- College of Horticulture, Nanjing Agricultural University, Nanjing, China
The family of DNA binding with one finger (DOF) transcription factors is plant specific, and these proteins contain a highly conserved domain (DOF domain) of 50-52 amino acids that includes a C2C2-type zinc finger motif at the N-terminus that is known to function in a number of plant processes. Here, we characterized 20 DOF genes in the important ornamental species chrysanthemum (Chrysanthemum morifolium) based on transcriptomic sequences. Phylogenetic analysis identified one pair of putative orthologous proteins in Arabidopsis and chrysanthemum and six pairs of paralogous proteins in chrysanthemum. Conserved motifs in the DOF proteins shared by Arabidopsis and chrysanthemum were analyzed using MEME. Bioinformatics analysis revealed that 13 CmDOFs could be targeted by 16 miRNA families. Moreover, we used 5' RLM-RACE to map the cleavage sites in CmDOF3, 15, and 21. The expression of these 20 genes in response to phytohormone treatments and abiotic stresses was characterized, and the expression patterns of six pairs of paralogous CmDOF genes were found to completely differ from one another, except for CmDOF6 and CmDOF7. This work will promote our research of the various functions of DOF gene family members in plant hormone and stress responses.
Introduction
Transcription factors (TFs) that determine gene transcription rates can be bound to cis-regulatory elements of promoters. Many TFs can be divided into different gene families according to their conserved domains. The DNA binding with one finger (DOF) TF family is plant specific and contains a conserved DOF domain (Yanagisawa, 2002). The DOF domain structure includes a C2C2 zinc finger that contains 50-52 amino acid residues; this zinc finger specifically binds to an element with the sequence 5′-AAAG-3′ (Yanagisawa, 2004). In addition to the DNA-binding domain, DOF TF proteins contain a bipartite nuclear localization signal (NLS) that partly overlaps with the conserved DOF DNA-binding domain (Krebs et al., 2010) and a C-terminal transcriptional activation domain (Yanagisawa, 2001). Moreno-Risueno et al. (2007) grouped the DOF family into seven subfamilies based on tree topology and corresponding phylogenetic relationships that probably originated from gene duplication events from a paraphyletic basal grade.
Since the first DOF protein, ZmDOF1, was identified in maize (Yanagisawa and Izui, 1993), additional DOF proteins have been found in many other plants (Cai et al., 2013; Negi et al., 2013). DOF TFs play multiple roles in different biological processes, such as flowering time (Fornara et al., 2009; Wei et al., 2010), seed protein content and color (Gupta et al., 2011), carbon and nitrogen metabolism (Yanagisawa et al., 2004; Tanaka et al., 2009), germination (Isabel-Lamoneda et al., 2003), light-mediated regulation (Park et al., 2003), vascular system development (Le Hir and Bellini, 2013), seed storage protein accumulation (Gaur et al., 2011), and hormone response (Gabriele et al., 2010), in various plants. For example, AtDOF4.1 overexpression lines showed severe growth retardation and delayed stem bolting and flowering, suggesting that AtDOF4.1 might act as a transcriptional repressor in the regulation of flowering time (Ahmad et al., 2013). AtDOF4.2 might regulate shoot branching through the up-regulation of three branching-related genes and seed epidermis development through the direct binding and activation of the cell wall loosening-related gene AtEXPA9 in Arabidopsis (Zou et al., 2013). In silico cis-regulatory element analysis indicated that SbDOF genes might be involved in light responsiveness, endosperm-specific gene expression, hormone responsiveness, meristem-specific expression and the stress response (Kushwaha et al., 2011). DOF TFs in Arabidopsis also play a unique role in vascular development and function (Le Hir and Bellini, 2013).
Recently, an increasing number of studies have shown that DOF TFs are also involved in the regulation of biotic and abiotic stress responses (Corrales et al., 2014; Ma et al., 2015; Sasaki et al., 2015). Thirty-five DOF full-length cDNAs were recently identified in the potato genome, and many StDOF genes were detected in various organs; several of these genes were up-regulated by abscisic acid (ABA) and abiotic stresses, such as drought and salinity (Venkatesh and Park, 2015). Similarly, several ZmDOF genes were up-regulated during salt treatment of seedlings (Chen and Cao, 2014). Overexpression of BBF1 from tobacco stimulates the transcription of the tobacco mosaic virus resistance gene N and defense-related responses, including ROS production (Takano et al., 2013). The identification of DOF genes associated with newer functions, such as abiotic stresses, needs to be explored for crop improvement (Gupta et al., 2015).
Chrysanthemum (Chrysanthemum morifolium), one of the four most famous cut flowers in the world, is susceptible to various biotic and abiotic stresses (An et al., 2014). To our knowledge, little information has been reported on the isolation and functional analysis of DOF TFs in chrysanthemum. Here, we isolated 20 DOF TFs in chrysanthemum based on a set of transcriptomic data. We performed a comparative phylogenetic analysis of chrysanthemum and Arabidopsis genes in silico and investigated the transcript levels in response to various phytohormones and abiotic stresses using qRT-PCR. Moreover, CmDOF3, 15, and 21 were confirmed as real targets of miRNA in plants by 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE). The results provide novel insights into the stress responses of CmDOF genes and promote a better understanding of the structure and function of DOFs in chrysanthemum.
Materials and Methods
Plant Materials and Growth Conditions
Cuttings of the cut flower chrysanthemum cultivar “Jinba,” maintained by the Chrysanthemum Germplasm Resource Preservation Centre (Nanjing Agricultural University, Nanjing, China), were rooted in vermiculite in the absence of fertilizer in a greenhouse. After 14 days, the cuttings were transplanted to their corresponding growth substrates and then subjected to a range of stress and phytohormone treatments.
Database Searches and Sequencing of Full-Length CmDOF cDNAs
All of the putative DOF proteins were retrieved from C. morifolium transcriptome data (Zhang et al., 2014). Arabidopsis DOF protein sequences were downloaded from The Arabidopsis Information Resource (TAIR) database. The DOF domain sequences of Arabidopsis were used as query sequences to identify CmDOF proteins. Multiple alignments among the identified CmDOF sequences were also performed to avoid repetition. Furthermore, the full open reading frames of CmDOFs were obtained via RACE PCR. The first cDNA strand was synthesized using the dT adaptor primer dT-AP and then subjected to nested PCR using the primer pair CmDOFx-3-F1/F2 and the adaptor primer AP (Table S1). Finally, twenty pairs of gene-specific primers (Table S2) were designed to amplify the full open reading frame sequences. The amplicons were purified using an AxyPrep DNA Gel Extraction Kit (Axygen, Hangzhou, China) and cloned into pMD19-T (TaKaRa, Tokyo, Japan) for sequencing.
Phylogenetic Tree Construction and Sequence Analysis
A phylogenetic tree was constructed with MEGA version 6.0 using the neighbor-joining method (Tamura et al., 2013). Multi-sequence alignments of DOF TFs were performed between Arabidopsis and C. morifolium using ClustalW software (Larkin et al., 2007). Computation of the theoretical isoelectric point (pI) and molecular weight (Mw) of CmDOF proteins was performed using the Compute pI/Mw online tool (http://web.expasy.org/compute_pi/), and ProtComp 9.0 and PSORT were used to predict their subcellular localization. The MEME v4.10.2 program (Bailey et al., 2015) was employed to identify the motifs present in the CmDOF proteins using the parameter settings suggested by Song et al. (2014b). Target prediction for miRNA was performed using the psRNATarget online tool (Dai and Zhao, 2011).
Target Validation by RLM-RACE
To confirm the predicted targets, RLM-RACE was performed using a FirstChoice RLM-RACE Kit (Ambion, Austin, TX, USA) following the methods described by Song et al. (2015). The RLM-RACE primer and gene-specific primers are shown in Table S5. The RLM-RACE products were purified using an Agarose Gel DNA Purification Kit (TaKaRa), ligated into the pMD19-T vector (TaKaRa), and sequenced.
Plant Treatments
The tissue-specific and treatment-induced transcription profiles of 20 CmDOF genes were explored in young seedling roots, stems and leaves as well as in the tube and ray florets of inflorescences at the bud stage and pollen. A variety of abiotic stresses was imposed, including high salinity (200 mM NaCl) and moisture deficit (20% w/v polyethylene glycol 6000, PEG 6000) (Song et al., 2012).
For the NaCl and PEG 6000 assays, young plants were transferred to liquid medium containing the stress agent, and the second true leaves were sampled at various time points (Song et al., 2014a). Other seedlings were subjected to a period of exposure at either 4°C or 40°C in a chamber providing a 16 h photoperiod and 50 μmol· m−2·s−1 of light, after which the second true leaves were sampled (Song et al., 2014c). The wounding treatment involved cutting the second true leaf. The phytohormone treatments involved spraying the leaves with either 50 μM ABA, 1 mM methyl jasmonate (MeJA) or 200 μM salicylic acid (SA) (Song et al., 2014b). Plants were sampled prior to stress treatment and then after 1, 4, 12, and 24 h.
After sampling, all of the collected material was snap frozen in liquid nitrogen and stored at −70°C. Each treatment was replicated three times.
Real-Time Quantitative PCR (qPCR)
Total RNA was isolated from samples using the RNAiso reagent (TaKaRa), according to the manufacturer's instructions; the RNA was then treated with RNase-free DNase I (TaKaRa) to remove potential genomic DNA contamination. The first cDNA strand was synthesized from 1 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The qPCR was performed using a Mastercycler ep realplex instrument (Eppendorf, Hamburg, Germany). Each 20 μL amplification reaction contained 10 μL of SYBR® Premix Ex Taq™ II (TaKaRa), 0.4 μL of each primer (10 μM), 4.2 μL of H2O and 5 μL of cDNA template. The PCR cycling regime consisted of an initial denaturation (95°C/2 min) followed by 40 cycles of 95°C/10, 55°C/15, and 72°C/20 s. A melting curve analysis was conducted following each assay to confirm the specificity of the amplicons. Gene-specific primers (sequences shown in Table S3) were designed using Primer3 Release 2.3.4 (Rozen and Skaletsky, 2000), and the EF1α gene was employed as a reference sequence. Relative transcript abundances were calculated via the 2−ΔΔCT method (Livak and Schmittgen, 2001). Three independent experiments were conducted.
Data Analysis
The relative transcript expression levels of each CmDOF were log2 transformed. The profiles were compared using Cluster v3.0 software (De Hoon et al., 2004) and visualized using Treeview (Eisen et al., 1998). SPSS v17.0 software (SPSS Inc., Chicago, IL, USA) was employed for all statistical analyses.
Results
Phylogenetic Relationships Among DOF Proteins of Chrysanthemum
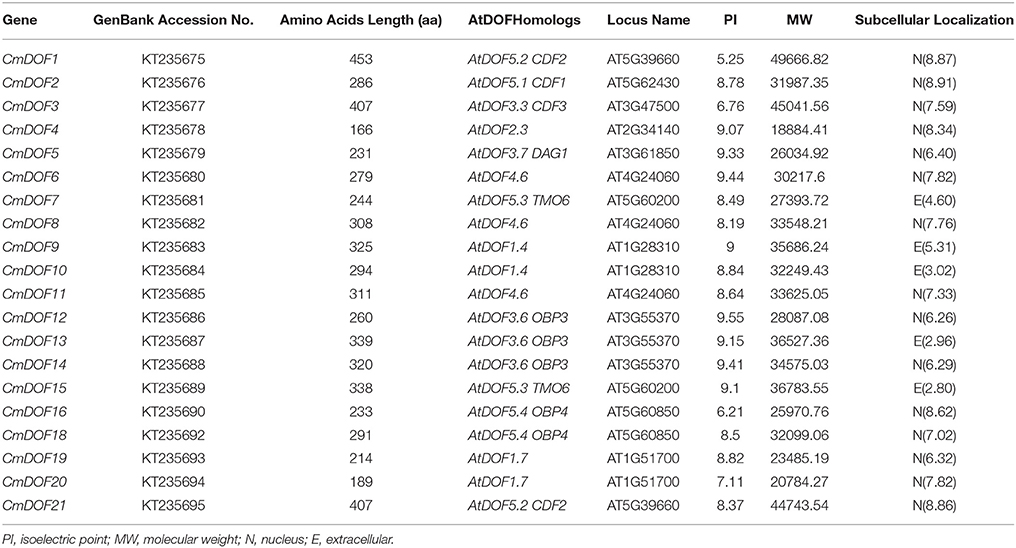
The 20 isolated DOF sequences were designated as CmDOF1 through CmDOF21, except CmDOF17, whose DOF domain is lost. The full-length cDNAs varied in length from 645 to 1817 bp, and their predicted protein products were composed of between 166 and 453 amino acid residues. Details regarding the CmDOF sequences are given in Table 1. Fifteen CmDOF proteins were predicted to show nuclear localization, excluding CmDOF7, 9, 10, 13, and 15. The conserved bipartite NLS was not found in the latter six proteins, which were predicted as being localized to the cytoplasm based on PSORT analysis.
To evaluate the evolutionary relationship between Arabidopsis and chrysanthemum DOF proteins, the deduced amino acid sequences of the DOF genes identified in Arabidopsis and chrysanthemum were completely aligned. A combined phylogenetic tree (Figure 1) was then constructed using the neighbor-joining method and bootstrap analysis (1000 reiterations). Twenty CmDOF genes were distributed across five of seven DOF groups: Group II, III, IV, V, and VI. Furthermore, one pair of putative orthologous proteins was identified in Arabidopsis and chrysanthemum: AtDOF5.4 with CmDOF18. In contrast, six pairs of paralogous DOF family proteins were identified in chrysanthemum: CmDOF2 and CmDOF3, CmDOF6 and CmDOF7, CmDOF8 and CmDOF11, CmDOF9 and CmDOF10, CmDOF13 and CmDOF14, and CmDOF19 and CmDOF20.
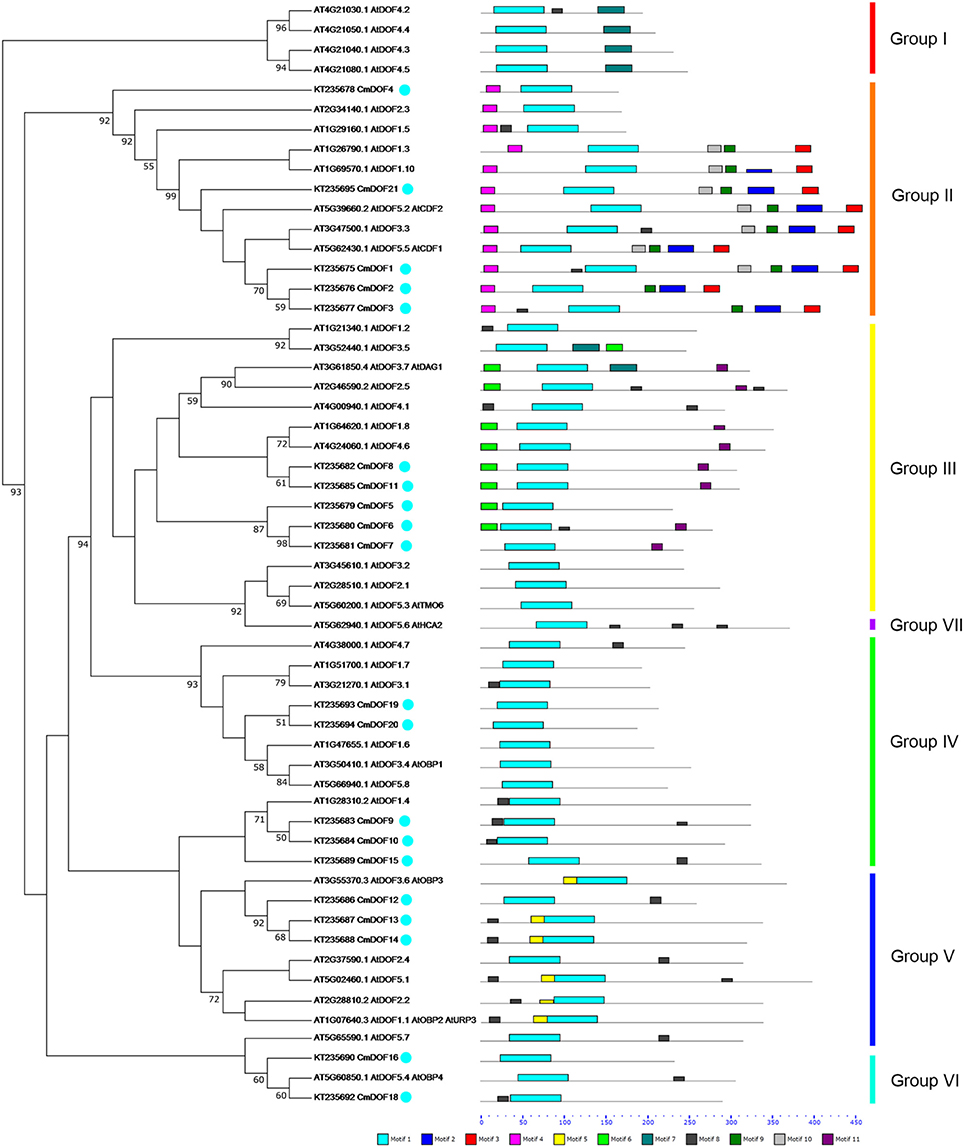
Figure 1. Phylogenetic tree and distribution of conserved motifs for the Arabidopsis and chrysanthemum DOF proteins. The tree was constructed from a complete alignment of 36 Arabidopsis and 20 chrysanthemum DOF proteins using the neighbor-joining method, bootstrap values of greater than 50% are shown at the nodal branches. The right portion shows the distribution of conserved motifs in Arabidopsis and chrysanthemum DOF proteins.
Conserved Sequences in DOF Proteins
DOF TFs contain the DOF DNA-binding domain that is usually located near the N-terminal region of the protein. DOF domains were present in all of the deduced DOF proteins in Arabidopsis and chrysanthemum based on MEME analysis, and each domain sequence contained ~56 amino acid residues (Motif 1, as shown in Figures 1, 2).
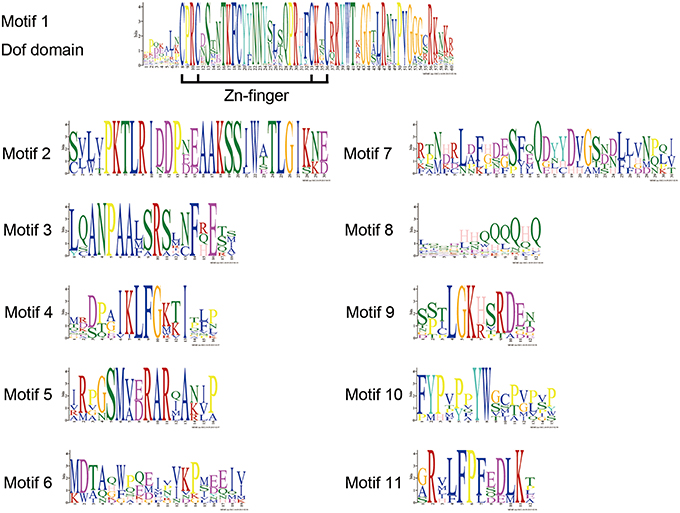
Figure 2. DOF protein motifs as derived by MEME analysis. The sequences of motifs of DOF proteins shown in Figure 1.
We also identified 10 other motifs shared by Arabidopsis and chrysanthemum; for example, Motifs 2/3/4/9 were only shared in Group II, Motif 5 was only present in Group V, and Group III had one specific motif, Motif 6 (Figure 1). Details on these motif features are shown in Figure 2. The Zn finger-like structure is the string CX2CX21CX2C type, which binds zinc (Zn2+) (Figure 2).
miRNA Target Site Prediction and Validation
All plant miRNA data were used to predict target transcript candidates of CmDOFs. As shown in Table S4, 13 CmDOFs can be targeted by 16 miRNA families. CmDOF1 has three target sites and CmDOF21 has two target sites, whereas the other 11 CmDOFs (3, 5, 6, 8, 9, 11, 15, 16, 18, 19, and 20) have only one target site. Moreover, we used 5′ RLM-RACE to map the cleavage sites in three predicted target genes. CmDOF3, 15 and 21 were confirmed as real targets of miRNA, as all of the 5′ ends of the mRNA fragments mapped to the nucleotide that paired to the tenth nucleotide of each miRNA with higher frequencies than depicted for each pairing oligo (Figure 3).
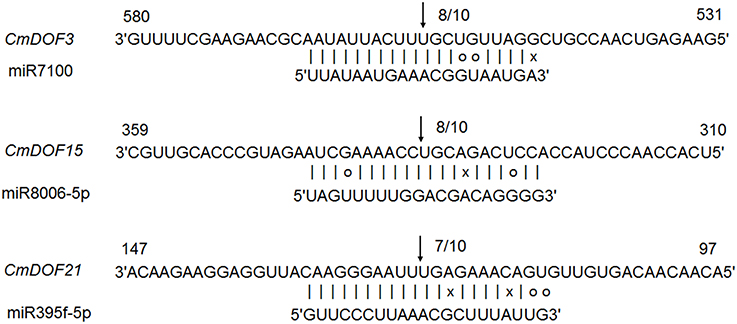
Figure 3. Mapping of mRNA cleavage sites confirmed by 5′ RLM-RACE. Arrows indicate the 5′ ends of mRNA fragments, as identified by cloned 5' RLM-RACE products, with the frequency of clones shown.
Transcription Profiling of CmDOF Genes
The 20 CmDOF genes were differentially expressed throughout the plant (Figure 4). The expression of CmDOF13 in ray florets was more than four orders of magnitude higher than that of CmDOF5 in the roots, whereas the CmDOF9 transcript was not detectable in the ray florets. The expression of CmDOF20 and CmDOF21 was significantly higher in reproductive organs than that in vegetative organs, whereas CmDOF16 was only highly expressed in roots. Interestingly, the expression patterns of six pairs of paralogous CmDOF genes were completely different from one another, with the exception CmDOF6 and CmDOF7, which exhibited similar expression patterns.
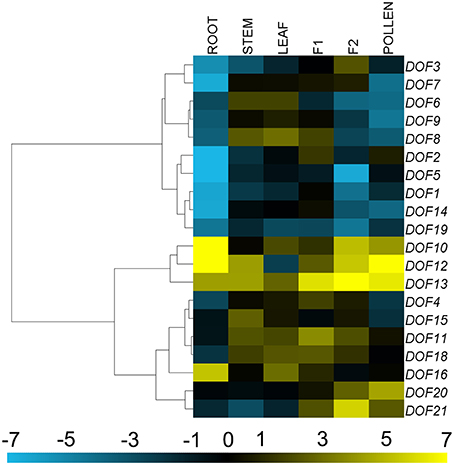
Figure 4. Differential transcription of CmDOF genes. F1, tubular florets; F2, ray florets at budding stage. Blue and yellow indicate lower and higher transcript abundance, respectively, compared to the relevant controls. Grey blocks indicate that transcription was not detected.
Expression of CmDOF Genes in Plants Challenged with Phytohormones
Seventeen of twenty CmDOF genes were significantly down-regulated by exogenous ABA, although CmDOF3, 4, 7, 8, 11, 19, and 21 were induced at 12 h. The CmDOF12 and CmDOF20 transcripts were increased at 4 and 24 h after ABA treatment, whereas CmDOF2 was only induced at 4 h (Figure 5A). The chrysanthemum DOF family genes exhibited three main expression patterns under MeJA treatment. CmDOF3, 4, 16, and 19 were induced by the treatment, whereas CmDOF1, 2, 5, 7, 8, 12, 14, 20, and 21 were repressed. The transcripts of the other seven CmDOFs were decreased at 1 h, increased at 4 h, and decreased at 12 h (Figure 5B). Eighteen of the genes were significantly repressed after 24 h of exposure to SA, whereas the expression of CmDOF9 and 19 was not significantly influenced by SA. Five genes (CmDOF2, 5, 6, 10, and 12) were induced at 1 and 12 h. The degree of inhibition of CmDOF1 increased with a longer processing time (Figure 5C).
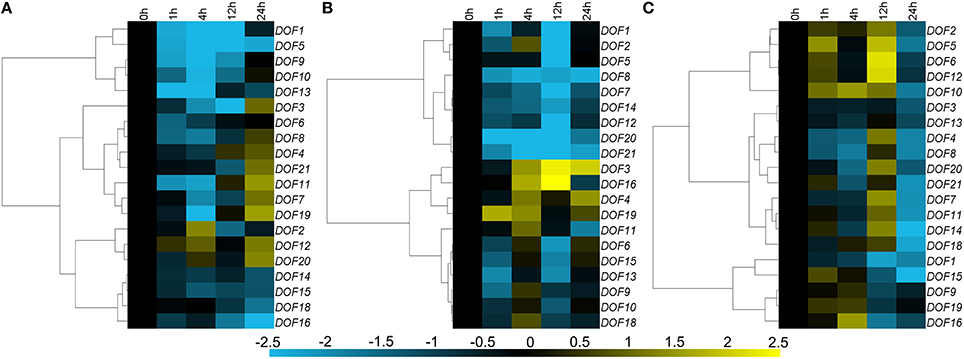
Figure 5. Differential transcription of CmDOF genes in leaves as induced by the exogenous supply of (A) abscisic acid (ABA), (B) methyl jasmonate (MeJA), and (C) salicylic acid (SA) treatments. Blue and yellow indicate lower and higher transcript abundance, respectively, compared to the relevant controls.
Differential Responses of the CmDOF Genes to Abiotic Stress
Three main expression patterns of CmDOFs were observed under salinity stress treatment. Eleven CmDOFs (1, 3, 6, 7, 8, 9, 11, 13, 14, 15, and 21) were suppressed, whereas five CmDOFs (5, 10, 12, 16, and 20) were weakly regulated by salinity stress, with a range of variation of less than 2-fold. Furthermore, CmDOF2, 4, 18, and 19 were up-regulated (Figure 6A). The expression of CmDOF5, 6, 8, 10, 12, 13, and 20 was not significantly altered by PEG treatment. Moisture stress up-regulated CmDOF2, 16, and 18 and markedly suppressed the transcription of 10 other CmDOFs (Figure 6B). Twelve of the 20 CmDOF genes (1, 2, 3, 7, 8, 9, 10, 11, 15, 19, 20, and 21) were suppressed by exposure to low temperature at 24 h. The transcript abundance of CmDOF7 and 19 was increased at 1 h and that of CmDOF1, 2, 3, and 10 was increased at 4 h. CmDOF16 was induced at 12 and 24 h, whereas the other seven CmDOFs (4, 5, 6, 12, 12, 14, and 18) were not significantly affected by low temperature (Figure 6C). Five CmDOF genes (1, 3, 7, 8, and 21) were down-regulated by high temperature, whereas seven CmDOF genes (2, 4, 5, 6, 16, 18, and 19) were up-regulated. CmDOF12, 13, and 14 were induced by high temperature treatment at 1 h but were then repressed thereafter. The other five CmDOFs (9, 10, 11, 15, and 20) were not significantly affected by low temperature (Figure 6D). All of the genes, with the exception of CmDOF2, 5, 16, 18, and 20, were down-regulated by mechanical damage (Figure 6E).
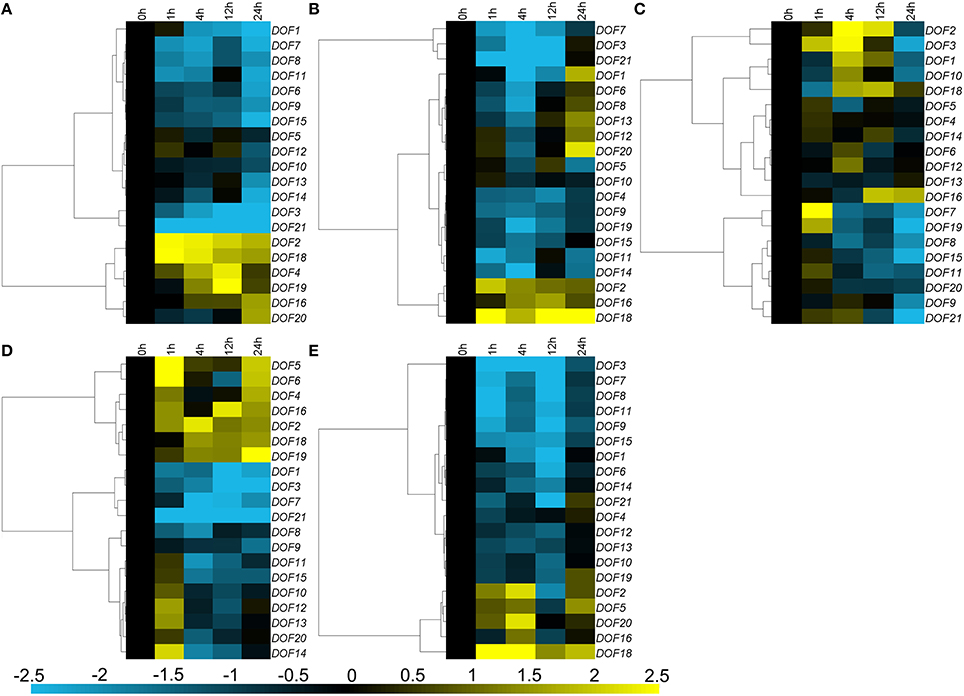
Figure 6. Differential transcription of CmDOF genes in leaves as induced by (A) salinity stress, (B) moisture stress, (C) low temperature (4°C), (D) high temperature (40°C), and (E) wound treatments. Blue and yellow indicate lower and higher transcript abundance, respectively, compared to the relevant controls.
Discussion
The DOF genes, members of the plant-specific TF family, are ubiquitous in photosynthetic organisms, ranging from green unicellular algae to vascular plants, and are implicated in important biological processes in plants. The function and evolution of DOF genes have been identified in Arabidopsis (Gupta et al., 2015), rice (Gaur et al., 2011), maize (Chen and Cao, 2014), poplar (Yang and Tuskan, 2006), Brachypodium distachyon (Hernando-Amado et al., 2012), bread wheat (Dong et al., 2007), and sorghum (Kushwaha et al., 2011). Nevertheless, little is known about the chrysanthemum DOF family. In this study, comparative analysis of the DOF family between Arabidopsis and chrysanthemum allowed for the prediction of various functions of the chrysanthemum DOF family members and helped to facilitate further gene function analysis.
Comparative Analysis of the Chrysanthemum and Arabidopsis DOF Gene Families
In this study, 20 CmDOF genes were identified in chrysanthemum based on transcriptome data and were classified by the presence of a highly conserved DOF domain. Gene duplication and differentiation have long been viewed as the major pathways of origin for new genes and for the differentiation of gene function. Therefore, to clarify the phylogenetic relationships among the CmDOF genes and infer the evolutionary history of this gene family, a combined phylogenetic tree was constructed based on the alignment of Arabidopsis and chrysanthemum DOF sequences (Figure 1). Phylogenetic analysis indicated that there are five subgroups in the transcriptome data. The results suggested that chrysanthemum has other unknown DOF genes, which may not have been identified here due to the limited available transcriptome data for chrysanthemum. Here, we also detected six pairs of paralogous CmDOF genes and one pair of putative orthologues based on phylogenetic analysis (Figure 1). Orthologues are defined as genes in different genomes that have been created by the splitting of taxonomic lineages, and paralogs are genes in the same genome created by gene duplication events (Thornton and DeSalle, 2000). Paralogs usually display different functions, whereas orthologues may retain the same function (Tatusov et al., 1997).
Motifs Analyses of the DOF Family in Chrysanthemum
We further analyzed the conserved motifs in the chrysanthemum DOF family using the MEME program. The majority of the CmDOFs in the same group shared similar motifs, suggesting that these conserved motifs play crucial roles in group-specific functions. However, high divergence in their structures was found between the different groups. For example, Group II contains Motifs 2, 3, 4, and 9, whereas Group III contains Motifs 6 and 11 (Figure 1), reflecting the complex nature of the function of DOF proteins in chrysanthemum. We also found motifs conserved in certain groups, e.g., Motif 7 in Group I, Motif 11 in Group III, and Motif 5 in Group V. The motif distribution indicated that the genes containing the same motifs were likely produced via gene expansion within the same groups. In total, 41 conserved motifs were identified in poplar, Arabidopsis, and rice DOF protein sequences (Yang and Tuskan, 2006). After comparison, we found that most motifs (except Motif 8) are shared by chrysanthemum, poplar, Arabidopsis, and rice.
The DNA-binding domain in CmDOF includes a C2C2-type zinc-finger like motif, although the amino acid sequence of this domain is largely different from those of other zinc-finger domains (Figure 2). The cysteine residues for putative coordination of zinc are shown in the DOF domain amino acid sequence. TFs sometimes contain multiple DNA-binding domains. For example, plant-specific WRKY TFs possess different numbers of WRKY DNA-binding domains, which allows the proteins to be classified into subgroups (Song et al., 2014b). However, in the case of DOF proteins, only a single copy of the DOF domain can consistently be found in their N-terminal regions (Figure 1).
miRNA Target Site Prediction and Validation
To our knowledge, reports on miRNA-DOF interactions have been rare. We predicted that 13 CmDOFs could be targeted by 16 miRNA families, respectively (Table S4). We also confirmed that CmDOF3, 15, and 21 were real targets of the miRNA in plants (Figure 3). However, the function of these interaction regulatory networks in plants should be determined by further research.
Organ-Preferential Expression of CmDOF Genes
Because gene expression patterns can provide important clues for gene function, we used qRT-PCR to examine the expression of CmDOF genes in young seedling roots, stems and leaves as well as in the tube and ray florets of inflorescences at the bud stage and in pollen (Figure 4). The expression profiles reveal spatial variations in the expression of CmDOFs in different organs. Five pairs of paralogous genes, except for CmDOF6 plus CmDOF7, showed distinct expression patterns, suggesting that significant functional divergence might occur after duplication events.
CmDOF10, 12, 13, and 16 showed relatively high expression levels in roots. Among them, only CmDOF16 was highly expressed in the root, indicating that it could play a role in the development of the plant root. These expression patterns were similar to those of their homolog in Arabidopsis, AtOBP3, which regulates phytochrome and cryptochrome signaling (Ward et al., 2005). Expression of CmDOF20 and CmDOF21 was significantly higher in reproductive organs than that in vegetative organs, indicating that they could play key roles in reproductive development. The CmDOF21 homolog AtCDF2 regulates the timing of transition from the vegetative to reproductive phase (Fornara et al., 2009). The expression level of CmDOF2 was higher in tube florets, whereas that of the paralogous gene CmDOF3 was higher in ray florets, suggesting that these genes could play different roles in chrysanthemum flower development. The highly expressed or differentially expressed CmDOF genes reported in this study may play a regulatory role in chrysanthemum plant development. However, additional research is needed to determine the functions of the CmDOF genes.
The Expression Profiles of CmDOF Genes Under Phytohormone and Abiotic Stress Treatments
Some plant hormonal signals, such as ABA, SA, and MeJA, are involved in the response to various stresses through activation of the transcription of several defense-related genes. For example, SA and MeJA co-ordinately play a critical role in biotic stress signaling upon pathogen infection (Vos et al., 2015), while ABA is extensively involved in the response to various biotic and abiotic stresses, including pathogen infection, cold, and osmotic stress (Lim et al., 2015). Therefore, in this study, we investigated the responses of CmDOFs to different plant hormone signals and abiotic stress treatments. The results showed that CmDOFs were both up-regulated and down-regulated by the treatments (Figures 4, 5), indicating that CmDOFs might be involved in the responses to various plant hormones that signal a stress response.
In previous reports, DOF proteins have been shown to be regulators of plant hormone-responsive genes and have been shown to mediate the response of gibberellins and auxins (Gupta et al., 2015). The rice DOF protein OsDof3 might be a mediator of GA signaling during germination (Washio, 2001). NtBBF1, which is a DOF protein known to play a pivotal role in regulating rolB expression, might provide the possible mechanism of auxin induction (Baumann et al., 1999). OBP3 (AtDof3.6) is induced by SA (Kang and Singh, 2000), although its homologs in chrysanthemum exhibited a different expression pattern. CmDOF12 was induced by SA, whereas CmDOF13 and CmDOF14 were not (Figure 5C). However, little is known about the role of the DOF gene family in ABA and JA hormonal signaling pathways. Our results may provide the basis for advancing research on DOF family genes in stress phytohormone signaling.
A group of five tomato DOF genes that are homologous to Arabidopsis Cycling DOF Factors (CDFs) function as transcriptional regulators involved in responses to drought and salt stress and flowering-time control in a gene-specific manner (Corrales et al., 2014). SlCDF1–5 genes were differentially induced in response to osmotic, salt, heat, and low-temperature stresses (Corrales et al., 2014). In chrysanthemum, CmDOF1 is homologous to AtCDF2, CmDOF2, and CmDOF21 are homologous to AtCDF1, and CmDOF3 is homologous to AtCDF3 (Table 1). However, they have different expression patterns in the presence of abiotic stress. CmDOF2 was induced by stress, whereas CmDOF1, 3, and 21 were repressed. This result indicated that chrysanthemum homologs of Arabidopsis CDF might have various roles in abiotic stress. As very few studies have investigated the role of DOF genes in the plant stress response, this work will lay the foundation for further investigations regarding the role of DOF in the stress response.
Conclusions
This study is the first transcriptome-wide analysis of the DOF TF family in chrysanthemum. The expression of 20 CmDOFs in response to a range of phytohormones and abiotic stress treatments was characterized. In addition, CmDOF3, 15, and 21 were confirmed as real targets of miRNA in plants. These findings lay the foundation for future research on the function of CmDOF genes in the plant stress response, which will promote their application in chrysanthemum breeding.
Author Contributions
Conceived and designed the experiments: AS, SC, and FC. Performed the experiments: AS, TG, JX, QF, and KZ. Analyzed the data: AS, SC, and FC. Contributed reagents/materials/analysis tools: ZG and FC. Wrote the paper: AS, PL, and DW. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by the National Science Fund for Distinguished Young Scholars (31425022), the Fundamental Research Funds for the Central Universities (KJQN201658, KYTZ201401), National Natural Science Foundation of China (31501792), the Natural Science Fund of Jiangsu Province (BK20150657), the China Postdoctoral Science Foundation (2014M561673, 2015T80564), the New Agricultural Varieties, Technology and New Model Update Project of Jiangsu Province [SXGC(2015)322], and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00199
References
Ahmad, M., Rim, Y., Chen, H., and Kim, J. (2013). Functional characterization of Arabidopsis Dof transcription factor AtDof4. 1. Russ. J. Plant Physiol. 60, 116–123. doi: 10.1134/S1021443712060027
An, J., Song, A., Guan, Z., Jiang, J., Chen, F., Lou, W., et al. (2014). The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 41, 4155–4162. doi: 10.1007/s11033-014-3287-2
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Baumann, K., De Paolis, A., Costantino, P., and Gualberti, G. (1999). The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11, 323–333. doi: 10.1105/tpc.11.3.323
Cai, X., Zhang, Y., Zhang, C., Zhang, T., Hu, T., Ye, J., et al. (2013). Genome-wide analysis of Plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 55, 552–566. doi: 10.1111/jipb.12043
Chen, Y., and Cao, J. (2014). Comparative analysis of Dof transcription factor family in maize. Plant Mol. Biol. Rep. 33, 1245–1258. doi: 10.1007/s11105-014-0835-9
Corrales, A.-R., Nebauer, S. G., Carrillo, L., Fernández-Nohales, P., Marqués, J., Renau-Morata, B., et al. (2014). Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
De Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20, 1453–1454. doi: 10.1093/bioinformatics/bth078
Dong, G., Ni, Z., Yao, Y., Nie, X., and Sun, Q. (2007). Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol. Biol. 63, 73–84. doi: 10.1007/s11103-006-9073-3
Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868. doi: 10.1073/pnas.95.25.14863
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Rühl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Gabriele, S., Rizza, A., Martone, J., Circelli, P., Costantino, P., and Vittorioso, P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61, 312–323. doi: 10.1111/j.1365-313X.2009.04055.x
Gaur, V. S., Singh, U., and Kumar, A. (2011). Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol. Biol. Rep. 38, 2827–2848. doi: 10.1007/s11033-010-0429-z
Gupta, N., Gupta, A. K., Singh, N., and Kumar, A. (2011). Differential expression of PBF Dof transcription factor in different tissues of three finger millet genotypes differing in seed protein content and color. Plant Mol. Biol. Rep. 29, 69–76. doi: 10.1007/s11105-010-0208-y
Gupta, S., Malviya, N., Kushwaha, H., Nasim, J., Bisht, N. C., Singh, V. K., et al. (2015). Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 241, 549–562. doi: 10.1007/s00425-014-2239-3
Hernando-Amado, S., González-Calle, V., Carbonero, P., and Barrero-Sicilia, C. (2012). The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol. 12:202. doi: 10.1186/1471-2229-12-202
Isabel-Lamoneda, I., Diaz, I., Martinez, M., Mena, M., and Carbonero, P. (2003). SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J. 33, 329–340. doi: 10.1046/j.1365-313X.2003.01628.x
Kang, H. G., and Singh, K. B. (2000). Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J. 21, 329–339. doi: 10.1046/j.1365-313x.2000.00678.x
Krebs, J., Mueller-Roeber, B., and Ruzicic, S. (2010). A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. J. Plant Physiol. 167, 583–586. doi: 10.1016/j.jplph.2009.11.016
Kushwaha, H., Gupta, S., Singh, V. K., Rastogi, S., and Yadav, D. (2011). Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 38, 5037–5053. doi: 10.1007/s11033-010-0650-9
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Le Hir, R., and Bellini, C. (2013). The plant-specific Dof transcription factors family: new players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 4:164. doi: 10.3389/fpls.2013.00164
Lim, C. W., Baek, W., Jung, J., Kim, J.-H., and Lee, S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16, 15251–15270. doi: 10.3390/ijms160715251
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J., Li, M.-Y., Wang, F., Tang, J., and Xiong, A.-S. (2015). Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16:33. doi: 10.1186/s12864-015-1242-9
Moreno-Risueno, M., Martínez, M., Vicente-Carbajosa, J., and Carbonero, P. (2007). The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol. Genet. Genomics 277, 379–390. doi: 10.1007/s00438-006-0186-9
Negi, J., Moriwaki, K., Konishi, M., Yokoyama, R., Nakano, T., Kusumi, K., et al. (2013). A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 23, 479–484. doi: 10.1016/j.cub.2013.02.001
Park, D. H., Lim, P. O., Kim, J. S., Cho, D. S., Hong, S. H., and Nam, H. G. (2003). The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 34, 161–171. doi: 10.1046/j.1365-313X.2003.01710.x
Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Sasaki, N., Matsumaru, M., Odaira, S., Nakata, A., Nakata, K., Nakayama, I., et al. (2015). Transient expression of tobacco BBF1-related Dof proteins, BBF2 and BBF3, upregulates genes involved in virus resistance and pathogen defense. Physiol. Mol. Plant Pathol. 89, 70–77. doi: 10.1016/j.pmpp.2014.12.005
Song, A., An, J., Guan, Z., Jiang, J., Chen, F., Lou, W., et al. (2014a). The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 80, 114–120. doi: 10.1016/j.plaphy.2014.03.030
Song, A., Li, P., Jiang, J., Chen, S., Li, H., Zeng, J., et al. (2014b). Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 15, 14442–14455. doi: 10.3390/ijms150814442
Song, A., Lu, J., Jiang, J., Chen, S., Guan, Z., Fang, W., et al. (2012). Isolation and characterisation of Chrysanthemum crassum SOS1, encoding a putative plasma membrane Na+ /H+ antiporter. Plant Biol. (Stuttg). 14, 706–713. doi: 10.1111/j.1438-8677.2011.00560.x
Song, A., Wang, L., Chen, S., Jiang, J., Guan, Z., Li, P., et al. (2015). Identification of nitrogen starvation-responsive microRNAs in Chrysanthemum nankingense. Plant Physiol. Biochem. 91, 41–48. doi: 10.1016/j.plaphy.2015.04.003
Song, A., Zhu, X., Chen, F., Gao, H., Jiang, J., and Chen, S. (2014c). A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 15, 5063–5078. doi: 10.3390/ijms15035063
Takano, M., Haque, M. A., Odaira, S., Nakata, K., Sasaki, N., and Nyunoya, H. (2013). Overexpression of a tobacco Dof transcription factor BBF1 stimulates the transcription of the tobacco mosaic virus resistance gene N and defense-related responses including ROS production. Plant Biotechnol. 30, 37–46. doi: 10.5511/plantbiotechnology.12.1207a
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, M., Takahata, Y., Nakayama, H., Nakatani, M., and Tahara, M. (2009). Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 230, 737–746. doi: 10.1007/s00425-009-0979-2
Tatusov, R. L., Koonin, E. V., and Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
Thornton, J. W., and DeSalle, R. (2000). Gene family evolution and homology: genomics meets phylogenetics. Annu. Rev. Genomics Hum. Genet. 1, 41–73. doi: 10.1146/annurev.genom.1.1.41
Venkatesh, J., and Park, S. W. (2015). Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 94, 73–85. doi: 10.1016/j.plaphy.2015.05.010
Vos, I. A., Moritz, L., Pieterse, C. M., and Van Wees, S. C. (2015). Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 6:639. doi: 10.3389/fpls.2015.00639
Ward, J. M., Cufr, C. A., Denzel, M. A., and Neff, M. M. (2005). The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17, 475–485. doi: 10.1105/tpc.104.027722
Washio, K. (2001). Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochem. Biophys. Acta 1520, 54–62. doi: 10.1016/S0167-4781(01)00251-2
Wei, P.-C., Tan, F., Gao, X.-Q., Zhang, X.-Q., Wang, G.-Q., Xu, H., et al. (2010). Overexpression of AtDOF4. 7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol. 153, 1031–1045. doi: 10.1104/pp.110.153247
Yanagisawa, S. (2001). The transcriptional activation domain of the plant-specific Dof1 factor functions in plant, animal, and yeast cells. Plant Cell Physiol. 42, 813–822. doi: 10.1093/pcp/pce105
Yanagisawa, S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. doi: 10.1016/S1360-1385(02)02362-2
Yanagisawa, S. (2004). Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. doi: 10.1093/pcp/pch055
Yanagisawa, S., Akiyama, A., Kisaka, H., Uchimiya, H., and Miwa, T. (2004). Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. U.S.A. 101, 7833–7838. doi: 10.1073/pnas.0402267101
Yanagisawa, S., and Izui, K. (1993). Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 268, 16028–16036.
Yang, X., and Tuskan, G. A. (2006). Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142, 820–830. doi: 10.1104/pp.106.083642
Zhang, F., Wang, Z., Dong, W., Sun, C., Wang, H., Song, A., et al. (2014). Transcriptomic and proteomic analysis reveals mechanisms of embryo abortion during chrysanthemum cross breeding. Sci. Rep. 4:6536. doi: 10.1038/srep06536
Keywords: Chrysanthemum morifolium, DNA binding with one finger, phylogenetic analysis, stress response, transcription factors
Citation: Song A, Gao T, Li P, Chen S, Guan Z, Wu D, Xin J, Fan Q, Zhao K and Chen F (2016) Transcriptome-Wide Identification and Expression Profiling of the DOF Transcription Factor Gene Family in Chrysanthemum morifolium. Front. Plant Sci. 7:199. doi: 10.3389/fpls.2016.00199
Received: 11 November 2015; Accepted: 05 February 2016;
Published: 23 February 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Dinesh Yadav, DDU Gorakhpur University, IndiaPaola Vittorioso, Sapienza University of Rome, Italy
Copyright © 2016 Song, Gao, Li, Chen, Guan, Wu, Xin, Fan, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fadi Chen, Y2hlbmZkQG5qYXUuZWR1LmNu
 Aiping Song
Aiping Song Tianwei Gao
Tianwei Gao