- 1State Key Laboratory of Tree Genetics and Breeding, Chinese Academy of Forestry, Beijing, China
- 2Key Laboratory of Tree Breeding of Zhejiang Province, The Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou, China
- 3Institute of Leisure Agriculture, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 4National Engineering Laboratory of Soil Pollution Control and Remediation Technologies, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
The heat shock transcription factor (Hsf) family, an important member in plant stress response, affects cadmium (Cd) tolerance in plants. In this study, we identified and functionally characterized a transcript of the Hsf A4 subgroup from Sedum alfredii. Designated as SaHsfA4c, the open reading frame was 1,302 bp long and encoded a putative protein of 433 amino acids containing a complete DNA-binding domain (DBD). Heterologous expression of SaHsfA4c in yeast enhanced Cd stress tolerance and accumulation, whereas expression of the alternatively spliced transcript InSaHsfA4c which contained an intron and harbored an incomplete DBD, resulted in relatively poor Cd stress tolerance and low Cd accumulation in transgenic yeast. The function of SaHsfA4c under Cd stress was characterized in transgenic Arabidopsis and non-hyperaccumulation ecotype S. alfredii. SaHsfA4c was able to rescue the Cd sensitivity of the Arabidopsis athsfa4c mutant. SaHsfA4c reduced reactive oxygen species (ROS) accumulation and increased the expression of ROS-scavenging enzyme genes and Hsps in transgenic lines. The present results suggest that SaHsfA4c increases plant resistance to stress by up-regulating the activities of ROS-scavenging enzyme and the expression of Hsps.
Introduction
Heavy metal pollution from industrial and agricultural activities has become a worldwide concern (Akesson et al., 2014). Given that heavy metals are persistent and non-degradable, their contamination of soil is irreversible and leads to a decline in soil fertility as well as in crop yield and quality (Luo et al., 2015). Heavy metals accumulated in plants and animals can enter the food chain and subsequently harm human health (Iqbal et al., 2015). Thus, the remediation of heavy metal-contaminated soil has long been a challenge and a popular research focus. Phytoremediation, in which hyperaccumulator plants remove contaminants from polluted soil, is an eco-friendly and cost-effective remediation measure (Lee, 2013).
Heavy metal hyperaccumulators are species of plants that can naturally accumulate substantial amounts of heavy metals in their aerial organs without showing apparent signs of toxicity (Verbruggen et al., 2013). Sedum alfredii Hance (Crassulaceae), a species first discovered in mining regions in China (Yang et al., 2002), is a highly valuable material for phytoremediation because of its propensity to hyperaccumulate cadmium (Cd), zinc (Zn) and lead (Pb). Recently, the allied species Sedum plumbizincicola was described (Wu et al., 2013). The physiological mechanisms underlying this species’ resistance and hyperaccumulation of different metal ions have been investigated (Lu et al., 2008; Jin et al., 2008; Li et al., 2016; Tian et al., 2016; Tian et al., 2017). Analyses of the transcriptome, small RNAs, degradome and proteome have been carried out under Cd stress (Li et al., 2016; Han et al., 2016; Peng et al., 2017). Furthermore, many genes associated with Cd uptake, resistance and hyperaccumulation, such as SpHMA3, SpMTL, SaREF, SaMT2, SaCAX2 and SaCu/ZnSOD, have been characterized from S. plumbizincicola and S. alfredii (Zhang et al., 2014; Liu et al., 2016; Zhang et al., 2016; Li et al., 2017; Liu et al., 2017; Peng et al., 2017). Despite these findings, transcription factors, which play an important role in abiotic stress, remain largely unknown in S. alfredii.
Heat shock transcription factors (Hsfs) comprise an important family of transcription factors involved in plant growth, development, signal transduction and stress response. Numerous studies have reported that the Hsfs of diverse plant species show different characteristics in response to abiotic stresses. Hsfs respond to external heat stress through activating the expression of heat shock proteins (Hsps) and other downstream genes by binding specifically the heat shock element (HSE) in the promoters of Hsf-responsive genes (Yamaguchi-Shinozaki and Shinozaki, 2006; Scharf et al., 2012; Wang et al., 2016). Hsfs are also involved in heavy metal tolerance. For instance, HsfA1a confers Cd tolerance in tomato (Solanum lycopersicum L. cv Ailsa Craig) by partially upregulating Hsps expression (Cai et al., 2017). In HsfA1a-silenced tomato plants, melatonin levels were down-regulated, whereas in HsfA1a-expressing plants the melatonin biosynthetic gene caffeic acid O-methyltransferase 1 (COMT1) was up-regulated and melatonin accumulation was induced under Cd treatment (Cai et al., 2017). The downstream gene metallothionein of HsfA4a is critical to enhance Cd tolerance in wheat (Triticum aestivum) and rice (Oryza sativa) (Shim et al., 2009). These findings demonstrate that Hsfs can bind to HSEs in the promoters of heavy metal stress-responsive genes to regulate the expression of downstream genes.
We previously observed that 18 SaHsfs showed a high expression level among the 22 members of the Hsf gene family identified in hyperaccumulation ecotype (HE) S. alfredii after Cd treatment. Furthermore, SaHsfA4c improves the Cd tolerance and accumulation in transgenic yeast (Chen et al., 2018). The function of SaHsfA4c in the regulation of Cd tolerance, however, remains unknown. In the present study, we analyzed the functions and subcellular location of SaHsfA4c from the HE S. alfredii. We concluded that overexpression of SaHsfA4c in Arabidopsis and non-hyperaccumulation ecotype (NHE) S. alfredii enhanced Cd tolerance by regulating the activities of reactive oxygen species (ROS)–scavenging enzymes and Hsps expression. SaHsfA4c is thus a potentially valuable gene for phytoremediation.
Materials and Methods
Plant Materials and Growth Conditions
Hyperaccumulation and non-hyperaccumulation ecotype S. alfredii were collected from mining regions in Quzhou City, China, and a tea plantation in Hangzhou City, China, respectively. The shoots were excised and cultivated in half-strength Hoagland-Arnon solution (pH 5.8). The nutrient solution was renewed every 3 days. After three weeks, robust and uniform plantlets (approximately 10 cm in height) were selected and cultured in half-strength Hoagland-Arnon nutrient solution containing 400 μM CdCl2. Plants were harvested and separated into roots, stems and leaves after 0, 0.5, 1, 6 and 12 h of treatment. The plants were grown in an artificial climate chamber under controlled conditions with a long-day photoperiod (16 h light/8 h dark) at 25°C.
Arabidopsis thaliana (Col-0) plants and transgenic Arabidopsis were grown under sterile conditions using half-strength Murashige and Skoog medium under the same conditions as S. alfredii. Seven-day-old plants were then incubated in Hoagland-Arnon nutrient solution for one month. Root and leaf samples were obtained after treatment with nutrient solution supplemented with or without 30 µM CdCl2 for one week. All experiments were performed in triplicate.
Gene Isolated, Identification and Plasmid Construction
To clone SaHsfA4c, total RNA was isolated from HE S. alfredii using the Total RNA Purification Kit (Aidlab, Beijing, China). cDNA was synthesized from 1 µg total RNA using the SuperScript® III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The full-length SaHsfA4c cDNA was isolated from HE S. alfredii using the species-specific primers SaHsfA4c-F/R designed based on the transcriptome sequences. In addition, the genomic sequence of SaHsfA4c was PCR-amplified from genomic DNA, which was isolated from the leaf of HE S. alfredii using the cetyltrimethyl ammonium bromide method. The obtained sequences were confirmed by sequencing and Blast analysis against the NCBI database (http://www.ncbi.nlm.nih.gov/). The Heatster platform (https://applbio.biologie.uni-frankfurt.de/hsf/heatster/) was used for sequence identification and domain analysis.
The purified PCR products were first ligated into the pDONR222 vector (Invitrogen, Carlsbad, USA) using the Gateway® BP Clonase reaction. This construct was then sequenced and recombined with the destination vectors pK7WGF2.0, pYES-DEST52 and pH2GW7.0 using the Gateway® LP Clonase reaction to generate subcellular localization, yeast, and plant expression vectors, respectively.
Phylogenetic Analysis
Deduced amino acid sequences containing the N-terminal region, DNA binding domain (DBD), and oligomerization domain (HR-A/B regions) of SaHsfA4c and other HsfA4 members from different plant species were used to compile a multiple sequence alignment. The sequence alignment was generated using ClustalX software and visualized with GeneDoc. Neighbor-joining trees were constructed using MEGA 6.0 with 1000 bootstrap repetitions performed to evaluate statistical support for the tree topology. Information on the sequences analyzed is given in Supplementary Table S1.
Quantitative Real-Time-PCR, Western Blotting and Immunolocalization
SYBR-based Quantitative real-time PCR (qRT-PCR) mixtures (TaKaRa, Da Lian, China) were prepared according to the manufacturer’s instructions and analyzed on a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). At least three biological replicates, with three technical replicates, were performed per sample. UBC9 and TUB served as internal controls for HE and NHE S. alfredii (Sang et al., 2013), whereas AtActin was used for Arabidopsis. Relative expression levels were calculated using the 2−ΔΔCt method. To construct a heat-map, the original expression levels of the analyzed genes were first normalized by the z-score method. And then, the normalized mRNA levels of the wild type (WT) under the non-stress condition (Control) were set arbitrarily to 1. Finally, the results were displayed using HemI software (The CUCKOO Workgroup, Wuhan, China). Values ranging from −2 to +2 were used to indicate the absolute signal intensity, and red and green colors were used to represent high and low expression values, respectively. All primers used in this analysis are listed in Supplementary Table S2.
A specific peptide (KEDNIKYDGLLTMH) for the SaHsfA4c protein was synthesized and used to produce polyclonal antibodies in rabbits (Abmart, Shanghai, China). The root, stem, and leaf from HE S. alfredii were ground into powder in liquid nitrogen and incubated with extraction buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1 mM PMSF, and 0.5% NP-40) at room temperature for 5 min. The lysate was centrifuged at 12,000 rpm for 15 min and the supernatant was retained. The protein concentration was quantified with the Enhanced BCA Protein Assay Kit (Beyotime, Shanghai, China). A total of 30 μg of protein per sample was used for SDS–PAGE and western blotting. Western blotting and immunolocalization were performed in accordance with methods detailed in previous studies (Han et al., 2017).
Subcellular Localization
The subcellular location of SaHsfA4c protein was determined by polyethylene glycol-mediated transformation of Arabidopsis protoplasts according the description by Yoo et al. (2007). The empty vector p35S-GFP was used as control. Arabidopsis protoplasts transformed with pK7WGF2.0-SaHsfA4c were observed under a LSM510 confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany).
Cadmium Tolerance Analysis in Yeast
The lithium acetate method was used to introduce expression constructs into Saccharomyces cerevisiae mutant strain Δycf1 (Szczypka et al., 1994). The transformants pYES-DEST52-SaHsfA4c and pYES-DEST52-InSaHsfA4c and yeast containing the empty vector pYES2.0, were cultured in liquid synthetic dextrose−uracil medium until OD600 up to the logarithmic phase. The bacteria solutions were then diluted serially (OD600 = 100, 10−1, 10−2, 10−3, 10−4 and 10−5) and spotted on synthetic galactose–uracil agar plates supplemented with CdCl2 (0, 15, 30, or 50 µM). Photographs were taken after incubation at 28°C for 3 days. To evaluate the growth status of yeast transformants in SG-U liquid medium containing 15 µM CdCl2, the OD600 was measured every 12 h for 96 h. The Cd content was measured as described by Li et al. (2017). The empty vector pYES2.0 was used as the control in each experiment. At least three biological replicates, with three technical replicates, were performed per analysis.
Plant Transformation and Physiological Analysis
The floral dip method (Clough and Bent, 1998) and a previously reported protocol (Liu et al., 2017) were applied for transformation of Arabidopsis and NHE S. alfredii, respectively, using Agrobacterium tumefaciens strain EHA105 carrying pH2GW7.0-SaHsfA4c. An Arabidopsis AtHsfA4c T-DNA insertion line (SALK_086202; athsfa4c) was obtained from the SALK collection, and homozygous mutants were used for the phenotype rescue experiment. The following transgenic plant lines with high transcript levels of SaHsfA4c were selected for subsequent experiments. oxSaHsfA4c#1 and oxSaHsfA4c#2 for overexpression in Arabidopsis, athsfa4c/SaHsfA4c#1 and athsfa4c/SaHsfA4c#2 for rescue of the Arabidopsis mutant athsfa4c, and oxSaHsfA4c#4′, oxSaHsfA4c#7′ and oxSaHsfA4c#10′ for overexpression in NHE S. alfredii.
To investigate the potential effects of SaHsfA4c in plants, leaves of Arabidopsis were stained with 3,3′diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) to reveal in situ accumulation of two ROS indices, namely hydrogen peroxide (H2O2) and superoxide anion (O2−), respectively. Leaves from the same location in each plant were harvested and immersed in DAB (1 mg∙ml−1 DAB in ddH2O, pH 3.8) or NBT solution (1 mg∙ml−1 NBT in 10 mM phosphate buffer, pH 7.8), respectively. The samples were then incubated at room temperature for 12 h in darkness. Finally, the leaves were bleached by boiling in ethanol for 20 min to remove chlorophyll. Brown and blue spots respectively indicated the presence of H2O2 and O2− in situ (Thordal-Christensen et al., 1997; Wohlgemuth et al., 2002). The activities of ROS-scavenging enzymes, including ascorbate peroxidase (APX), catalase (CAT) and peroxidase (POD), were measured. Ice-cold phosphate buffer was added to one gram of frozen, pulverized leaf samples. The homogenate was centrifuged with 12,000 rpm at 4°C for 10 min, and the supernatant was used for enzyme activity determination. The activities of APX, CAT, and POD were measured by absorption photometry as described by Aebi (1984); Ullah et al. (2016) and Polle et al. (1994), respectively. All experiments were repeated three times independently.
Co-Expression Network Construction
Gene co-expression relationships in HE S. alfredii were determined from published transcriptome data under Cd stress (Han et al., 2016). To obtain insights into the functions of gene co-expressed with SaHsfA4c, gene ontology (GO) terms were analyzed using osgo (http://www.omicshare.com/tools/Home/Soft/osgo). Finally, Hsfs, Hsps and antioxidant activity-related genes were selected to construct the co-expression subnetwork of SaHsfA4c (Supplementary Table S3). The subnetwork was visualized using Cytoscape software (Smoot et al., 2011).
Statistical Analyses
For a given biological sample, three or more representative technical repeats were analyzed and the mean value for the biological sample was determined. Data were calculated as the means ± standard deviation (SD) of three or more independent biological repeats of each sample. SPSS package 20.0 (IBM Corporation, Armonk, NY, USA) and Origin 8.5 (OriginLab, Northampton, MA, USA) software were used to perform the statistical analyses and display the results, respectively. The significance of differences among multiple groups was evaluated by one-way ANOVA followed by Tukey’s multiple range test (p = 0.05).
Results
Gene Isolation and Bioinformatic Analysis
The full-length coding sequence of SaHsfA4c was cloned and confirmed by sequencing. Interestingly, two PCR products were detected by gel electrophoresis (Figure 1A). DNA sequencing revealed that SaHsfA4c was alternatively spliced into two distinct transcripts of 1,302 and 1,503 bp length. The longer transcript, with a 201 bp intron, was present in the SaHsfA4c genomic DNA sequence; thus, the two transcripts were designated SaHsfA4c and InSaHsfA4c. The SaHsfA4c transcript encoded 433 amino acids and included a conserved DBD domain, an intermediate HR-A/B region, a nuclear location signal (NLS), two transcriptional activation domains (AHA motifs) and one nuclear export signal (NES) predicted using the Heatster online tool (Figure 1B). The second transcript, InSaHsfA4c, contains an advanced stop codon; it encoded a truncated protein variant of 109 amino acids, harbored an incomplete DBD, and lacked both the HR-A/B and AHA motifs (Figure 1A). Phylogenetic analysis revealed that SaHsfA4c belongs to the HsfA4c subgroup, as it showed a high similarity to other known HsfA4c proteins (Figure 1C).
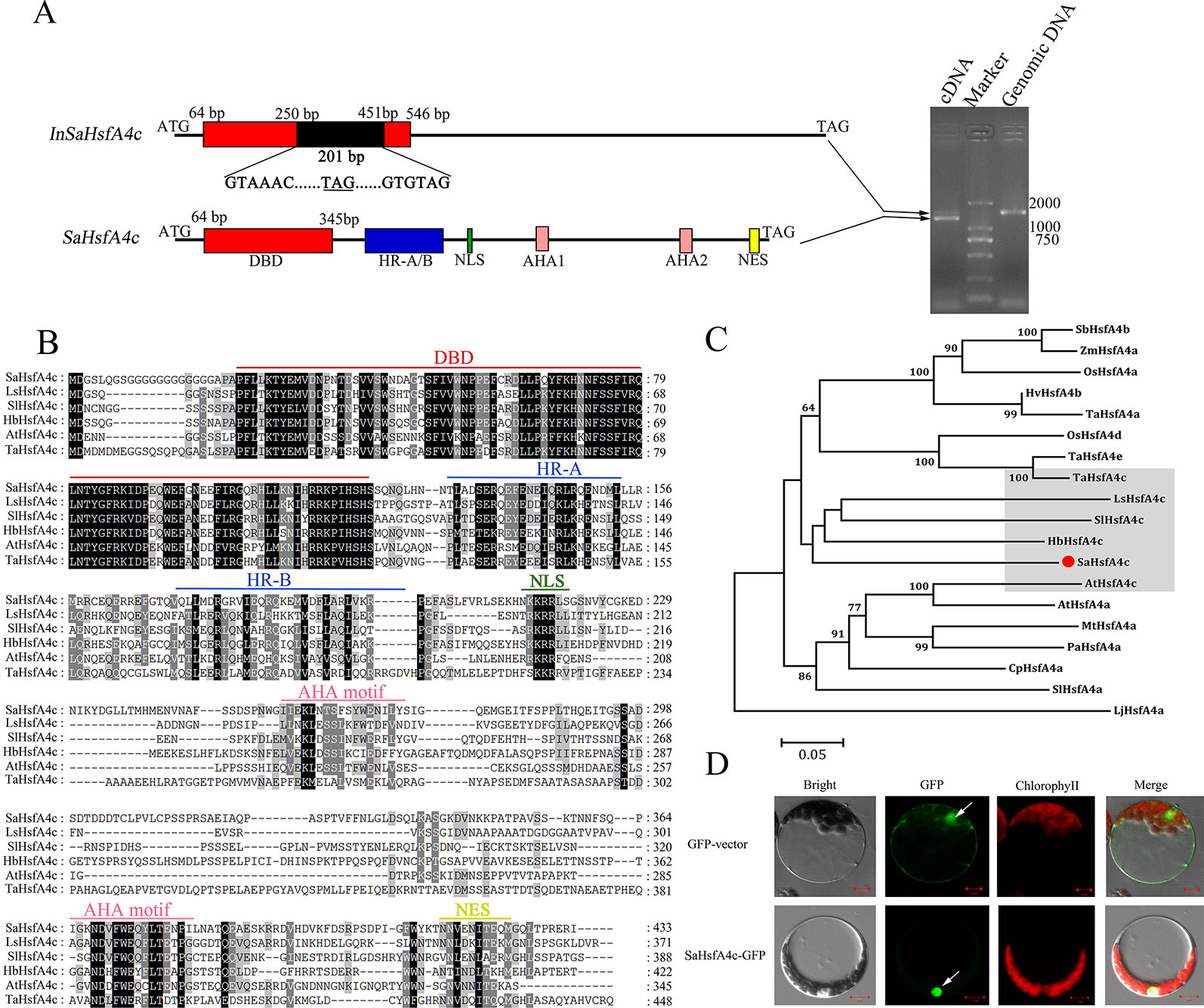
Figure 1 Identification of SaHsfA4c. (A) Schematic diagram of the SaHsfA4c and its two transcripts (Left). A 201 bp intron existed in the InSaHsfA4c mRNA, introducing a new stop codon. The new stop codons are underlined. Genomic DNA or cDNA was used as the template for PCR amplification of SaHsfA4c gene (Right). (B) Comparing amino acids sequence from SaHsfA4c and its orthologs. (C) Phylogenetic analysis of SaHsfA4c and other Hsf proteins. The species of origin of the HsfA4 s are indicated by the abbreviation before the gene names: SbHsfA4b, XP_002456359.1, Sorghum bicolor; ZmHsfA4a, CAA58117.1,Zea mays; OsHsfA4a, AK109856, Oryza sativa; OsHsfA4d, AK100412, Oryza sativa; HvHsfA4b, AEB26589.1, Hordeum vulgare subsp. Vulgare; TaHsfA4a, ACN93796.2, Triticum aestivum; TaHsfA4c, AHZ44767, Triticum aestivum; TaHsfA4e, AHZ44768, Triticum aestivum; LsHsfA4c, XP_023766107.1, Lactuca sativa; SlHsfA4a, XP_010317423.1, Solanum lycopersicum; SlHsfA4c, XP_004243245.1, Solanum lycopersicum; HbHsfA4c, XP_021689590.1, Hevea brasiliensis; AtHsfA4a, NP_193623.1, Arabidopsis thaliana; AtHsfA4c, NP_199383.1, Arabidopsis thaliana; MtHsfA4a, XP_003629847.1, Medicago truncatula; PaHsfA4a, AAL12248.1, Phaseolus acutifolius; CpHsfA4a, XP_021890826.1, Carica papaya. (D) Subcellular localization of SaHsfA4c. White arrows indicate the nucleus of cell. Control, p35S-GFP vector; SaHsfA4c-GFP, SaHsfA4c fused with GFP. Scale bar = 10 μm.
The predicted NLS domain located between the HR-A/B region and the AHA1 motif suggested a putative nuclear targeting of SaHsfA4c, and the SaHsfA4c protein contained NLS sequences (KKRR). To test this hypothesis, transient expression analysis in Arabidopsis protoplasts was conducted to verify the subcellular localization of the SaHsfA4c protein. An expression construct containing SaHsfA4c fused with GFP was introduced into the protoplasts. Green fluorescence signals were mainly detected in the nucleus of cells. p35S-GFP, used as the control vector, was expressed throughout the cells (Figure 1D).
Expression Patterns and Immunolocalization
On the basis of qRT-PCR analysis, SaHsfA4c was expressed under the non-stress condition in all three tissues analyzed (root, stem and leaf) of HE S. alfredii. The highest expression level was detected in the stem, followed by the root (Figure 2A). Under Cd stress, SaHsfA4c transcript and protein levels increased in all tissues. SaHsfA4c expression attained a peak of 2.15- and 1.82-fold at 1 h and 0.5 h after Cd stress in the root and leaf, respectively, and then gradually decreased (Figures 2B, C). In contrast, SaHsfA4c expression in the stem increased gradually, and the SaHsfA4c transcript level after 12 h treatment was 1.68-fold higher than that before treatment (Figure 2D). Similar trends in SaHsfA4c protein levels with or without exposure to Cd stress were observed.
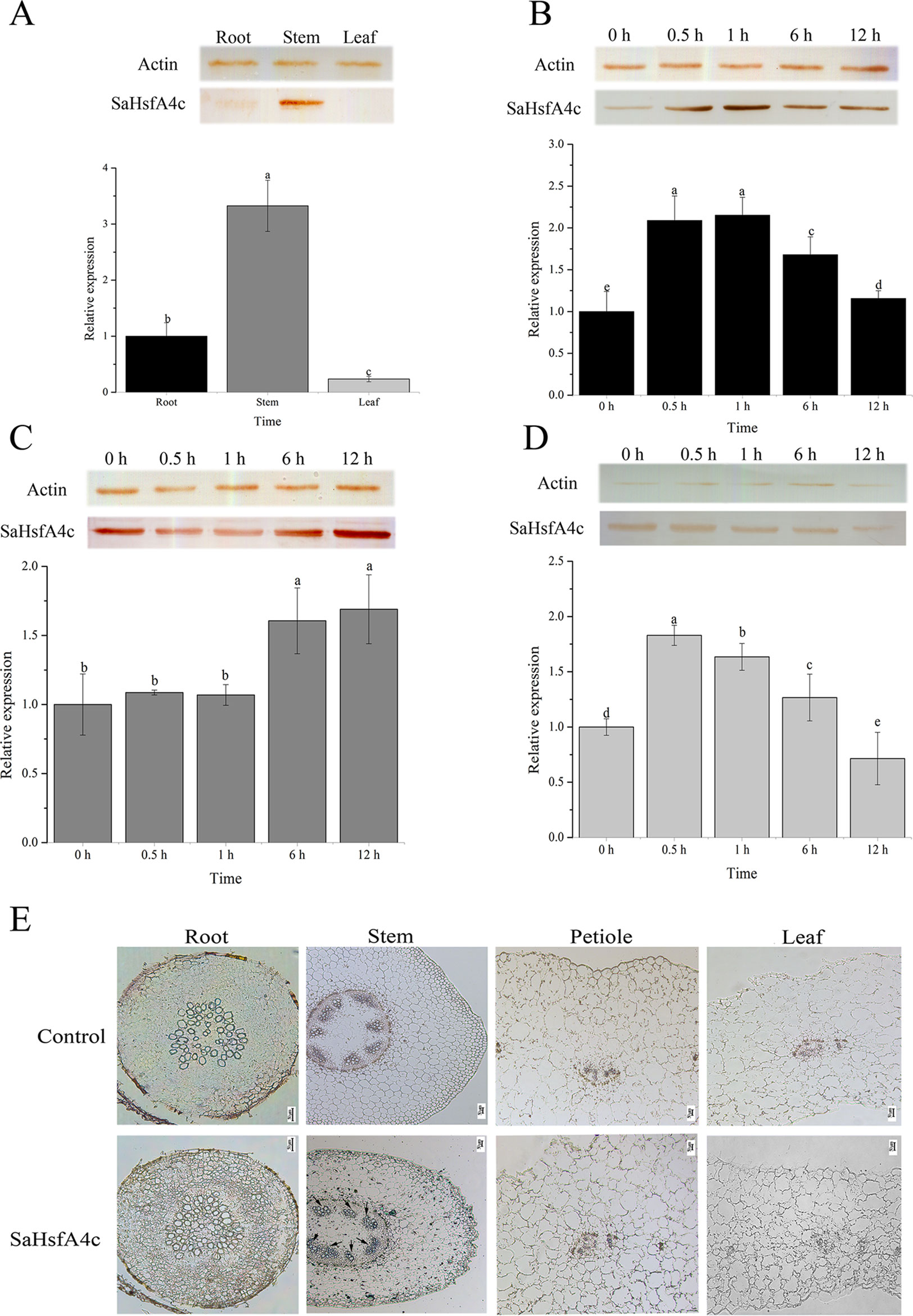
Figure 2 Expression patterns and immunolocalization of SaHsfA4c. (A–D) Expression patterns of SaHsfA4c under Cd treatment. Western blotting to quantify SaHsfA4c protein accumulation in treated tissues was present as SDS-PAGE gel, and the expression profiles of SaHsfA4c in S. alfredii treated with CdCl2 for different amounts of time were displayed in histogram. The normalized mRNA levels of the root (A) and tissues (B–D) without treatment were set arbitrarily to 1, respectively. (A) Without Cd treatment. (E) Immunolocalization of SaHsfA4c. Black arrows indicate the hybridization signals. A total of 30 μg of protein per sample was used for SDS–PAGE and western blotting. Different letters on the bars indicate significant difference among multiple groups according to Tukey’s multiple range test (p = 0.05).
To determine the tissue specificity of SaHsfA4c, immunolocalization was carried out using root, stem, leaf and petiole sections. Highly specific SaHsfA4c signals were mainly detected in stem xylem cells, with weaker signals observed in the root, petiole, and leaf. No signals were detected in the control (non-hybridization with SaHsfA4c) (Figure 2E). These results were generally consistent with the expression patterns of SaHsfA4c obtained by qRT-PCR and western blotting, which suggested that the SaHsfA4c protein specifically localized to the stem in HE S. alfredii.
The DBD of SaHsfA4c Is Vital for Cd Tolerance in Yeast
To determine their influence on Cd tolerance, SaHsfA4c and InSaHsfA4c transcripts were recombined in the yeast expression vector pYES-DEST52. Interestingly, only SaHsfA4c substantially enhanced Cd tolerance in Δycf1. In the absence of CdCl2, no significant difference in colony number or size was observed between the empty vector (EV) and Δycf1 harboring SaHsfA4c or InSaHsfA4c. On SG-U medium supplemented with 15, 30, or 50 μM CdCl2, the growth of the three transgenic yeast strains was inhibited; however, growth of the Δycf1 strain expressing SaHsfA4c was superior to that of the Δycf1 strain transfected with the EV (Figure 3A). In liquid media supplemented with 15 μM CdCl2, the growth of yeast cells expressing SaHsfA4c was higher than that of the EV and InSaHsfA4c (Figure 3B). Furthermore, the Cd content of the strain overexpressing SaHsfA4c was significantly higher than that of the other two strains. The Cd contents of yeast overexpressing SaHsfA4c and InSaHsfA4c were respectively 2.8- and 0.24-fold higher than that of the EV-transfected strain (Figure 3C). These results indicated that the yeast expressing SaHsfA4c showed increased Cd uptake and accumulation, and that growth of transgenic yeast was promoted by SaHsfA4c under Cd stress.
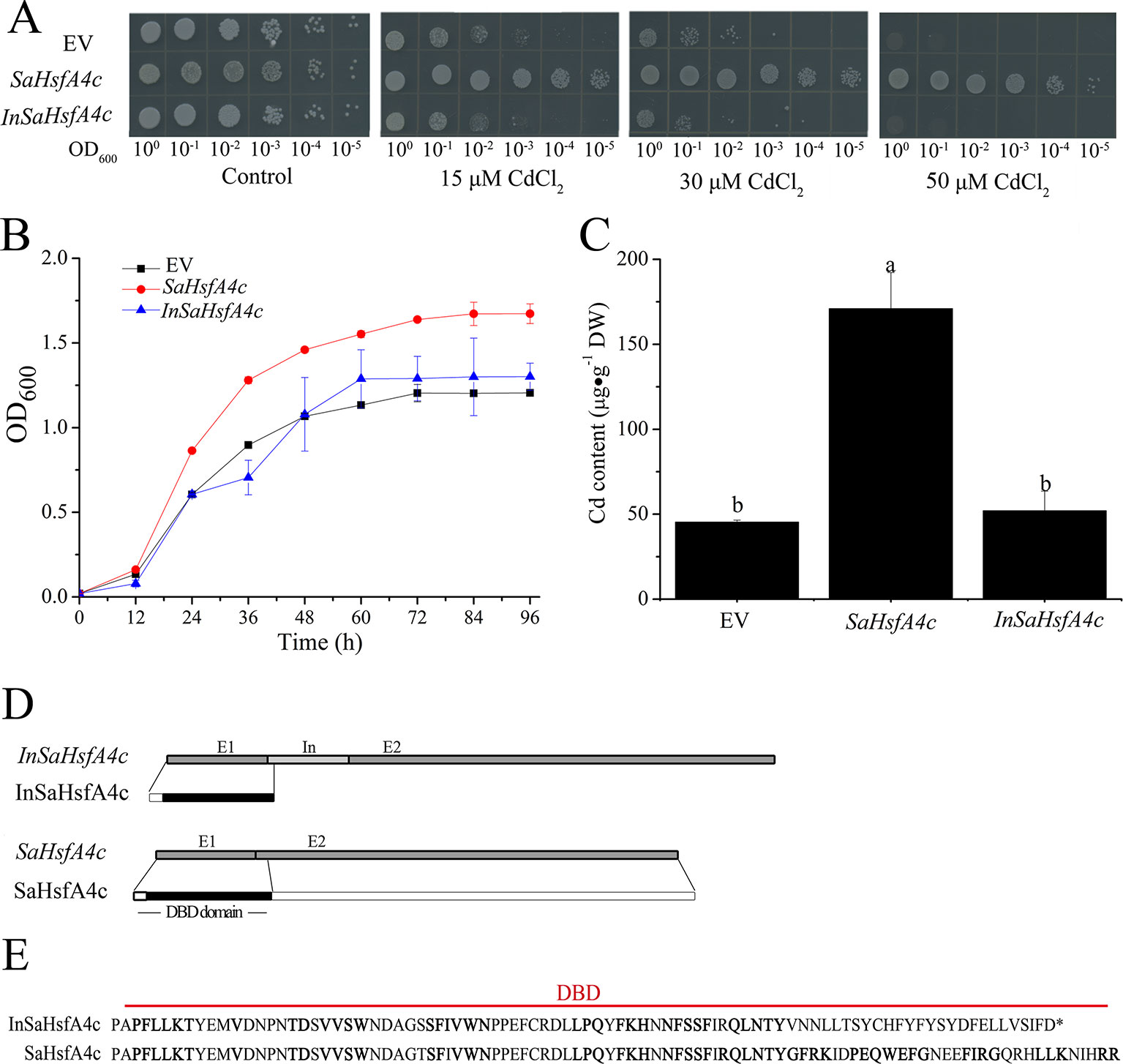
Figure 3 SaHsfA4c increased Cd tolerance in transgenic yeast. (A) Cd tolerance assay. (B) The time course growth of each yeast strains was measured by OD600. (C) Cd content. (D) Transcript and protein structures of SaHsfA4c and InSaHsfA4c. E1, E2 and In represent exon 1, 2 and intron. (E) The sequences of DBD in SaHsfA4c and InSaHsfA4c.
Comparison of the nucleotide sequences of the two transcripts (SaHsfA4c and InSaHsfA4c) revealed the presence of an intron between exons 1 and exons 2 of InSaHsfA4c (Figure 3D). Heatster predicted the DBD of SaHsfA4c to contain a complete exon 1 and partial exon 2, whereas InSaHsfA4c was predicted to harbor a premature termination codon in the intron and contain an incomplete DBD domain (Figures 3D, E). These results suggested that the DBD of SaHsfA4c is critical for Cd resistance.
SaHsfA4c Enhances Cd Tolerance in Transgenic Arabidopsis and Rescues Cd-Sensitive Defects of the athsfa4c Mutant
Ten transgenic Arabidopsis lines were obtained, among which two lines (oxSaHsfA4c#1 and oxSaHsfA4c#2) expressing high levels of SaHsfA4c were selected for subsequent experiments (Supplementary Figure S1A). Higher expression levels of SaHsfA4c were observed in oxSaHsfA4c#1 and oxSaHsfA4c#2 under the non-stress condition (Supplementary Figure S1B). After Cd treatment for 7 days, the uppermost leaves of oxSaHsfA4c#1 and oxSaHsfA4c#2 plants remained green with only the basal leaves appearing slightly yellow, whereas most leaves of WT plants became severely wilted (Figure 4A). In addition, Cd stress up-regulated SaHsfA4c expression in the transgenic lines oxSaHsfA4c#1 and oxSaHsfA4c#2 (Figure 4B).
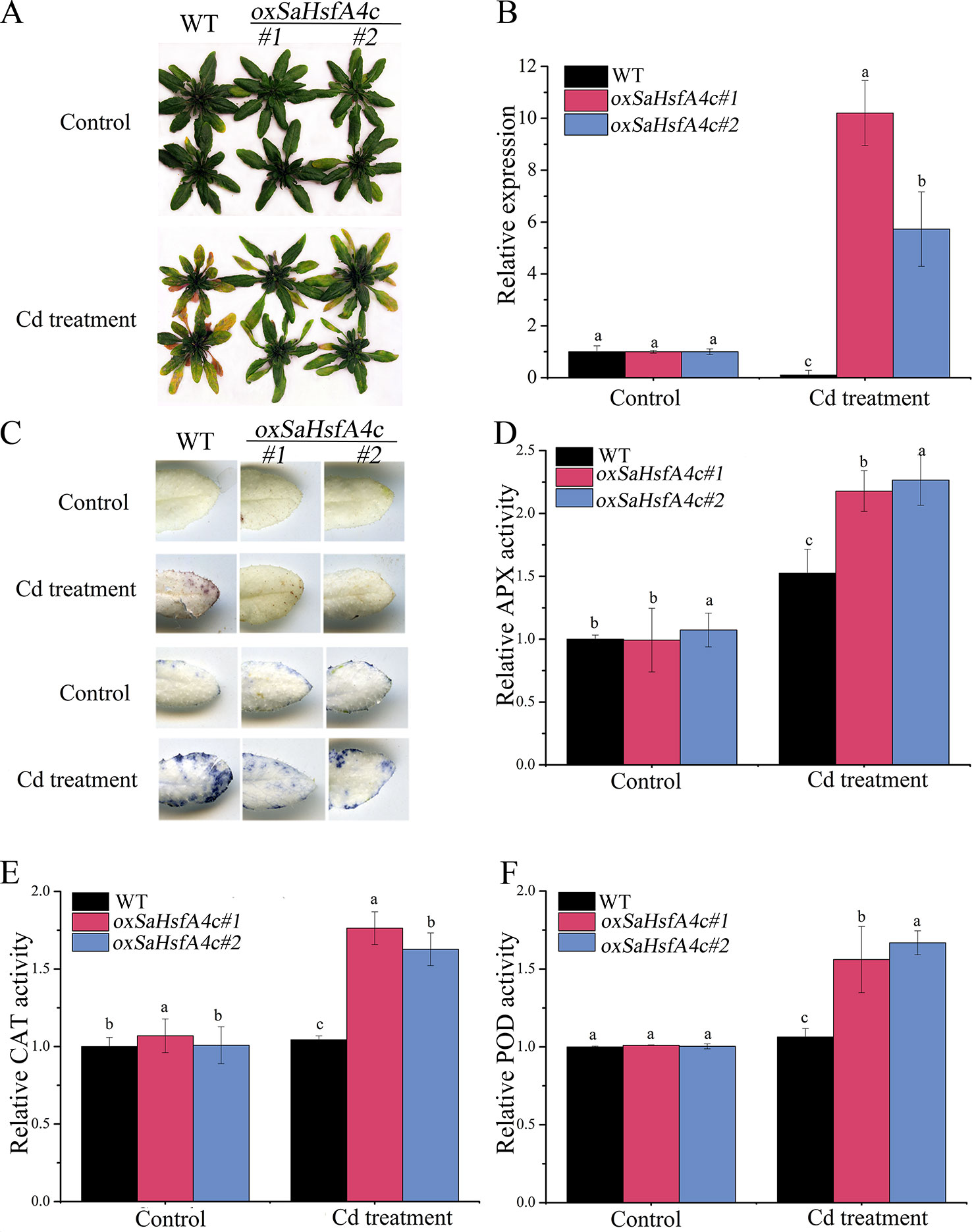
Figure 4 SaHsfA4c exhibits resistance to CdCl2 stress in transgenic Arabidopsis. (A) Growth status. (B) Relative expression level of SaHsfA4c in transgenic Arabidopsis. (C) DAB staining (upper) and NBT staining (lower). (D) Relative APX activity in the leaf. (E) Relative CAT activity in the leaf. (F) Relative POD activity in the leaf. Control, without Cd treatment; Cd treatment, 30 µM CdCl2 treatment for one week. Different letters on the bars indicate significant difference between WT and other lines. Bars indicate means ± standard deviations (SDs) of at least three independent biological experiments. Different letters on the bars indicate significant difference among multiple groups according to Tukey’s multiple range test (p = 0.05).
Physiological responses to Cd stress were compared between the WT and the two transgenic Arabidopsis lines (oxSaHsfA4c#1 and oxSaHsfA4c#2). Histochemical staining revealed that significantly less H2O2 and O2− accumulated in the transgenic Arabidopsis lines (oxSaHsfA4c#1 and oxSaHsfA4c#2) than in the WT under Cd treatment (Figure 4C). The activities of ROS-scavenging enzymes in the WT and transgenic lines were assayed to elucidate the role of SaHsfA4c in ROS homeostasis. Under the non-stress condition, APX, CAT, and POD activities were slightly higher in the transgenic Arabidopsis lines than those in the WT. Furthermore, the transgenic plants showed 1.2- to 3.0-fold higher APX, CAT and POD activities than the WT plants after Cd treatment (Figures 4D–F, Supplementary Figure S2). Next, the expression levels of ROS homeostasis-related genes (encoding ROS -scavenging enzymes), including AtAPX, AtCAT and AtPOD, and the downstream target genes of Hsfs, AtHsp18.1, AtHsp22, AtHsp23.6, AtHsp70, AtHsp90 and AtHsp101, were analyzed by qRT-PCR. Although, ROS scavenging related genes in leaves of all tested lines were significantly up-regulated after Cd treatment, the expression levels of oxSaHsfA4c#1 and oxSaHsfA4c#2 lines were markedly higher than those of the WT (Figure 5).
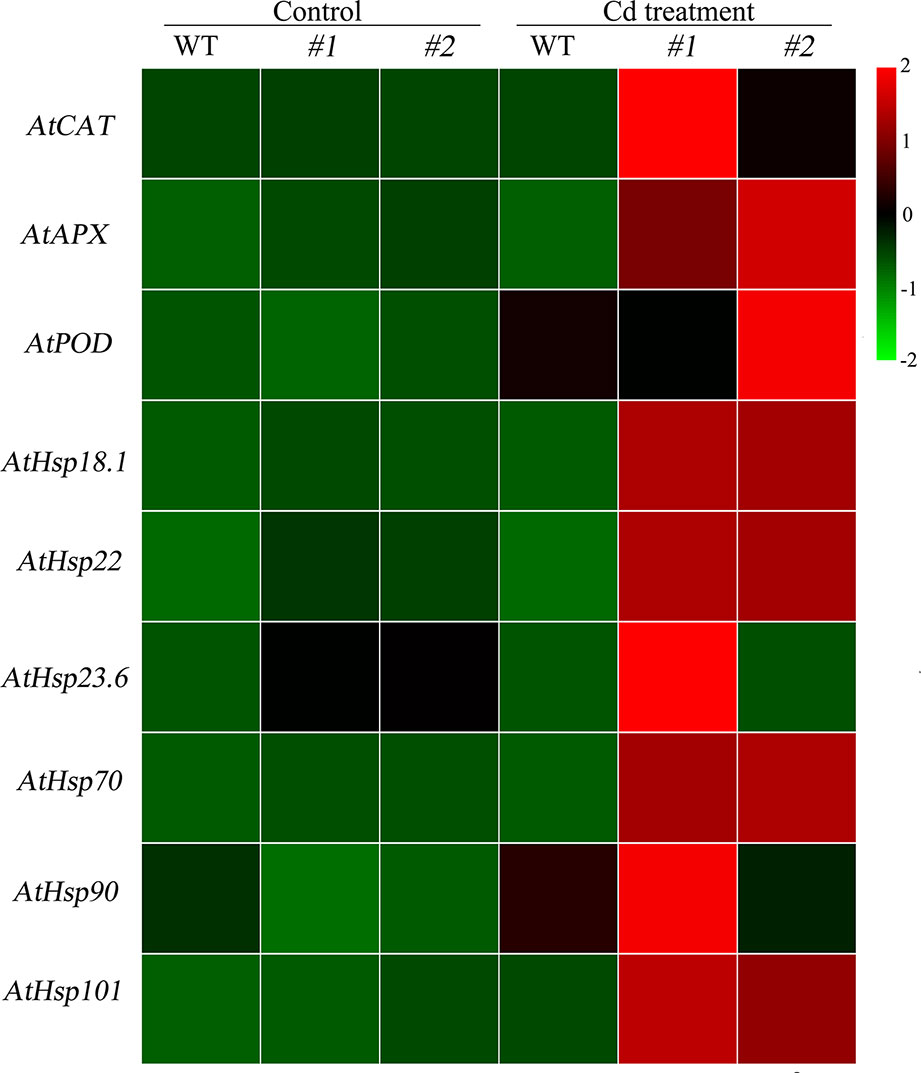
Figure 5 Expression patterns of ROS related genes and Hsps in transgenic Arabidopsis expressing SaHsfA4c. #1 and #2 represent the transgenic lines oxSaHsfA4c#1 and oxSaHsfA4c#2, respectively. Control, without Cd treatment; Cd treatment, 30 µM CdCl2 treatment for one week. The red and green colors were used to represent high and low expression values, respectively.
To further elucidate the functions of SaHsfA4c in regulating Cd tolerance, the plant expression vector harboring SaHsfA4c was also introduced into the Cd-stress-sensitive Arabidopsis athsfa4c mutant. Two transgenic lines (athsfa4c/SaHsfA4c#1 and athsfa4c/SaHsfA4c#2) showed significantly higher SaHsfA4c expression levels than the WT and athsfa4c mutant (Supplementary Figure S1C). Under the control condition, growth status was not significantly different among the four lines (WT, athsfa4c, athsfa4c/SaHsfA4c#1 and athsfa4c/SaHsfA4c#2). After Cd treatment for 7 days, both transgenic lines exhibited less yellowing compared with that of the WT (Figure 6A). In addition, SaHsfA4c was up-regulated in the transgenic lines (athsfa4c/SaHsfA4c#1 and athsfa4c/SaHsfA4c#2) by Cd treatment, with its expression increasing by 2.05- and 2.83-fold compared with that of the control (Figure 6B).
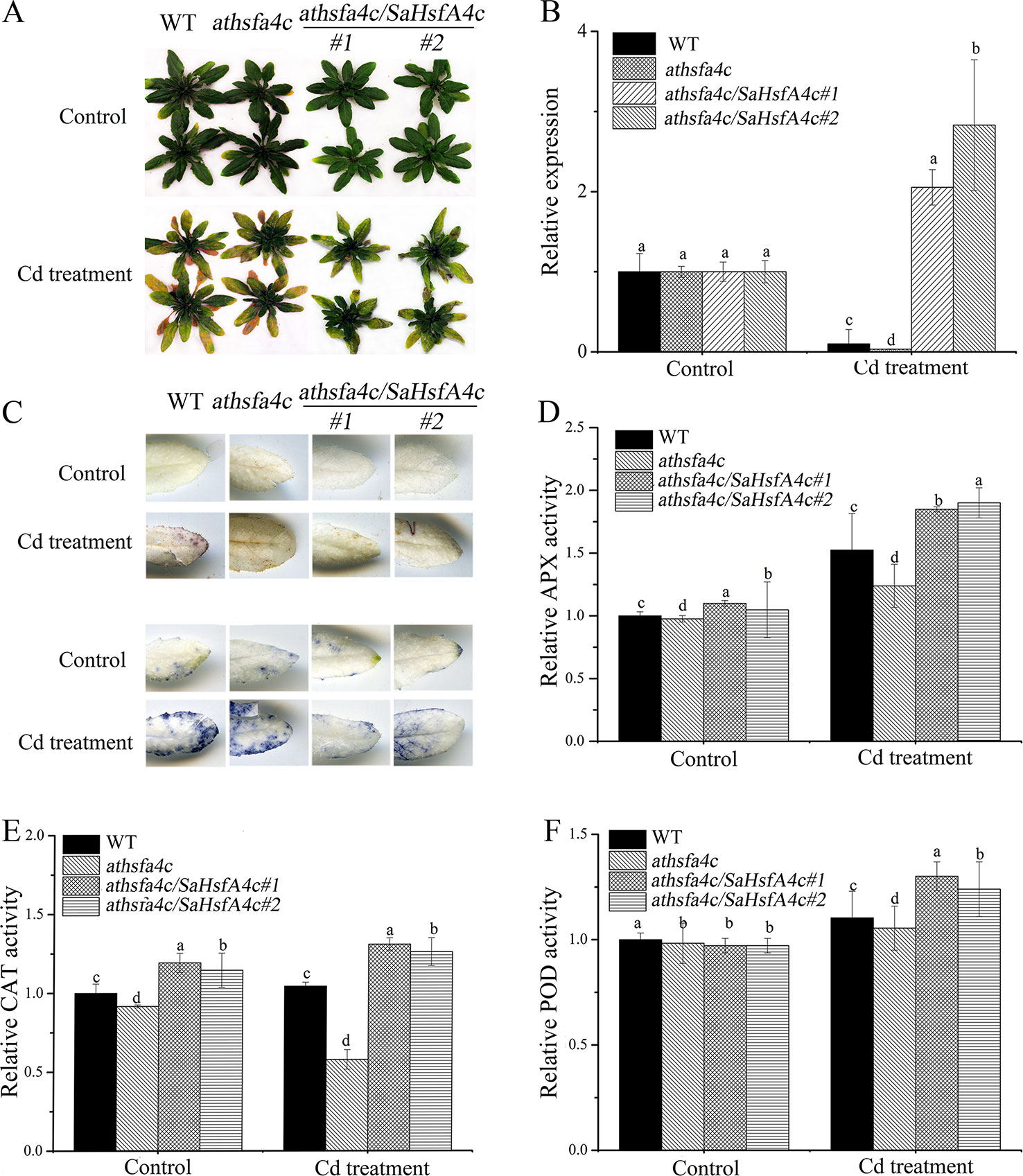
Figure 6 Characterization of Arabidopsis athsfa4c mutant transforming SaHsfA4c. (A) Growth status. (B) Relative expression level of SaHsfA4c. (C) DAB staining (upper) and NBT staining (lower). (D) Relative APX activity in the leaf. (E) Relative CAT activity in the leaf. (F) Relative POD activity in the leaf. Control, without Cd treatment; Cd treatment, 30 µM CdCl2 treatment for one week. Bars indicate means ± standard deviations (SDs) of at least three independent biological experiments. Different letters on the bars indicate significant difference among multiple groups according to Tukey’s multiple range test (p = 0.05).
Histochemical staining revealed that markedly less H2O2 and O2− accumulated in rescue Arabidopsis lines (athsfa4c/SaHsfA4c#1 and athsfa4c/SaHsfA4c#2) under Cd treatment, whereas ROS accumulation was detected in the leaves of both the WT and the mutant line (Figure 6C). We also measured the activities of ROS-scavenging enzyme activities. Even under the non-stress condition, APX activity in the root and CAT activity level in the leaf of rescued plants were 1.15- to 1.3-fold higher than those of WT plants, which suggested that the increased SaHsfA4c expression of rescued plants resulted in higher ROS-scavenging enzyme activities. Furthermore, the rescue lines showed 1.25- to 2.2-fold higher APX and CAT activities than those of WT plants after Cd treatment (Figures 6D, E, Supplementary Figure S3). Activities of POD in the transgenic Arabidopsis lines were not significantly different from those of the WT under the non-stress condition but were 1.2- to 1.3-fold higher than those in the WT after Cd treatment (Figure 6F, Supplementary Figure S3). In summary, APX, CAT, and POD activities of athsfa4c/SaHsfA4c#1 and athsfa4c/SaHsfA4c#2 markedly increased in both the leaf and root under Cd treatment, and were higher than those detected in the WT and athsfa4c mutant.
Overexpression of SaHsfA4c Confers Cd Tolerance in NHE S. alfredii
After validating their higher expression levels of SaHsfA4c under the control condition (Supplementary Figure S1D), three independent SaHsfA4c-overexpressing transgenic NHE S. alfredii lines (oxSaHsfA4c#4′, oxSaHsfA4c#7′ and oxSaHsfA4c#10′) were selected for further study. After 400 µM CdCl2 treatment, SaHsfA4c expression was significantly increased in these three transgenic lines but was slightly reduced in the WT (Figure 7A).
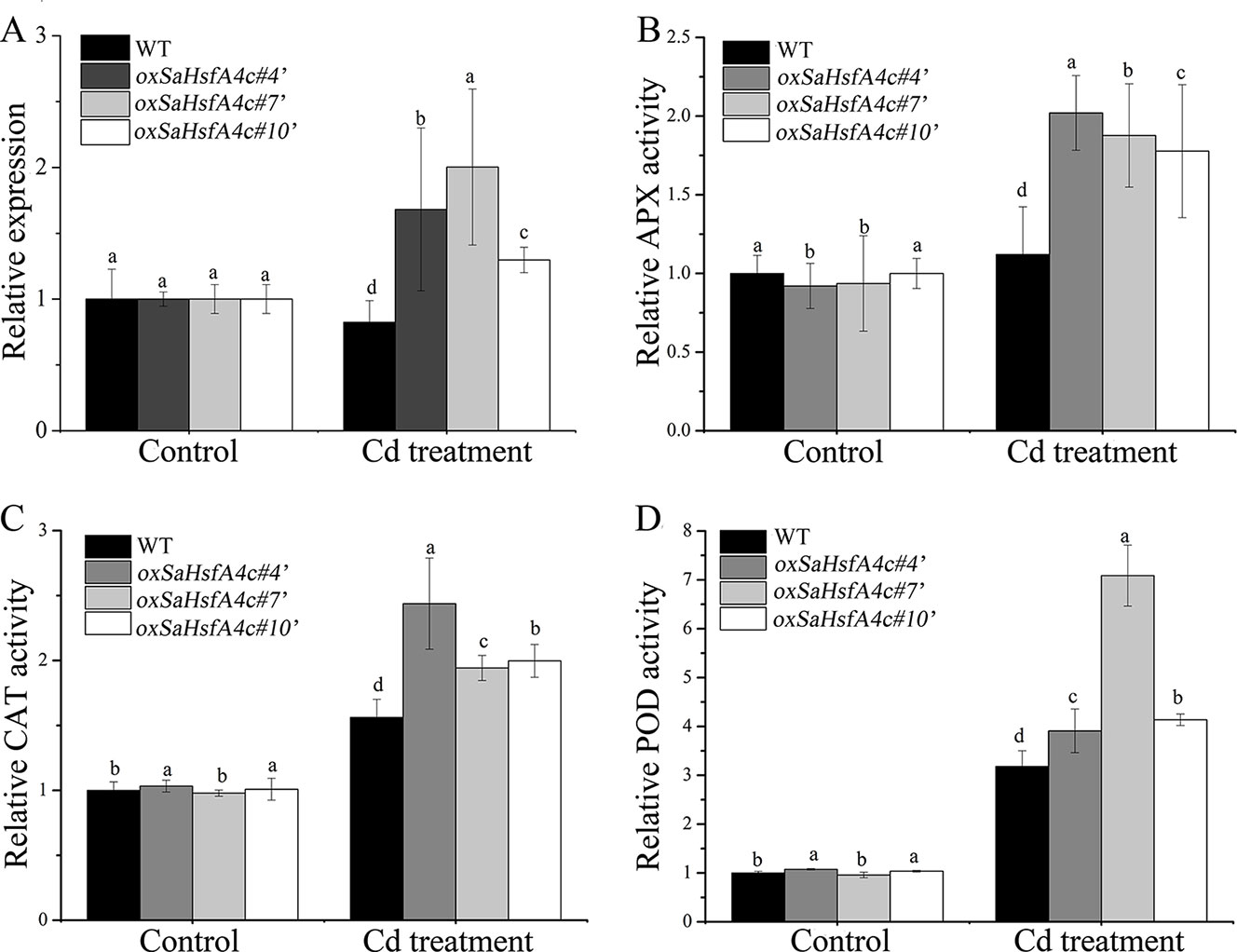
Figure 7 SaHsfA4c exhibits resistance to CdCl2 stress in transgenic NHE S. alfredii lines. (A) Relative expression level. (B) Relative APX activity in the leaf. (C) Relative CAT activity in the leaf. (D) Relative POD activity in the leaf. Control, without Cd treatment; Cd treatment, 400 µM CdCl2 treatment for one week. Bars indicate means ± standard deviations (SDs) of at least three independent biological experiments. Different letters on the bars indicate significant difference among multiple groups according to Tukey’s multiple range test (p = 0.05).
Plant physiological responses to Cd stress were also analyzed. As in Arabidopsis, activities of the ROS-scavenging enzymes APX, CAT, and POD in the leaf and root of the transgenic lines were higher than those of the WT, both under the control condition and under Cd stress (Figures 7B–D, Supplementary Figure S4).
To evaluate the regulatory roles of SaHsfA4c in NHE S. alfredii, the homologs of Arabidopsis’ ROS-scavenging related genes and Hsps were examined. All of these genes were up-regulated under Cd stress. Transcript levels of the tested genes were significantly higher in oxSaHsfA4c#4′, oxSaHsfA4c#7′ and oxSaHsfA4c#10′ than in the WT under Cd stress (Figure 8).
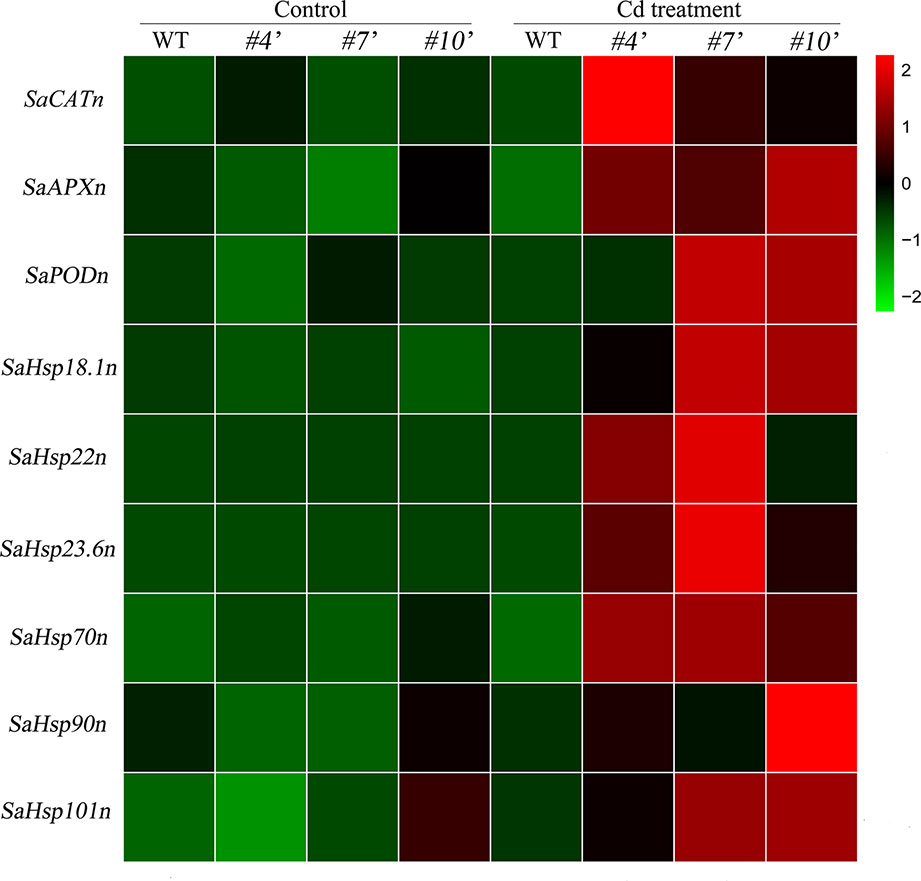
Figure 8 Expression patterns of candidate genes were performed in transgenic NHE S. alfredii overexpressing SaHsfA4c. #4′, #7′ and #10′ represent the transgenic lines oxSaHsfA4c#4′, oxSaHsfA4c#7′ and oxSaHsfA4c#10′, respectively. Control, without Cd treatment; Cd treatment, 400 µM CdCl2 treatment for one week. The red and green colors were used to represent high and low expression values, respectively.
Co-Expression Network of SaHsfA4c in HE S. alfredii
To provide additional context for the proposed function of SaHsfA4c, a gene co-expression analysis and GO functional classification were conducted using the transcriptome database obtained from HE S. alfredii under Cd stress (Han et al., 2016; Supplementary Figure S5). On the basis of the above mentioned results, which suggested that SaHsfA4c enhanced Cd tolerance by regulating ROS homeostasis and Hsp’ expression, we searched for relevant genes in the transcriptome data. A total of 45 genes were identified in the co-expression network (Figure 9). Notably, 18 genes in the co-expression network have been shown to be associated with ROS: 11 PODs, two APXs and two SODs. In addition, 19 Hsps with different molecular weight and eight Hsfs, which mainly belonged to the A and B subclasses, were identified as co-expressed genes.
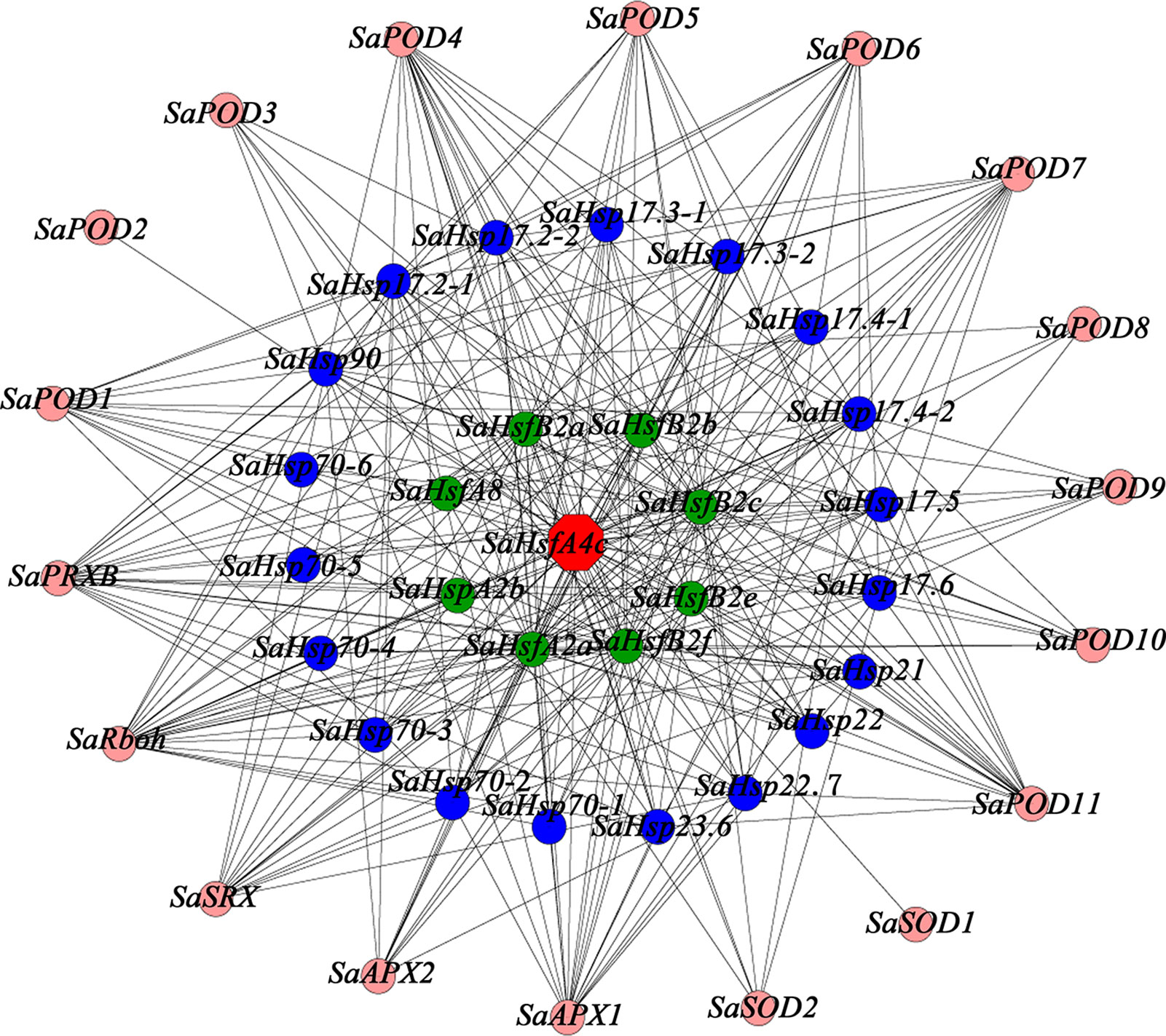
Figure 9 Co-expression network for SaHsfA4c. The red node is SaHsfA4c, pink nodes are genes related to encode ROS-scavenging enzymes, blue nodes are Hsps and green nodes are other Hsfs.
Discussion
In this study, SaHsfA4c, an ortholog of Hsf, was cloned from HE S. alfredii, and the functional properties and potential role of the gene in yeast and plants were characterized. The results showed that SaHsfA4c plays a role in Cd tolerance. It is proposed that this tolerance is mediated by target genes (ROS-related genes and Hsps) on the basis of two observations. First, SaHsfA4c was up-regulated under Cd stress, with higher transcript levels detected in the root, stem, and leaf (Figure 2). Second, ROS-related genes and Hsps were up-regulated in transgenic plants overexpressing SaHsfA4c (Figures 5 and 8).
In a previous study, TaHsfA4a and OsHsfA4a transgenic yeast exhibited Cd tolerance, and the growth status of the yeast strain overexpressing TaHsfA4a was superior to that of the control in liquid culture with CdCl2 (Shim et al., 2009). Similarly, the heterologous expression of SaHsfA4c improved Cd tolerance and accumulation in yeast in the current study. Compared with the control (EV), Δycf1 harboring a fusion vector (pYES-DEST52-SaHsfA4c) showed enhanced growth (Figure 3A), whereas InSaHsfA4c did not. Our detailed analyses of the functional domains of the two different transcripts of SaHsfA4c revealed that SaHsfA4c containing the completed DBD enhanced Cd resistance in yeast; in contrast, InSaHsfA4c with a truncated DBD did not improved Cd resistance, which indicated that the DBD is critical for Cd tolerance (Figure 3). We therefore speculate that the DBD activates or enhances other factors, thereby promoting the expression of target genes. These results suggested that SaHsfA4c can compensate for the Cd sensitivity of yeast mutants, and that the DBD is essential for Cd resistance and accumulation.
Reactive oxygen species, which are induced by abiotic stresses, are harmful to plant growth and development (Li et al., 2017). In the athsfa4a mutant, the H2O2 content is much higher than that of the WT, and the accumulation of H2O2 in atHsfA4a-overexpressing plants is lower than that in the WT under salinity stress (Guo et al., 2016). Plant response to stress-induced ROS production is important to protect the stability of intracellular ROS levels (Azarabadi et al., 2017). To protect plant cells from oxidative damage, plants use a high effective enzymatic antioxidant defense system involving ROS scavenging enzymes, such as APX, POD and CAT (Gill and Tuteja, 2010). Many studies have revealed crosstalk between Hsfs, ROS and ROS-scavenging enzymes. In Populus ussuriensis, PuHSFA4a can increase glutathione S-transferase activity to reduce ROS production and improve Zn resistance of roots by directly regulating the target gene PuGSTU17 (Zhang et al., 2019). Transgenic Arabidopsis overexpressing maize ZmHsf06 can enhance salt tolerance by increases POD and SOD activities (Li et al., 2015). AtHsfA2 regulated the ability of Arabidopsis to cope with heat and oxidative stress by inducing APX expression (Li et al., 2005). Transgenic tobacco overexpressing PeHsf produced more CAT under salt stress (Shen et al., 2013). Activities of SOD, APX and CAT in transgenic chrysanthemum overexpressing CmHsfA4, were higher than those of WT lines under salt stress (Li et al., 2017). In addition, Hsf activates the expression of downstream genes (including Hsps) and shows a similar pattern of expression to protect plants from external stress (Snyman and Cronjé, 2008; Xue et al., 2014; Xue et al., 2015; Chen et al., 2018). In the present study, we also observed that transgenic Arabidopsis and NHE S. alfredii expressing SaHsfA4c showed higher CAT, POD, and APX activities than those of the WT after Cd stress, and were able to induce the expression of ROS-scavenging system related genes (POD, CAT and APX) and Hsps. These results indicate that SaHsfA4c enhances the Cd tolerance of transgenic plants by activating ROS-scavenging enzyme activities and up-regulating Hsps.
Heavy metal tolerance and accumulation in plants is associated with a highly complex regulatory network system involving multiple genes. This system includes genes that are involved in diverse functions, such as metal ion absorption and transport, sequestration, chelation, detoxification and signal transduction (Luo et al., 2016). In the present study, the expression level of SaHsfA4c was high in the stem under the non-stress condition, and the expression levels in all tested tissues were also increased after Cd treatment. Although we observed that ROS-scavenging enzyme-related genes and Hsps may be the downstream target genes of SaHsfs, these genes are not the only target of SaHsfs in the regulation of Cd tolerance, which is similar to the finding that overexpression of CUP1 in yeast does not satisfy the Cd tolerance of TaHsfA4a (Shim et al., 2009). A co-expression subnetwork with SaHsfA4c as a hub gene was also constructed using a transcriptome database of HE S. alfredii under Cd stress, from which 45 genes, including Hsfs, Hsps and ROS scavenging genes, were detected (Figure 9). Multiple major pathways and numerous target genes may participate in the Cd tolerance mechanism induced by SaHsfA4c. Further identification of the target genes of SaHsfA4c may provide new insights into the mechanism of Cd tolerance.
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
SC, XH, and RZ planned and designed the research. SC performed the experiments. MY, HL, YW, and ZL helped the plant culture and assisted some experiments. LW and YZ contributed analytical tools, LW contributed the design of the work. ML and GQ helped to modify the manuscript, and SC, XH and RZ wrote the manuscript and coordinated its revision. RZ contributed reagents/materials/funds support. MY helped to proof the final version. All authors read and provided helpful discussions, and approved the final version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31872168); the National Nonprofit Institute Research Grant of Chinese Academy of Forestry (CAFYBB2017ZY007 and CAFYBB2016SY008); and the National Key R&D Program of China (No. 2016YFD0800801).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00142/full#supplementary-material
References
Aebi, H. (1984). Catalase in vitro. Methods In Enzymol. 105, 121–126. doi: 10.1016/S0076-6879(84)05016-3
Akesson, A., Barregard, L., Bergdahl, I. A., Nordberg, G. F., Nordberg, M., Skerfving, S. (2014). Non-renal effects and the risk assessment of environmental cadmium exposure. Environ. Health Perspect. 122 (5), 431–438. doi: 10.1289/ehp.1307110
Azarabadi, S., Abdollahi, H., Torabi, M., Salehi, Z., Nasiri, J. (2017). ROS generation, oxidative burst and dynamic expression profiles of ROS-scavenging enzymes of superoxide dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX) in response to Erwinia amylovora in pear (Pyrus communis L). Eur. J. Plant Pathol. 147 (2), 279–294. doi: 10.1007/s10658-016-1000-0
Cai, S. Y., Zhang, Y., Xu, Y. P., Qi, Z. Y., Li, M. Q., Ahammed, G. J., et al. (2017). HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal. Res. 62 (2), e12387. doi: 10.1111/jpi.12387
Chen, S. S., Jiang, J., Han, X. J., Zhang, Y. X., Zhuo, R. Y. (2018). Identification, expression analysis of the Hsf family, and characterization of Class A4 in Sedum alfredii Hance under cadmium stress. Int. J. Mol. Sci. 19 (4), 1216–1233. doi: 10.3390/ijms19041216
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 (12), 909–930. doi: 10.1016/j.plaphy.2010.08.016
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. In Plant Sci. 7, 114–127. doi: 10.3389/fpls.2016.00114
Han, X. J., Yin, H. F., Song, X. X., Zhang, Y. X., Liu, M. Y., Sang, J., et al. (2016). Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation. Plant Biotechnol. J. 14 (6), 1470–1483. doi: 10.1111/pbi.12512
Han, X. J., He, X. L., Qiu, W. M., Lu, Z. C., Zhang, Y. X., Chen, S. S., et al. (2017). Pathogenesis-related protein PR10 from Salix matsudana Koidz exhibits resistance to salt stress in transgenic Arabidopsis thaliana. Environ. Exp. Bot. 141, 74–82. doi: 10.1016/j.envexpbot.2017.07.008
Iqbal, M., Ahmad, A., Ansari, M. K. A., Qureshi, M. I., Aref, I. M., Khan, P. R., et al. (2015). Improving the phytoextraction capacity of plants to scavenge metal(loid)-contaminated sites. Environ. Rev. 23 (1), 44–65. doi: 10.1139/er-2014-0043
Jin, X. F., Yang, X. E., Islam, E., Liu, D., Mahmood, Q. (2008). Effects of cadmium on ultrastructure and antioxidative defense system in hyperaccumulator and non-hyperaccumulator ecotypes of Sedum alfredii Hance. J. Hazard. Mater. 156, 387–397. doi: 10.1016/j.jhazmat.2007.12.064
Lee, J. H. (2013). An overview of phytoremediation as a potentially promising technology for environmental pollution control. Biotechnol. Bioprocess. Eng. 18, 431–439. doi: 10.1007/s12257-013-0193-8
Li, C. G., Chen, Q. J., Gao, X. Q., Qi, B. S., Chen, N. Z., Xu, S. M., et al. (2005). AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci. In China Ser. C: Life Sci. 48, 540–550. doi: 10.1360/062005-119
Li, H. C., Zhang, H. R., Li, G. L., Liu, Z. H., Zhang, Y. M., Zhang, H. M., et al. (2015). Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Funct. Plant Biol. 42 (11), 1080–1091. doi: 10.1071/FP15080
Li, L. Z., Tu, C., Wu, L. H., Peijnenburg, W. J. G. M., Ebbs, S., Luo, Y. M. (2016). Pathways of root uptake and membrane transport of Cd2+ in the Zn/Cd hyperaccumulating plant Sedum plumbizincicola. Environ. Toxicol. Chem. 36, 1038–1046. doi: 10.1002/etc.3625
Li, F., Zhang, H. R., Zhao, H. S., Gao, T. W., Song, A. P., Jiang, J. D., et al. (2017). Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 16, 1311–1321. doi: 10.1111/pbi.12871
Li, Z., Han, X. J., Song, X. X., Zhang, Y. X., Jiang, J., Han, Q., et al. (2017). Overexpressing the Sedum alfredii Cu/Zn superoxide dismutase increased resistance to oxidative stress in transgenic Arabidopsis. Front. In Plant Sci. 8, 1010–1023. doi: 10.3389/fpls.2017.01010
Liu, M. Y., Qiu, W. M., He, X. L., Zheng, L., Song, X. X., Han, X. J., et al. (2016). Functional characterization of a gene in Sedum alfredii Hance resembling rubber elongation factor endowed with functions associated with cadmium tolerance. Front. In Plant Sci. 7, 965–981. doi: 10.3389/fpls.2016.00965
Liu, H., Zhao, H. X., Wu, L. H., Xu, W. Z. (2017). A genetic transformation method for cadmium hyperaccumulator Sedum plumbizincicola and non-hyperaccumulating ecotype of Sedum alfredii. Front. In Plant Sci. 8, 1047–1057. doi: 10.3389/fpls.2017.01047
Liu, H., Zhao, H. X., Wu, L. H., Liu, A. N., Zhao, F. J., Xu, W. Z. (2017). Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 215 (2), 687–698. doi: 10.1111/nph.14622
Lu, L. L., Tian, S. K., Yang, X. E., Wang, X. C., Brown, P., Li, T. Q., et al. (2008). Enhanced root-to-shoot translocation of cadmium in the hyperaccumulating ecotype of Sedum alfredii. J. Exp. Bot. 59 (11), 3203–3213. doi: 10.1093/jxb/ern174
Luo, K., Ma, T. T., Liu, H. Y., Wu, L. H., Ren, J., Nai, F. J., et al. (2015). Efficiency of repeated phytoextraction of cadmium and zinc from an agricultural soil contaminated with sewage sludge. Int. J. Phytorem. 17 (6), 575–582. doi: 10.1080/15226514.2014.935286
Luo, Z. B., He, J. L., Polle, A., Rennenberg, H. (2016). Heavy metal accumulation and signal transduction in herbaceous and woody plants: Paving the way for enhancing phytoremediation efficiency. Biotechnol. Adv. 34 (6), 1131–1148. doi: 10.1016/j.biotechadv.2016.07.003
Peng, J. S., Ding, G., Meng, S., Yi, H. Y., Gong, J. M. (2017). Enhanced metal tolerance correlates with heterotypic variation in SpMTL, a metallothionein-like protein from the hyperaccumulator Sedum plumbizincicola. Plant Cell Environ. 40 (8), 1368–1378. doi: 10.1111/pce.12929
Peng, J. S., Wang, Y. J., Ding, G., Ma, H. L., Zhang, Y. J., Gong, J. M. (2017). A pivotal role of cell wall in cadmium accumulation in the crassulaceae hyperaccumulator Sedum plumbizincicola. Mol. Plant 10, 771–774. doi: 10.1016/j.molp.2016.12.007
Polle, A., Otter, T., Seifert, F. (1994). Apoplastic peroxidases and lignification in needles of Norway Spruce (Picea abies L.). Plant Physiol. 106 (1), 53–60. doi: 10.1104/pp.106.1.53
Sang, J., Han, X. J., Liu, M. Y., Qiao, G. R., Jiang, J., Zhuo, R. (2013). Selection and validation of reference genes for real-time quantitative pcr in hyperaccumulating ecotype of Sedum alfredii under different heavy metals stresses. PloS One. 8 (12), e82927. doi: 10.1371/journal.pone.0082927
Scharf, K. D., Berberich, T., Ebersberger, I., Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 1819 (2), 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Shen, Z. D., Ding, M. Q., Sun, J., Deng, S. R., Zhao, R., Wang, M. J., et al. (2013). Overexpression of PeHSF mediates leaf ROS homeostasis in transgenic tobacco lines grown under salt stress conditions. Plant Cell Tissue Organ Culture. 115 (3), 299–308. doi: 10.1007/s11240-013-0362-7
Shim, D., Hwang, J. U., Lee, J., Lee, S., Choi, Y., An, G., et al. (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell. 21 (12), 4031–4043. doi: 10.1105/tpc.109.066902
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., Ideker, T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27 (3), 431–432. doi: 10.1093/bioinformatics/btq675
Snyman, M., Cronjé, M. J. (2008). Modulation of heat shock factors accompanies salicylic acid-mediated potentiation of Hsp70 in tomato seedlings. J. Exp. Bot. 59 (8), 2125–2132. doi: 10.1093/jxb/ern075
Szczypka, M. S., Wemmie, J. A., Moye-Rowley, W. S., Thiele, D. J. (1994). A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269 (36), 22853–22857. doi: 10.1016/S0092-8674(94)90702-1
Thordal-Christensen, H., Zhang, Z. G., Wei, Y. D., Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 (6), 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tian, S. K., Xie, R. H., Wang, H. X., Hu, Y., Ge, J., Liao, X. X., et al. (2016). Calcium deficiency triggers phloem remobilization of cadmium in a hyperaccumulating species. Plant Physiol. 172, 2300–2313. doi: 10.1104/pp.16.01348
Tian, S. K., Xie, R. H., Wang, H. X., Hu, Y., Hou, D. D., Liao, X. C., et al. (2017). Uptake, sequestration and tolerance of cadmium at cellular levels in the hyperaccumulator plant species Sedum alfredii. J. Exp. Bot. 68 (9), 2387–2398. doi: 10.1093/jxb/erx112
Ullah, S., Kolo, Z., Egbichi, I., Keyster, M., Ludidi, N. (2016). Nitric oxide influences glycine betaine content and ascorbate peroxidase activity in maize. South Afr. J. Bot. 105, 218–225. doi: 10.1016/j.sajb.2016.04.003
Verbruggen, N., Juraniec, M., Baliardini, C., Meyer, C. L. (2013). Tolerance to cadmium in plants: the special case of hyperaccumulators. Biometals 26 (4), 633–638. doi: 10.1007/s10534-013-9659-6
Wang, X. Y., Huang, W. L., Yang, Z. M., Liu, J., Huang, B. R. (2016). Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci. Rep. 6, 28021. doi: 10.1038/srep28021
Wohlgemuth, H., Mittelstrass, K., Kschieschan, S., Bender, J., Weigel, H. J., Overmyer, K., et al. (2002). Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell Environ. 25 (6), 717–726. doi: 10.1046/j.1365-3040.2002.00859.x
Wu, L. H., Liu, Y. J., Zhou, S. B., Guo, F. G., Bi, D., Guo, X. H., et al. (2013). Sedum plumbizincicola X.H. Guo et S.B. Zhou ex L.H. Wu (Crassulaceae): a new species from Zhejiang Province, China. Plant Syst. Evol. 299 (3), 487–498. doi: 10.1007/s00606-012-0738-x
Xue, G. P., Sadat, S., Drenth, J., McIntyre, C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65 (2), 539–557. doi: 10.1093/jxb/ert399
Xue, G. P., Drenth, J., McIntyre, C. L. (2015). TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 66 (3), 1025–1039. doi: 10.1093/jxb/eru462
Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 (1), 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, X. E., Long, X. X., Ni, W. Z., Fu, C. X. (2002). Sedum alfredii H: A new Zn hyperaccumulating plant first found in China. Chin. Sci. Bull. 47 (19), 1634–1637. doi: 10.1007/bf03184113
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 (7), 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, J., Zhang, M., Tian, S. K., Lu, L. L., Shohag, M. J. I., Yang, X. E. (2014). Metallothionein 2 (SaMT2) from Sedum alfredii Hance confers increased cd tolerance and accumulation in yeast and tobacco. PloS One 9 (7). doi: 10.1371/journal.pone.0102750
Zhang, M., Zhang, J., Lu, L. L., Zhu, Z. Q., Yang, X. E. (2016). Functional analysis of CAX2-like transporters isolated from two ecotypes of Sedum alfredii. Biol. Plant. 60 (1), 37–47. doi: 10.1007/s10535-015-0557-3
Keywords: Sedum alfredii Hance, heat shock transcription factor, cadmium stress, overexpression, ROS-scavenging enzyme
Citation: Chen S, Yu M, Li H, Wang Y, Lu Z, Zhang Y, Liu M, Qiao G, Wu L, Han X and Zhuo R (2020) SaHsfA4c From Sedum alfredii Hance Enhances Cadmium Tolerance by Regulating ROS-Scavenger Activities and Heat Shock Proteins Expression. Front. Plant Sci. 11:142. doi: 10.3389/fpls.2020.00142
Received: 18 July 2019; Accepted: 29 January 2020;
Published: 28 February 2020.
Edited by:
Ruth Grene, Virginia Tech, United StatesReviewed by:
Toshiro Shigaki, The University of Tokyo, JapanCharles L. Guy, University of Florida, United States
Copyright © 2020 Chen, Yu, Li, Wang, Lu, Zhang, Liu, Qiao, Wu, Han and Zhuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renying Zhuo, emh1b3J5QGdtYWlsLmNvbQ==; Xiaojiao Han, aGFueGlhb2ppYW8xMDA0QDE2My5jb20=
 Shuangshuang Chen
Shuangshuang Chen Miao Yu1,2
Miao Yu1,2 Yunxing Zhang
Yunxing Zhang Renying Zhuo
Renying Zhuo