- 1Department of Plant Sciences, School of Life Sciences, University of Hyderabad, Hyderabad, Telangana, India
- 2School of Agriculture and Biosciences, Karunya Institute of Technology and Sciences, Coimbatore, Tamil Nadu, India
- 3National Institute of Plant Genome Research (NIPGR), New Delhi, India
Millets constitute a significant proportion of underutilized grasses and are well known for their climate resilience as well as excellent nutritional profiles. Among millets, foxtail millet (Setaria italica) and its wild relative green foxtail (S. viridis) are collectively regarded as models for studying broad-spectrum traits, including abiotic stress tolerance, C4 photosynthesis, biofuel, and nutritional traits. Since the genome sequence release, the crop has seen an exponential increase in omics studies to dissect agronomic, nutritional, biofuel, and climate-resilience traits. These studies have provided first-hand information on the structure, organization, evolution, and expression of several genes; however, knowledge of the precise roles of such genes and their products remains elusive. Several open-access databases have also been instituted to enable advanced scientific research on these important crops. In this context, the current review enumerates the contemporary trend of research on understanding the climate resilience and other essential traits in Setaria, the knowledge gap, and how the information could be translated for the crop improvement of related millets, biofuel crops, and cereals. Also, the review provides a roadmap for studying other underutilized crop species using Setaria as a model.
Introduction
Crop productivity, limited by changing climatic conditions such as increasing temperature, drought, soil salinity, etc., imposes a severe threat on food security worldwide. Evidence suggests that expanding our study and utilizing neglected and underutilized cereals for sustainable agricultural production is imperative. Millets collectively form a group of small-grained cereals, including several distantly related species. This group of crops is cultivated in adverse climatic conditions, such as high temperature and drought, with low agricultural inputs. The most widely grown small millets are finger millet (Eleusine coracana), pearl millet (Pennisetum glaucum), proso millet (Panicum miliaceum), foxtail millet (Setaria italica), and barnyard millet (Echinochloa crus-galli). Although less popular than major cereal crops, millets are a rich source of protein, resistant starch, micronutrients, antioxidants, and bioactive compounds (Kumar et al., 2018). The water use efficiency (WUE) and Nitrogen-use efficiency (NUE) of millets are also better than popular cereals. Also, the millets are gluten-free and have a low glycemic index (Muthamilarasan and Prasad, 2021). Foxtail millet is considered an alternative sustainable protein source compared to other cereals and millets (Sachdev et al., 2021). Being a rich source of zinc and iron, the consumption of millets imparts immunity (Kumar et al., 2018). Altogether, small millets are promising cereals with the capability to ensure food security in the future. In the past 2 years, amid the recent COVID-19 pandemic, major emphasis has been shifted to the regular consumption of food that boosts immunity. Millets contain easily digestible proteins and a better essential amino acid profile than other cereal crops. These characteristics make millets nutritionally superior to other major cereals and desirable for ensuring food security amidst pandemics (Muthamilarasan and Prasad, 2021).
The genus Setaria, a group of panicoid grasses, belongs to the tribe Paniceae and is characterized by sterile bristles. This genus comprises more than 125 species distributed in temperate regions worldwide (Kellogg, 2017). However, foxtail millet (S. italica) and its wild progenitor green foxtail (Setaria viridis), remains the most studied millets in this genus. S. viridis is one of the most widely spread weeds. Setaria italica was domesticated from S. viridis ~9–11,000 years back in China near the Yellow River valley. Further, it has been widely cultivated in arid and semi-arid regions (Pant et al., 2016). Tsang et al. (2017) have reported that foxtail millet was domesticated ~5,000 years back in Taiwan. Both S. viridis and S. italica (henceforth, S. italica and S. viridis will be collectively called Setaria) offer several advantages as compared to the established models viz. rice and maize, such as short life cycle (5–6 weeks), small diploid genome (395–500 Mb), short stature (10–30 cm), C4 photosynthesis. Since several millet species are shown to be resilient to adverse climatic conditions and biotic and abiotic stresses (Lata et al., 2013; Muthamilarasan and Prasad, 2015), the study of the Setaria genus has gained popularity in the past decade. In particular, foxtail, pearl, and proso millet have been considered appealing substitutes for small grain production (Pant et al., 2016). Further, C4 metabolism actuates food productivity by efficiently utilizing water, carbon, and nitrogen; therefore, elucidating the C4 metabolic pathways is important. Considering that Setaria is proven valuable for C4 photosynthesis study, it might enable the long-term goal of engineering C4 traits into staple crops, rice and wheat. A rapid-cycling mini foxtail millet mutant, xiaomi, was recently presented as a model system to study the C4 mechanism (Yang et al., 2020).
Altogether, the physiological as well as genetic, and climate resilience attributes of Setaria present it as a valuable model system for research (Peng and Zhang, 2021). Although there have been considerable advances in understanding the unique traits of Setaria, our interpretation of the underlying mechanism of its climate resilience is still in its infancy. Recent advances in genome sequencing, RNA-seq, and the discovery of unique trait-related QTLs have further provided momentum to millet research. Analysis of the available information for Setaria would lead to applying current knowledge to enhance our understanding of other crop species. The present review provides a comprehensive view of genome sequencing of Setaria, transcriptome and proteome analysis, publicly available databases, agronomically important trait-linked markers, and characterization of genes predicted from various platforms.
Setaria omics during the pre-genome sequencing era
Earlier foxtail millet was predominantly cultivated in the Chinese belts. Before the green revolution, the farmers repeatedly cultivated trait-specific landraces. Despite the consumption shift toward rice and wheat, foxtail millet was recognized for its climate resilience among marginal farmers. This led to the development of improved varieties and pure lines of foxtail millet from the “All India Coordinated Improvement Project for small millets” during the 1960s. Preceding the advent of molecular markers, the research in foxtail millet began with morphological markers. Initially, Restriction fragment length polymorphisms (RFLP) markers were used in foxtail millet to group accessions from various countries. These RFLP codominant markers detected the ribosomal DNA variability and grouped the foxtail millet genotypes into European, western European, and Asiatic lines (Schontz and Rether, 1998). The RFLP markers also successfully identified the historical recombination events with three types of length polymorphisms by atp6 probes in the mitochondrial sequences of foxtail millet (Fukunaga and Kato, 2003). Besides RFLP, the most prominent marker utilized in foxtail millet was Simple-sequence repeats (SSR). These markers were implied in varying populations to enumerate the Linkage disequilibrium (LD) values. The association analysis in S. viridis with SSR markers projected a higher LD decay than other cereals. Thus, the SSR markers exhibited novel marker-trait associations in foxtail millet (Wang et al., 2010). A higher diversity with a maximum LD value in various landraces in China’s yellow river detected the genetic richness in landraces than the cultivated species. Hence, these markers helped in understanding the effect of domestication and artificial selection in the cultivated lines. In this series, randomly amplified polymorphic DNA (RAPD) and Inter Simple Sequence Repeats (ISSR) markers were also utilized by Kumari et al. (2011) in analyzing the higher genetic variance in South Indian foxtail millet collections.
In addition to diversity analysis, the molecular markers also resolved the ambiguities in the phylogenetic relationship of foxtail millet. In this aspect, a set of transposon display markers were utilized in the international collection. Transposon display is a modified form of AFLP that focuses on the long terminal repeats of retrotransposons (LTRs) and miniature inverted-repeat transposable elements (MITE). These LTRs are concentrated in the centromeric and peri-centromeric regions. While MITEs are in the euchromatic regions. In foxtail millet, these LTRs and MITEs are identified in the mutant alleles of the Waxy gene (GBSS 1). These mutants have a TE insertion to produce a sticky endosperm and are found in the Asian collections. These TD markers identified in the waxy alleles were grouped into eight clusters by Hirano et al. (2011). In addition, 98 novel intron length polymorphic markers (ILP) from rice were used in the molecular diversity analysis. Of these, 26 ILPs successfully classified 45 accessions in Setaria (Gupta et al., 2011). Further, molecular diversity with 45 SSR markers having di- and tri-nucleotide repeat motifs in 12 populations of Taiwan was shown by Lin et al. (2011). The genetic maps in foxtail millet have been constructed with RFLP, SSR, and SNP markers in trisomics, RILs, and F2 populations. The RFLP-based map with 160 loci was generated from the trisomic lines in S. italica × S. viridis. The total span length of this map was 964 cM. This was the first successful attempt with 80% coverage across the chromosomes, revealing that S. viridis carried a gene for gamete fertility (Wang et al., 1998). After this, SSR markers were used to develop a linkage map, and their presence in the genome was predominant in the intergenic regions. With 100 SSR markers in the F2 population between S. italica × S. viridis, this map was developed with a span length of 1,645 cM. There was an uneven distribution of markers in this map, with three on chromosome 3 and 18 on chromosome 9. The distribution pattern was due to the variations in high and low copy numbers across the genome and constructed in mapmaker version 3 using the kosambi mapping function (Jia et al., 2009).
The morphological traits, namely, anthocyanin pigmentation, length of bristles, plant height, non-glutinous seeds, seed size, and seed color, were used to distinguish the interspecific hybrids (Figure 1). The distribution of these traits in the F2 and their subsequent generations were involved in determining the nature of genes (dominance/recessive). Later, the cytological and isozyme markers were utilized in identifying their homologous pairing and the phylogeny in the related species. Using such an approach, S. pyconoma was found to have eight intermediate and 14 viridis type chromosomes, which provided information about its ancestry. Subsequently, molecular cytogenetics involving, Genomic in situ hybridization (GISH) and Fluorescence in situ hybridization (FISH) elaborated the similarities among the chromosomal structures in S. viridis and S. italica. Although the cultivated and wild species were morphologically different, they had similar cytological features. These molecular cytology tools also successfully distinguished the interspecific hybrids of italica x verticillata complex and facilitated grouping of the species in their respective gene pools (Benabdelmouna et al., 2001; Pallares et al., 2004; Brenner et al., 2015).

Figure 1. Genetic diversity of various seed-related traits in foxtail millet. Variation in anther color (A–D), panicle density (E–H), husk color (I–N), bristles (O–R), and panicle attitude (S–V) are shown. The names of the accessions are listed in Supplementary Table 1. figure not to scale.
Further, screening of the germplasm based on their seed morphology using seed fluorescence imaging as per ISTA for varietal identification was adopted in the classical genetics’ characterization. Other essential biochemical markers utilized in the classical diversity analysis were based on the phenol test, ferrous sulfate test, and SDS-protein analysis. In the cluster analysis, the proteins quantified through SDS PAGE were implied as isozyme markers to group the accessions. Among all, the enzymes peroxidase and esterase were the crucially utilized isozymes for detecting polymorphism in foxtail millet. Thus, the pre-sequencing era in foxtail millet focused on documenting the wild species and sub-races of S. italica. Overall, these studies laid a foundation for understanding the allelic richness and the course of historical domestication events in the related species (Willweber-Kishiomoto, 1962; Khan, 1997; Haroun, 2002; Upadhyaya et al., 2009).
Genome sequencing and resequencing efforts in Setaria
The foxtail genome was decoded by Bennetzen et al. (2012) and Zhang et al. (2012). These two findings revealed the overall genetic potential of S. italica. The first draft by Bennetzen et al. (2012) comprised 400 Mb across nine chromosomes with a 992-locus genetic map. The sequencing was performed by Sanger sequencing analysis in the seedlings of the cultivar Yugu 1 by Joint Genome Institute, United States. The genetic sequence revealed 40% transposable elements in which long terminal repeat retrotransposons were the most abundant, and there were around 24,000 protein-encoding genes. In addition, a genetic map based on these sequences representing 1,416 cM was constructed from the F2 of B100 × S. viridis. This map had 992 SNP markers identified by sequencing the 247 RIL progenies.
The second high-quality draft genome sequence of S. italica was reported by Zhang et al. (2012). The draft genome of 423 Mb across the nine chromosomes was sequenced by a whole-genome shotgun next-generation sequencing in the foxtail millet cultivar “Zhang gu.” The total number of genes annotated was 38,801, of which ~81% were expressed, and ~46% were transposable repeats (TE: 31.60% and DNA Transposon: 9.40%). Sequencing analysis revealed the presence of chromosome reshuffling events, where the Chr 7, Chr 9, Chr 3, and 10 of rice shared similarity with Chr 2 and Chr 9 of foxtail millet. These results represented the course of domestication events and the phylogenetic relationships of foxtail millet with rice and other cereals. In addition, A2, a photo-thermosensitive male sterile line, was also sequenced by Illumina GA II, and this was used to construct a genetic map. The comparative alignment of this sequence with Zhan gu identified 542,322 SNPs, 33,587 indels, and 10,839 structural variants (Zhang et al., 2012). Later, the draft genome sequence was utilized to tap the stress-regulated gene families in foxtail millet.
With the advancement in sequencing technologies, the resequenced genome assembly of foxtail millet was completed by Ni et al. (2017). Resequencing of 184 foxtail millet recombinant inbred lines updated the Zhang gu reference genome sequence. Approximately 16 Mb was added to the reference genome assembly as unanchored scaffolds, and the error correction with 3,158 gaps in Yugu 1 reference was completed by a bin map construction. The study further determined QTLs for nine agronomic traits with two major QTLs for plant height. These QTLs exhibited 89% similarity to the sd1 gene in rice (Ni et al., 2017). Further, QTL-seq analysis was performed involving whole-genome resequencing using pair-end sequencing with bulk segregants of F2 obtained from Shinanotsubuhime × Yuikogne for heading date (Yoshitsu et al., 2017). Genome-wide comparison of three bulks, viz., early, late, and extremely late heading, revealed two associated QTLs for heading. The first QTL, qDTH2, was identified on chromosome 2 for late heading, whereas the second QTL, qDTH7, was identified on chromosome 7 for extremely late heading (Yoshitsu et al., 2017). Next, a map-based cloning and high-throughput sequencing of loose panicle 1 mutant with increased panicle lengths and grain size encoded a novel WRKY transcription factor. A single shift in the G-A transition of the fifth intron in LP1 resulted in three disorganized splicing events owing to an enlarged panicle attitude which could be explored in developing bold grains in foxtail millet` (Xiang et al., 2017).
Furthermore, the sequencing effort involving dd-RAD sequencing of 142 foxtail millet core collection identified 844 SNPs on chromosome 5–2,153 on chromosome 8 with an average SNP frequency of 25.90 per Mb. The genome-wide association study (GWAS) of this data revealed 81 trait-associated markers for 10 traits across the genome (Jaiswal et al., 2019). Recently, resequencing of 164 RILs generated from Longgu7 × Yugu 1 was also conducted to identify QTLs and SNPs. Approximately 1,047,978 SNPs were detected between the parents. Three thousand four hundred thirteen bin markers were then used to generate a map having 3,963 recombinant breakpoints. Mapping and sequencing analysis depicted 47 QTLs for four traits: straw weight, panicle weight, grain weight per plant, and 1,000-grain weight. It also detected nine stable QTL clusters mapped on chromosomes 3, 6, 7, and 9 (Liu et al., 2020b). Deep sequencing was used to develop a high-density bin map with a 3,477-marker bin, which identified 26 QTLs associated with plant height (He et al., 2021).
Owing to the innovative sequencing technologies, several cost-effective genotyping by sequencing and RNA-seq have been carried out in this era (Supplementary Table 2). Such genotyping by sequencing 328 foxtail millet landraces and 12 green foxtail accessions presented a total of 5,677 single nucleotide polymorphisms for phylogenetic construction (Hunt et al., 2021). Another deep sequencing of long non-coding RNAs (lnRNA) in foxtail millet revealed their responses toward herbicides. Two thousand five hundred forty-seven lncRNAs were identified, of which 787 were known, whereas 1760 were novel. These lncRNAs were differentially expressed across the genotypes and could be utilized in developing herbicide tolerance in foxtail millet (Wang et al., 2020).
Foxtail millet is known as a hardy crop. Deep resequencing of 312 local landraces was performed to dissect its adaptation mechanism to various environmental conditions. Approximately 3.02 million SNPs were detected in this genome-wide scan, and a pseudogene regulator viz., SiPRR37, for heading date was identified. This was produced from a Tc1-Mariner transposon insertion in the first intron, and it was also discovered to be responsible for adaptability to harsh climates in north-eastern ecoregions (Li et al., 2021a). These novel discoveries in foxtail millet accessions enhanced the curiosity to sequence new cultivars for dissecting key traits for manipulation by breeding. In this direction, the de novo genome assembly of Huagu 11 compared to Yugu 1 depicted the nature of imazethapyr tolerance in foxtail millet. The assembly size was 408.37 Mb with a scaffold N50 size of 45.89 Mb. About 627 protein-encoding transcripts were identified in Huagu11 compared to 723 in Yugu 1. Here, Ser-626-Aln substitution was identified in acetohydroxy acid synthase gene to confer imazethapyr tolerance in Hugua 1. The intraspecies comparison revealed 969,596 SNPs and 156,282 indels with four chromosomal inversions (Wang et al., 2021c). The comprehensiveness and accurateness of genome sequence assemblies depend greatly on the techniques used for the task. Currently, Arabidopsis and rice genome sequence is the gold standard for other plant species and crops. Availability of gold standard genome sequence helps generate proper genotype and phenotype information for target gene discovery and prioritization, efficiently facilitating genome editing—however, most crops, including millets, lack gold standard genome assembly. The advancement of NGS technologies has greatly improved the closeness and correctness of genome assemblies (Kersey, 2019). In this context, the contiguity of the existing genome sequence of many crops has been improved through these NGS approaches (Belser et al., 2018). Similarly, applying advanced sequencing techniques might provide an improved genome sequence of foxtail millet, which will give a comprehensive understanding of genome architecture to discover agronomically important genes in this crop. Thus, the transition of sequencing technologies has paved the way to unravel the genomics of complex traits in foxtail millet which could be utilized in developing improved varieties (Figure 2).
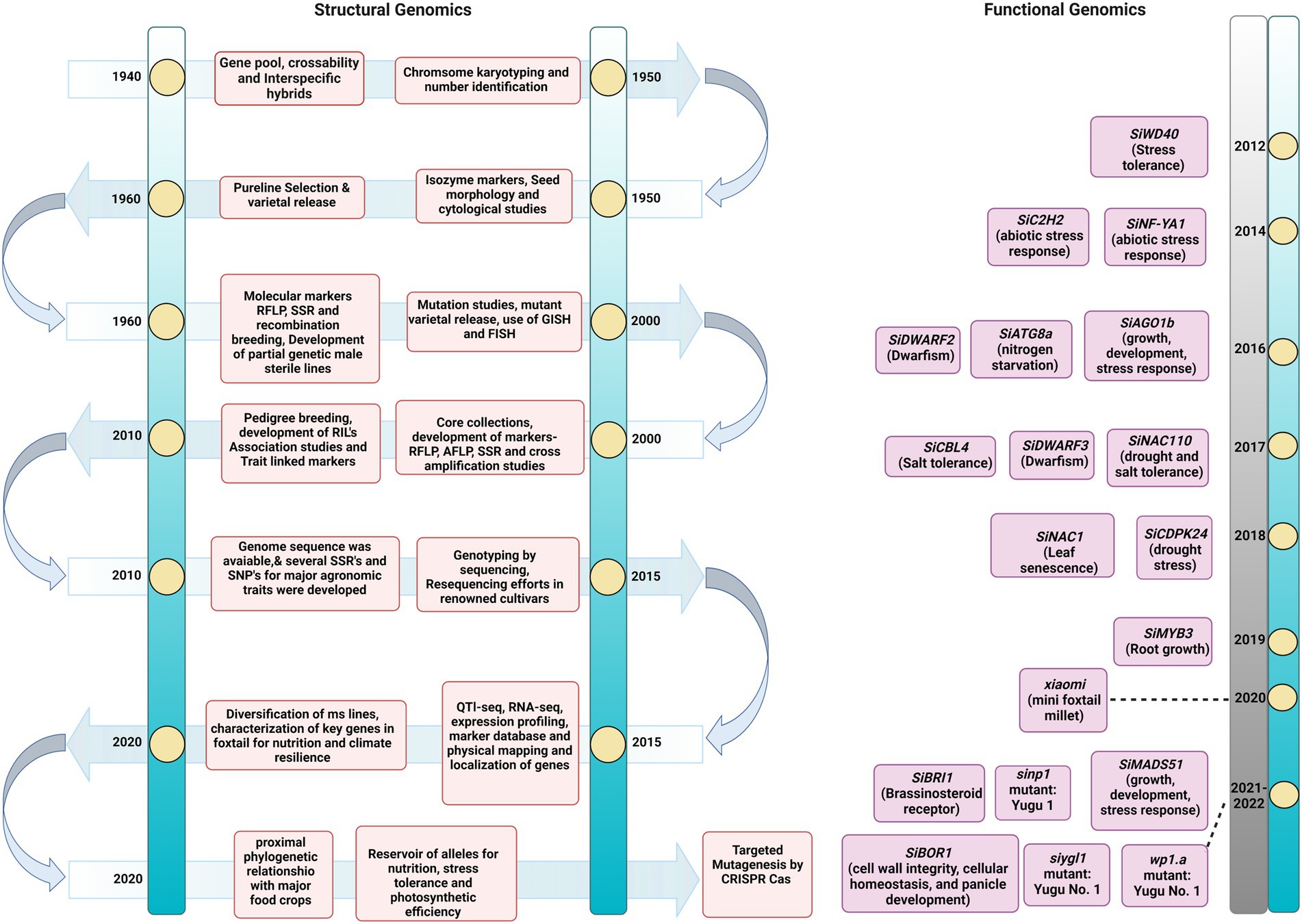
Figure 2. Timeline of advances in Setaria omics pre- and post-genome sequence release. The timeline of events in the breeding and genomics efforts completed for Setaria. The figure describes the significant achievements in classical and modern genetics.
Omics resources in Setaria
Genetics and genomics
Molecular markers are the key tools required for assisting the breeding programs in crops. There are studies on molecular diversity with 40 SSR markers in Taiwan landraces (Lin et al., 2012) and 77 SSR markers in green foxtail accessions (Jia et al., 2013). These programs recorded the advantageous utility of SSRs in marker-assisted breeding of foxtail millet. To facilitate the breeders of foxtail millet, a database of markers in foxtail millet was also developed by Bonthala et al. (2014). This database is a repository for genic SSRs, genomic SSRs, and ILP markers. From the reference genome sequence of Yugu 1, 5,020 highly repetitive microsatellite motifs were successfully utilized to design 788 SSR primers. These primers were analyzed in S. italica and S. viridis, resulting in successful amplicons in 733 primers. These findings were used to analyze the presence of SSRs throughout its genome (Zhang et al., 2014).
Other than SSRs, studies on developing functional miRNA-based markers from the conserved regions of foxtail millet and its related species were also developed with 100% amplification potential (Yadav et al., 2014). Later, implying ISSR markers in Coix and Setaria, their relatedness in cluster analysis was analyzed, suggesting that Setaria could be utilized as a model crop to tap the genetic potential of the non-sequenced crops (Dvořáková et al., 2015). After the accessibility of the whole-genome sequence in S. italica, the GWAS in several accessions was performed to formulate a haplotype map (Bennetzen et al., 2012; Zhang et al., 2012; Jia et al., 2013). Recent studies with SNP markers and GBS were predominantly used in association analysis. Population structure analysis among the races by Upadhyaya et al. (2015) exhibited less diversity in the indica variety. This race was comparatively unique to Moharia and maxima from the international core collection. Chander et al. (2017) incorporated 27 SSR and 4 EST-SSR in foxtail millet core collections.
Apart from analyzing the diversity for general traits, the CNL disease-resistant genes from a group of foxtail millet accessions were also used as markers in rice and Arabidopsis to identify their syntenic relationships. Among them, Si08 was similar to Os11 in rice for disease resistance (Andersen and Nepal, 2017a). Therefore, markers starting from RFLP to SNP were deployed in foxtail millet (Table 1), rendering it a model crop in cereals due to its higher transferability across species (Yadav et al., 2014). Among all the markers that were used in association analysis, it was found that the SNP markers resulted in a higher LD in germplasm. The association analysis with ddRAD approach in GWAS detected 844 SNPs in chromosome 5–2,153 SNPs in chromosome 8 (Jaiswal et al., 2019). Along with SNPs, segregation patterns and polymorphism in M2 mutants were analyzed with RAPD dominant markers (Anittha and Mullainathan, 2019). In addition, novel marker-trait analysis with ISSR markers for chlorogenic acid, catechin naringin, and myricetin concentrations presented a higher diversity for radical scavenging activity in foxtail millet, which could act as a base for stress breeding programs (Ghimire et al., 2019). The current focus with recent studies in hybrid production was also conducted to diversify the male sterile cytoplasm in foxtail millet with cpDNA markers. Four different cytoplasmic groups among the populations from cytoplasmic genetic sequences were identified, which have to be further increased in the near future to overcome cytoplasmic-specific pathogens (Liu et al., 2019a). Further, fingerprinting foxtail millet varieties using SSR and RAPD markers helped assess the genetic purity between CO (Te) 7 and the newly released variety, ATL1 (Natesan et al., 2020).
A genetic map with 128 SSR markers was also successfully developed with a span length of 1239.90 cM in RILs (Qie et al., 2014). Further, the reference genome sequence of Yugu 1 was used to develop 1,013 SSR markers. These markers were implied in F2 to construct a high-density linkage map. This map was refined and produced maximum coverage of 1,035 loci across nine chromosomes. The total span length of the map was 1318.80 cM and mapped the position of 29 QTLs for 11 traits in join-map 4.3 (Fang et al., 2016). In addition to SSRs, SNP for linkage map construction in foxtail millet was used in various studies. Initially, with 992 SNP markers in RILs with a span of 1,416 cM, a linkage map was developed by Bennetzen et al. (2012). Successively, another linkage map with SNP markers in RILs identified around 59 QTLs. The total span length of the SNP formulated map was 1934.60 cM. Through multiplexed shotgun sequencing, Zhang et al. (2017a) performed a genome-wide marker discovery genotyping with MSTmap. This study also reported the location of the hybrid sterility gene on chromosome 6. Following this, next-generation sequencing methods were utilized in the reference genome sequence, which detected 2,668, 587 SNP loci across the genome. Using this data, 9,968 SNP markers in F2 were deployed to form a high-density SNP linkage map. Eleven major QTLs for eight agronomic traits were mapped on the chromosomes with MSTmap. The total span length in 9 linkage groups was 1648.80 cM (Wang et al., 2017). These constructed genetic maps could be used to identify candidate genes underlying key traits to carry out the effective marker-assisted selection in foxtail millet.
Further, map-based cloning for SiAGO1b in bulk segregation analysis and fine mapping with SSR and SNP markers refined its location in 46.30 kb region between the SNP markers SNP027326466 and SNP27372797 on chromosome 7 (Liu et al., 2016a). Following this, the positional cloning of SiYGL1 for leaf color was performed by Li et al. (2016a,b). Bulk segregation analysis in F2 for markers on chromosome 9 was focused, and it was mapped to a 288.50 kb region between the SSR markers CAAS9005 and b248. The candidate gene was refined to a 77.10 kb region between CAPS697 and InDel3595. Another approach for SisTL1 locus on chromosome 9 was performed in F2 populations with CAPS marker. This gene was refined to be located at a 91 kb interval between CAPS-8 (4,339,573 on chromosome 4) and CAPS-7 (4,430,449 on chromosome 4). The resequencing of this locus revealed three SNPs, with one in the 15th exon, the second in the intergenic region of chromosome 4 (4412036), and the third in the first exon (Tang et al., 2018). Similar studies with locus SisTL2 were fine mapped to a 12 Mb region from 46.80 Mb to 58.90 Mb on chromosome 9. This gene led to the striped leaves, and it was located in a 1 Mb region with four putative mutants. On chromosome 9, this phenotype was caused due to an alteration in G to A and found to alter a splicing site (Zhang et al., 2017a). Recent studies in S. viridis located the reduced leaf angle phenotype in a homozygous locus at the 800 kb region on chromosome 5. This region contained 104 disruptive SNPs and 687 indels. Also, it was found that the insertion of G at this locus Chr_5:41489494 controlled the ligule development in S. viridis (Mamidi et al., 2020). These gene localizations in maps could be used for determining the key factors in the expression of a phenotype in genomics-assisted breeding (Supplementary Figure 1).
Following genome sequencing and annotation, identifying genes and their functional characterization is imperative to provide researchers with tools for biological research. The summary of gene families examined in Setaria is given in Supplementary Table 3. A study of transcription factor families in Setaria led to the identification of 147 NAC (Puranik et al., 2013), 171 AP2/ERF (Lata et al., 2014), 209 MYB (Muthamilarasan et al., 2014b), 149 bHLH (Wang et al., 2018b), 124 C2H2-ZF (Muthamilarasan et al., 2014a), 110 WRKY (Muthamilarasan et al., 2015a), 44 SCL (Liu et al., 2017a,b), 35 Dof (Zhang et al., 2017b), 47 HD-ZIP (Chai et al., 2018) and 27 Trihelix transcription factors (Wang et al., 2018a). In addition to TFs, 53 members of SET domain-containing proteins known to catalyze histone lysine methylation were also identified (Yadav et al., 2016). Pentatricopeptide repeat (PPR) protein and ADP-ribosylation factors family connected to post-transcriptional processes comprised 486 and 25 members, respectively (Muthamilarasan et al., 2016; Liu et al., 2016a). Further, gene families associated with RNA silencing complex viz. Dicer-like (8), Argonaute (19), and RNA-dependent RNA polymerase (11) were also studied (Yadav et al., 2015). Few of these genes showed altered expression in response to abiotic stress. Another genome-wide study identified 39 Nuclear Factor Y (NF-Y) genes in foxtail millet. Of these, SiNF-YA1 and SiNF-YB8 were responsive to salt and drought stresses (Feng et al., 2015). Genome-wide analysis of the 14–3-3 family in foxtail millet and its downstream characterization revealed that Si14-3-3 proteins interact with SiRSZ21A (a nucleoplasmic shuttling protein) in a phosphorylation-dependent manner and are involved in multiple abiotic stress responses (Kumar et al., 2015). Analysis of different heat shock proteins, such as HSP100, HSP90, HSP70, HSP60, and sHSP, demonstrated that many of the genes were responsive during abiotic stress. In particular, expression of SisHSP-27 was significantly increased in the foxtail millet tolerant cultivar., suggesting its importance for further characterization (Singh et al., 2016). Also, foxtail millet aquaporins viz. 12 plasma membrane intrinsic proteins (PIPs), 11 tonoplast intrinsic proteins (TIPs), 13 NOD26-like intrinsic proteins (NIPs), and 3 small basic intrinsic proteins (SIPs) were classified, and determination of their expression in response to various abiotic stresses highlighted the role of SiPIP3;1 and SiSIP1;1 in stress response (Singh et al., 2019). In addition, 11 Cytokinin oxidase/dehydrogenases linked to hormone metabolism (Wang et al., 2014) and 20 Aldehyde dehydrogenases (Zhu et al., 2014) were also identified through genome-wide analysis in Setaria.
Setaria is considered a valuable experimental system for gene discovery related to biofuel traits. In this regard, Ferreira et al. (2019) reported the analysis of gene families encoding for crucial enzymes linked to various bioenergy generation processes. Muthamilarasan et al. (2015b) described 13 gene families involved in secondary cell wall biosynthesis. In Setaria, 10 Phenylalanine ammonia lyase, 3 Cinnamate 4-hydroxylase, 5 4-Coumarate: CoA ligase, 2 Hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase, 1 p-coumarate 3-hydroxylase, 6 Caffeoyl-CoA 3-O-methyltransferase, 10 Cinnamoyl-CoA reductase, 2 Ferulate 5-hydroxylase, 6 Caffeic acid O-methyltransferase and 11 Cinnamyl alcohol dehydrogenase gene family members were investigated (Ferreira et al., 2019). Furthermore, a recent study identified 56 Laccase family members linked to lignin metabolism (Simões et al., 2020). In addition, various other gene families involved in multiple stress responses, disease resistance, seed storage or transport, such as WD40 (Mishra et al., 2014), 14–3-3 (Kumar et al., 2015), ZIP (Alagarasan et al., 2017), SOD (Wang et al., 2018c), OASTL (Liu et al., 2019b), ATG (Li et al., 2016a,b), LIM (Yang et al., 2019), CDPK (Yu et al., 2018), NBS-LRR (Zhao et al., 2016), CNL (Andersen and Nepal, 2017b), SSP (Gaur et al., 2018), MADS-box (Zhao et al., 2021a), BES/BZR (Liu et al., 2021), SNARE (Wang et al., 2021a), and PTI1 (Huangfu et al., 2021; Supplementary Table 3). Genome-wide identification and in silico analysis of phosphate transporter 1 gene (PHT 1) in Setaria viridis depicted 12 PHT 1 gene families for phosphorous uptake (Ceasar, 2019). Karunanithi et al. (2020) identified 32 terpene synthase (TPS) gene family members in S. italica genome. It was also found that cytochrome P450 monooxygenase (CYP99A17) catalyzes the C19 hydroxylation of SiTPS8 to generate the corresponding diterpene alcohols. Recently, 94 amino acid transporters (AATs) have been identified and divided into 12 subfamilies in the foxtail millet (Yang et al., 2021b). Further, characterization of the chitinase gene family in foxtail millet has identified 40 genes. Of these, a few genes might have a potential role in defense mechanisms under low temperature, drought, and osmotic stress conditions (Motukuri et al., 2021). Genome-wide identification and evolutionary analysis of the ARF gene family in foxtail millet revealed that duplication and purifying selection contribute to functional redundancy (Chen et al., 2021; Supplementary Table 3).
Transcriptomics
Studies examining the dynamics of the whole transcriptome have hinted at the differential expression of several classes of genes playing roles during condition-dependent experimental systems (Figure 3). Earlier, suppression subtractive hybridization (SSH) based transcriptome analysis of S. italica led to the identification of dehydration-responsive transcripts in susceptible and tolerant cultivars (Lata et al., 2010). Another SSH-based differential gene expression analysis identified a Lipid transfer protein, SiLTP, that showed significant involvement in drought and salt response in foxtail millet (Pan et al., 2016). Using reference-based and de novo assembly, various transcripts and simple sequence repeats (SSR) involved in C4 photosynthesis were identified in S. viridis (Xu et al., 2013). In addition, transcriptome analysis of different zones of S. viridis internode uncovered various genes and primary metabolites involved in the transition from primary to secondary cell wall synthesis (Martin et al., 2016). Another transcriptome study of drought-tolerant S. italica cultivar “Yugu1” and drought-sensitive “An04” revealed the altered expression of genes involved in phytohormone metabolism and signaling, detoxification, transcription factors, and stress-related proteins (Tang et al., 2017). RNA-seq was carried out for drought-resistant M79 and its parental lines to understand the relationship between drought stress and photosynthesis. The analysis showed that photosynthetic pathway-related genes were highly expressed in M79 (Shi et al., 2018). Further, spatiotemporal transcriptome analysis of S. viridis inflorescence provided an overview of the regulatory network underlining sequential developmental stages (Zhu et al., 2018). Next, RNA-seq analysis of S. italica during water-deficit stress induced by PEG-6000 identified biological pathways involving ABA-response, proline and soluble sugar synthesis, reactive oxygen species (ROS) metabolism, channel protein genes, and transcription factors (Xu et al., 2019). Leaf transcriptome of S. viridis during high light and low light conditions revealed that HL and LL affect sugar accumulation and photosynthesis in a contrasting manner (Henry et al., 2020). Furthermore, differential gene expression profiling identified various genes and pathways involved in the stress response mechanism during Uromyces setariae-italicae, and Sclerospora graminicola infection in S. italica (Li et al., 2015, 2020).
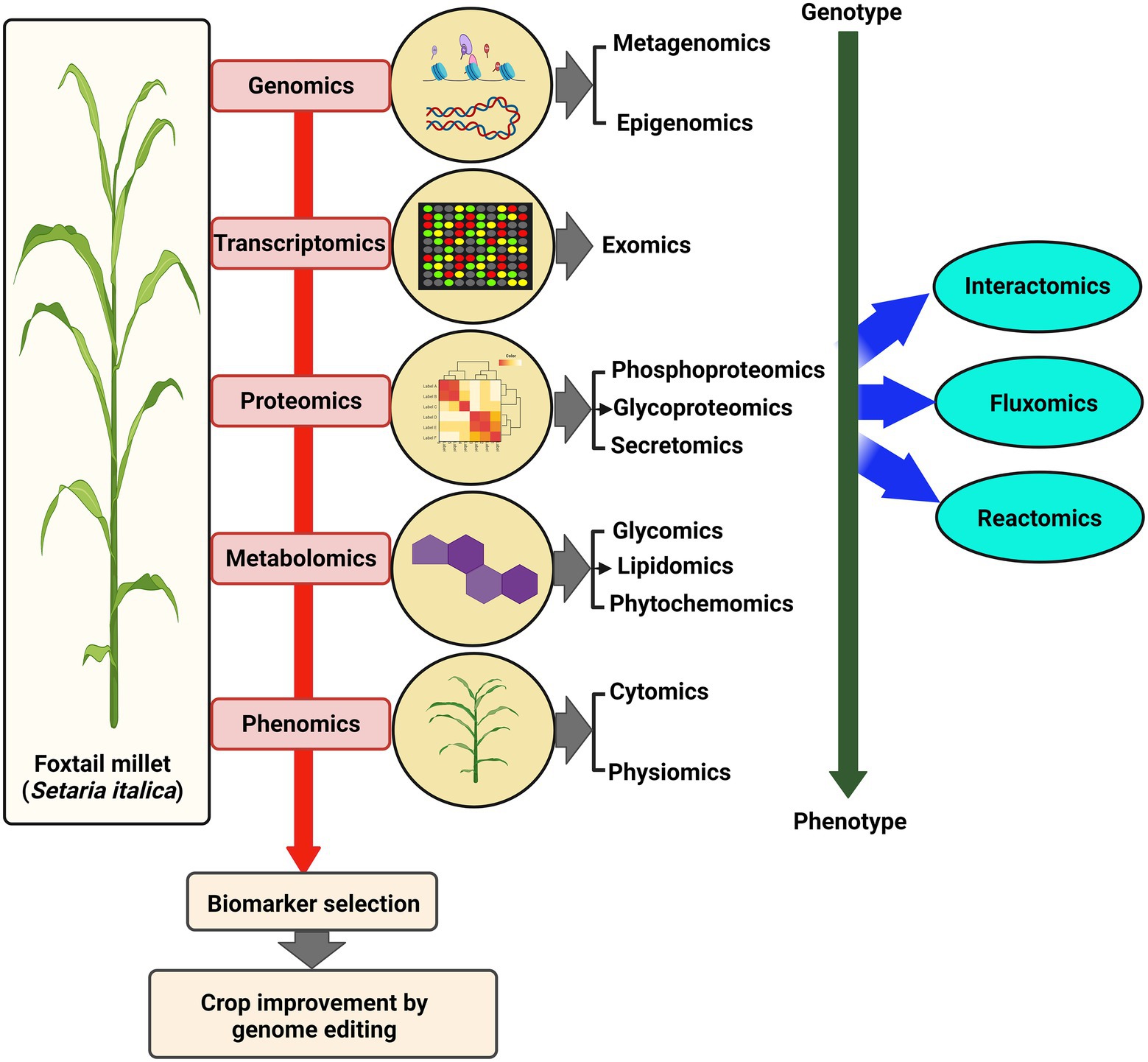
Figure 3. Multi-OMICS approach for crop improvement. Various advanced “OMICS” approaches used to understand different biological aspects in foxtail millet, from genotype to phenotype. These large-scale studies lead to the identification of biomarkers that paves the way for OMICS-assisted crop improvement.
Genome-wide gene annotation and non-coding RNA studies for drought responses by deep sequencing exhibited the role of small interfering RNAs and long non-coding RNAs. The transcriptomic sequencing explored the role of 24-nt siRNA flanking genes beneath the upregulated genes under drought stress. The PEG stimulated drought samples were initially subjected to Illumina pair-end sequencing technology. Further RNA seq analysis identified 2,824 genes responsive to drought expression patterns, and among them, 48.23% were upregulated, and 51.77% were downregulated. The key genes expressed encoded the late embryogenesis abundant protein (LEA), heat shock proteins (HSP), and aquaporin phosphatase 2C (PP2c; Qi et al., 2013). Parallel Illumina sequencing was utilized in another study to decode the role of miRNA targets toward drought stress. Degradome sequencing of the An04-4783 inbred line established 81 miRNAs belonging to 28 families. Among these, 14miRNAs were upregulated, and four were downregulated. The sequence information of the miRNAs revealed the role of 56 known genes and 26 novel, unknown genes in response to drought (Wang et al., 2016a,b).
Subsequently transcriptome analysis of mutants such as no pollen 1(NP1) characterized by defective pollen exine revealed that SiNP1 encodes a protein involved in carbohydrate metabolism and fatty acid biosynthesis (Zhang et al., 2021a,b,c). Similarly, the characterization of SiBOR 1 for boron accumulation in foxtail millet revealed a G-A transition at the seventh exon, and it is dominantly expressed in panicles. This mutant resulted in reduced boron with thicker cell walls in Setaria (Wang et al., 2021b). DNA methylation studies and transcriptome analysis revealed the role of epigenetic modifications during grain filling (Wang et al., 2021d). Recently, RNA-seq analysis of two mutants siaux1-1 and siaux-1-2 was performed to identify auxin-responsive factors beneath C4 root system architecture in shaping the root apical meristem (Tang et al., 2022). The findings pave a domain for screening the mutants to understand the C4 mechanism. Transcriptional profiling of foxtail millet seed at different developmental stages depicted dynamic changes in fatty acid and phytosterol content (Yuan et al., 2021).
Proteomics and metabolomics
Proteomics and metabolomics have been employed to elucidate the protein and metabolite complement of a particular molecular network that regulates the biosynthetic, regulatory, and signaling pathways in various plant species (Figure 3). In Setaria, the accountability of global studies based on proteome and metabolome analyses is significantly less. Veeranagamallaiah et al. (2008) reported a 2-dimensional gel electrophoresis proteome analysis of S. italica during the salt stress response. Differential response of green foxtail and yellow foxtail has been studied during the application of Pyroxsulam using metabolite analysis (Satchivi et al., 2017). A proteomic study of Zhangzagu3 was performed, which is known for its climate adaptability and disease tolerance, to evaluate the parental contribution to elite traits (derived from Zhangzagu3fu and A2). This study provided valuable information related to the molecular mechanism of heterosis in hybrid millets (Song et al., 2018). Further, Tandem mass tags (TMT) followed by liquid chromatography coupled mass spectrometry (LC–MS/MS) analysis identified 321 drought-responsive proteins in foxtail millet (Pan et al., 2018). In two recent studies, changes in the proteome of foxtail millet grains during drought stress led to identifying 104 and 83 differentially abundant proteins using 2-DE (Li et al., 2019a; Xu et al., 2020). Comparative proteome profiling after the foliar application of sodium selenite has identified 123 differentially expressed proteins in foxtail millet (Liang et al., 2020). Primary and secondary metabolic profiling of PTGMS and hybrid foxtail millet lines and their comparison with rice led to identifying several species-specific metabolites accumulated during different developmental stages (Li et al., 2018). Recently, metabolome analysis of S. viridis roots colonized with symbiotic bacteria, Herbaspirillum seropedicae revealed that nitrogen, starch, and sucrose metabolism linked metabolites were reduced compared to the uninoculated roots. In contrast, metabolites involved in purine, zeatin, and riboflavin pathways were significantly enriched (Agtuca et al., 2020). Using iTRAQ, 610 and 276 differentially expressed proteins (DEPs) in foxtail millet, Zhangzagu10 and the female/male parent lines were identified (Weng et al., 2020).
Using a system biology-based approach, foxtail millet has also been chosen as a model to gain a deeper insight into the C4 photosynthetic pathway. Metabolic reconstruction, combined with transcriptome and proteome analysis, unraveled similarities and differences in the central metabolism of mature and immature tissues in S. italica (de Oliveira Dal’Molin et al., 2016). Integrated transcriptome and metabolome analysis explained the comprehensive regulatory network underlying drought and salinity stress response in foxtail millet (Pan et al., 2020; Yu et al., 2020). Quantitative determination of 62 volatile compounds has been performed using simultaneous distillation extraction (SDE) and gas chromatography–mass spectrometry (GC–MS) in four foxtail millet varieties, namely Jigu 42, Henggu13, Henglvgu1, and Heinuogu (Li et al., 2021b). Metabolome-based GWAS identified the basis of natural genetic variations in foxtail millet germplasm (Wei et al., 2021).
Functional databases and online resources
After decoding the whole-genome sequence of S. viridis and S. italica (Bennetzen et al., 2012; Zhang et al., 2012), advances were made to generate large-scale genomic resources. The first-ever database for Setaria, the Foxtail millet Marker Database (FmMDb; http://www.nipgr.res.in/foxtail.html), was developed by Dr. Manoj Prasad Laboratory at the National Institute of Plant Genome Research (NIPGR), New Delhi, India (Bonthala et al., 2013). It provides access to genomic-, genic-SSRs, and ILP markers linking basic and applied sciences in foxtail millet. Further, the Foxtail Millet Transcription Factor Database (FmTFDb; http://59.163.192.91/FmTFDb/index.html) comprised 2,297 putative TFs belonging to 55 families (Bonthala et al., 2014). This database encompasses genomic location, sequence features, phylogeny, gene ontology (GO), and tissue-specific expression for all the TFs. In addition, the foxtail millet microRNA Database (FmMiRNADb: http://59.163.192.91/FmMiRNADb/index.html) provided marker information on 355 mature miRNAs, their secondary structure, and putative targets (Khan et al., 2014). Setaria italica Functional Genomics Database, SIFGD1 was established to predict gene function, motif analysis, regulatory modules, and gene family (You et al., 2015). This database represents genome, transcript, and protein sequence information combined with data sources like Beijing Genomics Institute, NCBI, and Phytozome. Furthermore, considering the significance of transposable elements (TE) based markers, Yadav et al. (2015) constructed the Foxtail millet Transposable Element-based Marker Database (FmTEMDb; http://59.163.192.83/ltrdb/index.html) from 30,706 TEs and 20,278 TE-based markers. In addition to the databases solely designed for Setaria, other public data sources, such as Plantgbd, Phytozome, and Gramene, also contain information for genome mining in millets (Duvick et al., 2008; Goodstein et al., 2012; Gupta et al., 2016).
Functional studies on stress tolerance traits
Numerous studies have reported millets adaptation and its molecular mechanism during the abiotic stress response. Investigation of foxtail millet varieties, Zhangzagu 10 and Jingu 21 after herbicide tribenuron-methyl (TBM) treatment revealed that the grain yield decreased with increasing TBM concentration (Ning et al., 2015). In foxtail millet, high-temperature stress was linked to root-shoot growth and metabolism (Aidoo et al., 2016). Using parallel analysis of RNA ends (PARE) approach during drought stress, gene degradation and gene transcription data were combined to study uncapped mRNAs in foxtail millet (Yi et al., 2015). Gene family identification and expression analysis of WRKY transcription factors indicated the putative role of SiWRKY066 and SiWRKY082 in dehydration and salinity stress response (Muthamilarasan et al., 2015a). Drought tolerance of foxtail millet was linked to plant growth-promoting rhizobacteria (PGPR), namely, Pseudomonas fluorescens, Enterobacter hormaechei, and Pseudomonas migulae (Niu et al., 2018). During low-nitrogen conditions, the root system was found to be decreased compared to increased biomass in foxtail millet. Expression of transporters, SiNRT1.1, SiNRT2.1, and SiNAR2.1 in root and SiNRT1.11 and SiNRT1.12 in the shoot was also increased, resulting in enhanced nitrate uptake and remobilization (Nadeem et al., 2018). Recently, an interesting study reported that 30 mg/m3 sulfur dioxide (SO2) application leads to increased tolerance to drought stress by decreased stomatal apertures and a reduced leaf transpiration rate (Han et al., 2019a,b). Also, applying exogenous SO2 derivatives sodium sulfite and sodium bisulfite alleviates heavy metal stress in foxtail millet (Han et al., 2018). Sodium hydrogen sulfide (NaHS) treatment changes the proline content by modulating the activity of proline-5-carboxylate reductase (P5CR) and proline dehydrogenase (PDH) enzymes while combating cadmium (Cd) stress in foxtail millet (Tian et al., 2016). In another study, adding 10 mM Ca2+ to the growth medium regulated superoxide dismutase and catalase expression, thereby increasing tolerance to salt stress (Han et al., 2019a,b). Quantification of polyamines in the salt-tolerant cultivar., Prasad, and susceptible cultivar., Lepakshi revealed that increased level of polyamine during salinity stress was due to enhanced activity of spermidine synthase and S-adenosyl methionine decarboxylase enzymes in the cultivar Prasad (Sudhakar et al., 2015). Plant treatment with polyamines such as putrescine (Put) and spermidine (Spd) revealed their protective effect during salinity stress in foxtail millet (Rathinapriya et al., 2020). The HAK/KUP/KT transporter family protein in S. italica, SiHAK1, was overexpressed in Arabidopsis to understand the K homeostasis during K+ deficiency and salt stress which provided the clue of the potassium homeostasis mechanism in foxtail millet (Zhang et al., 2018a). Also, a study has shown the relation between an enlarged root system, increased expression of phosphate transporters SiPHT1.1, SiPHT1.4, and reduced expression of nitrate transporters SiNRT2.1, SiNAR2.1 in roots during phosphate limitation (Ahmad et al., 2018).
Transcriptome analysis of low potassium stress-tolerant variety Longgu 25 led to identifying 1982 DEGs and 18 candidate genes. Further, heterologous expression of one of the candidate genes, SiMYB3, in Arabidopsis promoted elongation of primary roots and K+ deficiency tolerance (Cao et al., 2019). Overexpression of SiMYB3 also increased low-nitrogen stress tolerance by regulating root growth in rice and Arabidopsis (Ge et al., 2019). Genome-wide identification of core ABA signaling components SvPYL1 to SvPYL8, SvPP2C1 to SvPP2C12, SvSnRK2.1 to SvSnRK2.11 and their expression profiling during salt, drought, and cold stresses have provided targets for C4 plant engineering (Duarte et al., 2019). Overexpression of the SET domain-containing protein SiSET14 in yeast unfolded its role in cold stress response (Yadav et al., 2016). Liu et al. (2019c) showed the relationship between stimulating rhizosheath (layer of soil particles adhered with root surface by root hairs and mucilage) formation and declining soil water content. Recently, conserved miR394 targeted F-box gene SiFBP6 was identified using RLM-RACE (RNA ligase mediated rapid amplification of 5′ cDNA ends), which positively regulated drought resistance in foxtail millet (Geng et al., 2021). Interestingly, P-solubilizing microbes Acinetobacter calcoaceticus EU- LRNA-72 and Penicillium sp. EU-FTF-6 has been shown to mitigate the effect of drought stress by inducing the accumulation of glycine betaine, proline, and sugars, and decreasing lipid peroxidation in foxtail millet (Kour et al., 2020). A recent study discussed various genomic designing approaches for foxtail millet for abiotic stress tolerance (Rana et al., 2021).
Among biotic stress responses, Digital gene expression (DGE) constructed from S. italica cultivar Shilixiang during rust pathogen Uromyces setariae-italicae infection shed light on the mechanism of rust-response in foxtail millet (Li et al., 2015). The transcriptional study of foxtail millet recently revealed various stress-responsive pathways during the early stage of S. graminicola infection (Li et al., 2020). Transcriptional profiling of foxtail millet infected with Ustilago crameri showed that SiCDPK and SiRboh might be positive regulators during stress response (Hao et al., 2020). In another study, two maize insect pests, namely Spodoptera exigua (beet armyworm; BAW) and Spodoptera fugiperda (fall armyworm; FAW), were found to feed on Setaria plants as well. The study further showed that while JA-induced response was similar in both maize and Setaria, secondary metabolites such as benzoxazinoids and volatiles production showed a significant difference in both the plants (Hunter et al., 2020). Foxtail millet varieties exhibit resistance against blight disease caused by Rhizoctonia solani Kuhn (Patro et al., 2020). An ultra-density genetic linkage map identified QTLs linked to blast resistance in foxtail millet (Tian et al., 2021). Further, the current status of blast disease in foxtail millet caused by Pyricularia setariae Nishikado has been shown by Das et al. (2021). Taken together, various studies have been performed elucidating the regulatory mechanisms involved in biotic and abiotic stress response in Setaria. A combination of OMICS-based tools can be employed in the future to decipher the key factors contributing to the stress tolerance trait in foxtail millet, drought stress in particular. This information would help researchers globally to understand the basis of stress response in plants and utilize the knowledge further to inculcate stress tolerance in other important cereal crops.
Functional studies on biofuel traits
The Panicoideae clade comprises commercially important fuel stock grasses, such as maize, sugarcane, sorghum, and switchgrass (Lawrence and Walbot, 2007). However, the polyploid genome is one of the major limiting factors for genetic studies and manipulation of these crops. In the past decade, being genetically closer to the popular fuel stock grasses, S. viridis and S. italica are widely accepted as a promising model for biofuel research (Li and Brutnell, 2011; Muthamilarasan and Prasad, 2015; Muthamilarasan et al., 2015b; Mauro-Herrera and Doust, 2016). Petti et al. (2013) performed a deep analysis of S. viridis cellulose, neutral sugars, lignin, cellulose biosynthesis inhibitor response, and phylogenetic analysis of CESA genes and compared it with sorghum, maize, and switchgrass. Overexpression of PvMYB4 in switchgrass (Panicum virgatum), led to increased cellulosic ethanol yield by suppressing the lignin deposition and phenolic fermentation inhibitors and balancing soluble sugars and pectic polysaccharides (Shen et al., 2013). In S. viridis, silencing of acyl-CoA transferase SvBAHD01 caused enhanced biomass digestibility; therefore, it was found to be a suitable candidate for further investigation (De Souza et al., 2018). Several gene families linked to biofuel traits, namely, PAL, C4H, 4CL, HCT, C3′H, CCoAOMT, CCR, F5H, COMT, and CAD have been identified in Setaria (Ferreira et al., 2019). Due to its small and diploid genome, Setaria, particularly green foxtail, appears to be a promising model to conduct future research deciphering its potential as a biofuel crop. Further, identifying regulatory pathways underlying the potential biofuel trait and an in-depth analysis of candidate genes using reverse genetics approaches have to be done to explore the genetic determinant for the biofuel attribute regulation in Setaria, which can be utilized further for translational research.
Functional studies on photosynthetic traits
Fahlgren et al. (2015) developed the Bellwether Phenotyping Platform for automated recording of plant growth and multimodel phenotyping. Further, an open-source Plant Computer Vision (PlantCV) was developed for qualitative image analysis and used to detect the genotypic and environmental effect of plant height, biomass, water use efficiency (WUE), and architecture (Fahlgren et al., 2015). Various accessions have been studied to determine the WUE and heat stress tolerance in S. viridis (Saha et al., 2016). Thermal imaging and visible–near infrared spectroscopy were proven helpful in evaluating WUE and other physiological responses in S. italica (Wang et al., 2016a,b). The relationship between WUE and plant growth was also determined by Feldman et al. (2018), and the loci linked to pleiotropic components of WUE were identified through linkage mapping. Forward genetics study in S. viridis led to the identification of the SvAUX1 (AUXIN1) gene responsible for sparse panicle1 (spp1) phenotype sharing significant homology with ZmAUX1 in maize (Huang et al., 2017). Depletion of β-carbonic anhydrase (β-CA) in S. viridis indicated that photosynthesis was not affected at normal CO2 partial pressure even after the decreased level of CA. Also, mesophyll conductance was the limiting factor with CA only during low CO2 partial pressure (Osborn et al., 2017). In a similar report, CO2 assimilation was enhanced, and photosynthetic efficiency was increased during low partial pressure due to high mesophyll conductance (Ubierna et al., 2018). In S. viridis and S. italica, the correlation between photoperiod changes and flowering time was studied, indicating that flowering in Setaria is quicker during short-day conditions. Also, long-day conditions employ secondary genetic regulation additional to those employed by short-day photoperiod (Doust et al., 2017).
Carbon isotopic signature (δ13C) from leaf was found to be strongly linked to water use, transpiration rate, biomass, stomatal conductance, and transpiration efficiency in both S. viridis and S. italica (Ellsworth et al., 2017). Further, as an extension to this study, genetic link elucidation of this correlation identified three QTLs for δ13Cleaf (Ellsworth et al., 2020). In S. italica cultivar Yugu1 549 ethyl methane sulfonate-induced mutants were generated and screened for the disruption of Kranz anatomy, associated with C4 photosynthesis in many plants. About 14 mutants with abnormal Kranz structures were identified, which will help elucidate genes involved in the development of the Kranz structure (Luo et al., 2018). SiYGL2, the homolog of AtEGY1, was shown to be involved in the leaf senescence and Photosystem II function in S. italica (Zhang et al., 2018c). The genome-scale metabolic model was constructed in S. viridis that revealed the role of NH+4 and NO−3 ratio in balancing charges and 3-PGA/triosephosphate shuttle in proton balancing (Shaw and Maurice Cheung, 2019). Ermakova et al. (2019) have reported that the overexpression of Rieske FeS protein results in higher Cytochrome b6f content leading to the removal of electron transportation limit in mesophyll and bundle sheath cells. Overall, the C4 photosynthesis and CO2 assimilation level were markedly increased (Ermakova et al., 2019). Higher photosynthetic rates were also observed at elevated CO2 levels, and the response was analogous to C3 plants (Li et al., 2019b). Foxtail millet mutant library was generated, proving a valuable genetic resource for future functional studies. Further, map-based cloning identified deletion mutation in the phytoene synthase encoding WP1 exhibiting reduced chlorophyll and carotenoid in leaf and panicles (Sun et al., 2019). Transcriptome study of S. viridis leaves that serve as a source revealed that high light and low light exposure to a plant causes the contrasting deregulation of sugar sensors HXK and SnRK1 and trehalose pathway genes (Henry et al., 2020). White light exposure increases H2S generation, leading to hypocotyl elongation in foxtail millet (Liu et al., 2019d). Due to C4 photosynthesis, and high water and nitrogen use efficiency, foxtail millet is been a focus of researchers for years. However, genetic dissection of the photosynthetic traits to establish an in-depth understanding of the effect of environmental factors on yield is still in infancy. Advancement in the phenomics tools has provided high-throughput, non-destructive phenotyping platforms that hold the potential to be used for studying yield-related traits more efficiently. Further, identifying and validating molecular players linked to the photosynthetic traits and WUE/NUE is imperative to reduce the environmental impact on foxtail millet production.
Functional studies on nutritional traits
Foliar application of selenite in S. italica exhibited the potential of increasing Selenium and yellow pigment, thus increasing its dietary advantages (Ning et al., 2016). Numerous reports have shown the health benefits of foxtail millet, such as antiproliferative activity, lowering blood pressure, low glycemic index, glucose-lowering effect, multi-drug resistance in human HCT-8/Fu colorectal cancer, antioxidant properties, immunostimulatory activity, attenuation of Atherosclerosis (Zhang and Liu, 2015; Chen et al., 2017; Lestari et al., 2017; Ren et al., 2018a,b; Lu et al., 2018b; Marak et al., 2019; Srinivasan et al., 2020; Liu et al., 2020a). Evaluation of phenolic antioxidants of foxtail millet indicated its potential in regulating postprandial hyperglycemia (Xiang et al., 2019). Characteristics of various elements, GABA and polyphenols, β-glucan (Sharma et al., 2018b,c), and the effect of storage conditions on germination (Sharma et al., 2018a) were analyzed in S. italica. Gel forming ability of foxtail millet was increased significantly with the addition of CaCl2 and FeSO4, adding to its nutraceutical property (Nagaprabha and Bhattacharya, 2016). Further, physiochemical characterization of starch gel from green gram and foxtail millet was also performed by Nagaprabha et al. (2018). Terahertz time-domain spectroscopy (THz-TDS) was employed for qualitative and quantitative analyses of (a) glutamic acid and glutamine from yellow foxtail millet, and (b) ternary amino acid from foxtail millet, highlighting the application of this technique for binary and ternary amino acids measurement as compared to other methods (Lu et al., 2016, 2018a). Soluble dietary fiber content analysis from foxtail millet bran showed several health beneficial properties (Dong et al., 2019; Ji et al., 2019). Aroma compound isolation from foxtail millet grain after boiling, freeze-drying after boiling, or roasting exhibited that unsaturated aldehydes, benzene, and alcohols surged after boiling. At the same time, freeze-drying lowered the content of volatile compounds linked to the aroma. On the other hand, roasting enhanced pyrazine content (Bi et al., 2019).
In a previous study, cooked millet was also shown as promising nutraceutical food for delaying type 2 diabetes (Ren et al., 2016). Nutritional quality comparison elucidated that protein, fat, and fiber were higher in millet food products than in traditional rice products (Verma et al., 2015). Setaria italica resistant starch content and effect of different treatment was determined to analyze the structure and digestibility (Babu et al., 2019). In addition, the difference at the genetic level in starch physiochemical properties was determined in different accessions of Chinese foxtail millet and landraces found in Taiwan (Qi et al., 2019; Yin et al., 2019). Further, foxtail millet has been assessed for yield, quality, and morpho-nutritional traits to explore the genetic variability (Srilatha et al., 2020; Karvar et al., 2021). Sharma and Sharma (2021) showed that bioprocessing, including soaking, germination, fermentation, and a combination of aforesaid treatments, reduced the antinutrients while enhancing the bioactive profile in foxtail millet grains. Also, different milling fractions of foxtail millet were analyzed for their phenolic profiles, antioxidant properties, and α-glucosidase inhibitory effects (Zhang et al., 2021a,b,c). Further, it has been shown that the unsaturated fatty acids are responsible for forming volatiles, thus leading to an unpleasant aroma during germination in foxtail millet (Li et al., 2021b). A recent study identified 18 long non-coding RNAs (lncRNAs) in relation to grain yield and predicted them to function as miRNA target mimics (Zhao et al., 2020). The nutritional quality of foxtail millet was analyzed in response to elevated CO2 levels and found that eCO2 significantly enhanced the accumulation of K, Mn, Zn, and starch and promoted P accumulation. These findings advocate that foxtail millet holds great potential to provide food security and nutrition under eCO2 (Gong et al., 2021). Collectively, these studies emphasize an array of health benefits and nutritional qualities of foxtail millet. However, most of these are only limited to demonstrating the effect of genetic variability, storage, and method of cooking on nutritional qualities. Metabolomics can be employed further to illustrate the primary and secondary metabolic compounds associated with these qualities. In addition, multi-OMICs would help to understand the key players attributing to the health benefits of foxtail millet and other millets.
Functional studies on other unique traits
In situ hybridization mapping revealed that the apospory-specific genomic region (ASGR) of Pennisetum squamulatum has collinearity with chromosome 2 of S. italica (Sapkota et al., 2016). The anatomical study of the abscission zone (AZ) was performed to understand the difference in AZ development in the two closely related species, S. viridis and its domesticate, S. italica (Hodge and Kellogg, 2016). Further, the anatomy of early shoot development was investigated, specifically targeting Kranz anatomy initiation (Junqueira et al., 2018). Phenotyping assays were optimized for S. viridis at three developmental stages, viz. seed germination, early seedling, and adult plant growth, which will be helpful for physiological analysis of Setaria species and other panicoid species grasses (Acharya et al., 2017). Carbon and nitrogen isotope ratios in different tissues (leaves and grain) were determined to implicate the necessity of accounting for it further during paleodietary reconstructions (Lightfoot et al., 2016). Interestingly, the property of the foxtail millet shell as an efficient bio-sorbent was highlighted for the absorption of Cu, Zn, Cd, and Cr ions (Peng et al., 2018). Recently, variation in the utilization of N was shown at the seedling stage in different foxtail millet varieties (Erying et al., 2020). Desai et al. (2018) established S. viridis as a model system to uncover the mechanism underlining the Time of Day of Anther Appearance (TAA) regulation. In situ localization study of phytoliths exhibited species-specific morphotypes in different tissues of Setaria species that could be used for taxonomic characterization of Setaria (Bhat et al., 2018). Root microbiome examination of foxtail millet plants sampled from two geographically isolated locations indicated that the host plant assists the growth of specific bacteria in the rhizosphere (Jin et al., 2017). Metagenomic analysis of rhizoplane and endophytic bacteria in wild S. viridis and domesticated S. italica also classified species-associated microbiome (Chaluvadi and Bennetzen, 2018). While studying the rhizome of dried foxtail millet, a gram-positive bacterial strain, Amnibacterium setariae was isolated and characterized (Kim et al., 2019).
Endophyte isolation from both root and panicle of S. viridis and S. pumila affirmed distinct and conserved microbial taxa across genotypes and geographical distribution (Rodríguez et al., 2018). Aihemaiti et al. (2019) studied the effect of Vanadium (V) concentration on plant growth and the uptake of essential elements. At a concentration lower than 47.4 mg/L, V positively affected the accumulation of elements, such as P, Fe, Cu, Zn, and Mo, while at a higher concentration, accumulation was reduced. Root growth was more susceptible to increasing V concentration than shoot growth in S. viridis (Aihemaiti et al., 2019). In foxtail millet, it was found that Iso-potentials of PEG (used for drought treatment) and laundry detergent had a more adverse effect on seed germination and overall plant growth than PEG (Heidari et al., 2019). In a recent study, iron plaque (IP) formation was observed in the less studied grass S. parviflora which helps plants adapt to an iron-rich environment (Oliveira de Araujo et al., 2020). Transient expression in tobacco leaves revealed that foxtail millet PPLS1, a bHLH transcription factor associated with SiMYB85 controlled the purple color of pulvinus and leaf sheath (PPLS) trait used as an indicative characteristic of the authentic hybrids (Bai et al., 2020). Recently, the extract from foxtail millet leaves and the stem has shown an allelopathic effect on three different weeds (Dong et al., 2019). Interestingly, the peptides derived from foxtail millet have demonstrated antioxidant and anti-inflammatory activity in HaCaT cells and RAW264.7 murine macrophages (Ji et al., 2020).
Breeding, molecular breeding, and speed breeding in Setaria
Foxtail millet is a diploid cereal free of gluten and has several nutritional emoluments. Breeding for foxtail millet to develop elite cultivars began in the 1950s (Paroda and Mal, 1989). Foxtail millet, being a self-pollinated crop with minute inflorescence, the pure-line selection was the most predominantly used method for developing superior cultivars in foxtail millet (Lata et al., 2013). This remained a barrier to exploiting the prevailing genetic richness in its gene pool. Recently, CO 6 and CO (Te) 7 were released as hybrid derivatives by overcoming these constraints by crossing using a standard hot water method of emasculation and approach method for dusting (Pramitha Lydia et al., 2021). Now, advanced molecular breeding tools are being employed to utilize this prevailing diversity in its gene pool. Several genetic diversity analyses represent the variability for all morphological, biochemical, and nutritional traits in its germplasm, proving a way to perform effective selection and hybridization techniques in foxtail millet to meet the key breeding objectives (Ghimire et al., 2019). In addition, the availability of gene-specific and trait-linked markers from association analysis is an essential tool in developing high-yielding resilient varieties in foxtail millet (Vetriventhan et al., 2014).
After the availability of a complete genome sequence (Bennetzen et al., 2012; Jia et al., 2013), the breeding of foxtail millet has accelerated to inducing targeted mutation. Foxtail millet being a diploid crop, is feasible for dissecting observable mutants. Its smaller growth duration (95 DAS) also prefers a favorable condition to raise subsequent generations to develop a homozygous line within a year. Mutation in foxtail millet began in the mid of 70s (Gupta and Yashvir, 1975) and was initially used to develop elite cultivars of the previously released version. Recently with the joined venture of omics, the mutation breeding in foxtail millet has seen a different phase of success (Mamidi et al., 2020). Anittha and Mullainathan (2019) treated CO (Te) 7 with EMS and DES in various concentrations and visualized different chlorophyll and morphological mutations in M2 generation. This study confronted that EMS was more effective in attaining desirable variants in M2 generation (Anittha and Mullainathan, 2019). Generating mutant sources in foxtail millet serves as a new source for functional genomics. Recently, a library of mutants with 1,353 independent M2 lines exhibited varying chlorophyll and morphological types developed from EMS. Of these, 16 M2 lines were resequenced, and by map-based cloning, Wp 1 gene was identified to have a significant role in chlorophyll accumulation. The results depicted an eight-base pair deletion located at the 6th exon in LOC101786849. The wild Wp1 gene was studied to code phytoene synthase; therefore, the wp1 mutants exhibited reduced chlorophyll and carotenoid contents in leaves and panicles due to premature termination (Sun et al., 2019). Later, advanced approaches as a beginning of reverse genetics were initiated by analyzing de novo assemblies of S. viridis. This study identified loci for three traits, viz., response to climate, non-shattering, and leaf angle, which is a major factor for yield. By incorporating CRISPR-Cas 9, the Less shattering 1 (SvLes 1) was validated for its role in controlling seed shattering. Comparative analysis of this gene in S. italica revealed that the same gene was non-functional due to an insertion of retrotransposon on domestication (SiLes1-TE). Thus, this trait could be manipulated in cultivars to prevent shattering losses in foxtail millet.
Developing mutants by spontaneous and induced mutations also serves to study male sterility. Although several Genic Male sterility (GMS), Photoperiod Genetic Male Sterile (PGMS), and Cytoplasmic Genetic Male Sterile (CGMS) were screened in China, the successful male sterile system that is still used in hybrid seed production is Partial Genetic male sterility (PAGMS). Using this, two superior cultivars, Yugu 1 and Zhaogu 1 were developed (Diao and Jia, 2017). Recent studies on mutagenic agents also revealed that lower doses of EMS were more effective in yielding desirable mutants, whereas higher doses resulted in meiotic abnormalities (Kumar and Pandey, 2021). Hence, breeding for desirable traits by a mutation in foxtail millet requires properly handling the mutagenic agents.
The recent avenue of breeding focuses on reducing the cycle of selection in developing superior varieties, reducing the time span of releasing cultivars to the farmers. Speed breeding focuses on inducing haploids, which could be essential in achieving rapid homozygosity. Such double haploid standard and haploid induction are yet to be manifested in foxtail millet. Overcoming this, an initiative for editing of SiMTL gene, which is orthologous to the maize haploid inducer gene, was prompted by Cheng et al. (2021). The study successfully achieved an average haploid induction rate of 2.8%. It was also found that this could be enhanced by developing knock-out mutants of SiMTL. Future approaches to inducing haploids and DH techniques will be more rewarding to millet breeders in developing genetic resources that could alter the phase of foxtail millet cultivation.
Hotspots in Setaria research
In recent years, Setaria has been considered an ideal model system for expanding the identification and characterization of the underlying mechanism of important agronomical traits (Figure 4). The shift in the dietary habits toward foxtail millet further focuses on improving its palatability and consumer preference for marketability. The consumer prefers a yellow-colored grain with a higher concentration of aldehydes which offer a sweet aroma. The yellow-colored grain also has a higher resistant starch content than other varieties. Molecular markers for selecting foxtail millet in rendering a preferable quality have a major thrust. Three genes control the color of seeds in foxtail millet, viz., B, I, and K. The genes B and I are mapped on chromosomes 7 and 9, while K is not mapped yet (He et al., 2015). In addition, a significant focus on reducing the amylose content to improve the cooking quality also plays a significant role in consumers’ choices. Following this, the second primary preference of the consumers is the consumption of unbroken grains in diets. Breeding for bold seeded types in foxtail millet with a maximum grain recovery during threshing is a major issue that must be resolved in the upcoming years. Setaria is highly nutritious and rich in protein and minerals, including iron and zinc. Inclusively, research on improving its folate and selenium content has been initiated to enrich its overall value (He et al., 2015). The Second International Setaria Genetics Conference, held at the Donald Danforth Plant Science Center, St. Louis, MO, United States, has highlighted the progress of research work in Setaria till March 2017 (Zhu et al., 2017). Research in the past decade has uncovered several aspects of Setaria with a major focus on the domesticated variety, S. italica. However, considering the importance and present demand of foxtail millet, extensive work is needed to elucidate the basis of genetic variation among their germplasm and exploit this information further for crop improvement. Precisely, identification of foxtail millet cultivars exhibiting tolerance to multiple stress needs to be conducted. Evaluation of these cultivars holds the potential of using this trait in climate-smart agriculture. In addition to the current reports presenting the transcriptome analysis of Setaria, proteomic and metabolomic investigation should be performed to complement the current information and finally draw a hypothesis. Further, identifying and characterizing genes involved in the biotic, abiotic, and combined stress response in Setaria is important to find the biomarker linked to stress tolerance.
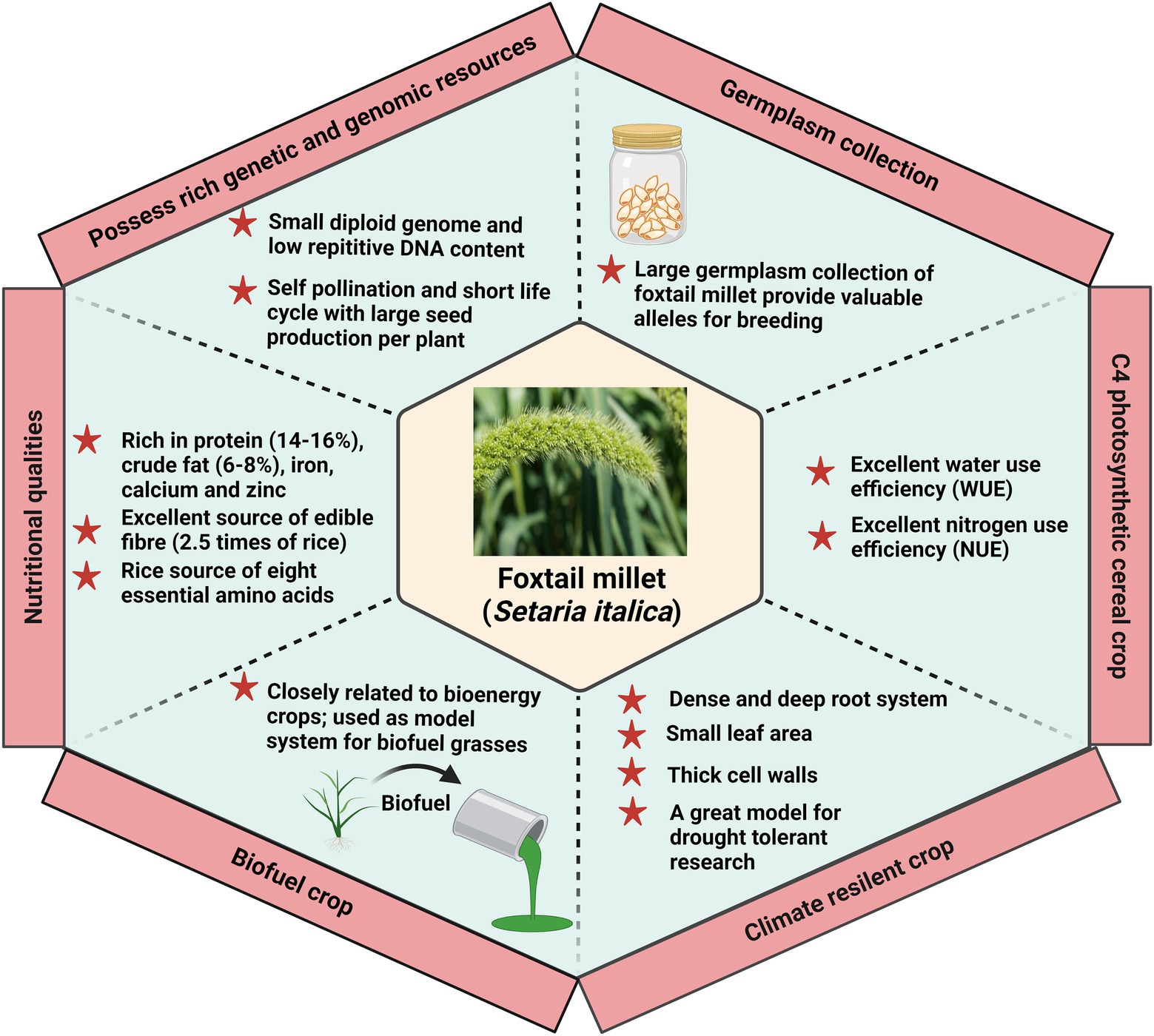
Figure 4. Foxtail millet as a model crop to translate the information in other crops. Foxtail millet is considered an ideal model system due to its physiological, genetic, and climate resilience attributes.
Exploring the correlation between photosynthetic traits and different climatic conditions with the crop yield is also necessary for nutrient management and yield enhancement. High-resistant starch content is another quality that needs to be investigated in detail to understand how this can be utilized in the regular diet for health benefits and disease prevention. In a previous study, biofortification in millets was proposed as a promising approach for nutritional security (Vinoth and Ravindhran, 2017). However, due to the antinutrients such as phytic acid, tannins, and polyphenols, the bioavailability of important nutrients becomes a limiting factor. Therefore, reducing antinutrients using RNAi or genome editing tools (CRISPER, TALENs, ZFNs, etc.) and/or enhancing the bioavailability of nutrients with the targeted expression of specific promoters can be focused as the area of research to utilize the nutritional potential of foxtail millet efficiently. A major issue with millet flour is rancidity, which persists because of high-fat content and lipase activity. An effective approach should be standardized to decrease the activity of rancidity-causing enzymes, leading to improved shelf life and long-term use of millet flour. Furthermore, our understanding of the biofuel traits of Setaria species has to be expanded to make this trait useful while engineering other crops.
Enhancing experimental design for the improvement of crops under ever-changing climatic conditions
Several studies have been conducted at different developmental stages or in response to multiple stresses and have identified candidate genes, which could be considered markers for crop improvement. Overexpression and knockdown of these genes have been performed in the native or heterologous system to comprehend their function. Methods for stable and efficient transformation of Setaria have been established for reverse genetics analysis of the genes identified from various experiments (Martins et al., 2015a,b; Saha and Blumwald, 2016; Van Eck, 2018; Rathinapriya et al., 2019; Nguyen et al., 2020; Santos et al., 2020; Sood et al., 2020). The shoot tip-based genetic transformation method has also been optimized for foxtail millet (Yang et al., 2021a). Also, the virus-mediated overexpression (VOX) vector based on Foxtail mosaic virus (genus Potexvirus) has been developed for protein expression in Setaria (Bouton et al., 2018). Earlier, foxtail mosaic virus (FoMV)-induced gene silencing (VIGS) was also established for functional genomics studies (Liu et al., 2016b). Further, successful attempts for gene editing and virus-induced flowering (VIF) have been made using the foxtail mosaic virus (Mei et al., 2019). Few studies have also optimized the reference gene for RT-PCR during developmental stages and/or stress responses in Setaria that can be used further for expression analyses (Lambret-Frotté et al., 2015; Martins et al., 2016; Nguyen et al., 2018).
Pan et al. (2014) identified and characterized a seed-specific promoter pF128 in foxtail millet. During genome-wide analysis of the NF-Y gene family and following transcriptome study identified two genes (SiNF-YA1 and SiNF-YB8) were highly responsive to salt and drought stresses. Overexpression of SiNF-YA1 imparted drought and salt tolerance in Arabidopsis plants by regulating the expression of stress-related genes (Feng et al., 2015). Foxtail millet Abscisic acid stress ripening protein, SiASR1 overexpression, leads to enhanced tolerance in tobacco by modulating the expression of oxidation-related genes, viz. NtRbohA, NtRbohB, NtCAT, NtSOD and NtAPX (Feng et al., 2016). Functional characterization of SiASR4 elucidated that ABA-responsive DRE-binding protein (SiARDP) was found to be upstream of the promoter region. Also, overexpression of SiASR4 in Arabidopsis and foxtail millet imparts drought and salt tolerance via an ABA-dependent pathway (Li et al., 2017). Similarly, SiLTP identified through Suppression subtractive hybridization (SSH) analysis was also regulated by SiARDP. Plants overexpressing SiLTP showed enhanced resistance, while RNAi plants exhibited sensitivity to drought and salt stress compared to wild-type foxtail millet plants (Pan et al., 2016). SiCDPK24 overexpression in Arabidopsis improved plant survival rate under drought stress by modulating the expression of genes, viz. RD29A, RD29B, RD22, KIN1, COR15, COR47, LEA14, CBF3/DREB1A, and DREB2A (Yu et al., 2018). Double zinc finger sequence harboring LIM domain-containing protein analysis pointed toward the presence of multiple stress-related cis-elements in the promoter of SiWLIM2b. Overexpression of SiWLIM2b in rice further advocated its possible role in multiple stress responses (Yang et al., 2019), making it an interesting candidate for future studies. In contrast, Kaur et al. (2018) showed the effect of overexpression of Arabidopsis AGG3 gene in S. viridis that positively mediate stress response and yield. During the genome-wide study of autophagy-associated genes gene family, a novel gene SiATG8a was identified whose heterologous expression led to improved drought and nitrogen starvation stress in Arabidopsis and rice (Li et al., 2016a). Expressing SiMYB3 in the heterologous system caused enhanced low-nitrogen stress tolerance by regulating root growth (Ge et al., 2019). Transgenic S. viridis plants overexpressing Brachypodium distachyon MATE gene showed increased tolerance to aluminum stress and higher root citrate exudation (Ribeiro et al., 2017). Overexpression of SiMYB3 in Arabidopsis promoted elongation of primary roots and K+ deficiency tolerance (Cao et al., 2019).
Aquaporins, SvPIP2;1 and SvNIP2;2, were expressed in Xenopus laevis oocyte to determine their water permeability. It was observed that during cell expansion in the stem, SvPIP2;1 serves as a water channel, and SvNIP2;2 mobilizes water and solutes from mature internodes (McGaughey et al., 2016). Map-based cloning identified a gene SiYGL1 linked to yellow leaf mutation that regulates a set of genes involved in photosynthesis, thylakoid development, and chloroplast signaling in S. italica (Li et al., 2016b). Characterization of Argonaute mutant, siago1b, followed by RNA-seq based comparison of wild-type and mutant plants, highlighted SiAGO1b function in energy metabolism, cell growth, programmed death, and abiotic stress responses in foxtail millet (Liu et al., 2016c). Another gene involved in the developmental process, SiTTG1, was also found to regulate salinity and high glucose stress response in foxtail millet (Liu et al., 2017a,b). Mutation in the Loose panicle 1 (LP1) gene in S. italica exhibited aberrant branch morphology, semi-dwarfism, and enlarged seed size (Xiang et al., 2017). Interestingly, brassinosteroid biosynthesis plays an essential role in the underlining alteration in inflorescence architecture (Yang et al., 2018). The transcription factor, SiNAC1, positively regulates leaf senescence in Arabidopsis in an ABA-dependent manner (Ren et al., 2018a,b). Loss of function mutation in both SiSTL1 and SiSTL2 genes leads to growth retardation, reduced chloroplast biogenesis, and leaf vein distances in an irregular manner in foxtail millet (Zhang et al., 2018b; Tang et al., 2019). Phosphate transporter SiPHT1 silencing in foxtail millet promoted several lateral roots and root hair, whereas decreased total and inorganic P accumulation in root and shoot (Ceasar et al., 2017). Further, the expression of PHT1 family genes leads to low phosphate stress tolerance in foxtail millet (Roch et al., 2020). RNAi of SvBAHD01 causes reduced feruloylation of the cell wall in the stem, thus increasing biomass digestibility in S. viridis (De Souza et al., 2018). Identification and characterization of BRASSINOSTEROID INSENSITIVE 1 (BRI1) showed the conserved role of SiBR1 in BR signaling in foxtail millet (Zhao et al., 2021b).
Conclusion
Most of the current research in Setaria focuses on genetic analysis-based QTL identification and transcriptome studies. However, proteome and metabolome analyses, key gene identification, and their characterization are still in infancy. Setaria, an annual diploid, offers scientists amenable features to explore as a model crop for tapping the hidden potential in underexplored crops. Current trends in foxtail millet also focus on its C4 mechanism. Identifying the regulatory genes beneath the C4 system establishes foxtail as a mini C4 model species. A collaborative effort with advanced breeding and phenotyping approaches for various agronomically essential traits is required to develop novel varieties of foxtail millet. There is also a crucial need to further explore the genetic resources of foxtail millet to identify several key molecular markers for performing trait genetics and association mapping. These findings would lead to marker-associated crop improvement in foxtail millet, and analogous markers can be studied in other related crops. Thus, applying advanced omics approaches will unravel the natural genetic variation in foxtail millet germplasm collection and open new opportunities for crop improvement in other crops. Also, foxtail millet could be used as a reference crop to understand the evolutionary relationship among other millet crops. The advancement of research and generated information helps study other panicoid grasses and related food and bioenergy crops.
Author contributions
PA and MM conceived and outlined the review. PA, LP, and PC wrote the manuscript. PA, LP, RS, and PS prepared tables and figures. MP and MM revised the manuscript and provided additional inputs. All authors contributed to the article and approved the submitted version.
Funding
Authors’ work in the area of millet genomics is funded by the Science and Engineering Research Board, Department of Science and Technology, Govt. of India (file no.: ECR/2017/001526), and Institute of Eminence grant (project no.: UoH-IoE-RC2-21-014) awarded to the University of Hyderabad by Ministry of Education, Govt. of India [Ref. no.: F11/9/2019-U3(A)].
Acknowledgments
The figures are created in BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.892736/full#supplementary-material
Footnotes
References
Acharya, B. R., Roy Choudhury, S., Estelle, A. B., Vijayakumar, A., Zhu, C., Hovis, L., et al. (2017). Optimization of phenotyping assays for the model monocot Setaria viridis. Front. Plant Sci. 8:2172. doi: 10.3389/fpls.2017.02172
Agtuca, B. J., Stopka, S. A., Tuleski, T. R., and Fernanda, P. (2020). In-situ metabolomic analysis of Setaria viridis roots colonized by beneficial endophytic bacteria. Mol. Plant-Microbe Interact. 33, 272–283. doi: 10.1094/MPMI-06-19-0174-R
Ahmad, Z., Nadeem, F., Wang, R., Diao, X., Han, Y., Xingchun, W., et al. (2018). A larger root system is coupled with contrasting expression patterns of phosphate and nitrate transporters in foxtail millet [Setaria italica (L.) beauv.] under phosphate limitation. Front. Plant Sci. 9:1367. doi: 10.3389/fpls.2018.01367
Aidoo, M. K., Bdolach, E., Fait, A., Lazarovitch, N., and Rachmilevitch, S. (2016). Tolerance to high soil temperature in foxtail millet (Setaria italica L.) is related to shoot and root growth and metabolism. Plant Physiol. Biochem. 106, 73–81. doi: 10.1016/j.plaphy.2016.04.038
Aihemaiti, A., Jiang, J., Gao, Y., Meng, Y., Zou, Q., Yang, M., et al. (2019). The effect of vanadium on essential element uptake of Setaria viridis’ seedlings. J. Environ. Manag. 237, 399–407. doi: 10.1016/j.jenvman.2019.02.054
Alagarasan, G., Dubey, M., Aswathy, K. S., and Chandel, G. (2017). Genome wide identification of orthologous ZIP genes associated with zinc and iron translocation in Setaria italica. Front. Plant Sci. 8:775. doi: 10.3389/fpls.2017.00775
Ali, A., Choi, Y. M., Do, Y. H., Lee, S., Oh, S., Park, H. J., et al. (2016). EST-SSR based genetic diversity and population structure among Korean landraces of foxtail millet (Setaria italica L.). Korean J. Plant Reso. 29, 322–330. doi: 10.7732/kjpr.2016.29.3.322
Andersen, E. J., and Nepal, M. P. (2017a). Genetic diversity of disease resistance genes in foxtail millet (Setaria italica L.). Plant Gene 10, 8–16. doi: 10.1016/j.plgene.2017.03.002
Andersen, E. J., and Nepal, M. P. (2017b). Data on the genome-wide identification of CNL R-genes in Setaria italica (L.) P. Beauv. Data Br. 13, 259–273. doi: 10.1016/j.dib.2017.05.035
Anittha, I., and Mullainathan, L. (2019). Molecular markers in determining phylogenetic relationship in foxtail millet (Setaria italica (L.) P. Beauv.) Var. co (Te) 7. J. Adv. Lab. Res. Biol. 10, 73–78.
Babu, A. S., Mohan, R. J., and Parimalavalli, R. (2019). Effect of single and dual-modifications on stability and structural characteristics of foxtail millet starch. Food Chem. 271, 457–465. doi: 10.1016/j.foodchem.2018.07.197
Bai, H., Cao, Y., Quan, J., Dong, L., Li, Z., Zhu, Y., et al. (2013). Identifying the genome-wide sequence variations and developing new molecular markers for genetics research by re-sequencing a landrace cultivar of foxtail millet. PLoS One 8:e73514. doi: 10.1371/journal.pone.0073514
Bai, H., Song, Z., Zhang, Y., and Li, Z. (2020). The bHLH transcription factor PPLS1 regulates the color of pulvinus and leaf sheath in foxtail millet (Setaria italica). Theor. Appl. Genet. 133, 1911–1926. doi: 10.1007/s00122-020-03566-4
Belser, C., Istace, B., Denis, E., Dubarry, M., Baurens, F. C., Falentin, C., et al. (2018). Chromosome-scale assemblies of plant genomes using nanopore long. Nat. Plants 4, 879–887. doi: 10.1038/s41477-018-0289-4
Benabdelmouna, A., Abirached-Darmency, M., and Darmency, H. (2001). Phylogenetic and genomic relationships in Setaria italica and its close relatives based on the molecular diversity and chromosomal organization of 5S and 18S-58S-25S rDNA genes. Theor. Appl. Genet. 103, 668–677. doi: 10.1007/s001220100596
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. doi: 10.1038/nbt.2196
Bhat, M. A., Shakoor, S. A., Badgal, P., and Soodan, A. S. (2018). Taxonomic demarcation of Setaria pumila (Poir.) Roem. & Schult., S. verticillata (L.) P. Beauv., and S. viridis (L.) P. Beauv. (cenchrinae, paniceae, panicoideae, poaceae) from phytolith signatures. Front. Plant Sci. 9:864. doi: 10.3389/fpls.2018.00864
Bi, S., Wang, A., Wang, Y., Xu, X., Luo, D., Shen, Q., et al. (2019). Effect of cooking on aroma profiles of Chinese foxtail millet (Setaria italica) and correlation with sensory quality. Food Chem. 289, 680–692. doi: 10.1016/j.foodchem.2019.03.108
Bonthala, V. S., Muthamilarasan, M., Misra, G., and Prasad, M. (2013). FmMDb: a versatile database of foxtail millet markers for millets and bioenergy grasses research. PLoS One 8:e71418. doi: 10.1371/journal.pone.0071418
Bonthala, V. S., Muthamilarasan, M., Roy, R., and Prasad, M. (2014). FmTFDb: a foxtail millet transcription factors database for expediting functional genomics in millets. Mol. Biol. Rep. 41, 6343–6348. doi: 10.1007/s11033-014-3574-y
Bouton, C., King, R. C., Chen, H., Azhakanandam, K., Bieri, S., Hammond-Kosack, K. E., et al. (2018). Foxtail mosaic virus: a viral vector for protein expression in cereals. Plant Physiol. 177, 1352–1367. doi: 10.1104/pp.17.01679
Brenner, D. M., Dekker, J., Niemi, J., and Pfiffner, L. (2015). Medical oxygen concentrators for releasing seed dormancy. Crop Sci. 55, 2291–2293. doi: 10.2135/cropsci2014.11.0783
Cao, X., Hu, L., Chen, X., Zhang, R., Cheng, D., Li, H., et al. (2019). Genome-wide analysis and identification of the low potassium stress responsive gene SiMYB3 in foxtail millet (Setaria italica L.). BMC Genomics 20:136. doi: 10.1186/s12864-019-5519-2
Ceasar, S. A. (2019). Genome-wide identification and in silico analysis of pht1 family genes and proteins in Setaria viridis: the best model to study nutrient transport in millets. Plant Genome 12:180019. doi: 10.3835/plantgenome2018.04.0019
Ceasar, S. A., Baker, A., and Ignacimuthu, S. (2017). Functional characterization of the PHT1 family transporters of foxtail millet with development of a novel agrobacterium-mediated transformation procedure. Sci. Rep. 7:14064. doi: 10.1038/s41598-017-14447-0
Chai, W., Si, W., Ji, W., Qin, Q., Zhao, M., and Jiang, H. (2018). Genome-wide investigation and expression profiling of HD-zip transcription factors in foxtail millet (Setaria italica L.). Biomed. Res. Int. 2018:8457614. doi: 10.1155/2018/8457614
Chaluvadi, S., and Bennetzen, J. L. (2018). Species-associated differences in the below-ground microbiomes of wild and domesticated Setaria. Front. Plant Sci. 9:1183. doi: 10.3389/fpls.2018.01183
Chander, S., Bhat, K. V., Kumari, R., Sen, S., Gaikwad, A. B., Gowda, M. V. C., et al. (2017). Analysis of spatial distribution of genetic diversity and validation of Indian foxtail millet core collection. Physiol. Mol. Biol. Plants 23, 663–673. doi: 10.1007/s12298-017-0448-5
Chen, J., Duan, W., Ren, X., Wang, C., Pan, Z., Diao, X., et al. (2017). Effect of foxtail millet protein hydrolysates on lowering blood pressure in spontaneously hypertensive rats. Eur. J. Nutr. 56, 2129–2138. doi: 10.1007/s00394-016-1252-7
Chen, Y., Liu, B., Zhao, Y., Yu, W., and Si, W. (2021). Whole-genome duplication and purifying selection contributes to the functional redundancy of auxin response factor (ARF) genes in foxtail millet (Setaria italica L.). Int. J. Genomics 2021:2590665. doi: 10.1155/2021/2590665
Cheng, Z., Sun, Y., Yang, S., Zhi, H., Yin, T., Ma, X., et al. (2021). Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J. 19, 1089–1091. doi: 10.1111/pbi.13584
Das, I. K., Krishnan, S., and Tonapi, V. (2021). Current Status and Management of foxtail millet [Setaria italica (L.) Beauv.] Blast Disease. Fungal Biology. Springer, Cham, 201–209.
De Souza, W. R., Martins, P. K., Freeman, J., Pellny, T. K., Michaelson, L. V., Sampaio, B. L., et al. (2018). Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytol. 218, 81–93. doi: 10.1111/nph.14970
de Oliveira Dal’Molin, C. G., Orellana, C., Gebbie, L., Steen, J., Hodson, M. P., Chrysanthopoulos, P., et al. (2016). Metabolic reconstruction of Setaria italica: A systems biology approach for integrating tissue-specific omics and pathway analysis of bioenergy grasses. Front. Plant Sci. 7:1138. doi: 10.3389/fpls.2016.01138
Desai, J. S., Slabaugh, E., Liebelt, D. J., Fredenberg, J. D., Gray, B. N., Jagadish, S. V. K., et al. (2018). Neural net classification combined with movement analysis to evaluate Setaria viridis as a model system for time of day of anther appearance. Front. Plant Sci. 9:5815. doi: 10.3389/fpls.2018.01585
Devos, K., Wang, Z., Beales, J., Sasaki, T., and Gale, M. D. (1998). Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theor. Appl. Genet. 96, 63–68. doi: 10.1007/s001220050709
Diao, X., and Jia, G. (2017). “Foxtail millet breeding in China,” in Genetics and Genomics of Setaria. Plant Genetics and Genomics: Crops and Models. eds. A. Doust and X. Diao (Cham: Springer).
Dong, J. L., Wang, L., Jing, L., Zhu, Y. Y., and Shen, R. L. (2019). Structural, antioxidant and adsorption properties of dietary fiber from foxtail millet (Setaria italica) bran. J. Sci. Food Agric. 99, 3886–3894. doi: 10.1002/jsfa.9611
Doust, A. N., Mauro-Herrera, M., Hodge, J. G., and Stromski, J. (2017). The C4 model grass Setaria is a short day plant with secondary long day genetic regulation. Front. Plant Sci. 8:1062. doi: 10.3389/fpls.2017.01062
Duarte, K. E., de Souza, W. R., Santiago, T. R., Sampaio, B. R., Ribeiro, P. A., Cotta, M. G., et al. (2019). Identification and characterization of core abscisic acid (ABA) signalling components and their gene expression profile in response to abiotic stresses in Setaria viridis. Sci. Rep. 9:4028. doi: 10.1038/s41598-019-40623-5
Duvick, J., Fu, A., Muppirala, U., Sabharwal, M., Mathew, D. W., Lawrence, J. C., et al. (2008). Plant GDB: a resource for comparative plant genomics. Nucleic Acids Res. 36, D959–D965. doi: 10.1093/nar/gkm1041
Dvořáková, Z., Čepková, P. H., Janovská, D., Viehmannov, I., Svobodová, E., Cusimamani, E. F., et al. (2015). Comparative analysis of genetic diversity of 8 millet genera revealed by ISSR markers. Emir. J. Food Agric. 28, 617–628. doi: 10.9755/ejfa.2015.04.077
Ellsworth, P. Z., Ellsworth, P. V., and Cousins, A. B. (2017). Relationship of leaf oxygen and carbon isotopic composition with transpiration efficiency in the C4 grasses Setaria viridis and Setaria italica. J. Exp. Bot. 68, 3513–3528. doi: 10.1093/jxb/erx185
Ellsworth, P. Z., Feldman, M. J., Baxter, I., and Cousins, A. B. (2020). A genetic link between leaf carbon isotope composition and whole-plant water use efficiency in the C4 grass Setaria. Plant J. 102, 1234–1248. doi: 10.1111/tpj.14696
Ermakova, M., Lopez-Calcagno, P. E., Raines, C. A., Furbank, R. T., and von Caemmerer, S. (2019). Overexpression of the Rieske FeS protein of the cytochrome b 6 f complex increases C4 photosynthesis in Setaria viridis. Commun. Biol. 2:314. doi: 10.1038/s42003-019-0561-9
Erying, C., Ling, Q., Yanbing, Y., and Huawen, Z. (2020). Variability of nitrogen use efficiency by foxtail millet cultivars at the seedling stage. Pesqui. Agropecu. Bras. 55:55. doi: 10.1590/s1678-3921.pab2020.v55.00832
Fahlgren, N., Feldman, M., Gehan, M. A., Wilson, M. S., Shyu, C., Bryant, D. W., et al. (2015). Versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant 8, 1520–1535. doi: 10.1016/j.molp.2015.06.005
Fang, X., Dong, K., Wang, X., Liu, T., He, J., Ren, R., et al. (2016). A high density genetic map and QTL for agronomic and yield traits in foxtail millet [Setaria italica (L.) P. Beauv]. BMC Genomics 17:336. doi: 10.1186/s12864-016-2628-z
Feldman, M. J., Ellsworth, P. Z., Fahlgren, N., Gehan, M. A., Cousins, A. B., and Baxter, I. (2018). Components of water use efficiency have unique genetic signatures in the model C4 grass Setaria. Plant Physiol. 178, 699–715. doi: 10.1104/pp.18.00146
Feng, Z. J., He, G. H., Zheng, W. J., Lu, P. P., Chen, M., Gong, Y. M., et al. (2015). Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 6:1142. doi: 10.3389/fpls.2015.01142
Feng, Z. J., Xu, Z. S., Sun, J., Li, L. C., Chen, M., Yang, G. X., et al. (2016). Investigation of the ASR family in foxtail millet and the role of ASR1 in drought/oxidative stress tolerance. Plant Cell Rep. 35, 115–128. doi: 10.1007/s00299-015-1873-y
Ferreira, S. S., Simões, M. S., Carvalho, G. G., de Lima, L. G. A., de Svartman, R. M., and Cesarino, I. (2019). The lignin toolbox of the model grass Setaria viridis. Plant Mol. Biol. 101, 235–255. doi: 10.1007/s11103-019-00897-9
Fukunaga, K., and Kato, K. (2003). Mitochondrial DNA variation in foxtail millet, Setaria italica (L) P Beauv. Euphytica 129, 7–13. doi: 10.1023/A:1021589019323
Gaur, V. S., Sood, S., Tiwari, S., and Kumar, A. (2018). Genome-wide identification and characterization of seed storage proteins (SSPs) of foxtail millet (Setaria italica (L.) P. Beauv.). 3 Biotech 8, 1–16. doi: 10.1007/s13205-018-1431-8
Ge, L., Dou, Y., Li, M., Qu, P., Zhang, H., Liu, Y., et al. (2019). SiMYB3 in foxtail millet (Setaria italica) confers tolerance to low-nitrogen stress by regulating root growth in transgenic plants. Int. J. Mol. Sci. 20:5741. doi: 10.3390/ijms20225741
Geng, Z., Liu, J., Li, D., Zhao, G., Liu, X., Dou, H., et al. (2021). A conserved miR394-targeted F-box gene positively regulates drought resistance in foxtail millet. J. Plant Biol. 64, 243–252. doi: 10.1007/s12374-021-09303-8
Ghimire, B. K., Yu, C. Y., Kim, S. H., and Chung, I. M. (2019). Assessment of diversity in the accessions of Setaria italica L based on phytochemical and morphological traits and ISSR markers. Molecules 24:1486. doi: 10.3390/molecules24081486
Gong, Z., Dong, L., Lam, S., Zhang, D., Zong, Y., Hao, X., et al. (2021). Nutritional quality in response to elevated CO2 concentration in foxtail millet (Setaria italica). J. Cereal Sci. 102:103318. doi: 10.1016/j.jcs.2021.103318
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, D. R., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gupta, S., Kumari, K., Das, J., Lata, C., Puranik, S., and Prasad, M. (2011). Development and utilization of novel intron length polymorphic markers in foxtail millet (Setaria italica (L.) P. Beauv.). Genome 54, 586–602. doi: 10.1139/g11-020
Gupta, P., Naithani, S., Tello-Ruiz, M. K., Chougule, K., Eustachio, P. D., Antonio, F., et al. (2016). Gramene database: navigating plant comparative genomics resources. Curr. Plant Biol. 7-8, 10–15. doi: 10.1016/j.cpb.2016.12.005
Gupta, P. K., and Yashvir, (1975). Induced mutations in foxtail millet (Setaria italica Beauv.). Theor. Appl. Genet. 45, 242–249. doi: 10.1007/BF00831895
Han, F., Sun, M., He, W., Cui, X., Pan, H., Wang, H., et al. (2019a). Ameliorating effects of exogenous Ca2+ on foxtail millet seedlings under salt stress. Funct. Plant Biol. 46, 407–416. doi: 10.1071/FP18314
Han, Y., Wu, M., Hao, L., and Yi, H. (2018). Sulphur dioxide derivatives alleviate cadmium toxicity by enhancing antioxidant defence and reducing Cd2+ uptake and translocation in foxtail millet seedlings. Ecotoxicol. Environ. Saf. 157, 207–215. doi: 10.1016/j.ecoenv.2018.03.084
Han, Y., Yang, H., Wu, M., and Yi, H. (2019b). Enhanced drought tolerance of foxtail millet seedlings by Sulphur dioxide fumigation. Ecotoxicol. Environ. Saf. 178, 9–16. doi: 10.1016/j.ecoenv.2019.04.006
Hao, L., Liu, J., Zhang, A., Han, Y., Guo, E., and Yi, H. (2020). Differential gene expression in foxtail millet during interaction with the smut fungus Ustilago crameri. Physiol. Mol. Plant Pathol. 110:101459. doi: 10.1016/j.pmpp.2020.101459
Haroun, S. (2002). Cytogenetic relationships between some species of Setaria L (Poaceae). Taeckholmia 22, 55–63. doi: 10.21608/taec.2002.12392
He, L., Zhang, B., Wang, X., Li, H., and Han, Y. (2015). Foxtail millet: nutritional and eating quality, and prospects for genetic improvement. Front. Agri. Sci. Eng. 2, 124–133. doi: 10.15302/J-FASE-2015054
He, Q., Zhi, H., Tang, S., Xing, L., Wang, S., Wang, H., et al. (2021). QTL mapping for foxtail millet plant height in multi-environment using an ultra-high density bin map. Theor. Appl. Genet. 134, 557–572. doi: 10.1007/s00122-020-03714-w
Heidari, H., Yosefi, M., Sasani, S., and Nosratti, I. (2019). Effect of irrigation with detergent-containing water on foxtail millet shoot biomass and ion accumulation. Environ. Sci. Pollut. Res. 26, 6328–6335. doi: 10.1007/s11356-018-3966-y
Henry, C., Watson-Lazowski, A., Oszvald, M., Griffiths, C., Paul, M. J., Furbank, R. T., et al. (2020). Sugar sensing responses to low and high light in leaves of the C4 model grass Setaria viridis. J. Exp. Bot. 71, 1039–1052. doi: 10.1093/jxb/erz495
Hirano, R., Naito, K., Fukunaga, K., Watanabe, K. N., Ohsawa, R., and Kawase, M. (2011). Genetic structure of landraces in foxtail millet (Setaria italica (L.) P. Beauv.) revealed with transposon display and interpretation to crop evolution of foxtail millet. Genome 54, 498–506. doi: 10.1139/g11-015
Hodge, J. G., and Kellogg, E. A. (2016). Abscission zone development in Setaria viridis and its domesticated relative, Setaria italica. Am. J. Bot. 103, 998–1005. doi: 10.3732/ajb.1500499
Huang, P., Jiang, H., Zhu, C., Barry, K., Jenkins, J., Sandor, L., et al. (2017). Sparse panicle1 is required for inflorescence development in Setaria viridis and maize. Nat. Plants. 3:17054. doi: 10.1038/nplants.2017.54
Huangfu, Y., Pan, J., Li, Z., Wang, Q., Mastouri, F., Li, Y., et al. (2021). Genome-wide identification of PTI1 family in Setaria italica and salinity-responsive functional analysis of SiPTI1-5. BMC Plant Biol. 21:319. doi: 10.1186/s12870-021-03077-4
Hunt, H. V., Przelomska, N., Campana, M. G., Cockram, J., Bligh, H., Kneale, C. J., et al. (2021). Population genomic structure of Eurasian and African foxtail millet landrace accessions inferred from genotyping-by-sequencing. Plant Genome 14:e20081. doi: 10.1002/tpg2.20081
Hunter, C. T., Block, A. K., Christensen, S. A., Li, Q. B., Rering, C., and Alborn, H. T. (2020). Setaria viridis as a model for translational genetic studies of jasmonic acid-related insect defenses in Zea mays. Plant Sci. 291:110329. doi: 10.1016/j.plantsci.2019.110329
Jaiswal, V., Gupta, S., Gahlaut, V., Muthamilarasan, M., Bandyopadhyay, T., Ramchiary, N., et al. (2019). Genome-wide association study of major agronomic traits in foxtail millet (Setaria italica L) using ddRAD sequencing. Sci. Rep. 9:5020. doi: 10.1038/s41598-019-41602-6
Ji, J., Liu, Y., Ge, Z., Zhang, Y., and Wang, X. (2019). Oleochemical properties for different fractions of foxtail millet bran. J. Oleo Sci. 68, 709–718. doi: 10.5650/jos.ess19063
Ji, Z., Mao, J., Chen, S., and Mao, J. (2020). Antioxidant and anti-inflammatory activity of peptides from foxtail millet (Setaria italica) prolamins in HaCaT cells and RAW264.7 murine macrophages. Food Biosci. 36:100636. doi: 10.1016/j.fbio.2020.100636
Jia, X. P., Shi, Y. S., Song, Y. C., Wang, G. Y., Wang, T. Y., and Li, Y. (2007). Development of EST-SSR in foxtail millet (Setaria italica). J. Genet. Resour. Crop Evol. 54, 233–236. doi: 10.1007/s10722-006-9139-8
Jia, G., Shi, S., Wang, C., Niu, Z., Chai, Y., Zhi, H., et al. (2013). Molecular diversity and population structure of Chinese green foxtail [Setaria viridis (L.) Beauv.] revealed by microsatellite analysis. J. Exp. Bot. 64, 3645–3656. doi: 10.1093/jxb/ert198
Jia, X., Zhang, Z., Liu, Y., Zhang, C., Shi, Y., Song, Y., et al. (2009). Development and genetic mapping of SSR markers in foxtail millet [Setaria italica (L.) P. Beauv.]. Theor. Appl. Genet. 118, 821–829. doi: 10.1007/s00122-008-0942-9
Jin, T., Wang, Y., Huang, Y., Xu, J., Zhang, P., Wang, N., et al. (2017). Taxonomic structure and functional association of foxtail millet root microbiome. Gigascience. 6, 1–12. doi: 10.1093/gigascience/gix089
Junqueira, N. E. G., Ortiz-Silva, B., Leal-Costa, M. V., Alves-Ferreira, M., Dickinson, H. G., Langdale, J. A., et al. (2018). Anatomy and ultrastructure of embryonic leaves of the C4 species Setaria viridis. Ann. Bot. 121, 1163–1172. doi: 10.1093/aob/mcx217
Karunanithi, P. S., Berrios, D. I., Wang, S., Davis, J., Shen, T., Fiehn, O., et al. (2020). The foxtail millet (Setaria italica) terpene synthase gene family. Plant J. 103, 781–800. doi: 10.1111/tpj.14771
Karvar, S. H., Vaidya, E. R., Ghorade, R. B., Gomashe, S. S., Kohakade, S. N., and Rajane, A. R. (2021). Extent of genetic variability for morpho-nutritional traits in foxtail millet [Setaria italica (L.) P. Beauv]. Pharma Innova. J. 10, 576–579.
Kaur, J., Choudhury, S. R., Vijayakumar, A., Hovis, L., Rhodes, Z., Polzin, R., et al. (2018). Arabidopsis type III Gγ protein AGG3 is a positive regulator of yield and stress responses in the model monocot Setaria viridis. Front. Plant Sci. 9:109. doi: 10.3389/fpls.2018.00109
Kellogg, E. A. (2017). Evolution of Setaria, in Genetics and Genomics of Setaria. A. Doust and Diao, X. Cham: Springer International Publishing, 3–27.
Kersey, P. J. (2019). Plant genome sequences: past, present, future. Curr. Opin. Plant Biol. 48, 1–8. doi: 10.1016/j.pbi.2018.11.001
Khan, M. F. (1997). Cytological characterisation of Setaria pallide-fusca (Schumach) Stapf and Hubb of district Rawalakot, Azad Kashmir-Pakistan. Cytologia 62, 225–229. doi: 10.1508/cytologia.62.225
Khan, Y., Yadav, A., Bonthala, V. S., Muthamilarasan, M., Yadav, C. B., and Prasad, M. (2014). Comprehensive genome-wide identification and expression profiling of foxtail millet [Setaria italica (L.)] miRNAs in response to abiotic stress and development of miRNA database. Plant Cell Tissue Organ Cult. 118, 279–292. doi: 10.1007/s11240-014-0480-x
Kim, I., Chhetri, G., Kim, J., and Seo, T. (2019). Amnibacterium Setariae Sp. nov., an endophytic actinobacterium isolated from dried foxtail. Antonie Van Leeuwenhoek 112, 1731–1738. doi: 10.1007/s10482-019-01302-7
Kour, D., Rana, K. L., Yadav, A. N., Sheikh, I., Kumar, V., Dhaliwal, H. L., et al. (2020). Amelioration of drought stress in foxtail millet (Setaria italica L.) by P-solubilizing drought-tolerant microbes with multifarious plant growth promoting attributes. Environ. Sustainability 3, 23–34. doi: 10.1007/s42398-020-00094-1
Kumar, K., Muthamilarasan, M., Bonthala, V. S., Roy, R., and Prasad, M. (2015). Unraveling 14-3-3 proteins in C4 panicoids with emphasis on model plant Setaria italica reveals phosphorylation-dependent subcellular localization of RS splicing factor. PLoS One 10, 1–22. doi: 10.1371/journal.pone.0123236
Kumar, G., and Pandey, I. K. (2021). EMS induced mutagenic effects on meiotic behaviour and photosynthetic pigment content in Setaria italica (L.) Beauv. Bull. Env. Pharmacol. Life Sci. 10, 127–132.
Kumar, A., Tomer, V., Kaur, A., Kumar, V., and Gupta, K. (2018). Millets: a solution to agrarian and nutritional challenges. Agric. Food Secur. 7:31. doi: 10.1186/s40066-018-0183-3
Kumari, R., Dikshit, N., Sharma, D., and Bhat, K. V. (2011). Analysis of molecular genetic diversity in a representative collection of foxtail millet [Setaria italica (L.) P. Beauv.] from different agro-ecological regions of India. Physiol. Mol. Biol. Plants 17, 363–374. doi: 10.1007/s12298-011-0085-3
Lambret-Frotté, J., De Almeida, L. C. S., De Moura, S. M., Souza, F. L. F., Linhares, F. S., and Alves-Ferreira, M. (2015). Validating internal control genes for the accurate normalization of qPCR expression analysis of the novel model plant Setaria viridis. PLoS One 10:e0135006. doi: 10.1371/journal.pone.0135006
Lata, C., Gupta, S., and Prasad, M. (2013). Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 33, 328–343. doi: 10.3109/07388551.2012.716809
Lata, C., Mishra, A. K., Muthamilarasan, M., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS One 9:e113092. doi: 10.1371/journal.pone.0113092
Lata, C., Sahu, P. P., and Prasad, M. (2010). Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem. Biophy. Research Comm. 393, 720–727. doi: 10.1016/j.bbrc.2010.02.068
Lawrence, C. J., and Walbot, V. (2007). Translational genomics for bioenergy production from fuel stock grasses: maize as the model species. Plant Cell 19, 2091–2094. doi: 10.1105/tpc.107.053660
Lestari, L. A., Huriyati, E., and Marsono, Y. (2017). The development of low glycemic index cookie bars from foxtail millet (Setaria italica), arrowroot (Maranta arundinacea) flour, and kidney beans (Phaseolus vulgaris). J. Food Sci. Technol. 54, 1406–1413. doi: 10.1007/s13197-017-2552-5
Li, P., and Brutnell, T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J. Exp. Bot. 62, 3031–3037. doi: 10.1093/jxb/err096
Li, W., Chen, M., Wang, E., Hu, L., Hawkesford, M. J., Zhong, L., et al. (2016a). Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics 17:797. doi: 10.1186/s12864-016-3113-4
Li, S., Dong, X., Fan, G., Yang, Q., Shi, J., Wei, W., et al. (2018). Comprehensive profiling and inheritance patterns of metabolites in foxtail millet. Front. Plant Sci. 9:1716. doi: 10.3389/fpls.2018.01716
Li, J., Dong, Y., Li, C., Pan, Y., and Yu, J. (2017). SiASR4, the target gene of SiARDP from Setaria italica, improves abiotic stress adaption in plants. Front. Plant Sci. 7:2053. doi: 10.3389/fpls.2016.02053
Li, R., Han, Y., Zhang, Q., Chang, G., Han, Y., Li, X., et al. (2020). Transcriptome profiling analysis reveals co-regulation of hormone pathways in foxtail millet during Sclerospora graminicola infection. Int. J. Mol. Sci. 21:1226. doi: 10.3390/ijms21041226
Li, P., Li, B., Seneweera, S., Zong, Y., Li, F. Y., Han, Y., et al. (2019b). Photosynthesis and yield response to elevated CO2, C4 plant foxtail millet behaves similarly to C3 species. Plant Sci. 285, 239–247. doi: 10.1016/j.plantsci.2019.05.006
Li, J., Li, X., Yang, Q., Luo, Y., Gong, X., Zhang, W., et al. (2019a). Proteomic changes in the grains of foxtail millet (Setaria italica (L.) beau) under drought stress. Spanish J. Agri. Res. 17:e0802. doi: 10.5424/sjar/2019172-14300
Li, W., Tang, S., Zhang, S., Shan, J., Tang, C., Chen, Q., et al. (2016b). Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet [Setaria italica (L.) P. Beauv]. Physiol. Plant. 157, 24–37. doi: 10.1111/ppl.12405
Li, Z. Y., Wang, N., Dong, L., Bai, H., Quan, Z. J., Liu, L., et al. (2015). Differential gene expression in foxtail millet during incompatible interaction with Uromyces setariae-italicae. PLoS One 10:e0123825. doi: 10.1371/journal.pone.0123825
Li, C., Wang, G., Li, H., Wang, G., Ma, J., Zhao, X., et al. (2021a). High-depth resequencing of 312 accessions reveals the local adaptation of foxtail millet. Theor. Appl. Genet. 134, 1303–1317. doi: 10.1007/s00122-020-03760-4
Li, S., Zhao, W., Liu, S., Li, P., Zhang, A., Zhang, J., et al. (2021b). Characterization of nutritional properties and aroma compounds in different colored kernel varieties of foxtail millet (Setaria italica). J. Cereal Sci. 100:103248. doi: 10.1016/j.jcs.2021.103248
Liang, K., Liang, S., and Zhu, H. (2020). Comparative proteomics analysis of the effect of selenium treatment on the quality of foxtail millet. LWT. 131:109691. doi: 10.1016/j.lwt.2020.109691
Lightfoot, E., Przelomska, N., Craven, M., Connell, T. C. O., He, L., Hunt, H. V., et al. (2016). Intraspecific carbon and nitrogen isotopic variability in foxtail millet (Setaria italica). Rapid Commun. Mass Spectrom. 30, 1475–1487. doi: 10.1002/rcm.7583
Lin, H. S., Chiang, C. Y., Chang, S. B., and Kuoh, C. S. (2011). Development of simple sequence repeats (SSR) markers in Setaria italica (Poaceae) and cross-amplification in related species. Int. J. Mol. Sci. 12, 7835–7845. doi: 10.3390/ijms12117835
Lin, H. S., Chiang, C. Y., Chang, S. B., Liao, G. I., and Kuoh, C. S. (2012). Genetic diversity in the foxtail millet ('Setaria italica') germplasm as determined by agronomic traits and microsatellite markers. Aust. J. Crop. Sci. 6, 342–349.
Liu, Z., Cao, C., Li, Y., Yang, G., and Pei, Y. (2019d). Light regulates hydrogen sulfide signalling during skoto- and photo-morphogenesis in foxtail millet. Funct. Plant Biol. 46, 916–924. doi: 10.1071/FP19079
Liu, D., Cui, Y., He, J., Li, S., Li, Q., Liang, D., et al. (2019a). Genetic diversity and classification of the cytoplasm of Chinese elite foxtail millet [Setaria italica (L.) P. Beauv.] parental lines revealed by chloroplast deoxyribonucleic acid variation. Front. Genet. 10:1198. doi: 10.3389/fgene.2019.01198
Liu, D., Cui, Y., Zhao, Z., Li, S., Liang, D., Wang, C., et al. (2021). Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genomics 22:682. doi: 10.1186/s12864-021-08002-5
Liu, T., He, J., Dong, K., Wang, X., Wang, W., Yang, P., et al. (2020b). QTL mapping of yield component traits on bin map generated from resequencing a RIL population of foxtail millet (Setaria italica). BMC Genomics 21:141. doi: 10.1186/s12864-020-6553-9
Liu, D., Li, J., Lu, J., Tian, B., Liu, X., Yang, G., et al. (2019b). Cloning and functional analysis of four O-Acetylserine (thiol) lyase family genes from foxtail millet. Plant Physiol. Biochem. 139, 325–332. doi: 10.1016/j.plaphy.2019.03.032
Liu, K., Qi, S., Li, D., Jin, C., Gao, C., Duan, S., et al. (2017b). TRANSPARENT TESTA GLABRA 1 ubiquitously regulates plant growth and development from Arabidopsis to foxtail millet (Setaria italica). Plant Sci. 254, 60–69. doi: 10.1016/j.plantsci.2016.10.010
Liu, H., Qin, J., Fan, H., Cheng, J., Li, L., and Liu, Z. (2017a). Genome-wide identification, phylogeny and expression analyses of SCARECROW-LIKE(SCL) genes in millet (Setaria italica). Physiol. Mol. Biol. Plants 23, 629–640. doi: 10.1007/s12298-017-0455-6
Liu, F., Shan, S., Li, H., and Li, Z. (2020a). Treatment of peroxidase derived from foxtail millet bran attenuates atherosclerosis by inhibition of CD36 and STAT3 in vitro and in vivo. J. Agric. Food Chem. 68, 1276–1285. doi: 10.1021/acs.jafc.9b06963
Liu, X., Tang, S., Jia, G., Schnable, J. C., Su, H., Tang, C., et al. (2016c). The C-terminal motif of SiAGO1b is required for the regulation of growth, development and stress responses in foxtail millet (Setaria italica (L.) P. Beauv). J. Exp. Bot. 67, 3237–3249. doi: 10.1093/jxb/erw135
Liu, N., Xie, K., Jia, Q., Zhao, J., Chen, T., Li, H., et al. (2016b). Foxtail mosaic virus-induced gene silencing in monocot plants. Plant Physiol. 171, 1801–1807. doi: 10.1104/pp.16.00010
Liu, J. M., Xu, Z. S., Lu, P. P., Li, W. W., Chen, M., Guo, C. H., et al. (2016a). Genome-wide investigation and expression analyses of the pentatricopeptide repeat protein gene family in foxtail millet. BMC Genomics 17, 1–16. doi: 10.1186/s12864-016-3184-2
Liu, T., Ye, N., Song, T., Cao, Y., Gao, B., Zhang, D., et al. (2019c). Rhizo-sheath formation and involvement in foxtail millet (Setaria italica) root growth under drought stress. J. Integr. Plant Biol. 61, 449–462. doi: 10.1111/jipb.12716
Lu, S. H., Li, B. Q., Zhai, H. L., Zhang, X., and Zhang, Z. Y. (2018a). An effective approach to quantitative analysis of ternary amino acids in foxtail millet substrate based on terahertz spectroscopy. Food Chem. 246, 220–227. doi: 10.1016/j.foodchem.2017.11.016
Lu, Y., Shan, S., Li, H., Shi, J., Zhang, X., and Li, Z. (2018b). Reversal effects of bound polyphenol from foxtail millet bran on multidrug resistance in human HCT-8/Fu colorectal cancer cell. J. Agric. Food Chem. 66, 5190–5199. doi: 10.1021/acs.jafc.8b01659
Lu, S., Zhang, X., Zhang, Z., Yang, Y., and Xiang, Y. (2016). Quantitative measurements of binary amino acids mixtures in yellow foxtail millet by terahertz time domain spectroscopy. Food Chem. 211, 494–501. doi: 10.1016/j.foodchem.2016.05.079
Luo, M., Zhang, S., Tang, C., Jia, G., Tang, S., Zhi, H., et al. (2018). Screening of mutants related to the C4 photosynthetic kranz structure in foxtail millet. Front. Plant Sci. 9:1650. doi: 10.3389/fpls.2018.01650
Mamidi, S., Healey, A., Huang, P., Grimwood, J., Jenkins, J., Barry, K., et al. (2020). A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat. Biotechnol. 38, 1203–1210. doi: 10.1038/s41587-020-0681-2
Marak, N. R., Malemnganbi, C. C., Marak, C. R., and Mishra, L. K. (2019). Functional and antioxidant properties of cookies incorporated with foxtail millet and ginger powder. J. Food Sci. Technol. 56, 5087–5096. doi: 10.1007/s13197-019-03981-6
Martin, A. P., Palmer, W. M., Brown, C., Abel, C., Lunn, J. E., Furbank, R. T., et al. (2016). A developing Setaria viridis internode: an experimental system for the study of biomass generation in a C4 model species. Biotechnol. Biofuels 9:45. doi: 10.1186/s13068-016-0457-6
Martins, P. K., Mafra, V., De Souza, W. R., Ribeiro, A. P., Vinecky, F., Basso, M. F., et al. (2016). Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 6:28348. doi: 10.1038/srep28348
Martins, P. K., Nakayama, T. J., Ribeiro, A. P., da Cunha, B. A. D. B., Nepomuceno, A. L., Harmon, F. G., et al. (2015b). Setaria viridis floral-dip: a simple and rapid agrobacterium-mediated transformation method. Biotechnol. Rep. 6, 61–63. doi: 10.1016/j.btre.2015.02.006
Martins, P. K., Ribeiro, A. P., da Cunha, B. A. D. B., Kobayashi, A. K., and Molinari, H. B. C. (2015a). A simple and highly efficient agrobacterium-mediated transformation protocol for Setaria viridis. Biotechnol. Rep. 6, 41–44. doi: 10.1016/j.btre.2015.02.002
Mauro-Herrera, M., and Doust, A. N. (2016). Development and genetic control of plant architecture and biomass in the panicoid grass, Setaria. PLoS One 11:e0151346. doi: 10.1371/journal.pone.0151346
McGaughey, S. A., Osborn, H. L., Chen, L., Pegler, J. L., Tyerman, S. D., Furbank, R. T., et al. (2016). Roles of aquaporins in Setaria viridis stem development and sugar storage. Front. Plant Sci. 7:1815. doi: 10.3389/fpls.2016.01815
Mei, Y., Beernink, B. M., Ellison, E. E., Konečná, E., Neelakandan, A. K., Voytas, D. F., et al. (2019). Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct 3:e00181. doi: 10.1002/pld3.181
Mishra, A. K., Muthamilarasan, M., Khan, Y., Parida, S. K., and Prasad, M. (2014). Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS One 9:e86852. doi: 10.1371/journal.pone.0086852
Motukuri, S. R. K., Nerella, D., Bathuru, J., Chodisetty, B., and Nallamothu, J. (2021). Genome-wide identification and in silico characterization of chitinase gene family in foxtail millet (Setaria italica). J. Appl. Biol. Biotechnol. 9, 19–30. doi: 10.7324/JABB.2021.9403
Muthamilarasan, M., Bonthala, V. S., Khandelwal, R., Jaishankar, J., Shweta, S., Nawaz, K., et al. (2015a). Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front. Plant Sci. 6:910. doi: 10.3389/fpls.2015.00910
Muthamilarasan, M., Bonthala, V. S., Mishra, A. K., Khandelwal, R., Khan, Y., Roy, R., et al. (2014a). C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct. Integr. Genomics 14, 531–543. doi: 10.1007/s10142-014-0383-2
Muthamilarasan, M., Khan, Y., Jaishankar, J., Shweta, S., Lata, C., and Prasad, M. (2015b). Integrative analysis and expression profiling of secondary cell wall genes in C4 biofuel model Setaria italica reveals targets for lignocellulose bioengineering. Front. Plant Sci. 6:965. doi: 10.3389/fpls.2015.00965
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., and Prasad, M. (2014b). Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS One 9:e109920. doi: 10.1371/journal.pone.0109920
Muthamilarasan, M., Mangu, V. R., Zandkarimi, H., Prasad, M., and Baisakh, N. (2016). Structure, organization and evolution of ADP-ribosylation factors in rice and foxtail millet, and their expression in rice. Sci. Rep. 6, 1–13. doi: 10.1038/srep24008
Muthamilarasan, M., and Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi: 10.1007/s00122-014-2399-3
Muthamilarasan, M., and Prasad, M. (2021). Small millets for enduring food security amidst pandemics. Trends Plant Sci. 26, 33–40. doi: 10.1016/j.tplants.2020.08.008
Nadeem, F., Ahmad, Z., Wang, R., Han, J., Shen, Q., Chang, F., et al. (2018). Foxtail millet [Setaria italica (L.) beauv.] grown under low nitrogen shows a smaller root system, enhanced biomass accumulation, and nitrate transporter expression. Front. Plant Sci. 9:205. doi: 10.3389/fpls.2018.00205
Nagaprabha, P., and Bhattacharya, S. (2016). Textural characterization of foxtail millet gels: effect of cations and hydrocolloids. J. Food Sci. Technol. 53, 257–268. doi: 10.1007/s13197-015-2046-2
Nagaprabha, P., Devisetti, R., and Bhattacharya, S. (2018). Physicochemical and microstructural characterisation of green gram and foxtail millet starch gels. J. Food Sci. Technol. 55, 782–791. doi: 10.1007/s13197-017-2991-z
Nakhoda, B., Mohammadi-Nejad, G., and Aminizadeh, A. (2020). Study of genetic diversity and identification of informative molecular markers related to drought tolerance in foxtail millet [Setaria italica (L.) P. Beauv.]. Agrl. Biotech. J. 11, 193–218. doi: 10.22103/JAB.2020.2530
Natesan, S., Murugesan, G., Murugan, N., Chandran, S., and Angamuthu, N. (2020). DNA fingerprinting of foxtail millet (Setaria italica L.) variety ATL 1 using SSR and RAPD markers along with morphological descriptors. Trop. Plant Res. 7, 587–593. doi: 10.22271/tpr.2020.v7.i3.072
Nguyen, D. Q., Eamens, A. L., and Grof, C. P. L. (2018). Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods 14:24. doi: 10.1186/s13007-018-0293-8
Nguyen, D. Q., Van Eck, J., Eamens, A. L., and Grof, C. P. L. (2020). Robust and reproducible agrobacterium-mediated transformation system of the C4 genetic model species Setaria viridis. Front. Plant Sci. 11:281. doi: 10.3389/fpls.2020.00281
Ni, X., Xia, Q., Zhang, H., Cheng, S., Li, H., Fan, G., et al. (2017). Updated foxtail millet genome assembly and gene mapping of nine key agronomic traits by resequencing a RIL population. GigaScience 6, 1–8. doi: 10.1093/gigascience/giw005
Ning, N., Yuan, X., Dong, S., Wen, Y., Gao, Z., Guo, M., et al. (2015). Grain yield and quality of foxtail millet (Setaria italica L.) in response to tribenuron-methyl. PLoS One 10:e0142557. doi: 10.1371/journal.pone.0142557
Ning, N., Yuan, X. Y., Dong, S. Q., Wen, Y. Y., Gao, Z. P., Guo, M. J., et al. (2016). Increasing selenium and yellow pigment concentrations in foxtail millet (Setaria italica L.) grain with foliar application of selenite. Biol. Trace Elem. Res. 170, 245–252. doi: 10.1007/s12011-015-0440-2
Niu, X., Song, L., Xiao, Y., and Ge, W. (2018). Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid and their potential in alleviating drought stress. Front. Microbiol. 8:2580. doi: 10.3389/fmicb.2017.02580
Oliveira de Araujo, T. O., Isaure, M. P., Alchoubassi, G., Bierla, K., Szpanur, J., Trcera, N., et al. (2020). Paspalum urvillei and Setaria parviflora, two grasses naturally adapted to extreme iron-rich environments. Plant Physiol. Biochem. 151, 144–156. doi: 10.1016/j.plaphy.2020.03.014
Osborn, H. L., Alonso-Cantabrana, H., Sharwood, R. E., Covsholf, S., Evans, J. R., Furbank, R. T., et al. (2017). Effects of reduced carbonic anhydrase activity on CO2 assimilation rates in Setaria viridis: a transgenic analysis. J. Exp. Bot. 68, 299–310. doi: 10.1093/jxb/erw357
Pallares, I. N., Ferrari, L., Postulka, E. B., and Ritta, M. N. (2004). Identification of cultivars of foxtail millet (Setaria italica (L) Beauv) from morphological and biochemical traits of their seed. Seed Sci. Technol. 32, 187–196. doi: 10.15258/sst.2004.32.1.19
Pan, J., Li, Z., and Dai, S. (2020). Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep. 10:13660. doi: 10.1038/s41598-020-70520-1
Pan, Y., Li, J., Jiao, L., Li, C., Zhu, D., and Yu, J. (2016). A non-specific Setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front. Plant Sci. 7:1752. doi: 10.3389/fpls.2016.01752
Pan, J., Li, Z., Wang, Q., Garrell, A. K., Liu, M., Guan, Y., et al. (2018). Comparative proteomic investigation of drought responses in foxtail millet. BMC Plant Biol. 18:315. doi: 10.1186/s12870-018-1533-9
Pan, Y., Ma, X., Liang, H., Zhao, Q., Zhu, D., and Yu, J. (2014). Spatial and temporal activity of the foxtail millet (Setaria italica) seed-specific promoter pF128. Planta 241, 57–67. doi: 10.1007/s00425-014-2164-5
Pandey, G., Misra, G., Kumari, K., Gupta, S., Parida, S. K., Chattopadhyay, D., et al. (2013). Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. 20, 197–207. doi: 10.1093/dnares/dst002
Pant, S. R., Irigoyen, S., Doust, A. N., Scholthof, K. B., and Mandadi, K. K. (2016). Setaria: a food crop and translational research model for C4 grasses. Front. Plant Sci. 7:1885. doi: 10.3389/fpls.2016.01885
Paroda, R. S., and Mal, B. (1989). “New plant Sources for Food and Industry in India,” in New Crops for Food and Industry. eds. G. E. Wickens, N. Haq, and P. Day (London: Chapman and Hall), 135–149.
Patro, T. S. S. K., Georgia, K. E., Raj Kumar, S., Anuradha, N., Rani, Y. S., Triveni, U., et al. (2020). Identification of foxtail millet varieties against banded blight disease incited by Rhizoctonia solani Kuhn. J Pharmacogn. Phytochem. 9, 1265–1267.
Peng, S. H., Wang, R., Yang, L. Z., He, L., He, X., and Liu, X. (2018). Biosorption of copper, zinc, cadmium and chromium ions from aqueous solution by natural foxtail millet shell. Ecotoxicol. Environ. Saf. 165, 61–69. doi: 10.1016/j.ecoenv.2018.08.084
Peng, R., and Zhang, B. (2021). Foxtail millet: a new model for C4 plants. Trends Plant Sci. 26, 199–201. doi: 10.1016/j.tplants.2020.12.003
Petti, C., Shearer, A., Tateno, M., Ruwaya, M., Nokes, S., Brutnell, T., et al. (2013). Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and panicoid crop species. Front. Plant Sci. 4:181. doi: 10.3389/fpls.2013.00181
Pramitha Lydia, J., Subalakshmi, S. V. K. I., Jeeva, G., and Surendhar, A. (2021). Small Millets Exploring the Hidden Potential. New Delhi, India: Scripown Publications.
Puranik, S., Sahu, P. P., Mandal, S. N., Venkata Suresh, B., Parida, S. K., and Prasad, M. (2013). Comprehensive genome-wide survey, genomic constitution and expression profiling of the nac transcription factor family in foxtail millet (Setaria italica L.). PLoS One 8:e64594. doi: 10.1371/journal.pone.0064594
Qi, Y., Wang, N., Yu, J., Wang, S., Wang, S., and Copeland, L. (2019). Insights into structure-function relationships of starch from foxtail millet cultivars grown in China. Int. J. Biol. Macromol. 155, 1176–1183. doi: 10.1016/j.ijbiomac.2019.11.085
Qi, X., Xie, S., Liu, Y., Yi, F., and Yu, J. (2013). Genome-wide annotation of genes and noncoding RNAs of foxtail millet in response to simulated drought stress by deep sequencing. Plant Mol. Biol. 83, 459–473. doi: 10.1007/s11103-013-0104-6
Qie, L., Jia, G., Zhang, W., Schnable, J., Shang, Z., Li, W., et al. (2014). Mapping of quantitative trait locus (QTLs) that contribute to germination and early seedling drought tolerance in the interspecific cross Setaria italica×Setaria viridis. PLoS One 9:e101868. doi: 10.1371/journal.pone.0101868
Rana, S., Pramitha, L., and Muthamilarasan, M. (2021). “Genomic designing for abiotic stress tolerance in foxtail millet (Setaria Italica L.),” in Genomic Designing for Abiotic Stress Resistant Cereal Crops. ed. C. Kole (Cham: Springer), 255–289.
Rathinapriya, P., Pandian, S., Rakkammal, K., Balasangeetha, M., Alexpandi, R., Satish, L., et al. (2020). The protective effects of polyamines on salinity stress tolerance in foxtail millet (Setaria italica L.), an important C4 model crop. Physiol. Mol. Biol. Plants 26, 1815–1829. doi: 10.1007/s12298-020-00869-0
Rathinapriya, P., Satish, L., Rameshkumar, R., Pandian, S., Rency, A. S., and Ramesh, M. (2019). Role of activated charcoal and amino acids in developing an efficient regeneration system for foxtail millet (Setaria italica (L.) Beauv.) using leaf base segments. Physiol. Mol. Biol. Plants 25, 533–548. doi: 10.1007/s12298-018-0619-z
Ren, X., Chen, J., Molla, M. M., Wang, C., Diao, X., and Shen, Q. (2016). In vitro starch digestibility and in vivo glycemic response of foxtail millet and its products. Food Funct. 7, 372–379. doi: 10.1039/C5FO01074H
Ren, T., Wang, J., Zhao, M., Gong, X., Wang, S., Wang, G., et al. (2018a). Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 247, 53–68. doi: 10.1007/s00425-017-2770-0
Ren, X., Yin, R., Hou, D., Zhang, M., Diao, X., Zhang, Y., et al. (2018b). The glucose-lowering effect of foxtail millet in subjects with impaired glucose tolerance: a self-controlled clinical trial. Nutrients 10:1509. doi: 10.3390/nu10101509
Ribeiro, A. P., de Souza, W. R., Martins, P. K., Vinecky, F., Duarte, K. E., Basso, M. F., et al. (2017). Overexpression of BdMATE gene improves aluminum tolerance in Setaria viridis. Front. Plant Sci. 8:865. doi: 10.3389/fpls.2017.00865
Roch, G. V., Maharajan, T., Krishna, T. P. A., Ignacimuthu, S., and Ceasar, S. A. (2020). Expression of PHT1 family transporter genes contributes for low phosphate stress tolerance in foxtail millet (Setaria italica) genotypes. Planta 252:98. doi: 10.1007/s00425-020-03503-1
Rodríguez, C. E., Mitter, B., Antonielli, L., Trognitz, F., Compant, S., and Sessitsch, A. (2018). Roots and panicles of the C4 model grasses Setaria viridis (L). and S. pumila host distinct bacterial assemblages with core taxa conserved across host genotypes and sampling sites. Front. Microbiol. 9:2708. doi: 10.3389/fmicb.2018.02708
Sachdev, N., Goomer, S., and Singh, L. R. (2021). Foxtail millet: a potential crop to meet future demand scenario for alternative sustainable protein. J. Sci. Food Agric. 101, 831–842. doi: 10.1002/jsfa.10716
Saha, P., and Blumwald, E. (2016). Spike-dip transformation of Setaria viridis. Plant J. 86, 89–101. doi: 10.1111/tpj.13148
Saha, P., Sade, N., Arzani, A., Wilhelmi, M. D. R., Coe, K. M., Li, B., et al. (2016). Effects of abiotic stress on physiological plasticity and water use of Setaria viridis (L.). Plant Sci. 251, 128–138. doi: 10.1016/j.plantsci.2016.06.011
Santos, C. M., Romeiro, D., Silva, J. P., Basso, M. F., Molinari, H. B. C., and Centeno, D. C. (2020). An improved protocol for efficient transformation and regeneration of Setaria italica. Plant Cell Rep. 39, 501–510. doi: 10.1007/s00299-019-02505-y
Sapkota, S., Conner, J. A., Hanna, W. W., Simon, B., Fengler, K., Deschamps, S., et al. (2016). In silico and fluorescence in situ hybridization mapping reveals collinearity between the Pennisetum squamulatum apomixis carrier-chromosome and chromosome 2 of sorghum and foxtail millet. PLoS One 11:e0152411. doi: 10.1371/journal.pone.0152411
Satchivi, N. M., Deboer, G. J., and Bell, J. L. (2017). Understanding the differential response of Setaria viridis L. (green foxtail) and Setaria pumila Poir. (yellow foxtail) to pyroxsulam. J. Agric. Food Chem. 65, 7328–7336. doi: 10.1021/acs.jafc.7b01453
Schontz, D., and Rether, B. (1998). Genetic variability in foxtail millet, Setaria italica (L) P Beauv-RFLP using a heterologous rDNA probe. Plant Breed. 117, 231–234. doi: 10.1111/j.1439-0523.1998.tb01931.x
Sharma, N., Alam, T., Goyal, S. K., Fatma, S., Pathania, S., and Niranajan, K. (2018a). Effect of different storage conditions on analytical and sensory quality of thermally processed, milk-based germinated foxtail millet porridge. J. Food Sci. 83, 3076–3084. doi: 10.1111/1750-3841.14371
Sharma, S., Saxena, D. C., and Riar, C. S. (2018b). Characteristics of β-glucan extracted from raw and germinated foxtail (Setaria italica) and kodo (Paspalum scrobiculatum) millets. Int. J. Biol. Macromol. 118, 141–148. doi: 10.1016/j.ijbiomac.2018.06.064
Sharma, S., Saxena, D. C., and Riar, C. S. (2018c). Changes in the GABA and polyphenols contents of foxtail millet on germination and their relationship with in vitro antioxidant activity. Food Chem. 245, 863–870. doi: 10.1016/j.foodchem.2017.11.093
Sharma, R., and Sharma, S. (2021). Anti-nutrient & bioactive profile, in vitro nutrient digestibility, techno-functionality, molecular and structural interactions of foxtail millet (Setaria italica L.) as influenced by biological processing techniques. Food Chem. 368:130815. doi: 10.1016/j.foodchem.2021.130815
Shaw, R., and Maurice Cheung, C. Y. (2019). A mass and charge balanced metabolic model of Setaria viridis revealed mechanisms of proton balancing in C4 plants. BMC Bioinformatics 20:357. doi: 10.1186/s12859-019-2941-z
Shen, H., Poovaiah, C. R., Ziebell, A., Tschaplinkski, T. J., Pattathil, S., Gjersing, E., et al. (2013). Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol. Biofuels. 6:71. doi: 10.1186/1754-6834-6-71
Shi, W., Cheng, J., Wen, X., Wang, J., Shi, G., Yao, J., et al. (2018). Transcriptomic studies reveal a key metabolic pathway contributing to a well-maintained photosynthetic system under drought stress in foxtail millet (Setaria italica L.). Peer J. 6:e4752. doi: 10.7717/peerj.4752
Simões, M. S., Carvalho, G. G., Ferreira, S. S., Hernandes-Lopes, J., de Setta, N., and Cesarino, I. (2020). Genome-wide characterization of the laccase gene family in Setaria viridis reveals members potentially involved in lignification. Planta 251, 1–18. doi: 10.1007/s00425-020-03337-x
Singh, R. K., Jaishankar, J., Muthamilarasan, M., Shweta, S., Dangi, A., and Prasad, M. (2016). Genome-wide analysis of heat shock proteins in C4 model, foxtail millet identifies potential candidates for crop improvement under abiotic stress. Sci. Rep. 6:32641. doi: 10.1038/srep32641
Singh, R. K., Shweta, S., Muthamilarasan, M., Rani, R., and Prasad, M. (2019). Study on aquaporins of Setaria italica suggests the involvement of SiPIP3;1 and SiSIP1;1 in abiotic stress response. Funct. Integr. Genomics 19, 587–596. doi: 10.1007/s10142-018-00653-0
Song, X. Q., Zhao, Y., Weng, Q. Y., Yuan, J. C., Dong, Z. P., and Zhao, Z. H. (2018). Proteomic analysis of Zhangzagu3 (Setaria italica) and its parents based on iTRAQ technique. Biotechnol. Equip. 32, 1407–1417. doi: 10.1080/13102818.2018.1528179
Sood, P., Singh, R. K., and Prasad, M. (2020). An efficient agrobacterium-mediated genetic transformation method for foxtail millet (Setaria italica L.). Plant Cell Rep. 39, 511–525. doi: 10.1007/s00299-019-02507-w
Srilatha, J., Haritha, T., Reddy, C. V. C. M., and Ramesh, D. (2020). Genetic variability studies for yield and nutritional traits in foxtail millet (Setaria italica (L.) Beauv.). J. Pharmacogn. Phytochem. 9, 596–600.
Srinivasan, A., Ekambaram, S. P., Perumal, S. S., Aruldhas, J., and Erusappan, T. (2020). Chemical characterization and immunostimulatory activity of phenolic acid bound arabinoxylans derived from foxtail and barnyard millets. J. Food Biochem. 44:e13116. doi: 10.1111/jfbc.13116
Sudhakar, C., Veeranagamallaiah, G., Nareshkumar, A., Sudhakarbabu, O., Sivakumar, M., Pandurangaiah, M., et al. (2015). Polyamine metabolism influences antioxidant defense mechanism in foxtail millet (Setaria italica L.) cultivars with different salinity tolerance. Plant Cell Rep. 34, 141–156. doi: 10.1007/s00299-014-1695-3
Sun, J., Luu, N. S., Chen, Z., Cui, X., Wu, J., Zhang, Z., et al. (2019). Generation and characterization of a foxtail millet (Setaria italica) mutant library. Front. Plant Sci. 10:369. doi: 10.3389/fpls.2019.00369
Tang, S., Li, L., Wang, Y., Chen, Q., Zhang, W., Jia, J., et al. (2017). Genotype-specific physiological and transcriptomic responses to drought stress in Setaria italica (an emerging model for Panicoideae grasses). Sci. Rep. 7:10009. doi: 10.1038/s41598-017-08854-6
Tang, S., Shahriari, M., Xiang, J., Pasternak, T., Igolkina, A., Aminizade, S., et al. (2022). The role of AUX1 during lateral root development in the domestication of the model C4 grass Setaria italica. J. Exp. Bot. 73, 2021–2034. doi: 10.1093/jxb/erab556
Tang, C., Tang, S., Zhang, S., Luo, M., Jia, G., Zhi, H., et al. (2019). SiSTL1, encoding a large subunit of ribonucleotide reductase, is crucial for plant growth, chloroplast biogenesis, and cell cycle progression in Setaria italica. J. Exp. Bot. 70, 1167–1182. doi: 10.1093/jxb/ery429
Tang, S., Zhang, H., Li, L., Liu, X., Chen, L., Chen, W., et al. (2018). Exogenous spermidine enhances the photosynthetic and antioxidant capacity of rice under heat stress during early grain-filling period. Funct. Plant Biol. 45, 911–921. doi: 10.1071/FP17149
Tian, B., Qiao, Z., Zhang, L., Li, H., and Pei, Y. (2016). Hydrogen sulphide and proline cooperate to alleviate cadmium stress in foxtail millet seedlings. Plant Physiol. Biochem. 109, 293–299. doi: 10.1016/j.plaphy.2016.10.006
Tian, B., Zhang, L., Liu, Y., Wu, P., Wang, W., Zhang, Y., et al. (2021). Identification of QTL for resistance to leaf blast in foxtail millet by genome re-sequencing analysis. Theor. Appl. Genet. 134, 743–754. doi: 10.1007/s00122-020-03730-w
Trivedi, A. K., Arya, L., Verma, S. K., Tyagi, R. K., Hemantaranjan, A., Verma, M., et al. (2018). Molecular profiling of foxtail millet (Setaria italica (L.) P. Beauv) from central himalayan region for genetic variability and nutritional quality. J. Agric. Sci. 156, 333–341. doi: 10.1017/S0021859618000382
Tsang, C. H., Li, K. T., Hsu, T. F., Tsai, Y. C., Fang, P. H., and Hsing, Y. I. C. (2017). Broomcorn and foxtail millet were cultivated in Taiwan about 5000 years ago. Bot. Stud. 58:3. doi: 10.1186/s40529-016-0158-2
Ubierna, N., Gandin, A., and Cousins, A. B. (2018). The response of mesophyll conductance to short-term variation in CO2 in the C4 plants Setaria viridis and Zea mays. J. Exp. Bot. 69, 1159–1170. doi: 10.1093/jxb/erx464
Upadhyaya, H. D., Pundir, R. P. S., Gowda, C. L. L., Reddy, V. G., and Singh, S. (2009). Establishing a core collection of foxtail millet to enhance the utilization of germplasm of an underutilized crop. Plant Genet. 7, 177–184. doi: 10.1017/S1479262108178042
Upadhyaya, H. D., Vetriventhan, M., Deshpande, S. P., Sivasubramani, S., Wallace, J. G., Buckler, E. S., et al. (2015). Population genetics and structure of a global foxtail millet germplasm collection. The Plant Genome 8, 1–13. doi: 10.3835/plantgenome2015.07.0054
Van Eck, J. (2018). The status of Setaria viridis transformation: agrobacterium-mediated to floral dip. Front. Plant Sci. 9:652. doi: 10.3389/fpls.2018.00652
Veeranagamallaiah, G., Jyothsnakumari, G., Thippeswamy, M., Reddy, P. C. O., Surabhi, G. K., Sriranganayakalu, G., et al. (2008). Proteomic analysis of salt stress responses in foxtail millet (Setaria italica L. cv. Prasad) seedlings. Plant Sci. 175, 631–641. doi: 10.1016/j.plantsci.2008.06.017
Verma, S., Srivastava, S., and Tiwari, N. (2015). Comparative study on nutritional and sensory quality of barnyard and foxtail millet food products with traditional rice products. J. Food Sci. Technol. 52, 5147–5155. doi: 10.1007/s13197-014-1617-y
Vetriventhan, M., Upadhyaya, H. D., Anandakumar, C. R., Senthilvel, S., Varshney, R. K., and Parzies, H. K. (2014). Population structure and linkage disequilibrium of ICRISAT foxtail millet (Setaria italica (L) P. Beauv.) core collection. Euphytica 196, 423–435. doi: 10.1007/s10681-013-1044-6
Vinoth, A., and Ravindhran, R. (2017). Biofortification in millets: a sustainable approach for nutritional security. Front. Plant Sci. 8:29. doi: 10.3389/fpls.2017.00029
Wang, C., Chen, J., Zhi, H., Yang, L., Li, W., Wang, Y., et al. (2010). Population genetics of foxtail millet and its wild ancestor. BMC Genet. 11:90. doi: 10.1186/1471-2156-11-90
Wang, Z. M., Devos, K. M., Liu, C. J., Wang, R. Q., and Gale, M. D. (1998). Construction of RFLP-based maps of foxtail millet, Setaria italica (L) P Beauv. Theor. Appl. Genet. 96, 31–36. doi: 10.1007/s001220050705
Wang, M., Ellsworth, P. Z., Zhou, J., Cousins, A. B., and Sankaran, S. (2016a). Evaluation of water-use efficiency in foxtail millet (Setaria italica) using visible-near infrared and thermal spectral sensing techniques. Talanta 152, 531–539. doi: 10.1016/j.talanta.2016.01.062
Wang, H., Hao, D., Wang, X., Zhang, H., Yang, P., Zhang, L., et al. (2021a). Genome-wide identification and expression analysis of the SNARE genes in foxtail millet (Setaria italica) reveals its roles in drought stress. Plant Growth Regul. 95, 355–369. doi: 10.1007/s10725-021-00746-0
Wang, C., Jia, G., Zhi, H., Niu, Z., Chai, Y., Li, W., et al. (2012). Genetic diversity and population structure of Chinese foxtail millet [Setaria italica (L.) Beauv.] landraces. Genes, Genomes, Genetics 2, 769–777. doi: 10.1534/g3.112.002907
Wang, J., Li, S., Lan, L., Xie, M., Cheng, S., Gan, X., et al. (2021c). De novo genome assembly of a foxtail millet cultivar Huagu11 uncovered the genetic difference to the cultivar Yugu1, and the genetic mechanism of imazethapyr tolerance. BMC Plant Biol. 21:271. doi: 10.1186/s12870-021-03003-8
Wang, Y., Li, L., Tang, S., Liu, J., Zhang, H., Zhi, H., et al. (2016b). Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genet. 17:57. doi: 10.1186/s12863-016-0364-7
Wang, Y., Liu, H., and Xin, Q. (2014). Genome-wide analysis and identification of cytokinin oxidase/dehydrogenase (CKX) gene family in foxtail millet (Setaria italica). J. Crop. Prod. 2, 244–254. doi: 10.1016/j.cj.2014.05.001
Wang, T., Lu, Q., Song, H., Hu, N., Wei, Y., Li, P., et al. (2021d). DNA methylation and RNA-sequencing analysis show epigenetic function during grain filling in foxtail millet (Setaria italica L.). Front. Plant Sci. 12:741415. doi: 10.3389/fpls.2021.741415
Wang, T., Song, H., Wei, Y., Li, P., Hu, N., Liu, J., et al. (2020). High throughput deep sequencing elucidates the important role of lncRNAs in foxtail millet response to herbicides. Genomics 112, 4463–4473. doi: 10.1016/j.ygeno.2020.07.045
Wang, T., Song, H., Zhang, B., Lu, Q., Liu, Z., Zhang, S., et al. (2018c). Genome-wide identification, characterization, and expression analysis of superoxide dismutase (SOD) genes in foxtail millet (Setaria italica L.). 3. Biotech 8:486. doi: 10.1007/s13205-018-1502-x
Wang, H., Tang, S., Zhi, H., Xing, L., Zhang, H., Tang, C., et al. (2021b). The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet. Crop J. 10, 342–353. doi: 10.1016/j.cj.2021.05.002
Wang, J., Wang, Z., Du, X., Yang, H., Han, F., Han, Y., et al. (2017). A high-density genetic map and QTL analysis of agronomic traits in foxtail millet [Setaria italica (L.) P. Beauv.] using RAD-seq. PLoS One 12:e0179717. doi: 10.1371/journal.pone.0179717
Wang, P., Wang, H., Wang, Y., Ren, F., and Liu, W. (2018b). Analysis of bHLH genes from foxtail millet (Setaria italica) and their potential relevance to drought stress. PLoS One 13:e0207344. doi: 10.1371/journal.pone.0207344
Wang, Z., Zhao, K., Pan, Y., Wang, J., Song, X., Ge, W., et al. (2018a). Genomic, expressional, protein-protein interactional analysis of Trihelix transcription factor genes in Setaria italica and inference of their evolutionary trajectory. BMC Genomics 19, 1–12. doi: 10.1186/s12864-018-5051-9
Wei, W., Li, S., Wang, Y., Wang, B., Fan, G., Zeng, Q., et al. (2021). Metabolome-based genome-wide association study provides genetic insights into the natural variation of foxtail millet. Front. Plant Sci. 12:665530. doi: 10.3389/fpls.2021.665530
Weng, Q., Song, X., Zhao, Y., Yuan, J., Dong, Z., Zhao, Z., et al. (2020). Proteomic profiling of foxtail millet hybrid Zhangzagu10 and its parent lines using iTRAQ-based technique. J. Plant Biochem. Biotechnol. 29, 439–449. doi: 10.1007/s13562-020-00551-2
Willweber-Kishiomoto, E. (1962). Interspecific Relationships in the Genus Setaria Contributions from the Biological Laboratory, vol. 14 Kyoto University, 1–41 Available at: http://hdlhandlenet/2433/155927
Xiang, J., Tang, S., Zhi, H., Jia, G., Wang, H., and DIao, X. (2017). Loose Panicle1 encoding a novel WRKY transcription factor, regulates panicle development, stem elongation, and seed size in foxtail millet [Setaria italica (L.) P. Beauv.]. PLoS One 12:e0178730. doi: 10.1371/journal.pone.0178730
Xiang, J., Zhang, M., Apea-Bah, F. B., and Beta, T. (2019). Hydroxycinnamic acid amide (HCAA) derivatives, flavonoid C-glycosides, phenolic acids and antioxidant properties of foxtail millet. Food Chem. 295, 214–223. doi: 10.1016/j.foodchem.2019.05.058
Xu, B., Gao, X., Dong, K., Li, X., Yang, P., Yang, T., et al. (2020). Grain protein content comparison and proteomic analysis of foxtail millet (Setaria italica L.) seed response to different drought stress levels. Acta Physiol. Plant. 42, 1–11. doi: 10.1007/s11738-019-2999-2
Xu, B., Gao, X., Gao, J., Li, J., Yang, P., and Feng, B. (2019). Transcriptome profiling using RNA-seq to provide insights into foxtail millet seedling tolerance to short-term water deficit stress induced by PEG-6000. J. Integr. Agric. 18, 2457–2471. doi: 10.1016/S2095-3119(19)62576-1
Xu, J., Li, Y., Ma, X., Ding, J., Wang, K., Wang, S., et al. (2013). Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol. Biol. 83, 77–87. doi: 10.1007/s11103-013-0025-4
Yadav, C. B., Bonthala, V. S., Muthamilarasan, M., Pandey, G., Khan, Y., and Prasad, M. (2015). Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res. 22, 79–90. doi: 10.1093/dnares/dsu039
Yadav, C., Muthamilarasan, M., Dangi, A., Shweta, S., and Prasad, M. (2016). Comprehensive analysis of SET domain gene family in foxtail millet identifies the putative role of SiSET14 in abiotic stress tolerance. Sci. Rep. 6:32621. doi: 10.1038/srep32621
Yadav, C. B., Muthamilarasan, M., Pandey, G., Khan, Y., and Prasad, M. (2014). Development of novel microRNA-based genetic markers in foxtail millet for genotyping applications in related grass species. Mol. Breed. 34, 2219–2224. doi: 10.1007/s11032-014-0137-9
Yang, Y., Chai, Y., Liu, J., Zheng, J., Zhao, Z., Amo, A., et al. (2021b). Amino acid transporter (AAT) gene family in foxtail millet (Setaria italica L.): widespread family expansion, functional differentiation, roles in quality formation and response to abiotic stresses. BMC Genomics 22:519. doi: 10.1186/s12864-021-07779-9
Yang, R., Chen, M., Sun, J. C., Yu, Y., Min, D. H., Chen, J., et al. (2019). Genome-wide analysis of LIM family genes in foxtail millet (Setaria italica L.) and characterization of the role of siwlim2b in drought tolerance. Int. J. Mol. Sci. 20:1303. doi: 10.3390/ijms20061303
Yang, L., Liu, Y., Xiang, Y., Sun, X., Yan, J., and Zhang, A. (2021a). Establishment and optimization of a shoot tip-based genetic transformation system for foxtail millet. Chinese Bull. Bot. 56, 71–79. doi: 10.11983/CBB20119
Yang, J., Thames, S., Best, N. B., Jiang, H., Huang, P., Dilkes, B. P., et al. (2018). Brassinosteroids modulate meristem fate and differentiation of unique inflorescence morphology in Setaria viridis. Plant Cell 30, 48–66. doi: 10.1105/tpc.17.00816
Yang, Z., Zhang, H., Li, X., Shen, H., Gao, J., Hou, S., et al. (2020). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants. 6, 1167–1178. doi: 10.1038/s41477-020-0747-7
Yazdizadeh, M., Mohammadi-Nejad, G., Fahmideh, L., Ebrahimi, F., Solouki, M., and Nakhoda, B. (2021). Association analysis of molecular markers with essential agronomic traits under normal and salt stress in wide germplasm of foxtail millet (Setaria italica L.). Res. Square. doi: 10.21203/rs.3.rs-739296/v1
Yi, F., Chen, J., and Yu, J. (2015). Global analysis of uncapped mRNA changes under drought stress and microRNA-dependent endonucleolytic cleavages in foxtail millet. BMC Plant Biol. 15:241. doi: 10.1186/s12870-015-0632-0
Yin, S. Y., Kuo, S. M., Chen, Y. R., Tsai, Y. C., Wu, Y. P., and Lin, Y. R. (2019). Genetic variation of physicochemical properties and digestibility of foxtail millet (Setaria italica) landraces of Taiwan. Molecules 24:4323. doi: 10.3390/molecules24234323
Yoshitsu, Y., Takakusagi, M., Abe, A., Takagi, H., Uemura, A., Yaegashi, H., et al. (2017). QTL-seq analysis identifies two genomic regions determining the heading date of foxtail millet, Setaria italica (L) P Beauv. Breed. Sci. 67, 518–527. doi: 10.1270/jsbbs.17061
You, Q., Zhang, L., Yi, X., Zhang, Z., Xu, W., and Su, Z. (2015). SIFGD: Setaria italica functional genomics database. Mol. Plant 8, 967–970. doi: 10.1016/j.molp.2015.02.001
Yu, T. F., Zhao, W. Y., Fu, J. D., Liu, Y. W., Chen, M., Zhou, Y. B., et al. (2018). Genome-wide analysis of CDPK family in foxtail millet and determination of SiCDPK24 functions in drought stress. Front. Plant Sci. 9:651. doi: 10.3389/fpls.2018.00651
Yu, A., Zhao, J., Wang, Z., Cheng, K., Zhang, P., Tian, G., et al. (2020). Transcriptome and metabolite analysis reveal the drought tolerance of foxtail millet significantly correlated with phenylpropanoids-related pathways during germination process under PEG stress. BMC Plant Biol. 20:274. doi: 10.1186/s12870-020-02483-4
Yuan, Y., Liu, C., Zhao, G., Gong, X., Dang, K., Yang, Q., et al. (2021). Transcriptome analysis reveals the mechanism associated with dynamic changes in fatty acid and phytosterol content in foxtail millet (Setaria italica) during seed development. Food Res. Int. 145:110429. doi: 10.1016/j.foodres.2021.110429
Zhang, K., Fan, G., Zhang, X., Zhao, F., Wei, W., Du, G., et al. (2017a). Identification of QTLs for 14 agronomically important traits in Setaria italica based on SNPs generated from high-throughput sequencing. G3: Genes Genomes Genetics 7, 1587–1594. doi: 10.1534/g3.117.041517
Zhang, L. Z., and Liu, R. H. (2015). Phenolic and carotenoid profiles and antiproliferative activity of foxtail millet. Food Chem. 174, 495–501. doi: 10.1016/j.foodchem.2014.09.089
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. doi: 10.1038/nbt.2195
Zhang, L., Liu, B., Zheng, G., Zhang, A., and Li, R. (2017b). Genome-wide characterization of the SiDof gene family in foxtail millet (Setaria italica). Biosystems 151, 27–33. doi: 10.1016/j.biosystems.2016.11.007
Zhang, H., Tang, S., Schnable, J. C., He, Q., Gao, Y., Luo, M., et al. (2021a). Genome-wide DNA polymorphism analysis and molecular marker development for the Setaria italica variety "SSR41" and positional cloning of the Setaria white leaf sheath gene SiWLS1. Front. Plant Sci. 12:743782. doi: 10.3389/fpls.2021.743782
Zhang, S., Tang, S., Tang, C., Luo, M., Jia, G., Zhi, H., et al. (2018b). SiSTL2 is required for cell cycle, leaf organ development, chloroplast biogenesis, and has effects on C4 photosynthesis in Setaria italica (L.) P. Beauv. Front. Plant Sci. 9:1103. doi: 10.3389/fpls.2018.01103
Zhang, S., Tang, C., Zhao, Q., Li, J., Yang, L., Qie, L., et al. (2014). Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in foxtail millet [Setaria italica (L.) P. Beauv]. BMC Genomics 15:78. doi: 10.1186/1471-2164-15-78
Zhang, H., Xiao, W., Yu, W., Yao, L., Li, L., Wei, J., et al. (2018a). Foxtail millet SiHAK1 excites extreme high-affinity K+ uptake to maintain K+ homeostasis under low K+ or salt stress. Plant Cell Rep. 37, 1533–1546. doi: 10.1007/s00299-018-2325-2
Zhang, M., Xu, Y., Xiang, J., Zheng, B., Yuan, Y., Luo, D., et al. (2021b). Comparative evaluation on phenolic profiles, antioxidant properties and α-glucosidase inhibitory effects of different milling fractions of foxtail millet. J. Cereal Sci. 99:103217. doi: 10.1016/j.jcs.2021.103217
Zhang, S., Zhi, H., Li, W., Shan, J., Tang, C., Jia, G., et al. (2018c). SiYGL2 is involved in the regulation of leaf senescence and photosystem II efficiency in Setaria italica (L.) P. Beauv. Front Plant Sci. 9:1308. doi: 10.3389/fpls.2018.01308
Zhang, W., Zhi, H., Tang, S., Zhang, H., Sui, Y., Jia, G., et al. (2021c). Identification of no pollen 1 provides a candidate gene for heterosis utilization in foxtail millet (Setaria italica L.). Crop J. 9, 1309–1319. doi: 10.1016/j.cj.2021.03.018
Zhao, Z., Liu, D., Cui, Y., Li, S., Liang, D., Sun, D., et al. (2020). Genome-wide identification and characterization of long non-coding RNAs related to grain yield in foxtail millet [Setaria italica (L.) P. Beauv.]. BMC Genomics 21:853. doi: 10.1186/s12864-020-07272-9
Zhao, Z., Tang, S., Zhang, Y., Yue, J., Xu, J., Tang, W., et al. (2021b). Evolutionary analysis and functional characterization of SiBRI1 as a Brassinosteroid receptor gene in foxtail millet. BMC Plant Biol. 21:291. doi: 10.1186/s12870-021-03081-8
Zhao, Y., Weng, Q., Song, J., Ma, H., Yuan, J., Dong, Z., et al. (2016). Bioinformatics analysis of NBS-LRR encoding resistance genes in Setaria italica. Biochem. Genet. 54, 232–248. doi: 10.1007/s10528-016-9715-3
Zhao, Q., Zhang, R., Wang, R., Gao, J., Han, Y., Yang, Z., et al. (2018). Genome-wide identification of molecular markers based on genomic re-sequencing of foxtail millet elite cultivar Jingu 21. Acta Agron. Sin. 44, 686–696. doi: 10.3724/SP.J.1006.2018.00686
Zhao, W., Zhang, L. L., Xu, Z. S., Fu, L., Pang, H. X., Ma, Y. Z., et al. (2021a). Genome-wide analysis of MADS-Box genes in foxtail millet (Setaria italica L.) and functional assessment of the role of SiMADS51 in the drought stress response. Front. Plant Sci. 12:659474. doi: 10.3389/fpls.2021.659474
Zhu, C., Ming, C., Zhao-shi, X., Lian-cheng, L., Xue-ping, C., and You-zhi, M. (2014). Characteristics and expression patterns of the aldehyde dehydrogenase (ALDH) gene superfamily of foxtail millet (Setaria italica L.). PLoS One 9:e101136. doi: 10.1371/journal.pone.0101136
Zhu, C., Yang, J., Box, M. S., Kellogg, E. A., and Eveland, A. L. (2018). A dynamic co-expression map of early inflorescence development in Setaria viridis provides a resource for gene discovery and comparative genomics. Front. Plant Sci. 9:1309. doi: 10.3389/fpls.2018.01309
Keywords: foxtail millet, climate resilience, C4 photosynthesis, nutrition, stress tolerance, integrated omics, breeding
Citation: Aggarwal PR, Pramitha L, Choudhary P, Singh RK, Shukla P, Prasad M and Muthamilarasan M (2022) Multi-omics intervention in Setaria to dissect climate-resilient traits: Progress and prospects. Front. Plant Sci. 13:892736. doi: 10.3389/fpls.2022.892736
Edited by:
Andy Pereira, University of Arkansas, United StatesReviewed by:
Babu Valliyodan, Lincoln University, United StatesPranav Pankaj Sahu, Czech Academy of Sciences, Czechia
Sha Tang, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Aggarwal, Pramitha, Choudhary, Singh, Shukla, Prasad and Muthamilarasan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehanathan Muthamilarasan, bXV0aHVAdW9oeWQuYWMuaW4=
†ORCID: Mehanathan Muthamilarasan https://orcid.org/0000-0001-7589-6372
 Pooja Rani Aggarwal1
Pooja Rani Aggarwal1 Roshan Kumar Singh
Roshan Kumar Singh Manoj Prasad
Manoj Prasad Mehanathan Muthamilarasan
Mehanathan Muthamilarasan