- 1Integrated Molecular Plant Physiology Research, Department of Biology, University of Antwerp, Antwerp, Belgium
- 2Department of Botany, Faculty of Science, University of Beni-Suef, Beni-Suef, Egypt
- 3Centre of Excellence Plant and Vegetation Ecology, Department of Biology, University of Antwerp, Antwerp, Belgium
- 4Division of Plant Physiology, Indian Agricultural Research Institute, New Delhi, India
- 5Department of Botany and Microbiology, Faculty of Science, Cairo University, Giza, Egypt
Salinity negatively affects plant growth and causes significant crop yield losses world-wide. Maize is an economically important cereal crop affected by high salinity. In this study, maize seedlings were subjected to 75 mM and 150 mM NaCl, to emulate high soil salinity. Roots, mature leaves (basal leaf-pair 1,2) and young leaves (distal leaf-pair 3,4) were harvested after 3 weeks of sowing. Roots showed the highest reduction in biomass, followed by mature and young leaves in the salt-stressed plants. Concomitant with the pattern of growth reduction, roots accumulated the highest levels of Na+ followed by mature and young leaves. High salinity induced oxidative stress in the roots and mature leaves, but to a lesser extent in younger leaves. The younger leaves showed increased electrolyte leakage (EL), malondialdehyde (MDA), and hydrogen peroxide (H2O2) concentrations only at 150 mM NaCl. Total antioxidant capacity (TAC) and polyphenol content increased with the increase in salinity levels in roots and mature leaves, but showed no changes in the young leaves. Under salinity stress, reduced ascorbate (ASC) and glutathione (GSH) content increased in roots, while total tocopherol levels increased specifically in the shoot tissues. Similarly, redox changes estimated by the ratio of redox couples (ASC/total ascorbate and GSH/total glutathione) showed significant decreases in the roots. Activities of enzymatic antioxidants, catalase (CAT, EC 1.11.1.6) and dehydroascorbate reductase (DHAR, EC 1.8.5.1), increased in all organs of salt-treated plants, while superoxide dismutase (SOD, EC 1.15.1.1), ascorbate peroxidase (APX, EC 1.11.1.11), glutathione-s-transferase (GST, EC 2.5.1.18) and glutathione reductase (GR, EC 1.6.4.2) increased specifically in the roots. Overall, these results suggest that Na+ is retained and detoxified mainly in roots, and less stress impact is observed in mature and younger leaves. This study also indicates a possible role of ROS in the systemic signaling from roots to leaves, allowing leaves to activate their defense mechanisms for better protection against salt stress.
Introduction
Soil salinization is a serious threat to crop productivity and predicted to increase in the face of global climate change (FAO, 2011)1. Estimations show that as much as 12 billion US$ will be lost globally, each year, due to the reduction in agricultural production from salt-affected areas (Qadir et al., 2008; Flowers et al., 2010). The situation is worst in arid and semi-arid regions, which are deficient in water and face high temperatures, resulting in more water-loss from plants due to higher evapotranspiration rates, aggravating the effects of salinity.
Salinity stress induces a multitude of responses in plants including morphological, physiological, biochemical, and molecular changes (Ambede et al., 2012; Abreu et al., 2013). It causes ionic imbalance, which results in ionic toxicity, osmotic stress, and generation of reactive oxygen species (ROS; Chaparzadeh et al., 2004; Chawla et al., 2013). For instance, accumulation of Na+ under salinity stress competes with K+ binding in proteins, causing inhibition of protein synthesis and metabolic enzymes (Schachtman and Liu, 1999; Pardo and Quintero, 2002). High concentrations of NaCl outside the roots reduce the water potential, making it more difficult for plants to extract water, and results in osmotic stress. In leaves, high salt levels cause stomatal closure, impairment of electron transport and the photosynthetic apparatus, leading to reduced photosynthesis and productivity (Abreu et al., 2013; Deinlein et al., 2014). To cope with osmotic stress, salt-stressed plants tend to accumulate compatible solutes such as proline (Pro) and glycine betaine (Holmström et al., 2000; Qureshi et al., 2013), that decrease the cytoplasmic osmotic potential, enabling water absorption (Pottosin et al., 2014; Puniran-Hartley et al., 2014).
High salinity also induces the formation of ROS within plant cells, and its over accumulation results in oxidative damage of membrane lipids, proteins and nucleic acids (Pérez-López et al., 2009; Gill and Tuteja, 2010). To scavenge high ROS levels, an efficient system of non-enzymatic and enzymatic antioxidants is involved (Apel and Hirt, 2004; Gill and Tuteja, 2010; Karuppanapandian et al., 2011). Non-enzymatic antioxidants include phenolics, flavonoids, tocopherols, ASC, and GSH (Munné-Bosch, 2005; Rai et al., 2013; Gupta and Huang, 2014; Rakhmankulova et al., 2015; Talbi et al., 2015). Enzymatic antioxidants include superoxide dismutase (SOD), peroxidase (POX), catalase (CAT), as well as the enzymes of the ascorbate (ASC)–glutathione (GSH) cycle [GSH reductase (GR), ASC peroxidase (APX), monodehydroascorbate dehydrogenase (MDHAR), and dehydroascorbate reductase (DHAR)] that detoxify ROS (Gill and Tuteja, 2010; Foyer and Noctor, 2011; Chawla et al., 2013). Up-regulation of antioxidants has been observed in salt tolerant cultivars of tomato, pea, Jatropha, and Calendula (Hernandez et al., 2000; Chaparzadeh et al., 2004; Mittova et al., 2004; Gao et al., 2008), suggesting a pertinent role of antioxidants in alleviating salt stress-induced oxidative damage. So far, no salinity-specific ROS or salinity-specific antioxidants have been identified in plants in general, the same species of ROS are produced under different abiotic and some biotic stresses, in part as by products of the stress and to serve as signals to nearby tissues as well (reviewed by Apel and Hirt, 2004). However, some salinity-specific physiological and biochemical mechanisms are identified. For instance, the uptake, compartmentalization and/or excretion of ions from specialized cells under salinity stress is essential for normal growth (Gupta and Huang, 2014; Viehweger, 2014).
Roots are the first organs to undergo salinity stress, and show greater reduction in growth than shoots (Lazof and Bernstein, 1999). However, other plants, such as avocado, showed a higher root to shoot ratio under high salinity (Bernstein et al., 2004). Moreover, some developmental stages, including germination, seedling emergence, and flowering are more sensitive to salt stress, and therefore the effects of high salinity depend on the plant developmental stage (Houle et al., 2001). Interestingly, age-related differences in responses in plant cells, tissues and organs have been observed. For instance, mature maize leave cells (at distal leaf parts), are more sensitive to high salinity than younger cells (from actively expanding leaf parts) which have higher expression of genes of the antioxidant enzymes (Kravchik and Bernstein, 2013). Also, cotton and Fragaria virginiana leaves of different ages, under drought stress, showed different stomatal conductance and osmotic potential adjustment responses (Jordan et al., 1975). However, differences in oxidative stress and antioxidant defenses, in different organs and developmental stages, have been much less studied.
Maize is an economically important cereal crop and its productivity is affected by high soil salinity in various parts of the world. Therefore, this work aims to elucidate the effects of salinity on maize growth. Based on our previous observations, we hypothesized that different organs and/or leaf tissues of different developmental stages may respond differently to high salinity stress and trigger specific defense mechanisms. We therefore compared oxidative stress and antioxidant responses of roots, mature leaves (basal leaf-pair 1, 2) and young leaves (distal leaf-pair 3, 4) harvested from 3 weeks old, salt-treated and non-treated maize seedlings.
Materials and Methods
Plant Material and Growth Conditions
Maize (Zea mays L. cv Giza 119) seeds were obtained from Agricultural Research Center, Giza, Egypt. Giza 119 is a local commercial variety first developed in 1973. Seeds were soaked on filter paper saturated with distilled water, and incubated at 26°C in the dark. Three days later, seedlings selected for uniform growth were transplanted into sandy soil (90% sand, pH 7.6). The soil initially contained 1.2% carbon, 15 mg nitrate-nitrogen (N), 1 mg ammonium-N, 10 mg phosphorus (P)/g air dry soil at a humidity of 0.30 g water/g dry soil. Growth conditions were; 16/8 h day/night photoperiod, 26/22°C temperature, and 70/80% relative humidity. High salinity treatments started immediately after transplantation by irrigating soils daily with 25 ml of 75 mM or 150 mM NaCl. These treatments were selected based on a preliminary study, where 75 mM and 150 mM showed mild and sever effects on plant growth, respectively. After 3 weeks of growth, roots, mature leaf-pair 1, 2 (L1,2) and distal leaf-pair 3, 4 (L3,4), were harvested. Samples were immediately frozen in liquid nitrogen and stored at -80°C, for further biochemical analyses. Dry weight (DW) of the plant organs was measured after drying the tissues for 5 days at 70°C.
Na+, K+, and Cl- Content Determination
Na+, K+, and Cl- content was determined in 100 mg DW of plant material. Samples were digested in 5 ml of HNO3/H2O (5:1), and supernatants after clarifying the digestate were analyzed by mass spectrometry (ICP-MS, Finnigan Element XR, Scientific, Bremen, Germany) as described previously (Hamad et al., 2015).
Oxidative Stress Markers
Malondialdehyde (MDA)
Malondialdehyde content was assayed according to Hodges et al. (1999). Fifty milligram fresh weight (FW) of tissue was homogenized in 1 ml of 80% (v/v) ethanol using a MagNA Lyser (Roche, Vilvoorde, Belgium). After centrifugation, the supernatant reacted with thiobarbituric acid to produce pinkish red chromogen, thiobarbituric acid-malondialdehyde (TBA-MDA). Absorbance was measured at 440, 532, and 600 nm by using a micro-plate reader (Synergy Mx, Biotek Instruments Inc., Vermont, VT, USA). MDA content was calculated and expressed as nmol/g FW tissue.
H2O2 Concentration
Fifty milligram fresh tissue was homogenized in 0.5 ml 0.1% TCA, centrifuged and 50 μl was used in the assay. Hydrogen peroxide (H2O2) concentration was measured according to Jiang et al. (1990), based on the peroxide-mediated oxidation of Fe2+, followed by reaction of Fe3+, with xylenol orange. Specificity for H2O2 was tested by eliminating H2O2 from the reaction mixture with CAT.
Electrolyte Leakage
Membrane leakage of cells was measured by cutting 1 cm2 disks of leaf tissues and segments of root tissues, washing thoroughly, and incubation in 20 ml deionized water for 18 h at room temperature. After this incubation leaf disks were boiled for 30 min. The conductivity of the incubation solutions was measured at three time points using a conductivity meter (WTW GmbH, Weilheim, Germany; Lutts et al., 1995).
Antioxidant Molecules
Total Antioxidant Capacity (TAC)
Two hundred milligram (FW) frozen plant tissue was ground with mortar and pestle in liquid nitrogen, and antioxidants were extracted in 2 ml of ice-cold 80% ethanol. FRAP (ferric reducing antioxidant power) reagent (300 mM acetate buffer (pH 3.6), 0.01 mM 2,4,6-tripirydylo-S-triazine (TPTZ)) in 0.04 mM HCl and 20 mM FeCl3.6H2O), was mixed with the extract and measured at 600 nm using a microplate reader (Benzie and Strain, 1999). Trolox was used as a standard.
Polyphenols and Flavonoids
Polyphenols and flavonoids were extracted by homogenizing 50 mg tissues in 0.5 ml 80% ethanol (v/v), centrifuged, pellet was washed twice each with 0.5 ml 80% ethanol (v/v), supernatants were pooled. Total phenolic content was determined using a Folin–Ciocalteu assay according to Zhang et al. (2006). Gallic acid was used as a standard. Flavonoid content was estimated using the modified aluminum chloride calorimetric method (Chang et al., 2002), with quercetin as a standard.
Ascorbate and Glutathione
One hundred milligram (FW) of frozen tissues were ground in a Mag NALyser (Roche, Vilvoorde, Belgium), and extracted in ice-cold 6% (v/v) phosphoric acid. Reduced ASC and GSH contents were determined by HPLC analysis (Potters et al., 2004). The identity of the peaks was confirmed using an in-line diode array detector (DAD, SPD-M10AVP, Shimadzu). Total ASC and GSH concentration was determined after reducing the samples with 40 mM DTT and the ASC and GSH redox status are calculated as the ratio between reduced and total amount of ASC and GSH, respectively.
Tocopherols
One hundred milligram (FW) of frozen plant tissue was extracted in 6 ml of hexane, and centrifuged at 14,000 × g for 15 min. Extracts were dried (CentriVap concentrator, Labconco, Kansas City, MO, USA) and resuspended again in hexane. Tocopherols were separated and quantified by HPLC analysis (Shimadzu, Hertogenbosch, The Netherlands, normal phase conditions, Particil Pac 5 mm column material, length 250 mm, i.d. 4.6 mm). Dimethyl tocol (DMT) was used as internal standard (5 ppm). Data were analyzed with Shimadzu Class VP 6.14 software.
Enzyme Activities
Superoxide dismutase, POX (EC 1.11.1.7), CAT, GSH POX (GPX), APX, GR, glutathione-S-transferase (GST), monodehydroascorbate reductase (MDHAR, EC 1.6.5.4) and DHAR were determined in an homogenate of 100 mg (FW) of leaf tissues, prepared in 1 ml of 50 mM potassium phosphate buffer (pH 7.0), containing 10% (w/v) polyvinylpyrrolidone (PVP), 0.25% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride(PMSF) and 1 mM ASC, by using a MagNA Lyser (Roche, Vilvoorde, Belgium). SOD activity was determined according to Dhindsa et al. (1982) by measuring the inhibition of NBT (nitroblue tetrazolium) reduction at 560 nm. POX activity was determined by the oxidation of pyrogallol (ε430 = 2.47 mM-1 cm-1; Kumar and Khan, 1982). CAT activity was assayed according to the Aebi (1984) by monitoring the decomposition of H2O2 at 240 nm. APX, MDHAR, DHAR, and GR activities were measured by the methods of (Murshed et al., 2008). GST activity was estimated by measuring the conjugation of GSH with excess 1-chloro-2,4-dinitrobenzene (CDNB) at 340 nm (ε340 = 0.0096 μM-1 cm-1; Habig et al., 1974). GPX activity was assayed by measuring the decrease in NADPH absorbance measured at 340 nm (ε340 = 6.22 mM-1cm-1; Drotar et al., 1985). All activity measurements were scaled down for semi-high throughput using a micro-plate reader (Synergy Mx, Biotek Instruments Inc., Winooski, VT, USA), and optimized to obtain linear time and protein concentration dependence.
Protein Concentration
The soluble protein was extracted by homogenizing 100 mg fresh tissue in content was estimated according to the method of Lowry et al. (1951).
Proline concentration
Proline content was estimated by homogenizing 0.2 g fresh weight in 2 ml of 3% aqueous sulfosalicylic acid, centrifugation at 10,000 rpm for 30 min. The supernatant was decanted and pellet was washed twice with 3% aqueous sulfosalicylic acid. The supernatants were pooled and proline content was measured using ninhydrin reagent and toluene extraction (Bates et al., 1973)
Statistical Analysis
All results were analyzed by one-way ANOVA using SPSS 16.0 statistical software and significant differences between the means of parameters (n = 8) were determined by using the Duncan test (P < 0.05). The treatments of each organ were compared with their corresponding control where significant differences were given different letters.
Results
Biomass
To assess the effect of high salinity (NaCl) on maize growth, biomass (DW) of roots, L1,2, and L3,4, was measured. Increase in the NaCl concentration resulted in a significant decrease in the biomass of roots and L1,2, and larger decline was observed at the higher (150 mM) NaCl concentration (Figure 1). However, L3,4 harvested from NaCl-treated plants showed no statistical difference in the biomass compared to the controls. This suggests that severe salinity reduced growth progressively from roots along the vertical gradient of the maize plant, with the root found to be most sensitive (-40%), followed by L1,2 (-20%).
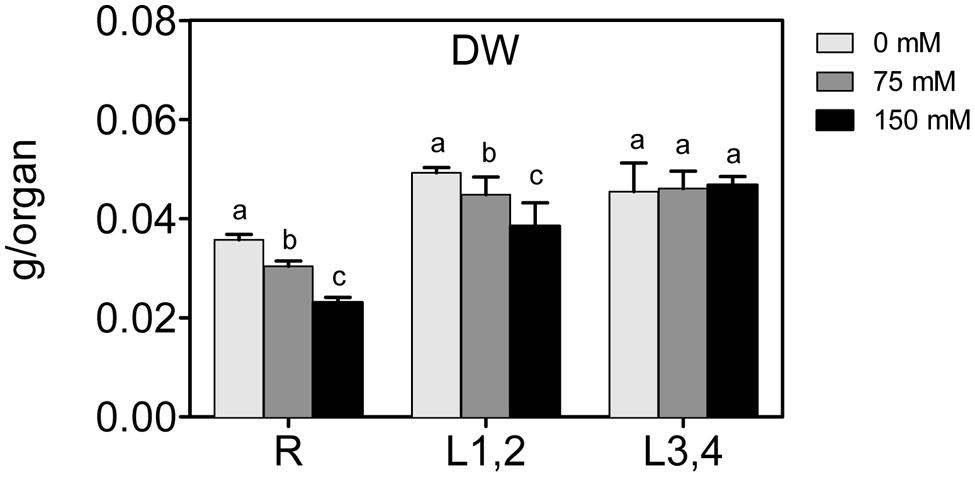
FIGURE 1. Effect of different salinity levels on dry weight (DW) of root (R), mature leaf pair (L1,2), and young leaf pair (L3,4) of 3 weeks old maize seedlings. Values are means of at least three replicates and significant differences between means, as determined by Duncan test (P < 0.05), are indicated by different letters.
Na+, K+, and Cl- Content
High soil salinity causes over-accumulation of Na+ inside the cell, which may result in ionic imbalances and metabolic toxicity. Na+ content increased significantly with the increase of applied NaCl concentration in root and L1,2 but Na+ accumulation was significant in L3,4 only under 75 mM treatment and not significant when further increased to 150 mM (Figure 2A). As with the effect of salt on biomass, there was an apparent gradient, with highest Na+ accumulation in roots (+404%), followed by L1,2 (+208%) and L3,4 (+137.3%), under severe salinity stress, compared to their respective controls. The change in Cl- content of different organs under salinity behaved similar to Na+ but the L1,2 content of Cl- increased with salinity in a concentration independent manner (Figure 2B). K+ content of roots decreased with the increase in salinity stress but in L1,2 and L3,4, no significant change was observed (Figure 2C).
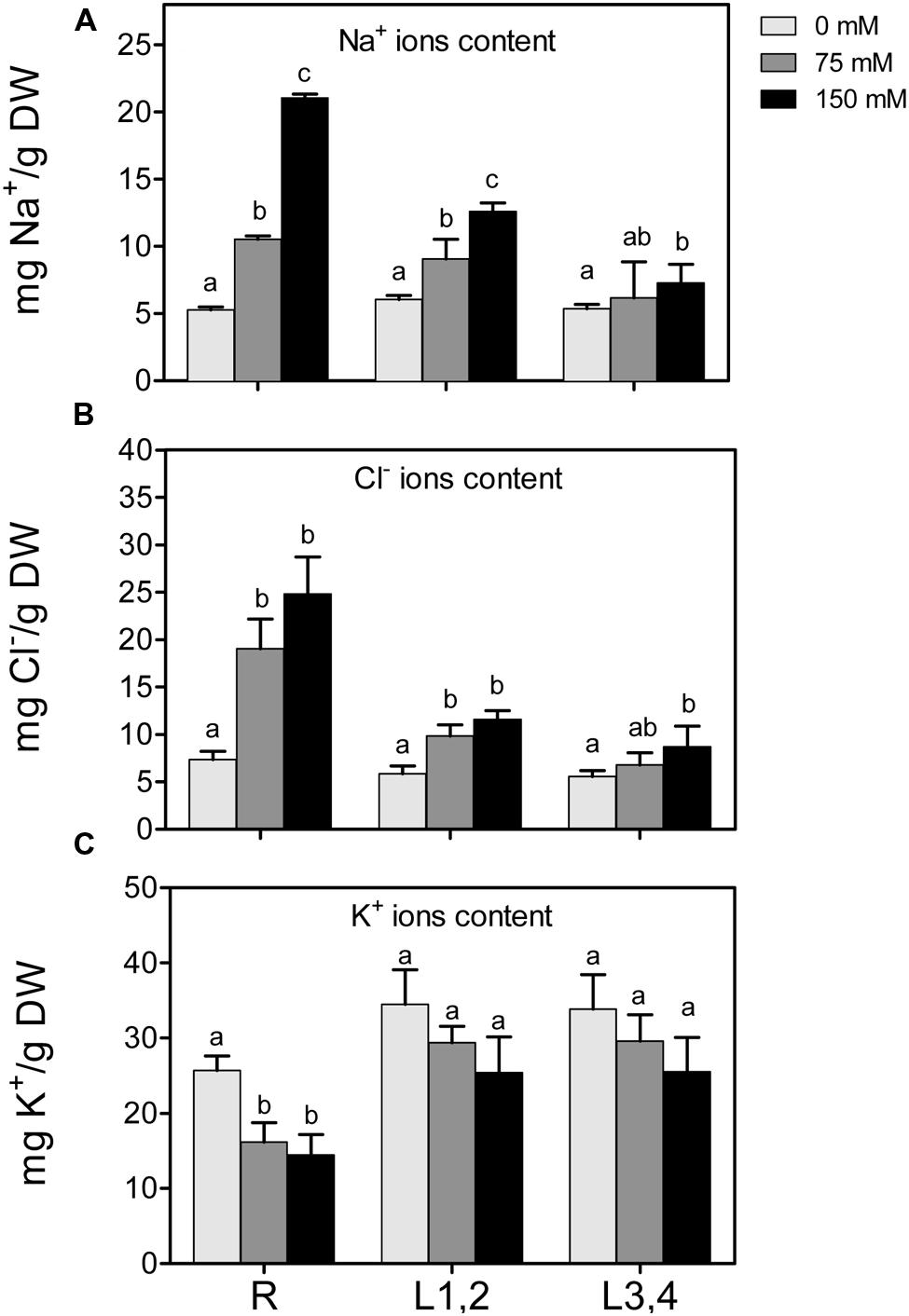
FIGURE 2. Effect of different salinity levels on Na+, Cl-, and K+ ion content of root (R), mature leaf pair (L1,2), and young leaf pair (L3,4) of 3 weeks old maize seedlings. Values are means of at least three replicates and significant differences between means, as determined by Duncan test (P < 0.05), are indicated by different letters.
Oxidative Stress Markers
Salt stress also leads to higher accumulation of ROS, which disturb cellular redox homeostasis, and result in oxidative damage. H2O2 content and markers of oxidative damage of cell membranes, i.e., electrolyte leakage (EL) and lipid peroxidation (MDA) were measured (Figure 3). Increase in the NaCl concentration resulted in a significant increase in the accumulation of H2O2 content in roots and L1,2. However, in L3,4, the increase was only observed at the highest NaCl (150 mM) concentration. Following the trend of H2O2, EL, and MDA also increased significantly (p < 0.05) in root and L1,2 but only at 150 mM NaCl in L3,4. Thus, oxidative stress parameters indicate that root tissues suffered the most from the salinity-induced oxidative stress, followed by L1,2 and L3,4 was the least affected.
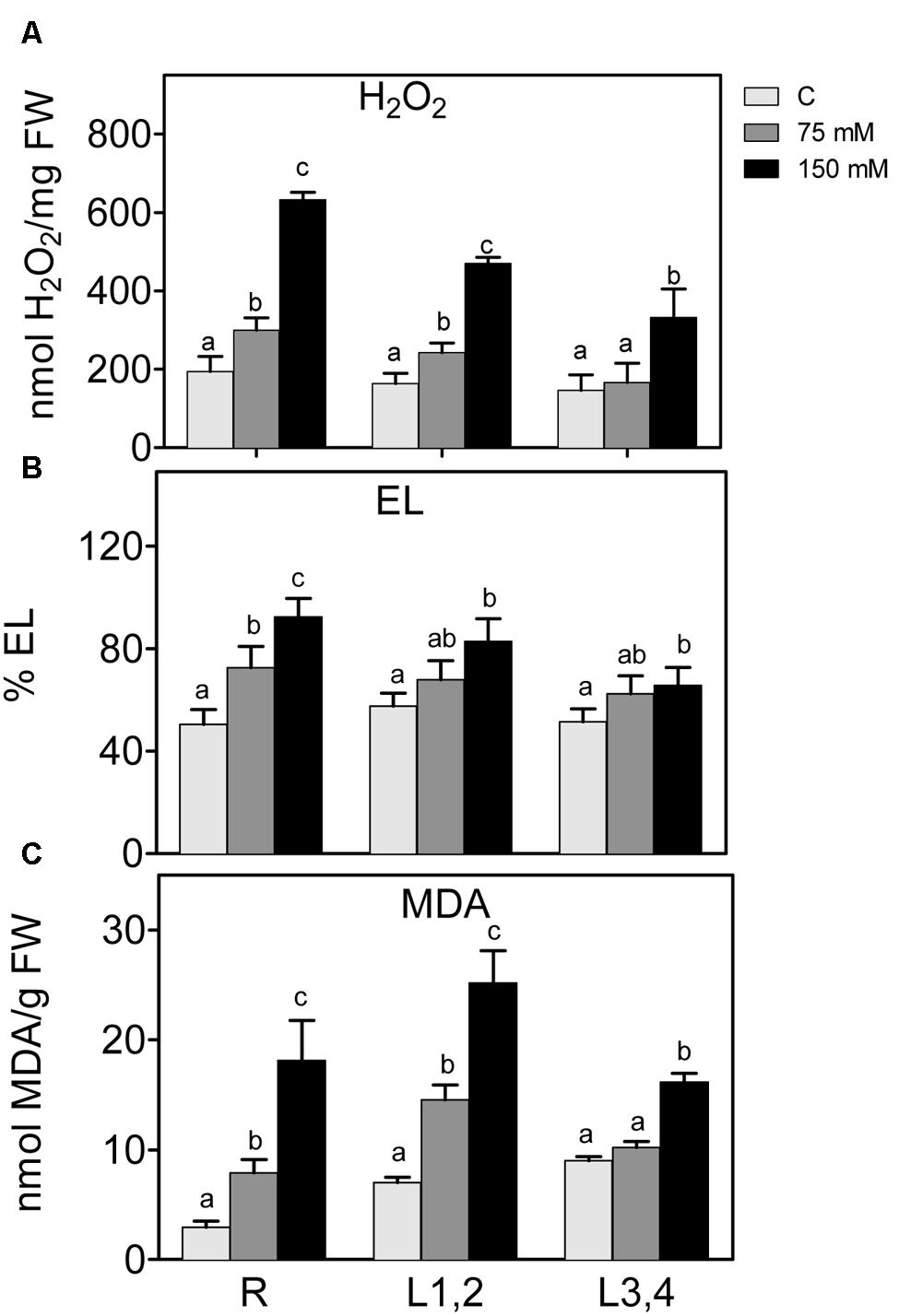
FIGURE 3. Effect of different salinity levels on H2O2 (A) relative electrolyte leakage (EL, B), and lipid peroxidation measured as malondialdehyde content (C) of root (R), mature leaf pair (L1,2), and young leaf pair (L3,4) of 3 weeks old maize seedlings. Values are means of at least three replicates and significant differences between means, as determined by Duncan test (P < 0.05), are indicated by different letters.
Antioxidant Defense Systems
Non-enzymatic Antioxidants
Changes in polyphenol and flavonoid content in roots, L1,2 and L3,4 under 75 mM and 150 mM NaCl, are shown in (Figures 4A,B). Polyphenol content of roots, increased with increasing salinity in a concentration dependent manner. However, its increment in L1,2 and L3,4 under salinity stress was concentration independent. High salinity did not affect levels of flavonoids in any of the tested organs. Increasing severity of salinity induced increases in the TAC, as determined by the FRAP-assay (Figure 4C), of L1,2 in a concentration-dependent manner. Root and L3,4 tissues were less responsive, their antioxidant capacity only slightly increased under mild or severe salinity stress. Slight increase in total tocopherol content was observed in L1,2 under mild salinity stress but not in root or L3,4 tissues. However, under severe salinity stress, the total tocopherol content increased significantly only in L1,2 and L3,4 (Figure 4D). The Pro content slightly decreased in L1,2 leaves, but did not show significant changes in maize root and L3,4 leaves (Figure 4E) under exposure to salinity stress.
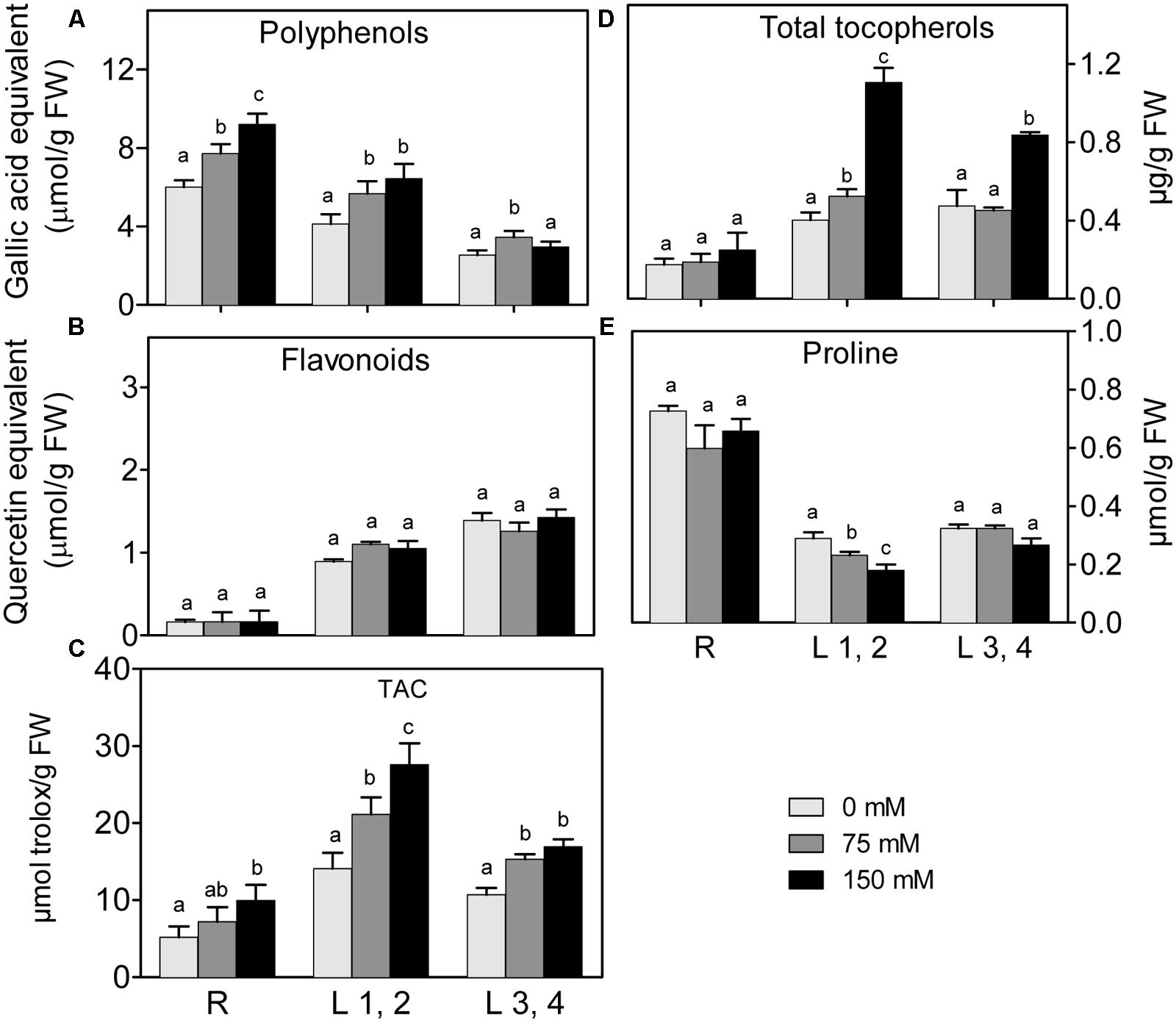
FIGURE 4. Effect of different salinity levels on polyphenols (A), flavonoids (B), total antioxidant capacity (TAC, C), total tocopherols (D), and Proline (E) content of root (R), mature leaf pair (L1,2) and young leaf pair (L3,4) of 3 weeks old maize seedlings. Values are means of at least three replicates and significant differences between means, as determined by Duncan test (P < 0.05), are indicated by different letters.
Enzymatic Antioxidants and ASC/GSH-Cycle Components
Mild or severe salinity stress increased SOD activity in root in a concentration independent manner. L1,2 and L3,4 content of SOD activity was not significantly affected by salinity stress (Figure 5A). POX did not show significant differences in activity upon exposure to mild or severe salinity in either roots, L1,2 or L3,4 leaves (Figure 5B). Only the highest salinity level resulted in a significant increase in CAT activity, and this was strongest in roots and L1,2 leaves (Figure 5C).
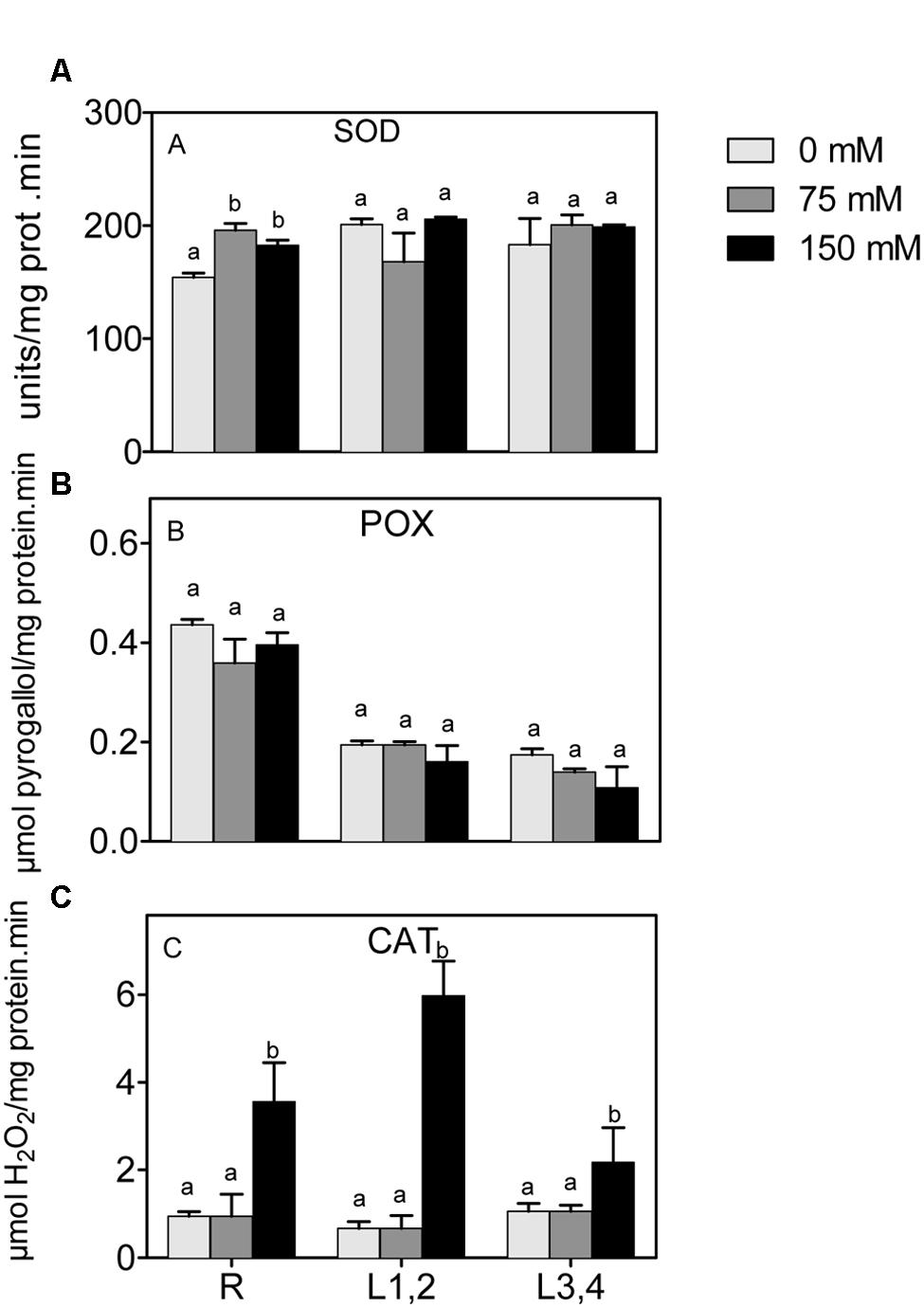
FIGURE 5. Effect of different salinity levels on superoxide dismutase (A), peroxidase (B), and catalase (C) activities of root (R), mature leaf pair (L1,2), and young leaf pair (L3,4) of 3 weeks old maize seedlings. Values are means of at least three replicates and significant differences between means, as determined by Duncan test (P < 0.05), are indicated by different letters.
In all plant parts, except L3,4, there was a significant increase of ASC under salinity stress (Figure 6A). However, the ascorbate redox status (ASC/total ascorbate, Figure 6B) decreased in root upon the increase in salinity. The L1,2 and L3,4 GSH content, and the GSH redox status (GSH/total glutathione), did not significantly change under any degree of salinity stress (Figures 6C,D). The, root GSH content increased, but the redox status decreased under salinity. APX activity (Figure 6E) in roots increased progressively with the increase in salinity strength, whereas in L1,2 and L3,4 tissues, its activity increased only slightly under mild but not severe salinity (Figure 6E). GR, MDHAR, and GST activities increased in root tissues only under severe salinity. Their activities in L1,2 and L3,4 was not affected by salinity (Figures 6F,G,I). DHAR activity (Figure 6H) in root tissues increased with increase in salinity in a concentration dependent manner. Only severe salinity increased DHAR activity in L1,2 and L3,4.
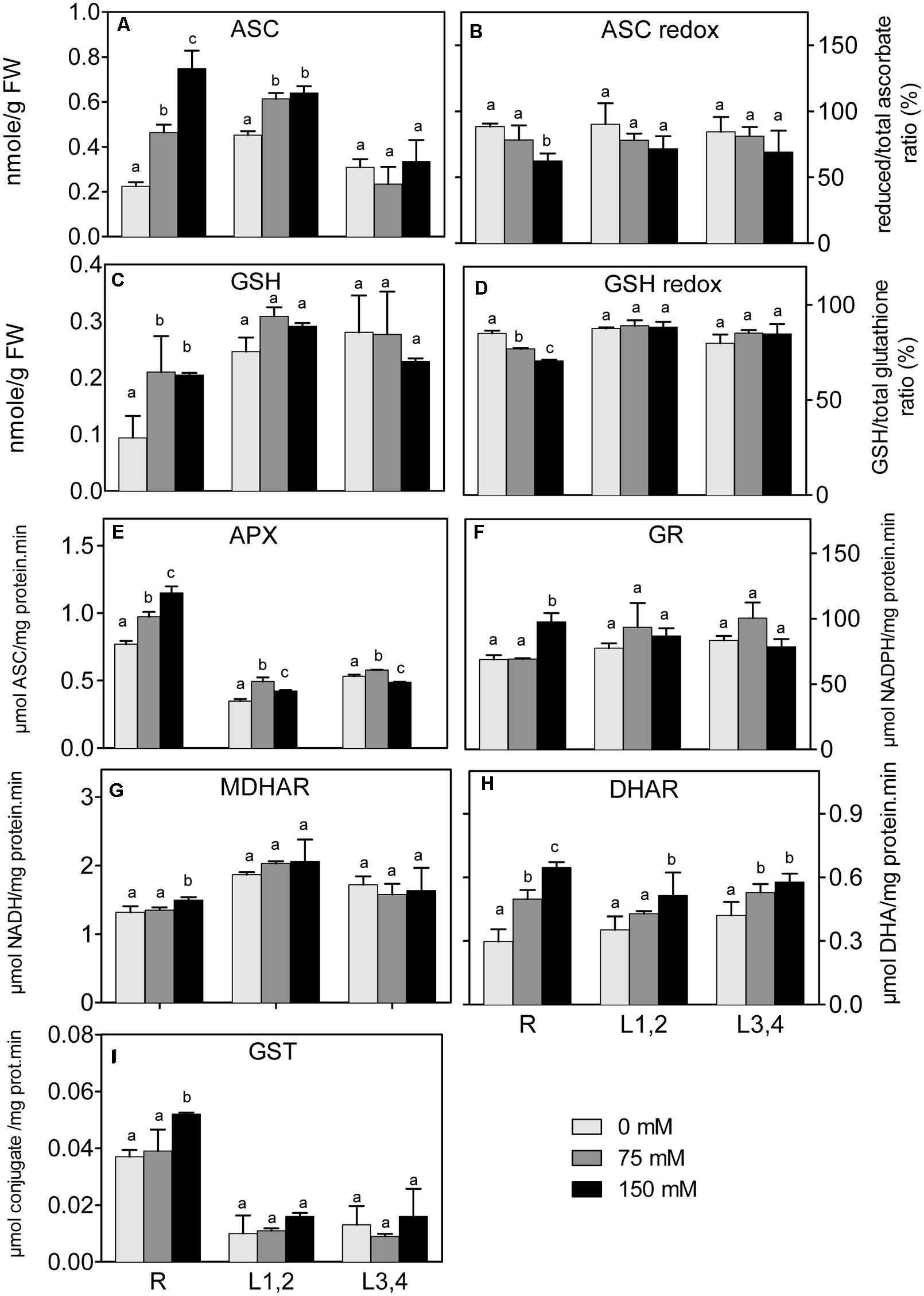
FIGURE 6. Effect of different salinity levels on reduced ascorbic acid (ASC, A), ascorbate redox status (reduced/total ascorbate, B), reduced glutathione (GSH, C), glutathione redox status (reduced/total glutathione ratio, D), ascorbate peroxidase (APX, E), glutathione reductase (GR, F), monodehydroascorbate reductase (MDHAR, G), dehydroascorbate reductase (DHAR, H), and glutathione-S-transferase (GST, I) activities of root (R), mature leaf pair (L1,2), and young leaf pair (L3,4) of 3 weeks old maize seedlings. Values are means of at least three replicates and significant differences between means, as determined by Duncan test (P < 0.05), are indicated by different letters.
Discussion
Soil salinity has emerged as a serious problem affecting crop productivity as well as distribution and survival of wild plants (Shabala et al., 2014). To contribute to our understanding of the mechanisms underlying salinity stress responses, we exposed maize seedlings to two different salinity levels, and compared responses in roots, mature leaves (L1,2) and young distal leaves (L3,4).
The results show that biomass (DW) of roots and older leaves (L1,2) decreased linearly with the increase in salinity, but biomass of young leaves (L3,4) was not affected by any level of salinity. This result may indicate lowered growth in roots and lowered CO2 assimilation in L1,2 but not L3,4 tissues was correlated to salinity. Growth inhibition in these organs also correlates linearly with increasing Na+ accumulation within tissues. Na+ and Cl- ions accumulation in leaf tissues could have resulted in stomatal closure and/or damage of photosynthetic machinery that in turn resulted in lower CO2 assimilation and lower growth and DW accumulation. A correlation between Na+ ion accumulation in root and shoot tissues and lowered DW was also previously observed. Pumpkin (Cucurbita pepo) and ground nut (Vigna subterranea) root and shoot length, FW and DW decreased in correlation to increasing salinity (Ambede et al., 2012; Kurum et al., 2013). Moreover, a decrease in chlorophyll content and photosynthetic efficiency under salinity stresses was observed in various plant species (Rout et al., 1997; Jamil et al., 2007; Ambede et al., 2012). Na+ ions accumulated in the roots of salt tolerant as well as salt sensitive green bean seedlings upon exposure to salinity stress. However, Na+ accumulated in the young leaves of salt sensitive genotype but in the older leaves of salt tolerant genotype (Yasar et al., 2006). The accumulation of Na+ in roots caused nutrient imbalance in the root tissues as indicated by decreased K+/Na+ ratio with the increase in salinity. The nutrient imbalance, in turn, contributed to lower growth and dry matter accumulation in root tissues. Similar results in maize has been found in salt sensitive maize (Hussain et al., 2014).
The accumulation of Na+ ions in roots and L1,2 was accompanied by increased levels of oxidative stress parameters, i.e., EL, MDA, and H2O2, as well as decreased GSH and ASC redox status in root tissues. High salinity has been reported to induce oxidative stress in different plants and tissues (Ashraf and Harris, 2004; Chawla et al., 2013). Under salinity stress, the level of ROS increases in the plant tissues as a result of irregularities in the electron transport chain and accumulation of photoreducing power. This excess of electrochemical energy can be dissipated through the Mehler reaction, resulting in ROS generation, including H2O2 (Asada, 1999), and damage of membranes, reflected in elevated EL and MDA levels (Pérez-López et al., 2009; Sharma et al., 2012; Chawla et al., 2013).
The results show that oxidative stress defense in maize under high salinity occurs through non-enzymatic and enzymatic mechanisms. Although there are common responses to salinity stress in different organs (e.g., CAT, DHAR, APX), there are also organ-specific reactions. Roots and older leaves (L1,2) responded by increasing polyphenol content and ASC, as non-enzymatic antioxidants, correlating with higher Na+ accumulation in these tissues. The accumulation of tocopherol in L1,2 and L3,4 but not in root tissues under salinity stress could be due to the specificity of tocopherols in scavenging singlet oxygen radicals in photosystem II (Kruk et al., 2005). Similarly, Hichem et al. (2009) found that salt-challenged maize plants accumulated higher polyphenols, but young leaves accumulated more polyphenols than older ones. ASC and tocopherols content have been shown to increase in leaves of tomato to protect them against oxidative stress under triazole treatment, as well as in wheat leaves under high salinity (Salama et al., 1994; Tuna, 2014). On the other hand, levels of ASC and tocopherols declined in de-etiolated rice leaves under salinity stress (Turan and Tripathy, 2013), these conflicting results could be due to plant/tissue specificity. At the level of organ specificity, our results also showed increased GSH level in response to high salinity that was root-specific. This could be ascribed to the increased demand and metabolism of sulfur under stress for biosynthesis of antioxidants such as GSH (Gill et al., 2013). Moreover, the root-located biosynthesis of the stress hormone ABA, which may act as a stress signal to the shoot (Bittner et al., 2001), increases under different abiotic stresses and requires sulfur supply. Upregulated expression of the sulfate transporter gene, AtSULTR3;1, and higher sulfate absorption are observed in salt-stressed Arabidopsis plants (Cao et al., 2014; Gallardo et al., 2014). The increased absorption of sulfate affects ABA and GSH levels in tissues under high salinity (Ruiz and Blumwald, 2002; Gill et al., 2013; Cao et al., 2014) and under drought stress (Chan et al., 2013; Gill et al., 2013). The increased content of ASC and GSH, accompanied by the reduced ASC and GSH redox status, indicates the crucial role of the ASC–GSH cycle for scavenging ROS especially in root tissues of maize seedlings. Similarly, Hernandez et al. (2000) have shown that the salinity tolerant, but not the sensitive variety, accumulated higher activities and transcripts of the ASC–GSH cycle.
At the level of enzymatic oxidative stress defenses, some responses to high salt appear to occur more general in roots and shoots (CAT, APX, and DHAR induction), whereas root-specific antioxidant enzymes (i.e., SOD, GR, and MDHAR) has been found. Yet other antioxidant enzymes, POX and MDHAR, were not affected by salinity stress in any of the tested organs. CAT, APX, and DHAR activities have been previously reported as general enzymatic antioxidant enzymes in different plant tissues and species, such as barley and rice (Pérez-López et al., 2009; Chawla et al., 2013). CAT has been shown by Gao et al. (2008) to be a major enzymatic antioxidant in radicles of Jatropha curcas L. challenged with salinity, especially under moderate salinity levels of 50 to 100 mM NaCl, compared to hypocotyl and cotyledons. Similarly, Chaparzadeh et al. (2004) showed increased CAT activity in leaves of Calendula offıcinalis under NaCl concentrations up to 100 mM, while MDHAR activity did not increase in either leaves or roots under salinity stress. Moreover, under sulfur deficiency, maize seedlings showed an increase in CAT activity in leaf sheaths and blades and decreased activity in roots while SOD activity decreased in roots and leaf sheaths but not in blades compared to their controls (Chorianopoulou et al., 2012) and these differential changes where ascribed to the differential ROS production in different organs under sulfur deficiency. Leaves of salinity tolerant rice cultivars showed higher activities of SOD and GR (Chawla et al., 2013). On the contrary, leaves of Calendula officinalis have lower SOD activity under salinity but enhanced GR activity compared to their control (Chaparzadeh et al., 2004).
Taken together, our results indicate that roots have a wide array of antioxidant metabolites and enzymes, mainly of the ASC–GSH cycle, that are upregulated in response to salinity stress. Tocopherol, on the other hand appears a more shoot-specific antioxidant in maize seedlings. Other antioxidant components did not show particular organ-specific increases, e.g., CAT, SOD, DHAR, and APX.
A root-specific increase in GST activities was observed under severe salinity. This increment in GST activity may be to detoxify secondary products of oxidative stress such as 4-hydroxynonenal, a toxic alkenal released due to oxidative damage of membranes (Edwards et al., 2000). Although GST is not strictly an antioxidant enzyme, rather it supports the detoxification of endobiotics and xenobiotics. Overexpression of a Tau-class GST in transgenic Arabidopsis and tobacco plants improved their oxidative and drought tolerance (Yu et al., 2003; Sharma et al., 2014).
Interestingly, Pro content did not change very much under any level of salinity in any tested organs. This is in contrast to many previous reports where Pro levels were increased under salinity stress to decrease the cellular water potential and improve water uptake, and possibly scavenge ROS molecules (Patel and Vora, 1985; Verslues and Sharma, 2010; Soshinkova et al., 2013). However, similar to our results Sorghum bicolor leaves showed no significant change in Pro content under salinity stress while in S. sudanense Pro decreased slightly with salinity (Pinho de Oliveira et al., 2013). Moreover, different maize varieties differed in Pro accumulation under salinity stress. Variety K3615.1 did not accumulate significant amounts of Pro under 50–100 mM NaCl, while K3653.2 showed decreased Pro content under elevated salinity, and other varieties accumulated large amounts of Pro upon NaCl treatment (Molazem and Bashirzadeh, 2015). The unchanged Pro levels under salinity stress in our study, could be due to the simultaneous upregulation of Pro synthesis and catabolizing enzymes (e.g., proline dehydrogenase) as a feedback response. Soshinkova et al. (2013) showed that upon addition of proline to the outer medium of cell suspensions, or to the nutrient solution of Thellungiella salsuginea, resulted in activation of proline dehydrogenase. It could also be ascribed to different thresholds of different maize varieties to synthesize proline under salinity stress. Moreover, the activation of enzymatic and non-enzymatic organic osmolytes/antioxidant defense molecules such as glycine betaine, ASC, GSH, SOD, CAT, etc. under salinity could be an alternative to proline to alleviate salt-induced damages.
These results indicate although there is universal antioxidant defense mechanisms apply for all tissues and developmental stages of the same plant. However, out of the arsenal of alternative antioxidant mechanisms available in all maize tissues, each organ uses mechanisms that are most efficient and in harmony with the other metabolic networks within that specific organ at the particular point of its age, to cope with the salinity and the resulting oxidative stress. Deciphering these specific alternatives could help developing more efficient metabolic engineering mechanisms specific to different organs and ages to cope with specific stress conditions. For example, engineering maize plants (var. Giza 119) with higher level of expression of key enzymes of ASC–GSH cycle in root, and synthesizing higher levels of tocopherols in shoots, and/or plants with reduced transport of Na+ from root to shoot, might help to cope with salinity stress at least at the seedling stage of the plant life.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abreu, I. A., Farinha, A. P., Negrão, S., Gonçalves, N., Fonseca, C., Rodrigues, M., et al. (2013). Coping with abiotic stress: proteome changes for crop improvement. J. Proteom. 93, 145–168. doi: 10.1016/j.jprot.2013.07.014
Aebi, H. (1984). “Catalase in vitro,” in Methods in Enzymology, ed. P. Lester (Cambridge, MA: Academic Press), 121–126.
Ambede, J. G., Netondo, G. W., Mwai, G. N., and Musyimi, D. M. (2012). NaCl salinity affects germination, growth, physiology, and biochemistry of bambara groundnut. Br. J. Plant Physiol. 24, 151–160. doi: 10.1590/S1677-04202012000300002
Apel, K., and Hirt, H. (2004). Reactive Oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Asada, K. (1999). THE water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. doi: 10.1146/annurev.arplant.50.1.601
Ashraf, M., and Harris, P. J. C. (2004). Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 166, 3–16. doi: 10.1016/j.plantsci.2003.10.024
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Benzie, I. F. F., and Strain, J. J. (1999). “Ferric reducing/antioxidant power assay: direct measure of total antioxidant activity of biological fluids and modified version for simultaneous measurement of total antioxidant power and ascorbic acid concentration,” in Methods in Enzymology, ed. P. Lester (Cambridge, CA: Academic Press), 15–27.
Bernstein, N., Meiri, A., and Zilberstaine, M. (2004). Root growth of avocado is more sensitive to salinity than shoot growth. J. Am. Soc. Hortic. Sci. 129, 188–192.
Bittner, F., Oreb, M., and Mendel, R. R. (2001). ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 276, 40381–40384. doi: 10.1074/jbc.C100472200
Cao, M.-J., Wang, Z., Zhao, Q., Mao, J.-L., Speiser, A., Wirtz, M., et al. (2014). Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J. 77, 604–615. doi: 10.1111/tpj.12407
Chan, K. X., Wirtz, M., Phua, S. Y., Estavillo, G. M., and Pogson, B. J. (2013). Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 18, 18–29. doi: 10.1016/j.tplants.2012.07.005
Chang, C., Yang, M., Wen, H., and Chern, J. (2002). Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug Anal. 10, 178–182.
Chaparzadeh, N., D’Amico, M. L., Khavari-Nejad, R. A., Izzo, R., and Navari-Izzo, F. (2004). Antioxidative responses of Calendula officinalis under salinity conditions. Plant Physiol. Biochem. 42, 695–701. doi: 10.1016/j.plaphy.2004.07.001
Chawla, S., Jain, S., and Jain, V. (2013). Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 22, 27–34. doi: 10.1007/s00709-011-0365-3
Chorianopoulou, S. N., Giamouroglou, M., and Bouranis, D. L. (2012). Differential early fluctuations in superoxide dismutase and catalase activities are included in the responses of young maize organs to s-deprivation. Am. J. Plant Sci. 3, 338–345. doi: 10.4236/ajps.2012.33040
de Oliveira, V. P., Marques, E. C., de Lacerda, C. F., Prisco, J. T., and Gomes-Filho, E. (2013). Physiological and biochemical characteristics of Sorghum bicolor and Sorghum sudanense subjected to salt stress in two stages of development. Afr. J. Agric. Res. 8, 660–670.
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Dhindsa, R. S., Plumb-Dhindsa, P. L., and Reid, D. M. (1982). Leaf senescence and lipid peroxidation: effects of some phytohormones, and scavengers of free radicals and singlet oxygen. Physiol. Plant. 56, 453–457. doi: 10.1111/j.1399-3054.1982.tb04539.x
Drotar, A., Phelps, P., and Fall, R. (1985). Evidence for glutathione peroxidase activities in cultured plant cells. Plant Sci. 42, 35–40. doi: 10.1016/0168-9452(85)90025-1
Edwards, R., Dixon, D. P., and Walbot, V. (2000). Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 5, 193–198. doi: 10.1016/S1360-1385(00)01601-0
Flowers, T. J., Gaur, P. M., Gowda, C. L., Krishnamurthy, L., Samineni, S., Siddique K. M., et al. (2010). Salt sensitivity in chickpea. Plant Cell Environ. 33, 490–509. doi: 10.1111/j.1365-3040.2009.02051.x
Foyer, C. H., and Noctor, G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18. doi: 10.1104/pp.110.167569
Gallardo, K., Courty, P.-E., Le Signor, C., Wipf, D., and Vernoud, V. (2014). Sulfate transporters in the plant’s response to drought and salinity: regulation and possible functions. Front. Plant Sci. 5:580. doi: 10.3389/fpls.2014.00580
Gao, S., Ouyang, C., Wang, S., Xu, Y., Tang, L., and Chen, F. (2008). Effects of salt stress on growth, antioxidant enzyme and phenylalanine ammonia-lyase activities in Jatropha curcas L. seedlings. Plant Soil Environ. 54, 374–381.
Gill, S. S., Anjum, N. A., Hasanuzzaman, M., Gill, R., Trivedi, D. K., Ahmad, I., et al. (2013). Glutathione and glutathione reductase: a boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 70, 204–212. doi: 10.1016/j.plaphy.2013.05.032
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J Genomics 2014:18. doi: 10.1155/2014/701596
Habig, W. H., Pabst, M. J., and Jakoby, W. B. (1974). Glutathione S-Transferases: the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249, 7130–7139.
Hamad, I., AbdElgawad, H., Al Jaouni, S., Zinta, G., Asard, H., Hassan, S., et al. (2015). Metabolic analysis of various date palm fruit (Phoenix dactylifera L.) cultivars from saudi arabia to assess their nutritional quality. Molecules 20:13620. doi: 10.3390/molecules200813620
Hernandez, J. A., Jimenez, A., Mullineaux, P., and Sevilla, F. (2000). Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell and Environ. 23, 853–862. doi: 10.1046/j.1365-3040.2000.00602.x
Hichem, H., Mounir, D., and Naceur, E. (2009). Differential responses of two maize (Zea mays L.) varieties to salt stress: changes on polyphenols composition of foliage and oxidative damages. Industrial Crops Prod. 30, 144–151. doi: 10.1016/j.indcrop.2009.03.003
Hodges, D. M., DeLong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1007/s004250050524
Holmström, K., Somersalo, S., Mandal, A., Palva, T., and Welin, B. (2000). Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J. Exp. Bot. 51, 177–185. doi: 10.1093/jexbot/51.343.177
Houle, G., Morel, L., Reynolds, C. E., and Siégel, J. (2001). The effect of salinity on different developmental stages of an endemic annual plant, Aster laurentianus (Asteraceae). Am. J. Bot. 88, 62–67. doi: 10.2307/2657127
Hussain, I., Ashraf, M. A., Anwar, F., Rasheed, R., Niaz, M., and Wahid, A. (2014). Biochemical characterization of maize (Zea mays L.) for salt tolerance. Plant Biosyst. 148, 1016–1026. doi: 10.1080/11263504.2013.798369
Jamil, M., Rehman, S., Lee, K. J., Kim, J. M., Kim, H., and Rha, E. S. (2007). Salinity reduced growth, PS2 photochemistry and chlorophyll content in radish. Sci. Agric. 64, 111–118. doi: 10.1590/S0103-90162007000200002
Jiang, Z.-Y., Woollard, A. C. S., and Wolff, S. P. (1990). Hydrogen peroxide production during experimental protein glycation. FEBS Lett. 268, 69–71. doi: 10.1016/0014-5793(90)80974-N
Jordan, W. R., Brown, K. W., and Thomas, J. C. (1975). Leaf age as a determinant in stomatal control of water loss from cotton during water stress. Plant Physiol. 56, 595–599. doi: 10.1104/pp.56.5.595
Karuppanapandian, T., Moon, J. C., Kim, C., Manoharan, K., and Kim, W. (2011). Reactive oxygen species in plants: their generation, signal transduction, and scavenging mechanisms. Austr. J. Crop Sci. 5, 709–725.
Kravchik, M., and Bernstein, N. (2013). Effects of salinity on the transcriptome of growing maize leaf cells point at cell-age specificity in the involvement of the antioxidative response in cell growth restriction. BMC Genomics 14:24. doi: 10.1186/1471-2164-14-24
Kruk, J., Holländer-Czytko, H., Oettmeier, W., and Trebst, A. (2005). Tocopherol as singlet oxygen scavenger in photosystem II. J. Plant Physiol. 162, 749–757. doi: 10.1016/j.jplph.2005.04.020
Kumar, K. B., and Khan, P. A. (1982). Peroxidase and polyphenol oxidase in excised ragi (Eleusine coracana cv PR 202) leaves during senescence [millets]. Ind. J. Exp. Bot. 20, e412–e416.
Kurum, R., Ulukapi, K., Aydinsakir, K., and Onus, A. N. (2013). The influence of salinity on seedling growth of some pumpkin varieties used as rootstock. Notulae Bot. Horti. Agrobot. Cluj Napoca 41, 219–225.
Lazof, D. B., and Bernstein, N. (1999). “The NaCl induced inhibition of shoot growth: the case for disturbed nutrition with special consideration of calcium,” in Advances in Botanical Research Incorporating Advances in Plant Pathology, Vol. 29, ed. J. A. Callow (Birmingham: University of Birmingham), 113–189.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the folin. Phenol Reagent J. Biol. Chem. 193, 265–275.
Lutts, S., Kinet, J. M., and Bouharmont, J. (1995). Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 46, 1843–1852. doi: 10.1093/jxb/46.12.1843
Mittova, V., Guy, M., Tal, M., and Volokita, M. (2004). Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J. Exp. Bot. 55, 1105–1113. doi: 10.1093/jxb/erh113
Molazem, D., and Bashirzadeh, A. (2015). Impact of salinity stress on proline reaction, peroxide activity, and antioxidant enzymes in maize (Zea mays L.). Polish J. Environ. Stud. 24, 597–603. doi: 10.1016/j.plaphy.2014.11.021
Munné-Bosch, S. (2005). The role of alpha-tocopherol in plant stress tolerance. J. Plant Physiol. 162, 743–748. doi: 10.1016/j.jplph.2005.04.022
Murshed, R., Lopez-Lauri, F., and Sallanon, H. (2008). Microplate quantification of enzymes of the plant ascorbate–glutathione cycle. Anal. Biochem. 383, 320–322. doi: 10.1016/j.ab.2008.07.020
Pardo, J. M., and Quintero, F. J. (2002). Plants and sodium ions: keeping company with the enemy. Genome Biol. 3, .1011–.1017. doi: 10.1186/gb-2002-3-6-reviews1017
Patel, J. A., and Vora, A. B. (1985). Free proline accumulation in drought-stressed plants. Plant Soil 84, 427–429. doi: 10.1093/treephys/tpv026
Pérez-López, U., Robredo, A., Lacuesta, M., Sgherri, C., Muñoz-Rueda, A., Navari-Izzo, F., et al. (2009). The oxidative stress caused by salinity in two barley cultivars is mitigated by elevated CO2. Physiol. Plant. 135, 29–42. doi: 10.1111/j.1399-3054.2008.01174.x
Potters, G., Horemans, N., Bellone, S., Caubergs, R., Trost, P., Guisez, Y., et al. (2004). Dehydroascorbate influences the plant cell cycle through a glutathione-independent reduction mechanism. Plant Physiol. 134, 1479–1487. doi: 10.1104/pp.103.033548
Pottosin, I., Bonales-Alatorre, E., and Shabala, S. (2014). Choline but not its derivative betaine blocks slow vacuolar channels in the halophyte Chenopodium quinoa: implications for salinity stress responses. FEBS Lett. 588, 3918–3923. doi: 10.1016/j.febslet.2014.09.003
Puniran-Hartley, N., Hartley, J., Shabala, L., and Shabala, S. (2014). Salinity-induced accumulation of organic osmolytes in barley and wheat leaves correlates with increased oxidative stress tolerance: in planta evidence for cross-tolerance. Plant Physiol. Biochem. 83, 32–39. doi: 10.1016/j.plaphy.2014.07.005
Qadir, M., Tubeileh, A., Akhtar, J., Larbi, A., Minhas, P. S., and Khan, M. A. (2008). Productivity enhancement of salt-affected environments through crop diversification. Land Degrad. Dev. 19, 429–453. doi: 10.1002/ldr.853
Qureshi, M., Abdin, M., Ahmad, J., and Iqbal, M. (2013). Effect of long-term salinity on cellular antioxidants, compatible solute and fatty acid profile of Sweet Annie (Artemisia annua L.). Phytochemistry 95, 215–223. doi: 10.1016/j.phytochem.2013.06.026
Rai, G. K., Rai, N. P., Rathaur, S., Kumar, S., and Singh, M. (2013). Expression of rd29A::AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol. Biochem. 69, 90–100. doi: 10.1016/j.plaphy.2013.05.002
Rakhmankulova, Z. F., Shuyskaya, E. V., Shcherbakov, A. V., Fedyaev, V. V., Biktimerova, G. Y., Khafisova, R. R., et al. (2015). Content of proline and flavonoids in the shoots of halophytes inhabiting the South Urals. Russian J. Plant Physiol. 62, 71–79. doi: 10.1134/S1021443715010112
Rout, N. P., Tripathi, S. B., and Shaw, B. P. (1997). Effect of salinity on chlorophyll and proline contents in three aquatic macrophytes. Biol. Plant. 40, 453–458. doi: 10.1023/A:1000984115298
Ruiz, J. M., and Blumwald, E. (2002). Salinity-induced glutathione synthesis in Brassica napus. Planta 214, 965–969. doi: 10.1007/s00425-002-0748-y
Salama, S., Trivedi, S., Busheva, M., Arafa, A. A., Garab, G., and Erdei, L. (1994). Effects of NaCl salinity on growth, cation accumulation, chloroplast structure and function in wheat cultivars differing in salt tolerance. J. Plant Physiol. 144, 241–247. doi: 10.1016/S0176-1617(11)80550-X
Schachtman, D., and Liu, W. (1999). Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 4, 281–287. doi: 10.1016/S1360-1385(99)01428-4
Shabala, S., Bose, J., and Hedrich, R. (2014). Salt bladders: do they matter? Trends Plant Sci. 19, 687–691. doi: 10.1016/j.tplants.2014.09.001
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012:26. doi: 10.1155/2012/217037
Sharma, R., Sahoo, A., Devendran, R., and Jain, M. (2014). Over-expression of a rice tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS ONE 9:e92900. doi: 10.1371/journal.pone.0092900
Soshinkova, T. N., Radyukina, N. L., Korolkova, D. V., and Nosov, A. V. (2013). Proline and functioning of the antioxidant system in Thellungiella salsuginea plants and cultured cells subjected to oxidative stress. Russian J. Plant Physiol. 60, 41–54. doi: 10.1134/S1021443713010093
Talbi, S., Romero-Puertas, M. C., Hernández, A., Terrón, L., Ferchichi, A., and Sandalio, L. M. (2015). Drought tolerance in a saharian plant oudneya africana: role of antioxidant defences. Environ. Exp. Bot. 111, 114–126. doi: 10.1016/j.envexpbot.2014.11.004
Tuna, A. L. (2014). Influence of foliarly applied different triazole compounds on growth, nutrition, and antioxidant enzyme activities in tomato (‘Solanum lycopersicum’ L.) under salt stress. Austr. J. Crop Sci. 8, 71–79.
Turan, S., and Tripathy, B. C. (2013). Salt and genotype impact on antioxidative enzymes and lipid peroxidation in two rice cultivars during de-etiolation. Protoplasma 250, 209–222. doi: 10.1007/s00709-012-0395-5
Verslues, P. E., and Sharma, S. (2010). Proline metabolism and its implications for plant-environment interaction. Arabidopsis Book 8:e0140. doi: 10.1199/tab.0140
Viehweger, K. (2014). How plants cope with heavy metals. Bot. Stud. 55, 1–12. doi: 10.1186/1999-3110-55-35
Yasar, F., Uzal, O., Tufenkci, S., and Yildiz, K. (2006). Ion accumulation in different organs of green bean genotypes grown under salt stress. Plant Soil Environ. 52, 476–480.
Yu, T. A. O., Li, Y. S., Chen, X. F., Hu, J., Chang, X. U. N., and Zhu, Y. G. (2003). Transgenic tobacco plants overexpressing cotton glutathione S-transferase (GST) show enhanced resistance to methyl viologen. J. Plant Physiol. 160, 1305–1311. doi: 10.1078/0176-1617-01205
Keywords: salinity, maize, biomass, reactive oxygen species (ROS), oxidative stress, antioxidants
Citation: AbdElgawad H, Zinta G, Hegab MM, Pandey R, Asard H and Abuelsoud W (2016) High Salinity Induces Different Oxidative Stress and Antioxidant Responses in Maize Seedlings Organs. Front. Plant Sci. 7:276. doi: 10.3389/fpls.2016.00276
Received: 27 November 2015; Accepted: 21 February 2016;
Published: 08 March 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Axel Tiessen, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoDevesh Shukla, Western Kentucky University, USA
Copyright © 2016 AbdElgawad, Zinta, Hegab, Pandey, Asard and Abuelsoud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hamada AbdElgawad, aGFtYWRhLmFiZGVsZ2F3YWRAdWFudHdlcnBlbi5iZQ==; Walid Abuelsoud, d2FsaWRhYmNAc2NpLmN1LmVkdS5lZw==
†Present address: Gaurav Zinta, Shanghai Center for Plant Stress Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
‡These authors are first authors.
 Hamada AbdElgawad
Hamada AbdElgawad Gaurav Zinta
Gaurav Zinta Momtaz M. Hegab
Momtaz M. Hegab Renu Pandey
Renu Pandey Han Asard
Han Asard Walid Abuelsoud
Walid Abuelsoud