- Departamento de Caracterización, Calidad y Seguridad, Instituto de Ciencia y Tecnología de Alimentos y Nutrición, ICTAN-CSIC, Ciudad Universitaria de Madrid, Madrid, Spain
C-repeat/dehydration-responsive element binding factors (CBF/DREB) are transcription factors which play a role in improving plant cold stress resistance and recognize the DRE/CRT element in the promoter of a set of cold regulated genes. Dehydrins (DHNs) are proteins that accumulate in plants in response to cold stress, which present, in some cases, CBF/DREB recognition sequences in their promoters and are activated by members of this transcription factor family. The application of a 3-day gaseous treatment with 20 kPa CO2 at 0∘C to table grapes cv. Autumn Royal maintained the quality of the bunches during postharvest storage at 0∘C, reducing weight loss and rachis browning. In order to determine the role of CBF/DREB genes in the beneficial effect of the gaseous treatment by regulating DHNs, we have analyzed the gene expression pattern of three VviDREBA1s (VviDREBA1-1, VviDREBA1-6, and VviDREBA1-7) as well as three VviDHNs (VviDHN1a, VviDHN2, and VviDHN4), in both alternative splicing forms. Results showed that the differences in VviDREBA1s expression were tissue and atmosphere composition dependent, although the application of high levels of CO2 caused a greater increase of VviDREBA1-1 in the skin, VviDREBA1-6 in the pulp and VviDREBA1-7 in the skin and pulp. Likewise, the application of high levels of CO2 regulated the retention of introns in the transcripts of the dehydrins studied in the different tissues analyzed. The DHNs promoter analysis showed that VviDHN2 presented the cis-acting DRE and CRT elements, whereas VviDHN1a presented only the DRE motif. Our electrophoretic mobility shift assays (EMSA) showed that VviDREBA1-1 was the only transcription factor that had in vitro binding capacity to the CRT element of the VviDHN2 promoter region, indicating that the transcriptional regulation of VviDHN1a and VviDHN4 would be carried out by activating other independent routes of these transcription factors. Our results suggest that the application of high CO2 levels to maintain table grape quality during storage at 0∘C, leads to an activation of CBF/DREBs transcription factors. Among these factors, VviDREBA1-1 seems to participate in the transcriptional activation of VviDHN2 via CRT binding, with the unspliced form of this DHN being activated by high CO2 levels in all the tissues analyzed.
Introduction
Storage temperature is one of the most important factors which affects postharvest fruit quality. Accordingly, low temperature is the most common abiotic stress applied to improve fruit quality during postharvest storage. To cope with low temperature, fruit have developed adaptive mechanisms, involving physiological and biochemical processes together with transcriptomic modifications, including regulatory and functional genes (Maul et al., 2008; Genero et al., 2016; Rosales et al., 2016). Among the regulatory genes, transcription factors play an important role in plant stress responses, acting as coordinators of stress signals and orchestrating the expression of functional genes (Singh et al., 2002; Wang et al., 2016). It is well known that C-repeat/dehydration-responsive element binding factors (CBF/DREB) are transcription factors which are involved in improving cold stress resistance (Stockinger et al., 1997; reviewed by Chew and Halliday, 2011). CBF/DREB proteins, which belong to the subgroup A1 of the DREB proteins and are members of the AP2/ERF (APETALA2/Ethylene-Responsive Factor) transcription factor superfamily, are able to recognize and bind to the DRE/CRT (Dehydration Responsive Element/C-Repeat) DNA regulatory motif in the promoters of many cold-responsive (COR) genes (Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997). In this respect, dehydrins (DHNs), a subgroup of LEA (Late Embryogenesis Abundant) proteins, are among the most commonly observed proteins which accumulate in plants in response to low temperature and environmental factors leading to cellular dehydration. The presence of DRE/CRT motifs in the promoter of several cold-regulated DHNs suggests that they play a role in the CBF/DREB-mediated signaling pathway (Gilmour et al., 2004; Wisniewski et al., 2006).
CBF/DREB transcription factors were first isolated from Arabidopsis (Stockinger et al., 1997; Medina et al., 1999; Haake et al., 2002) and thereafter they have been isolated from different woody species including almond (Barros et al., 2012), dwarf apple (Yang et al., 2011), sweet cherry (Kitashiba et al., 2004), and trifoliate orange (He et al., 2012). The overexpression of CBF/DREBs increased the tolerance to cold stress in Arabidopsis (Novillo et al., 2007), potato (Pino et al., 2008), and bilberry (Oakenfull et al., 2013). Thus, CBF/DREBs form part of the group of transcription factors identified as the first wave of cold-induced genes (Chinnusamy et al., 2007; Thomashow, 2010; Zhao and Zhu, 2016). The constitutive overexpression of CBFs in Arabidopsis activated the expression of DRE/CRT-containing target genes (including those that encode DHNs proteins such as COR47 and ERD10, or LEA proteins like COR15a) under normal growing conditions and enhanced freezing tolerance in the absence of a cold stimulus (Jaglo-Ottosen et al., 1998; Gilmour et al., 2004).
There is no doubt that CBF/DREB transcription factors are vital for plants to overcome cold stress, but little is known regarding how important they are for fruit. Storage of peach fruit at a chilling injury-delaying temperature (0°C) resulted in a greater accumulation of PpCBF1/5/6 transcripts than in fruit treated with a chilling injury-inducing temperature (5°C) (Liang et al., 2013). Zhang et al. (2017) indicated that several postharvest treatments, including ethylene (Zhao et al., 2009) and nitric oxide (Zhao et al., 2011), as well as hot water (Ma et al., 2014), triggered CBF expression in tomato fruit in the two first cases, and in kiwifruit in the latter, which enhanced cold tolerance during low temperature storage.
Grape (Vitis sp.) is one of the most economically important fruit crops in the world. Most of the studies on Vitis, which have deciphered the effect of CBFs on cold tolerance have been carried out with grapevine seedlings, and there is little information about their role in bunches. Thus, an increase in CBF3 and CBF4 expression was observed in the leaves of Vitis riparia and V. vinifera after 1–2 days at 4°C (Xiao et al., 2006, 2008), contrasting with the quick cold-induction observed in the case of CBF1 and CBF2 (Xiao et al., 2006). Karimi et al. (2015) compared the response of plants of one cultivar of V. riparia and two cultivars of V. vinifera to cold stress, and denoted that V. riparia which is endemic to cold regions, behaved stronger after 10 days of exposure to 4°C and showed higher expression for all the CBFs analyzed. Overexpression of VvCBF4 in V. vinifera cv. Freedom improved freezing survival and reduced freezing-induced electrolyte leakage by up to 2°C in non-cold-acclimated vines (Tillett et al., 2012). Likewise, overexpression of CBF transcription factors from V. vinifera cv. Koshu (Takuhara et al., 2011) or V. riparia (Siddiqua and Nassuth, 2011) in Arabidopsis improved the freezing tolerance of transgenic plants. Furthermore, the results from Siddiqua and Nassuth (2011) indicated that the Arabidopsis lines that overexpressed VrCBF1 or VrCBF4 showed an increase in the accumulation of COR genes, including dehydrin (COR47) and LEA_4 (COR15a), which have at least one DRE/CRT motif in their promoter (Kasuga et al., 1999).
The application of a 3-day gaseous treatment with 20 kPa CO2 at 0°C maintained the quality of red table grapes cv. Cardinal during postharvest storage at 0°C by reducing total decay and rachis browning (Sanchez-Ballesta et al., 2006; Rosales et al., 2016). Although table grapes are classified as chilling-tolerant, we have pointed out that the gaseous treatment help table grapes to face temperature shifts at 0°C (Sanchez-Ballesta et al., 2006) and thus CO2-treated berries reached a low ion leakage value during storage at 0°C in comparison to non-treated ones (Rosales et al., 2016). In a previous study, we observed that application of high CO2 levels at 0°C in table grapes cv. Cardinal for 3 days induced the expression of CBF1 and CBF4 (VviDREBA1-1 in this work) in the pulp and CBF4 in the rachis (Fernandez-Caballero et al., 2012). In a more recent work, we suggested that the beneficial effect of a high CO2 treatment to maintain Cardinal table grape quality seems to be mediated by the regulation of ERFs (Romero et al., 2016), which belong to the AP2/ERF transcription factor family. In the present study, we have analyzed the effect of a 3-day CO2 treatment in maintaining the table grape quality of a cultivar which is different from Cardinal, such as Autumn Royal. Unlike red-skinned Cardinal table grape cultivar which matures early (late spring to mid-summer) and presents very few seeds, Autumn Royal is a late-maturing seedless table grape cultivar (from autumn through early winter), with a purple-black to black berry skin. Furthermore, to investigate whether the role played by CBF/DREB transcription factors is cultivar dependent and/or a common feature of other DREBA1s, we have analyzed the transcript accumulation pattern of VviDREBA1-1, VviDREBA1-6, and VviDREBA1-7 in different tissues (skin, pulp, and rachis) of Autumn Royal bunches which were treated and non-treated with 20 kPa CO2 for 3 days at 0°C and then transferred to air for up to 13 days. Likewise, to study the role of CBF/DREBs in the regulation of DHNs in table grapes we first analyzed the pattern of expression of three dehydrins (VviDHN1a, VviDHN2, and VviDHN4) in different tissues of Autumn Royal bunches which were CO2-treated and non-treated; and we then analyzed their promoters so as to identify different cis-regulatory elements, including the DRE/CRT motifs. Finally, we examined the DNA-binding specificities of three CBF/DREBs, by using electrophoretic mobility shift assay (EMSA), showing that only VviDREBA1-1 was able to bind in vitro to the CRT element present in the VviDHN2 promoter.
Materials and Methods
Plant Material
Mature table grapes (V. vinifera L. cv. Autumn Royal) (12.87% total soluble solids; 0.46% tartaric acid) were harvested from a commercial orchard located in Abarán, Murcia, Spain (latitude: 38° 12′ 00′′ N; longitude: 01° 24′ 00′′ W) in November 2013. After random harvesting, field-packaged bunches were transported in the same day to the laboratory in Madrid (Spain), where bunches without physical and pathological defects were divided arbitrarily into two lots and stored at 0 ± 0.5°C with 95% relative humidity in two sealed methacrylate containers of 1 m3 capacity. One lot was stored under normal atmosphere for up to 41 days (non-treated fruit) and the second one was kept under a gas mixture containing 20 kPa CO2 + 20 kPa O2 + 60 kPa N2 (CO2-treated fruit) for 3 days. Thereafter, CO2-treated table bunches were transferred to air under the same conditions as non-treated ones until the end of the storage period. At time 0 and after 3 and 13 days of storage under air or CO2 conditions, skin, pulp and rachis from three biological replicates (each replicate consisting of 2 bunches) were collected independently, frozen in liquid nitrogen, grounded to a fine powder and stored at -80°C until analysis.
Quality Assessments
Berry quality assessment comprised soluble solids contents (SSC), titratable acidity (TA), pH, weight loss of bunches and rachis browning. SSC was determined using a digital refractometer Atago PR-101 (Atago Co. Ltd., Japan) at 20°C and expressed in °Brix. TA was determined by titration with 0.1 N NaOH up to pH 8.1 and results were expressed in % tartaric acid. The pH of the juice was measured using a pH meter with a glass electrode. The moisture content of berries was determined when a stable weight had been obtained after drying the fruit at 105°C, and it was expressed as g/100 g fresh weight (FW). The weight of the bunches was recorded on the day of harvest and after the different sampling dates. Cumulative weight losses were expressed as a percentage loss of the original weight. Rachis browning was determined using the following subjective scale: (0) none (rachis including pedicels, green-bright), slight (1) (rachis in good conditions and pedicels, green–gray), (2) moderate (secondary rachis and pedicels, green–brown), (3) intense (secondary rachis and pedicels, brown, and primary rachis, green with brown areas), (4) severe (pedicels, primary and secondary rachis, brown).
RNA Extraction, cDNA Synthesis and RT-PCR
For each sample, total RNA was extracted three times according to Zeng and Yang (2002), and treated with DNase I recombinant-RNase free (Roche) for genomic DNA removal. Concentration and purity of the total RNA samples were measured using the NanoDropTM 1000 Spectrophotometer (NanoDrop Technologies, Inc. Wilmington, DE, United States). Then, 1 μg of each extraction was used to synthesize cDNA by using the iScriptTM Reverse Transcription Supermix (Bio-Rad), according to the manufacturer’s instructions. VviDREBA1s full-length sequences were obtained by RT-PCR as described by Romero et al. (2008) using specific primers (Supplementary Table S1). Genomic DNA, obtained from leaves of V. vinifera cv. Autumn Royal as described by Lodhi et al. (1994) was used as template. PCR fragments were cloned and confirmed by sequencing as described by Romero et al. (2016).
Bioinformatic Tools
The V. vinifera CBF/DREBs previously identified (Xiao et al., 2006, 2008; Zhuang et al., 2009; Licausi et al., 2010; Cramer et al., 2014; Zhao et al., 2014; Carlow et al., 2017) were used to run a search in the 12X grape reference genome, V2.1 gene prediction hosted at Grape Genome Database (CRIBI1) (Vitulo et al., 2014). The BLASTP and BLASTX suites were used to perform similarity searches in the predicted proteome database. Protein sequence identity between the closest VviDREBA1s homologs were performed by using the LALIGN program2.
Multiple alignment analysis was performed at the Swiss EMBnet node web server using ClustalW and BoxShade tools, respectively. The prediction of ubiquitination and sumoylation sites was performed using, UbPred3 (Radivojac et al., 2010) and SUMOsp2.0 servers4 (Zhao et al., 2014), respectively. The proline (P) glutamic acid (E) serine (S) threonine (T) (PEST) regions in the proteins were found using the EMBOSS program pestfind5 with the default cut-off PEST score of 5.0 (Rice et al., 2000). Hydrophobic cluster analysis (HCA) was performed using online tools at http://bioserv.rpbs.univ-paris-diderot.fr/services/HCA/ (Gaboriaud et al., 1987).
Relative Gene Expression by Quantitative and Semi-Quantitative RT-PCR
Relative expression of VviDREBA1s as well as spliced and unspliced transcripts of VviDHN4 were assayed using quantitative RT-PCR (RT-qPCR) with samples of skin, pulp, and rachis from CO2-treated and non-treated bunches stored for 0, 3, and 13 days at 0°C. RT-qPCR was performed as described by Rosales et al. (2013), using gene-specific primer pairs (Supplementary Table S1). Actin1 gene from V. vinifera (ACT1: XM 002282480) was used as the internal reference gene for normalizing the transcript profiles following the 2-ΔΔCt method relative to a calibrator sample (day 0, bunches before storage). The specificity of products was validated by dissociation curve analysis and by agarose gel; and its sequences confirmed at the Genomic Department of the CIB-CSIC (Madrid, Spain).
Spliced and unspliced variants of VviDHN1a and VviDHN2 were evaluated by semi-quantitative RT-PCR as described by Navarro et al. (2015). Following amplification, products were visualized by electrophoresis in a 2% agarose gel stained with Goldview (Guangzhou Geneshun Biotech Ltd.). The identification of each PCR product was then confirmed by Sanger sequencing at the Genomic Department of the CIB-CSIC (Madrid, Spain).
Identification of Putative Cis-Regulatory Elements
Identification of the potential VviDHN2 and VviDHN4 promoter regions and transcription factor binding sites was conducted using the Genomatix suite of programs6 (Genomatix Software GmbH, Munich, Germany). The Gene2promoter program from the Genomatix software package was used to define 1500 bp upstream of the transcription start site of VviDHN2 and VviDHN4 promoter regions. The corresponding sequences were then used as the target sequences for putative transcription factor recognition site identification using the MatInspector Version 8.3 program (Cartharius et al., 2005) with the standard (0.75) core similarity and the optimized matrix similarity. DRE regulatory element from VviDHN1a promoter was previously identified (Rosales et al., 2014).
Production of the Recombinant VviDREBA1s Proteins in Escherichia coli
The full VviDREBA1s [GenBank Accession No. MF445007 (VviDREBA1-1), MF445008 (VviDREBA1-6) and MF445009 (VviDREBA1-7)] open reading frames (ORFs), including stop codons, were amplified by RT-PCR using the primers included in the Supplementary Table S1. The forward VviDREBA1-7 and VviDREBA1-1 primers contained a BamHI site, and the forward VviDREBA1-6 primer contained a XhoI site. The three reverse primers contained an EcoRI site. The resulting fragments digested with their respective restriction enzymes were cloned into the pTrcHisA vector, which contains an N-terminal His6 (Invitrogen, Carlsbad, CA, United States), previously digested with the same enzymes, and transformed into BL21-CodonPlus (DE3)-RIL competent cells. The induction and purification of recombinant proteins were performed according to Romero et al. (2008). The purified fusion proteins were concentrated as described by Romero et al. (2016). Protein analyses were performed on 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) using Mini-Protean II Cell (Bio-Rad) equipment as described by Rosales et al. (2014). Western blots were probed with antibodies and conditions previously described by Romero et al. (2016).
Electrophoretic Mobility Gel Shift Assay (EMSA) Assays
Purified VviDREBA1s recombinant proteins were used to determine DNA binding by EMSA as previously described by Romero et al. (2016). DRE/CRT motifs were synthetized and used as probes, which were biotin-labeled using the Biotin 3′ End DNA Labeling Kit (Thermo Scientific Pierce).
Statistical Analyses
The data were analyzed by ANOVA (one-way analysis of variance) and Duncan’s multiple range test was used (IBM Corp. SPSS Statistics version 22.0., Armonk, NY, United States). Statistical significance was assessed at the level P ≤ 0.05.
Results
Effect of a 3 Day-CO2 Treatment on the Quality of Table Grapes cv. Autumn Royal Stored at 0°C
The application of high CO2 levels for 3 days did not affect the SSC and TA values after 13 days of cold storage, in comparison to non-treated grapes (Table 1). Likewise, the maturity index (SSC/TA) in freshly harvested fruit (27.87) increased in both non-treated and CO2-treated grapes after 13 days of cold storage, due to the increase observed in the SSC content, reaching values of 29.63 and 29.06, respectively. By contrast, whereas the pH values did not change in non-treated fruit, a significant increase was observed in CO2-treated grapes, decreasing by day 13. Regarding the moisture content of berries, it decreased significantly during storage at 0°C in all the samples analyzed in comparison to freshly harvested fruit, although the major decrease was observed in non-treated samples after 13 days of cold storage. The CO2 treatment was effective in controlling the weight loss of bunches, as well as the rachis browning observed in non-treated table grapes. After 3 days, the percentage of weight loss and the rachis browning index were significantly lower in CO2-treated bunches, but the effect was only maintained after 13 days in the case of weight loss. By contrast, although the rachis browning index was also lower in CO2-treated bunches after 13 days at 0°C, the differences were not significant.
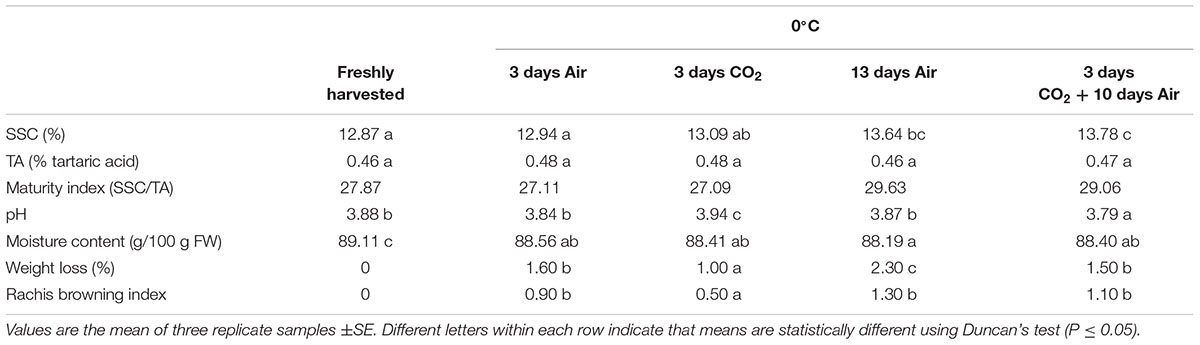
TABLE 1. Soluble solids content (SSC), titratable acidity (TA), pH, moisture content, weight loss and rachis browning index of table grapes cv. Autumn Royal treated with 20 kPa CO2 and stored during 13 days at 0°C.
Characterization of CBF/DREB Transcription Factors in Table Grapes
As a first step to characterize CBF/DREBs transcription factors, which belong to A1 subgroup, and because up to five different nomenclatures have been attributed (Table 2) for the different CBF/DREBs previously identified in V. vinifera (Xiao et al., 2006, 2008; Zhuang et al., 2009; Licausi et al., 2010; Cramer et al., 2014; Zhao et al., 2014; Carlow et al., 2017), we used the coding sequences of all of them to conduct a search in the 12X grape reference genome, V2.1 gene prediction (Vitulo et al., 2014). Six non-redundant CBF/DREBs transcription factors (VIT_216s0100g00380, VIT_202s0025g04460, VIT_211s0016g02140, VIT_206s0061g01440, VIT_206s0061g01390, VIT_208s0007g03790) were identified (Table 2). It is important to note that CBF1 (Xiao et al., 2006) was not found in the 12X grape genome database. According to the nomenclature system developed by the International Grape Genome Program (IGGP) Supernomenclature committee (Grimplet et al., 2014), we decided to rename the different CBF/DREBs in the same way as the one proposed by Zhuang et al. (2009), since it refers to the family (DREB) and subgroup (A1) (Table 2). Taking into account that CBF2 (VviDREBA1-7 in this work), CBF3 (VviDREBA1-6) and CBF4 (VviDREBA1-1) have been extensively studied in grapevine cuttings exposed to low temperature, and that CBF4 expression was modulated by high CO2 levels in Cardinal table grapes (Fernandez-Caballero et al., 2012), we have isolated them from table grapes cv. Autumn Royal using RT-PCR. VviDREBA1-1 consisted of 218 aa and shared 100% identity with VIT_216s0100g00380 and with VvCBF4 isolated from V. vinifera cv. Chardonnay (Xiao et al., 2008). VviDREBA1-6 (with 239 amino acids) was 83.3% identical with VIT_206s0061g01400 and the differences observed between them consisted of 10 variations of single amino acids and a protein fragment deletion of 28 amino acids in VIT_206s0061g01400. The percentage of identity of VviDREBA1-6 with VvCBF3 from V. vinifera cv. Chardonnay (Xiao et al., 2006) was 95.3, with differences in twelve amino acids. VviDREBA1-7 (253 amino acids) shared 99.6% identity with VIT_206s0061g01390, the difference being a variation of a single amino acid. Moreover, VviDREBA1-7 showed a 94.9% of identity with VvCBF2 (Xiao et al., 2006), presenting differences in 12 amino acids. It is important to point out that VviDREBA1-1 and VviDREBA1-6 presented an acidic predicted pI of 5.42 and 6.53, respectively, as occurs in dicot CBFs. However, VviDREBA1-7 showed a basic predicted pI of 9.71. Despite the pI difference, the putative activation domain at the C-terminus of the VviDREBA1-7 maintained a strongly acidic character with a pI of approximately 4.89, like the dicot CBFs.
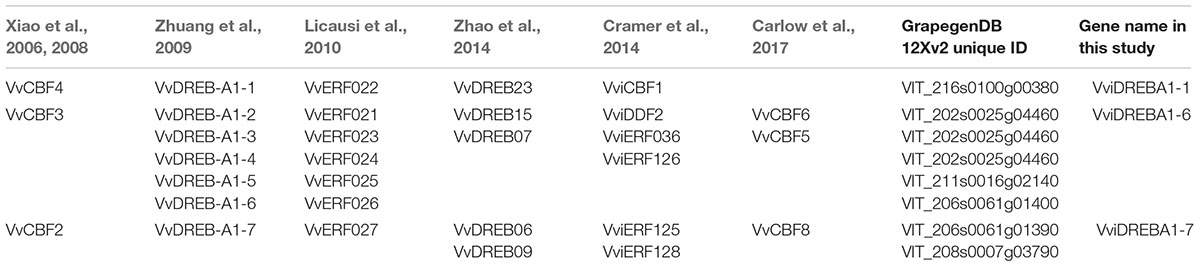
TABLE 2. Summary of the Vitis vinifera CBF/DREB gene names designated in this study and in previous works.
Multiple sequence analysis showed that all deduced proteins exhibited the typical features of DREBA1 proteins, including an AP2 domain with the YRG, WLG, and RAHD motifs (Supplementary Figure S1). Moreover, these VviDREBA1 proteins also had the conserved valine (V14) in the 14th position in the AP2 domain. However, the conserved glutamic acid (E19) in the 19th position was replaced in VviDREBA1-6 by aspartic acid (D), also negative charged. The AP2 domain displayed a higher degree of amino acid identity than the N-termini and C-termini of the VviDREBA1 proteins. PEST motifs were predicted in the three VviDREBA1s in the N-terminal end close to the AP2 domain (Figure 1). At the C-terminal, a relevant PEST motif (more than 20 residues) was also predicted for VviDREBA1-6.
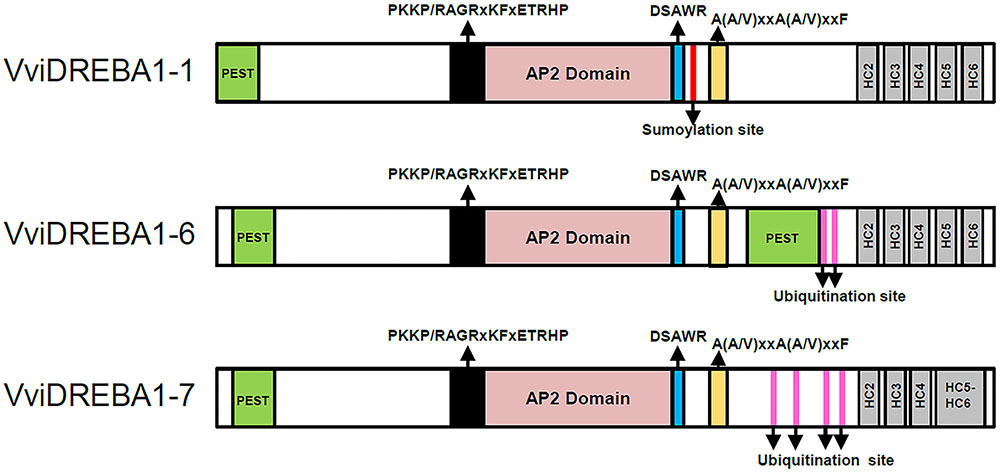
FIGURE 1. Schematic diagram of the three VviDREBA1s from Vitis vinifera cv. Autumn Royal showing conserved domains. The PEST sequences (green), the PKKPAGR motif (black), the AP2 domain (pink), the DSAWRL motif (blue), and the A(A/V)xxA(A/V)xxF motif (orange) are present in the three transcription factors. The hydrophobic clusters (HC2-HC6) (gray) which were present in the carboxy terminal region are indicated. A sumoylation target site (red line) was present in VviDREBA1-1 and ubiquitination sites (fuchsia lines) were present in VviDREBA1-6 and VviDREBA1-7.
The DREBA1 signature sequences PKKP/RAGRxKFxETRHP (abbreviated PKKPAGR) and DSAWR, located immediately upstream and downstream of the AP2 domain, respectively (Jaglo et al., 2001), were also observed in the VviDREBA1 proteins (Figure 1 and Supplementary Figure S1). However, only VviDREBA1-1 contained the conserved PKKPAGR consensus. Likewise, the domain A(A/V)xxA(A/V)xxF, with the underlined residues conserved in all known DREBA1 homologs (Xiong and Fei, 2006), was located in the downstream of the DSAWR motif in the three VviDREBA1s. However, the C-terminal LWSY motif (Dubouzet et al., 2003) was only conserved in the VviDREBA1-1 protein.
The hydrophobic cluster analysis of the C-terminus from VviDREBA1-1 and VviDREBA1-6 showed five hydrophobic clusters (HC2-HC6) (Supplementary Figure S2), which are known to be responsible for conferring trans-activation (Wang et al., 2005). By contrast, the analysis of VviDREBA1-7 revealed that HC5 and HC6 form a ‘mosaic cluster,’ which contained regular alternations of hydrophobic and non-hydrophobic residues, indicated as connecting lines.
The post-translational modifications were also evaluated through the prediction of ubiquitination and sumoylation sites associated with protein degradation and stability, respectively. A unique sumoylation site was predicted in VviDREBA1-1. However, two and four ubiquitination sites were predicted in VviDREBA1-6 and VviDREBA1-7, respectively.
Expression Profile of VviDREBA1s Transcription Factors in Different Tissues of Autumn Royal Bunches Exposed to Low Temperature and High CO2 Levels
In a previous work, we denoted that the application of CO2 levels at 0°C in table grapes cv. Cardinal for 3 days induced the transcript accumulation of VvcCBF1 in the pulp, and VvcCBF4 (VviDREBA1-1 in this work) in the pulp and rachis (Fernandez-Caballero et al., 2012). To study whether this effect is cultivar dependent and could be extensible to other VviDREBA1s, we analyzed the transcript accumulation of VviDREBA1-1, VviDREBA1-6, and VviDREBA1-7 in different tissues of cv. Autumn Royal bunches through RT-qPCR.
The storage at low temperature of table grapes cv. Autumn Royal induced different changes in VviDREBA1s expression according to the tissue analyzed and the atmosphere composition (Figure 2). Thus, in the case of the skin, only the application of a 3-day gaseous treatment at 0°C significantly induced the expression of VviDREBA1-1 and VviDREBA1-7, decreasing when table grapes were transferred to a normal atmosphere at 0°C. However, a slight but not significant accumulation of VviDREBA1-6 transcript was observed in this tissue at 0°C regardless of the atmosphere composition. In the case of the pulp, whereas VviDREBA1-6 and VviDREBA1-7 transcript levels were induced by the application of a 3-day gaseous treatment, in the case of VviDREBA1-1 an increase in the transcript accumulation was only observed after 13 days of storage of non-treated table grapes at 0°C. In the rachis, unlike in the skin and pulp, our results indicated that the gaseous treatment did not induce the expression of any VviDREBA1s, and only a significant accumulation of VviDREBA1-7 was observed after 13 days at 0°C in non-treated bunches. By contrast, a decrease in VviDREBA1-1 transcript levels was observed during the storage at 0°C regardless of the atmosphere composition, whereas VviDREBA1-6 expression only decreased significantly in 3-day CO2 treated samples.
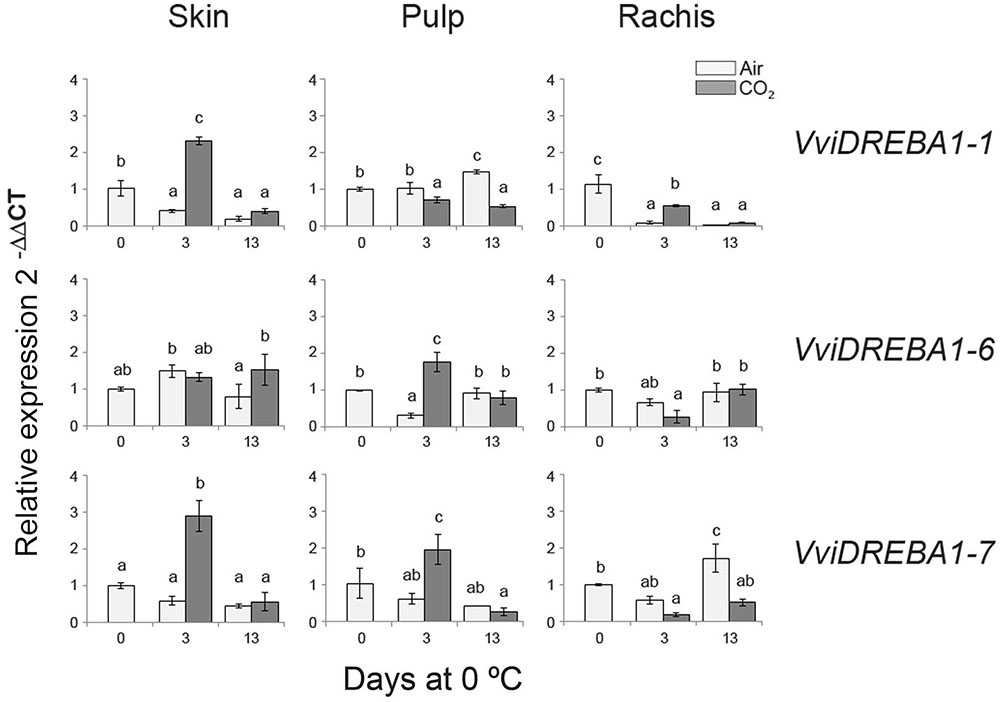
FIGURE 2. Expression pattern of VviDREBA1s in the skin, pulp and rachis of non-treated and CO2-treated bunches stored at 0°C up to 13 days. The relative quantification of transcripts was normalized to Actin1 and the results were calculated relative to a calibrator sample (day 0) using the formula 2-ΔΔCt. Values are the mean ± SD, n = 6. The different letters on the bars indicate that means are statistically different using Duncan’s test (P ≤ 0.05).
Modulation of VviDHNs Expression in Different Tissues of Autumn Royal Bunches by Low Temperature and High CO2 Levels
To analyze the putative role of VviDREBA1s in the regulation of DHNs, we first studied the changes of spliced and unspliced VviDHN1a, VviDHN2, and VviDHN4 mRNAs in fruit and non-fruit tissues of table grapes which were CO2-treated and non-treated and stored at 0°C. To this end, we performed semi-quantitative RT-PCR (VviDHN1a and VviDHN2) or RT-qPCR (VviDHN4). We observed that both VviDHN1a spliced and unspliced transcript levels were induced by storage at 0°C regardless of the atmosphere composition, although spliced transcripts were predominant in all the tissues analyzed (Figure 3). By contrast, differences between the spliced and unspliced transcript accumulation were observed in the case of VviDHN2 and VviDHN4 (Figures 3, 4). Thus, although VviDHN2 and VviDHN4 unspliced forms were activated by exposure to 0°C both in treated and non-treated samples in all the tissues analyzed, the accumulation observed by applying the gaseous treatment after 3 days was significantly higher. However, VviDHN2 spliced transcripts showed a tissue dependent regulation, remaining unchanged in the skin and increasing mainly by low temperature in the pulp of non-treated berries, whereas in the case of the rachis, higher levels of the spliced transcripts were reached in CO2-treated samples (Figure 3). By contrast, VviDHN4 spliced transcript accumulation remained constant in the skin and pulp of treated and non-treated berries until day 13 where the levels increased in the skin of CO2 treated fruit, decreasing in the pulp. In the rachis, an increase was observed after 13 days at 0°C regardless of a gaseous pretreatment, with it being significantly higher in non-treated samples (Figure 4).
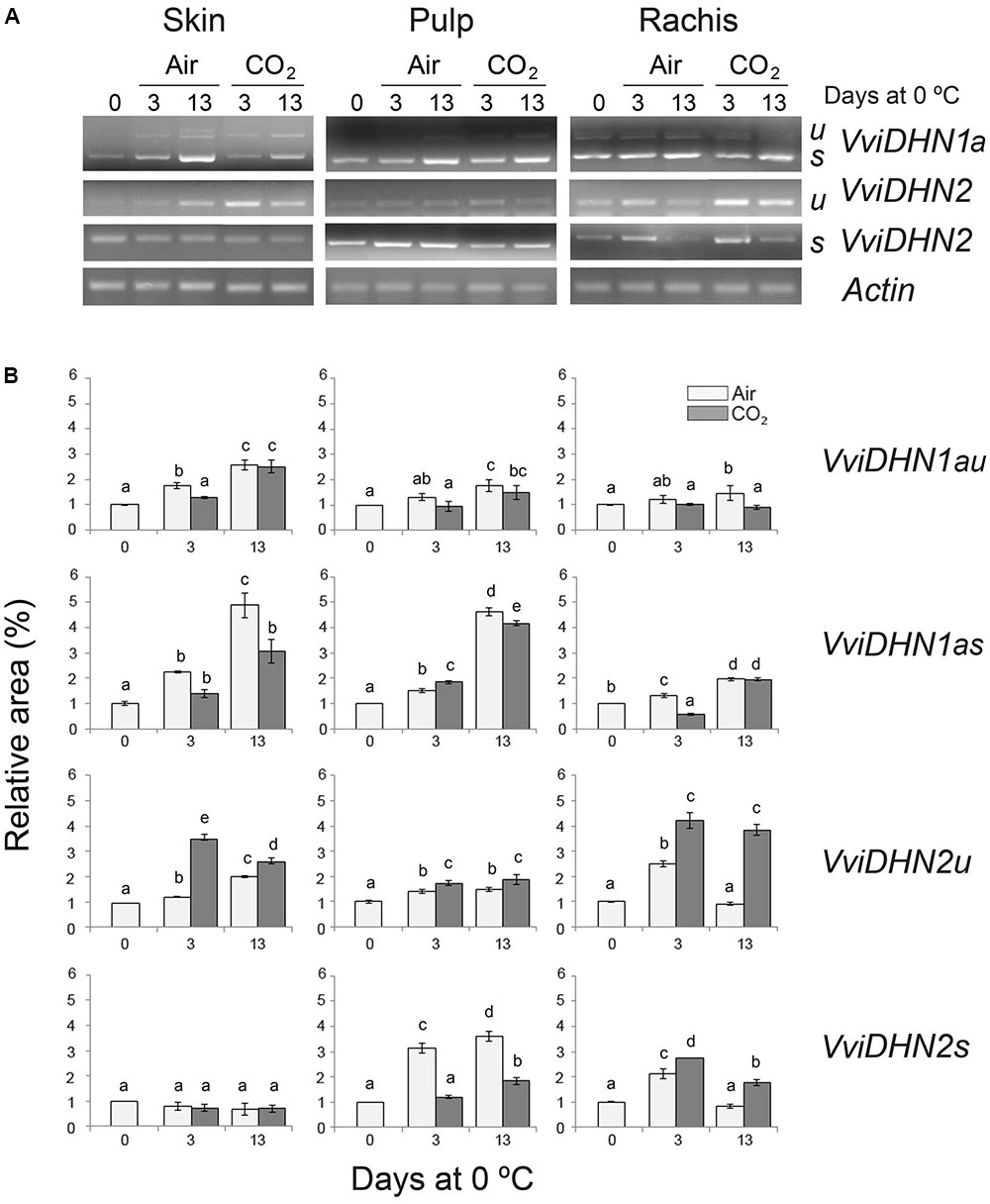
FIGURE 3. VviDHN1a and VviDHN2 gene expression in the skin, pulp and rachis of V. vinifera cv. Autumn Royal bunches after a 3-day high CO2 treatment and low temperature storage. (A) The levels of VviDHNs and Actin1a (control) transcripts were determined by semi-quantitative RT-PCR analysis. u: fragments produced in unspliced transcripts; s: fragments produced in spliced transcripts. (B) The results were calculated by densitometry quantification of the band intensity of each sample using Adobe Photoshop and were expressed as a relative fold-change with respect to bunches before storage (day 0; relative area). Values are the mean ± SD, n = 3. The different letters on the bars indicate that means are statistically different using Duncan’s test (P ≤ 0.05).
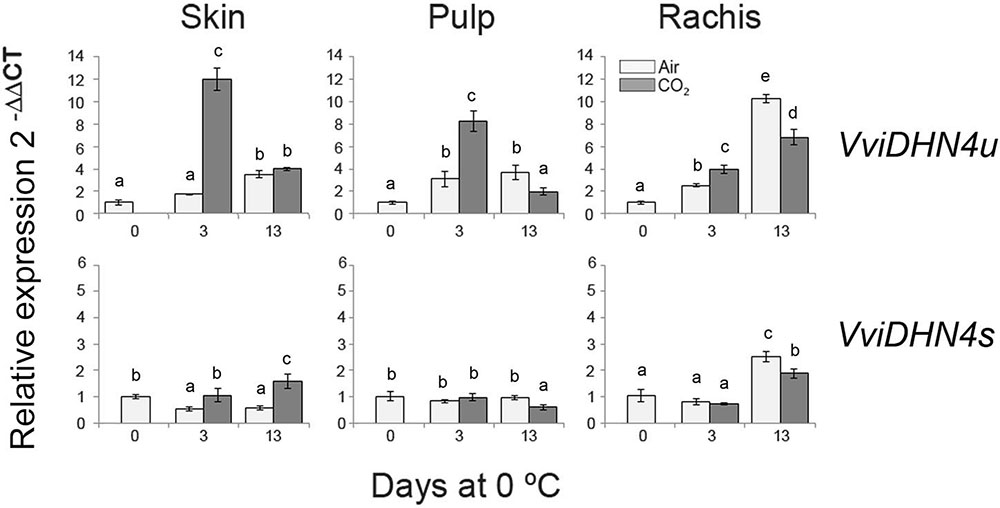
FIGURE 4. Relative gene expression of VviDHN4 spliced and unspliced (VviDHN4s and VviDHN4u) in the skin, pulp and rachis of V. vinifera cv. Autumn Royal bunches after a 3-day gaseous treatment and low temperature storage. The relative quantification of transcripts was normalized with Actin1 and the results were calculated relative to a calibrator sample (day 0) using the formula 2-ΔΔCt. Values are the mean ± SD, n = 6. The different letters on the bars indicate that means are statistically different using Duncan’s test (P ≤ 0.05).
Identification of Cis-Regulatory Elements in the Promoter Regions of VviDHN2 and VviDHN4
The identification and in silico analysis of 1500 bp of VviDHN2 and VviDHN4 promoter regions from V. vinifera using the Genomatix suite of programs led to the identification of several putative cis-acting regulatory elements associated with abiotic and biotic stress responses (Figure 5). The abiotic stress-related elements included ABA-responsive elements (ABRE), dehydration-responsive elements (DRE), and C-repeat elements (CRT). Identified within the biotic stress-related elements were methyl jasmonate-responsive elements (MeJa), ethylene-responsive elements (ERE), salicylic acid-responsive elements (TCA), and elicitor-responsive elements (W-box). VviDHN2 promoter harbored twelve cis-regulatory elements: eight related to abiotic stress and three related to biotic stress. Meanwhile, the VviDHN4 promoter presented ten cis-regulatory elements: two related to abiotic stress and seven related to biotic stress. Moreover, it is important to point out that VviDHN2 presented one DRE (ACCGAC core sequence) and one CRT (GCCGAC) motif, while VviDHN4 did not present any of them.
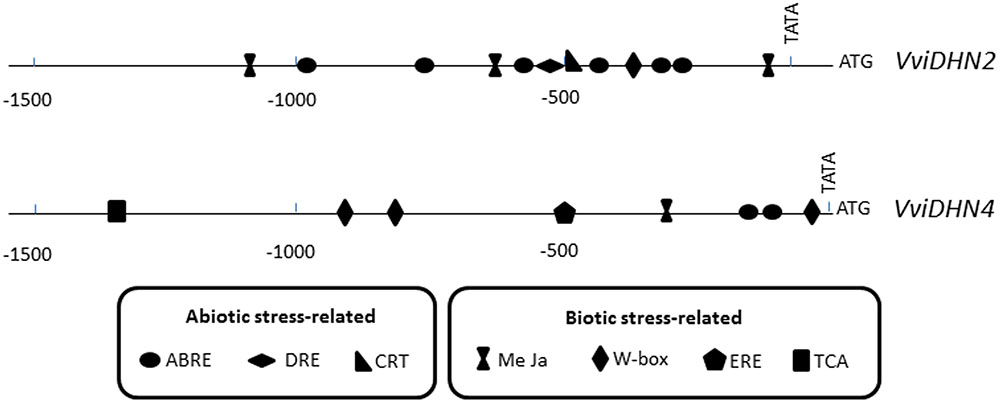
FIGURE 5. Schematic representation of biotic- and abiotic-related putative cis-acting regulatory elements in promoter regions of VviDHN2 and VviDHN4 genes.
Additionally, the promoter of VviDHN1a from table grapes cv. Cardinal was previously isolated (Rosales et al., 2014), and shared 98.4% identity with the corresponding sequence in the PN40024 grapevine reference genome. Among the ten cis-regulatory elements observed, a DRE motif was present.
VviDREBA1-1 Is a CRT Binding Protein
Purified VviDREBA1s recombinant proteins were obtained by Ni-NTA agarose and confirmed and visualized by Western Blot using the anti-6xHis monoclonal antibody (Figure 6A). EMSA analysis was performed to assess the capacity of the three VviDREBA1s to bind the DRE or CRT motifs. Only the probes from promoters of VviDHN1a and VviDHN2 were used since they contained the target motifs (DRE or CRT). However, mutated versions of the mentioned probes were included. Our results showed that only VviDREBA1-1 was able to bind specifically to the VviDHN2 CRT motif. In this case, the binding activity was abolished by competition with large amounts of unlabeled probe (Figure 6B). Likewise, we have observed that the change in the flanking regions of the CRT motif did not abolish the binding activity although the intensity was reduced (Supplementary Figure S3). By contrast, no binding or unspecific binding was observed in the assays performed with the VviDREBA1-1 and the probes harboring DRE motif; or with VviDREBA1-6 and VviDREBA1-7 proteins containing the DRE or CRT motifs; or with the mutated probes (data not shown). It is important to remark that only the results which show specific or unspecific binding have been included in Figure 6B.
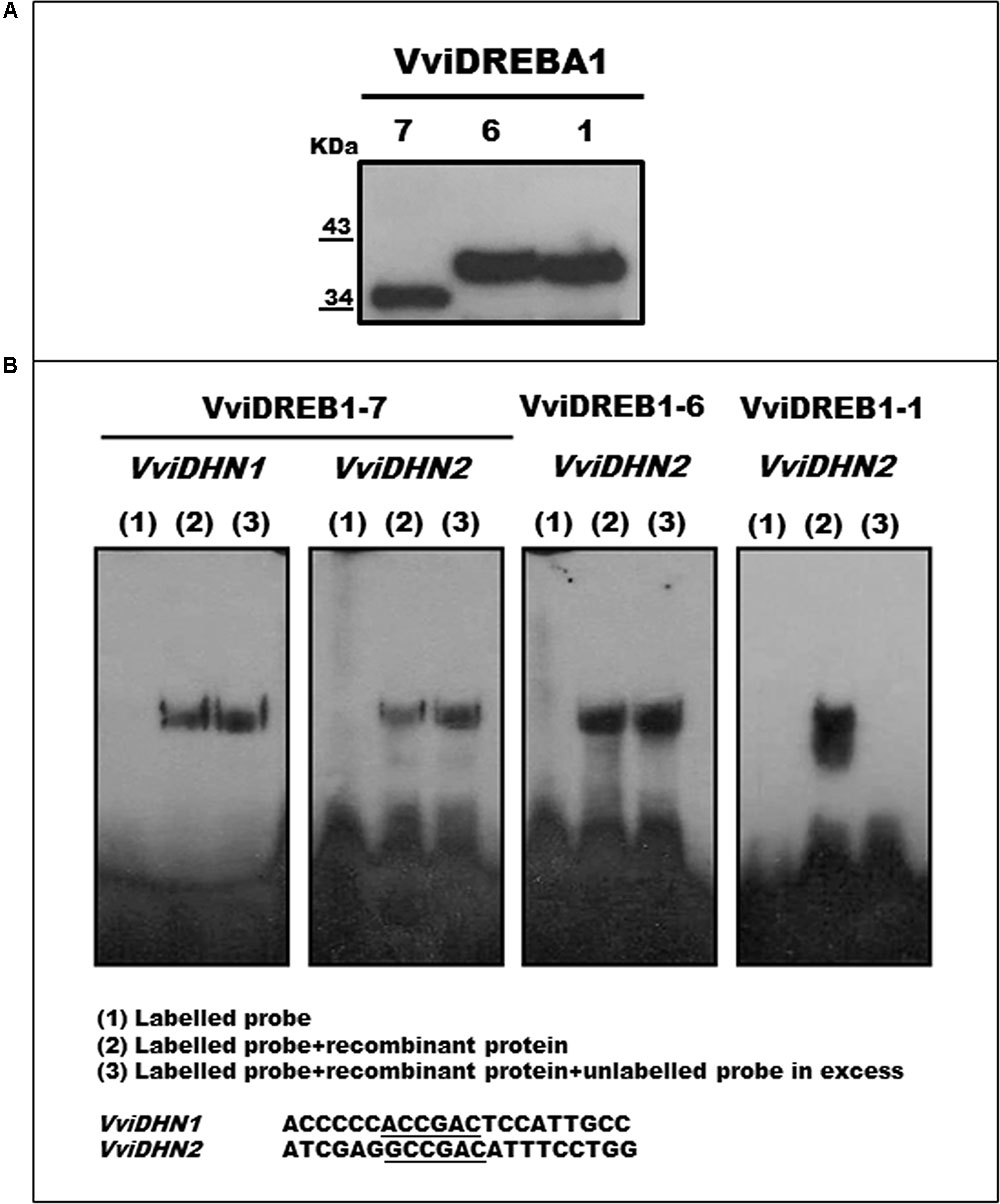
FIGURE 6. (A) Western blot of recombinant VviDREBA1s. (B) EMSA analyses were performed with recombinant VviDREBA1s proteins. DRE or CRT probes were labeled in the 3′end with biotin. For the competition assays, five hundred times of the unlabelled probe was added before the labeled probes. Sequences of the oligonucleotides containing the DRE or CRT motifs (underlined) used as probes are shown.
Discussion
Mature table grapes are non-climacteric fruit with a relatively low respiration rate, but are subject to serious water loss, softening and fungal attack after harvest. Grapes are classified as a non-chilling sensitive fruit and postharvest storage around 0°C is recommended to maintain their quality. However, we have observed that table grapes cv. Cardinal are sensitive to temperature shifts from the field to storage at 0°C with increasing rachis browning, water loss and ion leakage, as well as the activation of phenylpropanoid and pathogenesis related gene expression, whereas a 3-day high CO2 pretreatment at 0°C avoids and/or modifies these changes (Sanchez-Ballesta et al., 2006; Rosales et al., 2016). In the present study, we have observed that the application of high CO2 levels for 3 days at 0°C was effective in maintaining the quality of table grapes other than Cardinal, such as Autumn Royal. Previous results in Cardinal berries showed that the CO2 treatment significantly affected the moisture content of all tissues of the bunch, except for pulp tissues (Goñi et al., 2011). However, similar values of moisture content were quantified in 3-day CO2-treated and non-treated Autumn Royal berries stored at 0°C, as was observed in strawberries (Blanch et al., 2012). On the other hand, CO2-treated table grapes showed a low weight loss percentage in comparison to non-treated bunches during storage at 0°C. This effect has been also reported by Martínez-Romero et al. (2003) in table grapes, and in other types of fruit such as nectarines (Retamales et al., 2000), cucumbers (Wang and Qi, 1997), and cherries (Kappel et al., 2002) stored in modified atmosphere packaging at low temperature. Concerning the rachis browning index, it was lower in Autumn Royal bunches treated with CO2 for 3 days than in non-treated bunches, as has been observed in Cardinal table grapes (Rosales et al., 2013). However, this is not a common response to high CO2 levels. Thus, rachis appearance was adversely affected in Thompson Seedless and Red Globe bunches stored up to 40 or 45 days at low temperature under controlled atmosphere CO2 levels ≥15 kPa, respectively, and after 4 days of shelf-life (Retamales et al., 2003). The beneficial effect of the gaseous treatment on the rachis appearance of Autumn Royal bunches could be explained by lower water loss from the rachis under this treatment (Sanchez-Ballesta et al., 2006). Accordingly, in Cardinal bunches the relative water content of the rachis, an indicator of water balance status, decreased considerably throughout storage at 0°C in non-treated bunches whereas the application of 3-day high CO2 levels reduced this effect (Rosales et al., 2013).
In recent studies, we have shown that maintaining Cardinal table grape quality by the application of a 3-day CO2 treatment seems to be an active process which requires the activation of transcription factors, such as ERFs, belonging to the AP2/ERF transcription factor family (Romero et al., 2016; Rosales et al., 2016). In addition, we have observed that the gaseous treatment applied to Cardinal bunches induced the expression of other members of this transcription factor family such as CBF/DREB (Fernandez-Caballero et al., 2012). The CBF/DREB proteins play a crucial role in abiotic stress-mediated gene expression, representing one of the most attractive regulons for breeding programs (Zandkarimi et al., 2015) to cope with cold stress. Different studies have indeed shown the relationship between CBF/DREB expression and cold tolerance in different fruit such as tomato, peach, kiwifruit, and mango during low temperature storage in combination with different postharvest treatments (Liang et al., 2013; Ma et al., 2014; Zhang et al., 2017). However, with the exception of the study mentioned above, the relationship of these CBF/DREBs with the different sensitivity to temperature shifts induced in fruit by high CO2 levels is still unknown. Thus, we have isolated three DREBA1s from Autumn Royal table grapes and characterized their expression pattern in the skin, pulp and rachis during postharvest storage at low temperature with a 3-day high CO2 treatment. In this work, we observed that gene expression varied according to the VviDREBA1 analyzed and the tissue studied. The storage of Autumn Royal bunches at low temperature under normal atmosphere was enough to activate the expression of VviDREBA1-1 in the pulp and VviDREBA1-7 in the rachis. Likewise, as was observed in Cardinal grapes (Fernandez-Caballero et al., 2012), the 3-day high CO2 treatment at 0°C modulated the expression of VviDREBA1s by activating VviDREBA1-1 and VviDREBA1-7 in the skin, and VviDREBA1-6 and VviDREBA1-7 in the pulp. On the other hand, it is important to note that the effect of low temperature storage under normal atmosphere modulated VviDREBA1-1 and VviDREBA1-7 after 13 days of storage, while the three VviDREBA1s were induced at the end of the 3-day gaseous treatment at 0°C. This temporal difference could be important to help table grapes face temperature shifts at 0°C. Thus, Santiam tomato fruit showed less sensitivity to chilling than Lichum tomatoes, reflected in the lowest chilling injury index, ion leakage and malondialdehyde content. Meanwhile, LeCBF1 transcript accumulation was higher in Santiam tomatoes, indicating that there was a swift genetic response to chilling stress, which was positively correlated with cold tolerance of the cultivar (Zhao et al., 2009).
It is known that the activation of CBF/DREBs improves the expression of downstream target genes, especially those encoding LEA proteins, including DHNs, as these transcriptional activators can bind to the cis DRE/CRT motif present in the promoter regions of these genes. Overexpression of AtCBF1–AtCBF3 in transgenic Arabidopsis plants resulted in an increase in freeze tolerance and in the activation of different COR genes, including DHNs, at room temperature (Gilmour et al., 2004). A comparative study of two citrus species (Poncirus and Citrus) with different levels of freezing tolerance, showed a correlation between CBF1 expression and the degree of tolerance observed (Champ et al., 2007). These authors further demonstrated that CBF1 specifically recognized the consensus sequence (CCGAC) of the DRE/CRT elements from the dehydrin promoter of Poncirus trifoliata. To study the role of VviDREBA1s in the regulation of VviDHNs, we first studied the gene expression of spliced and unspliced VviDHN1, VviDHN2, and VviDHN4 mRNAs in different tissues of Autumn Royal bunches, both CO2-treated and non-treated with storage at 0°C. VviDHN1a, VvDHN2, and VviDHN4 are known to undergo alternative splicing (Fernandez-Caballero et al., 2012; Yang et al., 2012). Intron retention during VviDHNs pre-mRNA processing leads to the production of mRNAs with premature stop codons in the intron that give rise to truncated proteins without the K segment, which is characteristic of this protein family. Likewise, Lee et al. (2016) reported that OsCYP19-4 (the primary cyclophilin 19-4 transcript from rice) was able to generate up to eight alternative splice forms via two types of regulated splicing events, intron retention and exon skipping, under low temperatures. Thus, in the case of Arabidopsis, temperature stress (cold and heat) dramatically altered the alternative spliced of pre-mRNAs of several serine/arginine-rich proteins, a conserved family of splicing regulators in eukaryotes (Palusa et al., 2007). Although the functionality of unspliced DHNs is not well known, in a previous work we demonstrated that the unspliced DHN1a variant from V. vinifera cv. Cardinal slightly interacted with DNA (Rosales et al., 2014). In this work, we have observed that unspliced forms showed a higher regulation degree than spliced forms after the CO2 treatment. Therefore, with the exception of unspliced VviDHN1a and VviDHN4 in rachis, the 3-day CO2 treatment regulated the transcript levels in the unspliced forms in the different tissues of Autumn Royal.
The study of the VviDHNs promoter regions showed that, whereas the VviDHN2 promoter region showed the presence of one DRE element and one CRT element, VviDHN4 did not show any of them. Likewise, previous results showed the presence of a DRE element in the VviDHN1 promoter (Rosales et al., 2014). Through EMSA assays, our work provides evidence that the recombinant protein VviDREBA1-1 was the only one which showed specific binding to the CRT element (GCCGAC) presented in the VviDHN2 promoter, while no binding was observed to the DRE element (ACCGAC). In a recent work (Carlow et al., 2017), transient transactivation assays showed that all V. riparia CBFs except CBF5 activate via a CRT or DRE promoter element, whereby V. riparia CBF3 (homolog to Vv1DEBA1-6) and CBF4 (homolog to Vv1DEBA1-1) prefer a CRT element. It is known that not all CBF/DREBs have the same affinity and specificity for DRE/CRT elements. Thus, BNCBF17 from Brassica napus showed lower binding specificity than BNCBF5 to the core CCGAC sequence when this was mutated (Gao et al., 2002), while HvCBF1 from barley showed preference for binding to the sequence TTGCCGACAT, which contained the CRT motif instead of DRE (Xue, 2002). Likewise, the analysis of the promoters of COR genes induced in Arabidopsis plants which overexpressed CBFs demonstrated that variations in the sequences around the CRT element could affect the activation of the promoters (Maruyama et al., 2004). In this regard, our results showed that the region flanking the CRT element must have a role in the affinity of the VviDREBA1s, since changes in some nucleotides did not abolish the binding observed with VviDREBA1-1, but its intensity was reduced. Although more studies are needed to be able to understand the binding specificity of CBF/DREBs for target sequences, the results are in concordance with previous results reported by Nassuth et al. (2014) which suggest that CBF/DREBs paralogs in a plant, and possibly orthologs from different species, have a unique preference for DRE/CRT sequences. At this point it is important to remark that the three VviDREBA1s contained the signature sequences, PKKPAGR and DSAWR, flanking the AP2 domain which distinguishes this subgroup of transcription factors from the other AP2/ERF family members (Jaglo et al., 2001). Canella et al. (2010) showed that deletions or mutation in the PKKPAGR sequence greatly impaired the ability of AtCBF1 to induce expression of target COR genes because of its inability to bind to the DRE/CRT element indicating that amino acids beyond the AP2 domain are required. In the specific case of DREBA1s from Autumn Royal, we observed that whereas the PKKPAGR motif was well conserved in VviDREBA1-1 (PKKRAGRxKFxETRHP), there were three (HKRKAGRxKFxETRHP) and five (HKRKTGRxKFxKTRHP) amino acids which were different in VviDREBA1-6 and VviDREBA1-7, respectively. In Vitis, Xiao et al. (2006) observed that despite the three amino acid changes detected in CBF1, targeting and transactivation experiments denoted that CBF1 still functions. Nevertheless, transient activation experiments suggested that V. riparia CBF4 was a more effective activator than VrCBF1 (Xiao et al., 2008), and that Arabidopsis transgenic plants which overexpressed V. riparia CBF1 and CBF4 presented higher amounts of COR genes in the CBF4 lines (Siddiqua and Nassuth, 2011). Likewise, Nassuth et al. (2014) corroborated previous results and showed, with a new system of transactivation, that VrCBF1 and VrCBF4 transcription factors probably activate different overlapping sets of genes, and therefore play unique roles in cold acclimation. Thus, this evidence could be related to the changes observed in the PKKPAGR motif of Vitis CBFs.
Overall, our results demonstrated that the 3-day gaseous treatment was effective in maintaining table grape quality regardless of the cultivar and modulated the expression of VviDREBA1s transcription factors in a tissue dependent way. Furthermore, the application of high levels of CO2 regulated the retention of introns in the transcripts of the dehydrins in a different manner and in all the tissues analyzed. On the other hand, VviDREBA1-1 was the only transcription factor analyzed that presented in vitro binding capacity to the CRT element of the VviDHN2 promoter region, indicating that the transcriptional regulation of VviDHN1a and VviDHN4 would be carried out by activating other independent routes of these transcription factors (Figure 7). These results, together with the fact that VviDREBA1-1 gene expression was induced by high CO2 levels in the skin of Autumn Royal fruit and in the pulp and rachis of Cardinal bunches (Fernandez-Caballero et al., 2012), make this transcription factor a good candidate for further research with the aim of improving table grape quality.
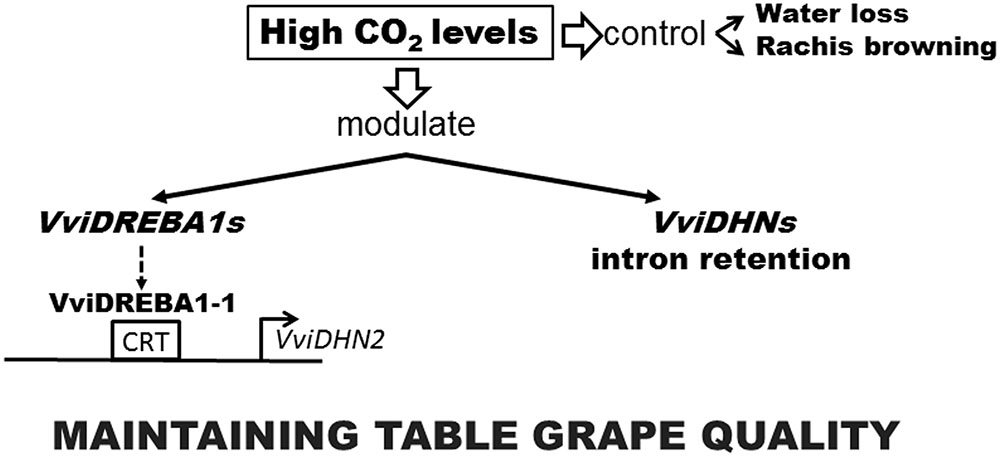
FIGURE 7. A scheme depicting events of Autumn Royal table grapes in response to high CO2 levels during postharvest storage at 0°C.
Author Contributions
MV-H and IR contributed equally to this work. MV-H: Participated in the table grape quality assessments, carried out the RNA extraction, RT-qPCR analysis, semiquantitative RT-PCR, statistical analyses. IR: Carried out the VviDREBA1s isolation and the production of recombinant proteins, the EMSA analysis, edited and collaborated in the first draft of the manuscript. ME: Conceived the postharvest table grapes storage experience, participated in the table grape quality assessments and critically revised the manuscript. CM: Participated in the table grape quality assessments and critically revised the manuscript. MS-B: conceived the VviDREBA1s and DHNs study, supervised and coordinated the experiments, interpreted the results and prepared the first draft of the manuscript. All authors have read and approved this manuscript.
Funding
This work was supported by the European Union under the 7th Framework Programme FP7-PEOPLE-2012-CIG no. 321694 and by CICYT projects AGL2011-26742 and AGL2014-53081-R. MV-H was supported by a predoctoral contract from the MEC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work benefited from the networking activities within the European COST Action FA1106-QualityFruit. Authors thank Dr. Carmen Lisset for her generosity in providing BL21-CodonPlus (DE3)-RIL competent cells. The authors are grateful to Frutas Esther S.A for providing tables grapes.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01591/full#supplementary-material
Footnotes
- ^http://genomes.cribi.unipd.it/grape/
- ^http://www.ch.embnet.org/software/LALIGN_form.html
- ^http://www.ubpred.org/
- ^http://sumosp.biocuckoo.org/online.php
- ^http://emboss.bioinformatics.nl/cgi-bin/emboss/epestfind
- ^http://www.genomatix.de
References
Barros, P. M., Gonçalves, N., Saibo, J. M. N., and Oliveira, M. M. (2012). Functional characterization of two almond C-repeat-binding factors involved in cold response. Tree Physiol. 32, 1113–1128. doi: 10.1093/treephys/tps067
Blanch, M., Sanchez-Ballesta, M. T., Escribano, M. I., and Merodio, C. (2012). Water distribution and ionic balance in response to high CO2 treatments in wild strawberries (Fragaria vesca cv. Mara de Bois). Postharvest Biol. Technol. 73, 63–71. doi: 10.1016/j.postharvbio.2012.06.003
Canella, D., Gilmour, S., Kuhn, L., and Thomashow, M. (2010). DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim. Biophys. Acta 1799, 454–462. doi: 10.1016/j.bbagrm.2009.11.017
Carlow, C. E., Faultless, J. T., Lee, C., Siddiqua, M., Edge, A., and Nassuth, A. (2017). Nuclear localization and transactivation by Vitis CBF transcription factors are regulated by combinations of conserved amino acid domains. Plant Physiol. Biochem. 118, 306–319. doi: 10.1016/j.plaphy.2017.06.027
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 13, 2933–2942. doi: 10.1093/bioinformatics/bti473
Champ, K. I., Febres, V. J., and Moore, G. A. (2007). The role of CBF transcriptional activators in two Citrus species (Poncirus and Citrus) with contrasting levels of freezing tolerance. Physiol. Plant 129, 529–541. doi: 10.1111/j.1399-3054.2006.00826.x
Chew, Y. H., and Halliday, K. (2011). A stress-free walk from Arabidopsis to crops. Curr. Opin. Biotechnol. 2, 281–286. doi: 10.1016/j.copbio.2010.11.011
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Cramer, G. R., Ghan, R., Schlauch, K. A., Tillett, R. L., Heymann, H., Ferrarini, A., et al. (2014). Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol. 14:370. doi: 10.1186/s12870-014-0370-8
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fernandez-Caballero, C., Rosales, R., Romero, I., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2012). Unraveling the roles of CBF1, CBF4 and dehydrin 1 genes in the response of table grapes to high CO2 levels and low temperature. J. Plant Physiol. 169, 744–748. doi: 10.1016/j.jplph.2011.12.018
Gaboriaud, C., Bissery, V., Benchetrit, T., and Mornon, J. P. (1987). Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 224, 149–155. doi: 10.1016/0014-5793(87)80439-8
Gao, M. J., Allard, G., Byass, L., Flanagan, A. M., and Singhet, J. (2002). Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol. Biol. 49, 459–471. doi: 10.1023/A:1015570308704
Genero, M., Gismondi, M., Monti, L. L., Gabilondo, J., Budde, C. O., Andreo, C. S., et al. (2016). Cell wall-related genes studies on peach cultivars with differential susceptibility to woolliness: looking for candidates as indicators of chilling tolerance. Plant Cell Rep. 35, 1235–1246. doi: 10.1007/s00299-016-1956-4
Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Goñi, O., Fernandez-Caballero, C., Sanchez-Ballesta, M. T., Escribano, M. I., and Merodio, C. (2011). Water status and quality improvement in high-CO2 treated table grapes. Food Chem. 128, 34–39. doi: 10.1016/j.foodchem.2011.02.073
Grimplet, J., Adam-Blondon, A.-F., Bert, P.-F., Bitz, O., Cantu, D., Davies, C., et al. (2014). The grapevine gene nomenclature system. BMC Genomics 15:1077. doi: 10.1186/1471-2164-15-1077
Haake, V., Cook, D., Riechmann, J. L., Pineda, O., Thomashow, M. F., and Zhang, J. Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130, 639–648. doi: 10.1104/pp.006478
He, L. G., Wang, H. L., Liu, D. C., Zhao, Y. J., Xu, M., Zhu, M., et al. (2012). Isolation and expression of a cold-responsive gene PtCBF in Poncirus trifoliate and isolation of citrus CBF promoters. Biol. Plant. 56, 484–492. doi: 10.1007/s10535-012-0059-5
Jaglo, K. R., Kleff, S., Amundsen, K. L., Zhang, X., Kaake, V., Xhan, J. Z., et al. (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127, 910–917. doi: 10.1104/pp.010548
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Kappel, F., Toivonen, P., MacKenzie, D. L., and Stam, S. (2002). Storage characteristics of new sweet cherry cultivars. Hortscience. 37, 139–143.
Karimi, M., Ebadi, A., Amir, S., Seyed, M., Salami, A., and Zareic, A. (2015). Comparison of CBF1, CBF2, CBF3 and CBF4 expression in some grapevine cultivars and species under cold stress. Sci. Hortic. 197, 521–526. doi: 10.1016/j.scienta.2015.10.011
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kitashiba, H., Ishizaka, T., Isuzugawa, K., Nishimura, K., and Suzuki, T. (2004). Expression of a sweet cherry DREB1/CBF ortholog in Arabidopsis confers salt and freezing tolerance. J. Plant Physiol. 161, 1171–1176. doi: 10.1016/j.jplph.2004.04.008
Lee, A., Lee, S. S., Jung, W. Y., Park, H. J., Lim, B. R., Kim, H. S., et al. (2016). The OsCYP19-4 gene is expressed as multiple alternatively spliced transcripts encoding isoforms with distinct cellular localizations and PPIase activities under cold stress. Int. J. Mol. Sci. 17:1154. doi: 10.3390/ijms17071154
Liang, L., Zhang, B., Yin, X. R., Xu, C. J., Sun, C. D., and Chen, K. S. (2013). Differential expression of the CBF gene family during postharvest cold storage and subsequent shelf-life of peach fruit. Plant Mol. Biol. Rep. 31, 1358–1367. doi: 10.1007/s11105-013-0600-5
Licausi, F., Giorgi, F. M., Zenoni, S., Osti, F., Pezzotti, M., and Perata, P. (2010). Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11:719. doi: 10.1186/1471-2164-11-719
Lodhi, M. A., Guang-Ning, Z., Weeden, F. N. F., and Reisch, B. I. (1994). A simple and efficient method for DNA extraction from grapevine cultivars, Vitis species and Ampelopsis. Plant Mol. Biol. Rep. 12, 6–13. doi: 10.1007/BF026686
Ma, Q., Suo, J., Huber, D. J., Dong, X., Han, Y., Zhang, Z., et al. (2014). Effect of hot water treatments on chilling injury and expression of a new C-repeat binding factor (CBF) in ‘Hongyang’ kiwifruit during low temperature storage. Postharvest Biol. Technol. 97, 102–110. doi: 10.1016/j.postharvbio.2014.05.018
Martínez-Romero, D., Guillén, F., Castillo, S., Valero, D., and Serrano, M. (2003). Modified atmosphere packaging maintains quality of table grapes. J. Food Sci. 68, 1838–1843. doi: 10.1111/j.1365-2621.2003.tb12339.x
Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., et al. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993. doi: 10.1111/j.1365-313X.2004.02100.x
Maul, P., McCollum, G. T., Popp, M., Guy, C. L., and Porat, R. (2008). Transcriptome profiling of grapefruit flavedo following exposure to low temperature and conditioning treatments uncovers principal molecular components involved in chilling tolerance and susceptibility. Plant Cell Environ. 31, 752–768. doi: 10.1111/j.1365-3040.2008.01793.x
Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–470. doi: 10.1104/pp.119.2.463
Nassuth, A., Siddiqua, M., Xiao, H., Moody, M. A., and Carlow, C. E. (2014). Newly developed quantitative transactivation system shows difference in activation by Vitis CBF transcription factors on DRE/CRT elements. Plant Methods 10:32. doi: 10.1186/1746-4811-10-32
Navarro, S., Vazquez-Hernandez, M., Rosales, R., Sanchez-Ballesta, M. T., Merodio, C., and Escribano, M. I. (2015). Differential regulation of dehydrin expression and trehalose levels in Cardinal table grape skin by low temperature and high CO2. J. Plant Physiol. 179, 1–11. doi: 10.1016/j.jplph.2015.02.007
Novillo, F., Medina, J., and Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. U.S.A. 104, 21002–21007. doi: 10.1073/pnas.0705639105
Oakenfull, R. J., Baxter, R., and Knight, M. R. (2013). A C-repeat binding factor transcriptional activator (CBF/DREB1) from European bilberry (Vaccinium myrtillus) induces freezing tolerance when expressed in Arabidopsis thaliana. PLOS ONE 8:e54119. doi: 10.1371/journal.pone.005411
Palusa, S. G., Ali, G. S., and Reddy, A. S. N. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49, 1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x
Pino, M. T., Skinner, J. S., Jeknic, Z., Hayes, P. M., Soeldner, A. H., Thomashow, M. F., et al. (2008). Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant Cell Environ. 31, 393–406. doi: 10.1111/j.1365-3040.2008.01776.x
Radivojac, P., Vacic, V., Haynes, C., Cocklin, R. R., Mohan, A., Heyen, J. W., et al. (2010). Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380. doi: 10.1002/prot.22555
Retamales, J., Defilippi, B., and Campos, R. (2000). Control of cold storage disorders in nectarines by modified atmosphere packaging. Fruits 55, 213–219.
Retamales, J., Defilippi, B., Castillo, P., and Manríquez, D. (2003). High-CO2 controlled atmospheres reduce decay incidence in Thompson Seedless & Red Globe table grapes. Postharvest Biol. Technol. 29, 177–182. doi: 10.1016/S0925-5214(03)00038-3
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Romero, I., Fernandez-Caballero, C., Goñi, O., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2008). Functionality of a class I beta-1,3-glucanase from skin of table grapes berries. Plant Sci. 174, 641–648. doi: 10.1016/j.plantsci.2008.03.019
Romero, I., Vazquez-Hernández, M., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2016). Expression profiles and DNA-binding affinity of five ERF genes in bunches of Vitis vinifera cv. Cardinal treated with high levels of CO2 at low temperature. Front. Plant Sci. 7:1748. doi: 10.3389/fpls.2016.01748
Rosales, R., Fernandez-Caballero, C., Romero, I., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2013). Molecular analysis of the improvement in rachis quality by high CO2 levels in table grapes stored at low temperature. Postharvest Biol. Technol. 77, 50–58. doi: 10.1016/j.postharvbio.2012.10.009
Rosales, R., Romero, I., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2014). The crucial role of Φ- and K-segments in the in vitro functionality of Vitis vinifera dehydrin DHN1a. Phytochemistry 108, 17–25. doi: 10.1016/j.phytochem.2014.10.006
Rosales, R., Romero, I., Fernandez-Caballero, C., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2016). Low temperature and short-term high-CO2 treatment in postharvest storage of table grapes at two maturity stages: effects on transcriptome profiling. Front. Plant Sci. 7:1020. doi: 10.3389/fpls.2016.01020
Sanchez-Ballesta, M. T., Jiménez, J. B., Romero, I., Orea, J. M., Maldonado, R., Ureña, A. G., et al. (2006). Effect of high CO2 pretreatment on quality, fungal decay and molecular regulation of stilbene phytoalexin biosynthesis in stored table grapes. Postharvest Biol. Technol. 42, 209–216. doi: 10.1016/j.postharvbio.2006.07.002
Siddiqua, M., and Nassuth, A. (2011). Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ. 34, 1345–1359. doi: 10.1111/j.1365-3040.2011.02334.x
Singh, K. B., Foley, R. C., and Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/S1369-5266(02)00289-3
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Takuhara, Y., Kobayashi, M., and Suzuki, S. (2011). Low-temperature-induced transcription factors in grapevine enhance cold tolerance in transgenic Arabidopsis plants. J. Plant Physiol. 168, 967–975. doi: 10.1016/j.jplph.2010.11.008
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Tillett, R. L., Wheatley, M. D., Tattersall, E. A. R., Schlauch, K. A., Cramer, G. R., and Cushman, J. C. (2012). The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotechnol. J. 10, 105–124. doi: 10.1111/j.1467-7652.2011.00648.x
Vitulo, N., Forcato, C., Carpinelli, E. C., Telatin, A., Campagna, D., D’Angelo, M., et al. (2014). A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 14:99. doi: 10.1186/1471-2229-14-99
Wang, C. Y., and Qi, L. (1997). Modified atmosphere packaging alleviates chilling injury in cucumbers. Postharvest Biol. Technol. 10, 195–200. doi: 10.1016/S0925-5214(97)01405-1
Wang, H., Wang, H., Shao, H., and Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67. doi: 10.3389/fpls.2016.00067
Wang, Z., Triezenberg, S. J., Thomashow, M. F., and Stockinger, E. J. (2005). Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Mol. Biol. 58, 543–559. doi: 10.1007/s11103-005-6760-4
Wisniewski, M. E., Bassett, C. L., Renaut, J., Farrell, R., Tworkoski, T., and Artlip, T. S. (2006). Differential regulation of two dehydrin genes from peach (Prunus persica) by photoperiod, low temperature and water deficit. Tree Physiol. 26, 575–584. doi: 10.1093/treephys/26.5.575
Xiao, H., Siddiqua, M., Braybrook, S., and Nassuth, A. (2006). Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 29, 1410–1421. doi: 10.1111/j.1365-3040.2006.01524.x
Xiao, H., Tattersall, E. A., Siddiqua, M. K., Cramer, G. R., and Nassuth, A. (2008). CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 31, 1–10. doi: 10.1111/j.1365-3040.2007.01741.x
Xiong, Y., and Fei, S. Z. (2006). Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 224, 878–888. doi: 10.1007/s00425-006-0273-5
Xue, G. P. (2002). The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J. 33, 373–383. doi: 10.1046/j.1365-313X.2003.01630.x
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yang, W., Liu, X.-D., Chi, X.-J., Wu, C.-A., Li, Y.-Z., Song, L.-L., et al. (2011). Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 233, 219–229. doi: 10.1007/s00425-010-1279-6
Yang, Y., He, M., Zhu, Z., Li, S., Xu, Y., Zhang, C., et al. (2012). Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 12:140. doi: 10.1186/1471-2229-12-140
Zandkarimi, H., Ebadi, A., Salami, S. A., Alizade, H., and Baisakh, N. (2015). Analyzing the expression profile of AREB/ABF and DREB/CBF genes under drought and salinity stresses in grape (Vitis vinifera L.). PLOS ONE 10:e0134288. doi: 10.1371/journal.pone.0134288
Zeng, Y., and Yang, T. (2002). RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol. Biol. Rep. 20:417. doi: 10.1007/BF02772130
Zhang, Z., Zhu, Q., Hu, M., Gao, Z., An, F., Li, M., et al. (2017). Low-temperature conditioning induces chilling tolerance in stored mango fruit. Food Chem. 219, 76–84. doi: 10.1016/j.foodchem.2016.09.123
Zhao, C., and Zhu, J. K. (2016). The broad roles of CBF genes: from development to abiotic stress. Plant Signal. Behav. 11:e1215794. doi: 10.1016/j.foodchem.2016.09.123
Zhao, D. Y., Shen, L., Fan, B., Liu, K. L., Yu, M. M., Zheng, Y., et al. (2009). Physiological and genetic properties of tomato fruits from 2 cultivars differing in chilling tolerance at cold storage. J. Food Sci. 74, 348–352. doi: 10.1111/j.1750-3841.2009.01156.x
Zhao, Q., Xie, Y., Zheng, Y., Jiang, S., Liu, W., Mu, W., et al. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, 325–330. doi: 10.1093/nar/gku383
Zhao, R., Sheng, J., Lv, S., Zheng, Y., Zhang, J., Yu, M., et al. (2011). Nitric oxide participates in the regulation of LeCBF1 gene expression and improves cold tolerance in harvested tomato fruit. Postharvest Biol. Technol. 62, 121–126. doi: 10.1016/j.postharvbio.2011.05.013
Keywords: CBF/DREB, dehydrins, transcription factors, Vitis vinifera, carbon dioxide, low temperature
Citation: Vazquez-Hernandez M, Romero I, Escribano MI, Merodio C and Sanchez-Ballesta MT (2017) Deciphering the Role of CBF/DREB Transcription Factors and Dehydrins in Maintaining the Quality of Table Grapes cv. Autumn Royal Treated with High CO2 Levels and Stored at 0°C. Front. Plant Sci. 8:1591. doi: 10.3389/fpls.2017.01591
Received: 10 July 2017; Accepted: 30 August 2017;
Published: 20 September 2017.
Edited by:
Michael Wisniewski, Appalachian Fruit Research Station (ARS-USDA), United StatesReviewed by:
Noam Alkan, Agricultural Research Organization, IsraelJason P. Londo, Grape Genetics Research Unit (USDA-ARS), United States
Copyright © 2017 Vazquez-Hernandez, Romero, Escribano, Merodio and Sanchez-Ballesta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. T. Sanchez-Ballesta, bWJhbGxlc3RhQGljdGFuLmNzaWMuZXM=
†These authors have contributed equally to this work.
 Maria Vazquez-Hernandez†
Maria Vazquez-Hernandez† Carmen Merodio
Carmen Merodio M. T. Sanchez-Ballesta
M. T. Sanchez-Ballesta