- 1Indian Council of Agricultural Research (ICAR) – Indian Institute of Pulses Research (IIPR), Kanpur, Uttar Pradesh, India
- 2Department of Agronomy, Feed the Future Innovation Lab for Collaborative Research on Sustainable Intensification, Kansas State University, Manhattan, KS, United States
- 3Department of Botany, Panjab University, Chandigarh, India
- 4College of Agriculture, Family Sciences and Technology, Fort Valley State University, Fort Valley, GA, United States
- 5Department of Plant Physiology, College of Agriculture, Vellayani, Kerala Agriculture University, Thiruvananthapuram, Kerala, India
- 6Institute of Agriculture, The University of Western Australia, Perth, Australia
Chickpea (Cicer arietinum L.) is a vital grain legume, offering an excellent balance of protein, carbohydrates, fats, fiber, essential micronutrients, and vitamins that can contribute to addressing the global population’s increasing food and nutritional demands. Chickpea protein offers a balanced source of amino acids with high bioavailability. Moreover, due to its balanced nutrients and affordable price, chickpea is an excellent alternative to animal protein, offering a formidable tool for combating hidden hunger and malnutrition, particularly prevalent in low-income countries. This review examines chickpea’s nutritional profile, encompassing protein, amino acids, carbohydrates, fatty acids, micronutrients, vitamins, antioxidant properties, and bioactive compounds of significance in health and pharmaceutical domains. Emphasis is placed on incorporating chickpeas into diets for their myriad health benefits and nutritional richness, aimed at enhancing human protein and micronutrient nutrition. We discuss advances in plant breeding and genomics that have facilitated the discovery of diverse genotypes and key genomic variants/regions/quantitative trait loci contributing to enhanced macro- and micronutrient contents and other quality parameters. Furthermore, we explore the potential of innovative breeding tools such as CRISPR/Cas9 in enhancing chickpea’s nutritional profile. Envisioning chickpea as a nutritionally smart crop, we endeavor to safeguard food security, combat hunger and malnutrition, and promote dietary diversity within sustainable agrifood systems.
Introduction
Chickpea (Cicer arietinum L.), a nutritionally dense pulse crop, is widely consumed by humans and cultivated annually, predominantly in semiarid and temperate climates under rainfed conditions (Gao et al., 2015). Global chickpea cultivation spans 14.56 million hectares (Mha), producing approximately 15 million tons (Mt) annually (FAOSTAT, 2021). Major producers include India, Australia, Pakistan, Central America, and East Africa (Knights and Hobson, 2016), with India leading production at 11.4 Mt from 9.9 Mha (FAOSTAT, 2021). Chickpea varieties are categorized as ‘desi’ and ‘kabuli’ based on seed shape and color (Knights and Hobson, 2016), with desi types predominant in Australia, Central America, East Africa, and India, while kabuli types thrive in the Mediterranean, Middle East, North Africa, and North America (Knights and Hobson, 2016; Grasso et al., 2022).
Global climate change and burgeoning human populations threaten food and nutritional security. Despite increased agricultural productivity, over 820 million people globally suffer from food insecurity, and at least 2 billion face nutritional insecurity (Jha et al., 2024). Approximately 3 billion people in Asia, Africa, and Latin America face micronutrient deficiencies, particularly zinc (Zn) and iron (Fe) (Welch and Graham, 2004; Darnton-Hill et al., 2006), crucial for optimal growth and development (Brown et al., 2002; Welch, 2002). Chickpea, inherently abundant in these micronutrients and vitamins, can help address ‘hidden hunger,’ particularly among infants and women of childbearing age in low-income countries (Broughton et al., 2003; Ufaz and Galili, 2008; Burchi et al., 2011). Moreover, chickpea contributes to human disease prevention, including diabetes, hyperlipidemia, kwashiorkor, and anemia (Younis et al., 2015).
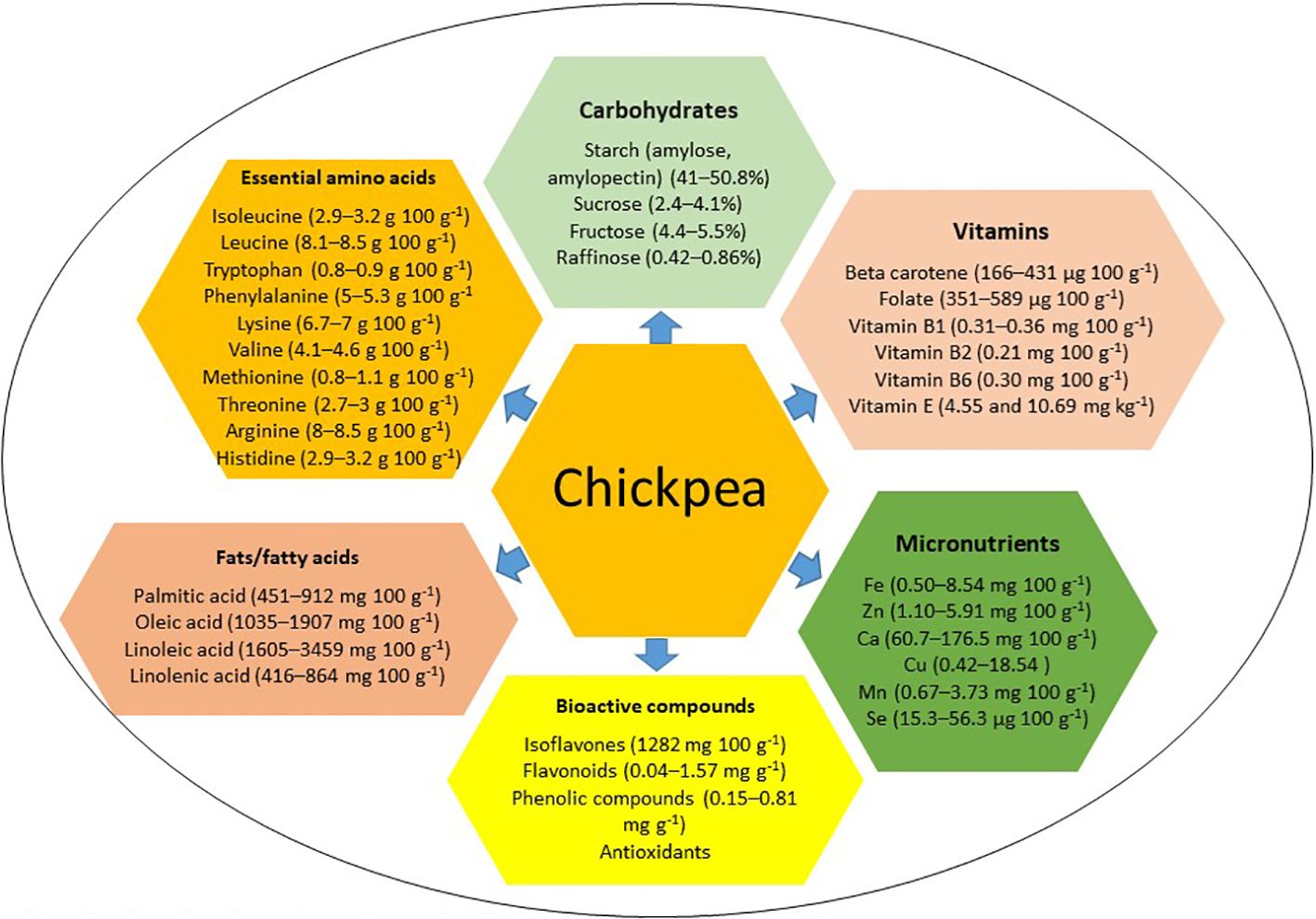
Chickpea, characterized by high dietary seed protein, abundant non-starch polysaccharides, low calorie content, low allergenicity, and high digestibility, offers a cost-effective protein source for low-income individuals and vegetarians (Kaur and Prasad, 2021; Yegrem, 2021; Begum et al., 2023). However, it lacks sulfur-containing essential amino acids methionine and cysteine (Jukanti et al., 2012; Grasso et al., 2022). Chickpea seeds are rich in carbohydrates, starch, fat, fiber, vitamins, and essential micronutrients (Heiras-Palazuelos et al., 2013; Kaur et al., 2019; Sharma et al., 2021; Grasso et al., 2022; Roorkiwal et al., 2022; Salaria et al., 2023) (see Figure 1). In Western countries, chickpea is mainly consumed in the form of ‘hummus’ (Wallace et al., 2016), with four tablespoons of chickpea-based hummus per day providing the equivalent of 2 cups of legumes per week and approximately 25 g of dietary fiber (Wallace et al., 2016). Approximately 1/3 cup of chickpea provides 5.4 g protein, 1.6 g fat, 16.7 g carbohydrates, 4.6 g dietary fiber, 29.9 mg calcium, 1.8 mg iron, 29.3 mg magnesium, 177.4 mg potassium, 104.9 µg folate, 0.6 mg vitamin A, and 0.2 mg vitamin C (Didinger and Thompson, 2021). Thus, regular chickpea consumption can meet the recommended daily allowance of secondary macronutrients and micronutrients (Thavarajah and Thavarajah, 2012). Chickpea can be used to make gluten-free bread (GFB), comprising 75% chickpea flour blended with 25% potato or cassava starch, for individuals with gluten-related disorders, enhancing nutritional quality, including dietary fiber (Santos et al., 2018). Moreover, chickpea harbors bioactive compounds like sterols, phenols, carotenoids, tannins, and isoflavones, with antioxidant properties and potential anti-glycemic and anti-cancer properties (Zhao et al., 2009; Tarzi et al., 2012; Rachwa-Rosiak et al., 2015; Domínguez et al., 2016; Wallace et al., 2016; Domínguez-Arispuro et al., 2018; Kaur and Prasad, 2021; Arora et al., 2023; Devi et al., 2023; Xiao et al., 2023). Recognizing that staple cereals alone cannot meet diverse micronutrient needs, supplementation with grain legumes like chickpea can provide essential micronutrients, fiber, and low glycemic index foods to combat nutritional insecurity and other health-related problems, particularly diabetes and obesity (Rebello et al., 2014; Ramani et al., 2021; Arya and Kumar, 2022; Gupta et al., 2023a). This review examines chickpea’s nutritional components, genetic determinants, and genomic regions contributing to improved nutrition. It also explores how emerging breeding tools and the CRISPR/Cas9 approach could enhance chickpea biofortification to help sustain global food security, address micronutrient deficiencies, mitigate malnutrition, and diversify food resources while supporting modern cropping systems and agricultural sustainability.
Harnessing genetic variability for improving seed protein
Chickpea contains high-quality protein with exceptional bioavailability (Sharma et al., 2016; Kaur et al., 2019; Kumar et al., 2021), making them a potential solution for addressing protein malnutrition in developing countries by providing a high protein percentage (Iqbal et al., 2006). While breeders have made significant strides in developing over 200 commercially viable chickpea cultivars with improved yield and resistance to biotic and abiotic stresses (Grewal et al., 2023), the focus on quality improvement, particularly seed protein content (SPC), has been somewhat overlooked. Consequently, information on SPC across different chickpea cultivars remains incomplete, necessitating focused breeding efforts and genomics research to develop superior chickpea genotypes capable of addressing the mounting challenges of malnutrition. Research on genetic variability has uncovered a wide spectrum of SPC in chickpea. Singh et al. (2010) noted SPC ranging from 15.7–31.5%, while Özer et al. (2010) reported 17.55–23.31% in 91 kabuli chickpea genotypes, highlighting high-protein genotypes like Hatay2 (23.3%), Hatay3 (23.9%), and Mersin 4 (23.23%). Gaur et al. (2016) explored the genetic control of SPC in an ICC5912 × ICC17109 F2 population, reporting substantial genetic variability in pink-flowered segregants (12.84–26.72%), blue-flowered segregants (17.10–26.08%), and white-flowered segregants (14.77–24.10%). Phenotypic assessments across diverse chickpea germplasm indicated significant genetic variability for SPC, ranging from 15.6–22.4% (Sharma et al., 2013; Misra et al., 2016; Upadhyaya et al., 2016b), 4.6–33.9% (Bhagyawant et al., 2018), 16.56–24.64% (Roorkiwal et al., 2022), 16.3–26.2% (Srungarapu et al., 2022a), and 20–24.8% (Onder et al., 2023). Similarly, Samineni et al. (2022) assessed SPC and micronutrients in 140 chickpea genotypes, revealing variability ranging from 11.6–24.8% under non-stress conditions, 15.7–26.2% under drought stress, and 15.9–24.7% under heat stress. A comprehensive analysis of 402 chickpea genotypes at two locations in India (Ludhiana and New Delhi) indicated SPC variability ranging from 18.19–33.56%, with the highest values in GNG 2144 (28.81%), H 82-2 (28.29%), Pusa 5023 (26.28%), and Pusa 417 (26.00%) (Grewal et al., 2023). Interestingly, protein contents among commercially cultivated varieties ranged from 18.19–30.44%, relatively lower than germplasm accessions (19.41–33.56%) (Grewal et al., 2023). Farida Traore et al. (2022) reported broad variability in SPC (18.9–32.4%) in 282 advanced chickpea breeding lines in Morocco. In another study, SPC varied between desi and kabuli types, with desi types generally lower (20.29%) than kabuli types (24.51%) (Sharma et al., 2013; Ghribi et al., 2015). Comparative studies have also revealed differences in SPC between kabuli (23.15–25.97%), desi (23.19–28.0%), and wild species (23.03–26.82%) (see Supplementary Table S1), with one study identifying GL 12021 and GNG 2171 (desi), L552 (kabuli), and C. pinnatifidum ILWC 261 (wild) as high-protein lines (Kaur et al., 2019).
Chickpea wild relatives are valuable reservoirs for agriculturally significant traits, including SPC (Ocampo et al., 1998; Sharma et al., 2021). Ocampo et al. (1998) assessed 228 chickpea accessions from eight annual wild species, reporting significant genetic variability for SPC, ranging from 16.8% (C. cuneatum) to 26.8% (C. pinnatifidum). Of the wild species, C. yamashitae exhibited the highest SPC (21.7%), while C. echinospermum had the lowest (19.2%) (Ocampo et al., 1998) (see Supplementary Table S1). A subsequent analysis of 41 cultivated chickpea accessions and the eight annual wild Cicer species unveiled significant variation in SPC (21–25.5%), with the highest in ICC17141 (C. chorassanicum), followed by ICC17124 (C. reticulatum; 25.1%), ICC17262 and ICC17261 (both C. reticulatum; 24.5%), and ICC20236 (C. chorassanicum; 24.3%) (Sharma et al., 2021). Despite identifying numerous chickpea genotypes with high protein content, it has been found that grain yield negatively correlates with seed protein (Srungarapu et al., 2022b), indicating a tradeoff between protein content, seed size, and grain yield (Gaur et al., 2016). Nonetheless, selecting transgressive segregants with high SPC and moderate grain yield holds promise for developing chickpea cultivars with improved SPC (Gaur et al., 2016). Commercially released cultivars with high SPC, viz., GNG 2144, H-82-2, L 144, Pusa 112, and Pusa 5023 (Grewal et al., 2023), could be potential donors in future chickpea breeding programs aimed at enhancing SPC content and yield.
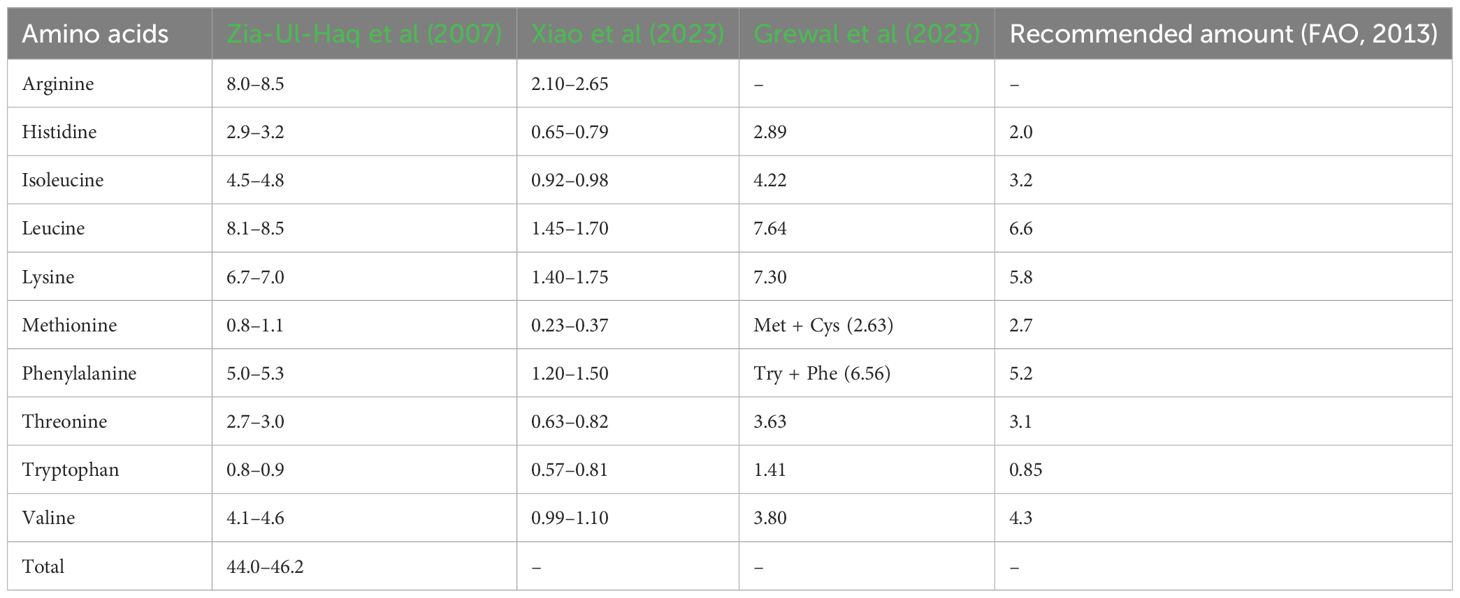
Chickpea proteins mainly comprise albumin (8–12%), globulin (53–60%), prolamin (3–7%), and glutelin (19–25%) (Day, 2013; Grasso et al., 2022), with significant variability observed between desi and kabuli types. Chickpea flour exhibits elevated levels of essential amino acids (39.89 g 100 g–1 protein) compared to wheat flour (32.20 g 100 g–1 protein), including arginine, aspartic acid, and glutamic acid, with a combined 36.85 g 100 g–1 protein for kabuli and 34.53 g 100 g–1 protein for desi varieties (Singh et al., 2010; Begum et al., 2023). Notably, chickpea contains essential amino acids above the recommended amounts by the WHO, including lysine (6.2–6.7 g 16 g–1 N; recommended 1.8), isoleucine (3.1–4.0 g 16 g–1 N; recommended 1.5), leucine (6.5–7.1 g 16 g–1 N; recommended 2.1), phenylalanine (5.1–5.8 g 16 g–1 N; recommended 2.1), threonine (3.1–3.4 g 16 g–1 N; recommended 1.1), valine (3.5–4.1 g 16 g–1 N; recommended 1.5), and histidine (2.3–2.8 g 16 g–1 N; recommended 1.5) (Onder et al., 2023). Moreover, selected commercially released chickpea cultivars exhibited higher essential amino acid contents than the WHO recommendations (see Table 1) (Grewal et al., 2023). Desi types have higher methionine (1.4%) than kabuli types (1.1%) (see Supplementary Table S1), with notable differences also reported for leucine, lysine, and serine (Ghribi et al., 2015; Ramani et al., 2021). Grewal et al. (2023) reported significant positive associations between SPC and aspartic acid, isoleucine, and phenylalanine but negative associations with methionine and cysteine. Therefore, cultivars with high amino acid content could be used to develop future biofortified chickpea cultivars to combat protein deficiency malnutrition. Among the major chickpea proteins, albumins (water-soluble proteins) are rich in sulfur-containing amino acids like methionine and cysteine (Bhatty, 1982), while globulins are salt-soluble proteins (Osborne, 1924), with legumin and vicilin the major globulins in chickpea (Boye et al., 2010; Day, 2013). Legumin contains higher methionine and cysteine levels than vicilin (Shevkani et al., 2019). Glutelins are soluble in dilute acid or alkali and contain methionine and cysteine (Singh and Jambunathan, 1982). Prolamins (alcohol-soluble) feature a high proportion of proline and glutamine (Osborne, 1924; Rachwa-Rosiak et al., 2015).
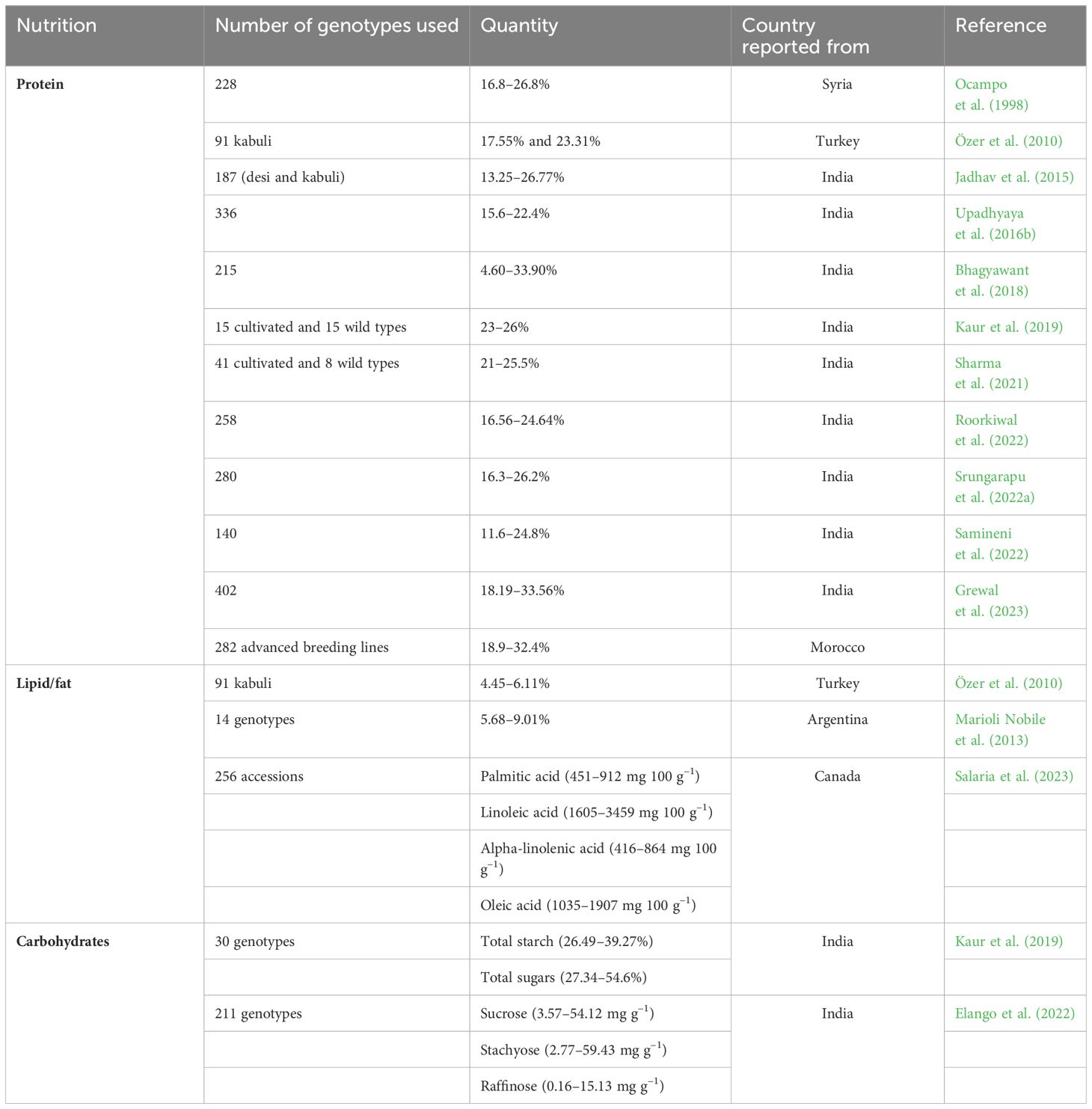
Table 1 Genetic variability for seed protein/lipid/carbohydrate content in chickpea from different countries.
Carbohydrate composition
Chickpea seed typically contains 50–58% carbohydrates (Jukanti et al., 2012), with starch being the primary component (41–50.8%) (Singh, 1985). Additionally, chickpea seed contains cellulose (4–13%), hemicellulose (3.5–8.8%), and pectin (1.5–3.8%) (Wood and Grusak, 2007). Kabuli and desi chickpea varieties contain amylose and amylopectin, with both types containing more amylopectin than amylose (Singh et al., 2004) and kabuli types typically exhibiting higher amylose contents than desi types (Grasso et al., 2022). Thus, chickpea starch has a low glycemic index, making it suitable for diabetic individuals (Kaur and Prasad, 2021). The characteristics of chickpea starch depend on factors like amylose content, swelling power, solubility, and water-binding capacity (Miao et al., 2009). Desi chickpea types typically exhibit total and apparent amylose contents of 35.24% and 31.11%, respectively, while kabuli types have 31.80% and 29.93%, respectively (Miao et al., 2009). Kabuli starch has a higher Mw (5.382×107 g mol–1) than desi starch (3.536×107 g mol–1) (Miao et al., 2009). Genetic variability in starch content among 91 kabuli chickpea landraces from Turkey ranged from 40.07–41.76% (Özer et al., 2010). An assessment of four selected chickpea genotypes revealed significant genetic variability for starch (30.6–49.9%), glucose (3.4–4.7%), fructose (4.4–5.5%), and sucrose (2.4–4.1%) (Ereifej et al., 2001). In addition to starch, chickpea carbohydrates comprise dietary fiber (18–22%), insoluble sugars (10–18% crude fiber), and soluble sugars (4–8%) (Tosh and Yada, 2010), with desi types exhibiting higher insoluble dietary fiber than kabuli types due to thicker seed coats (Rincon et al., 1998). Sánchez-Mata et al. (1998) reported various monosaccharide concentrations in chickpea, including ribose (0.11 g 100 g–1), galactose (0.7 g 100 g–1), glucose (0.05 g 100 g–1), and fructose (0.25 g 100 g–1). In one study, chickpea fiber ranged from 4.9–5.5% (Ereifej et al., 2001), while another reported 2.88% in chickpea genotypes originating from Sicily (Patane, 2006). In a study investigating soluble sugars, fructose content ranged from 3.66–4.33 mg g–1 in kabuli types, 2.0–5.33 mg g–1 in desi types, and 5.0–10.33 mg g–1 in wild chickpea species (Kaur et al., 2019). They also noted positive association of soluble sugar with starch content.
Sucrose, raffinose, verbascose, and stachyose form the primary soluble sugars present in chickpea seeds (Chibbar et al., 2010). Raffinose and stachyose are galactosyl derivatives of sucrose, with raffinose having one galactose moiety and stachyose having two galactose moieties attached to sucrose via α (1-6) glycosidic linkage (Chibbar et al., 2010; Kaur et al., 2019). These compounds are classified as raffinose family oligosaccharides (RFOs), which are non-digestible by humans and animals, often causing flatulence (Elango et al., 2022). Chickpea oligosaccharide levels varied among different cultivars, ranging from 5.54–8.82%, including 0.25–0.73% verbascose, 1.54–3.18% stachyose, 2.04–5.26% ciceritol, and 0.42–0.86% raffinose (Tosh and Yada, 2010; Singh and Kumari, 2011). Genetic variability for RFOs in chickpea has been noted, ranging from 10.33 mg g–1 in wild chickpea (accession EC 366342) to 0.66 mg g–1 in kabuli chickpea (GLK 14313) (Kaur et al., 2019). Taking into account the association of RFO with other sugars, RFO revealed a positive and significant correlation with raffinose, stachyose, and verbascose in both desi and kabuli types. Similarly, sucrose also exhibited a positive and significant correlation with raffinose, stachyose, and verbascose in both desi and kabuli types (Gangola et al., 2013).
Lipid/fat/oil composition
Fatty acids are classified into saturated fatty acids, monounsaturated fatty acids (MUFAs), and polyunsaturated fatty acids (PUFAs). Chickpea oil predominantly comprises unsaturated fatty acids (Gul and Egesel, 2008). Among grain legumes, chickpea has the highest fat (6.04%) (Allen et al., 2006). The lipid profile of chickpea includes storage lipids, primarily triacylglycerols, and membrane lipids like phospholipids, sphingolipids, glycolipids, and phytosterols (Madurapperumage et al., 2021). Both triacylglycerols and membrane lipids contribute to chickpea’s overall fat content (Zia-Ul-Haq et al., 2007; Madurapperumage et al., 2021). Chickpea seeds typically contain 56–67% triacylglycerols and 17–20% phospholipids (Zia-Ul-Haq et al., 2007). The recommended daily dietary allowance for infants and adults is 13.5 g palmitic acid, 7.5 g linoleic acid, 1.2 g alpha-linolenic acid, and 24 g oleic acid (Ervin et al., 2004). Daily consumption of chickpeas can provide 19.5 g of unsaturated fatty acids (Wallace et al., 2016). Interestingly, uncooked chickpeas contain more polyunsaturated fatty acids (2.73 g 100 g–1) than cooked chickpeas (1.15 g 100 g–1). Additionally, chickpea consumption contributes to lowering the intake of saturated fatty acids (22.4 g day–1), total fats (76.4 g day–1), and cholesterol (227 mg day–1) (Wallace et al., 2016). Various studies have reported significant variation in grain fat, ranging from 4.45–7.1% (Jood et al., 1998; Ereifej et al., 2001; Ravi and Harte, 2009; Shad et al., 2009; Özer et al., 2010). For instance, Özer et al. (2010) reported 4.45–6.11% grain fat in 91 Turkish kabuli chickpea samples, while Jood et al. (1998) reported 6–6.33% grain fat in Indian chickpea genotypes.
Chickpea oil content ranges from 3.8–10% (Gul and Egesel, 2008; Jukanti et al., 2012). The prominent fatty acids in chickpea oil are linolenic, oleic, and palmitic acids, with linoleic acid (18:2) the most abundant, ranging from 52.36–57.98% (Gul and Egesel, 2008). Omega-6 linoleic fatty acid constitutes the majority of chickpea oil (46–62%), followed by omega-9 oleic acid (24.12–29.48%) (Wood and Grusak, 2007; Gul and Egesel, 2008) and palmitic acid (16:0) (10.86–12.38%) (Gul and Egesel, 2008). Analysis of 14 Argentinean chickpea genotypes revealed oil contents ranging from 5.68–9.01%, with genotypes G101, P39, and L9 containing the most linoleic acid (mean 58.73%), linolenic acid (mean 3.06%), and oleic acid (18:1 omega-9) (mean 43.01%), respectively (Marioli Nobile et al., 2013). A fatty acid composition analysis in 256 chickpea accessions revealed wide genetic variability for palmitic acid (451–912 mg 100 g–1), linoleic acid (1605–3459 mg 100 g–1), alpha-linolenic acid (416–864 mg 100 g–1), and oleic acid (1035–1907 mg 100 g–1) (Salaria et al., 2023). A correlation study conducted by the same research group revealed a significant negative correlation between palmitic acid and linoleic acid, as well as between alphalinolenic acid and oleic acid. Further research on fatty acid composition in wild chickpea species could enhance our understanding and facilitate the development of biofortified chickpea varieties with improved fatty acid profiles to address nutritional needs among the human population.
Micronutrients
Chickpea seeds are a rich source of essential micronutrients like Fe, Zn, calcium (Ca), copper (Cu), and manganese (Mn) (Ibrikci et al., 2003; Jahan et al., 2019; Sharma et al., 2021; Roorkiwal et al., 2022; Srungarapu et al., 2022a). Feeding studies have indicated that daily chickpea consumption can provide 17.4 mg Fe (Wallace et al., 2016). Uncooked chickpea contains higher Zn (2.76 mg 100 g–1) than cooked chickpea (1.53 mg 100 g–1). Moreover, consuming 100 g of raw chickpeas can provide 160 mg Ca, 138 mg Mg, 4.1 mg Zn, and 5.0 mg Fe (Jukanti et al., 2012; Begum et al., 2023), highlighting the benefits of consuming uncooked chickpea for micronutrient uptake. The average Fe content in chickpea ranges from 3.0–14.3 mg 100 g–1 (Diapari et al., 2014). A recent study reported a wide range of genetic variability for grain Fe (0.50–8.54 mg 100 g–1) and Zn (1.10–5.91 mg 100 g–1) in 402 chickpea accessions across two locations, with Heera, H82-2, and H214 identified as promising genotypes for high Zn (>4 mg 100 g–1) and L550, KGD1168, PG114, JG74, and ICCV6 for high Fe (>6 mg 100 g–1) (Grewal et al., 2020). Samineni et al. (2022) investigated associations between marker traits and grain Fe and Zn contents in 140 chickpea accessions, revealing significant genetic variation for grain Fe (47.8–83.0, 49.4–86.2, and 41.4–77.6 mg kg–1 under non-stress, drought stress, and heat stress, respectively) and Zn (29.5–55.0, 28.1–63.1, 29.7–55.4 mg kg–1 under non-stress, drought stress, and heat stress, respectively). Similarly, Srungarapu et al. (2022a) noted wide genetic variability for grain Fe (44.1–76.7 mg kg–1) and Zn (36.3–56.2 mg kg–1) with high heritability (85–98%) in a two-year assessment of 280 chickpea accessions. However, grain Fe exhibited significant positive associations with Zn and 100-seed weight but a highly negative correlation with yield under all tested environments, consistent with findings by Samineni et al. (2022). Similarly, grain protein content negatively correlated with grain yield across all environments, suggesting that breeding programs should focus on selecting genotypes with moderate yield and high nutrient content, especially protein and micronutrients, rather than solely increasing nutrient content (Srungarapu et al., 2022a). Moreover, Roorkiwal et al. (2022) reported significant genetic variation in 258 chickpea accessions for various nutritional components: Fe (2.26–7.25 mg 100 g–1, mean: 4.36), Ca (60.7–176.5 mg 100 g–1, mean: 108.6), Mg (64.08–134.57 mg 100 g–1, mean: 100.35), Mn (0.67–3.73 mg 100 g–1, mean: 1.78), phytic acid (2.07–19.38 mg g–1, mean: 10.68), and Zn (1.15–4.59 mg 100 g–1, mean: 2.76. Similarly, Fayaz et al. (2022) reported significant genetic variation in various micronutrients in 147 chickpea genotypes over a two-year evaluation: Zn (1.45–20.49 and 0.936–20.58 mg 100 g–1, Fe (7.93–19.82 and 10.36–12.72 mg 100 g–1), Cu (0.42–18.54 and 0.366–19.03 mg 100 g–1), and Mn (0.907–5.43 and 0.48–12.28 mg 100 g–1) and Farida Traore et al. (2022) reported a wide range of genetic variability in 282 advanced chickpea breeding lines for Fe (3.12–8.1 mg 100 g–1) and Zn (3.21–8.61 mg 100 g–1). A correlation analysis revealed significant positive associations between grain protein content and Ca, Mn, Zn, β-carotene, and phytic acid contents but negative associations between β-carotene and Mn and between phytic acid and vitamin B1 (Roorkiwal et al., 2022). Another study reported that grain protein content negatively correlated with Zn content (Farida Traore et al. (2022), with Fe and Mn content (Sharma et al., 2021). These findings suggest an opportunity to target nutrient improvement in chickpea breeding programs.
A comprehensive analysis of 41 cultivated chickpea accessions and eight annual wild Cicer species revealed significant variation in Fe (48.6–166 mg kg–1) and Zn (35.3–47 mg kg–1) in six wild Cicer species—C. reticulatum, C. echinospermum, C. bijugum, C. pinnatifidum, C. chorassanicum, and C. yamashitae—compared to cultivated chickpeas (∼42 mg kg–1 for Fe and ∼28 mg kg–1 for Zn) (Sharma et al., 2021). The wild species also had significantly higher seed Mn (ranging from 57.9 mg kg–1 in C. chorassanicum to 162 mg kg–1 in C. pinnatifidum) than cultivated chickpea (37.1 mg kg–1). The wild species, except for C. echinospermum (3.1 mg kg–1) and C. bijugum (3.6 mg kg–1), also had significantly higher seed Cu, ranging from 4.3 mg kg–1 in C. reticulatum to 7.7 mg kg–1 in C. judaicum, than cultivated chickpea (3.4 mg kg–1). The eight wild Cicer species had significantly higher seed Ca, ranging from 3.02 g kg–1 in C. echinospermum to 6.09 g kg–1 in C. chorassanicum, than cultivated chickpea (2.22 g kg−1), and five wild Cicer species—C. bijugum, C. chorassanicum, C. judaicum, C. pinnatifidum, and C. reticulatum—had significantly higher seed Mg (1.65–1.83 g kg–1) than cultivated chickpea (1.41 g kg–1) (Sharma et al., 2021). The wide range of genetic variability in micronutrients found in various chickpea landraces and wild species presents opportunities for developing chickpea varieties to combat micronutrient-related ‘hidden hunger’.
Vitamins
Chickpea is recognized as an important source of various vitamins, including niacin, riboflavin, folic acid, thiamin, and β-carotene (Cabrera et al., 2003; Abbo et al., 2005; Jha et al., 2015; Rezaei et al., 2016).
β-carotene (precursor of vitamin A)
Chickpea contains significant amounts of β-carotene, the precursor of vitamin A, with reported wide ranges in carotenoid content from 22 μg g–1 (yellow cotyledon kabuli) to 44 μg g–1 (green cotyledon desi) (Rezaei et al., 2016). Lutein and zeaxanthin are the predominant components of carotenoids present in chickpea seeds, with green cotyledon chickpea cultivars exhibiting the highest provitamin A, including β-carotene and β-cryptoxanthin (Rezaei et al., 2016). Across five chickpea cultivars, CDC Jade had the highest lutein and β-cryptoxanthin in cotyledons at 16 days post-anthesis, while CDC Cory had the highest zeaxanthin and β-carotene (Rezaei et al., 2016). The same research group reported a wide range of total carotenoids in three segregating F2 populations (CDC Jade × CDC Frontier, CDC Cory × CDC Jade, and ICC4475 × CDC Jade), ranging from 10.6–40 µg g–1 in parental lines and 18.46–77.63 µg g–1 in segregating populations (Rezaei et al., 2019). Subsequently, Roorkiwal et al. (2022) identified wide genetic variation in β-carotene (0.003–0.104 mg 100 g–1) in 258 chickpea accessions using GWAS. Cooking chickpeas significantly reduces their vitamin A content from 67 IU 100 g–1 (uncooked) to 27 IU 100 g–1 (cooked) (Wallace et al., 2016).
Vitamin B complex
Chickpea exhibits a wide range of genetic variation for various B vitamins, including thiamine (B1), riboflavin (B2), and niacin (B3). They are rich sources of thiamin (453 μg) and riboflavin (173 μg) (Hall et al., 2017). A recent GWAS assessing 258 chickpea accessions revealed significant variability for these vitamins, ranging from 0.055–0.502 mg 100 g–1 thiamine (average 0.189), 0.011–0.638 mg100 g–1 riboflavin (average 0.111), and 0.116–1.57 mg100 g–1 niacin [average 0.411 (Roorkiwal et al., 2022)]. Moreover, a comparative analysis of four chickpea genotypes revealed significant variability for thiamine, ranging from 0.31–0.36 mg 100 g–1 (Xiao et al., 2023). Moreover, one study reported significant genetic variation in various B vitamins between desi and kabuli types, with 1.72 and 1.22 mg 100 g–1 niacin, 0.21 and 0.26 mg 100 g–1 riboflavin, 0.29 and 0.49 mg 100 g–1 thiamin, and 0.30 and 0.38 mg 100 g–1 pyridoxine, respectively (Jukanti et al., 2012). However, cooking chickpeas reduces the levels of these vitamins significantly from 1.54 to 0.52 mg 100 g–1 niacin, 0.2 to 0.063 mg 100 g–1 riboflavin, and 1.58 to 0.28 mg 100 g–1 pantothenic acid (Wallace et al., 2016).
Folic acid
Pulses are a significant source of dietary folate, with a wide range of folate levels documented across various pulse crops, including lentil (146–290 μg 100 g–1), yellow pea (50–202 μg 100 g–1), mung bean (141–169 μ 100 g–1), cowpea (96 μg 100 g–1), and common bean (103 μg 100 g–1) (Sen Gupta et al., 2013; Hefni and Witthoft, 2014). Folate content in chickpea ranges from 42–537 μg 100 g–1 (Hall et al., 2017), with cooking reducing the level from 557 μg 100 g–1 (uncooked) to 172 μg 100 g–1 (cooked) (Wallace et al., 2016). Moreover, chickpea consumers intake 627 µg day–1 folate compared to non-chickpea consumers (Wallace et al., 2016). Jha et al. (2015) used ultra-performance liquid chromatography and mass spectrometry to quantify six folate monoglutamates in four chickpea genotypes, revealing substantial genetic variability for folate (351–589 μg 100 g–1), with 5-methyltetrahydrofolate (5-MTHF) and 5-formyltetrahydrofolate (5-FTHF) the most abundant. Moreover, the study identified a significant effect of location and a location×cultivar interaction, suggesting that folate levels are responsive to genotype×environment interactions (Jha et al., 2015).
Vitamin E
Chickpea oil is particularly rich in tocopherols, with alpha-tocopherol the highest among pulses, reaching up to 13.7 mg 100 g–1 (Wood and Grusak, 2007; Pittaway et al., 2008). The oil comprises four different forms of tocopherols—alpha, beta, gamma, and delta (Zia-Ul-Haq et al., 2009), with gamma-tocopherol recognized as a natural seed antioxidant (Gul and Egesel, 2008; Boschin and Arnoldi, 2011). The total tocopherol in chickpea oil ranges from 20.69–52.44 mg kg–1 (Gul and Egesel, 2008), with higher amounts of vitamin E in uncooked chickpea (0.82 mg 100 g–1) than cooked chickpea (0.35 mg 100 g–1) (Wallace et al., 2016). The National Health and Nutrition Examination Survey reported that chickpea consumers intake 10.1 μg day–1 more vitamin E than non-chickpea consumers (Wallace et al., 2016). The significant genetic variability observed for these vitamins underscores the potential of chickpea for addressing nutritional deficiencies, especially in regions where vitamin-related deficiencies are prevalent.
Bioactive compounds
Chickpea is rich in bioactive compounds, including antioxidants, phenolic acids, flavonoids, and condensed tannins, which offer numerous health benefits (Heiras-Palazuelos et al., 2013). These compounds play significant roles in physiological and metabolic processes and contribute to reducing the risk of various diseases. Antioxidants in small quantities prevent the formation of free radicals or reactive oxygen species by retarding the oxidation of unsaturated fats, which are easily oxidized. Desi and kabuli chickpea exhibit varying antioxidant activities (Heiras-Palazuelos et al., 2013).
Phenols, essential bioactive compounds found in the chickpea seed coat (Xu et al., 2007), include phenolic acids, flavonoids, and condensed tannins (Singh et al., 2017; de Camargo et al., 2022), which offer various health benefits, including anti-carcinogenic, anti-thrombotic, anti-ulcer, anti-atherogenic, anti-allergenic, anti-inflammatory, antioxidant, and immune-modulating properties (de Camargo et al., 2022; Begum et al., 2023). Total phenolic contents in chickpea range from 27.48–48.01 mg 100 g–1 for kabuli types, 38.59–83.52 mg 100 g–1 for desi types, and 63.08–113.30 mg 100 g–1 for wild species (Kaur et al., 2019). They also observed that genotypes with higher phenol content exhibited greater antioxidant activity, suggesting a positive association between seed phenol content and antioxidant activity.A study using UPLC-MS/MS identified three specific phenolic acids in chickpea—taxifolin, biochanin, and m-hydroxybenzoic acid—that decrease oxidative damage in human HuH-7 cells induced by peroxy radicals, indicating hepatoprotective properties (de Camargo et al., 2022).
Anti-nutrients
Chickpea seeds also contain substances like tannins, protease inhibitors (such as trypsin and amylase inhibitors), phytic acid, and saponins (Soetan and Oyewole, 2009; Kaur et al., 2014, Kaur et al, 2019) that function as anti-nutrients by inhibiting the bioavailability of various nutrient components (Soetan and Oyewole, 2009). Tannins are recognized as significant anti-nutritional compounds, binding with enzyme proteins or minerals, resulting in the inactivation of digestive enzymes and reduced protein digestion (Kaur et al., 2019). Various studies have reported tannin contents in chickpea seeds ranging from 5.44–10.87 mg g–1 (Kaur et al., 2014), 17.52 mg g–1 (Bulbula and Urga, 2018), and 11.93 mg g–1 in desi types, 10.63 mg g–1 in kabuli types, and 16.38 mg g–1 in wild species (Kaur et al., 2019). Chickpea genotypes also varied in tannin contents, ranging from 9.08 mg g–1 in GL 14015 to 18.47 mg g–1 in C. judaicum ILWC 30 (Kaur et al., 2019).
Phytic acid forms complexes with proteins, impeding the absorption of micronutrients such as Fe, Ca, Zn, Cu, and Mg in the gastrointestinal tract (Tiwari and Singh, 2012). Studies have reported phytic acid levels ranging from 3.49–11.52 mg g–1 in desi types and 3.45–12.35 mg g–1 in kabuli types (Mondor et al., 2009), 11.33 mg g–1 in whole chickpeas, 11.53 mg g–1 in split chickpeas, and 14 mg g–1 in desi types (Shi et al., 2018), and 9.43–13.67 mg g–1 in kabuli types, 8.48–18.39 mg g–1 in desi types, and 4.24–8.48 mg g–1 in wild species (Kaur et al., 2019). Phytic acid has a negative association with most minerals except Zn; an increase in phytic acid suggests a negative impact on the absorption of these minerals (Kaur et al., 2019).
Saponins, naturally occurring surface-active glycosides, also inhibit nutrient absorption and bioavailability in chickpea (Choudhary et al., 2015; Katoch et al, 2016; Kaur et al., 2019; Singh et al., 2022). They contribute to enzyme inactivation, significantly impacting cellular metabolism and reducing nutrient absorption. Studies have reported saponin levels ranging from 4.98–12.23 mg g–1 in kabuli types (Choudhary et al., 2015) and 7.22 mg g–1 in desi types, 7.02 mg g–1 in kabuli types, and 8.38–9.68 mg g–1 in wild species (Kaur et al., 2019).
Trypsin inhibitors hinder digestive enzymes, specifically trypsin and chymotrypsin, affecting the utilization of sulfur amino acids in the body. Genetic variability for trypsin inhibitors in chickpea ranges from 111.5–218.4 trypsin inhibitor units (TIU) g–1 (Gupta et al., 2017), 38.53–64.47 TIU g–1 in kabuli types, 32.91–112.32 TIU g–1 in desi types, and 122.73–150.18 TIU g–1 in wild species (Kaur et al., 2019). Thus, minimizing anti-nutrient contents through breeding, genomics, and other innovative approaches could improve the bioavailability of essential micronutrients, vitamins, phosphorus, and other nutrients, enhancing their nutritional values and health benefits (Table 2).
Genomic resources for improving nutrients
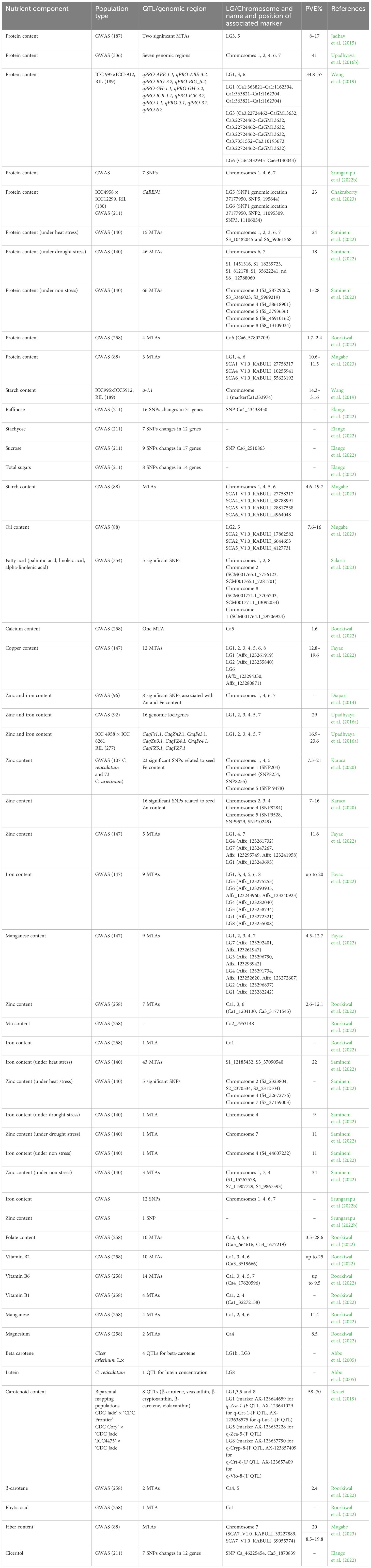
Advancements in genome sequencing technologies have revolutionized chickpea breeding by providing extensive genomic resources, including decoding the chickpea genome sequence, whole genome resequencing, pan-genome assembly, and transcriptome assembly for various traits (Varshney et al., 2013; Varshney et al, 2021). Leveraging these advanced genomic resources has made it possible to dissect various traits of agronomic importance, including nutrient components in chickpea. For example, Wang et al. (2019) genotyped a bi-parental mapping population developed from ICC5912 × ICC995 across four environments, identifying several SPC QTL explaining 34.8–57% phenotypic variation explained (PVE) with the quantitative trait loci (QTL) q-3.2 identified as the major seed protein QTL. Sab et al. (2020) identified 11 QTLs for seed Fe concentration on CaLG03, CaLG04, and CaLG05 using a F2:3 derived from MNK-1 and Annigeri 1, which explained 7.2% (CaqFe3.4) to 13.4% (CaqFe4.2) PVE. Further, eight QTLs for seed Zn, reported on CaLG04, CaLG05, and CaLG08, explained 5.7% (CaqZn8.1) to 13.7% PVE (CaqZn4.3) (Sab et al., 2020). Introgression of the QTL-hotspot region on CaLG04 that harbors several drought tolerance-related QTLs (Varshney et al., 2019) may increase seed Fe and Zn, with three QTLs for seed Fe and Zn (CaqFe4.4, CaqFe4.5, and CaqZn4.1) co-localized in this region (Sab et al., 2020). The authors identified genes in the QTL regions that encode Fe–S metabolism and Zn-dependent alcohol dehydrogenase activity on CaLG03, Fe ion binding oxidoreductase on CaLG04, and Zn-induced facilitator-like protein and ZIP zinc/iron transport family protein on CaLG05. Whole genome resequencing data identified 48 SNPs associated with targeted sugar types and nine genes “(Ca_06204, Ca_04353, and Ca_20828: Phosphatidylinositol N-acetylglucosaminyltransferase; Ca_17399 and Ca_22050: Remorin proteins; Ca_11152: Protein-serine/threonine phosphatase; Ca_10185, Ca_14209, and Ca_27229: UDP-glucose dehydrogenase)” as potential candidates for sugar metabolism and transport in chickpea (Elango et al., 2022). A GWAS using 16,376 single nucleotide polymorphisms (SNP) markers identified seven genomic regions associated with SPC in 336 chickpea accessions, explaining a combined 41% PVE (Upadhyaya et al., 2016b) (see Table 3).
Furthermore, a combined QTL-seq and GWAS approach identified two major QTLs that regulate SPC on LG5 and LG6 (Chakraborty et al., 2023). The highly significant SNPs and genomic region controlling SPC were validated in 211 chickpea accessions, indicating the tight association of an SNP marker with the “CaREN1 (ROP1 ENHANCER1)” genomic region, explaining 23% PVE. CaREN1 knockdown significantly reduced SPC, confirming the role of CaREN1 in governing SPC (Chakraborty et al., 2023).
Diapari et al. (2014) conducted GWAS using SNP markers to elucidate genomic regions associated with grain Zn and Fe in 96 chickpea genotypes, identifying eight SNPs contributing to Zn or Fe, with one SNP on chromosome 1 associated with both. Additionally, two SNPs related to Fe and Zn were identified on chromosomes 6 and 7, and three SNPs related to Zn and two SNPs related to Fe were identified on chromosome 4 (Diapari et al., 2014) (see Table 3).
A recombinant inbred lines-based bi-parental mapping population approach using an ICC 4958 × ICC 8261 population identified eight QTLs governing seed Fe and Zn on six chromosomes, explaining a combined 39.4% PVE (Upadhyaya et al., 2016a). Furthermore, genotyping 92 sequenced desi and kabuli accessions with 24,620 SNPs identified 16 genomic loci/genes contributing to seed Fe and Zn, accounting for a combined 29% PVE (Upadhyaya et al., 2016a). Subsequently, a GWAS on a diverse panel of 147 chickpea genotypes phenotyped for two years and genotyped with an “Axiom®50K CicerSNP array” identified 35 significant marker-trait associations (MTAs) contributing to grain Zn, Fe, Cu, and Mn, with five MTAs consistently identified in different environments (stable), six explaining more than 15% of the phenotypic variation (major), and three both stable and major MTAs (Fayaz et al., 2022). Likewise, over two years, SNP204 on LG1 and SNP9478 on LG5 showed significant MTAs for Fe content at Sanliurfa, and SNP8254 and SNP8255 on LG4 showed significant MTAs for Fe content at Bornova (Karaca et al., 2020). A GWAS on 258 chickpea genotypes using 318,644 SNPs derived from whole genome sequencing revealed 62 significant MTAs for 12 important nutritional traits, including crude protein, β-carotene, seed Ca, and folate content, on chromosomes Ca1, Ca3, Ca4, and Ca6, explaining up to 29% PVE (Roorkiwal et al., 2022). A GWAS on a reference set of 280 chickpea genotypes using a 5k SNP array and the FarmCPU and BLINK models identified seven significant SNPs for grain protein, 12 SNPs for Fe, and one SNP for Zn on chromosomes 1, 4, 6, and 7 (Srungarapu et al., 2022b). Another GWAS analysis identified 181 MTAs for grain protein, Zn, and Fe content in 140 diverse chickpea genotypes under non-stress, drought, and heat stress conditions, with 48 and 63 MTAs significantly associated with drought stress and heat stress, respectively (Samineni et al., 2022). Thus, targeting the identified overlapping/common genomic regions controlling these micronutrients for cloning could help elucidate the precise functions of candidate genes associated with nutrient content.
A GWAS on 354 kabuli and desi chickpea genotypes highlighted the significance of key grain fatty acids, including palmitic, linoleic, alpha-linolenic, and oleic acids, identifying five significant SNPs on chromosomes 1, 2, and 8 strongly associated with palmitic acid (Salaria et al., 2023). Another GWAS using 36,645 SNP markers derived from 88 chickpea accessions uncovered MTAs for starch on chromosome 2 (12% PVE), fiber content on chromosome 6 (20% PVE), oil content on chromosome 7 (11% PVE), and grain protein on chromosome 1 (11% PVE) (Mugabe et al., 2023).
Functional genomics approaches for discovering candidate gene(s) contributing to nutrients
Advances in functional genomics, particularly RNA-seq-based transcriptome assembly, have greatly enhanced our ability to identify trait-based candidate genes in chickpea (Kudapa et al., 2018). For instance, quantitative RT-PCR can be used to profile the expression of candidate genes for SPC. Notably, GWAS revealed higher differential upregulatory expression in high SPC-containing mapping individuals (21.5–22.4%) than in low SPC-containing mapping individuals (15.6–16.5%) during the seed development stage (Upadhyaya et al., 2016b). Similarly, functional analysis of the SPC gene ROP1ENHANCER1, identified through combined QTL-seq and candidate gene-based association mapping, demonstrated a significant reduction in SPC when this gene was knocked down in chickpea (Chakraborty et al., 2023). Similarly, Upadhyaya et al. (2016a) used qRT-PCR to explore the functional expression of candidate genes related to grain Fe and Zn, reporting high expression in the seeds of parental chickpea genotypes with high Fe and Zn compared to those with low Fe and Zn.
Furthermore, investigating the potential role of various transporter genes (“FRO2, IRT1, NRAMP3, V1T1, YSL1, FER3, GCN2, and WEE1”) in Fe metabolism, Jahan et al. (2023) validated their function in Fe uptake, root and stem translocation, and leaf tissue accumulation. Examining the expression patterns of identified genes related to carotenoid content, Rezaei et al. (2016) analyzed the expression of 19 selected genes associated with the carotenoid biosynthesis pathway in five chickpea cultivars, reporting up-regulatory expression in the CDC Jade cultivar.
Functional genomics advancements will potentially uncover more candidate genes associated with quality traits, offering insights into their precise functions. This knowledge could facilitate the cloning and transfer of these genes to elite chickpea cultivars.
Innovative breeding tools for improving nutritional components
Recent advances in breeding approaches, including genomic selection, speed breeding, and high-throughput phenotyping, offer promising avenues for improving the nutritional components of chickpea and developing nutritionally dense or biofortified genotypes. Genomic selection (GS) can harness high-throughput SNP markers derived from chickpea genomics resources to select progenies with superior genetic merit for various nutritional traits using prediction models trained on a large target population (Meuwissen et al., 2001). Speed breeding protocols can expedite the generation of mapping populations, such as recombinant lines and backcross populations, for mapping various nutritional component QTLs/genes (Watson et al., 2018). Advances in high-throughput phenotyping and non-destructive phenotyping, including hyperspectral imaging, Fourier transform near-infrared imaging, and micro-computed tomography imaging, offer efficient means of assessing nutritional components in chickpea (Hacisalihoglu and Armstrong, 2023). Emerging approaches like artificial intelligence and machine learning tools that use convolutional and deep neural networks could predict nutritional quality and the role of novel genes/pathways associated with various nutritional and anti-nutritional components in chickpea (Tachie et al., 2023). By integrating these innovative breeding tools into chickpea breeding programs, researchers can accelerate the development of nutritionally enhanced varieties, contributing to efforts to combat hunger and improve food security worldwide.
Scope of genome editing for improving nutrient bioavailability
Conventional breeding approaches have significantly increased the global yield and production of chickpea. Before the advent of CRISPR/Cas9 (Wang et al., 2016), other genome editing systems like zinc-finger nucleases (ZFNs) (Urnov et al., 2010), transcription activator-like effector nucleases (TALENs) (Joung and Sander, 2013), and homing endonucleases or meganucleases (Pâques and Duchateau, 2007; Daboussi et al., 2015) were used. Zinc-finger nucleases, comprising distinct DNA binding and FokI nuclease DNA cleavage domains, were among the earliest synthetic proteins used for targeted mutagenesis and gene replacement (Li et al., 1992; Carroll, 2011; Davies et al., 2017). However, they suffered from low specificity, limited efficacy, and inability to achieve gene knockout and RNA editing (Wang et al., 2016; Salsman and Dellaire, 2017; Kumar et al., 2022). Similarly, TALENs, which function as nonspecific DNA-cleaving nucleases, tether a restriction nuclease to a DNA-binding protein domain termed TAL effector (Hensel and Kumlehn, 2019) but faced similar drawbacks as ZFNs. Meganucleases, rare cutting enzymes also known as homing endonucleases (Pingoud and Silva, 2007), offered highly specific site cleavage (Khan, 2019) with low cytotoxicity. Despite their efficiency in excising large DNA sequences, challenges in manufacturing and potential off-targeting effects hindered their widespread use (Jin et al., 2016; Kumar et al., 2022).
In contrast, CRISPR/Cas9-based genome editing technology has revolutionized molecular biology by enabling precise and accurate targeted mutations in desired genomic regions (Wada et al., 2020). This breakthrough offers opportunities to improve functional quality traits, including nutrient-related parameters (Haun et al., 2014; Waltz, 2016; Sun et al., 2017; Li et al., 2018; Sánchez-León et al., 2018). Notably, a CRISPR/Cas9 protocol has been successfully established in chickpea (Badhan et al., 2021; Gupta et al., 2023a) and used to edit genes like 4-coumarate ligase (4CL) and Reveille 7 (RVE7) associated with drought tolerance (Badhan et al., 2021). Likewise, this technology has successfully knocked out chickpea phytoene desaturase (CaPDS), resulting in albino chickpea phenotypes (Gupta et al., 2023b).
Recent advances in genome editing, such as base editing and prime editing, further enhance the efficiency and precision of CRISPR/Cas9. Base editing introduces single nucleotide variants into DNA/RNA through programmable base editors (Porto et al., 2020), while prime editing enables insertions, deletions, or base conversions of up to 12 nucleotides without introducing double-strand breaks (Anzalone et al., 2019; Zong et al., 2022; Ahmad et al., 2023; Zhao et al., 2023). These technologies hold promise for addressing challenges in crop improvement, including developing biofortified chickpea varieties to combat malnutrition and promote global nutritional security. However, overcoming technical hurdles like reliable transformation and regeneration protocols, identifying genomic regions for target traits, and determining the precise metabolic pathways involved (Ku and Ha, 2020) remains crucial for realizing the full potential of genome editing in crop enhancement.
Conclusions and future perspectives
Addressing the challenges of global food security and malnutrition requires concerted efforts to enhance the nutritional quality of crops like chickpea. While progress has been made in increasing chickpea yield, there remains a need to balance this with improved nutritional traits. Thorough studies on the correlation of various quality traits, including carbohydrates, proteins, fats, and micronutrients, are crucial for integrating these factors into efforts aimed at enhancing nutritional characteristics. Recognizing the tradeoff between production and quality traits is crucial, and breeding programs should aim to find the optimal balance to meet both needs.
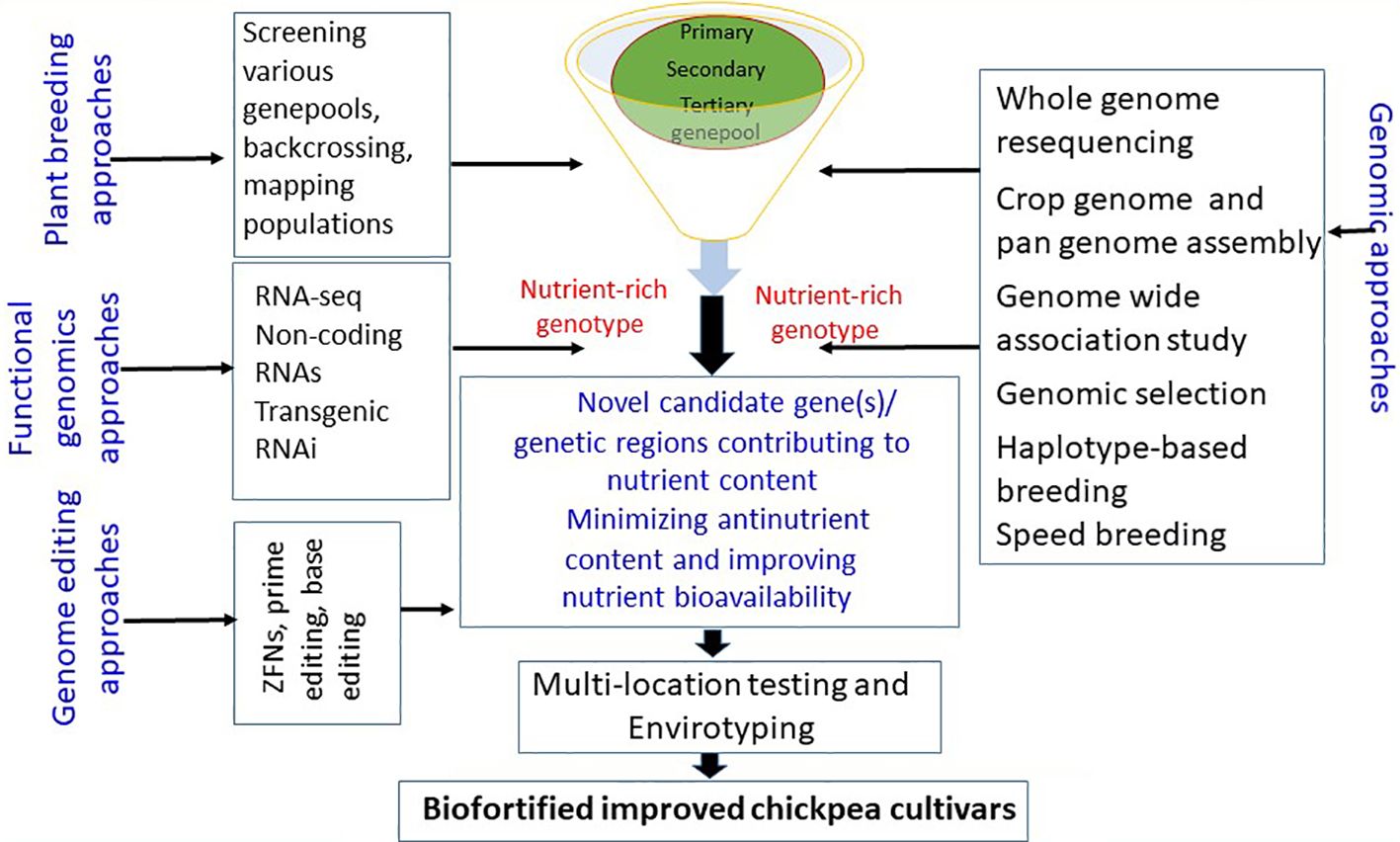
Using the genetic diversity in chickpea crop wild relatives, landraces, and germplasm resources and leveraging genomic resources such as the chickpea genome sequence and pan-genome assembly can help identify the key genetic determinants/gene(s)/QTL controlling nutritional traits. Marker-assisted selection can facilitate the transfer of nutrient-dense genomic regions to elite chickpea cultivars (see Figure 2). Furthermore, CRISPR/Cas9 genome editing offers precise editing of genomic regions related to anti-nutrients, enhancing nutrient bioavailability. Efforts to elucidate metabolic pathways associated with quality traits will deepen our understanding of the molecular mechanisms and gene(s) governing these quality traits. Developing improved chickpea cultivars with enhanced nutrition can help meet the rising demand for protein-rich diets and combat malnutrition, contributing to global food and nutrition security, modern cropping system diversity, and agricultural sustainability.
Author contributions
UJ: Writing – original draft, Writing – review & editing. HN: Writing – review & editing. MT: Writing – review & editing. BR: Writing – review & editing. PP: Writing – original draft, Writing – review & editing. KS: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Contribution number 24-241-J from Kansas Agricultural Experiment Station is gratefully acknowledged.
Acknowledgments
UJ and PP acknowledge Kansas State University for Support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1391496/full#supplementary-material
References
Abbo, S., Molina, C., Jungmann, R., Grusak, M. A., Berkovitch, Z., Reifen, R., et al. (2005). Quantitative trait loci governing carotenoid concentration and weight in seeds of chickpea (Cicer arietinum L.). Theor. Appl. Genet. 111, 185–195. doi: 10.1007/s00122-005-1930-y
Ahmad, N., Awan, M. J. A., Mansoor, S. (2023). Improving editing efficiency of prime editor in plants. Trends Plant Sci. 28, 1–3. doi: 10.1016/j.tplants.2022.09.001
Allen, L., Benoist, B., Dary, O., Hurrell, R. (2006). “Guidelines on food fortification with micronutrients,” in VWHO— Word Health Organization, FAO—Food and Agricultural Organization of the United Nations (Geneva: WHO).
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Arora, J., Kanthaliya, B., Joshi, A., Meena, M., Meena, S., Siddiqui, M. H., et al. (2023). Evaluation of total isoflavones in chickpea (Cicer arietinum L.) sprouts germinated under precursors (p-coumaric acid and L-phenylalanine) supplementation. Plants 12 (15), 2823. doi: 10.3390/plants12152823
Arya, N., Kumar, R. (2022). Utilizing chickpea isolates as a fortificant to develop protein dense yogurt: A review. Global J. Agric. Innov. Res. Dev. 9, 61–80. doi: 10.15377/2409-9813.2022.09.6
Badhan, S., Ball, A. S., Mantri, N. (2021). First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 22, 396. doi: 10.3390/ijms22010396
Begum, N., Khan, Q. U., Liu, L. G., Li, W., Liu, D., Haq, I. U. (2023). Nutritional composition, health benefits and bio-active compounds of chickpea (Cicer arietinum L.). Front. Nutr. 10, 1218468. doi: 10.3389/fnut.2023.1218468
Bhagyawant, S. S., Gautam, A. K., Narvekar, D. T., Gupta, N., Bhadkaria, A., Srivastava, N., et al. (2018). Biochemical diversity evaluation in chickpea accessions employing mini-core collection. Physiol. Mol. Biol. Plants. 24, 1165–1183. doi: 10.1007/s12298-018-0579-3
Bhatty, R. S. (1982). Albumin proteins of eight edible grain legume species: Electrophoretic patterns and amino acid composition. J. Agric. Food Chem. 30, 620–622. doi: 10.1021/jf00111a057
Boschin, G., Arnoldi, A. (2011). Legumes are valuable sources of tocopherols. Food Chem. 127, 1199–1203. doi: 10.1016/j.foodchem.2011.01.124
Boye, J. I., Zare, F., Pletch, A. (2010). Pulse proteins: Processing, characterization, functional properties and applications in food and feed. Food Res. Int. 43, 414–431. doi: 10.1016/j.foodres.2009.09.003
Broughton, W. J., Hernandez, G., Blair, M. W., Beebe, S. E., Gepts, P., Vanderleyden, J. (2003). Beans (Phaseolus spp.) model food legumes. Plant Soil 252, 55–128. doi: 10.1023/A:1024146710611
Brown, K. H., Peerson, J. M., Rivera, J., Allen, L. H. (2002). Effect of supplemental zinc on the growth and serum zinc concentrations of pre-pubertal children: A meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 75, 957–958. doi: 10.1093/ajcn/75.6.1062
Bulbula, D. D., Urga, K. (2018). Study on the effect of traditional processing methods on nutritional composition and antinutritional factors in chickpea (Cicer arietinum). Cogent Food Agric. 4, 1422370. doi: 10.1080/23311932.2017.1422370
Burchi, F., Fanzo, J., Frison, E. (2011). The role of food and nutrition system approaches in tackling hidden hunger. Int. J. Environ. Res. Public Health 8, 358–373. doi: 10.3390/ijerph8020358
Cabrera, C., Lloris, F., Gimeínez, R., Olalla, M., Loípez, M. C. (2003). Mineral content in legumes and nuts: contribution to the Spanish dietary intake. Sci. Total Environ. 308, 1–14. doi: 10.1016/S0048-9697(02)00611-3
Carroll, D. (2011). Genome engineering with zinc-finger nucleases. Genetics 188, 773–782. doi: 10.1534/genetics.111.131433
Chakraborty, A., Junaid, A., Parida, S. K., Bhatia, S. (2023). Integrated genomic approaches delineate a novel role of ROP1 ENHANCER1 in controlling seed protein content of chickpea. J. Expt. Bot. 74, 817–834. doi: 10.1093/jxb/erac452
Chibbar, R. N., Ambigaipalan, P., Hoover, R. (2010). Molecular diversity in pulse seed starch and complex carbohydrates and its role in human nutrition and health. Cereal Chem. 87, 342–352. doi: 10.1093/jxb/erac452
Choudhary, S., Kaur, J., Kaur, S., Kaur, S., Singh, I., Singh, S. (2015). Evaluation of antinutritional factors in kabuli chickpea cultivars differing in seed size. Indian J. Agric. Biochem. 28, 94–97.
Daboussi, F., Stoddard, T. J., Zhang, F. (2015). “Engineering meganuclease for precise plant genome modification. in Advances in new technology for targeted modification of plant genomes,”. Eds. Zhang, F., Puchta, H., Thomson, J. (New York, NY: Springer). doi: 10.1007/978-1-4939-2556-8_2
Darnton-Hill, I., Bloem, M., Chopra, M. (2006). Achieving the millennium development goals through mainstreaming nutrition: Speaking with one voice. Public Health Nutr. 9, 537–539. doi: 10.1079/PHN2006965
Davies, J. P., Kumar, S., Sastry-Dent, L. (2017). Use of zinc-finger nucleases for crop improvement. Prog. Mol. Biol. Transl. Sci. 149, 47–63. doi: 10.1016/bs.pmbts.2017.03.006
Day, L. (2013). Proteins from land plants—Potential resources for human nutrition and food security. Trends Food Sci. Technol. 32, 25–42. doi: 10.1016/j.tifs.2013.05.005
de Camargo, A. C., Concepcioín Alvarez, A., Arias-Santeí, M. F., Oyarzuín, J. E., Andia, M. E., Uribe, S., et al. (2022). Soluble free, esterified and insoluble-bound phenolic antioxidants from chickpeas prevent cytotoxicity in human hepatoma HuH-7 cells induced by peroxyl radicals. Antioxidants 11 (6), 1139. doi: 10.3390/antiox11061139
Devi, P., Awasthi, R., Jha, U., Sharma, K. D., Prasad, P. V. V., Siddique, K. H. M., et al. (2023). Understanding the effect of heat stress during seed filling on nutritional composition and seed yield in chickpea (Cicer arietinum L.). Sci. Reps. 13, 15450. doi: 10.1038/s41598-023-42586-0
Diapari, M., Sindhu, A., Bett, K., Deokar, A., Warkentin, T. D., Tar’an, B. (2014). Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.). Genome 57, 459–568. doi: 10.1139/gen-2014-0108
Didinger, C., Thompson, H. J. (2021). Defining nutritional and functional niches of legumes: A call for clarity to distinguish a future role for pulses in the dietary guidelines for Americans. Nutrients. 13, 1100. doi: 10.3390/nu13041100
Domínguez, D., Cuevas, E., Milan, J., Garzón, J. A., Canizalez, A., Gutirerrez, R., et al. (2016). Enhancement the antioxidant activity, total phenolic and flavonoid contents by optimizing the germination conditions of desi chickpea (Cicer arietinum L) seeds. FASEB J. 30, 1176.31.
Domínguez-Arispuro, D. M., Cuevas-Rodríguez, E. O., Milán-Carrillo, J., León-López, L., Gutiérrez-Dorado, R., Reyes-Moreno, C. (2018). Optimal germination condition impacts on the antioxidant activity and phenolic acids profile in pigmented desi chickpea (Cicer arietinum L.) seeds. J. Food Sci. Technol. 55, 638–647. doi: 10.1007/s13197-017-2973-1
Elango, D., Wang, W., Thudi, M., Sebastiar, S., Ramadoss, B. R., Varshney, R. K. (2022). Genome-wide association mapping of seed oligosaccharides in chickpea. Front. Plant Sci. 13,1024543. doi: 10.3389/fpls.2022.1024543
Ereifej, K. I., Al-Karaki, G. N., Hammouri, M. K. (2001). Seed chemical composition of improved chickpea cultivars grown under semiarid Mediterranean conditions. Int. J. Food Prop. 4, 239–246. doi: 10.1081/JFP-100105190
Ervin, R. B., Wright, J. D., Wang, C. Y., Kennedy-Stephenson, J. (2004). Dietary intake of fats and fatty acids for the United States population: 1999–2000. Adv. Data. 8 348), 1–6.
FAOSTAT (2021). Food and agriculture Organization of the United Nations, Statistics division (FAO). (FAOSTAT Database). Available at: http://www.fao.org/faostat/en/#data/QC.
Farida Traore, F., El-Baouchi, A., En-Nahli, Y., Hejjaoui, K., Metougui, M. L., Hamwieh, A., et al. (2022). Exploring the genetic variability and potential correlations between nutritional quality and agro-physiological traits in kabuli chickpea germplasm collection (Cicer arietinum L.). Front. Plant Sci. 13, 905320. doi: 10.3389/fpls.2022.905320
Fayaz, H., Tyagi, S., Wani, A. A., Pandey, R., Akhtar, S., Bhat, M. A., et al. (2022). Genome-wide association analysis to delineate high-quality SNPs for seed micronutrient density in chickpea (Cicer arietinum L.). Sci. Reps. 12, 11357. doi: 10.1038/s41598-022-14487-1
Gangola, M. P., Khedikar, Y. P., Gaur, P. M., Båga, M., Chibbar, R. N. (2013). Genotype and growing environment interaction shows a positive correlation between substrates of raffinose family oligosaccharides (RFO) biosynthesis and their accumulation in chickpea (Cicer arietinum L.) seeds. J. Agric. Food Chem. 61, 4943–4952. doi: 10.1021/jf3054033
Gao, Y., Yao, Y., Zhu, Y., Ren, G. (2015). Isoflavone content and com- position in chickpea (Cicer arietinum L.) sprouts germinated under different conditions. J. Agric. Food Chem. 63, 2701–2707. doi: 10.1021/jf5057524
Gaur, P. M., Singh, M. K., Samineni, S., Sajja, S. B., Jukanti, A. K., Kamatam, S., et al. (2016). Inheritance of protein content and its relationships with seed size, grain yield and other traits in chickpea. Euphytica 209, 253–260. doi: 10.1007/s10681-016-1678-2
Ghribi, A. M., Maklouf, I., Blecker, C., Attia, H., Besbes, S. (2015). Nutritional and compositional study of Desi and Kabuli chickpea (Cicer arietinum L.) flours from Tunisian cultivars. Adv. Food Technol. Nutr. Sci. 1, 38–47. doi: 10.17140/AFTNSOJ-1-107
Grasso, N., Lynch, N. L., Arendt, E. K., O’Mahony, J. A. (2022). Chickpea protein ingredients: A review of composition, functionality, and applications. Compr. Rev. Food Sci. Food Saf. 21, 435–452. doi: 10.1111/1541-4337.12878
Grewal, S. K., Sharma, K. P., Bharadwaj, R. D., Hegde, V., Sidhu, S. K., Singh, S., et al. (2023). Characterization of chickpea cultivars and trait specific germplasm for grain protein content and amino acids composition and identification of potential donors for genetic improvement of its nutritional quality. Plant Genet. Resour. 20, 383–393. doi: 10.1017/S147926212300028X
Grewal, S. K., Sharma, K. P., Bharadwaj, R. D., Hegde, V., Tripathi, S., Singh, S., et al. (2020). Understanding genotypic variation and identification of promising genotypes for iron and zinc content in chickpea (Cicer arietinum L.). J. Food Comps. Anal. 88, 103458. doi: 10.1016/j.jfca.2020.103458
Gul, M. K., Egesel, C. (2008). The effects of planting time on fatty acids and tocopherols in chickpea. Eur. Food Res. Technol. 226, 517–522. doi: 10.1007/s00217-007-0564-5
Gupta, N., Shrivastava, N., Bhagyawant, S. S. (2017). Multivariate analysis based on nutritional value, antinutritional profile and antioxidant capacity of forty chickpea genotypes grown in India. J. Nutr. Food Sci. 7, 3. doi: 10.4172/2155-9600
Gupta, A., Tripti, S., Singh, S. P., Bhardwaj, A., Srivastava, D., Kumar, R. (2023a). Prospects of microgreens as budding living functional food: Breeding and biofortification through OMICS and other approaches for nutritional security. Front. Genet. 14, 1053810. doi: 10.3389/fgene.2023.1053810
Gupta, S. K., Vishwakarma, N. K., Malakar, P., Vanspati, P., Sharma, N. K., Chattopadhyay, D. (2023b). Development of an Agrobacterium-delivered codon-optimized CRISPR/Cas9 system for chickpea genome editing. Protoplasma 260, 1437–1451. doi: 10.1007/s00709-023-01856-4
Hacisalihoglu, G., Armstrong, P. (2023). Crop seed phenomics: focus on non-destructive functional trait phenotyping methods and applications. Plants 12 (5), 1177. doi: 10.3390/plants12051177
Hall, C., Hillen, C., Garden Robinson, J. (2017). Composition, nutritional value, and health benefits of pulses. Cereal Chem. 94, 11–31. doi: 10.1094/CCHEM-03-16-0069-FI
Haun, W., Coffman, A., Clasen, B. M., Demorest, Z. L., Lowy, A., Ray, E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. doi: 10.1111/pbi.12201
Hefni, M., Witthoft, C. M. (2014). Folate content in processed legume foods commonly consumed in Egypt. LWT Food Sci. Technol. 57, 337–343. doi: 10.1016/j.lwt.2013.12.026
Heiras-Palazuelos, M. J., Ochoa-Lugo, M., Gutierrez-Dorado, R., LopezValenzuela, J. A., Mora, R. S., Milan-Carrillo, J., et al. (2013). Technological properties, antioxidant activity and phenolic and flavonoid content of pigmented chickpea (Cicer arietinum L.) cultivars. Int. J. Food Sci. Nutr. 64, 69–76. doi: 10.3109/09637486.2012.694854
Hensel, G., Kumlehn, J. (2019). Genome engineering using TALENs. Barley: Methods Mol. Biol. 1900, 195–215. doi: 10.1007/978-1-4939-8944-7_13
Ibrikci, H., Knewtson, S. J. B., Grusak, M. A. (2003). Chickpea leaves as a vegetble green for humans: Evaluation of mineral composition. J. Sci. Food Agric. 83, 945–950. doi: 10.1002/jsfa.1427
Iqbal, A., Khalil, I. A., Ateeq, N., Khan, M. S. (2006). Nutritional quality of important food legumes. Food Chem. 97, 331–335. doi: 10.1016/j.foodchem.2005.05.011
Jadhav, A. A., Rayate, S. J., Mhase, L. B., Thudi, M., Chitikineni, A., Harer, P. N., et al. (2015). Marker-trait association study for protein content in chickpea (Cicer arietinum L.). J. Genet. 94, 279–286. doi: 10.1007/s12041-015-0529-6
Jahan, T. A., Kalve, S., Belak, Z., Eskiw, C., Tar’an, B. (2023). Iron accumulation and partitioning in hydroponically grown wild and cultivated chickpea (Cicer arietinum L). Front. Plant Sci. 14, 1092493. doi: 10.3389/fpls.2023.1092493
Jahan, T. A., Vandenberg, A., Glahn, R. P., Tyler, R. T., Reaney, M. J. T., Tar’an, B. (2019). Iron fortification and bioavailability of chickpea (Cicer arietinum L.) seeds and flour. Nutrients, 11.
Jha, A. B., Ashokkumar, K., Diapari, M., Ambrose, S. J., Zhang, H., Tar’an, B., et al. (2015). Genetic diversity of folate profiles in seeds of common bean, lentil, chickpea and pea. J. Food Comp. Anal. 42, 134–140. doi: 10.1016/j.jfca.2015.03.006
Jha, R., Zhang, K., He, Y., Mendler-Drienyovszki, N., Magyar-Tábori, K., Quinet, M., et al. (2024). Global nutritional challenges and opportunities: Buckwheat, a potential bridge between nutrient deficiency and food security. Trends Food Sci. Technol. 145, 104365. doi: 10.1016/j.tifs.2024.104365
Jin, L., Lange, W., Kempmann, A., Maybeck, V., Günther, A., Gruteser, N., et al. (2016). High-efficiency transduction and specific expression of ChR2opt for optogenetic manipulation of primary cortical neurons mediated by recombinant adeno-associated viruses. J. Biotechnol. 233, 171–180. doi: 10.1016/j.jbiotec.2016.07.001
Jood, S., Bishnoi, S., Sharma, A. (1998). Chemical analysis and physico-chemical properties of chickpea and lentil cultivars. Food/Nahrung 42 (02), 71–74. doi: 10.1002/(SICI)1521-3803(199804)42:02<71::AID-FOOD71>3.3.CO;2-U
Joung, J. K., Sander, J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell. Biol. 14, 49–55. doi: 10.1038/nrm3486
Jukanti, A. K., Gaur, P. M., Gowda, C. L. L., Chibbar, R. N. (2012). Nutritional quality and health bene ts of chickpea (Cicer arietinum L.): A review. Br. J. Nutr. 108, S11–S26. doi: 10.1017/S0007114512000797
Karaca, N., Ates, D., Nemli, S., Ozkuru, E., Yilmaz, H., Yagmur, B., et al. (2020). Identification of SNP markers associated with iron and zinc concentrations in Cicer seeds. Curr. Genomics 21, 212–223. doi: 10.2174/1389202921666200413150951
Katoch, O., Chauhan, U. S., Yadav, R., Yadav, S. S., Kumar, A., Yadav, A., et al. (2016). Nitrate Reductase based phylogenetic analysis in chickpea. RJCE 20, 1–8.
Kaur, K., Grewal, S. K., Gill, P. S., Singh, S. (2019). Comparison of cultivated and wild chickpea genotypes for nutritional quality and antioxidant potential. J. Food Sci. Technol. 56, 1864–1876. doi: 10.1007/s13197-019-03646-4
Kaur, S., Kaur, S., Gupta, A. K., Kaur, N., Javed, M. (2014). Biochemical and nutritional characterization of chickpea (Cicer arietinum) genotypes. Indian J. Agric. Sci. 84, 479–486.
Kaur, R., Prasad, K. (2021). Technological, processing and nutritional aspects of chickpea (Cicer arietinum) - a review. Trends Food Sci. Technol. 109, 448–463. doi: 10.1016/j.tifs.2021.01.044
Khan, S. H. (2019). Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Therapy-Nucleic Acids 16, 326–334. doi: 10.1016/j.omtn.2019.02.027
Knights, E. J., Hobson, K. B. (2016). “Chickpea overview,” in Reference module in food science, 2nd Ed, Vol. 1. (Elsevier Ltd). doi: 10.1016/b978-0-08-100596-5.00035-4
Ku, H. K., Ha, S. H. (2020). Improving nutritional and functional quality by genome editing of crops: status and perspectives. Front. Plant Sci. 11, 577313. doi: 10.3389/fpls.2020.577313
Kudapa, H., Garg, V., Chitikineni, A., Varshney, R. K. (2018). The RNA-Seq-based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio-temporal changes associated with growth and development. Plant Cell Environ. 41, 2209–2225. doi: 10.1111/pce.13210
Kumar, R., Singh, R. K., Misra, J., Yadav, A., Kumar, A., Yadav, R., et al. (2021). Dissecting proteomic estimates for enhanced bioavailable nutrition during varied stages of germination and identification of potential genotypes in chickpea. Legume Res. 459, 1082–1087. doi: 10.18805/LR-4531
Kumar, D., Yadav, A., Ahmad, R., Dwivedi, U. N., Yadav, K. (2022). CRISPR-based genome editing for nutrient enrichment in crops: A promising approach toward global food security. Front. Genet. 13. doi: 10.3389/fgene.2022.932859
Li, L., Wu, L. P., Chandrasegaran, S. (1992). Functional domains inI restriction endonuclease. Proc. Natl. Acad. Sci. U.S.A. 89, 4275–4279. doi: 10.1073/pnas.89.10.4275
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9, 559. doi: 10.3389/fpls.2018.00559
Madurapperumage, A., Tang, L., Thavarajah, P., Bridges, W., Shipe, E., Vandemark, G., et al. (2021). Chickpea (Cicer arietinum L.) as a source of essential fatty acids–a biofortification approach. Front. Plant Sci. 12, 734980. doi: 10.3389/fpls.2021.734980
Marioli Nobile, C., Carreras, J., Grosso, R., Inga, C. M., Silva, M. P., Aguilar, R., et al. (2013). Proximate composition and seed lipid components of “kabuli”-type chickpea (Cicer arietinum L.) from Argentina (Scientific Research Publishing).
Meuwissen, T. H., Hayes, B. J., Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. doi: 10.1093/genetics/157.4.1819
Miao, M., Zhang, T., Jiang, B. (2009). Characterizations of kabuli and desi chickpea starches cultivated in China. Food Chem. 113, 1025–1032. doi: 10.1016/j.foodchem.2008.08.056
Misra, J. P., Yadav, A., Kumar, A., Yadav, R., Kumar, R. (2016). Bio-chemical characterization of chickpea genotypes with special reference to protein. RJCE 20, 38–43.
Mondor, M., Aksay, S., Drolet, H., Roufik, S., Farnworth, E., Boye, J. I. (2009). Influence of processing on composition and antinutritional factors of chickpea protein concentrates produced by isoelectric precipitation and ultrafiltration. Innov. Food Sci. Emerg. Technol. 10, 342–347. doi: 10.1016/j.ifset.2009.01.007
Mugabe, D., Frieszell, C. M., Warburton, M. L., Coyne, C. J., Sari, H., Uhdre, R., et al. (2023). Kabuli chickpea seed quality diversity and preliminary genome-wide association study identifies markers and potential candidate genes. Agrosystems Geosci. Environ. 6, 20437. doi: 10.1002/agg2.20437
Ocampo, B., Robertson, L. D., Singh, K. B. (1998). Variation in seed protein content in the annual wild Cicer species. J. Sci. Food Agric. 78, 220–224. doi: 10.1002/(ISSN)1097-0010
Onder, S., Can Karaca, A., Ozcelik, B., Alamri, A. S., Ibrahim, S. A., Galanakis, C. M. (2023). Exploring the amino-acid composition, secondary structure, and physicochemical and functional properties of chickpea protein isolates. ACS Omega 8, 1486–1495. doi: 10.1021/acsomega.2c06912
Özer, S., Karaköy, T., Toklu, F., Baloch, F. S., Kilian, B., Özkan, H. (2010). Nutritional and physicochemical variation in Turkish kabuli chickpea (Cicer arietinum L.) landraces. Euphytica 175, 237–249. doi: 10.1007/s10681-010-0174-3
Pâques, F., Duchateau, P. (2007). Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 7, 49–66. doi: 10.2174/156652307779940216
Patane, C. (2006). Variation and relationship among some nutritional traits in Sicilian genotypes of chickpea (Cicer arietinum L.). J. Food Qual. 29, 282–293.
Pingoud, A., Silva, G. H. (2007). Precision genome surgery. Nat. Biotechnol. 25, 743–744. doi: 10.1038/nbt0707-743
Pittaway, J. K., Robertson, I. K., Ball, M. J. (2008). Chickpeas may influence fatty acid and fiber intake in an ad libitum diet, leading to small improvements in serum lipid profile and glycemic control. J. Am. Diet. Assoc. 108 (6), 1009–1013. doi: 10.1016/j.jada.2008.03.009
Porto, E. M., Komor, A. C., Slaymaker, I. M., Yeo, G. W. (2020). Base editing: advances and therapeutic opportunities. Nat. Rev. Drug Discov. 19 (12), 839–859. doi: 10.1038/s41573-020-0084-6
Rachwa-Rosiak, D., Nebesny, E., Budryn, G. (2015). Chickpeas—composition, nutritional value, health benefits, application to bread and snacks: a review. Crit. Rev. Food Sci. Nutr. 55, 1137–1145. doi: 10.1080/10408398.2012.687418
Ramani, A., Kushwaha, R., Malaviya, R., Kumar, R., Yadav, N. (2021). Molecular, functional and nutritional properties of chickpea (Cicer arietinum L.) protein isolates prepared by modified solubilization methods. J. Food Measurement Characterization 15, 2352–2368. doi: 10.1007/s11694-020-00778-6
Ravi, R., Harte, J. (2009). Milling and physic-chemical properties of chickpea (Cicer arietinum L.) varieties. J. Sci. Food Agric. 89, 258–266. doi: 10.1002/jsfa.3435
Rebello, C. J., Greenway, F. L., Finley, J. W. (2014). A review of the nutritional value of legumes and their effects on obesity and its related co-morbidities. Obes. Rev. 15, 392–407. doi: 10.1111/obr.12144
Rezaei, M. K., Deokar, A. A., Arganosa, G., Roorkiwal, M., Pandey, S. K., Warkentin, T. D., et al. (2019). Mapping quantitative trait loci for carotenoid concentration in three F2 populations of chickpea. Plant Genome. 12, 1–12. doi: 10.3835/plantgenome2019.07.0067
Rezaei, M. K., Deokar, A., Tar’an, B. (2016). Identification and expression analysis of candidate genes involved in carotenoid biosynthesis in chickpea seeds. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01867
Rincon, F., Martinez, B., Ibanez, M. V. (1998). Proximate composition and antinutritive substances in chickpea (Cicer arietinum L) as affected by the biotype factor. J. Sci. Food Agric. 78, 382–388. doi: 10.1002/(ISSN)1097-0010
Roorkiwal, M., Bhandari, A., Barmukh, R., Bajaj, P., Valluri, V. K., Chitikineni, A., et al. (2022). Genome-wide association mapping of nutritional traits for designing superior chickpea varieties. Front. Plant Sci. 13, 843911. doi: 10.3389/fpls.2022.843911
Sab, S., Lokesha, R., Mannur, D. M., Jadhav, K., Mallikarjuna, B. P., Yeri, S., et al. (2020). Genome-wide SNP discovery and mapping QTLs for seed iron and zinc concentrations in chickpea (Cicer arietinum L.). Front. Nutr. 7,559120. doi: 10.3389/fnut.2020.559120
Salaria, S., Boatwright, J. L., Johnson, N., Madurapperumage, A., Joshi, P., Thavarajah, P., et al. (2023). Fatty acid composition and genome-wide associations of a chickpea (Cicer arietinum L.) diversity panel for biofortification efforts. Sci. Reps 13, 14002. doi: 10.1038/s41598-023-41274-3
Salsman, J., Dellaire, G. (2017). Precision genome editing in the CRISPR era. Biochem. Cell. Biol. 95, 187–201. doi: 10.1139/bcb-2016-0137
Samineni, S., Mahendrakar, M. D., Hotti, A., Chand, U., Rathore, A., Gaur, P. M. (2022). Impact of heat and drought stresses on grain nutrient content in chickpea: Genome-wide marker-trait associations for protein, Fe and Zn. Environ. Expt. Bot. 194, 104688. doi: 10.1016/j.envexpbot.2021.104688
Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi: 10.1111/pbi.12837
Sánchez-Mata, M. C., Peñuela-Teruel, M. J., Cámara-Hurtado, M., Díez-Marqués, C., Torija-Isasa, M. E. (1998). Determination of mono-, di-, and oligosaccharides in legumes by high performance liquid chromatography using an amino-bonded silica column. J. Agric. Food Chem. 46, 3648–3652.
Santos, F. G., Fratelli, C., Muniz, D. G., Capriles, V. D. (2018). Mixture design applied to the development of chickpea-based gluten-free bread with attractive technological, sensory, and nutritional quality. J. Food Sci. 83, 188–197. doi: 10.1111/1750-3841.14009
Sen Gupta, D., Thavarajah, D., Knutson, P., Thavarajah, P., McGee, R. J., Coyne, C. J., et al. (2013). Lentils (Lens culinaris L.), a rich source of folates. J. Agric. Food Chem. 61, 7794–7799. doi: 10.1021/jf401891p
Shad, M. A., Pervez, H., Zafar, Z. I., Zia-Ul-Haq, M., Nawaz, H. (2009). Evaluation of biochemical composition and physicochemical parameters of oil from seeds of desi chickpea varieties cultivated in arid zone of Pakistan. Pak. J. Bot. 41, 655–662.
Sharma, S., Lavale, S. A., Nimje, C., Singh, S. (2021). Characterization and identification of annual wild Cicer species for seed protein and mineral concentrations for chickpea improvement. Crop Sci. 61, 305–319. doi: 10.1002/csc2.20413
Sharma, S., Yadav, N., Singh, A., Kaur, D., Kumar, R. (2016). Impact of Thermal and Bioprocessing on Antioxidant and Functional Properties of nine Newly Developed Desi and Kabili Chickpea (Cicer arietinumL.) Cultivars. Vegetos 29, 78–86. doi: 10.5958/2229-4473.2016.00040.9
Sharma, S., Yadav, N., Singh, A., Kumar, R. (2013). Nutritional and antinutritional profile of newly developed chickpea (Cicer arietinum L.) varieties. Intl. Food Res. J. 20, 805–810.
Shevkani, K., Singh, N., Chen, Y., Kaur, A., Yu, L. (2019). Pulse proteins: Secondary structure, functionality and applications. J. Food Sci. Technol. 56, 2787–2798. doi: 10.1007/s13197-019-03723-8
Shi, L., Arntfield, S. D., Nickerson, M. (2018). Changes in level of phytic acid, lectins and oxalates during soaking and cooking of Canadian pulses. Food Res. Int. 107, 660–668. doi: 10.1016/j.foodres.2018.02.056
Singh, B., Singh, J. P., Kaur, A., Singh, N. (2017). Phenolic composition and antioxidant potential of grain legume seeds: a review. Food Res. Int. 101, 1–16. doi: 10.1016/j.foodres.2017.09.026
Singh, N., Kaur, S., Isono, N., Noda, T. (2010). Genotypic diversity in physico-chemical, pasting and gel textural properties of chickpea (Cicer arietinum L.). Food Chem. 122, 65–73. doi: 10.1016/j.foodchem.2010.02.015
Singh, N., Sandhu, K. S., Kaur, M. (2004). Characterization of starches separated from Indian chickpea (Cicer arietinum L.) cultivars. J. Food Eng. 63, 441–449. doi: 10.1016/j.jfoodeng.2003.09.003
Singh, U. (1985). Nutritional quality of chickpea (Cicer arietinum L.): Current status and future research needs. Plant Foods Hum. Nutr. 35, 339–351. doi: 10.1007/BF01091779
Singh, U., Jambunathan, R. (1982). Distribution of seed protein fractions and amino acids in different anatomical parts of chickpea (Cicer arietinum L.) and pigeonpea (Cajanus cajan L.). Plant Foods Hum. Nutr. 31, 347–354. doi: 10.1007/BF01094046
Singh, R., Kumari, N. (2011). HPLC based determination of oligosaccharides and diversity analysis in Chickpea (Cicer arietinum L.). Plant Arch. 11, 543–551.
Singh, A. P., Singh, R. K., Hegde, V. S., Shukla, N., Saini, P., Yadav, R., et al. (2022). Unclasping potential chickpea resources for the antioxidant enzyme Superoxide Dismutase. JSFA Rep. 2, 320–331. doi: 10.1002/jsf2.65
Soetan, K. O., Oyewole, O. E. (2009). The need for adequate processing to reduce the anti-nutritional factors in plants used as human foods and animal feeds: A review. Afr. J. Food Sci. 3, 223–232.
Srungarapu, R., Mahendrakar, M. D., Mohammad, L. A., Chand, U., Jagarlamudi, V. R., Kondamudi, K. P., et al. (2022b). Genome-wide association analysis reveals trait-linked markers for grain nutrient and agronomic traits in diverse set of chickpea germplasm. Cells 11 (15), 2457. doi: 10.3390/cells11152457
Srungarapu, R., Mohammad, L. A., Mahendrakar, M. D., Chand, U., Venkata, R. J., Kondamudi, K. P., et al. (2022a). Genetic variation for grain protein, Fe and Zn content traits in chickpea reference set. J. Food Comps. Anal. 114, 104774. doi: 10.1016/j.jfca.2022.104774
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8, 298. doi: 10.3389/fpls.2017.00298
Tachie, C., Tawiah, N. A., Aryee, A. N. (2023). Using machine learning models to predict the quality of plant-based foods. Curr. Res. Food Sci. 7, 100544. doi: 10.1016/j.crfs.2023.100544
Tarzi, B. G., Gharachorloo, M., Baharinia, M., Mortazavi, S. A. (2012). The effect of germination on phenolic content and antioxidant activity of chickpea. Iran. J. Pharm. Res. 11, 1137.
Thavarajah, P., Thavarajah, T. (2012). Evaluation of chickpea (Cicer arietinum L.) micronutrient composition: Biofortification opportunities to combat global micronutrient malnutrition. Food Res. Int. 49, 99–104. doi: 10.1016/j.foodres.2012.08.007
Tiwari, B. K., Singh, N. (2012). Pulse chemistry and technology (Cambridge, UK: Royal Society of Chemistry). doi: 10.1039/9781839169038
Tosh, S. M., Yada, S. (2010). Dietary fibres in pulse seeds and fractions: Characterization, functional attributes, and applications. Food Res. Int. 43, 450–460. doi: 10.1016/j.foodres.2009.09.005
Ufaz, S., Galili, G. (2008). Improving the content of essential amino acids in crop plants: Goals and opportunities. Plant Physiol. 147, 954–961. doi: 10.1104/pp.108.118091
Upadhyaya, H. D., Bajaj, D., Das, S., Kumar, V., Gowda, C. L. L., Sharma, S., et al. (2016a). Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci. Reps. 6 (1), 24050. doi: 10.1038/srep24050
Upadhyaya, H. D., Bajaj, D., Narnoliya, L., Das, S., Kumar, V., Gowda, C. L. L., et al. (2016b). Genome-wide scans for delineation of candidate genes regulating seed-protein content in chickpea. Front. Plant Sci. 7, 302. doi: 10.3389/fpls.2016.00302
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., Gregory, P. D. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. doi: 10.1038/nrg2842
Varshney, R. K., Roorkiwal, M., Sun, S., Bajaj, P., Chitikineni, A., Thudi, M., et al. (2021). A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature 599, 622–627. doi: 10.1038/s41586-021-04066-1
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246. doi: 10.1038/nbt.2491
Varshney, R. K., Thudi, M., Roorkiwal, M., He, W., Upadhyaya, H. D., Yang, W., et al. (2019). Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 51, 857–864. doi: 10.1038/s41588-019-0401-3
Wada, N., Ueta, R., Osakabe, Y., Osakabe, K. (2020). Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 20, 234. doi: 10.1186/s12870-020-02385-5
Wallace, T. C., Murray, R., Zelman, K. M. (2016). The nutritional value and health benefits of chickpeas and hummus. Nutrients 8, 766. doi: 10.3390/nu8120766
Waltz, E. (2016). CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 34, 582. doi: 10.1038/nbt0616-582
Wang, H., La Russa, M., Qi, L. S. (2016). CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wang, R., Gangola, M. P., Irvine, C., Gaur, P. M., Båga, M., Chibbar, R. N. (2019). Co-localization of genomic regions associated with seed morphology and composition in a desi chickpea (Cicer arietinum L.) population varying in seed protein concentration. Theor. Appl. Genet. 132, 1263–1281. doi: 10.1007/s00122-019-03277-5
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi: 10.1038/s41477-017-0083-8
Welch, R. M. (2002). The impact of mineral nutrients in food crops on global human health. Plant Soil. 247, 83–90. doi: 10.1023/A:1021140122921
Welch, R. M., Graham, R. D. (2004). Breeding for micronutrients in staple food crops from a human nutrition perspective. J. Expt. Bot. 55, 353–364. doi: 10.1093/jxb/erh064
Wood, J. A., Grusak, M. A. (2007). “Nutritional value of chickpea,” in Chickpea breeding and management. Eds. Yadav, S. S., Redden, B., Chen, W., Shar- ma, B. (CAB International, Wallingford), 101–142.
Xiao, S., Li, Z., Zhou, K., Fu, Y. (2023). Chemical composition of kabuli and desi chickpea (Cicer arietinum L.) cultivars grown in Xinjiang, China. Food Sci. Nutr. 11, 236–248. doi: 10.1002/fsn3.3056
Xu, B. J., Yuan, S. H., Chang, S. K. C. (2007). Comparative analyses of phenolic composition, antioxidant capacity, and colour of cool season legumes and other selected food legumes. J. Food Sci. 72, 167–177. doi: 10.1111/j.1750-3841.2006.00261.x
Yegrem, L. (2021). Nutritional composition, antinutritional factors, and utilization trends of Ethiopian chickpea (Cicer arietinum L.). Int. J. Food Sci. 2021, 5570753. doi: 10.1155/2021/5570753
Younis, K., Ahmad, S., Badpa, A. (2015). Malnutrition: causes and strategies. J. Food Process. Technol. 6, 434.
Zhao, S., Zhang, L., Gao, P., Shao, Z. (2009). Isolation and characterization of the isoflavones from sprouted chickpea seeds. Food Chem. 114, 869–873. doi: 10.1016/j.foodchem.2008.10.026
Zhao, Z., Shang, P., Mohanraju, P., Geijsen, N. (2023). Prime editing: advances and therapeutic applications. Trends Biotechnol. 41, 1000–1012. doi: 10.1016/j.tibtech.2023.03.004
Zia-Ul-Haq, M., Ahmad, S., Ahmad, M., Iqbal, S., Khawar, K. M. (2009). Effects of cultivar and row spacing on tocopherol and sterol composition of chickpea (Cicer arietinum L.) seed oil. J. Agric. Sci. 15, 25–30.
Zia-Ul-Haq, M., Iqbal, S., Ahmad, S., Imran, M., Niaz, A., Bhanger, M. I. (2007). Nutritional and compositional study of desi chickpea (Cicer arietinum L.) cultivars grown in Punjab, Pakistan. Food Chem. 105, 1357–1363. doi: 10.1016/j.foodchem.2007.05.004
Keywords: chickpea, protein, carbohydrate, fat, quality traits
Citation: Jha UC, Nayyar H, Thudi M, Beena R, Prasad PVV and Siddique KHM (2024) Unlocking the nutritional potential of chickpea: strategies for biofortification and enhanced multinutrient quality. Front. Plant Sci. 15:1391496. doi: 10.3389/fpls.2024.1391496
Received: 26 February 2024; Accepted: 20 May 2024;
Published: 07 June 2024.
Edited by:
Palak Chaturvedi, University of Vienna, AustriaReviewed by:
Lorenzo Rocchetti, NuCicer, Inc., United StatesRajendra Kumar, Indian Agricultural Research Institute (ICAR), India
Copyright © 2024 Jha, Nayyar, Thudi, Beena, Vara Prasad and Siddique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uday Chand Jha, dWNqQGtzdS5lZHU=; Kadambot H. M. Siddique, a2FkYW1ib3Quc2lkZGlxdWVAdXdhLmVkdS5hdQ==; P. V. Vara Prasad, dmFyYUBrc3UuZWR1
 Uday Chand Jha
Uday Chand Jha Harsh Nayyar
Harsh Nayyar Mahender Thudi
Mahender Thudi Radha Beena
Radha Beena P. V. Vara Prasad
P. V. Vara Prasad Kadambot H. M. Siddique
Kadambot H. M. Siddique