- 1Wheat Research Institute, School of Life Sciences and Engineering, Southwest University of Science and Technology, Mianyang, Sichuan, China
- 2Department of Plant Pathology, Washington State University, Pullman, WA, United States
- 3Wheat Health, Genetics, and Quality Research Unit, US Department of Agriculture-Agricultural Research Service (USDA-ARS), Pullman, WA, United States
- 4Crop Characteristic Resources Creation and Utilization Key Laboratory of Sichuan Province, Mianyang Institute of Agricultural Science, Mianyang, Sichuan, China
Introduction: Stripe rust is a global disease of wheat. Identification of new resistance genes is key to developing and growing resistant varieties for control of the disease. Wheat line PI 660122 has exhibited a high level of stripe rust resistance for over a decade. However, the genetics of stripe rust resistance in this line has not been studied. A set of 239 recombinant inbred lines (RILs) was developed from a cross between PI 660122 and an elite Chinese cultivar Zhengmai 9023.
Methods: The RIL population was phenotyped for stripe rust response in three field environments and genotyped with the Wheat 15K single-nucleotide polymorphism (SNP) array.
Results: A total of nine quantitative trait loci (QTLs) for stripe rust resistance were mapped to chromosomes 1B (one QTL), 2B (one QTL), 4B (two QTLs), 4D (two QTLs), 6A (one QTL), 6D (one QTL), and 7D (one QTL), of which seven QTLs were stable and designated as QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DS, QYrPI660122.swust-4DL, QYrZM9023.swust-6AS, QYrZM9023.swust-6DS, and QYrPI660122.swust-7DS. QYrPI660122.swust-4DS was a major all-stage resistance QTL explaining the highest percentage (10.67%–20.97%) of the total phenotypic variation and was mapped to a 12.15-cM interval flanked by SNP markers AX-110046962 and AX-111093894 on chromosome 4DS.
Discussion: The QTL and their linked SNP markers in this study can be used in wheat breeding to improve resistance to stripe rust. In addition, 26 lines were selected based on stripe rust resistance and agronomic traits in the field for further selection and release of new cultivars.
Introduction
Wheat stripe rust, caused by Puccinia striiformis Westend. f. sp. tritici Erikss. (Pst), is one of the most destructive diseases in the world (Chen, 2005; Milus et al., 2009). Losses from stripe rust typically range from 10% to 70% in commercial production environments, depending on the cultivar, prevailing climatic conditions, and inoculum pressure (Bariana et al., 2016; Zhou et al., 2022a). However, the disease can cause a 100% loss of yield for susceptible varieties (Bariana et al., 2016). Since 1950, the disease has occurred on an annual average of 4 million hectares in China. In particular, the five major outbreaks of wheat stripe rust in 1950, 1964, 1990, 2002, and 2017 all occurred on over 5.5 million hectares, resulting in a loss of 13.8 million tons of yield (Li and Zeng, 2000; Wan et al., 2004; Huang et al., 2018). Stripe rust can be controlled by resistant cultivars, fungicides, and some cultural practices. Compared to other approaches, planting resistant cultivars has been proven to be the most effective, easy-to-use, economical, and environmentally friendly way to control disease (Line, 2002; Chen, 2005).
Depending on phenotypic performance at different growth stages, wheat rust resistance can be classified into two types: all-stage resistance (ASR) and adult plant resistance (APR), sometimes also known as high-temperature adult plant (HTAP) resistance (Qayoum and Line, 1985; Chen, 2005; Lin and Chen, 2007; Rosewarne et al., 2013). All-stage resistance, also known as seedling resistance, can be detected at the seedling stage but is expressed at all growth stages. Such resistance is often race-specific and, thus, easily overcome by virulent races (Line, 2002; Chen, 2005; Chen, 2013). Due to race specificity, ASR often fails within 3–5 years of deployment (Jambuthenne et al., 2022). In contrast, HTAP resistance becomes more effective as plants grow older and the weather becomes warmer. It is usually non-race-specific, quantitatively inherited, and more likely to be durable. However, HTAP resistance is mostly incomplete, and the level is influenced by plant growth stage, temperature, and disease pressure (Chen, 2013).
To date, 84 permanently named and a large number of temporarily designated stripe rust resistance genes (Yr genes) and quantitative trait loci (QTLs) have been reported in wheat (Klymiuk et al., 2022). These resistance genes come mainly from common wheat cultivars, local germplasm, and wild relatives. Among them, Yr5/Yr7/YrSP, Yr10, Yr15, Yr18, Yr36, Yr46, YrU1, and YrAS2388 were cloned and characterized (Fu et al., 2009; Krattinger et al., 2009; Liu et al., 2014; Moore et al., 2015; Klymiuk et al., 2018; Marchal et al., 2018; Zhang et al., 2019; Wang et al., 2020). Most ASR genes have been overcome by virulent races. ASR genes Yr5 and Yr15 are still effective against all Pst races identified in the United States and many other countries (Wang and Chen, 2017). However, races virulent on Yr5 gene have been reported in Australia, India, China, and Turkey (Wellings and McIntosh, 1990; Zhang et al., 2020; Tekin et al., 2021), and the Yr15 virulence has been documented in Afghanistan (Gerechter-Amitai et al., 1989). Most wheat cultivars that have shown durable resistance to stripe rust have APR or HTAP resistance controlled by variable numbers of genes or QTL (Lin and Chen, 2007; Lin and Chen, 2008b; Santra et al., 2008; Carter et al., 2009; Paillard et al., 2012; Ren et al., 2012a; Lu et al., 2014; Zhou et al., 2014; Dong et al., 2017; Feng et al., 2018; Liu et al., 2018; Liu et al., 2019; Liu et al., 2020). In order to obtain a high degree of durable resistance, combining the two types of resistance types in the same background is considered a preferred method to improve the resistance to stripe rust in wheat breeding (Chen, 2007; Ren et al., 2012a; Chen, 2013; Liu et al., 2018).
The development of molecular markers, especially single-nucleotide polymorphism (SNP) markers, has revolutionized QTL analysis. A SNP is caused by a single-nucleotide mutation due to the insertion, deletion, and replacement of a single base segment in the genome. SNPs exist in the entire genomes of biological individuals and are the most abundant. SNP markers are now widely used in genetic analysis and breeding (Ma et al., 2019). Recent advances in sequencing technology have led to the availability of many SNP arrays in wheat (Rasheed et al., 2017). High-throughput genotyping techniques, including Wheat 9K (Cavanagh et al., 2013), 15K (Soleimani et al., 2020), 90K (Wang et al., 2014; Wu et al., 2018), 660K (Cui et al., 2017), and 820K SNP (Winfield et al., 2016) arrays, are now available. Among these SNP arrays, the 15K array is generally adequate and cost-effective for mapping traits of interest (Soleimani et al., 2020).
PI 660122, a spring wheat germplasm, was developed by the Wheat Health, Genetics, and Quality Research Unit of the US Department of Agriculture, Agricultural Research Service (USDA-ARS), and Washington State University and deposited in the USDA-ARS National Small Grains Collections (NSGC). In previous studies, the germplasm showed a high level of resistance in field tests over multiple years (Wang et al., 2012; Zhou et al., 2015b). At the seedling stage, it was resistant to US races PST-43 and PST-127 and Chinese races CYR29, CYR31, CYR32, and CYR33 and moderately resistant to US races PST-100 and PST-114 and Chinese race PST-HY8 of Pst (Wang et al., 2012; Zhou et al., 2015b). A comparison of greenhouse and field tests indicated that PI 660122 had effective ASR and possible HTAP resistance. The objectives of the present study were to further characterize the stripe rust resistance in PI 660122, map QTL for ASR and APR, and identify the QTL by comparing their chromosomal locations with previously reported stripe rust resistance QTL.
Materials and methods
Plant materials
To map the QTL for stripe rust resistance in PI 660122, we developed a mapping population from a cross between Zhengmai (ZM9023, as the female parent) and PI 660122 (as the male parent). PI 660122 was developed from cross Avocet S/PI 610755 (Wang et al., 2012). Avocet S (AvS), an Australian spring wheat selection, is highly susceptible to most Pst races in China and many other countries and has been used as a susceptible control in stripe rust tests. PI 610755 is a Mexico spring wheat variety, selected from the cross Altar 84/Aegilops tauschii (191)//Opata M85. ZM9023, a spring wheat cultivar developed by the Wheat Research Institute of Henan Academy of Agricultural Sciences, is moderately or highly susceptible to the currently predominant Pst races in China (Xue et al., 2014). We developed a total of 239 F5 and F6 recombinant inbred lines (RILs) from the ZM9023 × PI 660122 cross, using the single-seed descent method.
Greenhouse tests
Seedling tests were conducted in a greenhouse to evaluate the stripe rust responses of PI 660122 and Zhengmai 9023. For each genotype, 10–12 seeds were seeded in a 9 cm × 9 cm × 9 cm plot. At the one-leaf stage, seedlings were uniformly inoculated with fresh urediniospores of a Pst race mixed with talc at a ratio of 1:50. Three Chinese Pst races, CYR31, CYR32, and CYR34, were used in the seedling tests. Inoculated seedlings were kept in a dew chamber in the dark at 8°C and above 100% relative humidity for 24 h. The seedlings were then moved to a growth chamber at 16°C with a daily 16-h light for stripe rust development. The infection type (IT) data were recorded 18 days to 21 days after inoculation using the 0–9 scale (Line and Qayoum, 1992). Seedlings of AvS were included as the susceptibility check in each race test. Later, 15 RILs selected for each containing only one QTL were also tested together with the parents with the three races at the seedling stage in the greenhouse under the same conditions.
Field tests
The F5 and F6 RILs and their parents were tested for stripe rust responses to stripe rust in the experimental fields in Mianyang (MY; 31°33′N, 104°55′E) in 2021 (21) and both MY and Guangyuan (GY; 22:32°14′N, 106°17′E) in the Sichuan Province in 2022 (22). The field tests were conducted with one replicate at 21MY and 22GY and two replicates (completely randomized block design) at 22MY based on the available seed quantity. Each plot consisted of a single row, 1.0 m in length and with 25 cm between rows. Approximately 20 to 30 seeds were sown in each row. AvS was planted in a row every 20 rows as a susceptible check and spore spreader for increasing stripe rust pressure and uniformity in the nursery. To increase the Pst inoculum, AvS was also planted around the nursery. MY and GY are ideal regions for stripe rust, as Pst can over-winter and over-summer, and the nursery was naturally infected without artificial inoculation (Zhou et al., 2019).
The stripe rust IT of each parent or RIL was rated on a scale of 0–9 (Line and Qayoum, 1992). Disease severity (DS) was scored using a modified scale as previously described (Lin and Chen, 2007). Both IT and DS data were collected twice in each season. The first record was taken when susceptible AvS showed approximately 80% severity, and the second was approximately a week later (Nsabiyera et al., 2018). Agronomic traits such as plant height (PH), spike length (SL), productive tiller number (PTN), kernels per spike (KPS), and thousand-grain weight (TGW) were determined to select RILs. PH was measured from the ground to the top of the spike excluding awn after the milking stage; KPS, SL, and PTN of each plant were counted at maturity; TGW was measured after harvest.
DNA extraction and genotyping
Fresh young leaves of PI 660122, ZM9023, and 239 F5 RILs were harvested from the experimental field in January 2021. DNA from the fresh leaves was extracted using a modified cetyltrimethyl ammonium bromide (CTAB) method (Li et al., 2013). DNA was dissolved in ddH2O (100 μL), and DNA quality and concentration were determined by spectrophotometry (NanoDrop ND-1000, Thermo Scientific, Wilmington, DE, USA) after the DNA. DNA stock solutions were diluted with sterilized ddH2O to different concentrations according to individual experimental requirements for molecular analyses.
The parents and the 239 RILs were genotyped by China Golden Marker (Beijing) Biotech Co., Ltd. (http://www.cgmb.com.cn/) using the 15K SNP chip (Soleimani et al., 2020).
Statistical analysis, genetic map construction, and QTL mapping
Analysis of variance (ANOVA) and analysis of Pearson’s correlation coefficients were performed to analyze the stripe rust phenotypic data using the “AOV” tool in the QTL Ici Mapping V4.2 software (Wang, 2009; Meng et al., 2015). The same software was also used to analyze the genotypic data. After the genotypic data were scanned for missing and undetected data, redundant markers were deleted using the “Bin” function. Genetic maps were constructed using the Kosambi mapping function (Kosambi, 2016). QTL mapping was performed using the genetic maps and the IT and DS data based on inclusive composite interval mapping (ICIM) with preset parameters Step = 1 cM, value p for input variables (PIN) = 0.0001, and logarithm of odds (LOD) = 2.5. A QTL was identified when the logarithm of odds (LOD) score was greater than 2.5. To determine the additive effects of QTL, the effects of QTL combinations were demonstrated by plotting box plots for mean IT and mean DS of RILs sharing the same number of beneficial alleles.
Results
Stripe rust responses of the parents and RILs
In the greenhouse seedling tests, PI 660122 was highly resistant (IT of 2) to the tested three Chinese Pst races, whereas Zhengmai 9023 was highly susceptible (IT of 8–9) similar to the susceptible check AvS (Figure 1A). In the field tests under natural Pst infection, the final adult plant IT of PI 660122 was 2 across the two years and two locations, and its DS ranged from 5% to 10%, (Figure 1B). In contrast, Zhengmai 9023 was moderately resistant (IT of 5–6) with DS of 40%–50%. For comparison, AvS had IT of 9 and DS of 100%.
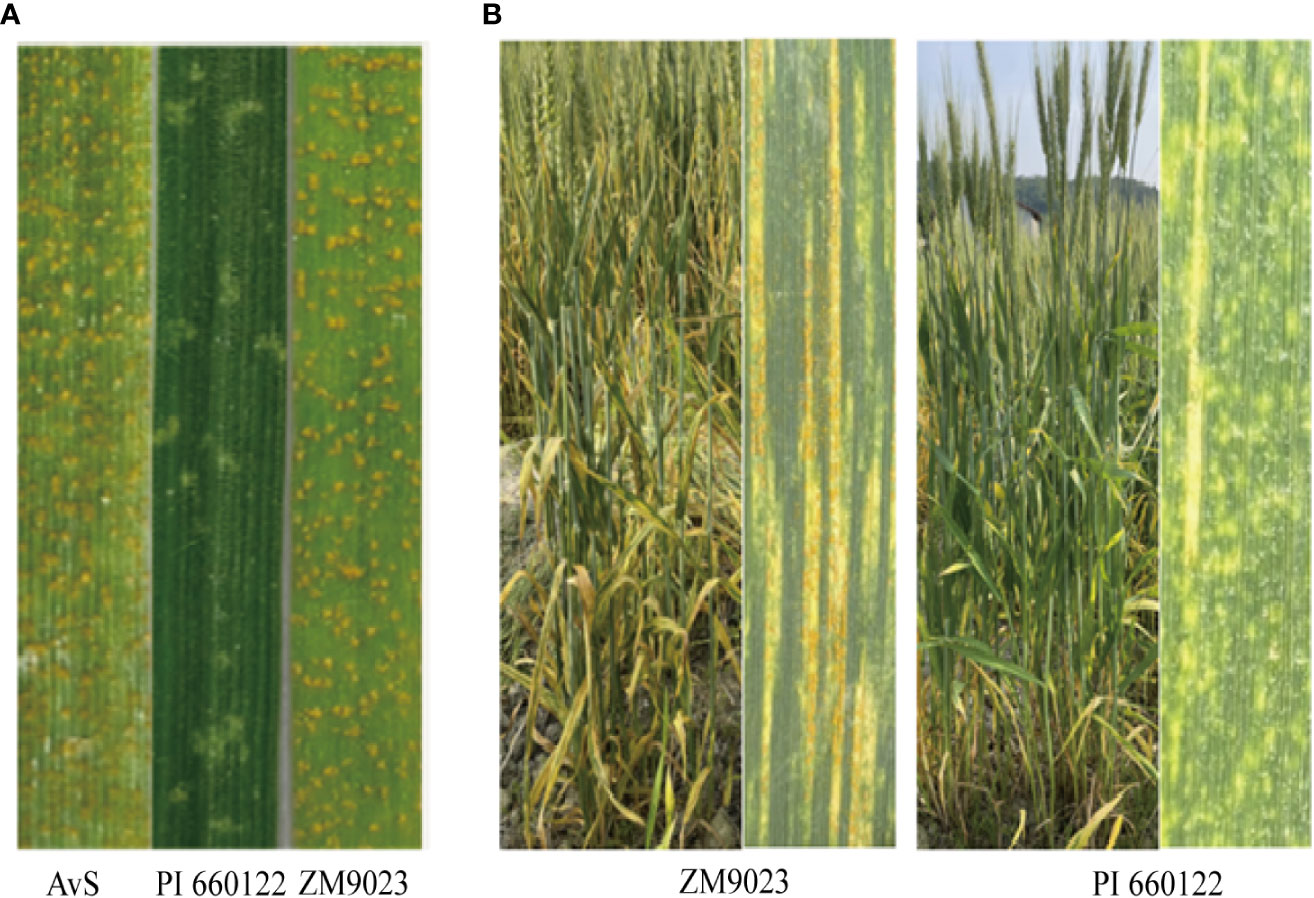
Figure 1 Stripe rust response of resistant parent PI 660122, susceptible parent Zhengmai 9023 (ZM9023), and susceptible check AvS with Chinese race CYR34 of Puccinia striiformis f sp. tritici at the seedling stage (A) and stripe rust reactions on flag leaves of ZM9023 and PI 660122 (B).
The RIL population had ITs ranging from 0 to 9 and DS from 0 to 90% across the years and locations (Figure 2). The IT and DS data from both sites and from both 2021 and 2022 at MY were each highly correlated (r = 0.76–0.81, p < 0.001 for IT; r = 0.61–0.75, p < 0.001 for DS) (Table 1). The ANOVA results showed significant variations (p < 0.001) among RILs, environments, and line × environment interactions for both IT and DS. The stripe rust phenotypes were influenced more by the environment than by the interaction of line and environment. The broad-sense heritability (h2) was estimated at 0.92 using the IT data and 0.86 based on the DS data across the two sites (Table 2).
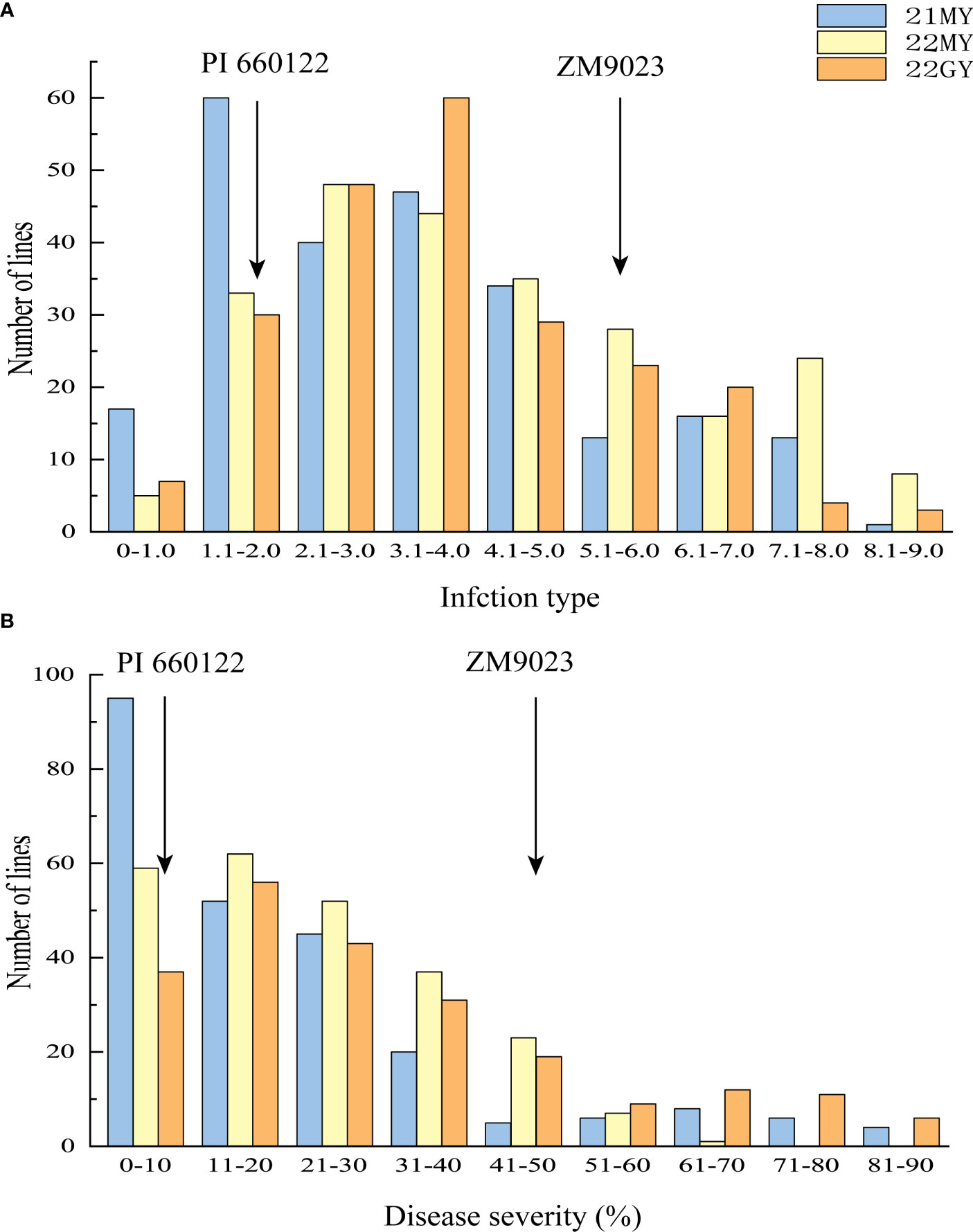
Figure 2 Frequency distributions of mean infection types (IT) (A) and disease severity (DS) (B) for 239 F6 RILs from cross Zhengmai 9023 × PI 660122 tested in Mianyang (MY) in 2021 (21) and 2022 (22) and Guangyuan (GY) in 2022. Arrows indicate the values of the parent lines. RILs, recombinant inbred lines.

Table 1 Correlation coefficients (r) of infection type (IT) and disease severity (DS) of the recombinant inbred lines Zhengmai 9023 × PI 660122 tested in different environments.
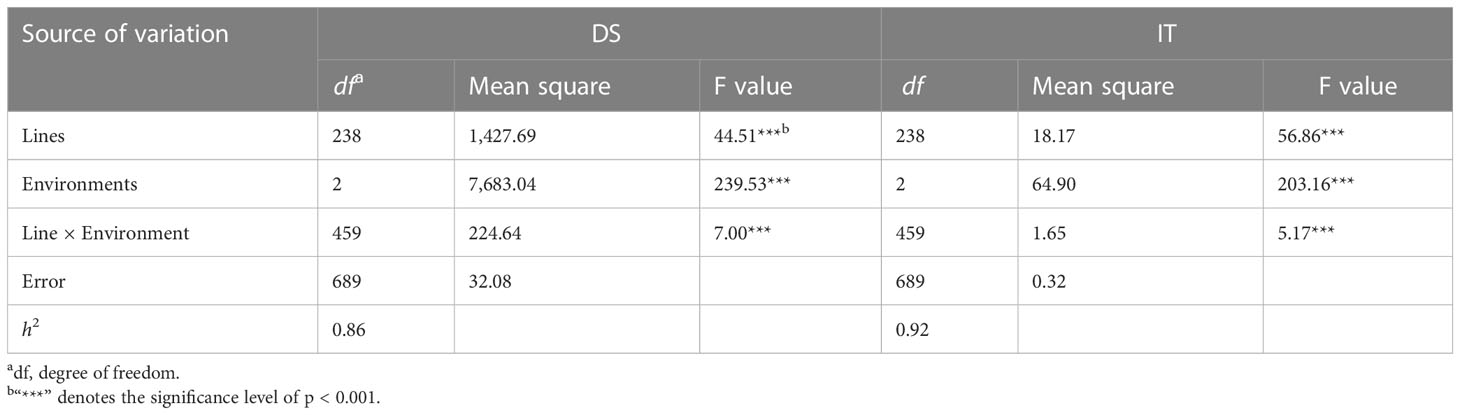
Table 2 Analysis of variance and estimate of broad-sense heritability (h2) of infection type (IT) and disease severity (DS) in the recombinant inbred line (RIL) population of Zhengmai 9023 × PI 660122 tested at Mianyang in 2021 and 2022 and Guangyuan in 2022.
Genetic linkage map construction
A total of 5,432 SNPs in the 15K SNP array showed homozygous polymorphisms between the two parents. After the redundant markers were filtered out, 4,102 SNPs with known chromosome locations were obtained and used as inputs in the linkage analysis using QTL Ici Mapping V4.2. The 4,102 SNPs covered a total map length of 7,937.6 cM, with the genetic length ranging from 135.6 cM for chromosome 1B to 635.5 cM for chromosome 5A (Table 3). The number of markers per chromosome ranged from 60 for chromosome 6A to 322 for chromosome 2A, with an average of 189 SNPs. The mean distance between adjacent SNP markers ranged from 0.5 cM for chromosome 1B to 7.3 cM for chromosome 2D, with an overall mean of 1.9 cM. Genomes A, B, and D included 1,374 (33.50%), 1,672 (40.76%), and 1,056 (25.74%) SNPs covering lengths of 2,630.1 cM, 2,144.1 cM, and 3,163.4 cM with mean marker distance of 1.91 cM, 1.28 cM, and 3.00 cM, respectively. The map was used to identify significant associations between SNPs and stripe rust resistance.
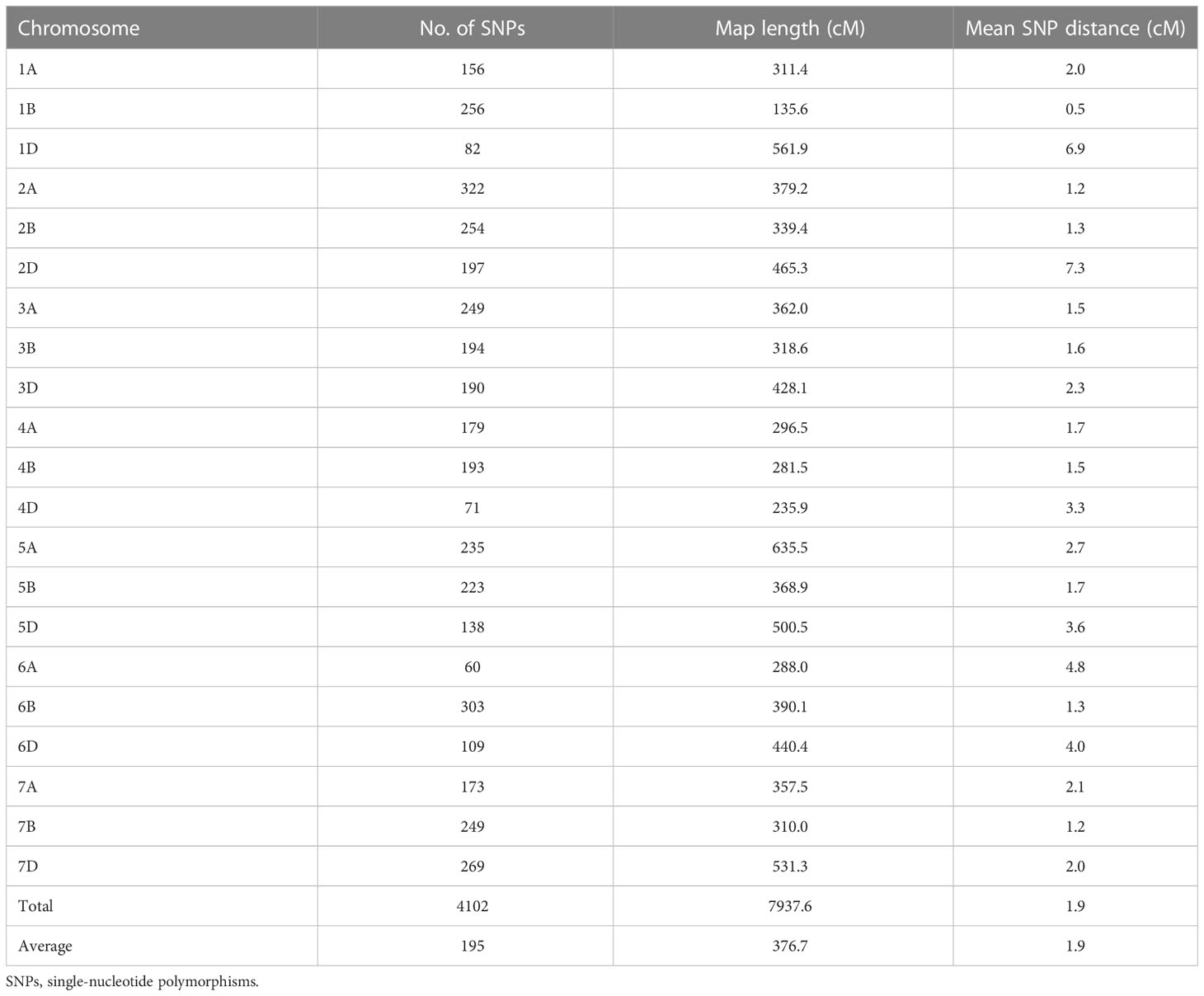
Table 3 Summary of chromosome assignment, number of SNPs, map length, and marker density of the genetic maps of the Zhengmai 9023 × PI 660122 recombinant inbred population.
QTL analysis of stripe rust resistance
QTL scans on all 21 chromosomes were performed using the ICIM method in the software QTL Ici Mapping V4.2. A total of nine QTLs contributing to stripe rust resistance in the Zhengmai 9023 × PI 660122 RIL population were identified with one QTL each on chromosomes 1B, 2B, 6A, 6D, and 7D and two QTLs each on 4B and 4D. Then, ICIM, single-marker analysis (SMA), and ICIM epistatic QTL (ICIM-EPI) for epistatic mapping were performed on QTL chromosome regions of these chromosomes. Among the QTL, seven (QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DS, QYrPI660122.swust-4DL, QYrZM9023.swust-6AS, QYrZM9023.swust-6DS, and QYrPI660122.swust-7DS) were detected in all environments, and two (QYrZM9023.swust-1BL and QYrPI660122.swust-2BL) were only detected in 22MY. Of the nine QTLs, six (QYrPI660122.swust-2BL, QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DS, QYrPI660122.swust-4DL, and QYrPI660122.swust-7DS) were from PI 660122 and three (QYrZM9023.swust-1BL, QYrZM9023.swust-6AS, and QYrZM9023.swust-6DS) from Zhengmai 9023.
QYrZM9023.swust-1BL, located at an 8.27-cM interval spanned by SNP markers AX-89763895 and AX-109273019, explained 7.41% and 7.29% of phenotypic variation in IT and DS, respectively, and was detected only in 22MY. QYrPI660122.swust-2BL, located at a 1.83-cM interval spanned by SNP markers AX-109849173 and AX-109349804, explained 4.65% and 5.57% of phenotypic variation in IT and DS, respectively, and was only detected in 22MY. QYrPI660122.swust-4BS, located at a 0.96-cM interval spanned by SNP markers AX-108767762 and AX-109309162, explained 10.94%–15.00% and 5.84%–12.93% of phenotypic variation in IT and DS, respectively, across all environments. QYrPI660122.swust-4BL, mapped to a 1.54-cM interval flanked by SNP markers AX-108935256 and AX-108984536, explained 8.25%–9.09% and 5.75%–13.51% phenotypic variation in IT and DS, respectively, and was detected in all environments. QYrPI660122.swust-4DS, located at a 12.15-cM interval spanned by SNP markers AX-110046962 and AX-111093894, explained 11.64%–17.20% and 13.22%–20.97% phenotypic variation in IT and DS, respectively, across all environments. QYrPI660122.swust-4DL, located at a 1.23-cM interval spanned by SNP markers AX-94560848 and AX-111557122, explained 11.18%–18.24% and 6.60%–17.37% phenotypic variation in IT and DS, respectively, across all environments. QYrZM9023.swust-6AS, located at a 10.37-cM interval spanned by SNP markers AX-95124889 and AX-110995858, explained 5.43%–9.11% and 6.23%–7.92% of phenotypic variation in IT and DS, respectively, across all environments. QYrZM9023.swust-6DS, located at a 2.58-cM interval spanned by SNP markers AX-11475193 and AX-109317417, explained 7.24%–13.33% and 7.25%–12.22% of phenotypic variation in IT and DS, respectively, across all environments. QYrPI660122.swust-7DS, located at a 4.75-cM interval spanned by SNP markers AX-110467729 and AX-89378255, explained 11.64%–17.20% and 13.22%–20.97% of phenotypic variation in IT and DS, respectively, and was detected in two environments, 21MY and 22GY (Figure 3, Table 4).
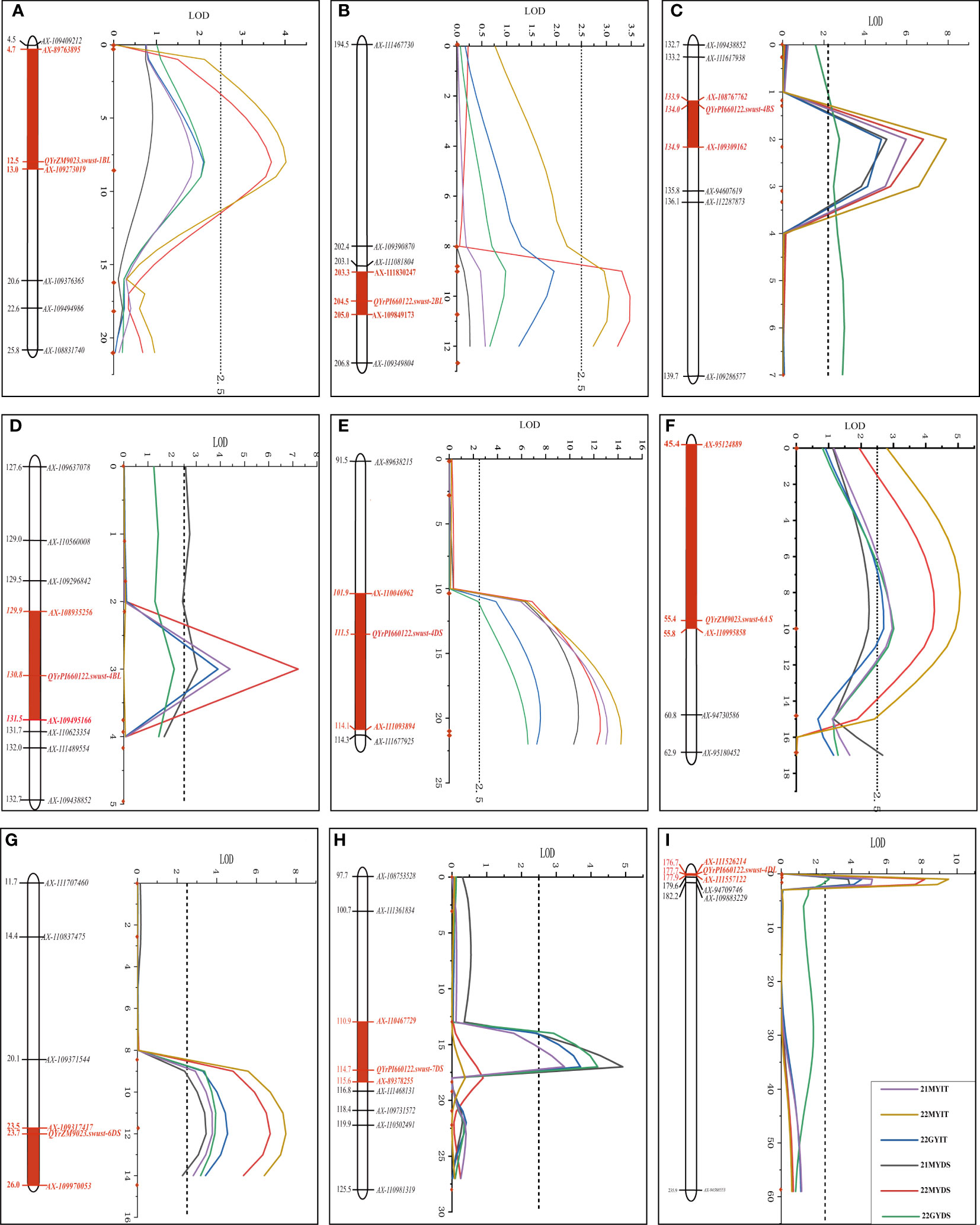
Figure 3 Stripe rust resistance QTLs on the genetic map of chromosomes 1BL (A), 2BL (B), 4BS (C), 4BL (D), 4DS (E), 6AS (F), 6DS (G), 7DS (H), and 4DS (I) based on infection type (IT) and disease severity (DS) data. The y-axis is in centimorgan (cM) distance, and the x-axis denotes LOD value. The red rectangle on the genetic map indicates the corresponding QTL region. QTLs, quantitative trait loci; LOD, limit of detection.
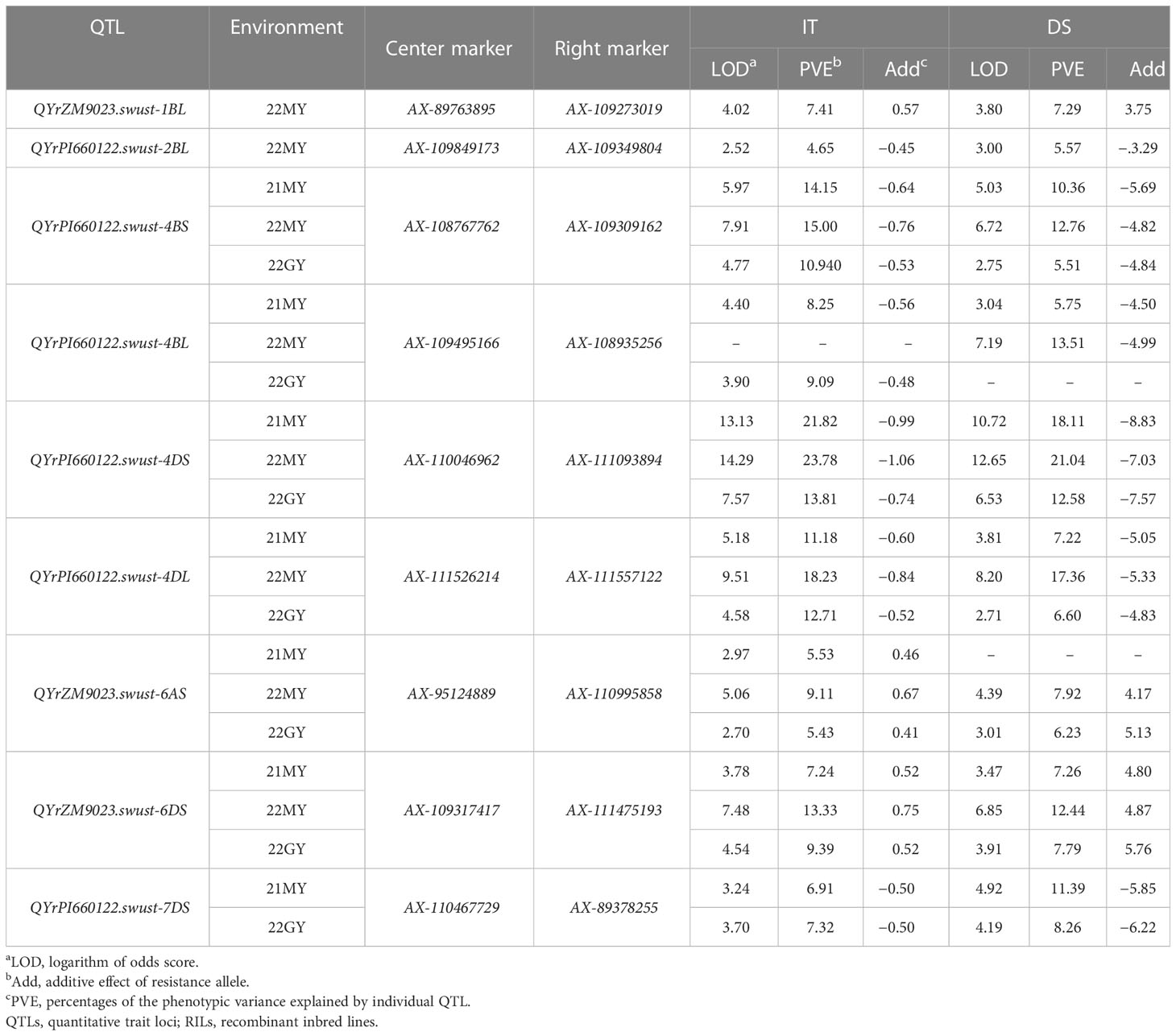
Table 4 Summary of nine stripe rust resistance QTLs identified based on mean disease severity (DS) and infection type (IT) of 239 RILs from Zhengmai 9023 × PI 660122 cross-tested in Mianyang 2021–2022 and Guangyuan 2022.
Identification of QTL resistance
Fifteen lines containing only one QTL were selected and tested for seedling reaction in the greenhouse using three Chinese Pst races (CYR31, CYR32, and CYR34). Among them, four lines contained QYrZM9023.swust-1BL, three lines contained QyrPI660122.swust-4DS, and eight lines contained QYrPI660122.swust-7DS. The lines containing QYrZM9023.swust-1BL or QYrPI660122.swust-7DS were susceptible (IT of 7–9) to all three races but showed moderate resistance at the adult-plant stage in the fields, indicating that these QTLs confer APR. In contrast, the lines containing QYrPI660122.swust-4DS were resistant (IT of 1–3) in the seedling tests, indicating that this QTL confers ASR (Figure 4, Table 5). The resistance types of QYrPI660122.swust-2BL, QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DL, QYrZM9023.swust-6AS, and QYrZM9023.swust-6DS were uncertain, as there were no single-QTL lines available from the RIL population.
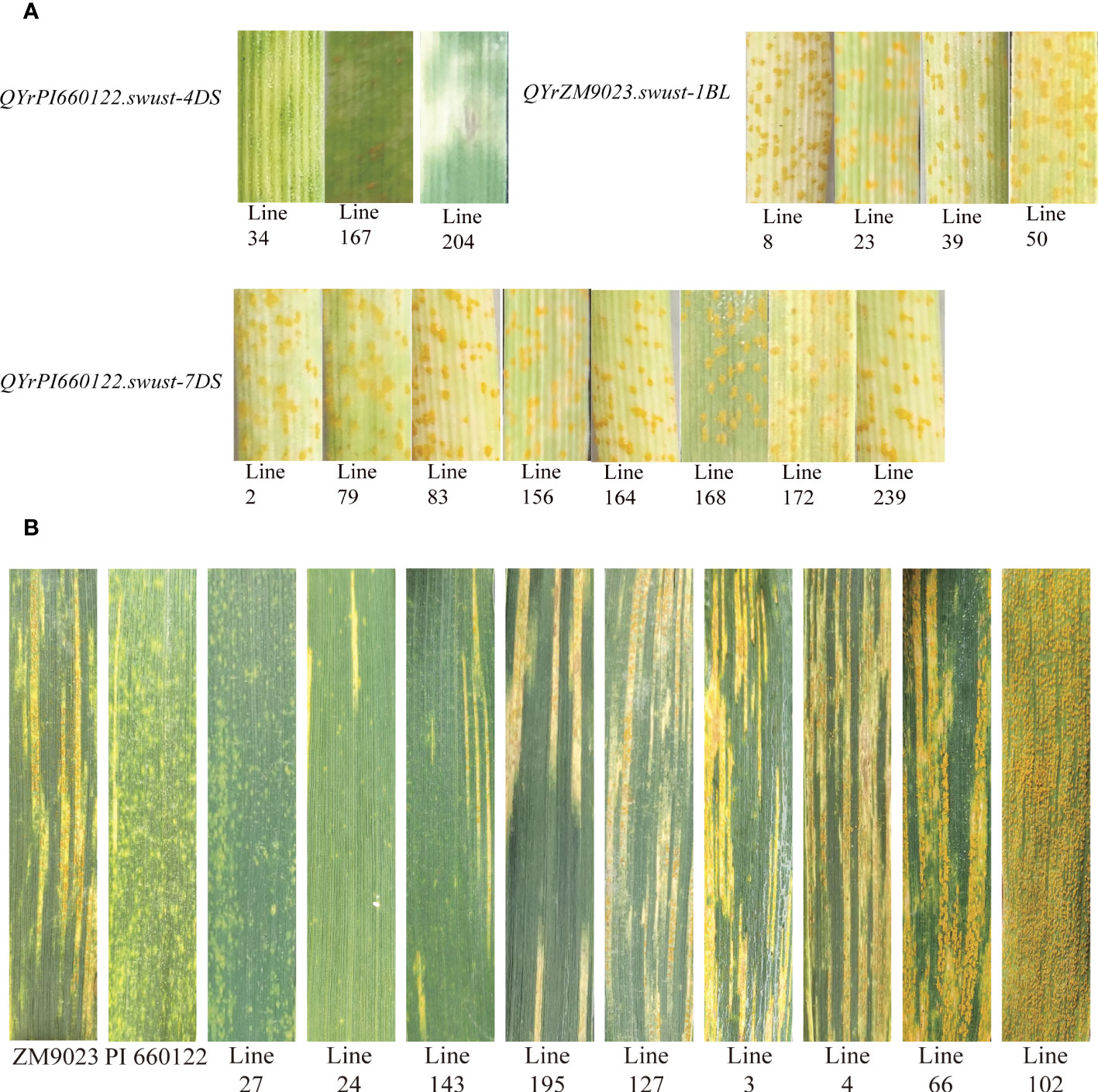
Figure 4 Stripe rust responses of lines containing only one QTL tested with Chinese race CYR32 of Puccinia striiformis f sp. tritici at the seedling stage (A) and on flag leaves of the resistant parent PI 660122, susceptible parent Zhengmai 9023 (ZM9023), and some selected recombinant inbred lines (RILs) (B). Line 27 containing QYrPI660122.swust-2BL, QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DS, QYrPI660122.swust-4DL, QYrZM9023.swust-6AS, QYrZM9023.swust-6DS, and QYrPI660122.swust-7DS; Line 24 containing QYrZM9023.swust-1BL, QYrPI660122.swust-2BL, QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DL, and QYrPI660122.swust-7DS; Line 143 containing QYrPI660122.swust-2BL, QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DL, and QYrPI660122.swust-7DS; Line 195 containing QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, QYrPI660122.swust-4DL, QYrZM9023.swust-6AS, and QYrZM9023.swust-6DS; Line 24 containing QYrPI660122.swust-2BL and QYrPI660122.swust-4DS; Line 3 containing QYrZM9023.swust-1BL, QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, and QYrPI660122.swust-4DL; Line 4 containing QYrPI660122.swust-2BL, QyrPI660122.swust-4BS, QyrPI660122.swust-4BL, and QyrPI660122.swust-4DL; Line 66 and Line 102 without any QTLs. QTL, quantitative trait locus.
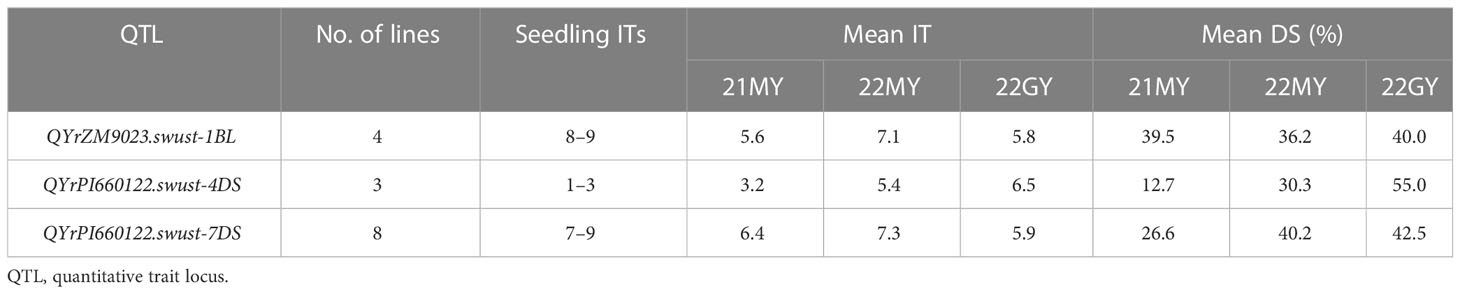
Table 5 Numbers of recombinant inbred lines from the Zhengmai 9023 × PI 660122 cross having only one stripe rust resistance and their infection types (ITs) at the seedling stage and mean IT and disease severity (DS) at the adult-plant stage in the fields of 2021 (21) and 2022 (22) at Mianyang (MY) and/or Guangyuan (GY).
QTL combinations
To determine the effects of the QTL in various combinations for Pst resistance, the 239 RILs were grouped into different genotypic groups based on the presence of markers closely associated with the nine QTL. These genotypes were further sorted into 10 groups based on the number of potential QTLs. Clearly, RILs carrying any number of QTL had lower mean DS than those without any of the QTL. Lines without any QTL had a mean IT of 6.6 and a mean DS of 46.91%. In comparison, when 0, 1, 2, 3, 4, 5, 6, and more than 6 QTLs were combined, the lines with one QTL had mean IT of 5.9 and mean DS of 36.04%, those with two QTLs had mean IT of 5.0 and mean DS of 31.81%, those with three QTLs had mean IT of 4.0 and mean DS of 23.25%, those with four QTLs had mean IT of 3.7 and mean DS of 19.52%, those with five QTLs had mean IT of 3.1 and mean DS of 14.76%, those with six QTLs had mean IT of 2.7 and mean DS of 10.45%, and those with seven or more QTL had mean IT of 2.4 and mean DS of 10.58%, close to the resistance level of PI 660122 (Figure 5).
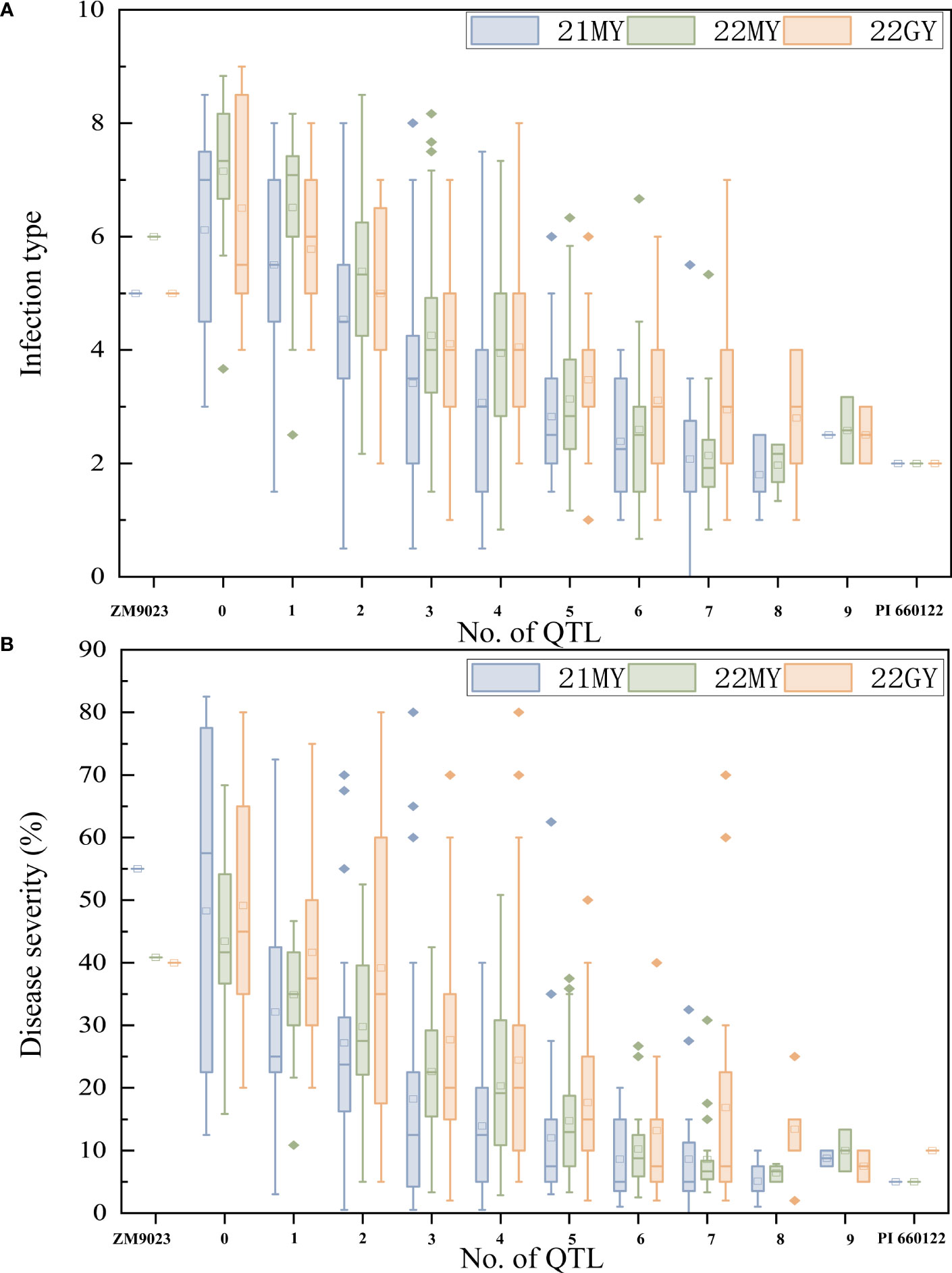
Figure 5 Effects of individual QTL and their combinations on stripe rust scores illustrated by mean infection type (IT) (A) and disease severity (DS) (B) scores of recombinant inbred lines from Zhengmai 9023 × PI 660122 (ZM9023 in three environments, 2021 Mianyang (21MY), 2022 Mianyang (22MY), and 2022 Guangyuan (22GY). Box plots indicate the infection type (IT) and disease severity (DS) associated with the identified QTL and their combination.
Selection of breeding lines
Various agronomic traits, including PH, PTN, SL, KPS, and TGW, of the parents and the 239 RILs were assessed in 2021 and 2022 in Mianyang and 2022 in Guangyuan. The mean PH values of PI 660122 and ZM9023 were 90.3 cm and 79.3 cm, respectively, and the RILs were mainly distributed in the range of 81–110 cm. The mean PTN values of PI 660122 and ZM9023 were 5 and 4, respectively, and the mean PTN values of RILs were between 4 and 10. The mean SL values of PI 660122 and ZM9023 were 9.8 cm and 8.4 cm, respectively, and the mean SL values of RILs were between 7.3 cm and 11.3 cm. The mean KPS values of PI 660122 and ZM9023 were 48 and 44, respectively, and the mean KPS values of RILs were between 33 and 58. The mean TGW values of PI 660122 and ZM9023 were 48 g and 44 g, respectively, and the mean TGW values of RILs were between 30.6 g and 58.4 g.
In order to select RILs with desirable agronomic traits, the following criteria were used: PH between 80 cm and 100 cm, PTN 5 or more, SL greater than 9 cm, KPS not less than 45, and TGW over 42 g, with stripe rust IT of 1–3 and DS < 20%. Based on these criteria, 26 lines were selected. The QTLs detected by their highly associated SNP markers in the selected lines are listed in Table 6. These lines had at least two QTLs, and three lines (F6-61, F6-78, and F6-86) had as many as seven QTLs. According to Table 7, the DS is negatively correlated with SL, PTN, KPS, and TGW, indicating that with the increase of DS, the SL, PTN, KPS, and TGW will decrease. The correlation coefficients between DS and PH, and PTN were 0.13 and 0.15, respectively, and the correlation was significant (p < 0.05). The correlation coefficients between DS and SL, KPS, and TGW were 0.05, 0.08, and 0.02, respectively, and the correlation was not significant (p > 0.05).
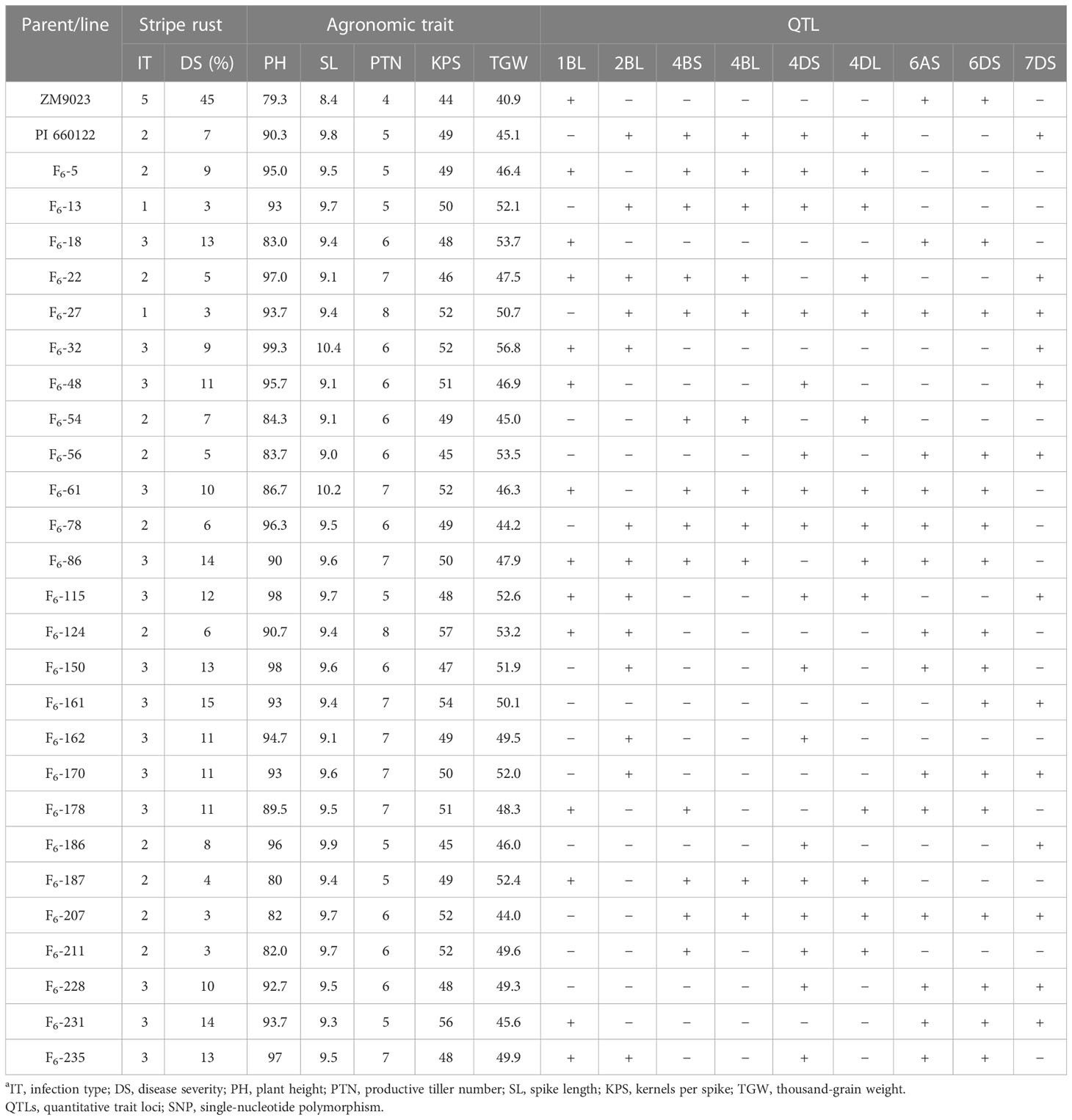
Table 6 Mean stripe rust response, agronomic traits, and presence (+) and absence (−) of resistant QTLs detected with SNP markers in Zhengmai (ZM) 9023, PI 661022, and selected recombinant inbred linesa.
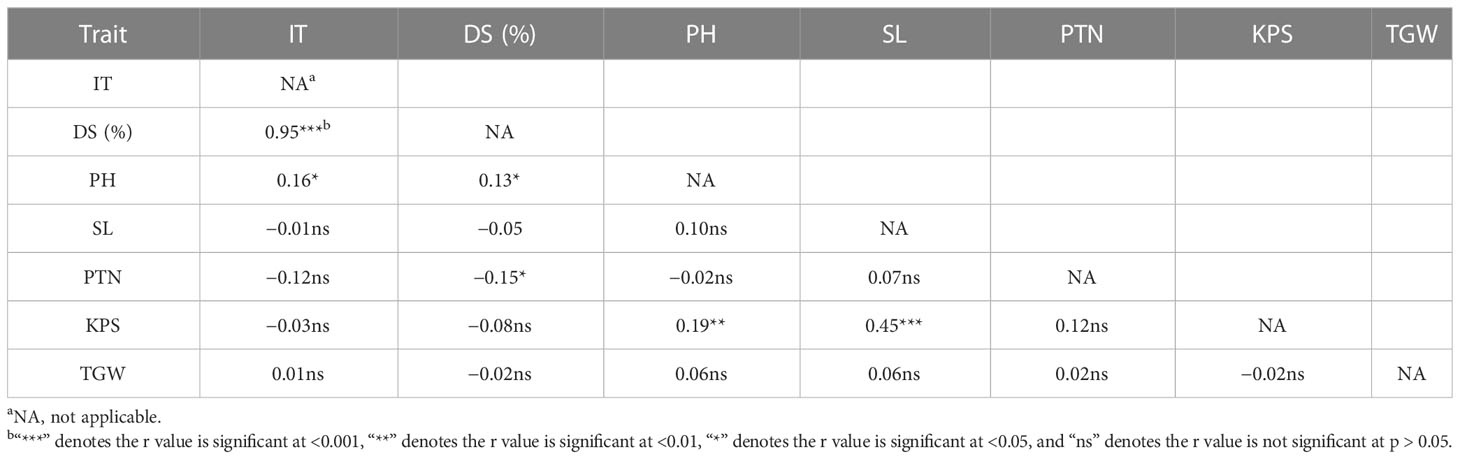
Table 7 Correlation coefficients (r) of important traits of the recombinant inbred lines Zhengmai 9023 × PI 660122.
Discussion
Developing durable resistance to stripe rust has been a top priority in wheat breeding over the past decade. Wheat line PI 660122 has exhibited high levels of stripe rust resistance for over a decade (Wang et al., 2012). In the present study, a RIL population containing 239 lines developed from a cross of PI 660122 with a Chinese elite cultivar, ZM9023, was phenotyped for stripe rust response in multiple environments and genotyped with the 15K wheat SNP array. We detected a total of nine QTLs, of which six (QYrPI660122.swust-2BL, QyrPI660122.swust-4BS, QyrPI660122.swust-4BL, QyrPI660122.swust-4DS, QyrPI660122.swust-4DL, and QyrPI660122.swust-7DS) came from PI 660122 and three (QYrZM9023.swust-1BL, QYrZM9023.swust-6AS, and QYrZM9023.swust-6DS) came from ZM9023.
QYrZM9023.swust-1BL, an APR QTL, was from Zhengmai 9023. This QTL was flanked by SNP markers AX-89763895 and AX-109273019, corresponding to the region from 670,429,611 bp to 681,685,826 bp of chromosome 1BL in the Chinese Spring (CS) genome (IWGSC RefSeq v1.0). Three permanently named stripe rust resistance genes, Yr21 (Chen and Line, 1995), Yr26 (Ma et al., 2001), and Yr29 (William et al., 2006), were mapped to 1BL. Among them, Yr21 and Yr26 confer ASR, while Yr29 is an APR gene linked with SSR markers Xwmc44 and Xwmc367 and located at the distal end of chromosome 1BL. The physical position of Xwmc367 is in the range of 678,736,681–678,736,834 bp, which is within the genome range of QYrZM9023.swust-1BL. According to a previous study, ZM9023 has Lr27/Yr30/Sr2 and Lr46/Yr29/Pm39/Sr58 (Li et al., 2020). As both Yr29 and QYrZM9023.swust-1BL confer APR, QYrZM9023.swust-1BL should be Yr29. Yr29 has been reported and deployed widely in wheat varieties around the world (William et al., 2006; Melichar et al., 2008; Lan et al., 2014; Hou et al., 2015; Maccaferri et al., 2015; Muleta et al., 2017; Wang and Chen, 2017; Cobo et al., 2018; Godoy et al., 2018; Liu et al., 2019; Rosa et al., 2019; Liu et al., 2020).
QYrPI660122.swust-2BL was derived from PI 660122, and it was flanked by SNP markers AX-109849173 and AX-109349804, corresponding to the region from 777,831,275 bp to 779,847,527 bp of chromosome 2BL in CS (IWGSC RefSeq v1.0). Seven permanently named stripe rust resistance genes, including Yr5 and Yr7 (Macer, 1963; Marchal et al., 2018), Yr43 (Cheng and Chen, 2010), Yr44 (Sui et al., 2009), Yr53 (Xu et al., 2013), Yr72 (McIntosh et al., 2016), and YrSP (Feng et al., 2015), were mapped to 2BL. Yr5/Yr7/YrSP (Marchal et al., 2018) had been cloned, and it encoded nucleotide-binding (NB) and leucine-rich repeat (LRR) proteins. The physical map position of Yr5/Yr7/YrSP was 685.265–685.27 Mb. Yr43, an ASR gene, was flanked by Xwgp110 and Xwgp103, but its physical map position is unknown. Yr44, an ASR gene, was derived from spring wheat cultivar Zak and flanked by XpWB5/N1R1 and Xwgp100, but its physical map position is unknown. Yr53, an ASR gene, was derived from PI 480148 and flanked by Xwmc441 and XLRRrev/NLRRrev350. The physical map position of Xwmc441 was 598,064,318–598,064,477 bp. Yr72, an ASR gene, was derived from AUS27507 and flanked by Xsun481 and IWB12294. The physical map position of IWB12294 was 767,171,587–767,171,587 bp. In addition, several major QTLs have been mapped to the long arm of chromosome 2B. QYr.hbaas-2BL was located at 453.3 Mb (Jia et al., 2020). Yr.niab-2B.1 was located at 683.05–750.12 Mb (Bouvet et al., 2022). QYrpd.Swust-2BL.1, QYrpd.Swust-2BL.2, QYrpd.Swust-2BL.3, and QYrpd.Swust-2BL.4 were located at 773.79–775.17 Mb, 753.37–777.52 Mb, 793.15–798.00 Mb, and 782.53–784.55 Mb, respectively (Zhou et al., 2022b). Qyr.gaas.2B.1 was located at 698.22–705.68 Mb (Cheng et al., 2022). YrQz was located at 557.37–630.40 Mb (Deng et al., 2004). QYr.nafu-2BL was located at 553.73–615.79 Mb (Zhou et al., 2015a; Hu et al., 2020). QYrww.wgp.2B-4 was located at 524 Mb (Mu et al., 2020). Yrdr.wgp-2BL was located at 709.84 Mb (Hou et al., 2015). QTL 2BL was located at 779.11–783.89 Mb (Somers et al., 2004). QYr.inra-2BL was located at 615.79–621.47 Mb (Mallard et al., 2005). QYraq.cau-2BL was located at 670.60–739.40 Mb (Ramburan et al., 2004). QYr.caas-2BL was located at 693.74–733.16 Mb (Ren et al., 2012b). Yrns.orz-2BL was located at 685.74 Mb (Vazquez et al., 2015). YrV23 is closely linked to Xwmc356 at position 796,684,893–796,685,357 bp (Wang et al., 2006). Based on the chromosomal positions, QYrPI660122.swust-2BL is likely different from Yr5/Yr7/YrSP and Yr53, but its relationships with other genes or QTL on 2BL need to be further studied.
QYrPI660122.swust-4BS was derived from PI 660122, and it was flanked by SNP markers AX-108767762 and AX-109309162, corresponding to the region from 32,961,964 bp to 36,395,734 bp of the CS (IWGSC RefSeq v1.0) chromosome 4BS. QYr.caas-4BS was located between markers Xwmc652 and Xgpw4388 (Wang et al., 2020). The physical map position of QYr.caas-4BS was 38.6–47.6 Mb. QYrcl.sicau-4B was located at the end of chromosome 4BS (Yao et al., 2020), which is different from the physical map position of QYrPI660122.swust-4BS. QYrPI660122.swust-4BS in PI 660122 is likely different from the previously mapped stripe rust resistance genes on chromosome 4BS.
QYrPI660122.swust-4BL was derived from PI 660122 and was flanked by SNP markers AX-109495166 and AX-108935256, corresponding to the position from 396,263,280 bp to 445,689,397 bp of the CS chromosome 4BL. Three permanently named stripe rust resistance genes, Yr50 (Liu et al., 2013), Yr62 (Lu et al., 2014), and Yr68 (McIntosh et al., 2016), were mapped to 4BL. Yr50, an ASR gene, was reported to be associated with Xbarc1096 and Xwmc47. The map position of Yr50 is 105.1 Mb–644.9 Mb. Yr62, a HTAP gene, was reported to be associated with Xgwm192 and Xgwm251. The physical map position of Yr62 was 482.8–568.6 Mb. Yr68, an APR gene, was reported to be associated with IWB74301 and IWB28394, and its physical map position was 575.04–600.66 Mb. In addition, several major QTLs were located on chromosome 4BL. QYrhm.nwafu-4B, Qyr.hbaas-4BL.1, and QYr.hbaas-4BL.2 overlapped Yr62 (Yuan et al., 2018; Jia et al., 2020). QYrhm.nwafu-4BL was derived from Humai 15 and flanked by AX-111150955 and Xgwm251, which was mapped to 523.4–568.6 Mb (Yuan et al., 2018). QYr.hbaas-4BL.1 was linked with IWB73717, and its physical map position was 531.3 Mb (Jia et al., 2020). QYr.hbaas-4BL.2 was linked with IWB63337 at the physical map position of 558.1 Mb. QYr.hbaas-4BL.3 was linked with IWB32927 at the physical map position of 579.4 Mb. QYr.sun-4B was derived from the Australian wheat cultivar Janz and exhibited minor variation (9.4%–16.8%) (Zwart et al., 2010). It was flanked by wPt-8543 and Xwmc238. The physical map position of Xwmc238 was 236,742,906–236,743,133 bp. QPst.jic-4B (Melichar et al., 2008) was derived from the UK winter wheat cultivar Guardian and mapped to the region between Xwmc652 and Xwmc692 with a PVE of 12%. QYr.crc-4BL was flanked by markers BS00048794_51 and RAC875_rep_c72961_977, and the physical map position of QYr.crc-4BL was 601.93–617.00 Mb (Rosa et al., 2019). YrBm, an APR QTL, was derived from Chinese winter wheat landrace Baimangmai (Hu et al., 2022). It was flanked by markers Xgpw7272 and Xwmc652. The physical map position of YrBm was 611.1–621.1 Mb. QYrPI660122.swust-4BL overlapped with Yr50, but further studies are needed to confirm the relationship between QYrPI660122.swust-4BL and Yr50 and determine the relationships with other QTLs on chromosome 4BL.
QYrPI660122.swust-4DS, an ASR QTL, was derived from PI 660122, and it was flanked by SNP markers AX-110046962 and AX-111093894 and corresponds to the region from 1,702,954 bp to 9,555,772 bp of the CS 4DS chromosome. Yr28 has been mapped to the short arm of chromosome 4D (Singh et al., 2000). Yr28 is a major ASR gene conferring stripe rust resistance from Ae. tauschii and located between SSR markers Xbcd265 and Xmwg634. Yr28 has been cloned and characterized, which encoded a typical nucleotide oligomerization domain-like receptor (NLR) (Zhang et al., 2019). The gene was further mapped between Xsdauw92 and Xsdauw96, approximately 0.13-cM interval, and its physical map position of Yr28 was 1.820–1.826 Mb. Based on the physical map position, the resistance type, and the Mexican wheat genotype PI 610755 that has Ae. tauschii in the pedigree as stripe rust resistance donor of PI 660122 (Wang et al., 2012), QYrPI660122.swust-4DS is highly likely Yr28.
QYrPI660122.swust-4DL was derived from PI 660122 and was flanked by SNP markers AX-94560848 and AX-111557122 corresponding to 288,430,275 bp to 310,458,135 bp of the CS chromosome 4DL. So far, only one permanently named Yr gene, Yr46 (Herrera-Foessel et al., 2011), has been reported on chromosome 4DL. Yr46 is an APR gene from wheat cultivar RL6007 and flanked by SSR markers Xgwm165 and Xgwm192, and its physical map position is approximately 417.2 Mb. Two QTLs, QYr.ucw-4DL (Cobo et al., 2018) and QYr.hbaas-4DL (Jia et al., 2020), have been also reported on chromosome 4DL. QYr.ucw-4DL was linked with the IWA2395, and its physical map position was 497.65 Mb (Cobo et al., 2018). QYr.hbaas-4DL is linked to SNP marker IWB44356 (Jia et al., 2020), with the physical map position of approximately 477.9 Mb. Based on the different physical map positions of QYrPI660122.swust-4DL from those of Yr46, QYr.ucw-4DL, and QYr.hbaas-4DL, QYrPI660122.swust-4DL is likely a new QTL for stripe rust resistance.
QYrZM9023.swust-6AS is derived from Zhengmai 9023 and flanked by SNP markers AX-95124889 and AX-110995858 corresponding to the 27,748,586–71,705,701-bp region of the CS chromosome 6AS. Numerous genes or QTLs for stripe rust resistance have been mapped to 6AS. Yr38 was mapped to 6AS (Marais et al., 2006), but its physical map position is unknown. Yr81 is flanked by KASP_3077 and Xgwm459 (Gessese et al., 2019), and the physical map position of Xgwm459 is within the 6,805,513–6,805,994-bp region. YrP10090 is flanked by AX-94460938 and AX-110585473 (Liu et al., 2021), and its physical map position is within 107.1–446.5 Mb. Qyr.gaas.6A was flanked by AX-109558600 and AX-109542604 (Cheng et al., 2022), and its physical map position is within 609.11–609.89 Mb. QYr-6A_Saar derived from the CIMMYT variety Saar (Lillemo et al., 2008) is flanked by XwPt-7063 and Xbarc3, and its physical map position is within 62.92–85.28 Mb. QYr.uaf-6A.1 and QYr.uaf-6A.4 were mapped with IWA8028 and IWB29623, respectively (Habib et al., 2020). The physical map position of QYr.uaf-6A.1 is at 105.02 Mb, and that of QYr.uaf-6A.4 is at 18.71 Mb. QYr.uga-6AS is flanked by wPt-671561 and wPt-7840 (Hao et al., 2011), and its physical map position is within 24.09–85.28 Mb. QYrex.wgp-6AS is flanked by markers Xgwm334 and Xwgp56 (Lin and Chen, 2008a), which indicate the QTL physical map position within 9.25–61.02 Mb. Yrq3 is flanked by SSR markers Xgwm334 and Xgwm169 (Cao et al., 2012), and its physical map position was found to be within 9.25–595.38 Mb. QYrcl.sicau-6A.1 was detected by two adjacent DArT-seq markers (3936688 and 1266956) on chromosome 6A between 5.14 and 5.83 Mb (Yao et al., 2020). QYr.sicau-6A is flanked by SNP markers AX-94548199 and AX-111101235 (Ma et al., 2019), and its physical map position of QYr.sicau-6A is within 90.32–97.52 Mb. QYrswp-6A is linked with IWA272 (Liu et al., 2019b), and its physical map position is 3.85 Mb. QYrZM9023.swust-6AS overlapped with QYr-6A_Saar, Yr.uga-6AS, and QYrex.wgp-6AS, but further studies are needed to determine if they are the same or different.
QYrZM9023.swust-6DS was also derived from Zhengmai 9023 and flanked by SNP markers AX-11475193 and AX-109317417 corresponding to the 35,630,857–44,498,347-bp region of the CS chromosome 6DS. Few genes or QTL for stripe rust resistance have been reported on 6DS. Yr77 is an APR gene flanked by Xbrac54 and Xcfd188, and the physical map position of Xcfd188 is within the 238,118,148–238,118,395-bp region (R. McIntosh, personal communication). QYr.ucw-6D is linked with IWA167 (Maccaferri et al., 2015), which is at the physical map position of 73.2 Mb. QYr.ufs-6D is flanked by Xgwm325 and Xbarc175 (Agenbag et al., 2012), and its physical map position is within the 79.96–411.88-Mb region. QYR7 is flanked by Xbcd1510 and XksuD27 (Boukhatem et al., 2002), and its physical map position is approximately 12 Mb. Based on the physical positions, QYrZM9023.swust-6DS is likely different from these genes or QTLs for stripe rust resistance genes previously mapped on chromosome 6DS.
QYrPI660122.swust-7DS, an APR QTL derived from PI 660122, is flanked by SNP markers AX-110467729 and AX-89378255 corresponding to the 43,386,933–47,379,368-bp region of the CS 7DS chromosome. Only the permanently named stripe rust resistance gene Yr18 has been mapped to chromosome 7DS. Yr18 was mapped to 7DS in a number of different wheat cultivars, such as Jupateco 73R and Opata 85 (Singh, 1992; Singh et al., 2000), Australian cultivar Cook (Bariana et al., 2001), and Fukuho-komugi (Suenaga et al., 2003). Yr18 is an APR gene flanked by Xgwm1220 and Xgwm29 and encodes a putative ATP-binding cassette transporter (Krattinger et al., 2009; Lagudah et al., 2009). The physical map position of Yr18 is from 47.412 Mb to 47.424 Mb, similar to QYrPI660122.swust-7DS. Yr18 is an important slow rusting gene and can confer high levels of resistance when combined with other minor genes (Singh and Rajaram, 1993; Navabi et al., 2004). Cultivars with Yr18 have been widely used in the International Maize and Wheat Improvement Center (CIMMYT) wheat breeding program (Singh et al., 2005). PI 660122 was developed from a cross of AvS with Mexican wheat genotype PI 610755 (Wang et al., 2012), PI 610755 has Opata 85 in its pedigree (Altar 84/Ae. tauschii (191)//Opata M85) (https://npgsweb.ars-grin.gov/gringlobal/accessiondetail?id=1580210), Yr18 was mapped in a RIL population for mapping Yr28 (Singh et al., 2000) as discussed above, and QYrPI660122.swust-7DS is most likely Yr18.
Conclusions
In the present study, we mapped nine QTLs conferring different types and levels of resistance to stripe rust. Among these QTLs, QYrZM9023.swust-1BL was identified as Yr29, QYrPI660122.swust-4DS as Yr28, and QYrPI660122.swust-7DS as Yr18, while QYrPI660122.swust-4BS, QYrPI660122.swust-4BL, and QYrZM9023.swust-6DS should be new. We demonstrated that combinations of different QTLs increased the levels of resistance. Furthermore, we selected lines from the RIL population with high adequate resistance to stripe rust combined with desirable agronomic traits, and these lines can be used in further evaluation for releasing commercial cultivars. The resistant lines and molecular markers for resistance QTL should be useful in developing wheat cultivars with high levels and durable resistance to stripe rust.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
QY and GJ detected the QTLs, analyzed the data, and prepared the first draft of the manuscript. QY, WT, RT, XQL, JF, XCZ and BS contributed to the collection of samples and phenotype data. XLZ contributed to the crosses and revised the manuscript. YR, XL, KH, and SY contributed to target line selection and population assessment. MW and XC provided seeds of the stripe rust-resistant parent. XLZ and XC conceived the project and generated the final version of the manuscript. All authors provided suggestions during the revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the Breakthrough in Wheat Breeding Material and Method Innovation and New Variety Breeding (Breeding Research Project, 2021YFYZ0002) and was partially funded by the Key Research and Development Program of the International Science and Technology Innovation Cooperation of Science and Technology Department of Sichuan Province, China (No. 2022YFH0032), National Natural Science Foundation of China (No. 32101707), PhD Foundation of Southwest University of Science and Technology (Nos. 19zx7116, 18zx7159, 16zx7162), and Longshan Academic Talent Research Support Program of SWUST (No. 17LZX5).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agenbag, G. M., Pretorius, Z. A., Boyd, L. A., Bender, C. M., Prins, R. (2012). Identification of adult plant resistance to stripe rust in the wheat cultivar Cappelle-Desprez. Theor. Appl. Genet. 125, 109–120. doi: 10.1007/s00122-012-1819-5
Bariana, H., Forrest, K., Qureshi, N., Miah, H., Hayden, M., Bansal, U. (2016). Adult plant stripe rust resistance gene Yr71 maps close to Lr24 in chromosome 3D of common wheat. Mol. Breed. 36, 98. doi: 10.1007/s11032-016-0528-1
Bariana, H. S., Hayden, M. J., Ahmed, N. U., Bell, J. A., McIntosh, R. A. (2001). Mapping of durable adult plant and seedling resistances to stripe rust and stem rust diseases in wheat. Aust. J. Agric. Res. 52, 1247–1255. doi: 10.1071/ar01040
Boukhatem, N., Baret, P. V., Mingeot, D., Jacquemin, J. M. (2002). Quantitative trait loci for resistance against yellow rust in two wheat-derived recombinant inbred line populations. Theor. Appl. Genet. 104, 111–118. doi: 10.1007/s001220200013
Bouvet, L., Percival-Alwyn, L., Berry, S., Fenwick, P., Mantello, C. C., Sharma, R., et al. (2022). Wheat genetic loci conferring resistance to stripe rust in the face of genetically diverse races of the fungus Puccinia striiformis f. sp. tritici. Theor. Appl. Genet. 135, 301–319. doi: 10.1007/s00122-021-03967-z
Cao, X., Zhou, J., Gong, X., Zhao, G., Jia, J., Qi, X. (2012). Identification and validation of a major quantitative trait locus for slow-rusting resistance to stripe rust in wheat. J. Integ. Plant Biol. 54, 330–344. doi: 10.1111/j.1744-7909.2012.01111.x
Carter, A. H., Chen, X. M., Garland-Campbell, K., Kidwell, K. K. (2009). Identifying QTL for high-temperature adult-plant resistance to stripe rust (Puccinia striiformis f. sp. tritici) in the spring wheat (Triticum aestivum L.) cultivar 'Louise'. Theor. Appl. Genet. 119, 1119–1128. doi: 10.1007/s00122-009-1114-2
Cavanagh, C. R., Chao, S., Wang, S., Huang, B. E., Stephen, S., Kiani, S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. U.S.A. 110, 8057–8062. doi: 10.1073/pnas.1217133110
Chen, X. M. (2005). Epidemiology and control of stripe rust (Puccinia striiformis f. sp. tritici) on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Chen, X. M. (2007). Challenges and solutions for stripe rust control in the United States. Aust. J. Agr. Res. 58, 648–655. doi: 10.1071/AR07045
Chen, X. M. (2013). High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4, 608–627. doi: 10.4236/ajps.2013.43080
Chen, X. M., Line, R. F. (1995). Gene number and heritability of wheat cultivars with durable, highr-temperature, adult-plantresistance and race-specific resistance to Puccinia striiformis. Phytopathology 85, 573–578. doi: 10.1094/Phyto-85-573
Cheng, P., Chen, X. M. (2010). Molecular mapping of a gene for stripe rust resistance in spring wheat cultivar IDO377s. Theor. Appl. Genet. 121, 195–204. doi: 10.1007/s00122-010-1302-0
Cheng, B., Gao, X., Cao, N., Ding, Y., Chen, T., Zhou, Q., et al. (2022). QTL mapping for adult plant resistance to wheat stripe rust in M96-5 × Guixie 3 wheat population. J. Appl. Genet. 63 (2), 265–279. doi: 10.1007/s13353-022-00686-z
Cobo, N., Pfluger, L., Chen, X. M., Dubcovsky, J. (2018). Mapping QTL for resistance to new virulent races of wheat stripe rust from two argentinean wheat cultivars. Crop Sci. 58, 2470–2483. doi: 10.2135/cropsci2018.04.0286
Cui, F., Zhang, N., Fan, X., Zhang, W., Zhao, C., Yang, L., et al. (2017). Utilization of a wheat 660K SNP array-derived high-density genetic map for high-resolution mapping of a major QTL for kernel number. Sci. Rep. 7, 3788. doi: 10.1038/s41598-017-04028-6
Deng, Z. Y., Zhang, X. Q., Wang, X. P., Jing, J. K., Wang, D. W. (2004). Identification and molecular mapping of a stripe rust resistance gene from a common wheat line Qz180. Acta Bot. Sin. 46, 236–241. doi: 10.1614/WS-03-091R
Dong, Z. Z., Hegarty, J. W., Zhang, J. L., Zhang, W. J., Chao, S. M., Chen, X. M., et al. (2017). Validation and characterization of a QTL for adult plant resistance to stripe rust on wheat chromosome arm 6BS (Yr78). Theor. Appl. Genet. 130, 2127–2137. doi: 10.1007/s00122-017-2946-9
Feng, J. Y., Wang, M. N., Chen, X. M., See, D. R., Zheng, Y. L., Chao, S. M., et al. (2015). Molecular mapping of YrSP and its relationship with other genes for stripe rust resistance in wheat chromosome 2BL. Phytopathology 105, 1206–1213. doi: 10.1094/PHYTO-03-15-0060-R
Feng, J. Y., Wang, M. N., See, D. R., Chao, S. M., Zheng, Y. L., Chen, X. M. (2018). Characterization of novel gene Yr79 and four additional QTL for all-stage and high-temperature adult-plant resistance to stripe rust in spring wheat PI 182103. Phytopathology 108, 737–747. doi: 10.1094/PHYTO-11-17-0375-R
Fu, D. L., Uauy, C., Distelfeld, A., Blechel, A., Epstein, L., Chen, X. M., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Gerechter-Amitai, Z. K., van Silfhout, C. H., Grama, A., Kleitman, F. (1989). Yr15 — a new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G-25. Euphytica 43, 187–190. doi: 10.1007/bf00037912
Gessese, M., Bariana, H., Wong, D., Hayden, M., Bansal, U. (2019). Molecular mapping of stripe rust resistance gene Yr81 in a common wheat landrace Aus27430. Plant Dis. 103, 1166–1171. doi: 10.1094/pdis-06-18-1055-re
Godoy, J., Rynearson, S., Chen, X. M., Pumphrey, M. (2018). Genome-wide association mapping of loci for resistance to stripe rust in North American elite spring wheat germplasm. Phytopathology 108, 234–245. doi: 10.1094/PHYTO-06-17-0195-R
Habib, M., Awan, F. S., Sadia, B., Zia, M. A. (2020). Genome-wide association mapping for stripe rust resistance in Pakistani spring wheat genotypes. Plants 9, 1056. doi: 10.3390/plants9091056
Hao, Y., Chen, Z., Wang, Y., Bland, D., Buck, J., Brown-Guedira, G., et al. (2011). Characterization of a major QTL for adult plant resistance to stripe rust in US soft red winter wheat. Theor. Appl. Genet. 123, 1401–1411. doi: 10.1007/s00122-011-1675-8
Herrera-Foessel, S. A., Lagudah, E. S., Huerta-Espino, J., Hayden, M. J., Bariana, H. S., Singh, D., et al. (2011). New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor. Appl. Genet. 122, 239–249. doi: 10.1007/s00122-010-1439-x
Hou, L., Chen, X. M., Wang, M. N., See, D. R., Chao, S., Bulli, P., et al. (2015). Mapping a large number of QTL for durable resistance to stripe rust in winter wheat Druchamp using SSR and SNP markers. PloS One 10, e0126794. doi: 10.1371/journal.pone.0126794
Hu, C. Y., Wang, F. T., Feng, J., Sun, C., Guo, J. Y., Lang, X. W., et al. (2022). Identification and molecular mapping of YrBm for adult plan resistance to stripe rust in Chinese wheat landrace Baimangmai. Theor. Appl. Genet. 135, 2655–2664. doi: 10.1007/s00122-022-04139-3
Hu, T., Zhong, X., Yang, Q., Zhou, X. L., Li, X., Yang, S. Z., et al. (2020). Introgression of two quantitative trait loci for stripe rust resistance into three Chinese wheat cultivars. Agronomy 10, 483. doi: 10.3390/agronomy10040483
Huang, C., Jiang, Y. Y., Li, P. L., Peng, H., Cui, Y., Yang, J. J., et al. (2018). Epidemics analysis of wheat stripe rust in China in 2017. Plant protection. 44, 162–166. doi: 10.16688/j.zwbh.2017268
Jambuthenne, D. T., Riaz, A., Athiyannan, N., Athiyannan, N., Ng, W. L., Ziems, L., et al. (2022). Mining the Vavilov wheat diversity panel for new sources of adult plant resistance to stripe rust. Theor. Appl. Genet. 135, 1355–1373. doi: 10.1007/s00122-022-04037-8
Jia, M., Yang, L., Zhang, W., Rosewarne, G., Li, J., Yang, E., et al. (2020). Genome-wide association analysis of stripe rust resistance in modern Chinese wheat. BMC Plant Biol. 20, 491. doi: 10.1186/s12870-020-02693-w
Klymiuk, V., Chawla, H. S., Wiebe, K., Ens, J., Fatiukha, A., Govta, L., et al. (2022). Discovery of stripe rust resistance with incomplete dominance in wild emmer wheat using bulked segregant analysis sequencing. Commun. Biol. 5, 826. doi: 10.1038/s42003-022-03773-3
Klymiuk, V., Yaniv, E., Huang, L., Raats, D., Fatiukha, A., Chen, S., et al. (2018). Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat. Commun. 9, 3735. doi: 10.1038/s41467-018-06138-9
Kosambi, D. D. (2016). The estimation of map distances from recombination values. Eds. Ramaswamy, R., Kosambi, D. D. (New Delhi: Springer), 125–130. doi: 10.1007/978-81-322-3676-4_16
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lagudah, E. S., Krattinger, S. G., Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., Spielmeyer, Q., et al. (2009). Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal disease. Theor. Appl. Genet. 119, 889–898. doi: 10.1007/s00122-009-1097-z
Lan, C. X., Rosewarne, G. M., Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Basnet, B. R., et al. (2014). QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin1. Mol. Breed. 34, 789–803. doi: 10.1007/s11032-014-0075-6
Li, W., Song, G. Q., Li, J. H., Li, Y. L., Zhang, S. J., Gao, J., et al. (2020). Molecular detection of four pleiotropic disease resistance genes in wheat. J. Triti. Crops 40, 395–400. doi: 10.7606/J.ISSN.1009-1041.2020.04.0
Li, J. L., Wang, S., Yu, J., Wang, L., Zhou, S. L. (2013). A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 48, 72–78. doi: 10.3724/sp.j.1259.2013.00072
Lillemo, M., Asalf, B., Singh, R. P., Huerta-Espino, J., Chen, X. M., He, Z. H., et al. (2008). The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116, 1155–1166. doi: 10.1007/s00122-008-0743-1
Lin, F., Chen, X. M. (2007). Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 114, 1277–1287. doi: 10.1007/s00122-007-0518-0
Lin, F., Chen, X. M. (2008a). Molecular mapping of genes for race-specific overall resistance to stripe rust in wheat cultivar Express. Theor. Appl. Genet. 116, 797–806. doi: 10.1007/s00122-008-0713-7
Lin, F., Chen, X. M. (2008b). Quantitative trait loci for non-race-specific, high-temperature adult-plant resistance to stripe rust in wheat cultivar Express. Theor. Appl. Genet. 118, 631–642. doi: 10.1007/s00122-008-0894-0
Line, R. F. (2002). Stripe rust of wheat and barley in North America: a retrospective historical review. Annu. Rev. Phytopathol. 40, 75–118. doi: 10.1146/annurev.phyto.40.020102.111645
Line, R. F., Qayoum, A. (1992). Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America 1968–87. Tech. Bulletin United States Department Agric. 1788, 1–44.
Liu, J., Chang, Z., Zhang, X., Yang, Z., Li, X., Jia, J., et al. (2013). Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor. Appl. Genet. 126, 265–274. doi: 10.1007/s00122-012-1979-3
Liu, W., Frick, M., Huel, R., Nykiforuk, C. L., Wang, X. M., Gaudet, D. A., et al. (2014). The stripe rust resistance gene Yr10 encodes an evolutionary-conserved and unique CC-NBS-LRR sequence in wheat. Mol. Plant 7, 1740–1755. doi: 10.1093/mp/ssu112
Liu, S., Huang, S., Zeng, Q., Wang, X., Yu, R., Wang, Q., et al. (2021). Refined mapping of stripe rust resistance gene YrP10090 within a desirable haplotype for wheat improvement on chromosome 6A. Theor. Appl. Genet. 134, 2005–2021. doi: 10.1007/s00122-021-03801-6
Liu, W., Kolmer, J., Rynearson, S., Chen, X., Gao, L., Anderson, J. A., et al. (2019b). Identifying loci conferring resistance to leaf and stripe rusts in a spring wheat population (Triticum aestivum L.) via genome-wide association mapping. Phytopathology 109, 1932–1940. doi: 10.1094/phyto-04-19-0143-r
Liu, L., Wang, M. N., Feng, J. Y., See, D. R., Chao, S. M., Chen, X. M. (2018). Combination of all-stage and high-temperature adult-plant resistance QTL confers high level, durable resistance to stripe rust in winter wheat cultivar Madsen. Theor. Appl. Genet. 131, 1835–1849. doi: 10.1007/s00122-018-3116-4
Liu, L., Yuan, C. Y., Wang, M. N., See, D. R., Chen, X. M. (2020). Mapping quantitative trait loci for high-temperature adult-plant resistance to stripe rust in spring wheat PI 197734 using a doubled haploid population and genotyping by multiplexed sequencing. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.596962
Liu, L., Yuan, C. Y., Wang, M. N., See, D. R., Zemetra, R. S., Chen, X. M. (2019). QTL analysis of durable stripe rust resistance in the North American winter wheat cultivar Skiles. Theor. Appl. Genet. 132, 1677–1691. doi: 10.1007/s00122-019-03307-2
Lu, Y., Wang, M., Chen, X., See, D., Chao, S., Jing, J. (2014). Mapping of Yr62 and a small-effect QTL for high-temperature adult-plant resistance to stripe rust in spring wheat PI 192252. Theor. Appl. Genet. 127, 1449–1459. doi: 10.1007/s00122-014-2312-0
Ma, J., Qin, N., Cai, B., Chen, G., Ding, P., Zhang, H., et al. (2019). Identification and validation of a novel major QTL for all-stage stripe rust resistance on 1BL in the winter wheat line 20828. Theor. Appl. Genet. 132, 1363–1373. doi: 10.1007/s00122-019-03283-7
Ma, J., Zhou, R., Dong, Y., Wang, L., Wang, X., Jia, J. (2001). Molecular mapping and detection of the yellow rust resistance gene Yr26 in wheat transferred from Triticum turgidum L. using microsatellite markers. Euphytica 120, 219–226. doi: 10.1023/a:1017510331721
Maccaferri, M., Zhang, J., Bulli, P., Abate, Z., Chao, S., Cantu, D., et al. (2015). A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3-Genes Genom. Genet. 5, 449–465. doi: 10.1534/g3.114.014563
Macer, R. (1963). The formal and monosomic genetic analysis of stripe rust (Puccinia striiformis) resistance in wheat, Hereditas. 2, 127–142
Mallard, S., Gaudet, D., Aldeia, A., Abelard, C., Besnard, A. L., Sourdille, P., et al. (2005). Genetic analysis of durable resistance to yellow rust in bread wheat. Theor. Appl. Genet. 110, 1401–1409. doi: 10.1007/s00122-005-1954-3
Marais, G. F., McCallum, B., Marais, A. S. (2006). Leaf rust and stripe rust resistance genes derived from Aegilops Sharonensis. Euphytica 149, 373–380. doi: 10.1007/s10681-006-9092-9
Marchal, C., Zhang, J., Zhang, P., Fenwick, P., Steuernagel, B., Adamski, N. M., et al. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plant 4, 662–668. doi: 10.1038/s41477-018-0236-4
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C., Xia, X. C. (2016) Catalogue of gene symbols for wheat: 2016 Supplement. Available at: http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015-2016.pdf (Accessed 15 May, 2023).
Melichar, J. P., Berry, S., Newell, C., MacCormack, R., Boyd, L. A. (2008). QTL identification and microphenotype characterisation of the developmentally regulated yellow rust resistance in the UK wheat cultivar Guardian. Theor. Appl. Genet. 117, 391–399. doi: 10.1007/s00122-008-0783-6
Meng, L., Li, H. H., Zhang, L. Y., Wang, J. K. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Milus, E., Kristensen, K., Hovmøller, M. S. (2009). Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99, 89–94. doi: 10.1094/phyto-99-1-0089
Moore, J. W., Herrera-Foessel, S., Lan, C. X., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Mu, J. M., Liu, L., Liu, Y., Wang, M. N., See, D. R., Han, D. J., et al. (2020). Genome-wide association study and gene specific markers identified 51 genes or QTL for resistance to stripe rust in US winter wheat cultivars and breeding lines. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00998
Muleta, K. T., Bulli, P., Rynearson, S., Chen, X. M., Pumphrey, M. (2017). Loci associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a core collection of spring wheat (Triticum aestivum). PloS One 12, e0179087. doi: 10.1371/journal.pone.0179087
Navabi, A., Singh, R. P., Tewari, J. P., Briggs, K. G. (2004). Inheritance of high levels of adult-plant resistance to stripe rust in five spring wheat genotypes. Crop Sci. 44, 1156–1162. doi: 10.2135/cropsci2004.1156
Nsabiyera, V., Bariana, H. S., Qureshi, W. D., Hayden, M. J., Bansal, U. K. (2018). Characterisation and mapping of adult plant stripe rust resistance in wheat accession Aus27284. Theor. Appl. Genet. 131, 1459–1467. doi: 10.1007/s00122-018-3090-x
Paillard, S., Trotoux-Verplancke, G., Perretant, M. R., Mohamadi, F., Leconte, M., Coëdel, S., et al. (2012). Durable resistance to stripe rust is due to three specific resistance genes in French bread wheat cultivar Apache. Theor. Appl. Genet. 125, 955–965. doi: 10.1007/s00122-012-1885-8
Qayoum, A., Line, R. F. (1985). High-temperature, adult-plant resistance to stripe rust of wheat. Phytopathology 75, 1121–1123. doi: 10.1094/phyto-75-1121
Ramburan, V. P., Pretorius, Z. A., Louw, J. H., Boyd, L. A., Smith, P. H., Boshoff, W. H. P., et al. (2004). A genetic analysis of adult plant resistance tostripe rust in the wheat cultivar Kariega. Theor. Appl. Genet. 108, 1426–1433. doi: 10.1007/s00122-003-1567-7
Rasheed, A., Hao, Y., Xia, X., Khan, A., Xu, Y., Varshney, R. K., et al. (2017). Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol. Plant 10, 1047–1064. doi: 10.1016/j.molp.2017.06.008
Ren, Y., He, Z. H., Li, J., Lillemo, M., Wu, L., Bai, B., et al. (2012b). QTL mapping of adult-plant resistance to stripe rust in a population derived from common wheat cultivars Naxos and Shanghai 3/Catbird. Theor. Appl. Genet. 125, 1211–1221. doi: 10.1007/s00122-012-1907-6
Ren, R. S., Wang, M. N., Chen, X. M., Zhang, Z. J. (2012a). Characterization and molecular mapping of Yr52 for high-temperature adult-plant resistance to stripe rust in spring wheat germplasm PI 183527. Theor. Appl. Genet. 125, 847–857. doi: 10.1007/s00122-012-1877-8
Rosa, S. B., Zanella, C. M., Hiebert, C. W., Brûlé-Babel, A. L., Randhawa, H. S., Shorter, S., et al. (2019). Genetic characterization of leaf and stripe rust resistance in the Brazilian wheat cultivar Toropi. Phytopathology 109, 1760–1768. doi: 10.1094/PHYTO-05-19-0159-R
Rosewarne, G. M., Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Lan, C. X., He, Z. H. (2013). Quantitative trait loci of stripe rust resistance in wheat. Theor. Appl. Genet. 126, 2427–2449. doi: 10.1007/s00122-013-2159-9
Santra, D. K., Chen, X. M., Santra, M., Campbell, K. G., Kidwell, K. K. (2008). Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum L.) cultivar 'Stephens'. Theor. Appl. Genet. 117, 793–802. doi: 10.1007/s00122-008-0820-5
Singh, R. P. (1992). Genetic association of leaf rust resistance gene Lr34 with adult plant resistance to stripe rust in bread wheat. Phytopathology 82, 835–838. doi: 10.1094/phyto-82-835
Singh, R. P., Huerta-Espino, J., William, H. M. (2005). Genetics and breeding for durable resistance to leaf and stripe rust of wheat. Turk. J. Agric. Forest. 29, 121–127.
Singh, R. P., Nelson, J. C., Sorrells, M. E. (2000). Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 40, 1148–1155. doi: 10.2135/cropsci2000.4041148x
Singh, R. P., Rajaram, S. (1993). Genetics of adult plant resistance to stripe rust in ten spring bread wheats. Euphytica 72, 1–7. doi: 10.1007/bf00023766
Soleimani, B., Lehnert, H., Keilwagen, J., Plieske, J., Ordon, F., Naseri, R. S., et al. (2020). Comparison between core set selection methods using different illumina marker platforms: a case study of assessment of diversity in wheat. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01040
Somers, D. J., Isaac, P., Edwards, K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114. doi: 10.1007/s00122-004-1740-7
Suenaga, K., Singh, R. P., Huerta-Espino, J., William, H. M. (2003). Microsatellite markers for genes Lr34/Yr18 and other quantitative trait Loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 93, 881–890. doi: 10.1094/phyto.2003.93.7.881
Sui, X. X., Wang, M. N., Chen, X. M. (2009). Molecular mapping of a stripe rust resistance gene in spring wheat cultivar Zak. Phytopathology 99, 1209–1215. doi: 10.1094/PHYTO-99-10-1209
Tekin, M., Cat, A., Akan, K., Catal, M., Akar, T. (2021). A new virulent race of wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) on the resistance gene Yr5 in Turkey. Plant Dis. 105, 3292. doi: 10.1094/pdis-03-21-0629-pdn
Vazquez, M. D., Zemetra, R., Peterson, C. J., Chen, X. M., Heesacker, A., Mundt, C. C. (2015). Multi-location wheat stripe rust QTL analysis: genetic background and epistatic interactions. Theor. Appl. Genet. 128, 1307–1318. doi: 10.1007/s00122-015-2507-z
Wan, A. M., Zhao, Z. H., Chen, X. M., He, Z. H., Jin, S. L., Jia, Q. Z., et al. (2004). Wheat stripe rust epidemic and virulence of Puccinia striiformis f.sp. tritici in China in 2002. Plant Dis. 88, 896–940. doi: 10.1094/PDIS.2004.88.8.896
Wang, J. K. (2009). Inclusive composite interval mapping of quantitative trait genes. Acta Agron. Sin. 35, 239–245. doi: 10.3724/SP.J.1006.2009.00239
Wang, M. N., Chen, X. M. (2017). Stripe rust resistance. Eds. Chen, X. M., Kang, Z. S. (Dordrecht: Springer Netherlands), 353–558.
Wang, M. N., Chen, X. M., Xu, L. S., Cheng, P., Bockelman, H. E. (2012). Registration of 70 common spring wheat germplasm lines resistant to stripe rust. J. Plant Regist. 6, 104–110. doi: 10.3198/jpr2011.05.0261crg
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Y. B., Xu, S. C., Xu, Z., Liu, T. G., Lin, R. M. (2006). A microsatellite marker linked to the stripe rust resistance gene YrV23 in the wheat variety Vilmorin23. Hereditas 28, 306–310. doi: 10.16288/j.yczz.2006.03.011
Wang, H., Zou, S. H., Li, Y. L., Lin, F. Y., Tang, D. Z. (2020). An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat. Commun. 11, 1353. doi: 10.1038/s41467-020-15139-6
Wellings, C. R., McIntosh, R. A. (1990). Puccinia striiformis f.sp. tritici in Australasia: pathogenic changes during the first 10 years. Plant Pathol. 39, 316–325. doi: 10.1111/j.1365-3059.1990.tb02509.x
William, H. M., Singh, R. P., Huerta-Espino, J., Palacios, G., Suenaga, K. (2006). Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49, 977–990. doi: 10.1139/g06-052
Winfield, M. O., Allen, A. M., Burridge, A. J., Barker, G. L. A., Benbow, H. R., Wilkinson, P. A., et al. (2016). High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206. doi: 10.1111/pbi.12485
Wu, J. H., Liu, S. J., Wang, Q. L., Zeng, Q. D., Mu, J. M., Huang, S., et al. (2018). Rapid identification of an adult plant stripe rust resistance gene in hexaploid wheat by high-throughput SNP array genotyping of pooled extremes. Theor. Appl. Genet. 131, 43–58. doi: 10.1007/s00122-017-2984-3
Xu, L. S., Wang, M. N., Cheng, P., Kang, Z. S., Hulbert, S. H., Chen, X. M. (2013). Molecular mapping of Yr53, a new gene for stripe rust resistance in durum wheat accession PI 480148 and its transfer to common wheat. Theor. Appl. Genet. 126, 523–533. doi: 10.1007/s00122-012-1998-0
Xue, W. B., Xu, X., Mu, J. M., Wang, Q. L., Wu, J. H., Huang, L. L., et al. (2014). Evaluation of stripe rust resistance and genes in Chinese elite wheat varieties. J. Tritic. Crops 34, 1054–1060. doi: 10.7606/j.issn.1009-1041.2014.08.06
Yao, F. J., Long, L., Wang, Y. Q., Duan, L. Y., Zhao, X. Y., Jiang, Y. F., et al. (2020). Population structure and genetic basis of the stripe rust resistance of 140 Chinese wheat landraces revealed by a genome-wide association study. Plant Sci. 301, 110688. doi: 10.1016/j.plantsci.2020.110688
Yuan, F. P., Zeng, Q. D., Wu, J. H., Wang, Q. L., Yang, Z. J., Liang, B. P., et al. (2018). QTL mapping and validation of adult plant resistance to stripe rust in Chinese wheat landrace Humai 15. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00968
Zhang, C. Z., Huang, L., Zhang, H. F., Hao, Q. Q., Lyu, B., Wang, M. N., et al. (2019). An ancestral NB-LRR with duplicated 3'UTRs confers stripe rust resistance in wheat and barley. Nat. Commun. 10, 4023. doi: 10.1038/s41467-019-11872-9
Zhang, G. S., Zhao, Y. Y., Kang, Z. S., Zhao, J. (2020). First report of a Puccinia striiformis f. sp. tritici race virulent to wheat stripe rust resistance gene Yr5 in China. Plant Dis. 104, 284. doi: 10.1094/pdis-05-19-0901-pdn
Zhou, X. L., Fang, T. H., Li, K. X., Huang, K. B., Ma, C. H., Zhang, M., et al. (2022a). Yield losses associated with different levels of stripe rust resistance of commercial wheat cultivars in China. Phytopathol 112, 1244–1251. doi: 10.1094/phyto-07-21-0286-r
Zhou, X. L., Han, D. J., Chen, X. M., Mu, J. M., Xue, W. B., Zeng, Q. D., et al. (2015a). QTL mapping of adult-plant resistance to stripe rust in wheat line P9897. Euphytica 205, 243–253. doi: 10.1007/s10681-015-1447-7
Zhou, X. L., Hu, T., Li, X., Yu, M., Li, Y., Yang, S., et al. (2019). Genome-wide mapping of adult plant stripe rust resistance in wheat cultivar Toni. Theor. Appl. Genet. 132, 1693–1704. doi: 10.1007/s00122-019-03308-1
Zhou, X. L., Li, X., Han, D. J., Yang, S. Z., Kang, Z. S., Ren, R. S. (2022b). Genome-wide QTL mapping for stripe rust resistance in winter wheat Pindong 34 using a 90K SNP array. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.932762
Zhou, X. L., Wang, M. N., Chen, X. M., Lu, Y., Kang, Z. S., Jing, J. X. (2014). Identification of Yr59 conferring high-temperature adult-plant resistance to stripe rust in wheat germplasm PI 178759. Theor. Appl. Genet. 127, 935–945. doi: 10.1007/s00122-014-2269-z
Zhou, X. L., Zhang, G. M., Huang, L. L., Han, D. J., Kang, Z. S. (2015b). Evaluation of resistance to stripe rust in eighty abroad spring wheat germplasms. Sci. Agric. Sin. 48, 1518–1526. doi: 10.3864/j.issn.0578-1752.2015.08.06
Keywords: stripe rust, wheat, resistance, QTL mapping, yellow rust
Citation: Yan Q, Jia G, Tan W, Tian R, Zheng X, Feng J, Luo X, Si B, Li X, Huang K, Wang M, Chen X, Ren Y, Yang S and Zhou X (2023) Genome-wide QTL mapping for stripe rust resistance in spring wheat line PI 660122 using the Wheat 15K SNP array. Front. Plant Sci. 14:1232897. doi: 10.3389/fpls.2023.1232897
Received: 01 June 2023; Accepted: 31 July 2023;
Published: 28 August 2023.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Hanif Khan, Indian Institute of Wheat and Barley Research (ICAR), IndiaYuefeng Ruan, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2023 Yan, Jia, Tan, Tian, Zheng, Feng, Luo, Si, Li, Huang, Wang, Chen, Ren, Yang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suizhuang Yang, eWFuZ3N6aEAxMjYuY29t; Xinli Zhou, ZWxpNjk1MUBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Qiong Yan1†
Qiong Yan1† Wenjing Tan
Wenjing Tan Xin Li
Xin Li Meinan Wang
Meinan Wang Xianming Chen
Xianming Chen Yong Ren
Yong Ren Suizhuang Yang
Suizhuang Yang Xinli Zhou
Xinli Zhou