Abstract
The increasing atmospheric nitrogen deposition, characterized by a rising proportion of nitrate nitrogen (NO3⁻-N), is exacerbating the spread of invasive plant species. Despite this trend, the response mechanisms of Mikania micrantha, a highly invasive plant, to NO3⁻-N remain poorly understood. This study investigates the unique adaptation strategies of M. micrantha to elevated NO3⁻-N levels, providing novel insights into its invasive success under changing nitrogen deposition patterns. Field experiments showed that M. micrantha rhizosphere soil contained higher NO3–N content and protease activity compared to companion plants (Paederia scandens, Ipomoea nil, and Ipomoea cairica). Both roots and stems of M. micrantha had higher NO3–N content and demonstrated stronger nitrogen metabolism capabilities. Pot experiments further showed that increasing NO3⁻-N concentrations (0 mM–40 mM) significantly promoted M. micrantha growth, with optimal phenotypic responses (main stem length, leaf number, branch number, and biomass) observed at 5 mM NO3⁻-N. Nitrogen metabolism enzyme assays revealed that nitrate reductase (NR), nitrite reductase (NiR), glutamate dehydrogenase (GDH), and free amino acid content increased progressively with NO3⁻-N concentration. Transcriptome sequencing and qPCR analyses identified upregulation of key genes related to transcription factors, nitrate transporter-related, nitrogen metabolism enzyme, and amino acid synthesis pathway. These findings demonstrate that M. micrantha employs a multifaceted strategy to exploit elevated NO3⁻-N conditions: enhanced NO3⁻-N uptake from soil, efficient transport to stems, and robust nitrogen metabolism facilitated by coordinated gene expression. This study reveals the adaptation mechanisms of M. micrantha to NO3⁻-N enrichment, offering critical insights for predicting and managing invasive species responses to global atmospheric nitrogen deposition changes. The results highlight the importance of considering nitrogen composition, rather than just quantity, in invasive species management strategies.
Introduction
Since the industrial revolution, human activities such as fertilizer application and fossil fuel combustion have significantly increased atmospheric nitrogen deposition (Stevens et al., 2011). China has become one of the three major high nitrogen deposition areas in the world. Data indicates that anthropogenic nitrogen production was 18.3 Tg N in 1980, which doubled by 2010 to 53.9 Tg N (Gu et al., 2015). Nitrogen is an essential nutrient for plant growth and development, with its demand significantly increasing compared to other elements (Toor et al., 2021). While high nitrogen deposition typically increases soil nitrogen availability and stimulates plant growth (Dong et al., 2022), excessive nitrogen can cause a range of ecological problems. These include impacts on human health, alterations in biogeochemical cycles, shifts in ecosystem structure and function, and even the potential extinction of species (Chen et al., 2020; Jia et al., 2020; Zhao et al., 2021). Ammonium-nitrogen (NH4+-N) and nitrate-nitrogen (NO3–N) are the two main inorganic nitrogen forms absorbed by plants (Wang et al., 2021). NO3–N deposition primarily originates from industrial and transportation fossil fuel emissions (Galloway et al., 2004), whereas NH4+-N deposition is largely derived from artificial fertilizers and livestock farming (Behera et al., 2013). Research indicates that the RNHx/NOy ratio in nitrogen deposition has been decreasing in China, due to an ongoing increase in NO3–N deposition alongside a decrease in NH4+-N deposition. This marks a transition from the previous NH4+-dominant nitrogen deposition mode to a new mode where NH4+-N and NO3–N deposition contribute equally (Zhu et al., 2015; Yu et al., 2019). Therefore, in the context of global nitrogen deposition, especially the alteration of nitrogen deposition components (RNHx/NOy), controlling the invasion of exotic plants has become an urgent priority (Shuvar and Korpita, 2021).
Biological invasions not only threaten the abundance and diversity of native species but also alters the carbon and nitrogen cycles in ecosystems, thereby affecting their structure and function. Increased nitrogen deposition significantly accelerates the growth and spread of nitrophilous plants, which can lead to the exclusion of plants with lower nitrogen requirements from the community, ultimately causing their decline or even extinction. Numerous studies indicated that nitrogen fertilization favored the growth and invasion of exotic species (Bobbink et al., 2010; Antonio and Mack, 2006). Eller and Oliveira (2017) found that the invasive plant Melinis minutiflora exhibited a stronger competitive advantages and interfered with the uptake of nitrogen by native specie Aristida adscensionis, making it more beneficial in high-nitrogen environments. Additionally, Peng et al. (2019) reported that nitrogen addition increased the leaf lifespan, plant height, and early flowering of the invasive plant Solidago canadensis. Invasive species typically exhibit excellent phenotypic plasticity and resource use efficiency compared to native species (Vaz-Pinto et al., 2014), enabling them to survive even under stressful conditions. Furthermore, studies suggested that different plants exhibit varied responses and preferences for different nitrogen forms (Yousaf et al., 2016; Qian et al., 2021; He et al., 2021). The invasive species Wedelia trilobata exhibited better adaptation to environmental conditions with an NH4+-N/NO3–N ratio of 2:1 through faster growth and antioxidant defense system compared to Wedelia chinensis (Huang et al., 2022). Under elevated NH4+-N levels, the invasive species Phyllostachys edulis demonstrated superior growth, nitrogen uptake and NH4+-N tolerance compared to Castanopsis fargesii, thereby facilitating its expansion (Zou et al., 2020). The invasive plant Flaveria bidentis exhibited an increase in plant height and branching under high ammonium cultivation conditions (Huangfu et al., 2016). Conversely, some studies suggested that invasive plants such as Amaranthus retroflexus and Bidens pilosa exhibit better growth advantages in habitats with higher NO3–N level (Wang et al., 2018; Chen and Chen, 2019). However, most researchers have primarily focused on the physiological and ecological responses of invasive plants to nitrogen deposition and its various forms, leaving the invasion mechanisms of exotic plants remain unclear.
Mikania micrantha (Asteraceae family), native to Central and South America, has become widespread in Asia and the Pacific Islands and is listed as one of the world’s top 100 most threatening alien invasive species. Due to its rapid growth and strong adaptability, it can quickly colonize invaded areas, causing significant damage to local ecosystems and severe economic losses (Day et al., 2016). The rapid growth of stem is an important characteristic of M. micrantha, with certain photosynthetic activity (Cai et al., 2023; Liu et al., 2020) and stress resistance (Chen et al., 2024; Zhang et al., 2019), playing an important role in its rapid invasion process. Fang et al. (2021) found that M. micrantha has expanded rapidly in terms of invaded area over the past 30 years. It is predicted that in the 2050s and 2070s, M. micrantha will continue to rapidly spread from Yunnan and Guangdong provinces towards the northern regions and inland areas. Therefore, understanding the mechanisms facilitating the rapid growth of M. micrantha is crucial for the effective control of invasive plant species. Studies have shown that as CO2 concentrations and nitrogen deposition rose, the invasive potential of M. micrantha increased (Zhang et al., 2016). Compared to native plants Polygonum chinense and Paederia scandens, M. micrantha demonstrated strong competitive resource utilization capabilities in terms of nitrogen acquisition and soil nitrogen mineralization (Yu et al., 2021). Liu et al. (2020) found that NH4+-N significantly increased in soil after the invasion of M. micrantha, but NO3−-N content significantly decreased. These results suggested that M. micrantha possesses a strong ability to acquire nitrogen, potentially exhibiting preferential selection for the NO3–N. Hence, we propose a hypothesis that, in the context of increasing global nitrogen deposition, particularly with the continuous increase of NO3−-N deposition, the rapid growth of M. micrantha may enhance nitrogen utilization efficiency in the main form of NO3− by regulating the expression levels of key genes or proteins involved in NO3−-N absorption, thereby accelerating its diffusion trend to the north and inland. Our study aims to improve the management of M. micrantha invasion control in the context of global change in the future.
Materials and methods
Plant collection and cultivation
The naturally growing M. micrantha and associated plants (P. scandens, Ipomoea nil, and Ipomoea cairica) were used for the identification of nitrogen absorption patterns. Samples were collected from the South China Normal University botanical garden in Guangzhou, China (23°10′N, 113°21′E) during growing season (July–August). Stems from the first to the fourth internodes of M. micrantha and its associated plants were collected for physiological data determination. The rhizosphere soil of plant was collected and stored at 4°C.
Control experiment starting from seed germination was carried out to observe the response of stems under different NO3−-N concentrations. Seeds of M. micrantha and P. scandens were placed in a constant temperature incubator (12h/12h light-dark cycle, light intensity of 100–120 mmol m-2 s-1, day/night temperature of 25 ± 1 °C). After 1–2 weeks of cultivation, healthy seedlings were selected and transplanted into pots. The cultivation substrate consisted of a mixture of Arabidopsis soil and vermiculite in a 3:1 ratio. After one month of cultivation, M. micrantha and P. scandens with consistent growth were selected for nutrient solution cultivation experiments. Samples with similar growth were divided into 6 groups, and were treated with modified Hoagland nutrient solutions with 0, 0.5, 5, 10, 20, and 40 mM NO3−-N respectively (Table 1). Fifteen repetitions were set for each group. Each group was treated for 30 days, with treatment every 3 days.
Table 1
| Nutrition | Reagent | Concentation (g L-1) | Dosage (mL) | |||||
|---|---|---|---|---|---|---|---|---|
| 0 mM | 0.5 mM | 5 mM | 10 mM | 20 mM | 40 mM | |||
| Macroelement | KNO3 | 50.55 | 0 | 1 | 10 | 20 | 40 | 80 |
| KCl | 37.28 | 80 | 79 | 70 | 60 | 40 | 0 | |
| Microelement | CaCl2 | 22.2 | 10 | |||||
| KH2PO4 | 5.444 | 10 | ||||||
| MgSO4·7H2O | 24.65 | 10 | ||||||
| H3BO3 | 0.185 | 1 | ||||||
| MnSO4·H2O | 0.0845 | 1 | ||||||
| CuSO4·5H2O | 0.0250 | 1 | ||||||
| ZnSO4·7H2O | 0.0288 | 1 | ||||||
| (NH4)6Mo7O24·4H2O | 0.124 | 1 | ||||||
| FeSO4·7H2O | 13.9 | 1 | ||||||
| EDTANa2·2H2O | 18.6 | 1 | ||||||
Modified hoagland nutrient solution.
Analysis of basic growth indicators
Plant main stem length, leaf number, branching number, and axillary bud number were measured using a ruler every 3 days. Phenotypic changes were documented using a camera. Subsequently, the roots, stems, and leaves of each plant were separated, dried at 75°C for 72 hours, and weighed to record the biomass.
Soil physicochemical properties
Soil physicochemical properties, including pH, moisture content, NH4+–N and NO3−–N levels, and protease enzyme activity, were measured as follows: (1) Measurement of pH value and moisture content: Following the method by Yu et al. (2021), 5 g of fresh rhizosphere soil was mixed with 0.01 M CaCl2 solution, shaken, and extracted for 30 minutes. Soil pH was measured directly measured using a pH meter (ST3100, Ohaus Instruments (China) Co., Ltd.), and moisture content was determined after drying at 60°C. (2) Nitrogen content determination: Inorganic nitrogen forms (NH4+ –N and NO3−–N) were extracted from fresh soil using 2 M KCl solution (Yu et al., 2021). NH4+–N was quantified using the indophenol blue colorimetric method, with absorbance measured at 625 nm. NO3−–N was measured directly via UV spectrophotometry. Standard curves were prepared using ammonium sulfate ((NH4)2SO4) for NH4+–N and potassium nitrate (KNO3) for NO3−–N. (3) Protease enzyme activity determination: Protease enzyme activity was determined using the sodium caseinate method (Ladd and Butler, 1972). Fresh soil (2.5 g) was incubated with Tris buffer (pH=8.1) and 2% sodium caseinate solution at 50°C for 2 hours. After adding 15% TCA solution and filtering, the supernatant was mixed with alkaline reagent and Folin’s reagent. Absorbance was measured at 700 nm, and a standard curve was prepared using sodium caseinate.
Determination of nitrogen content and nitrogen metabolism enzyme activities in plant tissues
The nitrogen content (NO3−–N and NH4+–N) and key enzymes activities related to nitrogen metabolism, including nitrate reductase (NR), nitrite reductase (NiR), glutamine synthetase (GS), glutamate synthase (GOGAT) in plant tissues, were measured using reagent kits from Suzhou Koming Biotechnology Co., Ltd. The activity of GDH was determined according to the method described by Zhou et al. (2015). Fresh stem segments (third or fourth) and lateral roots (0.1 g) were homogenized in 1 mL extraction buffer (Suzhou Koming Biotechnology Co., Ltd.) with quartz sand in an ice bath. The homogenate was centrifuged at 10,000 ×g and 4°C for 10 min, and the supernatant was stored at 4°C for analysis. The 3 mL reaction mixture consisted of 2.6 mL of stock solution (115.4 mmol L-1 pH=8 Tris-HCl buffer, 23.1 mmol L-1 a-Ketoglutaric acid, 231 mmol L-1 NH4Cl), 0.1 mL CaCl2, and 0.1 mL ddH2O. After incubating at 30°C for 10 minutes, 0.1 mL of NADH and 0.1 mL of the supernatant were added. Absorbance values at a wavelength of 340 nm were measured every minute using a UV-2450 spectrophotometer (Shimadzu, Tokyo, Japan). GDH activity were expressed as nmol min-1 g-1 FW.
Determination of free amino acid content
The free amino acid content was determined following Doi et al. (1981) with slight modifications.
Fresh stem segments (third or fourth) and lateral roots (0.1 g) were homogenized in 1.5 mL of 10% acetic acid with quartz sand in an ice bath. The homogenate was centrifuged at 12,000 × g and 4°C for 10 min to obtain the supernatant. A volume of 1 mL of the supernatant was mixed with 1 mL sodium acetate buffer (pH=5.4) and ninhydrin reagent, then heated in a boiling water bath at 100°C for 15 min. After cooling, 3 mL of 60% ethanol was added, followed by thorough mixing. The absorbance at OD570nm was measured using a spectrophotometer.
Analysis of transcriptome sequencing
To analyze the gene expression patterns of M. micrantha stems in response to different NO3−-N concentrations, the third to fourth internodes of stems at 0 mM and 5 mM were selected for transcriptomic sequencing. Total RNA extraction and library construction were conducted using the Spin Column Plant Total RNA Purification Kit (Sangon, Shanghai, China.) and the HiPure Total RNA Mini Kit (Magen), respectively. RNA integrity and purity were assessed using agarose gel electrophoresis and UV spectrophotometry. Qualified RNA were subjected to library construction using the Novogene NGS RNA Library Prep Kit. The quality of the constructed libraries was evaluated using the Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System, followed by sequencing on the IIIumina HiSeq 4000 platform after passing quality control. Clean reads were aligned to the reference genome using HISAT2 software. Differential gene expression analysis was performed using DESeq2 software, with the criteria for selecting differential genes set as an absolute fold change ≥2 and Padj value ≤0.05. DEGs were functionally annotated using databases such as Nonredundant protein (Nr), Nonredundant nucleotide (Nt), Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Ontology (GO).
Quantitative real time RT-PCR of key gene expression
Following the protocol of Cai et al. (2022), fresh stem segments (0.1 g) were ground in liquid nitrogen. Total RNA was isolated using a Quick RNA isolation Kit (Huayueyang) according to the manufacturer’s protocol and was quantified using a spectrometer (NanoDrop). cDNA was synthesized using the TopScript-RT-DryMIX (dT18) kit (Takara, Tokyo, Japan). The relative expression levels of key genes were analyzed was estimated by qRT-PCR with a SYBR Green master mix (SYBR Green Premix Ex Taq, Takara, Japan) in a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). The PCR program was run as follows: 95°C for 30 seconds, followed by 39 cycles of 95°C for 5 s, 60°C for 34 s, and 65°C for 5 s, with a final extension step at 95°C for 50 seconds. The reference gene used was 18S, and the relative expression of the candidate genes were calculated using the 2–ΔΔCT method. The specific primers for reference gene and candidate genes were listed in Table 2.
Table 2
| Gene name | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| 18s | GTCGGGGGCATTCGTATTTC | CGGCATCGTTTATGGTTGAG |
| NPF5 | TCAGCCGTCTTGACCACTTC | ACCTGAGCTTTCCTCTGCAC |
| NPF6 | GGGACAGGCACACTAGCATT | ATTTAAGACCGCCAGTCCCG |
| CLC-g | TGCTATCGTCGGCTCCAATC | CTGTTGCGCCAATCTTGCTT |
| SLAH3 | ATATGTCTCGGCGTCAGCAG | GGACGGAGATGCACCATAGG |
| HY5.1 | CCGAAGGTTCCCGGAGAAAA | TTGACTCGCACCTCCAACTC |
| LBD38 | GATACAGTCCTTCGAGGCGG | ACGGACTTCCCTGACGTAGA |
| NR1.1 | ACGACTGGTCCGTGGAGATA | GTTGCCGGGAACTCTCTTGA |
| NiR1.2 | AGTTCCACCAGGCTGTATGC | ATCGGTTCACCGCCAATCTT |
| GS | ATTTCCGCTGGTGACGAGTT | TCCAGCACCATTCCAGTCAC |
| GDH | ACGACTTGCTGGTTTCCGAT | GCAGCAGCATGCTTGTATCC |
Gene-specific primers used for qRT-PCR.
Data analysis
The statistical analyses, including significant difference analysis and regression analysis, were conducted using IBM SPSS Statistics 19.0 (IBM, NY, USA). The results were presented as mean ± standard error. Independent sample t-tests were utilized for the significant difference analysis between each concentration group (0.5–40 mM) and the control group (0 mM), with * indicating a significant difference (0.01<p<0.05), ** indicating an extremely significant difference (p<0.01), and ns indicating no significant difference (p>0.05). Data visualization was performed using SigmaPlot 14.0 (Systat Software, San Jose, CA, USA). Additionally, a correlational analysis (Pearson correlation) was applied to measure the correlations among between M. micrantha and P. scandens under different concentrations of NO3−–N (0–40 mM). Principal component analysis (PCA) was carried out to describe the degree of association and determine possible factors that affect the biomass in both plant under NO3−–N concentration. PCA analysis was performed using Origin 2018 software (OriginLab, Northampton, MA, USA).
Results
Differences in nitrogen absorption, utilization, and metabolism of M. micrantha and its companion plants
The analysis of rhizosphere soil properties (Table 3) showed that I. cairica exhibited significantly higher soil moisture content compared to M. micrantha, I. nil, and P. scandens (p<0.05). The NO3−–N content in M. micrantha was significantly higher compared to P. scandens, I. nil, and I. cairica by 2.3-, 2.5-, and 2-fold, respectively, as evidenced by the non-overlapping 95% confidence intervals (CI). Conversely, NH4+–N content in I. cairica and I. nil were notably higher than that in M. micrantha and P. scandens. The protease activity was significantly higher in M. micrantha, surpassing I. cairica and I. nil by approximately 1.3-fold but only slightly exceeding P. scandens.
Table 3
| Indices | M. micrantha | I. cairica | I. nil | P. scandens | |
|---|---|---|---|---|---|
| Soil | Moisture content(%) | 21.17±0.19b (20.68, 21.66) | 26.12±1.59a (22.04, 30.21) | 21.73±0.51b (20.53, 22.93) | 22.85±0.99b (20.31, 25.39) |
| NH4+–N (μg g-1 Fw) | 0.22±0.01bc (0.20, 0.25) | 0.34±0.02ab (0.23, 0.45) | 0.41±0.07a (0.18,0.64) | 0.16±0.08c (0.13, 0.19) | |
| NO3-–N (μg g-1 Fw) | 0.65±0.03a ( 0.57, 0.72 ) | 0.28±0.05b ( 0.13, 0.43) | 0.26±0.18b ( 0.20, 0.31) | 0.31±0.06b ( 0.13, 0.49) | |
| Protease (μg g-1 h Fw) | 26.88±0.40a ( 25.16, 28.59) | 20.36±2.02b (15.18, 25.55) | 20.58±1.10b (17.53, 23.63) | 25.37±0.93ab (21.36, 29.38) | |
| Root | NH4+–N (μg g-1 Fw) | 22.37±0.95b (19.33, 25.41) | 26.31±0.73ab (24.44, 28.17) | 25.23±0.91b (22.33, 28.13) | 32.05±3.73a (21.68, 42.42) |
| NO3-–N (μg g-1 Fw) | 123.14±8.35a (99.95, 146.32) | 118.71±9.04a (89.95, 147.47) | 53.97±3.06b (46.12, 61.83) | 62.41±3.99b (45.24, 79.59) | |
| NR (nmol min-1 g-1 Fw) | 225.05±11.74a (187.69, 262.41) | 233.09±15.39a (184.11, 282.07) | 106.90±3.56c (95.56, 118.24) | 172.32±6.56b (154.12, 190.53) | |
| GS (μmol min-1 g-1 Fw) | 5.59±0.09c (5.31, 5.87) | 7.80±0.30a (6.83, 8.76) | 6.33±0.08b (6.09, 6.56) | 6.77±0.21b (6.10, 7.44) | |
| GDH (nmol min-1 g-1 Fw) | 0.14±0.10a (0.10, 0.18) | 0.09±0.01b (0.07, 0.10) | 0.09±0.01b (0.07, 0.12) | 0.06±0.01c (0.03, 0.09) | |
| NH2-N content (mg g-1 Fw) | 0.22±0.05a (0.07, 0.37) | 0.14±0.03ab (0.03, 0.25) | 0.10±0.01b (0.07, 0.13) | 0.11±0.01ab (0.08, 0.15) | |
| Stem | NH4+–N (μg g-1 Fw) | 18.94±1.50d (14.16, 23.71) | 44.99±3.50b (35.27, 54.71) | 58.61±3.16a (48.56, 68.67) | 34.28±2.82c (25.31, 43.25) |
| NO3-–N (μg g-1 Fw) | 345.31±46.37a (145.79, 544.82) | 136.77±14.10b (76.13, 197.41) | 234.63±19.53b (150.59, 318.66) | 226.24±31.74b (125.24, 327.25) | |
| NR (nmol min-1 g-1 Fw) | 167.18±16.18a (97.56, 236.80) | 212.19±22.58a (115.03, 309.35) | 102.88±19.64b (18.36, 187.40) | 30.01±5.67c (5.61, 54.41) | |
| GS (μmol min-1 g-1 Fw) | 4.86±0.29c (4.12, 5.61) | 8.30±0.65b (6.64, 9.97) | 13.10±0.38a (12.12, 14.07) | 4.68±0.22c (4.06, 5.30) | |
| GDH (nmol min-1 g-1 Fw) | 0.11±0.01ab (0.07, 0.14) | 0.11±0.01a (0.08, 0.14) | 0.08±0.00b (0.07, 0.10) | 0.04±0.01c (0.03, 0.06) | |
| NH2-N content (mg g-1 Fw) | 4.35±0.73a (1.21, 7.49) | 3.62±0.37a (2.46, 4.78) | 1.60±0.11b (1.12, 2.08) | 2.21±0.28b (0.99, 3.44) |
Comparison of nitrogen absorption characteristics in soil, roots, and stems of M. micrantha and its associated plants under field conditions (mean ± standard error, n=5, numbers in parentheses are 95% CI).
Nitrogen nutrient analysis in the roots and stems (Table 3) of M. micrantha and its companion plants revealed that M. micrantha exhibited the highest NO3−–N content in both roots and stems compared to other plants. Specifically, the NO3−–N content in M. micrantha roots was twice with non-overlapping 95% CI as high as in I. nil and P. scandens, while in stems, it was 1.4- and 1.5-fold higher than I. nil and P. scandens, respectively. The NH4+–N content was highest in the roots of P. scandens, while I. nil had the highest NH4+–N content in the stems.
Further analysis of enzyme activities involved in nitrogen metabolism (Table 3) revealed that M. micrantha had significantly higher NR and GDH activity in roots and stem compared to other plants. Specifically, NR activity in M. micrantha roots and stem was 2.1-/1.6- and 1.3-/5.6-fold higher than I. nil and P. scandens, respectively. GDH activity in M. micrantha roots was 1.6-, 1.5-, and 2.3-fold higher than I. cairica, I. nil, and P. scandens. In the stem, GDH activity in M. micrantha was similar to I. cairica but significantly higher than P. scandens. However, GS activity in M. micrantha was lower than that of other plants in both roots and stems.
As products of nitrogen metabolism, the free amino acid content in M. micrantha was also higher than in other plants, with the root content significantly higher than I. nil, approximately 2.2-fold. In stems, free amino acids were 2.7- and 2.0-fold higher than in I. nil and P. scandens, respectively.
The change in phenotypic parameters of M. micrantha stems under different NO3−–N concentrations
The results showed that there was a significant increase in the main stem length, leaf number and branch numbers of M. micrantha in the 5, 10, 20, and 40 mM treatment groups compared to the 0 mM (p<0.05). The increments in stem length for the 0.5, 5, 10, 20, and 40 mM groups were respectively 1-, 1.4-, 1.3-, 1.3-, and 1.1-fold (Figure 1A). Notably, the 5 mM group exhibits a significantly highest mean value compared to the 0 mM (Supplementary Table S1; mean difference (Δ) (95% CI of Δ) = 26.733 (19.720, 33.747), p<0.001); The increases in leaf numbers were 1.3-, 2.1-, 2.3-, 2.1-, and 2.2-fold, respectively (Figure 2B), in which the 10 mM group has the highest mean increment compared to the 0 mM (Supplementary Table S1; Δ(95% CI of Δ) = 4.000 (3.274, 4.726), p<0.001); and the increases in branch numbers were 2.7-, 11.3-, 11.3-, 13-, and 12.7-fold higher than the 0 mM group, with the 20 mM group exhibiting the highest mean value (Supplementary Table S1; Δ(95% CI of Δ) = 13.000 (11.323, 14.677), p<0.001), while axillary buds numbers significantly decreased (Figure 1C). In contrast, P. scandens showed no significant changes in stem length across treatments, as their 95% confidence intervals of mean difference include zero (Supplementary Table S1), though branching and axillary bud numbers increased compared to the control, with the exception of axillary buds in the 20 mM group. Overall, M. micrantha exhibited greater increases in stem length, leaf number, branch, and axillary bud numbers than P. scandens at the same NO3−–N concentrations, consistent with the phenotypic results (Supplementary Figure S1).
Figure 1

Change in growth indicators of M. micrantha and P. scandens under different NO3⁻-N concentrations at 30 days. (A) is the increment of stem length, (B) is the increment of leaf number, (C) is the increment of axillary bud and branch number, (D) is the stem and leaf biomass ratio, and (E) is the plant biomass, including the stem, leaves, aboveground biomass, and total biomass. The results are the mean ± SEM of five biological replicates. Each concentration group (0.5–40 mM) was compared with the control group (0 mM) by independent sample t-tests. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001), and ns indicates no significant difference.
Figure 2

Changes in NO3−–N content and nitrogen metabolic enzyme Activity of M. micrantha and P. scandens under different NO3−–N concentrations at 30 days. (A) is the NO3−–N content in stem and leaf, (B–G) is the NR, NiR, GS, GDH, GOGAT activity and amino acid content in stem, respectively. The results are the mean ± SEM of five biological replicates. Each concentration group (0.5-40 mM) was compared with the control group (0 mM) by independent sample t-tests. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001), and ns indicates no significant difference.
The changes in biomass of different plant organs of M. micrantha stems under different NO3−–N concentrations
Further analysis of the biomass of the two plants showed that, compared to the 0 mM group, M. micrantha exhibited a significant increase in aboveground biomass and total biomass in the 5, 10, 20, and 40 mM treatment groups (Figures 1C, E), with the 10 mM group exhibiting the highest increment in aboveground biomass (Supplementary Table S1; Δ(95% CI of Δ) = 1.290 (1.046, 1.534), p<0.001). Specifically, the biomass values for stem, leaf, aboveground, and total biomass in M. micrantha were consistently higher across all treatments, with increases ranging from 1.1- to 2.8-fold for stem biomass, 1- to 3.5-fold for leaf biomass, 1.1- to 3-fold for aboveground biomass and 1.1- to 2.4-fold for total biomass. In contrast, P. scandens showed more moderate increases in biomass, with the highest increments in the 5 and 20 mM groups. Additionally, M. micrantha demonstrated an increase in leaf biomass ratio with the increase in NO3−–N concentration, while the stem biomass ratio remained relatively unchanged except for a 15.9% increase at 10 mM. Conversely, P. scandens showed no significant changes in stem and leaf biomass ratios under different treatments (Figure 1D).
The changes in NO3−–N content and the expression levels of key genes involved in NO3−–N absorption and transport in M. micrantha stems under different NO3−–N concentrations
The NO3−–N content in the stems of two plants was higher than that in the control group, with M. micrantha and P. scandens stems showing NO3−–N content 1.2-/1.9-fold, 3.1-/3.4-fold, 6.9-/4.9-fold, 15.7-/6.1-fold, and 21.9-/6.3-fold higher than the control group under 0.5, 5, 10, 20, and 40 mM treatments, respectively, suggesting that NO3−–N content of two species reached the highest level in the 40 mM group (Supplementary Table S2; Δ(95% CI of Δ) = 518.866 (398.579, 639.156), p<0.001/Δ(95% CI of Δ) = 296.782 (200.144, 393.421), p<0.001). Interestingly, the elevation in M. micrantha at 20 and 40 mM treatments were higher than that in P. scandens, as the higher mean values and mean differences of M. micrantha (Figure 2A; Supplementary Table S2). RNA-seq and qPCR results revealed that M. micrantha exhibited upregulation of genes related to NO3−–N absorption at 5 mM group. Notably, NPF6.3, CLCb1, CLCb2, and SLAH3 showed the most significant upregulation, with expression levels 3.8- to 9.3-fold higher than 0 mM group, respectively. However, two genes in the NPF family, NPF8.1 and NPF5.10, were significantly downregulated at 5 mM, with expression levels 0.48- and 0.17-fold of the control, respectively (Figure 3). QPCR results further demonstrated that under 0.5, 5, 10, 20, and 40 mM group, the expression levels of NPF5 in M. micrantha were 3.3- to 10-fold of the 0 mM group (Figure 4A). The expression levels of NPF6 in M. micrantha were 2.4- to 3.4-fold of the 0 mM group (Figure 4B). Further analysis showed varying expression patterns of transcription factors. Positive regulators, such as CLC-g, HY5.1, and SLAH3, exhibited the highest expression at different concentrations, with CLC-g (Figure 4C) and SLAH3 (Figure 4D) peaking at 5 mM (2.1- and 1.6-fold, respectively), while HY5.1 (Figure 4E) peaked at 20 mM (12-fold). The negative feedback factor LBD38 decreased in expression with higher NO3−–N concentrations (Figure 4F), especially at 10, 20, and 40 mM, where expression levels were 0.3-, 0.2-, and 0.2-fold of the 0 mM group.
Figure 3

Heat map of genes related to nitrogen metabolism in M. micrantha stem under 0 mM and 5 mM NO3−–N treatment.
Figure 4

Gene expression related to nitrogen absorption transporters in M. micrantha stem under different NO3−–N concentrations. (A, B) are E3N88_41881 (NPF5), E3N88_09314 (NPF6), respectively. (C–F) are CLC-g, SLAH3, HY5.1 and LBD38, respectively. (G–J) are NiR1.2, NR1.1, Gs and GDH, respectively. (K, L) are ALDO and CYSK, respectively. The results are the mean ± SEM of six biological replicates. Each concentration group (0.5-40 mM) was compared with the control group (0 mM) by independent sample t-tests. Asterisks indicate significant differences (*p < 0.05, **p < 0.01, ***p < 0.001), and ns indicates no significant difference.
The changes in nitrogen metabolism enzyme activities and the expression levels of key genes in nitrogen metabolism pathways in M. micrantha stems under different NO3−–N concentrations
In M. micrantha, NR activity significantly increased at 5, 10, and 20 mM NO3−–N concentrations compared to the 0 mM group, with 3.4-, 1.8-, and 1.9-fold increases, respectively, and a similar trend was observed in P. scandens (Figure 2B). NiR activity in M. micrantha was 1.8- and 1.5-fold higher in the 5 and 20 mM groups (Supplementary Table S2; Δ(95% CI of Δ) = 1.446 (1.051, 1.840), p<0.001/Δ(95% CI of Δ) = 0.853 (0.458, 1.247), p<0.001), while P. scandens showed a 16% increase at 5 mM (Supplementary Table S2; Δ(95% CI of Δ) = 1.296 (0.186, 2.406), p<0.05, Figure 2C). For the GS/GOGAT cycle, GS activity in M. micrantha significantly decreased in the 20 and 40 mM groups, while P. scandens showed no significant difference, except for a significant increase in the 20 mM group (Figure 2D). GOGAT activity in M. micrantha peaked at 5 mM, showing a 2.2-fold increase (Supplementary Table S2; Δ(95% CI of Δ) = 129.980 (79.608, 180.352), p<0.001), whereas P. scandens exhibited a significant decrease of 41.1% in the 5 mM group (Supplementary Table S2; Δ(95% CI of Δ) = -158.831 (-264.702, -52.960), p<0.01, Figure 2F). GDH activity in M. micrantha increased significantly at 5 mM (Supplementary Table S2; Δ(95% CI of Δ) = 10748.817 (7330.776, 14166.857), p<0.001), being 2.4-fold higher than in P. scandens (Figure 2D). Combining RNA-seq and qPCR results to further analyze genes related to nitrate assimilation, including NR, NiR, and GDH, showed high expression at 5 mM in M. micrantha. Notably, NiR1/2/3 expression increased 21.6-, 10.7-, and 11-fold, respectively (Figure 4G). In contrast, GS1 and GS2 expression was low at 5 mM. NR1.1 expression increased 11.8-fold at 10 mM (Figure 4H), while NiR1.2 expression was significantly higher in the 5, 10, and 20 mM groups (4.6-, 7.6-, and 6.6-fold, respectively). GS expression decreased by 62.4–71.0% at higher NO3−–N concentrations (Figure 4I), while GDH gene expression increased by 159.2% at 5 mM (Figure 4J).
The change in free amino acid content and the expression levels of key genes in amino acid synthesis pathways in M. micrantha stems under different NO3−–N concentrations
The free amino acid content in M. micrantha significantly higher under 5, 10, 20, and 40 mM treatments, with increases of 5.8-, 5.3-, 5.3-, and 6.3-fold, respectively. Similarly, P. scandens showed a significant increase in free amino acid content under 10, 20, and 40 mM treatments, with no significant difference under 0.5 and 5 mM treatments. Overall, M. micrantha exhibited higher free amino acid content than P. scandens under 5, 10, 20, and 40 mM treatments, as shown by the higher mean values and mean differences of M. micrantha (Figure 2G; Supplementary Table S2). Gene expression analysis revealed that the expression levels of ALDO and CYSK genes in M. micrantha was significantly higher at 5 mM group, approximately 2.5- and 3.8-fold of the 0 mM group, respectively. Conversely, PK gene expression was lower in the 5 mM group (Figure 5). qPCR results further demonstrated that under 0.5, 5, 10, 20, and 40 mM group, the expression levels of ALDO in M. micrantha were 0.8-, 2,4-, 2.0-, 4.9-and 0.2-fold of the 0 mM group, respectively (Figure 4K). The expression levels of CYSK in M. micrantha were 1.0-, 1.2-, 0.9-, 5.3- and 0.7-fold of the 0 mM group, respectively (Figure 4L).
Figure 5
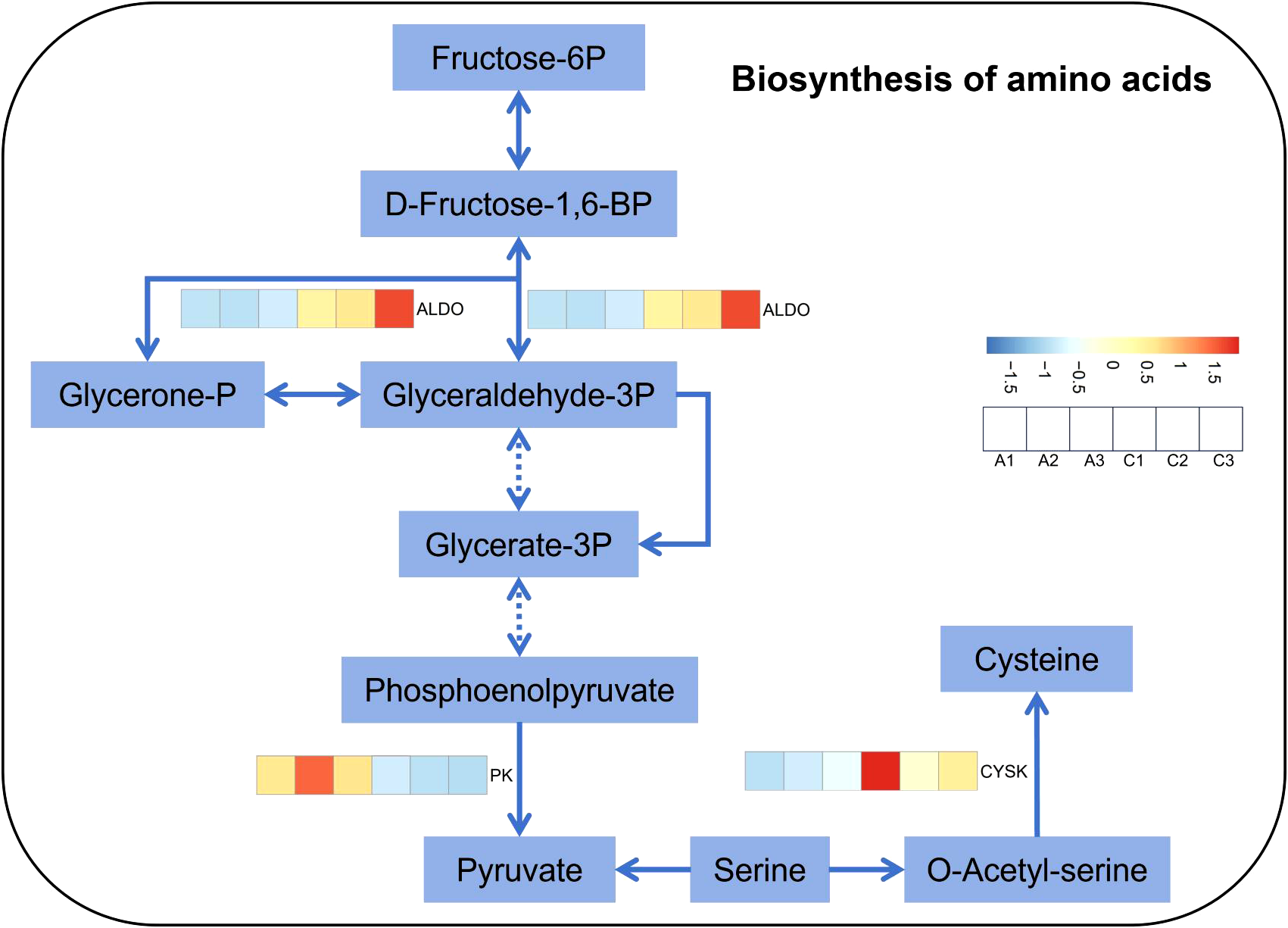
Heat map of genes related to biosynthesis of amino acids in M. micrantha stem under 0 mM and 5 mM NO3−–N treatment.
Correlation analysis between biomass and nitrogen metabolism-related enzyme activities
The correlation analysis revealed significant associations between biomass and nitrogen metabolism-related enzyme activities in two species. In M. micrantha, leaf biomass, stem biomass and total biomass displayed a highly positive correlation with NO3−–N content (Figure 6A). However, NO3−–N content had a relatively small impact on the stem biomass of P. scandens, and with no significant effect on leaf biomass (Figure 6B). Additionally, in M. micrantha, biomass demonstrated a significant positive correlation with GOGAT and FAA, a strong negative correlation with GS, and a positive correlation with NR and NiR but without statistical significance (Figure 6A). In P. scandens, biomass showed significant positive correlations with NR, Gs, GDH, and FAA, while presenting a negative correlation with NiR (Figure 6B).
Figure 6

Correlation analysis between biomass and nitrogen metabolism-related enzyme activities in M. micrantha(A) and P. scandens(B).
Principal component analysis
The scores plot showed that M. micrantha was significantly separated under different NO3−–N concentrations (Supplementary Figure S5A), suggesting its high sensitivity to changes in NO3−–N concentration and more variable metabolic response. The first two principal components (PC1 and PC2) accounted for 80.35% of the total variation. PC1 accounted for 52.21% of the total variance, with the highest contributions from Biomass (0.41), NiR (0.36), GOGAT (0.41) and FAA (0.44). These strongly correlated eigenvectors may affect stem growth by dynamically regulating nitrogen enzyme activities (NiR, GOGAT), thus explaining the rapid response to nitrate fluctuations. PC2 explained 28.14% of the total variance and was primarily influenced by Nitrate (-0.52), NR (0.46), GS (0.43), and GDH (0.40). The significant negative loading of NO3−–N and positive loadings of enzymes in PC2, potentially highlighting the influence of NO3−–N absorption and distribution on nitrogen metabolism (Supplementary Table S3-1). The PCA biplot reveals two distinct clustering groups. The 5 mM-40 mM groups were positively linked to NR, GDH, NiR, GOGAT, FAA, Biomass, and Nitrate, while the 0 mM-0.5 mM groups were positively associated with GS (Figure 7A). The diversity of biochemical traits in the 5 mM-40 mM group may suggest that M. micrantha responds to external nitrate fluctuations through flexible nitrogen assimilation strategy. Conversely, P. scandens clustered closely under different NO3−–N concentrations, indicating that it is insensitive to nitrate treatment and a relatively consistent response pattern (Supplementary Figures S5B, Figure 7B). PC1, which captures 55% of the variance, is largely influenced by Biomass (0.41), Nitrate (0.44), GS (0.35), GDH (0.37) and FAA (0.39), which may indicate that nitrogen metabolism (especially GS and GDH) possibly related to the insignificant changes in stem growth during increasing NO3−–N concentrations. PC2, which explains a smaller portion of the variance (21.59%), captures eigenvectors changes less directly related to responses under nitrate concentration (Supplementary Table S3-2).
Figure 7

The principal component analysis (PCA) of in M. micrantha(A) and P. scandens(B).
Discussion
M. micrantha prefers to absorb NO3−–N
The observed variations in soil properties among M. micrantha and its companion plants not only highlight the influence of plant species on rhizospheric characteristics but also raise intriguing questions about the underlying mechanisms. The significant variations in NO3−–N and NH4+–N levels in the rhizospheric soil among different plant species underscore the complexity of nitrogen cycling in the root zone. The preferential uptake of NO3−–N by M. micrantha, as evidenced by the higher NO3−–N content in its rhizospheric soil (Table 3), aligns with the well-documented preference of certain plant species for specific nitrogen forms (Wang et al., 2018;Chen and Chen, 2019). Previous studies had shown that blackberry plants preferentially take up NH4+–N (Duan et al., 2023). Sphagneticola canadensis was a NO3−-N-preferring plant (Wang et al., 2023). The invasive plant Xanthium strumarium preferred to use NO3−, while its native congener X. sibiricum preferred to use NH4+ (Zhang et al., 2022). The preference of Cymbidium tracyanum for NO3− as a nitrogen form may be a result of long-term adaptation to epiphytic habitat (Dong et al., 2024). The rhizospheric soil of Bidens pilosa (Spanish needle) exhibited a high concentration of NO3−–N, possibly attributed to the significant enhancement of soil nitrification by root exudates. This process converts more NH4+–N into absorbable NO3−–N, thereby increasing the root’s absorption efficiency of nitrogen nutrients and promoting the aboveground growth (Chen et al., 2009; Yu et al., 2021). Additionally, protease, as a crucial enzyme in the soil nitrogen cycle, influences the release of soil available nutrients. The rhizospheric soil of M. micrantha demonstrated elevated protease activity (Table 3), indicating an accelerated nitrogen metabolism process, thereby enhancing its efficiency in utilizing organic nitrogen (Li et al., 2006). Similar results have been observed in studies of other invasive plants such as I. cairica, Synedrella nodiflora, Lantana camara, and W. trilobata, where high protease activity in the rhizospheric soil was identified (Li et al., 2008).
High expression of transporter genes promotes the absorption of NO3−–N
Nitrate is one of the main forms of inorganic nitrogen absorbed by plants, and its concentration variation has a direct impact on plant growth. Currently, research on the influence of NO3−–N on plant growth primarily focuses on aspects such as leaf photosynthetic capacity, leaf morphology, or root architecture (Takei et al., 2001; Crawford and Forde, 2002; Guiboileau et al., 2012; Vidal et al., 2014; Chen and Chen, 2019). However, limited attention has been given to non-foliar organs such as stems. Most studies have observed changes in physiological characteristics such as increased plant height, stem thickness, and biomass under higher nitrate nitrogen levels (Song et al., 2021), consistent with the findings of our study (Figure 1). The rapid growth of stems is one of the most prominent characteristics of M. micrantha. Our study found that with increasing NO3−–N concentration, the main stem length (Figure 1A) and leaf number (Figure 1B) of M. micrantha significantly increased, indicating that the axillary buds continuously activated the development of growth component branches (Figure 1C). As a result, the number of axillary buds decreased, while the number of branches increased (Bennett and Leyser, 2006). Results from PCA (Figure 7) and correlation analysis (Figure 6) further indicated that, unlike P. scandens, M. micrantha was more significantly influenced by NO3−–N, and its biomass was positively correlated with NO3−–N, NR, NiR, GOGAT, and FAA. The transport and assimilation of NO3−–N within plants directly impact plant growth and development, linked to their nitrogen metabolism capabilities (Yang et al., 2023; Xu et al., 2023). Currently, there was limited research on the nitrogen metabolism capacity of plant stems. RNA-seq analysis was conducted on M. micrantha stems treated with 5mM and 0 mM NO3−–N. Using Fold Change ≥ 2 and FDR < 0.01 as criteria for differential gene selection, 345 differentially expressed genes were identified, with 181 upregulated and 164 downregulated (Supplementary Figure S2). These genes primarily enriched pathways related to plant hormone signal transduction, phenylpropanoid biosynthesis, pentose and glucuronate interconversions, and nitrogen metabolism (Supplementary Figure S4). The results suggested associations between these pathways and nitrogen nutrient responses, indicating the involvement of some components in multiple nitrogen-related metabolisms, consistent with Wei et al. (2016) findings in Barley. The primary processes for plants to acquire and utilize NO3−–N include the absorption and transport of NO3−, reduction of NO3− to NH4+, and assimilation of NH4+ into glutamic acid. Four gene families involved in NO3−–N absorption and transport have been discovered: NRT1/NPF, NRT2, CLC, and SLAH. These proteins are essential for the absorption, transport, and transfer of nitrate from the external environment to various cell types, tissues, and organs (Krapp et al., 2014). NO3− induced the upregulation of NPF6.3, a nitrate transporter, which was primarily involved in transporting nitrate from the root to aboveground organs (Tsay et al., 2007). The expression of NPF6.3 was significantly upregulated in the stem of M. micrantha (Figure 3), which played a role in transporting more NO3−–N for plant growth (Figure 4A) and as a signaling molecule regulating other physiological and ecological processes (Fan et al., 2017; Fredes et al., 2019). The high expression of NPF5 genes (Figure 4A) in M. micrantha stems can regulate NO3− balance between cytoplasm and vacuoles in response to changes in nitrogen supply (Wang et al., 2020), indicating that NPF5 gene significantly enhances the nitrogen redistribution ability in plants and improves nitrogen utilization efficiency. In contrast, the expression of NPF5.10 in M. micrantha stem was downregulated (Figure 3), which reduces the efflux of NO3−–N from the vacuole (Lu et al., 2022).
Additionally, the high expression of CLC and SLAC gene family members in M. micrantha (Figure 3) promotes NO3−–N transport within stems. Research suggested that these transporters also exhibited certain activity in NO3− transport, altering the balance and storage of NO3− and Cl− in cells (Chopin et al., 2007; O’Brien et al., 2016). M. micrantha absorbed NO3−, and its transcription factor expression levels were correlated with its involvement in regulating NO3−, uptake. Under NO3−–N treatment, the transcription factor HY5 in M. micrantha, involved in nitrate root signal regulation, showed increased expression levels at 10 mM and 20 mM NO3−–N treatments (Figure 4E). Previous research has highlighted the crucial role of HY5 in regulating genes associated with nitrogen uptake and assimilation in plants (Mankotia et al., 2024). HY5 positively regulates the expression of Nitrite reductase 1 (NiR1), known for converting nitrite into ammonium (Huang et al., 2015). Conversely, As a negative regulator, overexpression of LBD could have a certain inhibitory effect on nitrate responsive genes (NRT1.1, NRT2.5, etc.) (Rubin et al., 2009). Consistent with previous findings, the LBD gene family members in M. micrantha showed low expression levels under high NO3−–N treatment (Figure 4F). Therefore, the absorption and transport of NO3−–N are key steps in nitrogen absorption. The upregulation of key genes such as NPF6.3, NPF5, CLC, and SLAC, along with the regulatory roles of transcription factors like HY5 and LBD, underscores the complexity of nitrogen absorption in M. micrantha compared to native plant, which was more conducive to its absorption of NO3−–N.
Enhancing nitrogen metabolism capacity promotes the efficient assimilation of NO3−–N
The key enzymes involved in nitrogen metabolism play a crucial role in the nitrogen metabolism in plants. In M. micrantha, the expression levels of NR, NiR, and GDH were higher, while the expression of GS showed an opposite trend (Figures 4G–J, 3), which was consistent with physiological data (Figure 2). These results indicated that a portion of NO3− in the stem was being reduced, primarily through the GDH pathway for nitrogen assimilation, thus ensuring a higher nitrogen metabolism capacity. GDH and GS are key enzymes involved in ammonium assimilation in the nitrogen metabolism process. Previous studies have shown that nitrate can promote GS activity to some extent (Thu Hoai et al., 2003). However, under stress conditions, protein degradation produces a large amount of NH4+, which is prone to ammonia toxicity. The increase in GDH activity has a certain detoxification effect on NH4+ accumulation (Bittsánszky et al., 2015), which is similar to the opposite relationship with GS. With the increase of NO3−–N concentration, the NR (Figure 2B) and NiR (Figure 2C) activities in M. micrantha were higher, indicating that a large amount of NO3− was gradually reduced to NH4+. To avoid the toxic effects of NH4+, the plant upregulated the expression of GDH gene (Figure 4J), enhancing GDH enzyme activity (Figure 2E), converting free NH4+ into glutamic acid and further converts it into other forms of organic nitrogen, providing precursors for the biosynthesis of nitrogen-containing compounds in plants. Therefore, GDH is considered the main nitrogen assimilation pathway in the stems of M. micrantha. Under high nitrogen conditions, with the increasing concentration of nitrate, the expression level of GDH was upregulated, indicating that M. micrantha optimizes nitrogen utilization by adjusting nitrogen metabolism pathways, thus enhancing its biomass and adaptability. This finding provides new insights into how M. micrantha responds to high-nitrogen environments. Similar studies (Yang et al., 2010) found that in cucumber seedlings, root ammonia assimilation was primarily accomplished through the GDH-induced pathway, while leaf ammonia assimilation was achieved through the GS/GOGAT cycle under nitrate treatment. However, the interaction between GDH and GS in ammonia assimilation in M. micrantha and how they balance the response to external NO3− requires further research for in-depth exploration. GDH and GS play important roles in the assimilation process of M. micrantha, but further research is needed to balance the response to external NO3− and increase more nutrients for plant growth. Positive regulatory genes (ALDO and CYSK) play a role in enhancing amino acid synthesis. It was observed that M. micrantha upregulated the expression levels of these two genes (Figures 4KJ, 7), providing more precursors for amino acid synthesis, which was consistent with the significant increase in free amino acid content observed. Previous studies have suggested that tomato (Solanum lycopersicum L.) under low nitrogen conditions enhances the expression levels of ALDO gene to increase metabolism and obtain the substances or energy needed to adapt to environmental changes (Xu et al., 2023). Therefore, by regulating the expression levels of related transcription factors and nitrate transporter genes, M. micrantha enhances its ability to absorb nitrate nitrogen, improves nitrogen metabolism in the stem, and promotes the accumulation of a large number of amino acids, ensuring its higher biomass.
Conclusion
M. micrantha, as a rapidly growing invasive plant, has garnered widespread attention and research. Our study analyzed physicochemical properties in rhizosphere soils, identified relevant differentially expressed genes, and constructed key transcriptional regulatory pathways. The results revealed that NO3−–N effectively promoted the growth of M. micrantha, including an increase in plant height, branching and biomass. Two potential mechanisms underlie these phenotypic changes in M. micrantha for better NO3− acquisition. Firstly, compared to companion plants, M. micrantha exhibited higher NO3−–N content and protease activity in the soil. This process may accelerate nitrogen metabolism in the rhizosphere soil of M. micrantha, improving the efficiency of root utilization of nitrate nitrogen. Secondly, increased activities of NR, NiR, GS, and GOGAT in the stem enhanced nitrogen assimilation and amino acid biosynthesis, thereby promoting plant growth. In summary, under the backdrop of increasing global nitrogen deposition, particularly with the continued rise in NO3−–N deposition, the rapid growth of M. micrantha may be facilitated by the regulation of NO3−–N uptake transcription factors (HY5) and transport proteins (CLC, SCLA/C, NPF), as well as the expression regulation of key enzyme genes involved in nitrogen assimilation (NR, GS, GOGAT), thereby enhancing the nitrogen utilization efficiency of NO3−–N as the main form and accelerating the spread of M. micrantha. This study reveals the adaptation mechanisms of M. micrantha to NO3⁻-N enrichment, offering critical insights for predicting and managing invasive species responses to global atmospheric nitrogen deposition changes. The results highlight the importance of considering nitrogen composition, rather than just quantity, in invasive species management strategies.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MlC: Writing – original draft, Writing – review & editing. LC: Writing – original draft. MhC: Writing – original draft. WK: Writing – review & editing. DW: Writing – review & editing, Formal analysis. CP: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by National Natural Science Foundation of China (32401382, 3217130145, 31870374), Professorial and Doctoral Scientific Research Foundation of Huizhou University (2022JB086) and National College Students Innovation and Entrepreneurship Program Project (202310577015).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1525303/full#supplementary-material
References
1
Antonio C. M. D. Mack M. C. (2006). Nutrient limitation in a fire-derived, N rich Hawaiian grassland. Biotropica38, 458–467. doi: 10.1111/j.1744-7429.2006.00170.x
2
Behera S. N. Sharma M. Aneja V. P. Balasubramanian R. (2013). Ammonia in the atmosphere: a review on emission sources, atmospheric chemistry and deposition on terrestrial bodies. Environ. Sci. Pollution Res.20, 8092–8131. doi: 10.1007/s11356-013-2051-9
3
Bennett T. Leyser O. (2006). Something on the side: axillary meristems and plant development. Plant Mol. Biol.60, 843–854. doi: 10.1007/s11103-005-2763-4
4
Bittsánszky A. Pilinszky K. Gyulai G. Komives T. (2015). Overcoming ammonium toxicity. Plant Sci.231, 184–190. doi: 10.1016/j.plantsci.2014.12.005
5
Bobbink R. Hicks K. Galloway J. Spranger T. Alkemade R. Ashmore M. et al . (2010). Global assessment of N deposition effects on terrestrial plant diversity: a synthesis. Ecol. Appl.20, 30–59. doi: 10.1890/08-1140.1
6
Cai M. Chen L. Ke W. Chen M. Zhang J. Huang J. et al . (2023). Understanding the invasion mechanism of Malignant alien weed Mikania micrantha from the perspective of photosynthetic capacity of stems. Biol. Invasions25, 1181–1119. doi: 10.1007/s10530-022-02973-6
7
Chen W. B. Chen B. M. (2019). Considering the preferences for nitrogen forms by invasive plants: a case study from a hydroponic culture experiment. Weed Res.59, 49–57. doi: 10.1111/wre.2019.59.issue-1
8
Chen L. Cai M. Zhang Q. Pan Y. Chen M. Zhang X. et al . (2024). Why can Mikania micrantha cover trees quickly during invasion? BMC Plant Biol.24, 511.
9
Chen Q. Niu B. Hu Y. Wang J. Lei T. Zhou J. et al . (2020). Multilevel nitrogen additions alter chemical composition and turnover of the labile fraction soil organic matter via effects on vegetation and microorganisms. J. Geophysical Research: Biogeosciences125, e2019JG005316. doi: 10.1029/2019JG005316
10
Chen B. M. Peng S. L. Ni G. Y. (2009). Effects of the invasive plant Mikania micrantha HBK on soil nitrogen availability through allelopathy in South China. Biol. Invasions11, 1291–1299. doi: 10.1007/s10530-008-9336-9
11
Chopin F. Orsel M. Dorbe M. F. Chardon F. Truong H. N. Miller A. J. et al . (2007). The Arabidopsis AtNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell19, 1590–1602. doi: 10.1105/tpc.107.050542
12
Crawford N. M. Forde B. G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book1, e0011. doi: 10.1199/tab.0011
13
Day M. D. Clements D. R. Gile C. Senaratne W. K. Shen S. Weston L. A. et al . (2016). Biology and impacts of Pacific Islands invasive species. 13. Mikania micrantha Kunth (Asteraceae). Pacific Sci.70, 257–285. doi: 10.2984/70.3.1
14
Doi E. Shibata D. Matoba T. (1981). Modified colorimetric ninhydrin methods for peptidase assay. Analytical Biochem.118, 173–184. doi: 10.1016/0003-2697(81)90175-5
15
Dong S. Shang Z. Gao J. Boone R. (2022). Enhancing the ecological services of the Qinghai-Tibetan Plateau’s grasslands through sustainable restoration and management in era of global change. Agriculture Ecosystems Environ.326, 107756. doi: 10.1016/j.agee.2021.107756
16
Dong X. M. Zhang W. Hu H. Gao T. Y. Wang X. Q. Shi Q. et al . (2024). Physiological and transcriptome analysis reveal the nitrogen preference and regulatory pathways of nitrogen metabolism in an epiphytic orchid, Cymbidium tracyanum. Environ. Exp. Bot.219, 105618. doi: 10.1016/j.envexpbot.2023.105618
17
Duan Y. K. Yang H. Y. Yang H. Wu Y. Fan S. Wu W. et al . (2023). Integrative physiological, metabolomic and transcriptomic analysis reveals nitrogen preference and carbon and nitrogen metabolism in blackberry plants. J. Plant Physiol.280, 153888. doi: 10.1016/j.jplph.2022.153888
18
Eller C. B. Oliveira R. S. (2017). Effects of nitrogen availability on the competitive interactions between an invasive and a native grass from Brazilian cerrado. Plant Soil410, 63–72. doi: 10.1007/s11104-016-2984-0
19
Fan X. Naz M. Fan X. Xuan W. Miller A. J. Xu G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot.68, 2463–2475. doi: 10.1093/jxb/erx011
20
Fang Y. Zhang X. Wei H. Wang D. Chen R. Wang L. et al . (2021). Predicting the invasive trend of exotic plants in China based on the ensemble model under climate change: A case for three invasive plants of Asteraceae. Sci. Total Environ.756, 143841. doi: 10.1016/j.scitotenv.2020.143841
21
Fredes I. Moreno S. Díaz F. P. Gutiérrez R. A. (2019). Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol.47, 112–118. doi: 10.1016/j.pbi.2018.10.004
22
Galloway J. N. Dentener F. J. Capone D. G. Boyer E. W. Howarth R. W. Seitzinger S. P. et al . (2004). Nitrogen cycles: past, present, and future. Biogeochemistry70, 153–226. doi: 10.1007/s10533-004-0370-0
23
Gu B. Ju X. Chang J. Ge Y. Vitousek P. M. (2015). Integrated reactive nitrogen budgets and future trends in China. Proc. Natl. Acad. Sci.112, 8792–8797. doi: 10.1073/pnas.1510211112
24
Guiboileau A. Yoshimoto K. Soulay F. Bataillé M. P. Avice J. C. Masclaux‐Daubresse C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol.194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
25
He W. Zhang M. Jin G. Sui X. Zhang T. Song F. (2021). Effects of nitrogen deposition on nitrogen-mineralizing enzyme activity and soil microbial community structure in a Korean pine plantation. Microbial Ecol.81, 410–424. doi: 10.1007/s00248-020-01595-6
26
Huang P. Shen F. Abbas A. Wang H. Du Y. Du D. et al . (2022). Effects of different nitrogen forms and competitive treatments on the growth and antioxidant system of Wedelia trilobata and Wedelia chinensis under high nitrogen concentrations. Front. Plant Sci.13, 8510999. doi: 10.3389/fpls.2022.851099
27
Huang L. Zhang H. Zhang H. Deng X. W. Wei N. (2015). HY5 regulates nitrite reductase 1 (NIR1) and ammonium transporter1; 2 (AMT1; 2) in Arabidopsis seedlings. Plant Sci.238, 330–339.
28
Huangfu C. Li H. Chen X. Liu H. Wang H. Yang D. (2016). Response of an invasive plant, Flaveria bidentis, to nitrogen addition: a test of form-preference uptake. Biol. invasions18, 3365–3380. doi: 10.1007/s10530-016-1231-1
29
Jia X. Zhong Y. Liu J. Zhu G. Shangguan Z. Yan W. (2020). Effects of nitrogen enrichment on soil microbial characteristics: From biomass to enzyme activities. Geoderma366, 114256. doi: 10.1016/j.geoderma.2020.114256
30
Krapp A. David L. C. Chardin C. Girin T. Marmagne A. Leprince A. S. et al . (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot.65, 789–798. doi: 10.1093/jxb/eru001
31
Ladd J. N. Butler J. H. A. (1972). Short-term assays of soil proteolytic enzyme activities using proteins and dipeptide derivatives as substrates. Soil Biol. Biochem.4, 19–30. doi: 10.1016/0038-0717(72)90038-7
32
Li W. H. Zhang C. B. Jiang H. B. Xin G. R. Yang Z. Y. (2006). Changes in soil microbial community associated with invasion of the exotic weed, Mikania micrantha H.B.K. Plant Soil281, 309–324. doi: 10.1007/s11104-005-9641-3
33
Li W. H. Zhang C. B. Lin J. Y. Yang C. J. (2008). Characteristics of nitrogen metabolism and soil nitrogen of invasive plants. J. Trop. Subtropical Bot.16, 321–327. doi: 10.3969/j.issn.1005-3395.2008.04.005
34
Liu B. Yan J. Li W. H. Yin L. Li P. Yu H. et al . (2020). Mikania micrantha genome provides insights into the molecular mechanism of rapid growth. Nat. Commun.11, 340–353. doi: 10.1038/s41467-019-13926-4
35
Lu Y. T. Liu D. F. Wen T. T. Fang Z. J. Chen S. Y. Li H. et al . (2022). Vacuolar nitrate efflux requires multiple functional redundant nitrate transporter in Arabidopsis thaliana. Front. Plant Sci.13. doi: 10.3389/fpls.2022.926809
36
Mankotia S. Jakhar P. Satbhai S. B. (2024). HY5: a key regulator for light-mediated nutrient uptake and utilization by plants. New Phytol.241, 1929–1935. doi: 10.1111/nph.v241.5
37
O’Brien J. A. Vega A. Bouguyon E. Krouk G. Gojon A. Coruzzi G. et al . (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant9, 837–856. doi: 10.1016/j.molp.2016.05.004
38
Peng Y. Yang J. X. Zhou X. H. Peng P. H. Li J. J. Zhang S. M. et al . (2019). An invasive population of Solidago canadensis is less sensitive to warming and nitrogen-addition than its native population in an invaded range. Biol. Invasions21, 151–162. doi: 10.1007/s10530-018-1812-2
39
Qian J. Jin W. Hu J. Wang P. Wang C. Lu B. et al . (2021). Stable isotope analyses of nitrogen source and preference for ammonium versus nitrate of riparian plants during the plant growing season in Taihu Lake Basin. Sci. Total Environ.763, 143029. doi: 10.1016/j.scitotenv.2020.143029
40
Rubin G. Tohge T. Matsuda F. Saito K. Scheible W. R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell21, 3567–3584. doi: 10.1105/tpc.109.067041
41
Shuvar I. A. Korpita H. M. (2021). Invasion of rare weed species and its impact on natural biodiversity (Publishing House Baltija Publishing).
42
Song J. N. Yang J. L. Jeong B. R. (2021). Growth, quality, and nitrogen assimilation in response to high ammonium or nitrate supply in cabbage (Brassica campestris L.) and lettuce (Lactuca sativa L.). Agronomy11, 2556. doi: 10.3390/agronomy11122556
43
Stevens C. Dupre C. Gaudnik C. Dorland E. Dise N. Gowing D. et al . (2011). Changes in species composition of European acid grasslands observed along a gradient of nitrogen deposition. J. Vegetation Sci.22, 207–215. doi: 10.1111/j.1654-1103.2010.01254.x
44
Takei K. Sakakibara H. Taniguchi M. Sugiyama T. (2001). Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: Implication of cytokinin species that induces gene expression of maize response regulator. Plant Cell Physiol.42, 85–93. doi: 10.1093/pcp/pce009
45
Thu Hoai N. T. Shim I. S. Kobayashi K. Kenji U. (2003). Accumulation of some nitrogen compounds in response to salt stress and their relationships with salt tolerance in rice (Oryza sativa L.) seedlings. Plant Growth Regul.41, 159–164. doi: 10.1023/A:1027305522741
46
Toor M. D. Adnan M. Rehman F. U. Tahir R. Saeed M. S. Khan A. U. et al . (2021). Nutrients and their importance in agriculture crop production; A review. Indian J. Appl. Pure Biosci.9, 1–6. doi: 10.18782/2582-2845.8527
47
Tsay Y. F. Chiu C. C. Tsai C. B. Ho C. H. Hsu P. K. (2007). Nitrate transporters and peptide transporters. FEBS Lett.581, 2290–2300. doi: 10.1016/j.febslet.2007.04.047
48
Vaz-Pinto F. Martínez B. Olabarria C. Arenas F. (2014). Neighbourhood competition in coexisting species: the native Cystoseira humilis vs the invasive Sargassum muticum. J. Exp. Marine Biol. Ecol.454, 32–41. doi: 10.1016/j.jembe.2014.02.001
49
Vidal E. A. Moyano T. C. Canales J. Gutiérrez R. A. (2014). Nitrogen control of developmental phase transitions in Arabidopsis thaliana. J. Exp. Bot.65, 5611–5618. doi: 10.1093/jxb/eru326
50
Wang S. Liu Y. Chen L. Yang H. Wang G. Wang C. et al . (2021). Effects of excessive nitrogen on nitrogen uptake and transformation in the wetland soils of Liaohe estuary, northeast China. Sci. Total Environ.791, 148228. doi: 10.1016/j.scitotenv.2021.148228
51
Wang H. D. Wan Y. F. Buchner P. King R. Ma H. (2020). Phylogeny and gene expression of the complete NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in Triticum aestivum. J. Exp. Bot.71, 4531–4546. doi: 10.1093/jxb/eraa210
52
Wang C. Y. Wu B. D. Jiang K. Zhou J. (2018). Differences in functional traits between invasive and native Amaranthus species under simulated acid deposition with a gradient of pH levels. Acta Oecologica89, 32–37. doi: 10.1016/j.actao.2018.04.006
53
Wang W. J. Zhu Q. Y. Dai S. Y. Meng L. He M. Chen S. et al . (2023). Effects of Solidago canadensis L. @ on mineralization-immobilization turnover enhance its nitrogen competitiveness and invasiveness. Sci. Total Environ.882, 163641. doi: 10.1016/j.scitotenv.2023.163641
54
Wei Z. X. Zeng X. Q. Qin C. Wang Y. Bai L. Xu Q. et al . (2016). Comparative transcriptome analysis revealed genes commonly responsive to varied nitrate stress in leaves of Tibetan hulless barley. Front. Plant Sci.7, 1067. doi: 10.3389/fpls.2016.01067
55
Xu Y. Zhang K. Li S. Zhou Y. Ran S. Xu R. et al . (2023). Carbon and nitrogen metabolism in tomato (solanum lycopersicum L.) leaves response to nitrogen treatment. Plant Growth Regul.100, 747–756. doi: 10.1007/s10725-023-00969-3
56
Yang H. Y. Duan Y. K. Wu Y. Q. Zhang C. Wu W. Lyu L. et al . (2023). Physiological and transcriptional responses of carbohydrate and nitrogen metabolism and ion balance in blueberry plants under nitrogen deficiency. Plant Growth Regul.101, 519–535. doi: 10.1007/s10725-023-01038-5
57
Yang X. Y. Wang X. F. Wei M. Hikosaka S. Goto E. (2010). Response of ammonia assimilation in cucumber seedlings to nitrate stress. J. Plant Biol.53, 173–179. doi: 10.1007/s12374-010-9096-9
58
Yousaf M. Li X. Zhang Z. Ren T. Cong R. Ata-Ul-Karim S. T. et al . (2016). Nitrogen fertilizer management for enhancing crop productivity and nitrogen use efficiency in a rice-oilseed rape rotation system in China. Front. Plant Sci.7, 1496. doi: 10.3389/fpls.2016.01496
59
Yu G. Jia Y. He N. Zhu J. Chen Z. Wang Q. et al . (2019). Stabilization of atmospheric nitrogen deposition in China over the past decade. Nat. Geosci.12, 424–429. doi: 10.1038/s41561-019-0352-4
60
Yu H. X. Le Roux J. J. Jiang Z. Y. Sun F. Peng C. Li W. (2021). Soil nitrogen dynamics and competition during plant invasion: insights from Mikania micrantha invasions in China. New Phytol.229, 3440–3452. doi: 10.1111/nph.v229.6
61
Zhang L. Chen X. Wen D. (2016). Interactive effects of rising CO2 and elevated nitrogen and phosphorus on nitrogen allocation in invasive weeds Mikania micrantha and Chromolaena odorata. Biol. Invasions18, 1391–1407. doi: 10.1007/s10530-016-1089-2
62
Zhang Z. Zhang C. Zhang C. S. Wang W. B. Feng Y. L. (2022). Differences and related physiological mechanisms in effects of ammonium on the invasive plant Xanthium strumarium and its native congener X. sibiricum. Front. Plant Science.13, 999748. doi: 10.3389/fpls.2022.999748
63
Zhang Q. Zhai J. Shao L. Lin W. Peng C. (2019). Accumulation of anthocyanins: an adaptation strategy of Mikania micrantha to low temperature in winter. Front. Plant Sci.10, 1049.
64
Zhao N. Ze S. Z. Liu N. Y. Hu L. Ji M. Li Q. et al . (2021). Exogenous phytohormone application and transcriptome analysis of Mikania micrantha provides insights for a potential control strategy. Genomics113, 964–975. doi: 10.1016/j.ygeno.2021.02.008
65
Zhou Y. B. Zhang C. S. Lin C. S. Yang Y. Peng Y. Tang D. et al . (2015). Over-expression of a glutamate dehydrogenase gene, MgGDH, from Magnaporthe grisea confers tolerance to dehydration stress in transgenic rice. Planta241, 727–740. doi: 10.1007/s00425-014-2214-z
66
Zhu J. He N. Wang Q. Yuan G. Wen D. Yu G. et al . (2015). The composition, spatial patterns, and influencing factors of atmospheric wet nitrogen deposition in Chinese terrestrial ecosystems. Sci. Total Environ.511, 777–785. doi: 10.1016/j.scitotenv.2014.12.038
67
Zou N. Shi W. Hou L. Kronzucker H. J. Huang L. Gu H. et al . (2020). Superior growth, N uptake and NH4+ tolerance in the giant bamboo Phyllostachys edulis over the broad-leaved tree Castanopsis fargesii at elevated NH4+ may underlie community succession and favor the expansion of bamboo. Tree Physiol.40, 1606–1622. doi: 10.1093/treephys/tpaa086
Summary
Keywords
invasive plant, NO3⁻-N, RNA-seq, gene, stem
Citation
Cai M, Chen L, Chen M, Ke W, Wang D and Peng C (2025) Nitrate nitrogen uptake and metabolism in Mikania micrantha stem: insights into enhanced growth and invasiveness. Front. Plant Sci. 16:1525303. doi: 10.3389/fpls.2025.1525303
Received
09 November 2024
Accepted
11 April 2025
Published
02 May 2025
Volume
16 - 2025
Edited by
Nacer Bellaloui, United States Department of Agriculture, United States
Reviewed by
Yuping Hou, Ludong University, China
Lei Liu, Lanzhou University, China
Updates
Copyright
© 2025 Cai, Chen, Chen, Ke, Wang and Peng.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changlian Peng, pengchl@scib.ac.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.