- College of Horticulture, Shanxi Agricultural University, Jinzhong, Shanxi, China
The heat shock factors (HSFs) play important roles in activating heat stress responses in plants. Cerasus humilis (Ch) is a nutrient-rich fruit tree that can resist various abiotic and biotic stressors. However, the HSFs in C. humilis have not yet been characterized and their roles remain unclear. In this study, 21 ChHSF gene members were identified after searching the entire genome of C. humilis. Gene structure and motif composition analysis revealed that 16 ChHSF genes had only one intron and the motif3 was highly conserved in family of ChHSFs. Furthermore, the cis-acting elements analysis indicated that they most ChHSFs participate in plant growth and development, abiotic stress responses, and plant hormone regulations. By analyzing the tissue specific transcriptomes, it was found that most ChHSF genes had higher expression levels in leaves than in other tissues of C.humilis. Notably, the ChHSF04 gene exhibited a striking 115.5-, 14.4-, and 16.0-fold higher expression in leaves relative to seeds, roots, and fruits, respectively. The high temperature (40 °C) treated C. humilis seedlings quantitative real-time polymerase chain reaction (qRT-PCR) was conducted on all ChHSF gene members. The results show that the expression of most ChHSF genes in the leaves was significantly upregulated and peaked at 12 h under the heat stress and the expression levels of ChHSF04, ChHSF05, ChHSF12, ChHSF13, ChHSF15 and ChHSF16 exhibited 53-, 33-, 24-, 22-, 43- and 65-fold upregulation, indicating that these genes may play important roles in early response to heat stress in C. humilis. These results provide valuable insights into the evolutionary relationship of the ChHSF gene family and its role in high temperature stress responses.
1 Introduction
Plants are prone to various abiotic stresses, such as unfavorable temperatures, saline-alkali soil, drought, and other adverse environmental conditions (Liu et al., 2023). With the intensification of global climate change, high temperatures have become an important environmental factor restricting the growth and development of woody plants (Zhou et al., 2023). Therefore, plants must alleviate these adverse effects through self-regulation to enable them not only to survive but also to thrive in high-temperature environments (Song et al., 2014; Wan et al., 2019). Thank to evolution, plants have developed a set of mechanisms to respond to high-temperature stress including sensing external temperature changes and reacting quickly to alleviate damage by synthesizing osmotic regulators such as proline and up-regulating the expression of a series of stress response genes (Ahuja et al., 2010; Zanella et al., 2016; Gomez-Pastor et al., 2018; Zhou et al., 2019).
The heat shock transcription factors (HSF), an important transcription factor, was previously reported to participate in high temperature response and other stress responses in plants (Ohama et al., 2016; Wan et al., 2019; Wang et al., 2023). Based on the differences in gene structure, HSF genes could be divided into three subfamilies: types A, B, and C (Nover et al., 2001). Type A HSFs were proved to have transcriptional activation functions (Ren et al., 2023). Some studies showed that type B HSFs can assist type A HSFs in performing their functions, whereas few studies were focused on the gene function of type C HSFs (Andrási et al., 2021). In Arabidopsis thaliana, overexpression of the HSFB4 gene can shorten the length of the root, while overexpression of the HSFA2 gene not only improves the plant’s salt tolerance but also promotes the growth of callus tissue (Begum et al., 2013). In citrus fruits, CitHsfA7 was the only factor that resulted in a significant lowering of the citric acid content under hot air treatment (Li et al., 2022). In Poplar, stably overexpressing PeHSFA3 gene exhibited enhanced thermo-tolerance (Ren et al., 2024).
Cerasus humilis, a unique dwarfed deciduous fruit tree, belongs to the genus of Prunus in Rosaceae, which has a cultivation history of over 3000 years in China (Fu et al., 2021). It has become an important pioneer tree species in arid and semi-arid hilly areas because of its strong tolerance to various abiotic stresses, particularly drought and high temperatures (Wu et al., 2019). However, the heat tolerance mechanisms of C. humilis have not yet been studied. Based on the newly released whole genome of C. humilis, whole-genome identification of the HSF genes family of C. humilis was carried out in this study. The gene structure, physicochemical properties, conserved motifs, chromosome location, promoter elements, and phylogenetic relationships of C. humilis were analyzed using bioinformatics tools. In addition, qRT-PCR was employed to analyze the expression patterns of all ChHSF genes in C. humilis plants under high-temperature stress. The results of this study lay an important foundation for the functional identification ChHSF genes in the future and subsequent studies of the heat tolerance mechanism of C. humilis.
2 Materials and methods
2.1 High-temperature treatments of C. humilis
One-month-old cuttage seedlings of C. humilis cultivar ‘Nongda 4’ were taken from the resource nursery of Shanxi Agricultural University and then subjected to room-temperature (25 °C) and high-temperature (40 °C) treatments (Yang et al., 2024). Leaves from treated seedlings for 0 h, 12 h, 24 h, 36 h, and 48 h were collected, frozen in liquid nitrogen, and stored at -80 °C for later use.
2.2 Proline content determination
Determination of the proline contents were carried out followed the method described by Abd-ElGawad (Abd-ElGawad et al., 2020). An amount of 0.1 g of liquid nitrogen grinded sample was mixed with 5 mL of 3% sulfosalicylic acid solution, then the mixture was water-bath in boiled water for 15 min. The cooled mixture (1 mL) was transferred to a new test tube and mixed with 2 mL water and 2 mL acetic acid, then water-bathed in boiled water for 1 h to develop color. Subsequently, 5 mL of toluene was added and mixed thoroughly in the dark for 2–3 h. After the layers were completely separated, the absorbance was measured at 520 nm.Proline content per unit of fresh sample (%) = C×Vt/W×Vs×106·100%.
C value is the calculated proline weight from the standard curve (μg/mL), Vt value is the total volume of the extraction solution (mL), Vs value is the sample volume used for determination (mL), W value is the sample mass (g), and the factor 106 is used to convert g into μg.
2.3 Genome-wide identification of the ChHSF genes
The complete genome sequence of C. humilis was downloaded from https://doi.org/10.6084/m9.figshare.11669673. Two methods were used to identify the gene members of the ChHSF. First, the HSF protein sequences of Prunus persica obtained from the Gramene website (http://www.gramene.org/) were used to conduct local BLAST searches for homologous proteins in C. humilis. Secondly, the Hidden Markov Model (PF00447) was downloaded from the Pfam (http://pfam.xfam.org/) website. By running the local hmmsearch software (HMMER 2.3.2) in the local protein database of C. humilis, the sequences in that match PF00447 were selected. After removing repetitive sequences, DBDs and coiled-coil structures were studied using CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), finally, all HSF proteins with DBDs and coiled-coil structures were retained.
2.4 Physicochemical property prediction and phylogenetic analysis of ChHSFs
The number of amino acids, molecular weights, and isoelectric points of the ChHSFs were analyzed using the website ExPASy-ProtParam (https://web.expasy.org/protparam/). Subcellular localization prediction of ChHSFs was performed using the online software PSORT II (https://psort.hgc.jp/form2.html).
Sequences of Arabidopsis heat shock factors (AtHSFs) were obtained from the TAIR database (https://www.arabidopsis.org/) and then were aligned ChHSFs using MEGA software (version 7.0). A phylogenetic tree was constructed using the neighbor-joining method (bootstrap = 1000, other parameters set to default); and visualized using EvolView (https://evolgenius.info/evolview-v2/; Subramanian et al., 2019).
2.5 Chromosomal localization, gene structure, and conserved motif analysis of ChHSF genes
Based on the genome sequence and annotation information of C. humilis, TBtools was used to map the ChHSF genes onto the chromosomes. Collinearity and gene duplication events in the entire C. humilis genome were analyzed using Multiple Collinearity Scan Toolkit (MCScanX) software with default parameters. Gene structure analysis, domain analysis and visualization were performed using TBtools based on data from the CDD website. Conserved motifs in the ChHSFs were analyzed using MEME (https://meme-suite.org/meme/tools/meme).
2.6 Promoter analysis of ChHSF genes
The 2kb sequences upstream of the transcription start sites of all ChHSF genes were extracted and the cis-regulatory elements analyzed the online PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
2.7 Analysis of ChHSF proteins interaction network
The online website STRING (https://cn.string-db.org/) was used to construct a ChHSF protein interaction networks using P. persica as reference (Yang et al., 2023). GO enrichment analysis was performed using eggNOG-mapper (http://eggnog-mapper.embl.de/) and TBTools.
2.8 Expression patterns of ChHSF in different tissues of C. humilis
Transcriptome data for C. humilis roots (PRJNA683804), seeds (PRJNA420878), fruits (PRJNA417674), and leaves (PRJNA684437) were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/). Tissue specific expression heatmap of ChHSF genes were drawn using TBtools based on their FPKM values (Han et al., 2021).
2.9 Quantitative real-time polymerase chain reaction
Total RNA from the treated leaves samples was extracted using the CTAB method (Yang et al., 2018). The RNA concentration was determined using a nucleic acid analyzer (Thermo ND2000C) after gel electrophoresis. The cDNA was synthesized using the TaKaRa [PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time)] reverse transcription kit. QRT-PCR was performed on a 7500 real-time fluorescence quantitative PCR instrument using a SYBR Green (TaKaRa) kit. Each step of the reaction was performed in accordance with the SYBR Green manual. Primers were designed based on the gene sequences (ChHSF01-ChHSF21) using Vector NTI (Vector NT1 Advance 11.5). The primer information and gene accession numbers were shown in Supplementary Tables S1 and S2. The internal reference gene was ChActin (ouLi_007745). The 20 μL amplification system contained 2 μL cDNA, 0.8 μL of each of the forward and reverse primers, 0.4 μL of ROX Reference Dye II, 10 μL of SYBR, and 6 μL of ddH2O. The reaction procedure was as follows: pre-denaturation at 95°C for 30 s; denaturation at 95°C for 15 s, annealing at 58.5°C for 34 s, extension at 95°C for 15 s, 40 cycles, and each treatment was repeated three times. The relative expression of ChHSF was calculated with the 2-ΔΔCT method. Data analysis was performed using Excel 2007, Origin 9.0, and SAS 9.2.
3 Results
3.1 Identification and characterization analysis of ChHSFs in C. humulis
In total, 21 HSF gene members were identified in the whole genome of C. humilis and they were sequentially named ChHSF01 to ChHSF21 according to their positions on the chromosomes. The molecular weight (MW), theoretical isoelectric point (pI), subcellular localization, and other physicochemical properties were analyzed (Supplementary Table S2). As shown in Supplementary Table S2, the length of 21 ChHSF proteins varied from 194 aa to 534 aa and their MW ranged from 22.09 kDa to 59.38 kDa, whereas the pI values ranged from 4.67 to 8.53. Subcellular localization analysis indicated that most ChHSF proteins (18 members) were located in the nucleus while the rest (ChHSF04, ChHSF06, and ChHSF19) were located in the cytoplasm, indicating their important roles in the nucleus.
3.2 Phylogenetic analysis of ChHSFs in C. humulis
Based on the phylogenetic tree constructed by aligning 21 AtHSFs and 21 ChHSFs, the ChHSFs were divided into three subfamilies (Figure 1A). Type A was further divided into 9 subtypes (A1-A9) including 13 family members: ChHSF01, ChHSF02, ChHSF03, ChHSF04, ChHSF06, ChHSF07, ChHSF08, ChHSF09, ChHSF10, ChHSF15, ChHSF17, ChHSF19, and ChHSF21. Type B was divided into four subtypes (B1-B4) including seven family members: ChHSF05, ChHSF11, ChHSF12, ChHSF13, ChHSF14, ChHSF16, and ChHSF20. Type C included only one family member (ChHSF18). The evolutionary tree showed that multiple ChHSFs were tightly clustered with AtHSFs and no ChHSF subfamily members were identified in subtype A7.
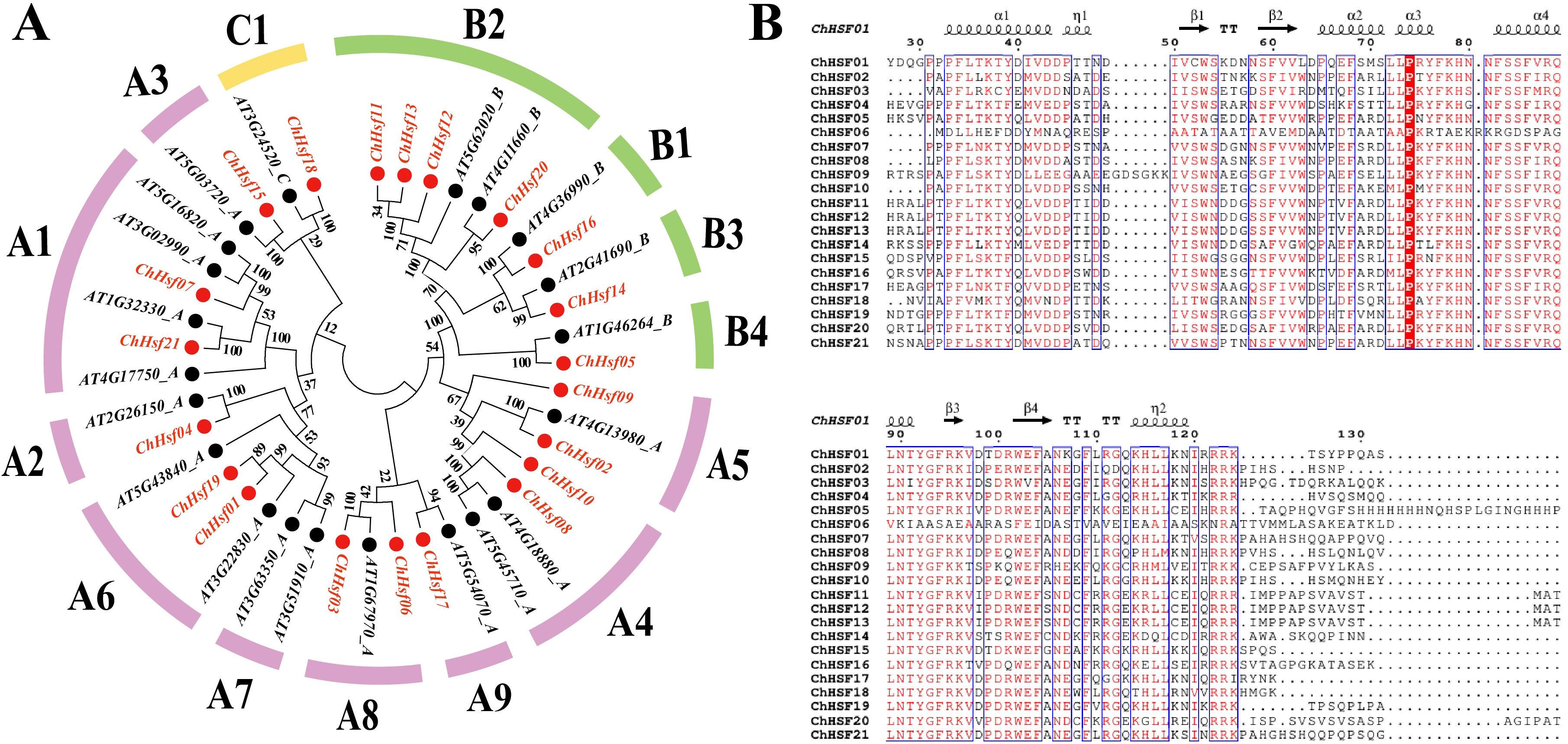
Figure 1. Evolutionary relationship and multiple sequence alignment of the ChHSF proteins. A phylogenetic tree of C. humilis and A. thaliana HSF proteins. C. humilis proteins are denoted in red, whereas A. thaliana proteins are marked in black (A). Multiple sequence alignment of the DBD domains of the ChHSF protein family (B).
The DBD (DNA binding domain) is the most conserved structural domain in HSFs comprising three helical bundles (α1, α2, and α3) and four antiparallel β sheets (β1, β2, β3, and β4) (Wang et al., 2024). To further investigate the evolutionary relationship between the structural domains of the ChHSF proteins in the three subfamilies, the amino acid sequences of the 21 ChHSF proteins were subjected to multiple sequence alignment (Figure 1B). Most ChHSFs had conserved structural domains and only minor variations were observed in the conserved region except ChHSF06. Interestingly, ChHSF09 has six additional amino acids between α1-β1, which may be related to its specific functional roles and needs to be explored further.
3.3 Gene structure and conserved motif analysis of the ChHSF family in C. humilis
To understand the structural composition of ChHSF genes, a structural map was constructed using their gene sequences (Figure 2A). The results indicated that most ChHSF genes have only one intron except the ChHSF03 (2 introns), ChHSF04 (2 introns), ChHSF06 (2 introns), ChHSF20 (2 introns) and ChHSF19 (4 introns) (Figure 2B). Based on the schematic diagram of the conserved motif of the ChHSFs, 10 conserved motifs were identified and the motif 3 was the most conserved motif in C. humilis which commonly present in all ChHSFs (Figures 2C, D). In group A, ChHSF11, ChHSF12 and ChHSF13 shared the same motif structure (motif 1, 2, 3, 4, 6, 7, 8, 9), while ChHSF05, ChHSF14 and ChHSF16 had the same motif structure (motif 1, 2, 3, 4). The HSF proteins in group B all contained motifs 1 to 4, and ChHSF02, ChHSF04, ChHSF07, ChHSF08, and ChHSF10 shared same motif structure (motif 1, 2, 3, 4, 5, 10). The analysis of conserved motifs revealed a conserved pattern in HSF family genes, and within the same group, similar motif compositions were observed.

Figure 2. Phylogenetic relationships, gene structure, and protein conservative motif analysis in the ChHSF genes family. ChHSF Phylogenetic tree (A). ChHSF genes structure (B). ChHSF protein conserved motif (C). Conservative motif logo (D).
3.4 Chromosome distribution and collinearity analysis of the ChHSFs in C. humilis
Twenty-one ChHSFs were unevenly distributed across the 8 chromosomes of C. humilis, and chromosome 1 contained most ChHSF gene members (ChHSF01-ChHSF06), while Chr2 contained 0 ChHSF genes. Three tandem duplication events involving seven ChHSF genes on chromosome 1 (ChHSF03/ChHSF04), chromosome 3 (ChHSF07/ChHSF08), and chromosome 5 (ChHSF11/ChHSF12/ChHSF13) were observed (Figure 3A). Moreover, six homologous sites and three segmental duplication events were identified: ChHSF01/ChHSF19, ChHSF04/ChHSF17, and ChHSF11/ChHSF20 (Figure 3B). These results suggest that ChHSF gene family may have expanded through both tandem and segmental duplication events.
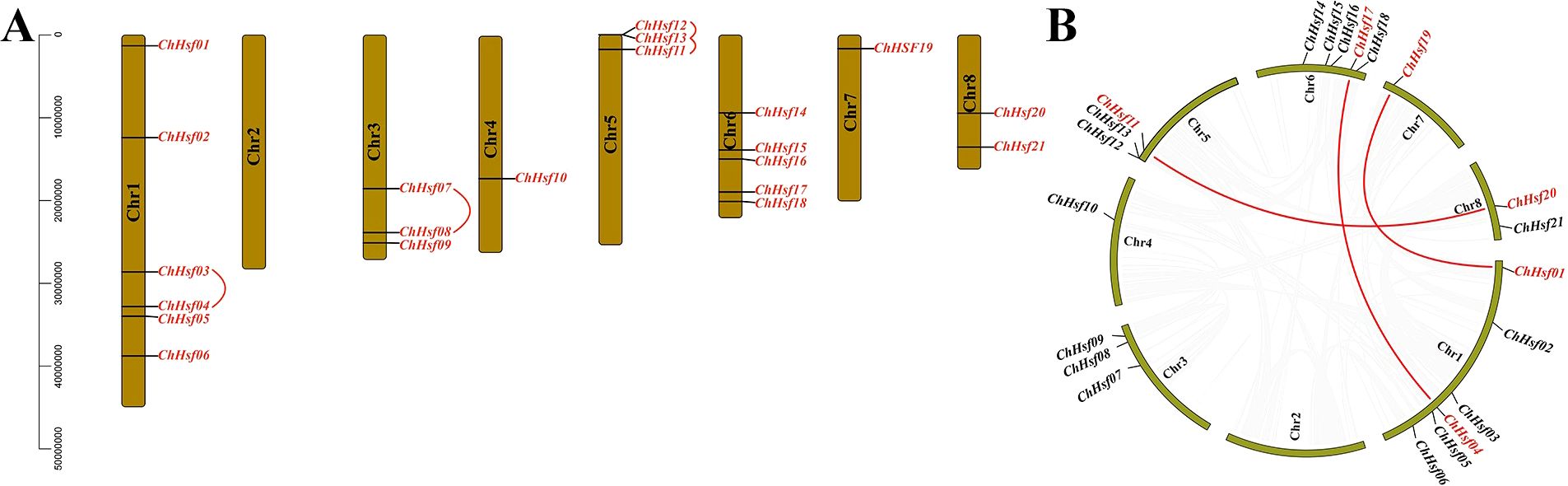
Figure 3. Chromosomal location of ChHSFs. Chr 1–8 represents 8 chromosomes respectively; Tandem duplicated of ChHSFs were marked with red curves (A); Synteny analysis of ChHSFs among chromosomes, red lines represent segmental duplication events (B).
3.5 Analysis of cis-acting elements in the promoter regions of the ChHSFs
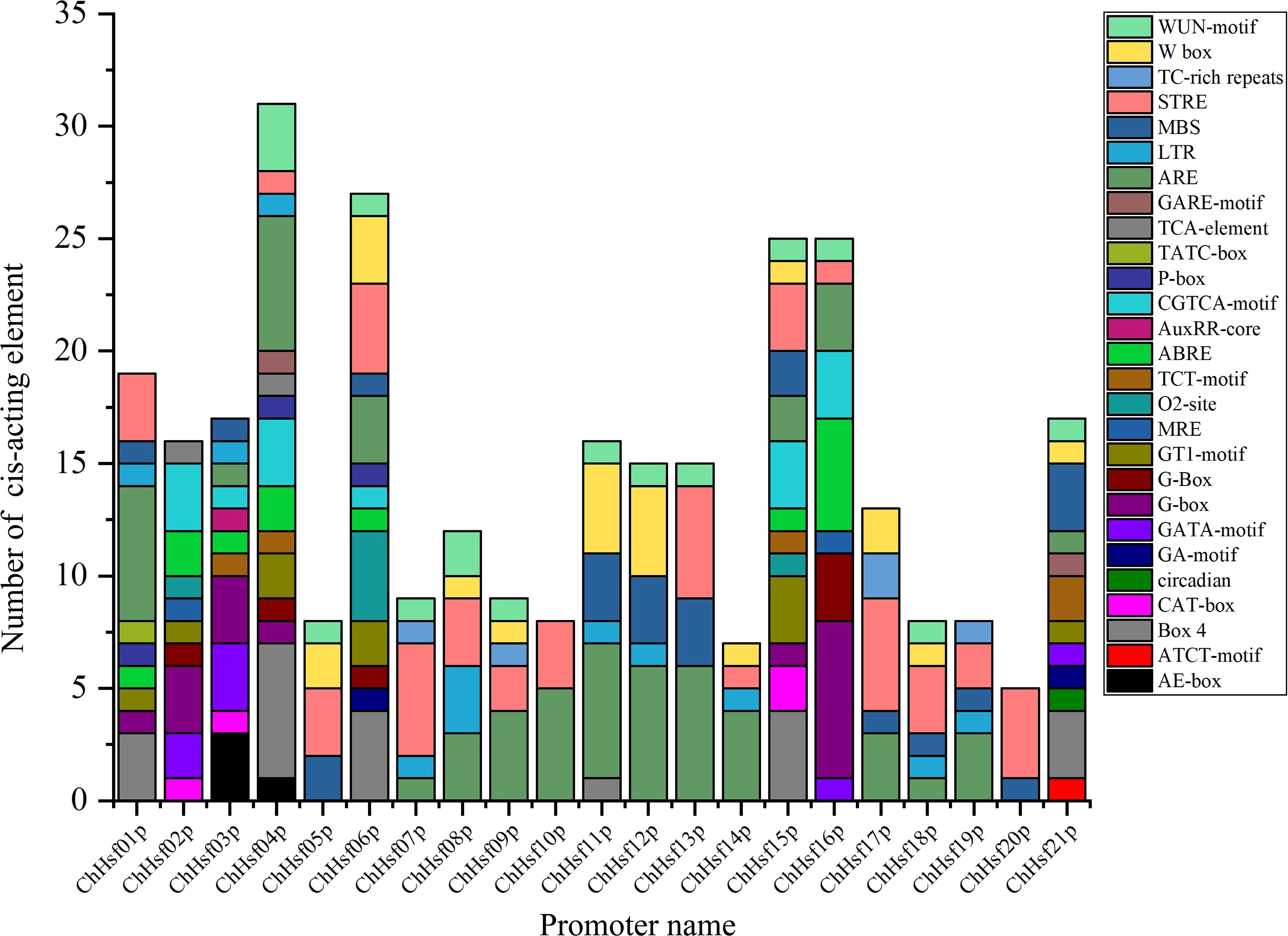
After predicting the cis-acting elements in the promoter regions of the ChHSFs by using the PlantCare online tool, a total of 27 cis-regulatory elements were detected (Figure 4), including 13 plant growth and development related elements, 7 hormone responsive elements, and 7 stress responsive elements (STREs). The cis-regulatory element number of ChHSFs ranged from 2 (ChHSF20) to 15 (ChHSF04). Interestingly, most ChHSF gene members had STREs in their promoter regions (Supplementary Table S3), indicating that ChHSFs may play important roles in stress responses in C. humilis. However, no heat stress-response cis-regulatory elements (HSE) were detected in the promoter regions of these ChHSF genes and it remains unclear whether the expression of the ChHSF genes is directly regulated by heat stress.
3.6 Protein interaction prediction of the ChHSFs
To obtain information on the possible interactions between ChHSF proteins in C. humilis, an interaction protein network was constructed using the STRING website (Figure 5A). The results showed that the three types of proteins had interactions with ChHSF proteins, including heat shock protein (HSP90), HSF-binding protein (HSBP1), and WD repeat protein (WD40). GO function analysis indicated that ChHSFs have the molecular function of transcription regulator activity and DNA-binding transcription factor activity, which are involved in the biological process of the nucleic acid-templated transcription, RNA biosynthetic and metabolic regulations (Figure 5B).
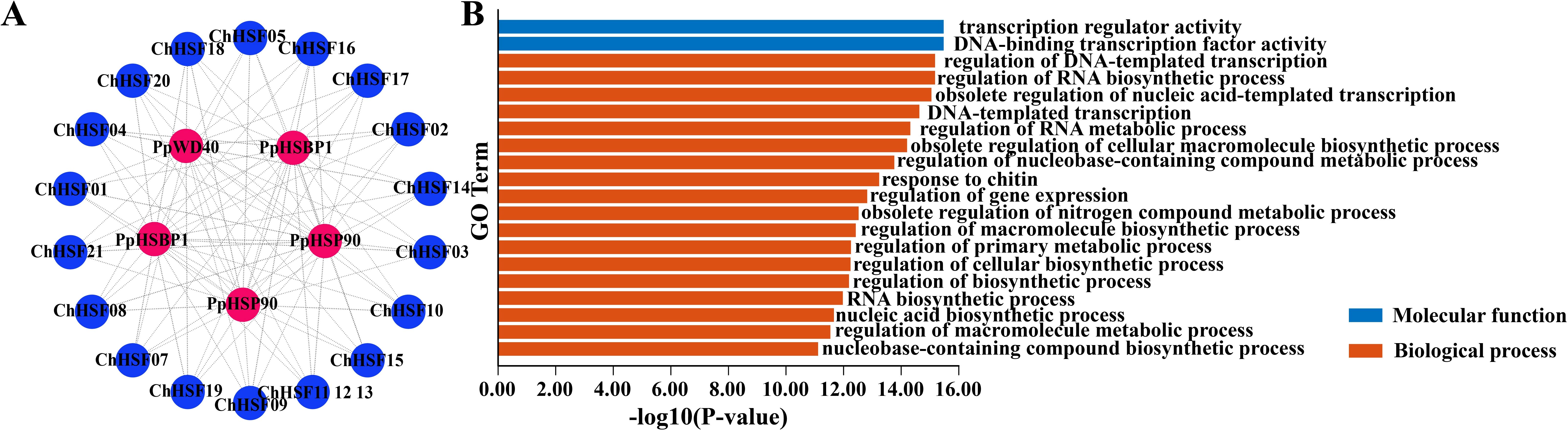
Figure 5. Protein interaction and GO function enrichment analysis of ChHSF genes (A). ChHSF protein-protein interaction (B). ChHSF gene function analysis, including biological process (BP) and molecular function (MF); biological only displays the top 20 items.
3.7 Tissue-specific expressions of ChHSF genes
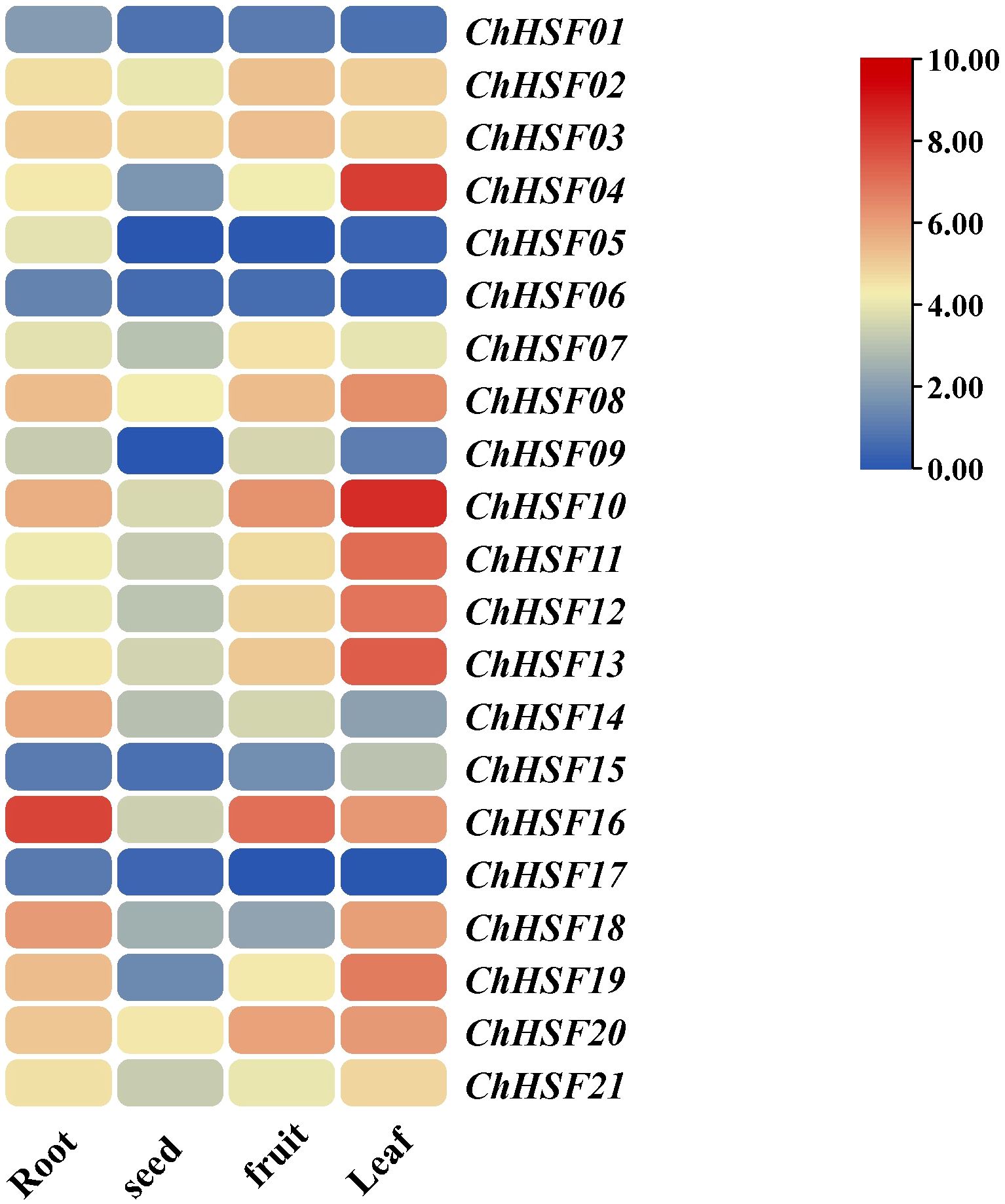
The expression patterns of 21 ChHSF genes were analyzed to elucidate their expression characteristics in four different tissues of C.humilis (roots, seeds, fruits, and leaves) (Figure 6). The results show that most ChHSF genes had higher expression levels in leaves than in the roots, fruits and seeds of C.humilis. Three ChHSF members (ChHSF01, ChHSF06, and ChHSF17) had low relatively low expression levels in all four tissues, indicating that they contribute less than other ChHSF genes in regulating the growth and development of these tissues in C.humilis. The ChHSF16 had the highest expression levels in roots and fruits of C.humilis, while ChHSF04 were only highly expressed in leaves. Tissue-specific expression results of the ChHSF family genes revealed their functional diversity and complexity.
3.8 Growth status and proline content of high temperature treated C. humilis cuttings
The growth status of C. humilis cuttings under normal temperature (25 °C) and high temperature (40 °C) treatments were documented (Figure 7A). The leaves of high temperature treated C. humilis cuttings begun to show signs of wilting after 12 h. From 12 to 24 h, the leaves started to turn yellow. After 48 h, the leaves became more fragile and are more likely to fall off. However, the leaves of normal temperature treated C. humilis cuttings remained healthy from 0–48 h.

Figure 7. Physiological state of C. humilis cutting seedlings under high-temperature stress. Growth of C. humilis cuttings at 25 and 40 °C for 0–48 h (A). Proline contents in leaves of C. humilis from 0–48 h under high-temperature stress (B).
To further verify the effects of high-temperature stress on C. humilis plants, the proline content in C. humilis leaves was determined (Figure 7B). The proline content in the leaves of C. humilis under 25 °C treatment kept stable from 0–48 h. However, the proline content in the leaves C. humilis under 40 °C treatment increased sharply with prolonged stress time, which peaked at 24 h, and then gradually decreases.
3.9 Expression analysis of ChHSF genes in high temperature treated C. humilis cuttings
The expression patterns of 21 ChHSF family genes in leaves of normal temperature and high temperature treated C. humilis cuttings were determined using qRT-PCR. In the early stage of high temperature treatment (0–12 h), 20 ChHSF family genes were up-regulated in leaves of high temperature treated C. humilis cuttings, while the ChHSF14 was significantly down-regulated (Figure 8). Noticeably, the expression levels of 10 ChHSF family members (ChHSF04, ChHSF05, ChHSF06, ChHSF12, ChHSF13, ChHSF15, ChHSF16, ChHSF17, ChHSF19, ChHSF20, and ChHSF21) exhibited more than ten-fold upregulation, indicating that these genes may play important roles in early response to heat stress in C. humilis. From 12–48 h, most ChHSF family members were significantly down-regulated, while the expression levels of ChHSF15 peaked at 24 h and then decreased in high temperature treated C. humilis cuttings.
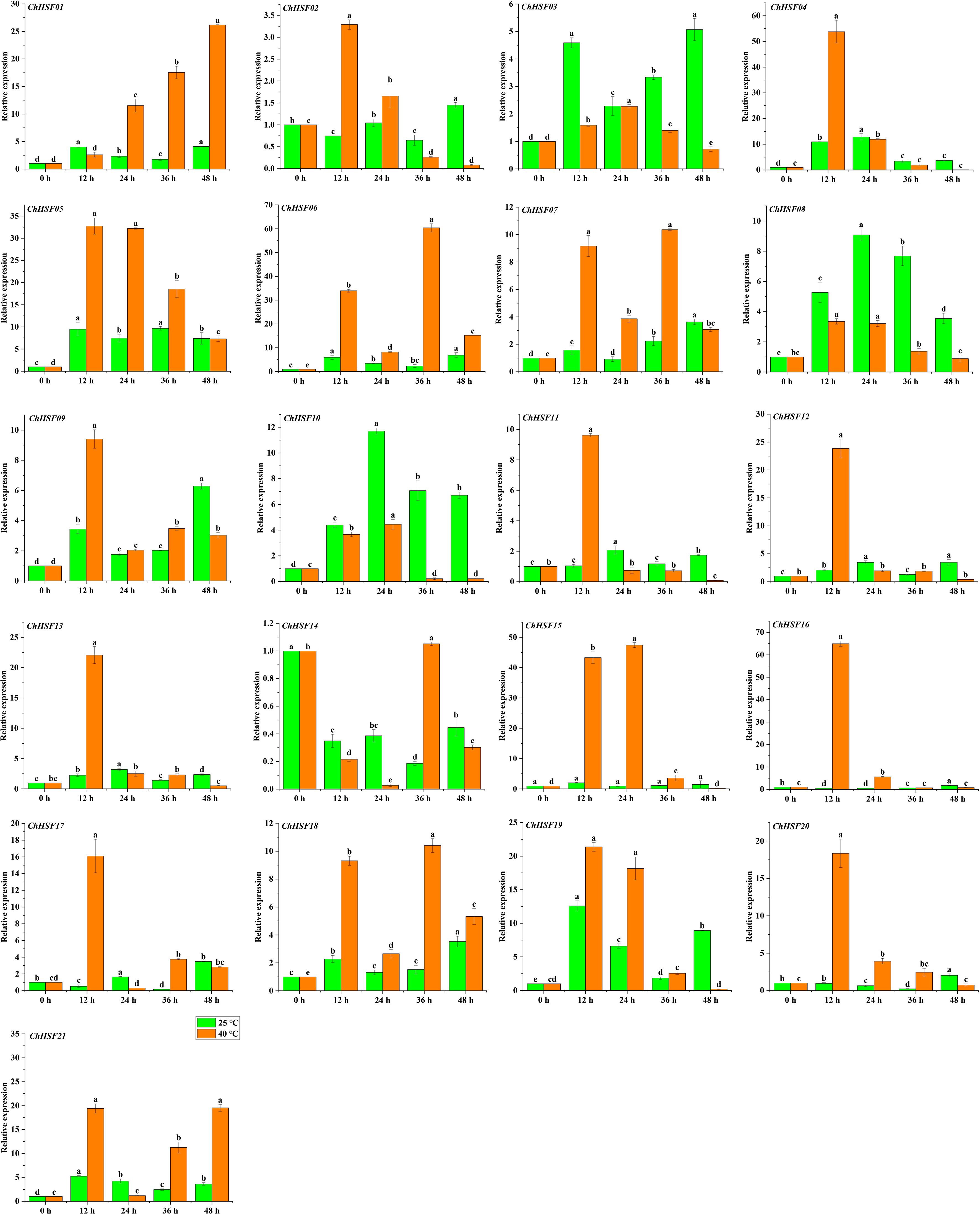
Figure 8. ChHSF genes’ expression in leaves of C. humilis cutting seedlings under high-temperature stress. Lowercase letters represents significance at p ≤ 0.05.
4 Discussion
HSFs are an important group of transcriptional regulatory factor widely present in plants (Wang et al., 2024). To date, HSF genes have been identified not only in A. thaliana (Nover et al., 2001), but also in many fruit species such as apple (Giorno et al., 2012), strawberry (Hu et al., 2015), Chinese white pear (Qiao et al., 2015), peach (Qiao et al., 2015), grape (Hu et al., 2016), citrus (Lin et al., 2015) and plum (Wan et al., 2019). Moreover, the number of HSF family genes varied from 17 (peach) to 35 (carrot) (Huang et al., 2015), the differences in the number of HSF family members among different plants may be due to the variations in the retention of HSF genes during the evolutionary process of adapting to the environment (Zheng et al., 2024). In this study, 21 ChHSF family members were identified in C. humilis and they were named as ChHSF01 to ChHSF21 according to their positions on the chromosomes. Interestingly, the size of HSF family C.humilis is bigger than in peach (Prunus persica) and plum (Prunus mume), indicating that the C.humilis is more ancient in evolution.
According to the conserved domains of Arabidopsis, the ChHSFs family members can be divided into three types: types A, B, and C. Type A includes 13 family members and subfamily A7 does not include C. humilis family members, it indicates that these genes are missing during evolution (Ren et al., 2023). Type B includes 7 family members, and type C includes one family member. Analysis of gene structure showed that 21 ChHSF genes contained different numbers of introns (1-4). As a part of plant evolution, introns may not only increase the length of genes and the frequency of recombination between genes, but also play an important role in regulation (Shabalina et al., 2010). In contrast, genes without introns have no advantage in the process of species evolution and will delay the regulatory response (Ren et al., 2023), therefore, many ChHSF members respond rapidly to stress treatment. Promoter analysis showed that the promoter region of ChHSF04 had the largest number of cis-regulatory elements among the ChHSF family, indicating that ChHSF04 may be a key gene in the defense against non-biological stress in C. humilis. Furthermore, no HSE elements were detected in ChHSFs promoter regions, suggesting that the expression of ChHSF genes related to high temperatures may not be directly induced by heat stress (Tang et al., 2016; Li et al., 2019; Zhang et al., 2020).
Research shows that the expression of the HSF gene varies in some species (Song et al., 2014; Liu et al., 2023). In C. humilis, several ChHSFs display tissue-specific expression patterns. For example, The ChHSF16 had the highest expression levels in roots of C.humilis, while ChHSF03 and ChHSF10 were highly expressed in seeds and fruits respectively, suggesting that HSFs play widespread roles in the growth and development of different organs and tissues. Studies have shown that HSF gene is involved in high temperature stress response (Fragkostefanakis et al., 2016), in this study, qRT-PCR was used to analyze the expression patterns of 21 HSF gene members in leaves of C.humilis under high temperature stress, the results showed that 20 ChHSF family genes were significantly up-regulated in leaves of high temperature treated C. humilis cuttings in the early stage of high temperature treatment (0–12 h). Interestingly, after 12 h of heat stress, the expression of 10 ChHSF family genes in leaves was significantly up-regulated in more than ten-fold, indicating that these genes warrant further investigation as promising candidate genes for genetic engineering strategies aimed at enhancing high-temperature resistance in C. humilis.
As described in the previous research, changes in proline content can serve as a good indicator of C.humilis stress response (Han et al., 2021). By measuring the changes of proline content in leaves at the same time, we found that the proline content in the leaves C. humilis under 40 °C treatment synchronously increased with expression levels of several ChHSF genes, and the functional connections between the proline accumulation and key ChHSFs’ expressions need to be further studied.
Analysis of the protein interaction network of ChHSF proteins in C. humilis shows that three proteins, that is HSP90, HSBP1, and WD40, interact with the HSF protein of C. humilis. HSP90 is a major member of HSP and is of great significance as a molecular chaperone (Wei et al., 2024). The HSF regulates the heat shock response of cells, it can bind to HSE under HS, activating the transcription and expression of downstream HSP genes (Li et al., 2020). Studies have shown that when plants are exposed to heat stress, the expression of heat shock genes increases, resulting in rapid accumulation of heat shock proteins (HSPs) (Aldubai et al., 2022). The HSF-mediated stress tolerance depends on the regulatory effect of plant HSBPs, which have inhibitory effects on the DNA-binding ability and transactivation activity of HSF (Muthusamy et al., 2023). Studies have shown that AtHSBP interacts with AtHSFs to reduce HSF transactivation and HSP gene protein levels (Hsu et al., 2010). In the study of Aldubai et al., it was found that several HSFs, including HsfA2, were expressed in tomato anthers at the early stage of pollen production, indicating that SlHSBP1 may play a regulatory role in HSF development (Aldubai et al., 2022). WD40, also known as the WD-repeat (WDR) protein, is widely present in eukaryotes and plays an important role in abiotic stress and other physiological and biochemical processes (Ke et al., 2023). In summary, the results of this study provide abundant resources for future research on protein interactions between HSF and HSP, WD40, and HSBP1 in C. humilis.
5 Conclusions
Altogether, we analyzed the gene structures, conserved motifs, cis-acting elements, and expression patterns of the ChHSF gene family under high temperature stress. The results provide valuable insights into evolutionary relationships of the ChHSF gene family and its role in high-temperature stress responses.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XM: Investigation, Writing – original draft, Writing – review & editing. JTZ: Investigation, Writing – original draft, Writing – review & editing. CW: Methodology, Writing – review & editing. LC: Methodology, Writing – review & editing. JYZ: Methodology, Writing – review & editing. PW: Software, Writing – review & editing. JCZ: Software, Writing – review & editing. BZ: Software, Writing – review & editing. JD: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Shanxi Agricultural University Science and Technology Innovation Doctoral Research Start-up Fund Project (grant numbers 2018YJ06 and 2016ZZ01).
Conflict of interest
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1553187/full#supplementary-material
Supplementary Table 1 | Primer sequences of quantitative RT-PCR.
Supplementary Table 2 | Identification of HSF gene family members in C. humilis.
Supplementary Table 3 | Distribution of the key cis-acting elements in promoters of ChHSFs.
References
Abd-ElGawad, A. M., Rashad, Y. M., Abdel-Azeem, A. M., Al-Barati, S. A., Assaeed, A. M., Mowafy, A. M. (2020). Calligonum polygonoides L. Shrubs provide species-specific facilitation for the understory plants in coastal ecosystem. Biol. (Basel). 9, 232. doi: 10.3390/biology9080232
Ahuja, I., de Vos, R. C., Bones, A. M., Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Science. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Aldubai, A. A., Alsadon, A. A., Migdadi, H. H., Alghamdi, S. S., Al-Faifi, S. A., Afzal, M. (2022). Response of tomato (Solanum lycopersicum L.) genotypes to heat stress using morphological and expression study. Plants (Basel). 11, 615. doi: 10.3390/plants11050615
Andrási, N., Pettkó-Szandtner, A., Szabados, L. (2021). Diversity of plant heat shock factors: regulation, interactions, and functions. J. Exp. Botany. 72, 1558–1575. doi: 10.1093/jxb/eraa576
Begum, T., Reuter, R., Schöffl, F. (2013). Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 130, 54–60. doi: 10.1016/j.mod.2012.05.008
Fragkostefanakis, S., Mesihovic, A., Simm, S., Paupière, M. J., Hu, Y., Paul, P., et al. (2016). HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol. 170, 2461–2477. doi: 10.1104/pp.15.01913
Fu, H., Qiao, Y., Wang, P., Mu, X., Zhang, J., Fu, B., et al. (2021). Changes of bioactive components and antioxidant potential during fruit development of Prunus humilis Bunge. PloS One 16, e0251300. doi: 10.1371/journal.pone.0251300
Giorno, F., Guerriero, G., Baric, S., Mariani, C. (2012). Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13, 639–651. doi: 10.1186/1471-2164-13-639
Gomez-Pastor, R., Burchfiel, E. T., Thiele, D. J. (2018). Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19. doi: 10.1038/nrm.2017.73
Han, H., Mu, X., Wang, P., Wang, Z., Fu, H., Gao, Y., et al. (2021). Identification of LecRLK gene family in Cerasus humilis through genomic-transcriptomic data mining and expression analyses. PloS One 16, e0254535. doi: 10.1371/journal.pone.0254535
Hsu, S. F., Lai, H. C., Jinn, T. L. (2010). Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant Physiol. 153, 773–784. doi: 10.1104/pp.109.151225
Hu, Y., Han, Y. T., Wei, W., Li, Y. J., Zhang, K., Gao, Y. R., et al. (2015). Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Science. 6. doi: 10.3389/fpls.2015.00736
Hu, Y., Han, Y. T., Zhang, K., Zhao, F. L., Li, Y. J., Zheng, Y., et al. (2016). Identification and expression analysis of heat shock transcription factors in the wild Chinese grapevine (Vitis pseudoreticulata). Plant Physiol. Biochem. 99, 1–10. doi: 10.1016/j.plaphy.2015.11.020
Huang, Y., Li, M. Y., Wang, F., Xu, Z. S., Huang, W., Wang, G. L., et al. (2015). Heat shock factors in carrot: genome-wide identification, classification, and expression profiles response to abiotic stress. Mol. Biol. Rep. 42, 893–905. doi: 10.1007/s11033-014-3826-x
Ke, S., Jiang, Y., Zhou, M., Li, Y. (2023). Genome-wide identification, evolution, and expression analysis of the WD40 subfamily in oryza genus. Int. J. Mol. Sci. 24, 15776. doi: 10.3390/ijms242115776
Li, S. J., Liu, S. C., Lin, X. H., Grierson, D., Yin, X. R., Chen, K. S. (2022). Citrus heat shock transcription factor CitHsfA7-mediated citric acid degradation in response to heat stress. Plant Cell Environ. 45, 95–104. doi: 10.1111/pce.14207
Li, M., Xie, F., Li, Y., Gong, L., Luo, Y., Zhang, Y., et al. (2020). Genome-wide analysis of the heat shock transcription factor gene family in brassica juncea: structure, evolution, and expression profiles. DNA Cell Biol. 39, 1990–2004. doi: 10.1089/dna.2020.5922
Li, W., Wan, X. L., Yu, J. Y., Wang, K. L., Zhang, J. (2019). Genome-wide identification, classification, and expression analysis of the Hsf gene family in carnation (Dianthus caryophyllus). Int. J. Mol. Sci. 20 (20), 5233. doi: 10.3390/ijms20205233
Lin, Q., Jiang, Q., Lin, J., Wang, D., Li, S., Liu, C., et al. (2015). Heat shock transcription factors expression during fruit development and under hot air stress in Ponkan (Citrus reticulata Blanco cv. Ponkan) fruit. Gene. 559, 129–136. doi: 10.1016/j.gene.2015.01.024
Liu, H., Li, X., Zi, Y., Zhao, G., Zhu, L., Hong, L., et al. (2023). Characterization of the heat shock transcription factor family in medicago sativa L. and its potential roles in response to abiotic stresses. Int. J. Mol. Sci. 24, 12683. doi: 10.3390/ijms241612683
Muthusamy, M., Son, S., Park, S. R., Lee, S. I. (2023). Heat shock factor binding protein BrHSBP1 regulates seed and pod development in Brassica rapa. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1232736
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones. 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2
Ohama, N., Kusakabe, K., Mizoi, J., Zhao, H., Kidokoro, S., Koizumi, S., et al. (2016). The transcriptional cascade in the heat stress response of arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell. 28, 181–201. doi: 10.1105/tpc.15.00435
Qiao, X., Li, M., Li, L., Yin, H., Wu, J., Zhang, S. (2015). Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 15, 12–27. doi: 10.1186/s12870-014-0401-5
Ren, Y., Ma, R., Xie, M., Fan, Y., Feng, L., Chen, L., et al. (2023). Genome-wide identification, phylogenetic and expression pattern analysis of HSF family genes in the Rye (Secale cereale L.). BMC Plant Biol. 23, 441. doi: 10.1186/s12870-023-04418-1
Ren, S.-X., Zou, H.-L., Cui, J.-W., Shen, N., Bao, H.-Y., Gan, Q., et al. (2024). PeHSFA3 is essential for the heat-stress response of Populus × euramericana. Industrial Crops Products. 219, 119054–119054. doi: 10.1016/j.indcrop.2024.119054
Shabalina, S. A., Ogurtsov, A. Y., Spiridonov, A. N., Novichkov, P. S., Spiridonov, N. A., Koonin, E. V. (2010). Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol. Biol. Evol. 27, 1745–1749. doi: 10.1093/molbev/msq086
Song, X., Liu, G., Duan, W., Liu, T., Huang, Z., Ren, J., et al. (2014). Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol. Genet. Genomics 289, 541–551. doi: 10.1007/s00438-014-0833-5
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Tang, R., Zhu, W., Song, X., Lin, X., Cai, J., Wang, M., et al. (2016). Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Science. 7. doi: 10.3389/fpls.2016.00490
Wan, X., Yang, J., Guo, C., Bao, M., Zhang, J. (2019). Genome-wide identification and classification of the Hsf and sHsp gene families in Prunus mume, and transcriptional analysis under heat stress. PeerJ. 7, e7312. doi: 10.7717/peerj.7312
Wang, Z., Wang, P., He, J., Kong, L., Zhang, W., Liu, W., et al. (2024). Genome-Wide Analysis of the HSF Gene Family Reveals Its Role in Astragalus mongholicus under Different Light Conditions. Biol. (Basel). 13, 280. doi: 10.3390/biology13040280
Wang, Q., Zhang, Z., Guo, C., Zhao, X., Li, Z., Mou, Y., et al. (2023). Hsf transcription factor gene family in peanut (Arachis hypogaea L.): genome-wide characterization and expression analysis under drought and salt stresses. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1214732
Wei, Y., Zhu, B., Liu, W., Cheng, X., Lin, D., He, C., et al. (2024). Heat shock protein 90 co-chaperone modules fine-tune the antagonistic interaction between salicylic acid and auxin biosynthesis in cassava. Cell Rep. 43, 114457. doi: 10.1016/j.celrep.2024.114457
Wu, Q., Yuan, R. Y., Feng, C. Y., Li, S. S., Wang, L. S. (2019). Analysis of polyphenols composition and antioxidant activity assessment of chinese dwarf cherry (Cerasus humilis (Bge.) sok.). Natural Product Communications. 14 (6), 1–10. doi: 10.1177/1934578X19856509
Yang, G., Gao, X., Ma, K., Li, D., Jia, C., Zhai, M., et al. (2018). The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol. 18, 367. doi: 10.1186/s12870-018-1568-y
Yang, R., Yang, Y., Hu, Y., Yin, L., Qu, P., Wang, P., et al. (2023). Comparison of bioactive compounds and antioxidant activities in differentially pigmented cerasus humilis fruits. Molecules. 28, 6272. doi: 10.3390/molecules28176272
Yang, Y., Yin, J., Zhu, L., Xu, L., Wu, W., Lu, Y., et al. (2024). Genome-Wide Analysis of the Liriodendron chinense Hsf Gene Family under Abiotic Stress and Characterization of the LcHsfA2a Gene. Int. J. Mol. Sci. 25, 2733. doi: 10.3390/ijms25052733
Zanella, M., Borghi, G. L., Pirone, C., Thalmann, M., Pazmino, D., Costa, A., et al. (2016). Beta-amylase 1 (bam1) degrades transitory starch to sustain proline biosynthesis during drought stress[J]. J Exp Bot. 67 (6), 1819–1826.
Zhang, X., Xu, W., Ni, D., Wang, M., Guo, G. (2020). Genome-wide characterization of tea plant (Camellia sinensis) Hsf transcription factor family and role of CsHsfA2 in heat tolerance. BMC Plant Biol. 20, 244. doi: 10.1186/s12870-020-02462-9
Zheng, R., Chen, J., Peng, Y., Zhu, X., Niu, M., Chen, X., et al. (2024). General Analysis of Heat Shock Factors in the Cymbidium ensifolium Genome Provided Insights into Their Evolution and Special Roles with Response to Temperature. Int. J. Mol. Sci. 25, 1002. doi: 10.3390/ijms25021002
Zhou, C., Wu, S., Li, C., Quan, W., Wang, A. (2023). Response mechanisms of woody plants to high-temperature stress. Plants (Basel). 12, 3643. doi: 10.3390/plants12203643
Keywords: Cerasus humilis, HSF, phylogenetic analysis, bioinformatics, gene expression
Citation: Mu X, Zhang J, Wang C, Chen L, Zhang J, Wang P, Zhang J, Zhang B and Du J (2025) Whole-genome identification of HSF family genes in Cerasus humilis and expression analysis under high-temperature stress. Front. Plant Sci. 16:1553187. doi: 10.3389/fpls.2025.1553187
Received: 30 December 2024; Accepted: 07 April 2025;
Published: 28 April 2025.
Edited by:
Christos Bazakos, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Ranjeet Ranjan Kumar, Indian Agricultural Research Institute (ICAR), IndiaMichail Michailidis, Aristotle University of Thessaloniki, Greece
Biao Jin, Yangzhou University, China
Copyright © 2025 Mu, Zhang, Wang, Chen, Zhang, Wang, Zhang, Zhang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junjie Du, ZGpqNzM4QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Xiaopeng Mu
Xiaopeng Mu Jiating Zhang†
Jiating Zhang† Chenyi Wang
Chenyi Wang Junjie Du
Junjie Du