- Key Laboratory of Biology and Genetic Improvement of Maize in Southwest Region, Ministry of Agriculture, Maize Research Institute, Sichuan Agricultural University, Chengdu, China
Zinc is an essential micronutrient for plant growth and development, and functions as a cofactor for hundreds of transcription factors and enzymes in numerous biological processes. Zinc deficiency is common abiotic stress resulting in yield loss and quality deterioration of crops, but zinc excess causes toxicity for biological systems. In plants, zinc homeostasis is tightly modulated by zinc transporters and binding compounds that uptake/release, transport, localize, and store zinc, as well as their upstream regulators. Lazarus 1 (LAZ1), a member of DUF300 protein family, functions as transmembrane organic solute transporter in vertebrates. However, the function of LAZ1 in plants is still obscure. In the present study, the ZmLAZ1-4 protein was confirmed to bind to zinc ions by bioinformatic prediction and thermal shift assay. Heterologous expression of ZmLAZ1-4 in the zinc-sensitive yeast mutant, Arabidopsis, and maize significantly facilitated the accumulation of Zn2+ in transgenic lines, respectively. The result of subcellular localization exhibited that ZmLAZ1-4 was localized on the plasma and vacuolar membrane, as well as chloroplast. Moreover, the ZmLAZ1-4 gene was negatively co-expressed with ZmBES1/BZR1-11 gene through co-expression and real-time quantitative PCR analysis. The results of yeast one-hybrid and dual-luciferase assay suggested that ZmBES1/BZR1-11 could bind to ZmLAZ1-4 promoter to inhibit its transcription. All results indicated that ZmLAZ1-4 was a novel zinc transporter on plasma and vacuolar membrane, and transported zinc under negative regulation of the ZmBES1/BZR1-11 transcription factor. The study provides insights into further underlying the mechanism of ZmLAZ1-4 regulating zinc homeostasis.
Introduction
Zinc (Zn) is an essential micronutrient for plant growth and development, and functions as a cofactor for hundreds of transcription factors and enzymes in numerous biological processes, such as chlorophyll biosynthesis, gene expression, signal transduction, and stress response (Palmer and Guerinot, 2009; Zlobin, 2021). Zn deficiency is common abiotic stress resulting in production loss and quality deterioration of crops, but Zn excess causes toxicity for biological systems (Grotz et al., 1998; Arrivault et al., 2006; Palmer and Guerinot, 2009). However, Zn deficiency is far more frequent than toxicity, because Zn content in the soils has low availability for plants (Alvarez and Rico, 2003; Cakmak, 2008). Zn toxicity only occurs on polluted soils containing excessive Zn in mining or industrial areas (Mossa et al., 2020). In plants, the excessive Zn is usually stored in the vacuole to avoid toxicity (Martinoia et al., 2007). Usually, Zn homeostasis is tightly modulated by Zn transporters and binding compounds that uptake/release, transport, localize, and store Zn within the whole plant as well as within individual tissues, cells, and cellular compartments (Palmer and Guerinot, 2009; Zlobin, 2021). Some metal tolerance proteins (MTPs) and heavy metal ATPases (HMAs) localize on the vacuolar membrane and modulate Zn homeostasis as a Zn sensor and transporter (Arrivault et al., 2006; Lan et al., 2013; Menguer et al., 2013; Tanaka et al., 2015).
Zn uptake from the soil, as well as transport in organs, tissues, cells, and intracellular compartments, is mediated by some members of the zinc–iron permease (ZIP) family on plasma and vacuolar membrane (Grotz et al., 1998; Milner et al., 2013). Overexpression of ZIP genes is responsive to Zn deficiency and restores Zn uptake in yeast mutants (Ishimaru et al., 2007; Evens et al., 2017; Yang et al., 2020). In addition, the Zn-regulated transporter (ZRT), iron-regulated transporter (IRT), and natural resistance-associated macrophage protein (NRAMP) have been reported to uptake and transport Zn (Wang et al., 2021). Furthermore, the expression of these Zn transporters is negatively regulated by upstream transcription factors such as members of the bZIP family, which directly bind to zinc deficiency response elements in the promoters of ZIP and other Zn transporter genes (Evens et al., 2017; Nazri et al., 2017; Pita-Barbosa et al., 2019; Lilay et al., 2021). Zinc in the cytoplasm is trapped by small cysteine-rich proteins (metallothioneins) and cysteine-containing peptides (phytochelatins). Consequently, the concentration of free Zn is kept at a low level in cytoplasm, thus protecting cells against Zn toxicity (Cobbett, 2000). The plasma membrane of plant cells contains at least two Zn extrusion transporters including HMA2 and HMA4 (Chong et al., 2009). In Arabidopsis, these ATPases release excessive Zn in the cytosol and mediate the intercellular and intertissue Zn transport (Hussain et al., 2004). ZIF1 (zinc-induced facilitator-1) also acts as a Zn transporter (Haydon and Cobbett, 2007). Zinc is also required in the chloroplast as cofactors for superoxide dismutase, which catalyze the conversion of superoxide to hydrogen peroxide, preventing cellular damage by the reactive hydroxyl radical species (Palmer and Guerinot, 2009). A possible candidate of Zn transporter across the chloroplast membrane is HMA1, which localizes to the chloroplast membrane and contributes to the detoxification of Zn excess (Moreno et al., 2008; Kim et al., 2009; Mikkelsen et al., 2012).
Maize is much more sensitive to Zn deficiency than other crops (Alvarez and Rico, 2003; Mattiello et al., 2015). So, Zn deficiency is recognized as one of the main limiting factors for maize yield. The application of Zn fertilizer achieves a yield gain of more than 18% (Potarzycki and Grzebisz, 2009; Xue et al., 2014; Hacisalihoglu, 2020). In maize, the expression of some ZmZIP genes is significantly increased under Zn deficiency (Mondal et al., 2014; Khatun et al., 2018; Mager et al., 2018). Likewise, overexpression of ZmZIP3, 5, 7, and ZmIRT1 genes increases Zn accumulation in transgenic maize and Arabidopsis (Li et al., 2015, 2016, 2019). Besides, little is known about other genes in the Zn regulation of maize.
Lazarus 1 (LAZ1) is a transmembrane protein with sequence homology and structural similarity to members of the DUF300 family (Malinovsky et al., 2010). DUF300 proteins function as transmembrane organic solute transporter in vertebrates (Wang et al., 2001). In Arabidopsis, two LAZ1 proteins are found to maintain vacuole integrity and mediate brassinosteroid (BR) signaling and localized on the vacuolar membrane (Liu et al., 2018). In our previous study, we cloned eight members of the ZmLAZ1 gene family from maize and found their differential expression among different organs, developmental stages, and under abiotic stresses, implying their functional diversity (Liu et al., 2020). In the present study, we demonstrated that the ZmLAZ1-4 protein was distinct from the other seven members, and it functioned as a zinc transporter on plasma and vacuolar membrane and modulated zinc homeostasis under the negative regulation of BR signaling transcription factor ZmBES1/BZR1-11.
Materials and Methods
Substrate Prediction and Thermal Shift Assay
To predict candidate substrates of ZmLAZ1 proteins, the amino acid sequences of eight ZmLAZ1 members were submitted to the SWISS-MODEL software1 to get a protein model structure file in PDB format. Then the PDB file of each ZmLAZ1 was searched against the RCSBPDB software2 to get putative substrates. The coding sequences (CDSs) of the ZmLAZ1-4 and ZmLAZ1-8 genes were amplified from pMD19-T-ZmLAZ1-4 and pMD19-T-ZmLAZ1-8 (Liu et al., 2020) by PCR primers CDS1-4F/CDS1-4R and CDS1-8F/CDS1-8R (Supplementary Table 1), respectively, and inserted into His-tagged prokaryotic expression vector pET-32a (Takara, Osaka, Japan) by using ClonExpress II One Step Cloning Kit (Vazyme Biotech, Nanjing, China). The construct was introduced into Escherichia coils BL21 (DE3), screened on Luria-Bertani (LB) plates containing 100 mg/ml ampicillin, and grown in LB medium at 37°C to OD600 = 0.6. The His-tagged proteins were induced by 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16°C overnight, purified by using Ni-TED 1 ml Sefinose (TM) Column (Sangon Biotech, Shanghai, China), detected by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), quantified in NanoDrop One Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States), and diluted to 1 μg/μl with 10% dimethyl sulfoxide (DMSO).
As described by Huynh and Partch (2015) with minor modification, 2 μl of 10% DMSO, 6 μg of the purified protein, 2 μl of 10 × SYPRO orange, and 0 (blank control) and 200 μM of each predicted substrate were added into each of three wells of a 96-well PCR plate. In CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States), the sampled plate was equilibrated at 25°C for 5 min and then ramped up to a final temperature of 95°C in increments of 1°C. Fluorescence was read every 0.2°C ramping up. The change rates of relative fluorescence units (RFUs) with time (T) [-d(RFU)/dT] were plotted vs. the temperature to generate melting curves of ZmLA1-4 incubated with each predicted substrate.
Zinc Transport Assay in Zinc-Sensitive Yeast Mutant
The CDS of ZmLAZ1-4 was amplified from pMD19-T-ZmLAZ1-4 plasmid (Liu et al., 2020) by using primers pYES2F/pYES2R (Supplementary Table 1) and used for construction of yeast expression vector pYES2-ZmLAZ1-4 by using ClonExpress® II One Step Cloning Kit (Vazyme Biotech, Nanjing, China). The pYES2-ZmLAZ1-4 and empty vector pYES2 were transformed into yeast Zn-sensitive mutant Δzrc1 (Miyabe et al., 2001) and wild-type (WT) strain BY4743, respectively. The positively transformed lines were selected on synthetic dropout medium (SD) plates containing 2% galactose (w/v) and without uracil (Ura), identified by PCR amplification with primers LAZ4F/LAZ4R (Supplementary Table 1), grown in SD medium at 30°C for 16 h, diluted to OD600 = 0.8, and then diluted by 10-fold serial to 1:104. Subsequently, 5 μl of each dilution were spotted on SD plates containing ZnSO4 (0 or 2 mM) and 2% galactose with three replicates and incubated at 30°C for 3 days. Meanwhile, 50 μl transformed lines were grown in a 10-ml SD liquid medium containing ZnSO4 (1 and 2 mM) and 2% galactose, and used for measurement OD600 at 0, 6, and 24 h. Then the cultures were washed with 10 μM ethylene diamine tetraacetic acid (EDTA) and used for the determination of Zn concentration by Inductively Coupled Plasma-Mass Spectrometry (Thermo Fisher Scientific, Waltham, MA, United States).
Transformation and Phenotyping of Arabidopsis and Maize
Overexpression vector (pC2300-35S-ZmLAZ1-4) of ZmLAZ1-4 was constructed as above and introduced into Agrobacterium tumefaciens strain GV3101. Positive strains were identified and used for transformation of WT Arabidopsis thaliana by floral dip. Transgenic lines were screened on kanamycin 1/2 MS plates and identified by PCR with primers LAZ4F1/LAZ4R1 (Supplementary Table 1). Referring to Kawachi et al. (2009), each homozygous line was grouped into three replicates and grown on 1/2 MS zinc deficiency plates (control) with 5 and 50 μM ZnSO4 at 22°C temperature, 50% humidity, and 16 h light of 120 μE m–2 s–1 illumination intensity/8 h dark period for 2 weeks. After photographing for the phenotype, the seedlings were dried at 60°C for 48 h, weighed for biomass, and digested in 80% nitric acid at 250°C overnight. The digested solution was diluted with ddH2O and used for measurement of Zn2+ content by Inductively Coupled Plasma-Mass Spectrometry (Thermo Fisher Scientific, Waltham, MA, United States).
Overexpression vector (pZZ00026-Ubi-ZmLAZ1-4-Tnos) of ZmLAZ1-4 was constructed as above and used to transform embryonic calli isolated from maize inbred line B73 by Agrobacterium mediation. Positive calli were screened on H6 medium with 0.06% (v/v, effective concentration) Basta herbicide. Regenerated plantlets were screened by PAT/bar EPSPS LFD Strip kit (Youlong, Shanghai, China) according to the manufacturer’s instruction and identified by PCR with primers LAZ4F2/LAZ4R2 (Supplementary Table 1) and real-time quantitative PCR (RT-qPCR) with primers LAZ4F3/LAZ4R3 (Supplementary Table 1). Homozygous T3 lines and WT were grown in vermiculite with Zn dropout Hoagland’s nutrient solution (Coolaber, Beijing, China) at 28°C and 300 μE m–2 s–1 illumination intensity for 16 h/20°C dark for 8 h. Referring to Kawachi et al. (2009), at three-leaf stage, the seedlings of each line were grouped into three replicates, treated with 5 and 50 μM ZnSO4 for 3 weeks, then photographed and dried at 60°C for 72 h, and weighed for biomass and used to measure Zn2+ content as above.
Subcellular Localization
The transmembrane domains of LAZ1-4 were predicted by the TMHMM v. 2.0 software.3 The CDS without termination codons of ZmLAZ1-4 and tonoplast maker gene AtTIP2 (Loque et al., 2005) was amplified from pMD19-T-ZmLAZ1-4 plasmid (Liu et al., 2020) and Arabidopsis cDNA using primers Non-Term1-4F/Non-Term1-4R and AtTIP2F/AtTIP2R (Supplementary Table 1) and used for construction of transient expression vector 35S-ZmLAZ1-4-eGFP and 35S-mCherry-AtTIP2 using ClonExpress® II One Step Cloning Kit (Vazyme Biotech, Nanjing, China), respectively. Subsequently, the 35S-ZmLAZ1-4-eGFP, 35S-mCherry-AtTIP2 of tonoplast maker, 35S-mCherry-OsRac3 of plasma membrane marker (donated by professor Shuangcheng Li, Tao et al., 2021), and empty vector 35S-eGFP (blank control) were introduced into Agrobacterium tumefaciens strain GV3101, respectively.
Maize mesophyll protoplasts were prepared with etiolated leaves and co-transfected with 35S-ZmLAZ1-4-eGFP and 35S-mCherry-OsRac3, as well as 35S-eGFP and 35S-mCherry-OsRac3 as blank control. After incubation at 25°C in dark for 12 h, the protoplasts were used to observe the fluorescence of eGFP and OsRac3 under laser scanning confocal microscope LSM 800 with 488 and 584 nm laser channel (Zeiss, Oberkochen, Germany), respectively.
The combination of 35S-ZmLAZ1-4-eGFP and 35S-mCherry-OsRac3, 35S-ZmLAZ1-4-eGFP and 35S-mCherry-AtTIP2, as well as 35S-eGFP and 35S-mCherry-OsRac3, and 35S-eGFP and 35S-mCherry-AtTIP2 plasmid was mixed with 0.1 M spermidine and 2.5 M CaCl2 and precipitated onto gold particles (φ = 60 μm), respectively. Onion bulbs were surface sterilized with 75% ethanol. The fifth scales without pigment were cut into 2 cm × 2 cm, incubated on MS medium for 4 h, and bombard in helium biolistic gun (Bio-Rad, Hercules, CA, United States) with above gold particles. After filtration at 28°C under dark for 24 h, the bombarded onion scales were used to observe the fluorescence of eGFP, OsRac3, and AtTIP2 under the same microscope.
The Agrobacterium strains harboring 35S-ZmLAZ1-4-eGFP and 35S-eGFP were infiltrated into the abaxial leaf surface of 3-week-old plants of Nicotiana benthamiana, respectively. After incubation at 22°C and 14 light/10 dark for 24 h, the infiltrated leaves were used to observe the fluorescence of eGFP and autofluorescence of chloroplasts under the same microscope.
Co-expression and Real-Time Quantitative PCR Analysis
The transcriptomic data of maize inbred line B73 were downloaded from MazieGDB4 and used for co-expression analysis with ZmLAZ1-4 by using a Perl script (Supplementary Data Set 1). The correlation coefficient was set as > 0.9 and < −0.9. Among the candidates, only the ZmBES1/BZR1-11 gene encoded transcription factor, co-expressed with ZmLAZ1-4 (correlation coefficient is −0.93), and used for RT-qPCR analysis. The seeds of inbred line B73 were surface sterilized with 30% H2O2, germinated in petri dish, and transplanted into a plastic mesh grid for hydroponic culture at 28°C under a photoperiod of 14 h light/10 h dark. At the three-leaf stage, the seedlings were subjected to the treatment of 5 μM ZnSO4. At the 0 (control), 1st, 2rd, and 3rd day of treatment, the whole plant was sampled, ground in liquid nitrogen, and used for total RNA extraction by using RNAiso plus kit (TaKaRa, Osaka, Japan). After removing probable genomic DNA contamination by using RNase-free DNase (TaKaRa, Osaka, Japan), these samples were quantified on NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States) and reverse transcribed into cDNA by using PrimeScript™ reagent kit (TaKaRa, Osaka, Japan). The RT-qPCR was performed as described by Sun et al. (2020). The ZmGAPDH was used as reference. The primers are listed in Supplementary Table 1.
Yeast One Hybrid and Dual Luciferase Assay
The CDS of ZmBES1/BZR1-11 was amplified with primers pGADT7F/pGADT7R (Supplementary Table 1) and used to construct vector pGADT7-ZmBES1/BZR1-11. The cis-acting elements bound by ZmBES1/BZR1-11 in ZmLAZ1-4 promoter were predicted by PlantCARE.5 The sequence (−1 to −1,100 bp) of ZmLAZ1-4 promoter (pZmLAZ1-4) containing cis-acting elements was amplified with primers pAbAiF/pAbAiR (Supplementary Table 1) and used to construct reporter vector pAbAi-pZmLAZ1-4. The pAbAi-pZmLAZ1-4 was restricted with BbsI and transformed into yeast Y1H gold by using a yeast transformation kit (Coolaber, Beijing, China). The transformant was plated onto Ura dropout SD medium and incubated at 30°C for 5 days. The positive clones were identified by PCR across the multiple cloning sites of the pAbAi vector and the ura3-52 gene of Y1H gold with primers Y1HF/Y1HR (Supplementary Table 1), diluted to 10–1, 10–2, 10–3, and 10–4 folds with 0.9% NaCl and plated onto Ura dropout SD medium containing either 50, 100, 200, or 400 ng/ml aureobasidin (AbA) to inhibit the Y1H gold background. Competent cells were prepared with the positive clones, transformed with prey vector pGADT7-ZmBES1/BZR1-11, plated onto the Leu dropout SD medium containing AbA at an optimal concentration, and incubated at 30°C for 5 days.
The promotor sequence (−1 to −1,100 bp) of ZmLAZ1-4 (pZmLAZ1-4) was amplified with specific primers pGreenIIF/pGreenIIR (Supplementary Table 1) and inserted into pGreenII-0800-LUC plasmid to drive firefly luciferase gene (LUC) and generate reporter vector pZmLAZ1-4-LUC. The Renilla luciferase gene REN driven by 35S promoter in pZmLAZ1-4-LUC plasmid was used as internal reference. The CDS of the ZmBES1/BZR1-11 gene was amplified with primers pCAMBIA2300F/pCAMBIA2300R (Supplementary Table 1) and inserted into pCAMBIA2300-35S-eGFP plasmid to create effector vector 35S-ZmBES1/BZR1-11. The reporter and the effector vectors were introduced into Agrobacterium strain GV3101, respectively, and used for co-infiltration of Nicotiana benthamiana leaves. After incubated at 22°C and 14 light/10 dark for 3 days, the leaves were visualized for LUC signal in ChemiDoc™ Imaging System (Bio-Rad, Hercules, CA, United States). The relative activities of LUC and REN were determined in a dual-luciferase reporter assay system (Thermo Fisher Scientific, Waltham, MA, United States) and used to calculate relative LUC activity (LUC/REN).
Statistical Analysis
All experiments were performed with three replicates. The data were shown as mean ± standard deviation and analyzed using Student’s t-test at *p < 0.05 and **p < 0.01 level.
Results
ZmLAZ1-4 Specifically Binds to Zinc
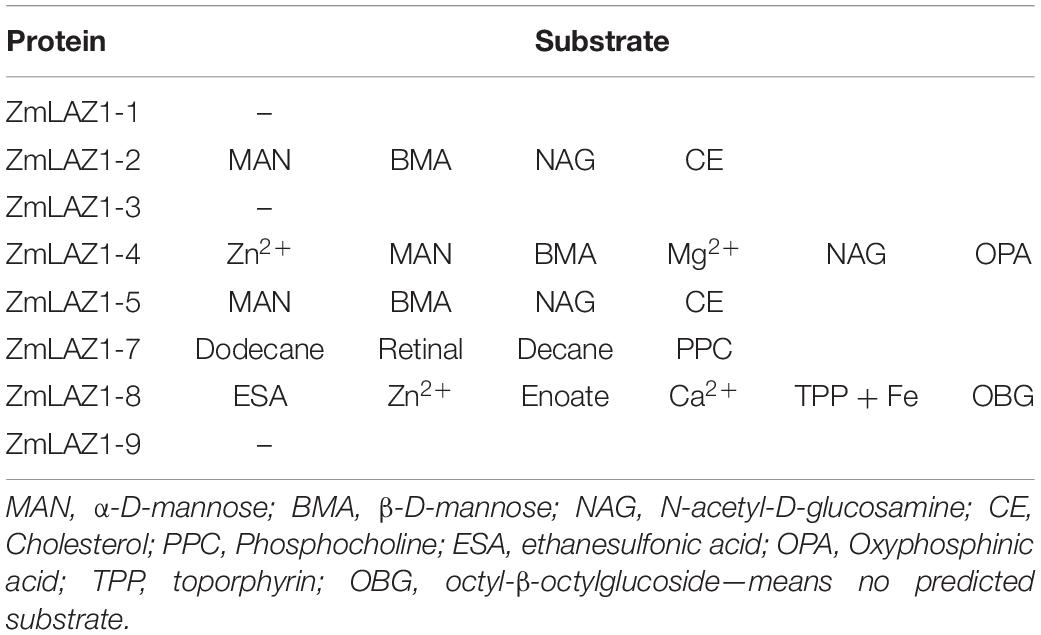
By the RCSBPDB software as described by Wang et al. (2001), substrates of eight ZmLAZ1 members were mainly predicted to be organic solutes including α-D-mannose, β-D-mannose, N-acetyl-D-glucosamine, cholesterol, phosphocholine, ethanesulfonic acid, phosphinic acid, toporphyrin, and octyl-β-octylglucoside. However, only ZmLAZ1-4 (Zm00001d012921) and ZmLAZ1-8 (Zm00001d036361) were predicted to combine inorganic ions containing zinc (Zn2+), magnesium (Mg2+), and calcium (Ca2+) (Table 1). During many times of prokaryotic expression, the ZmLAZ1-8 protein was not successfully purified (Supplementary Figure 1). Therefore, the ZmLAZ1-4 was used for further study.
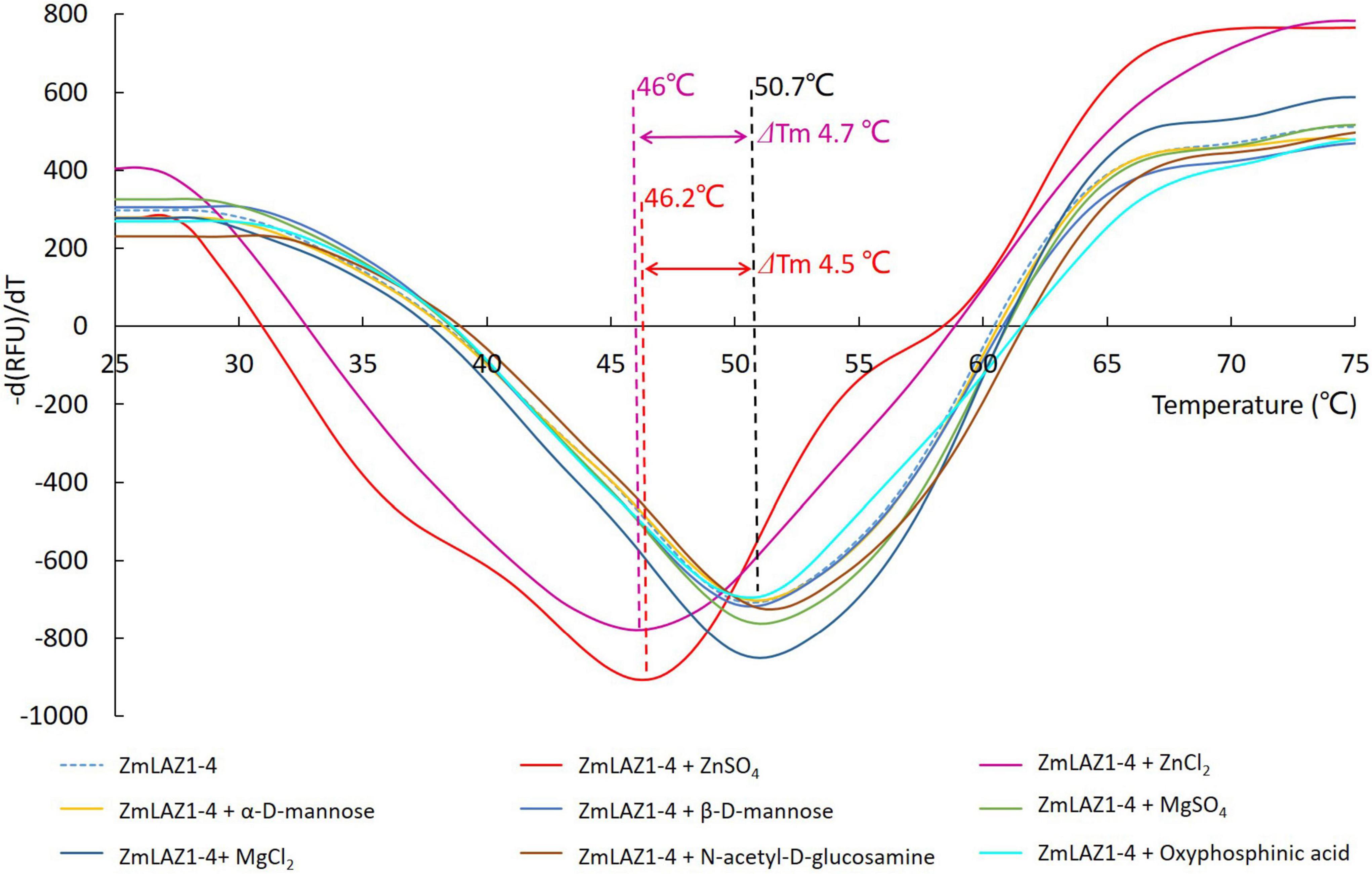
In the thermal shift assay, melting temperature (Tm) of the ZmLAZ1-4 protein incubated with ZnCl2 and ZnSO4 was 4.7 and 4.5°C lower than that of ZmLAZ1-4 incubated alone (blank control), respectively, whereas Tm of ZmLAZ1-4 incubated with other predicted substrates kept same value with blank control (Figure 1). This result suggested that zinc ion was candidate substrate of ZmLAZ1-4.
ZmLAZ1-4 Transports Zinc in Yeast, Arabidopsis, and Maize
Under non-stress (0 mM Zn2+), the diluted colonies and growth curves of OD600 showed no significant difference among Δzrc1 mutant transformed by the ZmLAZ1-4 gene, and Δzrc1 and WT transformed by empty vector pYES2. Under 2 mM Zn2+ stress, the difference was significant among these three lines (Figures 2A,B). The complementation of ZmLAZ1-4 significantly inhibited the growth of Zn-sensitive mutant Δzrc1. The Zn2+ concentration of Δzrc1 transformed by ZmLAZ1-4 was significantly higher than that of Δzrc1 and WT transformed by empty vector pYES2 (Figure 2C), suggesting that ZmLAZ1-4 could transport Zn2+ into cells.
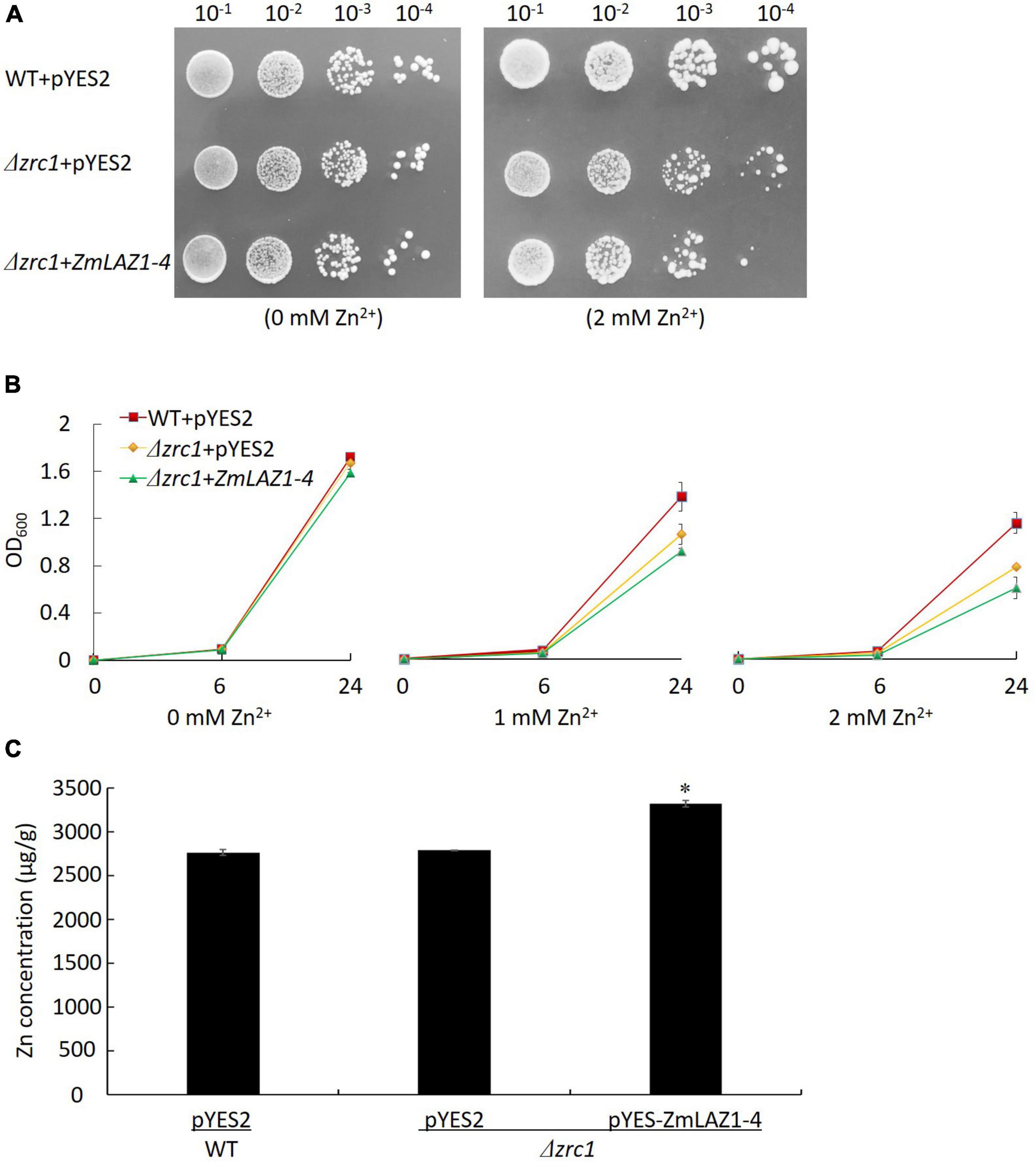
Figure 2. Zinc transport assay in yeast Zn-sensitive mutant. (A) Colonies of WT and Δzrc1 mutant transformed by empty vector and Δzrc1 transformed by ZmLA1-4 under 0 and 2 mM ZnSO4 stress. (B) OD600 curves of the three lines under 0, 1, and 2 mM ZnSO4 stress. (C) Zn concentration in yeast grown in synthetic dropout medium (SD) with 2 mM ZnSO4 for 48 h. *p < 0.05.
Two homozygous T3 Arabidopsis lines overexpressing ZmLAZ1-4 were screened on kanamycin 1/2 MS plates and identified by PCR amplification (Supplementary Figure 2). Under 0, 5, and 50 μM ZnSO4 treatments, the growth phenotype of T3 lines showed no obvious difference compared to WT (Figure 3A). However, Zn2+ content of transgenic lines was significantly higher than WT under 5 and 50 μM ZnSO4 treatments, while only trace content was measured under 0 μM ZnSO4 treatment (Figure 3B).
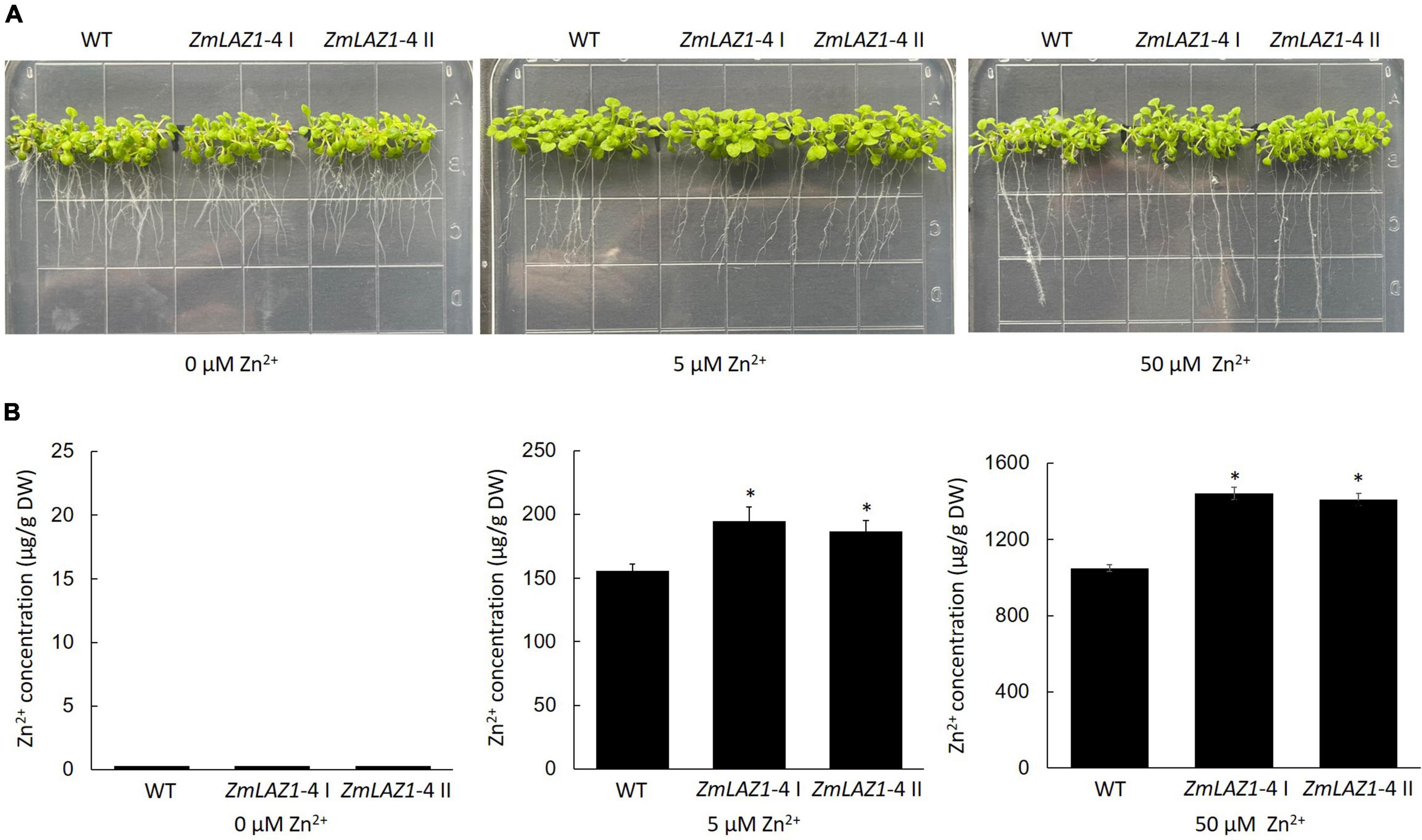
Figure 3. Phenotype and Zn2+ content of transgenic Arabidopsis. (A) Phenotype. (B) Zn2+ content. The seeds of every line were sterilized and grown on 1/2 MS zinc deficiency plates (control) with 5 and 50 μM ZnSO4 for 2 weeks, respectively. WT, wild type. ZmLA1-4 I and ZmLA1-4 II represent homozygous lines. *p < 0.05.
By A. tumefaciens-mediated embryonic calli transformation, from the positive transgenic calli harboring ZmLAZ1-4, ten plantlets were regenerated and four homozygous T3 maize lines overexpressing ZmLAZ1-4 were identified by PAT/bar EPSPS LFD Strips (Supplementary Figure 3), PCR amplification (Supplementary Figure 4), and RT-qPCR (Supplementary Figure 5). Four homozygous lines and WT were grown in vermiculite with Zn deficient Hoagland’s nutrient solution. After 3 weeks of 5 and 50 μM ZnSO4 treatments, the growth phenotype and biomass of all transgenic lines showed different compared with WT (Figures 4A,B). However, the Zn2+ content of all transgenic lines was significantly higher than WT under 5 and 50 μM ZnSO4 treatments (Figure 4C). The above results indicated that ZmLAZ1-4 functioned as a Zn transporter.
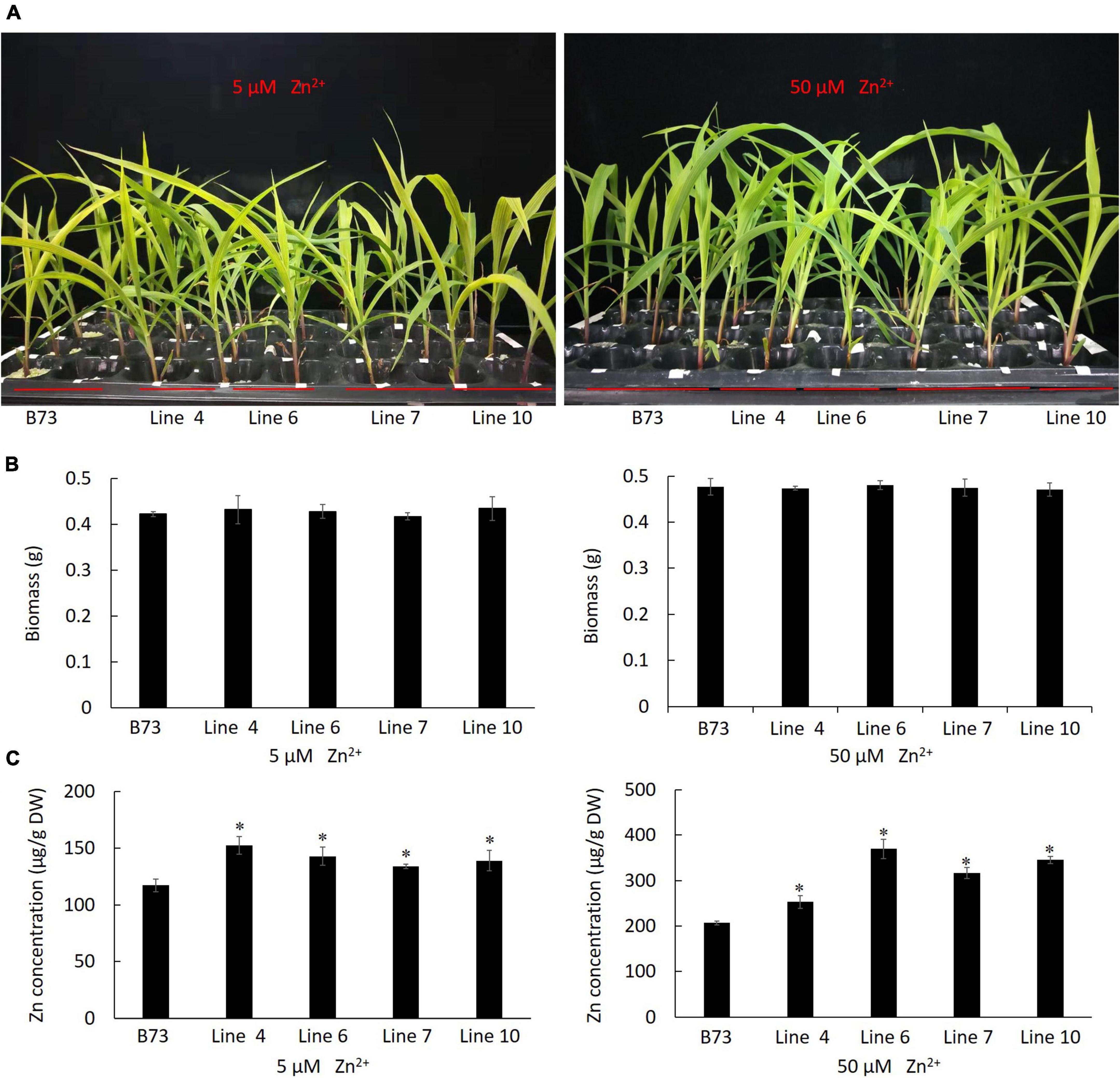
Figure 4. Phenotype of transgenic maize under ZnSO4 treatment. (A) Phenotype. (B) Biomass. (C) Zn2+ content. At three-leaf stage, the seedlings of each line were grouped into three replicates, treated with 5 and 50 μM ZnSO4 for 3 weeks, then photographed and dried at 60°C for 72 h, and weighed for biomass and used to measure Zn2+ content. The biomass of three seedlings of every line was shown. B73, the untransformed control. Line 4, 6, 7, and 10 are homozygous T3 lines. *p < 0.05.
ZmLAZ1-4 Localized on Plasma and Vacuolar Membranes
By the TMHMM software, seven transmembrane domains were predicted during ZmLA1-4 protein (Supplementary Figure 6). As shown in Figure 5, the GFP fluorescence was observed in the cytoplasm and nucleus in maize protoplasts, and onion cells transfected by empty vector 35S-eGFP. However, the GFP fluorescence from the fusion protein (35S-ZmLAZ1-4-GFP) was merged with red fluorescence of the plasma membrane marker OsRac3, tonoplast maker AtTIP2, and autofluorescence of chloroplasts. Especially when AtTIP2 was used as a maker, it could be clearly seen that ZmLAZ-4 was localized on the tonoplast. Furthermore, the ZmLAZ1-4 was also localized to chloroplast (Supplementary Figure 7). These results indicated the subcellular localization of the ZmLAZ1-4 protein on the plasma and vacuolar membrane.
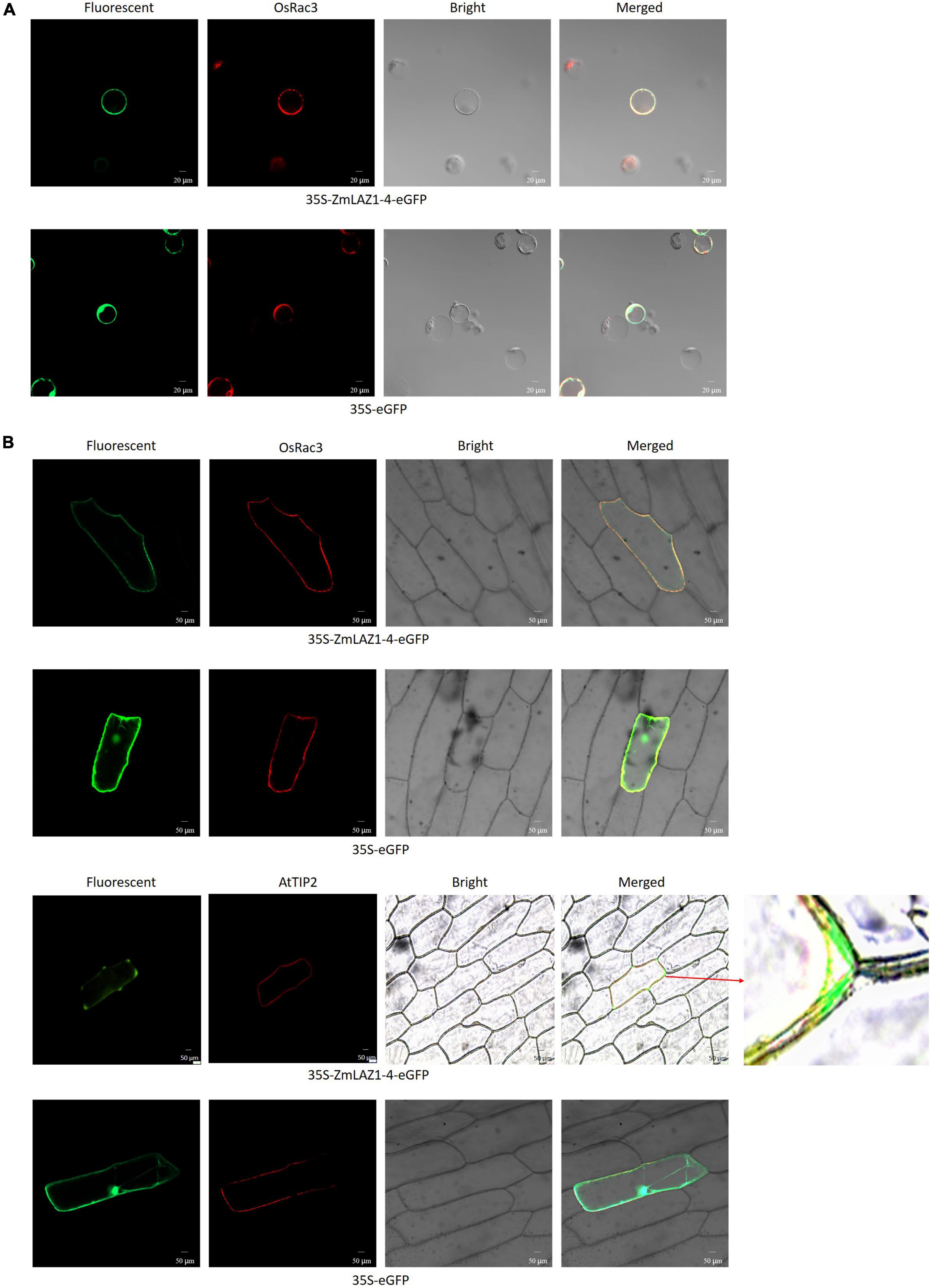
Figure 5. Subcellular localization of ZmLAZ1-4 in maize protoplasts (A) and onion cells (B). Scale bar is 50 μm.
ZmLAZ1-4 Is Negatively Regulated by ZmBES1/BZR1-11
In order to explore the mechanism of ZmLAZ1-4 regulating Zn transport, the co-expression analysis was conducted. The results showed that there were 27 genes co-expressed with ZmLAZ1-4 with correlation coefficient > 0.9 or < −0.9, and only ZmBES1/BZR1-11 among these candidates encoded transcription factor and negatively co-expressed with ZmLAZ1-4 (Supplementary Table 2). The result of RT-qPCR likewise showed that the expression of ZmLAZ1-4 and ZmBES1/BZR1-11 was significantly downregulated and upregulated by Zn deficiency (5 μM), respectively (Figure 6). It was predicted that there were six E-boxes (CAXXTG) of BES1/BZR1 binding element (Yin et al., 2005) during ZmLAZ1-4 promoter by PlantCARE. Hence, the yeast one-hybrid (Y1H) was performed. As shown in Figure 7, on Leu dropout SD medium, the growth of Y1H gold strain co-transformed by empty prey vector pGADT7 and pAbAi-pZmLAZ1-4 harboring six E-boxes elements of ZmLAZ1-4 promoter was inhibited by 200 ng/ml AbA, whereas the Y1H gold strain co-transformed by pGADT7-ZmBES1/BZR1-11 and pAbAi-pZmLAZ1-4 formed few colonies, indicating that the ZmBES1/BZR1-11 transcription factor could bind to ZmLAZ1-4 promoter. The result was further verified by dual-luciferase assay in vivo. The relative LUC activity (LUC/REN) of leaves co-infiltrated by reporter vector ZmLAZ1-4-LUC and effector vector 35S-ZmBES1/BZR1-11 was significantly lower than that of control (Figure 7B). These results indicate that the ZmBES1/BZR1-11 transcription factor binds to ZmLAZ1-4 promoter to inhibit ZmLAZ1-4 transcription.
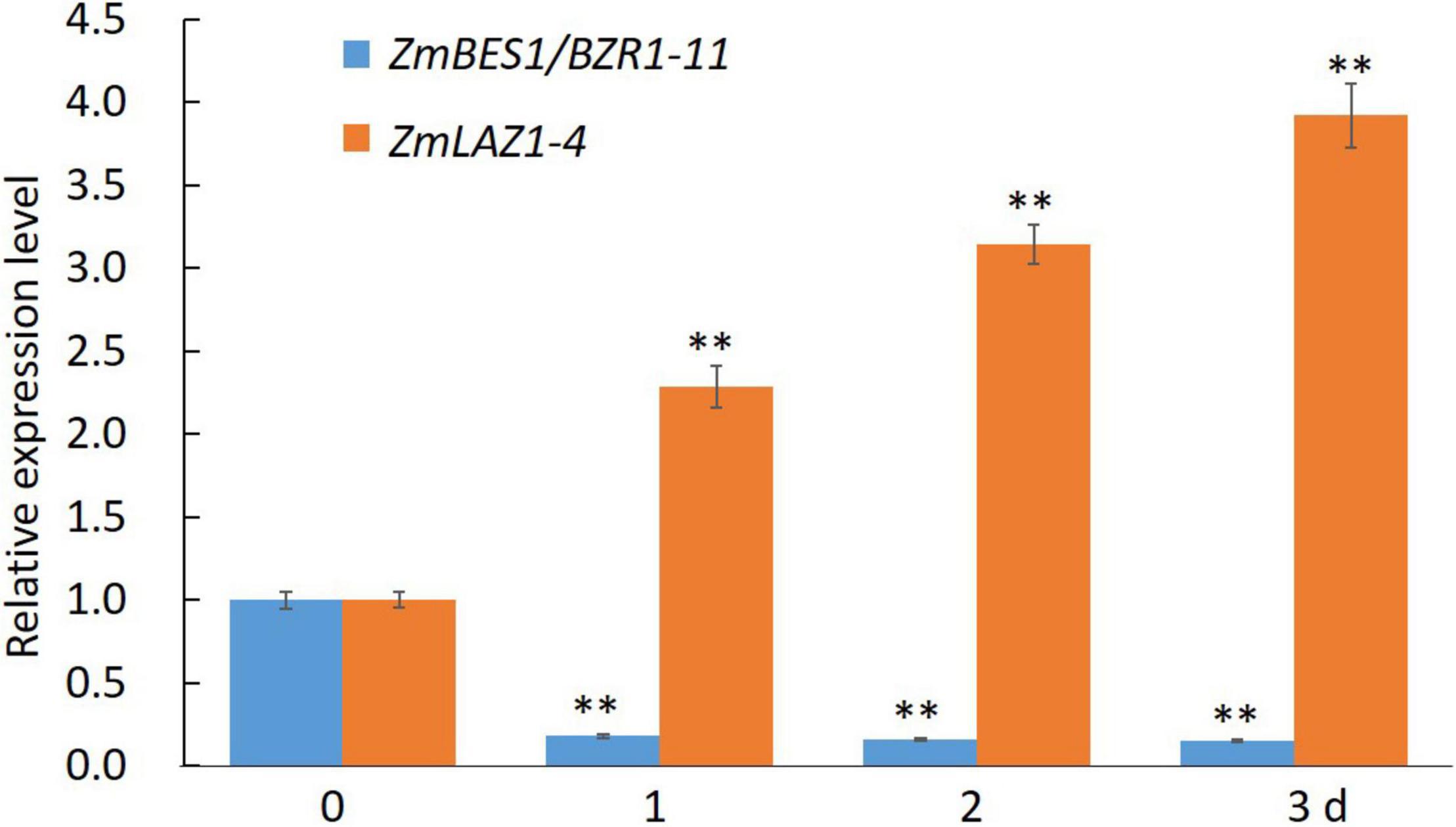
Figure 6. Relative expression level of ZmLAZ1-4 and ZmBES1/BZR1-11 in response to Zn deficiency. **p < 0.01.
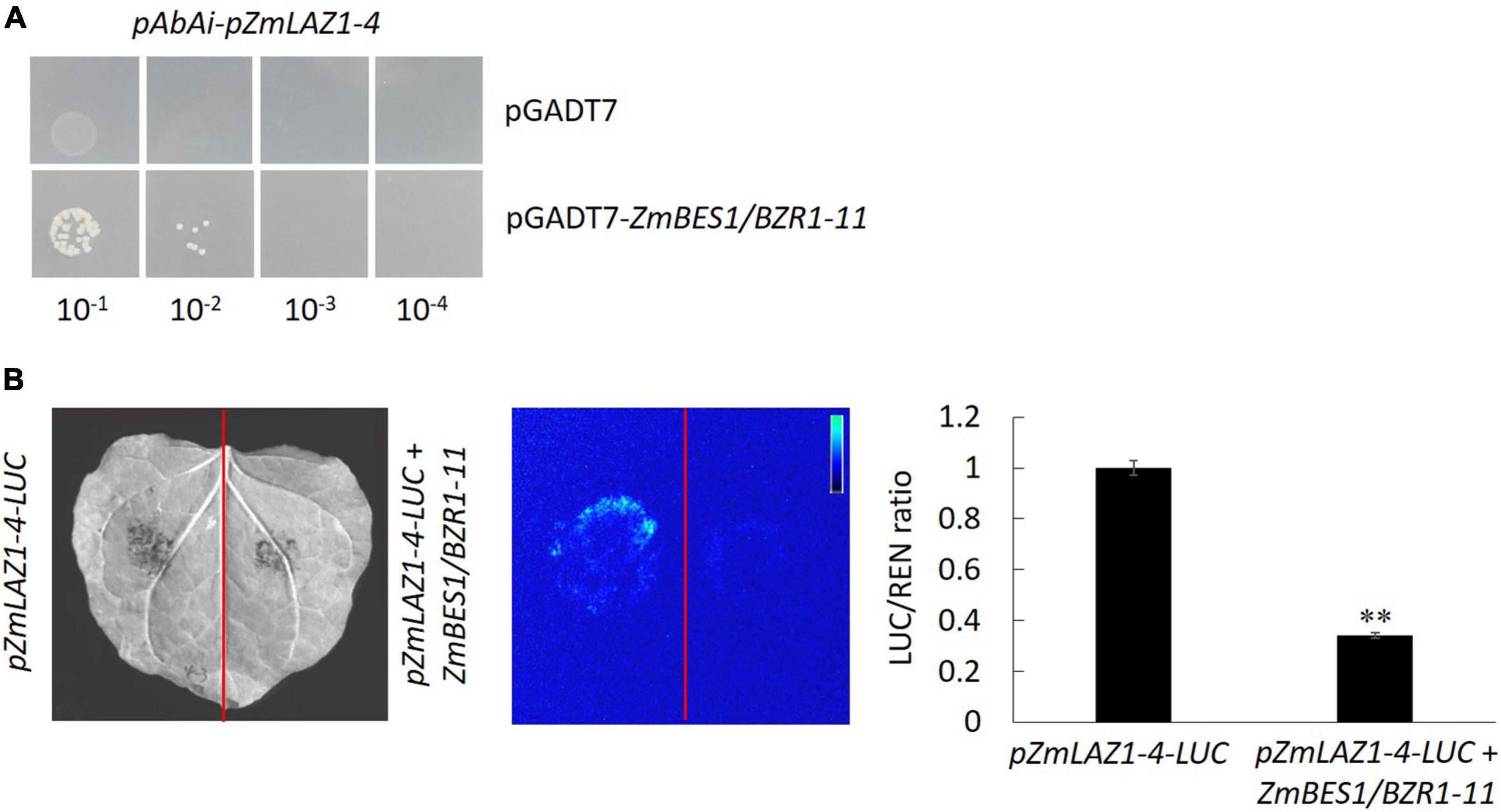
Figure 7. Confirmation of ZmBES1/BZR1-11 binding to ZmLAZ1-4 promoter. The Y1H (A) and dual-luciferase assay (B) were performed to verify the binding and negative regulation of ZmLAZ1-4 by ZmBES1/BZR1-11 transcription factor. The tobacco leaves were co-infiltrated by 35S-ZmBES1/BZR1-11 and pZmLAZ1-4-LUC, incubated at 22°C and 14 light/10 dark for 3 days, visualized for LUC signal, and used to measure activity LUC and REN. **p < 0.01.
Discussion
The eight ZmLAZ1 members were grouped into a family in phylogenetic analysis because of their sequence similarity, especially their conserved DUF300 domain (Malinovsky et al., 2010; Liu et al., 2020). In the present study, only ZmLAZ1-4 and ZmLAZ1-8 were predicted to combine metal ions including Zn2+, Mg2+, or Ca2+ (Table 1). During prokaryotic expression, ZmLAZ1-8 was not successfully purified (Supplementary Figure 1). Eukaryotic membrane proteins are often difficult to be purified (Newstead et al., 2007). Therefore, the combination of ZmLAZ1-4 to predicted substrates was verified by thermal shift assay (Figure 1). Even in ZIP family, only some members were identified as Zn transporters (Grotz et al., 1998; Ishimaru et al., 2007; Evens et al., 2017; Yang et al., 2020). The other members might function as transporters of other divalent ions (Milner et al., 2013). The overexpression of ZmLAZ1-4 in yeast mutant, Arabidopsis, and maize significantly increased Zn uptake (Figures 2–4), suggesting that the ZmLAZ1-4 protein was involved in Zn uptake in maize.
In our previous study, ZmLAZ1-4 was predicted to localize on chloroplast, plasmalemma, cytoplasm, and endoplasmic reticulum (Liu et al., 2020). The subcellular localization showed that ZmLAZ1-4 functioned on plasma and vacuolar membrane, as well as chloroplast using tonoplast maker AtTIP2 and plasma membrane marker OsRac3 (Figure 5 and Supplementary Figure 7), which well confirmed the tonoplast and plasma membrane localization (Loque et al., 2005; Tao et al., 2021). Hence, they could be used as a marker in our study. The phenomenon was similar with Mg2+ transporter AtMRS2 showing different intracellular localization patterns in yeast and chloroplast localization, and Pi transporter PHT2;1 localizing to mitochondria, plasma membrane, endoplasmic reticulum, and chloroplast (Wayne and Maria, 2002; Drummond et al., 2006). In Arabidopsis, two LAZ1 proteins were also localized on plasma and vacuolar membrane, but no specific marker was used for chloroplast localization (Malinovsky et al., 2010). Our result suggests that ZmLAZ1-4 functions on the plasma membrane and uptakes Zn from the soil, and transports Zn into vacuole. But the mechanism of ZmLAZ1-4 acting on chloroplast remains unclear. Before this study, Zn transport across chloroplast and vacuolar membrane was well documented to be mediated by HMA, MTP, and Oryza sativa Zn transporter (OTZ) proteins (Kobae et al., 2004; Arrivault et al., 2006; Martinoia et al., 2007; Moreno et al., 2008; Kim et al., 2009; Morel et al., 2009; Lan et al., 2013; Menguer et al., 2013; Tanaka et al., 2015). Some endoplasmic reticulum-localized and Golgi apparatus-localized zinc transporters were also involved in Zn homeostasis by controlling the release of zinc into cytosol (Fujiwara et al., 2015; Adulcikas et al., 2018; Wang et al., 2021). Our result of subcellular localization could not rule out the possibility of endoplasmic reticulum and Golgi apparatus localization of ZmLAZ1-4 (Supplementary Figure 7). This will be explored in further study.
Among the 27 genes co-expressing with ZmLAZ1-4, only ZmBES1/BZR1-11 encoded transcription factor and negatively co-expressed with ZmLAZ1-4 (Figure 6 and Supplementary Table 2). The ZmBES1/BZR1-11 can bind to E-boxes (CAXXTG) element in ZmLAZ1-4 promoter to inhibit ZmLAZ1-4 transcription (Yin et al., 2005), which was verified by Y1H and dual-luciferase assay (Figure 7). But previous studies exhibited that BES1/BZR1 transcription factor responds to BR induction and regulates the expression of BR-responsive genes (Yin et al., 2005; Yu et al., 2018), and Arabidopsis LAZ1 proteins localized on plasma and vacuolar membrane also mediated BR signaling (Malinovsky et al., 2010). It could be concluded that the ZmLAZ1-4 protein functioned as a Zn2+ transporter on plasma and vacuolar membrane, and chloroplast to modulate Zn homeostasis in maize. The expression of ZmLAZ1-4 was negatively regulated by ZmBES1/BZR1-11 transcription factor. The results of this study indicated that ZmLAZ1-4 was a novel zinc transporter distinct from the previously documented Zn transporters ZIP, ZRT, IRT, NRAMP, etc. (Grotz et al., 1998; Ishimaru et al., 2007; Milner et al., 2013; Evens et al., 2017; Yang et al., 2020), as plotted in a signaling diagram of zinc homeostasis together with the previously reported evidence (Figure 8).
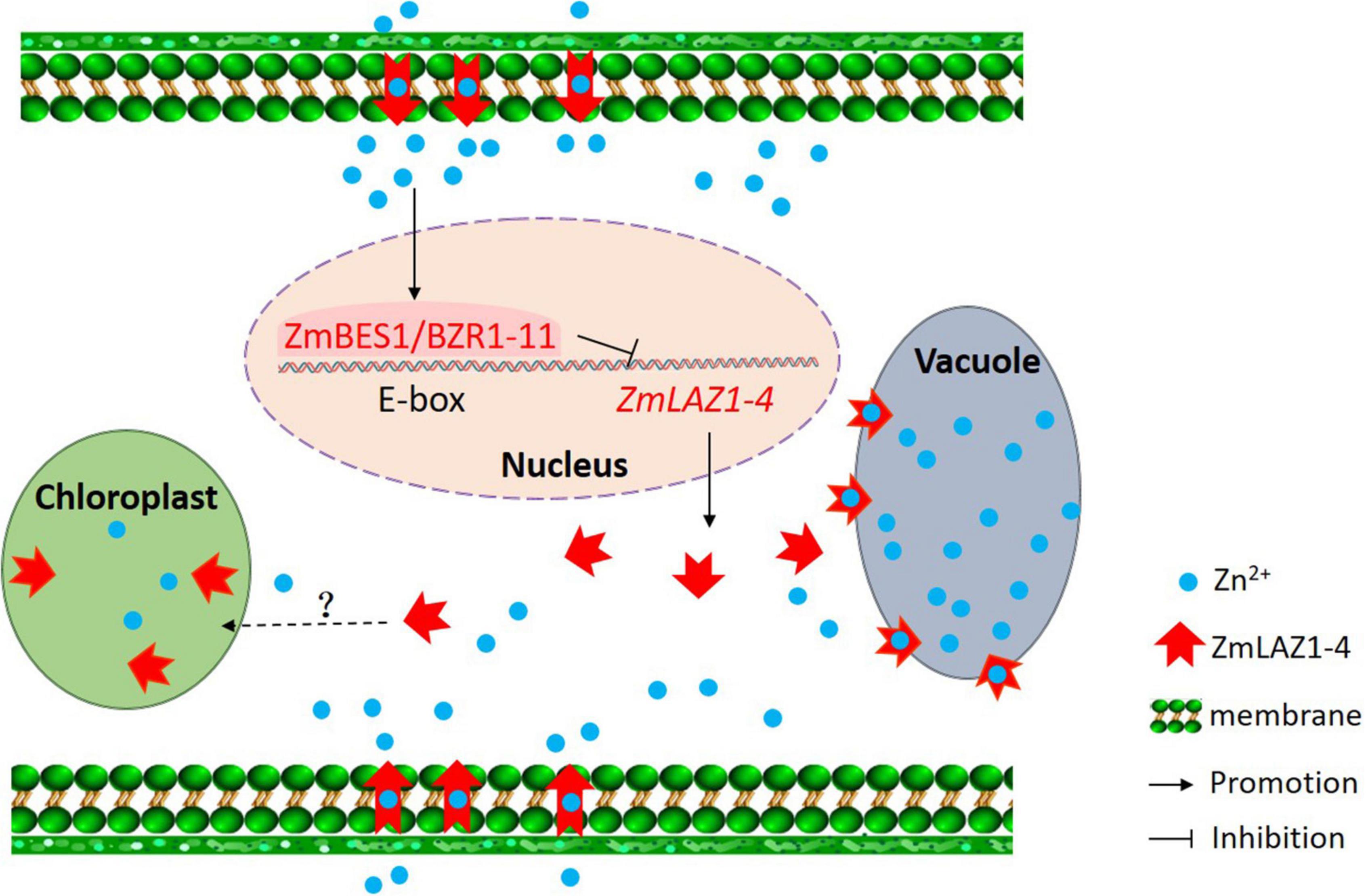
Figure 8. Signaling diagram of zinc homeostasis in maize. The black words, lines, and arrows indicate signaling pathways previously reported. The red words, lines, and arrows show signaling pathway demonstrated in the study.
Conclusion
The ZmLAZ1-4 protein is a novel zinc transporter that transports zinc ions across plasma and vacuolar membrane and modulates zinc homeostasis under the negative regulation of ZmBES1/BZR1-11 transcription factor.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: wap.maizegdb.org, Zea_mays.AGPv4.32gff3.
Author Contributions
FF and WL conceived and supervised the research. BL, HY, QyY, LD, FS, JQ, WF, and QqY performed the experiments. BL and WL drafted the manuscript. BL and HY revised the manuscript. All authors interpreted and discussed the data.
Funding
This work was supported by the National Key R&D Program of China (2021YFF1000303), the Science and Technology Program of Sichuan (2020YJ0353), and the Chengdu Science and Technology Bureau (2021-YF05-02024-SN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Shuangcheng Li (Rice Research Institute, Sichuan Agricultural University) for his donation of expression vector, Yi Wang (Wheat Research Institute, Sichuan Agricultural University) for his donation of yeast mutant and expression vector, and the Key Laboratory of Biology and Genetic Improvement of Maize in Southwest Region, Ministry of Agriculture, for its technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.881055/full#supplementary-material
Footnotes
- ^ https://swissmodel.expasy.org/
- ^ https://www.rcsb.org/
- ^ http://www.cbs.dtu.dk/services/TMHMM
- ^ https://www.maizegdb.org
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Adulcikas, J., Norouzi, S., Bretag, L., Sohal, S. S., and Myers, S. (2018). The zinc transporter SLC39A7 (ZIP7) harbours a highly-conserved histidine-rich N-terminal region that potentially contributes to zinc homeostasis in the endoplasmic reticulum. Comput. Biol. Med. 100, 196–202. doi: 10.1016/j.compbiomed.2018.07.007
Alvarez, J. M., and Rico, M. I. (2003). Effects of zinc complexes on the distribution of zinc in calcareous soil and zinc uptake by maize. J. Agric. Food Chem. 51, 5760–5767. doi: 10.1021/jf030092m
Arrivault, S., Senger, T., and Krämer, U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46, 861–879. doi: 10.1111/j.1365-313X.2006.02746.x
Cakmak, I. (2008). Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 302, 1–17. doi: 10.1007/s11104-007-9466-3
Chong, K., Jarvis, R. S., Sherson, S. M., and Cobbett, C. S. (2009). Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 181, 79–88. doi: 10.1111/j.1469-8137.2008.02637.x
Cobbett, C. S. (2000). Phytochelatins and their roles in heavy metal detoxication. Plant Physiol. 123, 825–832. doi: 10.1104/pp.104.4.1325
Drummond, R., Tutone, A., Li, Y. C., and Gardner, R. C. (2006). A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 170, 78–89. doi: 10.1016/j.plantsci.2005.08.018
Evens, N. P., Buchner, P., Williams, L. E., and Hawkesford, M. J. (2017). The role of ZIP transporters and group F bZIP transcription factors in the Zn-deficiency response of wheat (Triticum aestivum). Plant J. 92, 291–304. doi: 10.1111/tpj.13655
Fujiwara, T., Kawachi, M., Sato, Y., Mori, H., Kutsuna, N., Hasezawa, S., et al. (2015). A high molecular mass zinc transporter MTP12 forms a functional heteromeric complex with MTP5 in the Golgi in Arabidopsis thaliana. FEBS J. 283, 1965–1979. doi: 10.1111/febs.13252
Grotz, N., Fox, T., Connolly, E., Park, W., Guerinot, M. L., and Eide, D. (1998). Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. U.S.A. 95, 7220–7224. doi: 10.1073/pnas.95.12.7220
Hacisalihoglu, G. (2020). Zinc (Zn): the last nutrient in the alphabet and shedding light on Zn efficiency for the future of crop production under suboptimal Zn. Plants 9:1471. doi: 10.3390/plants9111471
Haydon, M. J., and Cobbett, C. S. (2007). A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143, 1705–1719. doi: 10.1104/pp.106.092015
Hussain, D., Haydon, M. J., Wang, Y., Wong, E., Sherson, S. M., Young, J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16, 1327–1339. doi: 10.1105/tpc.020487
Huynh, K., and Partch, C. L. (2015). Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79
Ishimaru, Y., Masuda, H., Suzuki, M., Bashir, K., Takahashi, M., Nakanishi, H., et al. (2007). Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J. Exp. Bot. 58, 2909–2915. doi: 10.1093/jxb/erm147
Kawachi, M., Kobae, Y., Mori, H., Tomioka, R., Lee, Y., and Maeshima, M. (2009). A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant Cell Physiol. 50, 1156–1170. doi: 10.1093/pcp/pcp067
Khatun, M. A., Hossain, M. M., Bari, M. A., Abdullahil, K. M., Parvez, M. S., Alam, M. F., et al. (2018). Zinc deficiency tolerance in maize is associated with the upregulation of Zn transporter genes and antioxidant activities. Plant Biol. 20, 765–770. doi: 10.1111/plb.12837
Kim, Y.-Y., Choi, H., Segami, S., Cho, H.-T., Martinoia, E., Maeshima, M., et al. (2009). AtHMA1 contributes to detoxification of excess Zn(II) in Arabidopsis. Plant J. 58, 737–753. doi: 10.1111/j.1365-313X.2009.03818.x
Kobae, Y., Uemura, T., Sato, M. H., Ohnishi, M., Mimura, T., Nakagawa, T., et al. (2004). Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 45, 1749–1758. doi: 10.1093/pcp/pci015
Lan, H. X., Wang, Z. F., Wang, Q. H., Wang, M. M., Bao, Y. M., Huang, J., et al. (2013). Characterization of a vacuolar zinc transporter OZT1 in rice (Oryza sativa L.). Mol. Biol. Rep. 40, 1201–1210. doi: 10.1007/s11033-012-2162-2
Li, S., Liu, X., Zhou, X., Li, Y., Yang, W., and Chen, R. (2019). Improving zinc and iron accumulation in maize grains using the zinc and iron transporter ZmZIP5. Plant Cell Physiol. 60, 2077–2085. doi: 10.1093/pcp/pcz104
Li, S., Zhou, X., Li, H., Liu, Y., Zhu, L., Guo, J., et al. (2015). Overexpression of ZmIRT1 and ZmZIP3 enhances iron and zinc accumulation in transgenic Arabidopsis. PLoS One 10:e0136647. doi: 10.1371/journal.pone.0136647
Li, S., Zhou, X., Zhao, Y., Li, H., Liu, Y., Zhu, L., et al. (2016). Constitutive expression of the ZmZIP7 in Arabidopsis alters metal homeostasis and increases Fe and Zn content. Plant Physiol. Biochem. 106, 1–10. doi: 10.1016/j.plaphy.2016.04.044
Lilay, G. H., Persson, D. P., Castro, P. H., Liao, F., Alexander, R. D., Aarts, M. G., et al. (2021). Arabidopsis bZIP19 and bZIP23 act as zinc sensors to control plant zinc status. Nat. Plant 7, 137–143. doi: 10.1038/s41477-021-00856-7
Liu, B. L., Yu, H. Q., Wen, Q., Fu, F. L., and Li, W. C. (2020). Genome-wide analysis of LAZ1 gene family from maize. J. Plant Growth Regul. 39, 656–668. doi: 10.1007/s00344-019-10008-z
Liu, Q., Vain, T., Viotti, C., Doyle, S. M., Tarkowska, D., Novak, O., et al. (2018). Vacuole integrity maintained by DUF300 proteins is required for brassinosteroid signaling regulation. Mol. Plant 11, 553–567. doi: 10.1016/j.molp.2017.12.015
Loque, D., Ludewig, U., Yuan, L., and von Wiren, N. (2005). Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137, 671–680. doi: 10.1104/pp.104.051268
Mager, S., Schönberger, B., and Ludewig, U. (2018). The transcriptome of zinc deficient maize roots and its relationship to DNA methylation loss. BMC Plant Biol. 18:372. doi: 10.1186/s12870-018-1603-z
Malinovsky, F. G., Brodersen, P., Fiil, B. K., McKinney, L. V., Thorgrimsen, S., Beck, M., et al. (2010). Lazarus1, a DUF300 protein, contributes to programmed cell death associated with Arabidopsis acd11 and the hypersensitive response. PLoS One 5:e12586. doi: 10.1371/journal.pone.0012586
Martinoia, E., Maeshima, M., and Neuhaus, E. (2007). Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 58, 83–102. doi: 10.1093/jxb/erl183
Mattiello, E. M., Ruiz, H. A., Neves, J. C., Ventrella, M. C., and Araújo, W. L. (2015). Zinc deficiency affects physiological and anatomical characteristics in maize leaves. J. Plant Physiol. 183, 138–143. doi: 10.1016/j.jplph.2015.05.014
Menguer, P. K., Farthing, E., Peaston, K. A., Ricachenevsky, F. K., Fett, J. P., and Williams, L. E. (2013). Functional analysis of the rice vacuolar zinc transporter OsMTP1. J. Exp. Bot. 64, 2871–2883. doi: 10.1093/jxb/ert136
Mikkelsen, M. D., Pedas, P., Schiller, M., Vincze, E., Mills, R., Borg, S., et al. (2012). Barley HvHMA1 is a heavy metal pump involved in mobilizing organellar Zn and Cu and plays a role in metal loading into grains. PLoS One 7:e49027.
Milner, M. J., Seamon, J., Craft, E., and Kochian, L. V. (2013). Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 64, 369–381. doi: 10.1093/jxb/ers315
Miyabe, S., Izawa, S., and Inoue, Y. (2001). The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 282, 79–83. doi: 10.1006/bbrc.2001.4522
Mondal, T. K., Ganie, S. A., Rana, M. K., and Sharma, T. R. (2014). Genome-wide analysis of zinc transporter genes of maize (Zea mays). Plant Mol. Biol. Rep. 32, 605–616. doi: 10.1007/s11105-013-0664-2
Morel, M., Crouzet, J., Gravot, A., Auroy, P., Leonhardt, N., Vavasseur, A., et al. (2009). AtHMA3, a P1B-ATPase allowing Cd/Zn/ Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 149, 894–904. doi: 10.1104/pp.108.130294
Moreno, I., Norambuena, L., Maturana, D., Toro, M., Vergara, C., Orellana, A., et al. (2008). AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J. Biol. Chem. 283, 9633–9641. doi: 10.1074/jbc.M800736200
Mossa, A. W., Young, S. D., and Crout, N. M. J. (2020). Zinc uptake and phyto-toxicity: comparing intensity- and capacity-based drivers. Sci. Total Environ. 699, 134314. doi: 10.1016/j.scitotenv.2019.134314
Nazri, A. Z., Griffin, J. H. C., Peaston, K. A., Alexander-Webber, D. G. A., and Williams, L. E. (2017). F-group bZIPs in barley-a role in Zn deficiency. Plant Cell Environ. 40, 2754–2770. doi: 10.1111/pce.13045
Newstead, S., Kim, H., von Heijne, G., Iwata, S., and Drew, D. (2007). High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 104, 13936–13941. doi: 10.1073/pnas.0704546104
Palmer, C., and Guerinot, M. L. (2009). A question of balance: facing the challenges of Cu, Fe and Zn homeostasis. Nat. Chem. Biol. 5, 333–340. doi: 10.1038/nchembio.166
Pita-Barbosa, A., Ricachenevsky, F. K., Wilson, M., Dottorini, T., and Salt, D. E. (2019). Transcriptional plasticity buffers genetic variation in zinc homeostasis. Sci. Rep. 9:19482. doi: 10.1038/s41598-019-55736-0
Potarzycki, J., and Grzebisz, W. (2009). Effect of zinc foliar application on grain yield of maize and its yielding components. Plant Soil Environ. 55, 519–527. doi: 10.3389/fpls.2018.00677
Sun, F. A., Yu, H. Q., Qu, J. T., Cao, Y., Ding, L., Feng, W. Q., et al. (2020). Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int. J. Mol. Sci. 21:996. doi: 10.3390/ijms21030996
Tanaka, N., Fujiwara, T., Tomioka, R., Krämer, U., Kawachi, M., and Maeshima, M. (2015). Characterization of the histidine-rich loop of Arabidopsis vacuolar membrane zinc transporter AtMTP1 as a sensor of zinc level in the cytosol. Plant Cell Physiol. 56, 510–519. doi: 10.1093/pcp/pcu194
Tao, Y., Zou, T., Zhang, X., Liu, R., Chen, H., Yuan, G., et al. (2021). Secretory lipid transfer protein OsLTPL94 acts as a target of EAT1 and is required for rice pollen wall development. Plant J. 108, 358–377. doi: 10.1111/tpj.15443
Wang, W., Seward, D. J., Li, L., Boyer, J. L., and Ballatori, N. (2001). Expression cloning of two genes that together mediate organic solute and steroid transport in the liver of a marine vertebrate. Proc. Natl. Acad. Sci. U.S.A. 98, 9431–9436. doi: 10.1073/pnas.161099898
Wang, Y., Yang, J., Miao, R., Kang, Y., and Qi, Z. (2021). A novel zinc transporter essential for Arabidopsis zinc and iron-dependent growth. J. Plant Physiol. 256:153296. doi: 10.1016/j.jplph.2020.153296
Wayne, K. V., and Maria, J. H. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14, 1751–1766. doi: 10.1105/tpc.002220
Xue, Y., Yue, S., Zhang, W., Liu, D., Cui, Z., Chen, X., et al. (2014). Zinc, iron, manganese and copper uptake requirement in response to nitrogen supply and the increased grain yield of summer maize. PLoS One 9:e93895. doi: 10.1371/journal.pone.0093895
Yang, M., Li, Y., Liu, Z., Tian, J., Liang, L., Qiu, Y., et al. (2020). A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 103, 1695–1709. doi: 10.1111/tpj.14855
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Yu, H., Feng, W., Sun, F., Zhang, Y., Qu, J., Liu, B., et al. (2018). Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Gow. Regul. 86, 235–249. doi: 10.1007/s10725-018-0424-2
Keywords: maize, ZmLAZ1-4, zinc transport, tonoplast, transcriptional regulation
Citation: Liu B, Yu H, Yang Q, Ding L, Sun F, Qu J, Feng W, Yang Q, Li W and Fu F (2022) Zinc Transporter ZmLAZ1-4 Modulates Zinc Homeostasis on Plasma and Vacuolar Membrane in Maize. Front. Plant Sci. 13:881055. doi: 10.3389/fpls.2022.881055
Received: 22 February 2022; Accepted: 04 April 2022;
Published: 02 May 2022.
Edited by:
Seçkin Eroğlu, Middle East Technical University, TurkeyReviewed by:
Babar Hussain, University of Central Punjab, PakistanAlexandra Lešková, CEA Cadarache, France
Bastian Meier, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2022 Liu, Yu, Yang, Ding, Sun, Qu, Feng, Yang, Li and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanchen Li, YXVtZHltc0BzaWNhdS5lZHUuY24=; Fengling Fu, ZmZsQHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Bingliang Liu
Bingliang Liu Haoqiang Yu
Haoqiang Yu Qinyu Yang
Qinyu Yang Lei Ding
Lei Ding Fuai Sun
Fuai Sun Jingtao Qu
Jingtao Qu Wenqi Feng
Wenqi Feng Qingqing Yang
Qingqing Yang Wanchen Li
Wanchen Li Fengling Fu
Fengling Fu