- 1National Nanfan Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Sanya, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China
- 3Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences/Key Laboratory of Ministry of Agriculture for Germplasm Resources, Conservation, and Utilization of Cassava, Danzhou, China
- 4Biotechnology Research Institute, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China
- 5State Key Laboratory of Crop Gene Resources and Breeding, Institute of Crop Sciences, CAAS, Beijing, China
- 6Chemistry Department, Tshwane University of Technology, Pretoria, South Africa
The phenylpropanoid pathway remains a key target for most climate-resilient crop development, owing to it being a precursor to over 8000 metabolites, including flavonoids and lignin compounds, including their derivatives. These metabolites are involved in biotic and abiotic stress tolerance, inviting several studies into their roles in plant defense, drought, temperature, UV, and nutrient stress tolerance. Literature is currently inundated with cutting-edge reports on the phenylpropanoid pathways and their functions. Here, we provide a comprehensive update on the biosynthesis of phenylpropanoids, mainly lignin and flavonoids, their roles in biotic and abiotic interaction, and transcending topics, including pest and diseases, drought, temperature, and UV stress tolerance. We further reviewed the post-transcriptional, post-translational, and epigenetic modifications regulating phenylpropanoid metabolism and highlighted their applications and optimization strategies for large-scale production. This review provides an all-inclusive update on recent reports on the metabolism of phenylpropanoids in plants.
1 Introduction
Phenylpropanoids are highly diverse secondary metabolites derived from the shikimate pathway, emanating from the glycolysis and the pentose phosphate pathways routes (Lehari and Kumar, 2024). The phenylpropanoid pathway branches into two, producing numerous lignin- and flavonoid-related metabolites, which are ubiquitous in the plant kingdom and greatly contribute to plant environmental interactions. Phenylpropanoids and other phenolic compounds formation commences with L-phenylalanine, an aromatic amino acid, and L-tyrosine in some grasses. An enormous array of plant self-serving metabolites are generated via the phenylpropanoid metabolic pathway through a few shikimate pathway intermediates (Siebeneichler et al., 2024). The resultant hydroxycinnamic acids and esters are converted by a series of oxygenases, reductases, and transferases, yielding developmental- and environmental cues-specific metabolites (Ninkuu et al., 2023c). Glycosides of phenylpropanoid exhibit a variety of bioactivity, including antioxidant effect, immunomodulatory effects, and enzyme-inhibitory effect (Pinar and Rodríguez-Couto, 2025).
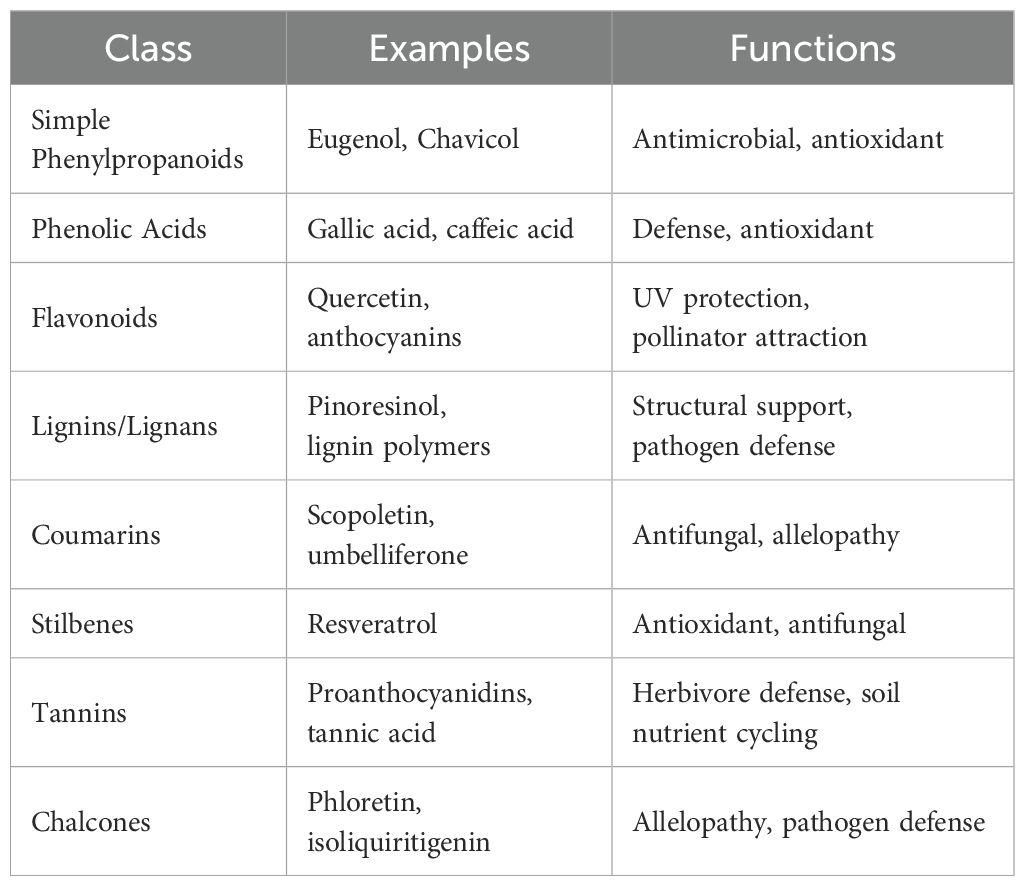
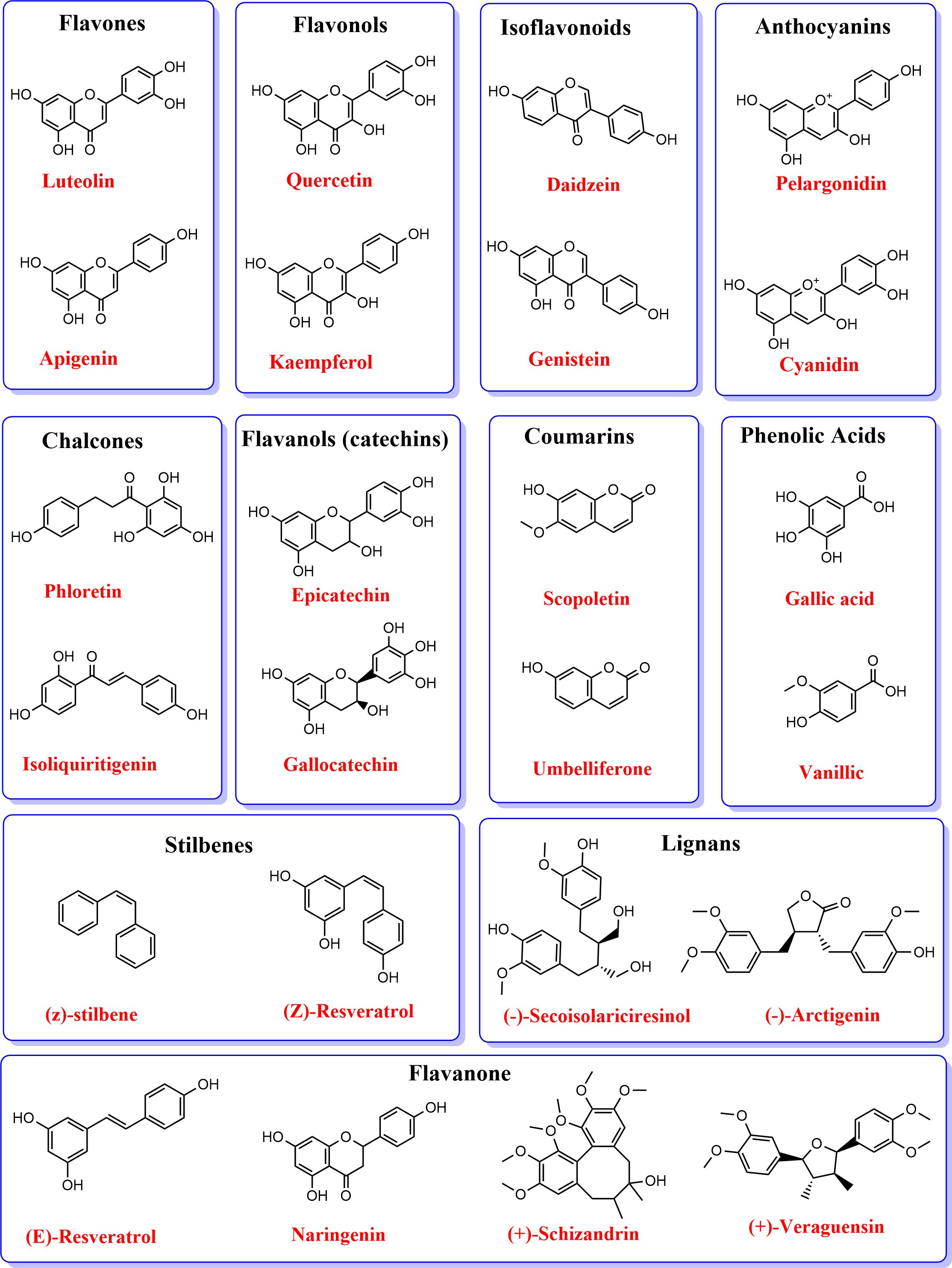
Phenylpropanoids are categorized into several classes, including simple phenylpropanoids such as cinnamic and p-coumaric acids, ferulic, caffeic, and sinapic acids; phenolic acids (hydrocinnamic and hydroxybenzoic acids); flavonoids (flavones, flavonols, flavanones, anthocyanins, isoflavonoids, etc.); lignin and lignans; coumarins, and stilbenoids (Dixon et al., 2002).
Recent studies have comprehensively elucidated the molecular regulation of phenylpropanoids, diversity, and plasticity. Additionally, the role of phenylpropanoid metabolites in biotic (plant diseases and pest control) and abiotic stress (drought, temperature, UV, nutrients, etc.) are interactions continuously changing the face of climate-resilient germplasm development in recent times. Moreover, phenylpropanoids such as lignin are required for mechanical support for plant growth and the promotion of water and mineral uptake and partitioning in plants (Uddin et al., 2024). The current article provides a comprehensive update on the biosynthesis of phenylpropanoids, mainly lignin and flavonoids, their roles in biotic and abiotic interaction, and topics, including pests and diseases tolerance, drought, temperature, nutrient signaling and uptake, and UV stress tolerance. We also examined post-transcriptional, post-translational, and epigenetic modifications involved in phenylpropanoid biosynthesis and highlighted their industrial applications as well as optimization strategies for large-scale production. This review provides an all-inclusive update on recent reports on the metabolism of phenylpropanoids in plants.
2 Overview of the phenylpropanoid pathway
The intracellular, plastidial localization, and the intricate regulation of the phenylpropanoid pathway have been explored for decades now, with almost all the pathway genes and proteins identified. Whereas tryptophan, phenylalanine, and tyrosine are useful aromatic amino acids synthesizing proteins, they are also precursors to several natural products, including hormones, pigments, alkaloids, and cell wall components. Intriguingly, all three are derivatives of the shikimate pathway, where approximately ≥30% of photosynthetic carbon is fixed on plants, providing essential diet components to humans and animals due to the loss of this pathway in their lineage (Maeda and Dudareva, 2012). The shikimate, which is a crucial precursor to the phenylpropanoids pathways, is driven by a seven-step pathway characterized by six enzymes and initiated via the condensation reaction of phosphoenolpyruvate and erythrose-4-phosphate. Notably, the phosphoenolpyruvate and erythrose-4-phosphate are also derivatives of glycolysis and the pentose phosphate pathways, respectively (Ren et al., 2024; Tzin and Galili, 2010). The formation of Arogenate from shikimate is the major biosynthetic route of phenylalanine and tyrosine, encoded by prephenate aminotransferase (PAT and CE) and arogenate dehydratase (ADT). However, recent advances have also linked phenylalanine biosynthesis to phenylpyruvate in microbes (Ren et al., 2024; Tzin and Galili, 2010) (Figure 1). Phenylalanine ammonia-lyase (PAL) is the gate opener for several glycosylation, acylation, hydroxylation, and methylation reactions, forming over 8000 metabolites in the phenylpropanoid pathway (Ninkuu et al., 2023a).
The phenylalanine and the tyrosine in some grasses diverge into different pathways, from Arogenate but reconverges, yielding p-coumarate, which is a precursor to coumaroyl CoA for the formation of an array of phenylpropanoid metabolites. Coumaroyl CoA is also the precursor for the lignin and flavonoid biosynthesis (Figure 1). Lignin is a heterogeneous phenolic polymer and the second most abundant polymer after cellulose, forming 30% of the earth’s organic carbons in the biosphere. The so-called heterogeneity of lignin results from its polymerization from various hydroxycinnamoyl alcohol derivatives. It is subsequently deposited in the cell walls of vascular plants, conferring many stress tolerance traits, including resistance to diseases and pests, drought, deterioration, heat stress, UV radiation, etc (de Oliveira et al., 2025; Ninkuu et al., 2022). Elsewhere, we comprehensively reviewed the 11 enzymes involved in lignin biosynthesis, the phytoalexins they produced, and their individual or collaborative roles in plant immunity induction (Ninkuu et al., 2023a).
Like lignin, flavonoid metabolism is the second branch of the phenylpropanoid pathways, producing over 6000 polyphenolic metabolites (Jie et al., 2023). Flavonoids are bioactive metabolites involved in plants’ biotic and abiotic interactions, including microbial signaling, allelopathy, and nutraceuticals for improved health (Oro et al., 2025; Zheng et al., 2025). Flavonoids are characterized by C6-C3-C6 diphenylpropane skeleton, where three carbon chains (C3) links the two aromatic rings (Shanker and Rana, 2025). Flavonoids are classified based on the heterocyclic C‐ring, such as chalcones, aurones, flavones, isoflavones, flavanones, dihydroflavonols, anthocyanidins, leucoanthocyanidins, flavonols, and flavan‐3‐ols (Chen et al., 2023). Table 1 and Figure 2 show the classifications of flavonoids and their structural forms, respectively. The first committed step in flavonoid biosynthesis is catalyzed by chalcone synthase (CHS), converting p‐coumaroyl‐CoA to chalcone, which directs the metabolic flux to flavonoid metabolism. Stilbene synthases (STS) also encode the formation of simple stilbenes from cinnamoly‐CoA and p‐coumaroyl‐CoA. Liu et al. (2021) review discusses the biosynthesis processes of flavonoids in plants, dissecting the various enzymes involved.
3 Biological functions of phenylpropanoid-derived metabolites
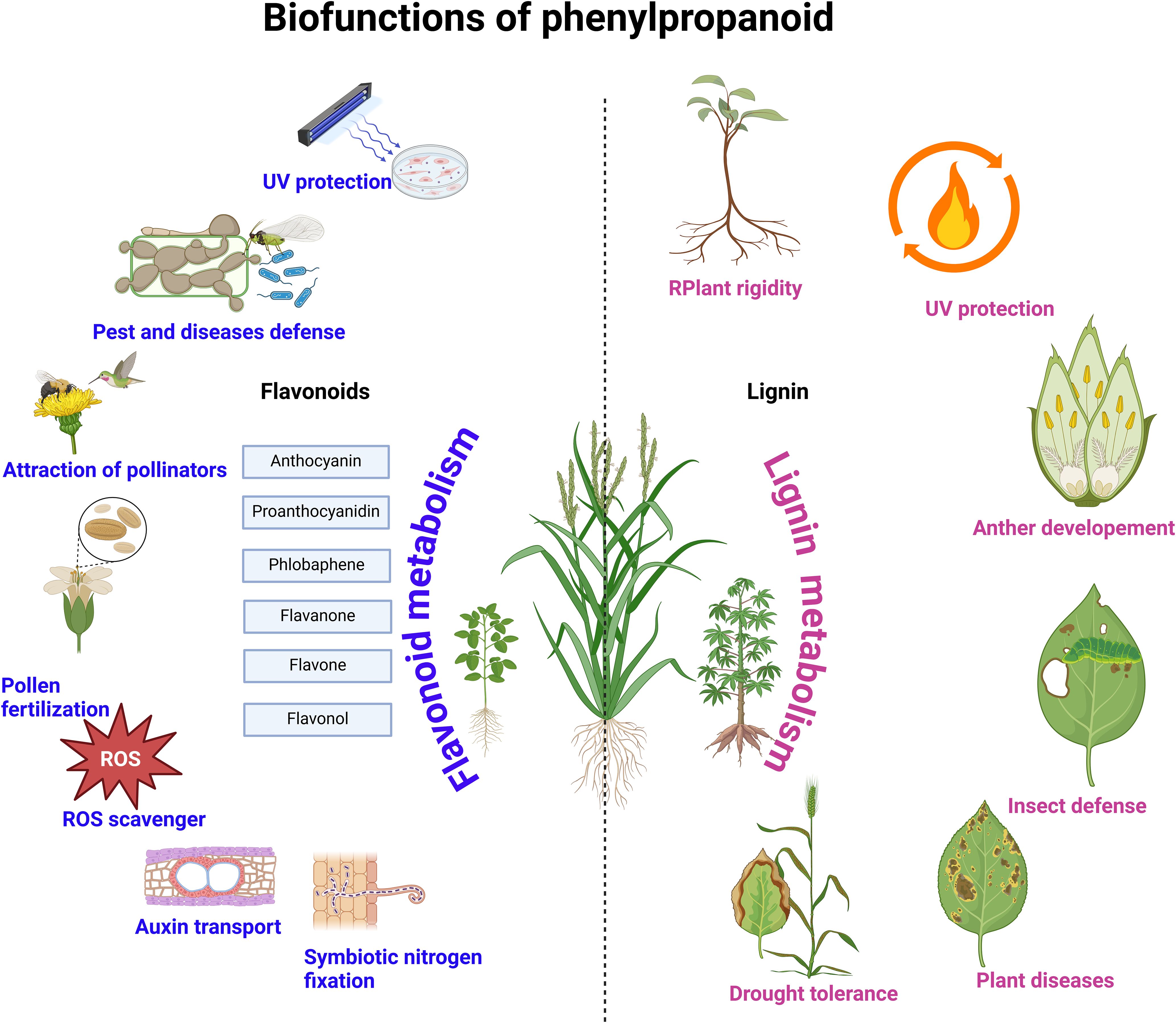
As a sessile land organism, plants are exposed to numerous but expected environmental hazards, including pathogens and insect infections, UV radiation, drought, heat, and cold stressors. The deterioration of crop products is also quite hastened by environmental influences. Notwithstanding these unavoidable stressors imposed partly due to climate change, studies have shown that phenylpropanoid metabolism can ameliorate these factors in plants (Figure 3). In the following sections, we highlight recent works elucidating the role of phenylpropanoid metabolism in resisting these stresses.
3.1 Phenylpropanoid metabolism enhances resistance to reactive oxygen species for stress tolerance
Reactive oxygen species (ROS), including superoxides (O2-), hydrogen peroxide (H2O2), hydroxyl radical (OH-), and singlet oxygen species (1O2) are by-products of cellular metabolism responsive to adverse environmental stressors in plants (Rabeh et al., 2025). ROS induction signals plant growth, differentiation, and immune responses. Moreover, ROS production under stressful conditions obstructs cellular functions, leading to oxidative damage and conferring biotic and abiotic stress responses in plants. However, plants adapt to excessive ROS induction using intricate ROS-scavenging mechanisms to offset damage to protein, lipids, and DNA (Rabeh et al., 2025; Wang et al., 2024; Gao et al., 2023; Yang et al., 2023). Moreover, plants have developed sophisticated mechanisms to cope with stressors, such as phenolic compound metabolism, to neutralize ROS DNA (Rabeh et al., 2025; Wang et al., 2024).
Meeting the growing food demand presents a significant challenge to global food security, as much of the world’s arable land remains vulnerable to abiotic stresses such as salinity, drought, extreme temperatures, UV radiation, and heavy metal toxicity. Phenylpropanoid biosynthesis becomes a crucial physiological need of abiotic-stressed plants. The surge in phenylpropanoid metabolism under abiotic stress detoxifies ROS and protects cellular components from oxidative damage. Hence, crucial genes encoding key phenolic enzymes, including PAL (phenylalanine ammonia-lyase), C4H (cinnamate 4-hydroxylase), 4CL (4-coumarate: CoA ligase), CHI (chalcone isomerase), and F3H (flavanone 3-hydroxylase) are predominantly upregulated in response to various abiotic stressors (Rabeh et al., 2025; Rao and Zheng, 2025; Sharma et al., 2019).
3.2 Phenylpropanoids enhance plants’ tolerance to UV-B radiation
Plants exposed to UV-B stress generate harmful ROS that severely damage their DNA and proteins (Naikoo et al., 2019; Singh et al., 2023). Nevertheless, such stresses can be mitigated by increasing cellular phenolic deposition, which shields the epidermal layers of the leaves (Olson and Ruhland, 2024; Xiao et al., 2023). Phenylpropanoids further plummet DNA damage by minimizing the photodamage of crucial enzymes such as NAD/NADP, while arresting thymine dimerization (Naikoo et al., 2019). Among these phenolics, flavonoids are considered effective UV-B screening filters deposited in leaf interiors and trichomes, for plant defense against harmful radiations (Choudhary et al., 2021; Singh et al., 2023). Several studies have affirmed that a spike in flavonoid biosynthesis promotes plant tolerance to UV- radiation (Table 3) (Hao et al., 2022; Rizi et al., 2021; Song et al., 2025). Hence, increased expression of flavonoid biosynthetic genes (F3H, CHS, CHI, and FLS) safeguards plants against UV-B stress. According to Zhao et al. (2020), revealed that the upregulation of FLS and FS’H in response to UV-B radiation promoted flavonoid biosynthesis in Ginkgo biloba leaves. Similar upregulation of the flavonoid-induced gene (F3H) has been reported in a desert plant, Reaumuria soongorica, indicating flavonoid regulates UV-B stress adaptation (Liu et al., 2013). Martínez-Silvestre et al. (2022) revealed a higher flavonoid content in the callus irradiated with UV-B, averting the harmful effects of UV-B radiation in Sideroxylon capiri. Thus, flavonoids function as a “signal trigger,” neutralizing the prospective effects of UV-B light.
3.3 Phenylpropanoids enhance plant responses to temperature stressors
Extremes of temperature retards plant growth and development (Aluko et al., 2021; Ma D. et al., 2025). Plants accumulate more phenolic compounds to detoxify ROS under temperature stress (Table 3). Hence, the increased expression of C3H and lignin levels in rhododendron contributes to cold tolerance (Wei et al., 2006). The crucial genes encoding lignin biosynthesis were highly expressed in cold-tolerant cultivars, indicating the contribution of lignin in peach adaptation to cold (Li et al., 2023b). Overexpressing CaPOA1 and CaCAD in Arabidopsis increases ROS scavenging and plant tolerance to cold injury (Xiao et al., 2025). A similar increase in phenolic compounds was observed in heat-stressed plants (Commisso et al., 2016; Wang J. et al., 2019; Yuan et al., 2025), indicating the crucial roles of phenylpropanoids in enhancing plants’ tolerance to temperature stress.
3.4 Transcriptional regulation of lignin and flavonoids roles in plant defense interactions
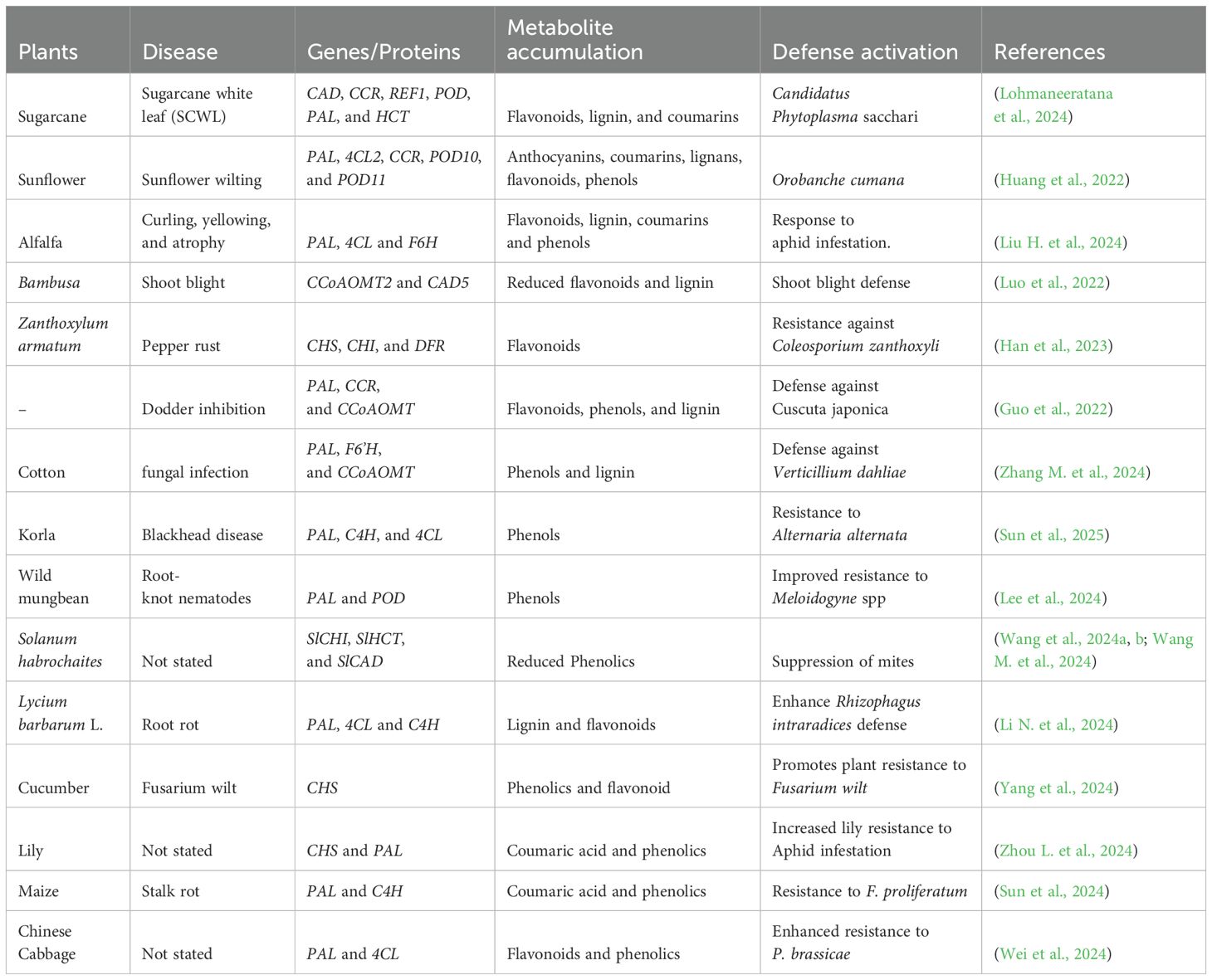
While lignin metabolism strengthens the cell wall, enhancing physical resistance to invasion, flavonoid biosynthesis produces essential phytoalexins that support plant immunity and serve as signaling molecules for microbial interactions. For example, the upregulation of the phenylpropanoid pathway under Hrip1 induction conferred resistance to rice blast fungi by reinforcing cell walls through extensive lignin deposition (Ninkuu et al., 2022; Zhang et al., 2021). Wang W. et al. (2025) also reported the enhanced accumulation of lignin against Tambocerus elongatus in Camellia sinensis. The oxidation of H2O2 promoted lignin accumulation by downregulating transcriptional inhibitors, including miR397b, that adversely regulate OsLAC7, OsLAC28, and OsLAC29, liberating Copalyl Diphosphate Synthase 2 (CPS2) for terpenoids metabolism (Cao and Dong, 2025; Ninkuu et al., 2021). Additionally, a pear plant over-expressing the PbrMYB14 enhanced lignin accumulation against Alternaria alternata and reduced leaf lesions by 68.95% (Yan et al., 2025). GhBGLU46 has been identified as a key activator of several lignin metabolism genes, including GhCCoAOMT2, GhCCR4, GhCAD6, and GhCAD. Thus the overexpressing GhBGLU46 increased lignin production against Verticillium wilt (Wang et al., 2025a). Li and Wang (2025) also found that CpVQ20-overexpressing lines in tobacco promoted flavonoid and lignin metabolism via upregulated NtF5H against powdery mildew.
Furthermore, lignin also mediates insect modulation. Recent literature has shown that the overexpressing lines of CCR in Populus enhanced lignin levels to mediate defense against L. dispar larvae (Li Y. et al., 2025). Sl4CLL6 mutant lines hampered the expression of genes downstream of the phenylpropanoid pathway, including SlHCT, SlCAD, and SlCHI, further compromising tomato resistance to mites (Wang et al., 2024).
Flavonoids such as anthocyanins, flavonols, and flavones are highly pigmented and contribute to the flower color of plants (Bisht and Gaikwad, 2025). Recent studies have revealed their novel roles in pest and disease mitigation (Tiwari et al., 2025). Chu et al. (2025) reported the role of NtWRKY28 in lignin and flavonoid metabolism against aphids in tobacco plants by inducing the upregulation of several phenylpropanoid biosynthetic genes (PAL, 4CL, CHI, CAD, HCT, CHS, C4H, and CCR). Additionally, VqWRKY56 enhances the transcription of VqbZIPC22, which activates salicylic acid and proanthocyanidin metabolism, strengthening resistance to powdery mildew in Vitis quinquangulari (Wang Y. et al., 2023). Quercetin accumulation in lima beans also enhances defense against Tetranychus urticae (Li F. et al., 2025), while Brown midrib 12 (BMR12) induction promoted COMT activity, increasing JA and flavonoids accumulation against fall armyworm (Kundu et al., 2025). In a study investigating the mechanism of phenylpropanoid’s defense against Alternaria alternata in korla fruits, Sun et al. (2025) reported high enzymatic activity of PAL, C4H, and 4CL resulting in significant accumulation of total phenolics, trans-cinnamic acid, ferulic acid, caffeic acid, p-coumaric acid, and sinapic acid. Notably, higher expression of CHS and CHI significantly improved flavonoid accumulation, including naringenin, rutin, apigenin, quercetin, and epicatechin in defense against A. alternata infection. Recent research has highlighted the role of phenylpropanoid metabolism in plant resistance to diseases and pests (Table 2).
3.5 Phenylpropanoids (Flavonoids) as signaling molecules for root nodulation in legumes
Flavonoids play a crucial role as signaling molecules and chemo-attractants in plant-microbe interactions, influencing organisms such as Fusarium spp., Rhizobium, and arbuscular mycorrhizal fungi. Additionally, they can activate virulence genes in Pseudomonas syringae and Agrobacterium tumefaciens (Falcone Ferreyra et al., 2012). Flavonoids also play a crucial role in the legume nodulation process (Figure 3). Thus, legume roots exudate flavonoids, which rhizobial nodulation (Nod) protein NodD detects, triggering the expression of nod genes and Nod factors (NF) (Ninkuu et al., 2025) (Figure 4). NFs induces legume responses for symbiotic interactions (Haskett et al., 2025). Evidence has shown that RNAi of chalcone synthase in legumes exhibited deficiency in nodulation due to the collapse of flavonoid biosynthesis (Abdel-Lateif et al., 2013; Das et al., 2024). Moreover, the Rlv3841 NodD regulatory domain deletion line activated NodDFI for transcript accumulation of NF genes (Haskett et al., 2025). Interestingly, flavonoid exudation into the rhizosphere to attract rhizobia spp. is complicated and involves several players. Elicitors have been implicated in inducing flavonoid exudation (Hassan and Mathesius, 2012). However, transgenic Arabidopsis harboring the mutant ABC transporter exhibited altered exudation of flavonoids. ABC transporters have been demonstrated to be involved in isoflavonoid genistein exudation in soybeans, and it has also been reported that flavonoids can be passively released by decomposing roots (Hassan and Mathesius, 2012).
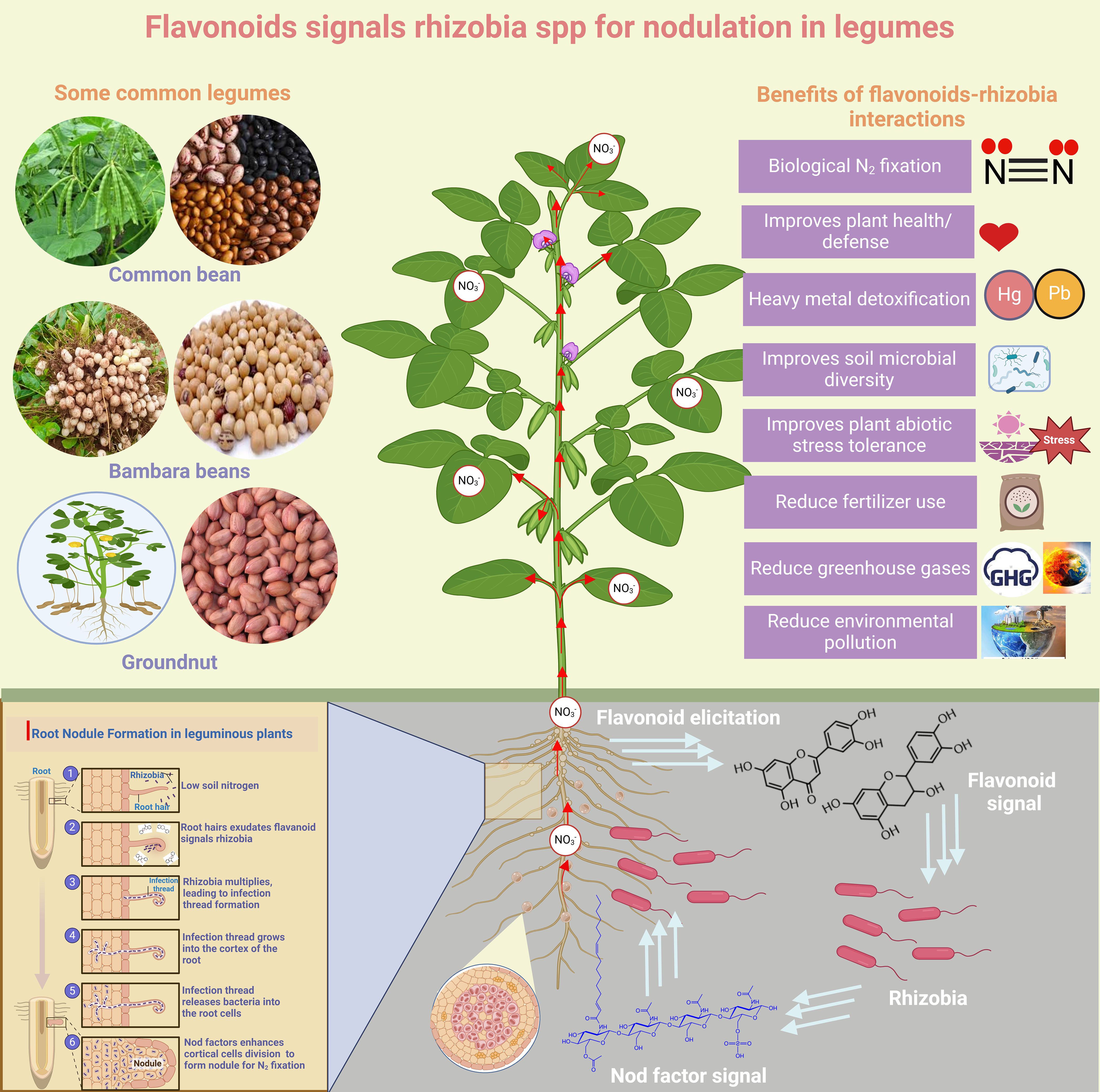
Figure 4. Phenylpropanoids (Flavonoids) signal rhizobia for root nodulation in legumes. The Figure provides an overview of the role of flavonoids as signaling molecules for rhizobia, which infects root hair legumes, leading to nodulation. This symbiotic relationship results in N2 fixation for crop growth. The Figure also highlighted the ecological benefits of nitrogen fixation.
Flavonoids-induced symbiotic interactions between roots of legumes and rhizobia spp. have several ecological benefits (Figure 4). Some of these include improvement of soil health, reduction of environmental pollution and GHG emissions from synthetic fertilizer use. Furthermore, ROS accumulation in legume roots upon detecting rhizobia spp. via nod factors can also initiate a crucial signaling cascade (Hérouart et al., 2002). Apart from coordinating symbiotic interaction, ROS production modifies the cell wall and modulates the expression of defense-related genes, positioning legumes’ defense machinery against pathogens. Interestingly, it is currently unknown how cell wall modification favors rhizobia infection but inhibits pathogens.
Two plant growth regulators, cytokinins and auxins, crucially enhance legumes nodulation process, promoting cell division and differentiation (Reid et al., 2017; Ryu et al., 2012). Additionally, cytokinins and auxins promote the growth of root primordia via cell elongation and division in the proliferating zone (Ryu et al., 2012).
4 Phenylpropanoids mediate osmotic stress adjustment
Osmotic stresses, such as drought and salinity, are major physiological factors that limit plant growth and yield improvement. The next sections discusses their impacts on plants and highlights the modulatory role of phenylpropanoids in stress response.
4.1 Phenylpropanoid biosynthesis is crucial for drought stress tolerance
Drought stress negates various plant physiological processes, ultimately retarding growth and development (Aluko et al., 2021; Jardim-Messeder et al., 2025). Nevertheless, plants have developed adaptative mechanisms for drought, specifically via phenylpropanoid biosynthesis (Rao and Zheng, 2025; Wagay et al., 2023). Earlier studies reported increases in the expression of flavanone-3-hydroxylase (F3H), PAL, 4CL, and flavonol synthase (FLS) enhanced plant tolerance to drought (Chen W. et al., 2025; Ghasemi et al., 2023; Park et al., 2023), perhaps because phenolic compounds mitigates ROS accumulation in the cells, preventing oxidative damage. It has been claimed that flavonoid deposition in the cytoplasm efficiently mitigates the harmful effect of the H2O2 molecule exerted by drought. However, La et al. (2023) detected a lesser content of flavonoids in soybean under drought stress conditions. Discrepancies in these findings may be influenced by factors including stages of seed development, tissue type, or drought severity (La et al., 2023). Ghasemi et al. (2023) reported a gradual decline in phenolic content, following an initial increase 7 days after drought treatment. Low phenolic formation during the later stages of stress indicates plants’ metabolic adjustment to prolonged stress (Ghasemi et al., 2023). Furthermore, Yan et al. (2023) reported the role of OsOLP1 in mediating rice tolerance to drought via lignin, proline, and abscisic acid accumulation. Elsewhere, Cao P. et al. (2024) identified BGC7 and BGC11 gene clusters consisting of 12 genes, including 4CLs, C3H, CPA, and SlMYB13 in phenolamide metabolism against drought stress tolerance in tomatoes, providing deeper insight into crop improvement techniques via genetic engineering and secondary metabolite elicitation. Detailed reports on crop drought-resistant mechanisms mediated by phenylpropanoids metabolism are highlighted in Table 3.
4.2 Salinity stress tolerance in plants under phenylpropanoid metabolism
Salinity stress is a crucial environmental constraint that halts plant growth and development (Ben Youssef et al., 2025; Safdar et al., 2019; Aluko et al., 2024). High soil salinity decreases leaf dry weight, plant height, photosynthesis, water, and nutrient uptake (Singh et al., 2025; Wang H. et al., 2025). Salt stress promotes the production of ROS, causing oxidative damage to plant cells (Jiang et al., 2025; Singh et al., 2025; Huang et al., 2024; Yang et al., 2023). Therefore, enhancing antioxidant defense systems could contribute to plant salinity tolerance (Ling et al., 2025). One of the most probable ways of improving plants defense system is by increasing the activities of antioxidant enzymes such as CAT and SOD, involved in the removal of H2O2 and O2-, safeguarding against cellular damage (Garcia-Caparros et al., 2021; Shomali et al., 2022). Cao Y. H. et al. (2024) reported a significant increase in SOD and CAT activities under salinity stress, particularly in salt-tolerant genotypes. The salt-tolerant genotype appears to have an in-built phenolic compound, acting as an antioxidant defense system, that scavenges harmful ROS (Cao Y. H. et al., 2024; Bistgani et al., 2019; Chen et al., 2019). Ample evidence revealed that increased expression of phenylpropanoid biosynthetic genes and their respective metabolites contributes to plant salt tolerance (Table 3). Increased expression of NtCHS1 facilitated tobacco tolerance to salt stress (Chen et al., 2019). Flavonoid biosynthetic genes, including LpFLS1 and LpCHI1, highly expressed in ryegrass, suggest their involvement in salt tolerance (Cao Y. H. et al., 2024). Overexpressing GmCHI4 in soybean enhanced isoflavones content in the salt-stressed root (Zhang J. et al., 2024). These and other findings suggest the contributions of phenylpropanoids in plant salt stress tolerance.
4.3 Phenylpropanoids role in postharvest deterioration
Postharvest physiological deterioration (PPD) severely threatens global food security, rendering crops unpalatable 1–3 days after harvest (Chang et al., 2024; Chen Z. et al., 2025; Ji et al., 2025). Different storage methods, including cellular storage, plastic bag wrapping, indoor sand storage, and paraffin wax coatings, have been previously used to improve plants’ postharvest quality. Yet, the interventions are time-consuming and labor-intensive (An et al., 2023; Chang et al., 2024; Chen Z. et al., 2025). Extending the postharvest shelf life is critical for sustainable crop productivity.
Attempts to extend postharvest shelf-life have been quite challenging due to the increased production of reactive oxygen species (ROS), which causes PPD. Phenylpropanoid metabolism has become a crucial defense mechanism to mitigate ROS-induced PPD and improve plant storage stability effectively (Liu Q. et al., 2024; Liu et al., 2017; Wahengbam et al., 2023). Specific phenylpropanoid-derived metabolites, such as phenolics, epicatechin, flavonoids, and ferulic acid, accumulate in stressed or injured plants during storage. Meanwhile, others, including 3,4-flavanone, coumarin, and isoflavone, decrease, suggesting changes in metabolite synthesis contribute to postharvest deterioration under stress. Zheng et al. (2022) recently revealed that changes in the synthesis of phenylpropanoid derivatives impact strawberry postharvest quality under temperature stress. Apple and bulb discoloration have also been attributed to phenylpropane biosynthesis, suggesting phenylpropanoids are crucial for fruit preservation (Chen Z. et al., 2025; Wang J. et al., 2023).
Studies have shown a significant increase in the expression of genes associated with phenolic biosynthesis and ROS turnover during storage, ultimately regulating PPD (An et al., 2023; Liu Q. et al., 2024; Vanderschuren et al., 2014; Wahengbam et al., 2023; Wang B. et al., 2019; Wang C. et al., 2023). Perhaps the reason why PAL expression, which was barely detectable in harvested cassava roots (0hr), increased by 70-fold 72hrs after wounding (Kumar and Knowles, 2003; Wang C. et al., 2023). The enhanced activity of PAL facilitates lignin biosynthesis (Liu et al., 2005); thus, the expression of cinnamate-4-hydroxylase (C4H), which synthesizes precursors of lignin biosynthesis, increased 72hrs after wound healing (Xu J. et al., 2019). Furthermore, 4-coumarate CoA ligase (4CL) facilitates the metabolic flux to flavonoids in PPD-susceptible plants (Wang C. et al., 2023; Wang et al., 2020), indicating the contributions of phenolic compounds in plant storage stability. Recent updates on the crucial roles of phenylpropanoid genes and the respective metabolites are indicated PPD (Tables 4, 5).
4.4 Interaction between plant growth regulators and phenylpropanoid metabolism
Phytohormones are natural signaling molecules that contribute to plants’ response to environmental cues (Samanta and Roychoudhury, 2025). Recent advances link these naturally synthesized and deployed molecules by plants to the modulatory activity of the phenylpropanoid pathway. For example, ethylene, auxin, strigolactone (SL), jasmonate (JA), and gibberellin are associated with the phenylpropanoid pathway (Silva et al., 2025), indicating the activities influencing phenylpropanoids intricately affect phytohormones. Shi et al. (2024) recent study reported the role of a novel phytohormone, 2,4-dichloroformamide cyclopropane acid (B2) in drought stress tolerance in Carex breviculmis. Transcriptome analysis of B2-treated plants activated the expression of drought stress-responsive transcription factors, including AP2/ERF-ERF, WRKY, and mTERF, which consequently upregulated the phenylpropanoid metabolism via the upregulation of HCT, COMT, and POD genes. B2 signaling modulated phytohormone-responsive genes, leading to abscisic acid accumulation for drought tolerance (Shi et al., 2024). Elsewhere, Dey and Sen Raychaudhuri (2024) reported that 1 μM MeJA treatment of Plantago ovata enriched the PAL and CHI for enhanced antioxidant defense through ROS signaling, activating significant metabolism of phenolic compounds, such as caffeic acid, chlorogenic acid, vanillic acid, coumaric acid and Luteoloside and PGRs including IAA and GA. Moreover, evidence indicates that FvTCP9 transcription factor regulates FaNCED1, which encodes 9-cis-epoxycarotenoid dioxygenase (NCED), a key enzyme in abscisic acid (ABA) biosynthesis. Activation of FaNCED1 leads to changes in ABA levels and may be involved in the PYR-PP2C-SnRK2 signaling pathway. Furthermore, FvTCP9 modulates the transcription of genes associated with anthocyanin biosynthesis (FaPAL, FaCHS, FaCHI, FaANS, FaUFGT), influencing strawberry coloration. Thus, exploring the intricate interaction of phenylpropanoids and phytohormones can enhance plants developmental cues and stress Reponses.
4.5 Phenylpropanoids regulate nutrient deficiency tolerance in plants
Nutrient deficiency stress is one of the leading causes of plant growth retardation and yield loss (Li et al., 2023a, b; Li C. et al., 2023; Ninkuu et al., 2023a). Nitrogen (N), phosphorus (P), and potassium (K+) deficiency stress, for instance, disrupt photosynthesis, nutrient uptake, and allocation. Nevertheless, phenylpropanoids mediate plant tolerance to nutrient stress (Table 6). Li J. et al. (2023) revealed that the upregulation of flavonoids under N deficiency stress maintains C/N balance of sugarbeet. Low N stress promotes flavonoid biosynthesis, increasing plant enzymatic activities in snow chrysanthemum (Li Z. et al., 2023). Similar reports on rapeseed and cassava have shown a significant boost in flavonoid content in response to N deficiency stress (Koeslin-Findeklee et al., 2015; Wang et al., 2025b). Wang et al. (2025b) affirm the upregulation of two CHI in response to low N stress, suggesting CHI is crucial for carbon flux redistribution. Evidence has shown the detrimental impact of N deficiency stress on carbon metabolism, redirecting photosynthetic carbon into the phenylpropanoid biosynthetic pathway (Aluko et al., 2021; Wang et al., 2025b). This shift promotes metabolic flux of the flavonoid downstream genes, resulting in increased flavonoid deposition (Xin et al., 2019). Samarina et al. (2024) reported that increases in lignin and flavonoids improved cell wall stability under N deficiency, suggesting their roles in tea adaptation to low N conditions. Lignin regulates root architecture and other plant physiological processes, and thus, lignin reprograms Neolamarckia cadamba root under N deficiency stress (Lu et al., 2021).
Increased activities of phenylpropanoid–derivatives under P deficiency have also been well-documented (Liu S. et al., 2024; Wu et al., 2022). Increased activities of PAL and 4CL suggest their enzymes are crucial downstream metabolites in response to P deficiency stress (Liu S. et al., 2024). Lignin, one of the vital downstream branches, was enhanced in response to P deficiency (Cesarino, 2019). More importantly, lignin biosynthetic genes, including CCR, CAD, and POD, were significantly upregulated in response to low P stress (Wu et al., 2022). Increasing lignin gene expressions may promote cell wall thickening, reduce permeability, and improve plant adaptation to low P stress (Cesarino, 2019).
The impact of phenylpropanoids on low K+ stress has been elucidated following the reports of excessive production of harmful ROS upon low K+ stress (Sun et al., 2023; Zeng et al., 2015, 2018). Potassium stress increases PAL deposition to detoxify ROS, which damages cell membrane stability (Sun et al., 2023). Moreover, UDP-glucosyl transferase activities have been demonstrated to regulate flavonoid-mediated auxin levels during grain development (Ninkuu et al., 2023b). Although the impact of phenylpropanoids in enhancing plant tolerance to individual stress has been harnessed, less is known under combined N, P, and K stressors (Table 6).
5 Post‐transcriptional regulation of phenylpropanoid metabolism
Plant cell retains their competitiveness to varying degrees of stress exposure by balancing growth and proliferation expenditures with the stress factors. Under such conditions, plants recruit different levels of gene regulatory activities, such as post-translational and post-transcriptional modification of mRNA, to respond to the stress factors and recovery processes (Hernández-Elvira and Sunnerhagen, 2022). Post-transcriptional gene modification is multi-layered, involving mRNA processing, stability, localization, and protein translations (Courtney, 2021).
The role of Micro RNAs (miRNAs) and small RNAs (RNAs) in targeting the structural genes regulating phenylpropanoid metabolism has been thoroughly studied in relation to plant stress responses (Nayak et al., 2025; Rosatti et al., 2024). MiRNAs modulate their target genes posttranscriptionally through mRNA cleavage or limiting its translation, which is critical in the downstream biochemical pathways and pigment synthesis (Ding et al., 2024). For instance, miRNA156 modulates flavonoid synthesis by targeting MYB TFs (Rosatti et al., 2024). Additionally, the loss of function of miR-858a liberated the targeting efficiency of flavonoid-specific transcriptional regulators, including AtMYB12 and AtCHS1 (Jiang et al., 2021). Moreover, miR-172, miR530, and miR157 have been demonstrated to regulate secondary metabolite accumulation in leaves and roots of rice, Arabidopsis, and Chlorophytum borivilianum (Jiang et al., 2021). Furthermore, miR-894, miR172, miR-9662, and miR-166 have also been reported to regulate phenylpropanoid metabolism in plants (Marcela et al., 2019). MiRNAs-TFs-target genes complex can upregulate or compromise phenylpropanoid metabolism. SPL9 and SPL13 are targeted explicitly by miR156 to stifle the mRNA level of DFR and inhibit anthocyanin accumulation in the process (Cui et al., 2014; Gou et al., 2011). Nevertheless, DFR expression is upregulated for anthocyanin accumulation via overexpressing miR156, which inhibits SPL13 in alfalfa (Feyissa et al., 2019). Our previous study showed that MiR396b/GRF module regulates Arabidopsis growth under low sulfur conditions (Ninkuu et al., 2024). Yuan et al. (2024) recent study showed that the miR396b/GRF6 module improved salt stress tolerance in rice by inhibiting H2O2 accumulation while elevating ROS-scavenging enzyme activities, including CAT, SOD and POD. Meanwhile, ZNF9 was identified as a negative regulator of salt stress tolerance by binding to the miR396b promoter region. In soybean, miR398b targets and represses the transcript level of GmCCS and GmCSD1b, compromising the defense prowess of the crop. Interestingly, the defense machinery of soybean against Heterodera glycines worsened when miR398b overexpressing levels were generated. However, silencing of miR398b in soybeans improved crop defense capabilities by modulating H2O2 and O2- levels (Zhang X. et al., 2024).
Plant pigmentation can also be regulated by miRNA in plants. Nayak et al. (2025) RNA sequencing identified 74 miRNA regulating white coloration and 61 responsible for brown color pigmentation in cotton by modulating flavonoid biosynthesis.
6 Post-translational modification of phenylpropanoid metabolism
Post-translational modifications (PTMs) play a significant role in protein functions, stability, localization, activity, structure, and molecular interactions. Post-translational modifications can also influence lignin biosynthesis and wood formation. Recent studies have demonstrated that PTMs of monolignol enzymes, such as phosphorylation and ubiquitination, inhibit enzymatic activity and stability of proteins (Sulis and Wang, 2020). It is worth noting that PTM of proteins are strongly associated with phenylpropanoid metabolism, including phosphorylation, ubiquitination, glycosylation, and S-nitrosylation. These PTMs are essential for biological processes in plants. For example, Kelch Domain F-Box (KFB) proteins (KFB1, KFB50, KFB20, and KFB39) inhibit phenylpropanoid metabolism via PAL ubiquitination and proteasome-mediated degradation. Moreover, MED5 mediates the activation of KFB39 and KFB50, while KFBCHS, which negatively regulates flavonoid biosynthesis, acts as the ubiquitination and degradation of CHS in A. thaliana (Kim et al., 2020). Additionally, the ubiquitination of PAL1–4 reduces KFB proteins, lowering their stability in Arabidopsis thaliana via the 26S proteasome. Similarly, the interaction of OsCCR with SCFOsFBK1 in rice decreases its stability through the 26S proteasome (Zhang et al., 2013). Zhang et al. also showed that MYB156 and MYB221 interaction with UBC34 diminishes their transactivation of lignin genes and may reduce their stability through the 26S proteasome in P. tomentosa (Zheng et al., 2019).
Phosphorylation has long been recognized as a key regulatory modification of proteins. Phosphorylation of PtrAldOMT2 by SDX deactivates its protein activity in Populus trichocarpa by ∼ 60% (Wang et al., 2015). Although R2R3-MYB family members are crucial regulators of gene expression, PtMYB4 is phosphorylated by PtMAPK6 during early xylem development (Morse et al., 2009).
7 Epigenetic regulation of phenylpropanoid metabolism
Epigenetic regulation, which modifies gene expression without altering DNA sequences, can influence phenylpropanoid metabolism, particularly lignin deposition in plants. Environmental factors can trigger epigenetic modification by altering plant gene expression, leading to phenylpropanoid metabolism as a response factor (Ma H. et al., 2025). Epigenetic regulatory mechanisms preceding lignin and flavonoid metabolism include histone modification, DNA methylation, and miRNA activity (Li W. et al., 2024). For instance, a histone deacetylase PtrHDA15, acting as an epigenetic inhibitor, relies on PtrbZIP44-A1 for chromatin histone modifications that repress PtrCCoAOMT2 and PtrCCR2 to inhibit lignin accumulation in P. trichocarpa (Li W. et al., 2024). Moreover, overexpressing PtrbZIP44-A1 or PtrHDA15 triggered the reduction of histone acetylation at PtrCCoAOMT2 and PtrCCR2 promoters, leading to reduced lignin accumulation. However, the ptrbzip44-a1 and ptrhda15 mutants detected higher histone acetylation levels at PtrCCoAOMT2 and PtrCCR2 promoters, triggering the expression of the target gene and lignin deposition (Li W. et al., 2024). The conserved histone H2 variant, H2A.Z, has been shown to negatively regulate anthocyanin biosynthesis in A. thaliana. Cai et al. (2019) reported that anthocyanin synthesis in H2A.Z deposition-deficient mutants is associated with increased levels of H3K4me3, which is upregulated by anthocyanin-related genes. Furthermore, Peng et al. (2020) demonstrated that virus-induced gene silencing of McHDA6 (Histone deacetylase 6) inhibited the transcriptional activity of methyltransferase 1 (McMET1), leading to enhanced expression of McMYB10 and increased anthocyanin accumulation in Malus crabapple.
8 Interaction between phenylpropanoid metabolism and plant signaling pathways
Phytohormones are naturally existing organic signaling molecules that crucially coordinate responses to plant biotic and abiotic interaction and developmental cues at lower concentrations. Plant phytohormones are highly diverse, fulfilling distinct regulatory roles or engaging in complex, multifunctional processes within the plant. They include auxins, cytokinins, Gibberellins (GA), Ascisic Acid (ABA), ethylene, Brassinosteroids, Salicylic Acid (SA), Jasmonates (JAs), and Strigolactones (Chakraborty et al., 2025; Iqbal et al., 2021). It is well-established that phytohormones can regulate phenylpropanoid metabolism, and NAC/MYB has been demonstrated to regulate these hierarchical interactions (Li C. et al., 2024; Li W. et al., 2024). For instance, PtoJAZ5 is a key regulator of JA-mediated lignin suppression in Populus, influencing secondary vascular development. Furthermore, transgenic lines overexpressing PtoJAZ5 in poplar and Arabidopsis exhibited collapsed secondary cell walls attributed to the downregulation of genes involved in SCW formation (Li C. et al., 2024; Li W. et al., 2024; Zhao et al., 2023). Overexpression of McMYB4 led to increased accumulation of flavonols and lignin in apples. Subsequent Y1H and electrophoretic mobility shift assays (EMSAs) demonstrated that McMYB4 directly binds to the promoter regions of McMYB4, CAD, and F5H, key genes involved in flavonoid and lignin biosynthesis. Additionally, McMYB4 was shown to interact with the promoters of AUX/ARF and BRI/BIN genes, thereby activating auxin and brassinosteroid signaling pathways to promote growth and reduce reactive oxygen species (ROS) (Hao et al., 2021). According to Xu C. et al. (2019), overexpression of PtoARF5.1 and PtoIAA9m, which encodes a stabilized form of the IAA9 protein, suppresses secondary xylem development by downregulating genes such as PAL4 and WND1B that are involved in lignin biosynthesis and xylem formation. This repression occurs through inhibiting their positive regulators, PtoHB7, PtoHB8, and two class III HD-ZIP transcription factors.
Exogenous application of benzylaminopurine and MeJA has also been shown to stimulate the accumulation of proline, ROS, and dehydrins, thereby enhancing antioxidant activity and reinforcing the cell wall with lignin as a physical defense barrier (Avalbaev et al., 2021). Similarly, the application of SA and JA enhanced resistance against drought stress in wheat and French bean via enhanced SOD and POD enzymatic activities, along with the accumulation of defense metabolites, such as anthocyanins, flavonoids, total phenolics, and saponin (Ilyas et al., 2017; Karamian et al., 2020; Mohi-Ud-Din et al., 2021).
9 Cutting-edge technologies for optimizing phenylpropanoids commercial production
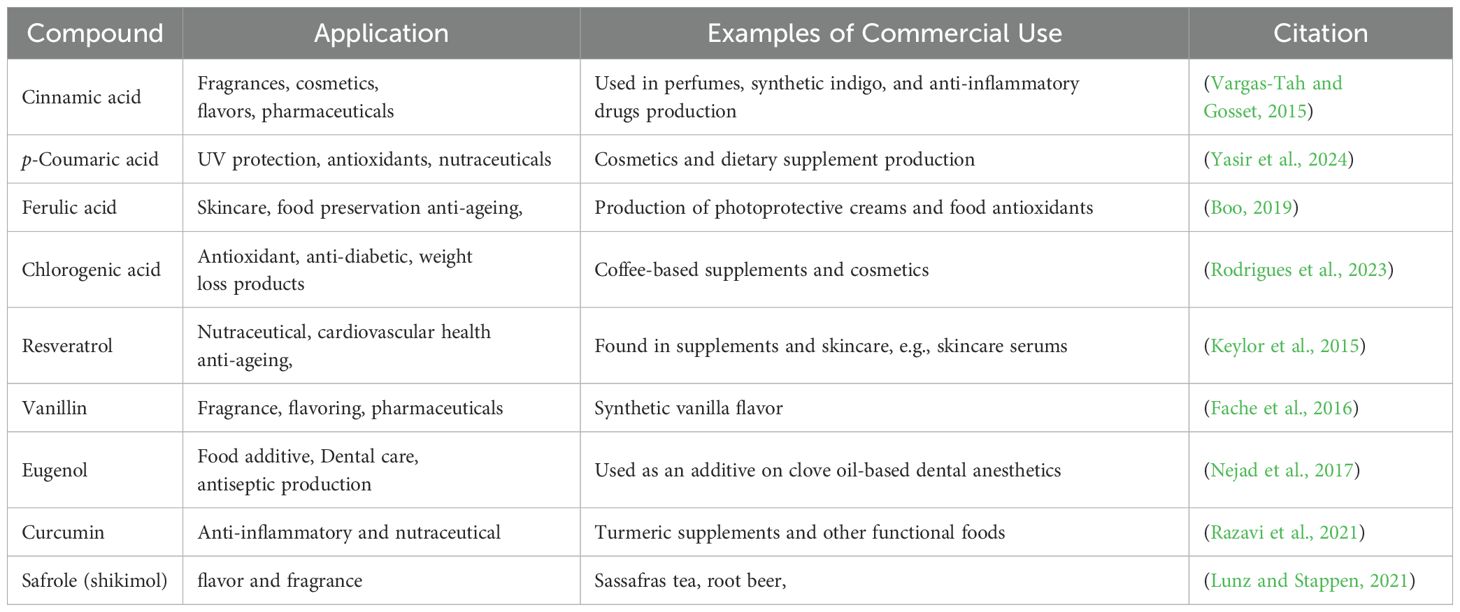
In recent times, the demand for green bioactive compounds has grown, driven by concerns over the environmental impact of synthetic alternatives. Additionally, the rising global population has stimulated growth in the pharmaceutical and food industries (Adetunde et al., 2025), creating a need for innovative methods to scale up the production of plant-based bioactive compounds. The phenylpropanoid pathway has generated several bioactive ingredients used in fragrance, flavor, food additives, neutraceuticals, and several other drugs (Table 7).
A range of methods has been used in the commercial production of phenylpropanoids. Traditional approaches, like solvent extraction for vanillin and related compounds, produce only minimal yields. As a result, modern high-yield techniques, such as microbial synthesis, have been developed and adopted. This approach depends on high-titre-tolerant microbes, such as Escherichia coli and Saccharomyces cerevisiae, as biofactories for the commercial production of phenylpropanoids (Ferulic acid, resveratrol, cinnamic acid) (Vargas-Tah and Gosset, 2015b). For example, heterologous expression PAL/TAL genes in microbes have been used to produce CA and pHCA strains. Under this condition, l-Tyr and l-Phe are transformed into pHCA and CA (Vargas-Tah and Gosset, 2015). Recently, Park et al. (2022) synthesized coniferyl alcohol (CA) and dihydroquercetin (DHQ) by reconstructing the phenylpropanoid pathway in E. coli. An E. coli strain that produces 187.7 mg/L was engineered to carry phenylpropanoid genes from A. thaliana, including 4CL4, OMT1, and CCR1. Similarly, naringenin was also produced via 239.4 mg/L of DHQ E. coli carrier, harboring A. thaliana genes (TT7, F3H, and CPR) (Park et al., 2022).
10 Conclusion
Phenylpropanoids are central to plant survival and environmental interactions, serving both structural and chemical roles, as well as biotic and abiotic stress resistance. This pathway has been a crucial target for climate-smart crop development due to the diverse metabolites’ functions in ROS scavenging, UV stress tolerance, salt stress resistance, and extreme temperature tolerance. Based on these functions, plant stress improvement techniques can be carried out to produce crop cultivars that can simultaneously exhibit these traits to enhance food production for the hungry world. Although lignin metabolism in crop plants has generated controversy over the end use of crop straws due to overly recalcitrant to chemical digestion, crop improvement techniques must sustainably engineer lignin pathways to meet crop resilience to stress and industrial application of crop straw. Furthermore, the rapid development of metabolic engineering techniques could benefit the engineering of most of these critical metabolites in the phenylpropanoids pathway for biopesticide development. Conclusively, our review provides a timely update of the current studies on phenylpropanoid metabolism and stress tolerance.
Author contributions
OA: Conceptualization, Writing – original draft. VN: Conceptualization, Writing – original draft. JY: Writing – review & editing. GL: Conceptualization, Supervision, Writing – review & editing. JZ: Conceptualization, Supervision, Writing – review & editing. HL: Conceptualization, Supervision, Writing – review & editing. SC: Writing – review & editing. HZ: Conceptualization, Supervision, Writing – review & editing. FD: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Hainan Provincial Sanya Yazhou Bay Science and Technology Innovation Joint Project (No: ZDYF2025GXJS150) and Key R&D Programs of Hainan Province (ZDYF2024XDNY210) to HL and The Nanfan Special Project, CAAS (YBXM2443), to Jun Zhao.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Lateif, K., Vaissayre, V., Gherbi, H., Verries, C., Meudec, E., Perrine-Walker, F., et al. (2013). Silencing of the chalcone synthase gene in C asuarina glauca highlights the important role of flavonoids during nodulation. New Phytol. 199, 1012–1021. doi: 10.1111/nph.12326
Adetunde, L. A., Osemwegie, O. O., Akinsanola, B. A., Odeyemi, A. T., and Ninkuu, V. (2025). Trend in pharmaceutical effluent discharge and management using microorganisms. Environ. Adv., 100617. doi: 10.1016/j.envadv.2025.100617
Aluko, O. O., Li, C., Wang, Q., and Liu, H. (2021). Sucrose utilization for improved crop yields: A review article. Int. J. Mol. Sci. 22, 4704. doi: 10.3390/ijms22094704
Aluko, O. O., Ninkuu, V., Ziemah, J., Jianpei, Y., Taiwo, E., Ninkuu, S. B., et al. (2024). Genome-wide identification and expression analysis of EIN3/EIL gene family in rice (Oryza sativa). Plant Stress 12, 100437. doi: 10.1016/j.stress.2024.100437
An, F., Cui, M., Chen, T., Cheng, C., Liu, Z., Luo, X., et al. (2023). Flavonoid accumulation modulates the responses of cassava tuberous roots to postharvest physiological deterioration. Postharvest Biol. Technol. 198, 112254. doi: 10.1016/j.postharvbio.2023.112254
An, Z., Yang, Z., Zhou, Y., Huo, S., Zhang, S., Wu, D., et al. (2024). OsJRL negatively regulates rice cold tolerance via interfering phenylalanine metabolism and flavonoid biosynthesis. Plant Cell Environ. 47, 4071–4085. doi: 10.1111/pce.v47.11
Avalbaev, A., Allagulova, C., Maslennikova, D., Fedorova, K., and Shakirova, F. (2021). Methyl jasmonate and cytokinin mitigate the salinity-induced oxidative injury in wheat seedlings. J. Plant Growth Regul. 40, 1741–1752. doi: 10.1007/s00344-020-10221-1
Ben Youssef, R., Jelali, N., Acosta Motos, J. R., Abdelly, C., and Albacete, A. (2025). Salicylic acid seed priming: A key frontier in conferring salt stress tolerance in barley seed germination and seedling growth. Agronomy 15, 154. doi: 10.3390/agronomy15010154
Bisht, S. and Gaikwad, K. K. (2025). Natural pigments or dyes for sustainable food packaging application. Food Biopro. Technol. 18, 4301–4325. doi: 10.1007/s11947-025-03756-2
Bistgani, Z. E., Hashemi, M., DaCosta, M., Craker, L., Maggi, F., and Morshedloo, M. R. (2019). Effect of salinity stress on the physiological characteristics, phenolic compounds and antioxidant activity of Thymus vulgaris L. and Thymus daenensis Celak. Indust. Crops Prod. 135, 311–320. doi: 10.1016/j.indcrop.2019.04.055
Boo, Y. C. (2019). p-Coumaric acid as an active ingredient in cosmetics: A review focusing on its antimelanogenic effects. Antioxidants 8, 275. doi: 10.3390/antiox8080275
Cai, H., Zhang, M., Chai, M., He, Q., Huang, X., Zhao, L., et al. (2019). Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol. 221, 295–308. doi: 10.1111/nph.15306
Cao, L. and Dong, X. (2025). H2O2 regulates rice defense via bHLH25 oxidation. Cell Res. 35. doi: 10.1038/s41422-025-01071-1
Cao, Y.-H., Lü, Z.-L., Li, Y.-H., Jiang, Y., and Zhang, J.-L. (2024). Integrated metabolomic and transcriptomic analysis reveals the role of root phenylpropanoid biosynthesis pathway in the salt tolerance of perennial ryegrass. BMC Plant Biol. 24, 1225. doi: 10.1186/s12870-024-05961-1
Cao, P., Yang, J., Xia, L., Zhang, Z., Wu, Z., Hao, Y., et al. (2024). Two gene clusters and their positive regulator SlMYB13 that have undergone domestication-associated negative selection control phenolamide accumulation and drought tolerance in tomato. Mol. Plant 17, 579–597. doi: 10.1016/j.molp.2024.02.003
Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56, 209–214. doi: 10.1016/j.copbio.2018.12.012
Chakraborty, R., Rehman, R. U., Siddiqui, M. W., Liu, H., and Seth, C. S. (2025). Phytohormones: Heart of plants’ signaling network under biotic, abiotic, and climate change stresses. Plant Physiol. Biochem. 223, 109839. doi: 10.1016/j.plaphy.2025.109839
Chang, N., Zheng, L., Xu, Y., Wang, C., Li, H., and Wang, Y. (2024). Integrated transcriptomic and metabolomic analysis reveals the molecular profiles of dynamic variation in Lilium brownii var. viridulum suffering from bulb rot. Front. Genet. 15, 1432997. doi: 10.3389/fgene.2024.1432997
Chen, W., Miao, Y., Ayyaz, A., Huang, Q., Hannan, F., Zou, H.-X., et al. (2025). Anthocyanin accumulation enhances drought tolerance in purple-leaf Brassica napus: Transcriptomic, metabolomic, and physiological evidence. Indust. Crops Prod. 223, 120149. doi: 10.1016/j.indcrop.2024.120149
Chen, S., Wang, X., Cheng, Y., Gao, H., and Chen, X. (2023). A review of classification, biosynthesis, biological activities and potential applications of flavonoids. Molecules 28, 4982. doi: 10.3390/molecules28134982
Chen, S., Wu, F., Li, Y., Qian, Y., Pan, X., Li, F., et al. (2019). NtMYB4 and NtCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness. Front. Plant Sci. 10, 178. doi: 10.3389/fpls.2019.00178
Chen, Z., Zhu, H., Zhang, K., and Cui, J. (2025). Starch–sucrose metabolism and phenylpropanoid biosynthesis pathways play crucial regulatory roles in extending the postharvest longevity of Lilium brownii var. viridulum bulbs. Postharvest Biol. Technol. 219, 113280. doi: 10.1016/j.postharvbio.2024.113280
Cheng, T., Lin, J., Zhou, X., Wang, H., Zhou, X., Huang, X., et al. (2025). Integrative metabolomics and transcriptomics profiling reveals differential expression of flavonoid synthesis in Ophiopogon japonicus (L. f.) Ker-Gawl. in adaptation to drought. PloS One 20, e0313580. doi: 10.1371/journal.pone.0313580
Choudhary, K. K., Singh, S., Agrawal, M., and Agrawal, S. (2021). Role of Jasmonic and salicylic acid signaling in plants under UV-B stress. Jasmonates Salicylates Signaling Plants, 45–63. doi: 10.1007/978-3-030-75805-9_3
Chu, L.-Y., Liu, T., Xia, P.-l., Li, J.-P., Tang, Z.-R., Zheng, Y.-L., et al. (2025). NtWRKY28 orchestrates flavonoid and lignin biosynthesis to defense aphid attack in tobacco plants. Plant Physiol. Biochem. 221, 109673. doi: 10.1016/j.plaphy.2025.109673
Commisso, M., Toffali, K., Strazzer, P., Stocchero, M., Ceoldo, S., Baldan, B., et al. (2016). Impact of phenylpropanoid compounds on heat stress tolerance in carrot cell cultures. Front. Plant Sci. 7, 1439. doi: 10.3389/fpls.2016.01439
Courtney, D. G. (2021). Post-transcriptional regulation of viral RNA through epitranscriptional modification. Cells 10, 1129. doi: 10.3390/cells10051129
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Das, A., Choudhury, S., Gopinath, V., Majeed, W., Chakraborty, S., Bhairavi, K. S., et al. (2024). “Functions of Flavonoids in Plant, Pathogen, and Opportunistic Fungal Interactions,” in Opportunistic Fungi, Nematode and Plant Interactions: Interplay and Mechanisms. Ed. Akhtar, M. S. (Singapore: Springer Nature Singapore), 91–123. doi: 10.1007/978-981-97-2045-3_6
Deng, Y., Zhu, Z., Chen, J., Kuang, L., Yan, T., li, L., et al. (2025). Comparative transcriptomics of indica and japonica rice roots under heat stress reveals the crucial role of OsMAPK3 in heat response. Plant Physiol. Biochem. 221, 109668. doi: 10.1016/j.plaphy.2025.109668
de Oliveira, D. R., Matos, M., Silva, K. T., Mazzetto, S. E., Magalhães, W. L., LoMonaco, D., et al. (2025). Unveiling the mechanistic aspects of different delignification methods and their effects on the structure of technical Eucalyptus lignins. Biomass Convers. Bioref., 1–15. doi: 10.1007/s13399-025-06502-y
Dey, S. and Sen Raychaudhuri, S. (2024). Methyl jasmonate improves selenium tolerance via regulating ROS signalling, hormonal crosstalk and phenylpropanoid pathway in Plantago ovata. Plant Physiol. Biochem. 209, 108533. doi: 10.1016/j.plaphy.2024.108533
Ding, T., Li, W., Li, F., Ren, M., and Wang, W. (2024). microRNAs: Key regulators in plant responses to abiotic and biotic stresses via endogenous and cross-kingdom mechanisms. Int. J. Mol. Sci. 25, 1154. doi: 10.3390/ijms25021154
Dixon, R. A., Achnine, L., Kota, P., Liu, C. J., Reddy, M. S., and Wang, L. (2002). The phenylpropanoid pathway and plant defence—a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Dong, B., Da, F., Chen, Y., and Ding, X. (2023). Melatonin treatment maintains the quality of fresh-cut gastrodia elata under low-temperature conditions by regulating reactive oxygen species metabolism and phenylpropanoid pathway. Int. J. Mol. Sci. 24, 14284. doi: 10.3390/ijms241814284
dos Santos Cotrim, G., da Silva, D. M., da Graça, J. P., de Oliveira Junior, A., de Castro, C., Zocolo, G. J., et al. (2023). Glycine max (L.) Merr.(Soybean) metabolome responses to potassium availability. Phytochemistry 205, 113472. doi: 10.1016/j.phytochem.2022.113472
Drapal, M., Rivera, T. M. O., Meléndez, J. L. L., Perez-Fons, L., Tran, T., Dufour, D., et al. (2024). Biochemical characterisation of a cassava (Manihot esculenta crantz) diversity panel for post-harvest physiological deterioration; metabolite involvement and environmental influence. J. Plant Physiol. 301, 154303. doi: 10.1016/j.jplph.2024.154303
Duan, H., Tiika, R. J., Tian, F., Lu, Y., Zhang, Q., Hu, Y., et al. (2023). Metabolomics analysis unveils important changes involved in the salt tolerance of Salicornia europaea. Front. Plant Sci. 13, 1097076. doi: 10.3389/fpls.2022.1097076
Eom, S. H., Ahn, M.-A., Kim, E., Lee, H. J., Lee, J. H., Wi, S. H., et al. (2022). Plant response to cold stress: Cold stress changes antioxidant metabolism in heading type kimchi cabbage (Brassica rapa L. ssp. Pekinensis). Antioxidants 11, 700. doi: 10.3390/antiox11040700
Fache, M., Boutevin, B., and Caillol, S. (2016). Vanillin production from lignin and its use as a renewable chemical. ACS Sustain. Chem. Eng. 4, 35–46. doi: 10.1021/acssuschemeng.5b01344
Falcone Ferreyra, M. L., Rius, S., and Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00222
Feng, X., Wang, D., and Li, D. (2025). Morphological, physiological and transcriptional analyses provide insights into the biosynthesis of phenolics in Juniperus rigida under UV-B treatment. Plant Physiol. Biochem. 220, 109534. doi: 10.1016/j.plaphy.2025.109534
Feyissa, B. A., Arshad, M., Gruber, M. Y., Kohalmi, S. E., and Hannoufa, A. (2019). The interplay between miR156/SPL13 and DFR/WD40–1 regulate drought tolerance in alfalfa. BMC Plant Biol. 19, 1–19. doi: 10.1186/s12870-019-2059-5
Gai, Q.-Y., Lu, Y., Jiao, J., Fu, J.-X., Xu, X.-J., Yao, L., et al. (2022). Application of UV-B radiation for enhancing the accumulation of bioactive phenolic compounds in pigeon pea [Cajanus cajan (L.) Millsp.] hairy root cultures. J. Photochem. Photobiol. B: Biol. 228, 112406. doi: 10.1016/j.jphotobiol.2022.112406
Gao, S., Chen, S., Yang, M., Wu, J., Chen, S., and Li, H. (2023). Mining salt stress-related genes in Spartina alterniflora via analyzing co-evolution signal across 365 plant species using phylogenetic profiling. aBIOTECH 4, 291–302. doi: 10.1007/s42994-023-00125-5
Gao, S., Wang, Y., Yu, S., Huang, Y., Liu, H., Chen, W., et al. (2020). Effects of drought stress on growth, physiology and secondary metabolites of Two Adonis species in Northeast China. Sci. Hortic. 259, 108795. doi: 10.1016/j.scienta.2019.108795
Garcia-Caparros, P., De Filippis, L., Gul, A., Hasanuzzaman, M., Ozturk, M., Altay, V., et al. (2021). Oxidative stress and antioxidant metabolism under adverse environmental conditions: a review. Bot. Rev. 87, 421–466. doi: 10.1007/s12229-020-09231-1
Ghasemi, S., Kumleh, H. H., Kordrostami, M., and Rezadoost, M. H. (2023). Drought stress-mediated alterations in secondary metabolites and biosynthetic gene expression in cumin plants: Insights from gene-specific and metabolite-level analyses. Plant Stress 10, 100241. doi: 10.1016/j.stress.2023.100241
Gong, F., Yu, W., Cao, K., Xu, H., and Zhou, X. (2024). RcTRP5 Transcription Factor Mediates the Molecular Mechanism of Lignin Biosynthesis Regulation in R. chrysanthum against UV-B Stress. Int. J. Mol. Sci. 25, 9205. doi: 10.3390/ijms25179205
Gong, F., Yu, W., Zeng, Q., Dong, J., Cao, K., Xu, H., et al. (2023). Rhododendron chrysanthum’s primary metabolites are converted to phenolics more quickly when exposed to UV-B radiation. Biomolecules 13, 1700. doi: 10.3390/biom13121700
Gou, J. Y., Felippes, F. F., Liu, C. J., Weigel, D., and Wang, J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. doi: 10.1105/tpc.111.084525
Gu, H., Liu, S., Li, G., Hou, L., Shen, T., Song, M., et al. (2024). Enhanced catalytic synthesis of flavonoid by UV-B radiation in Artemisia argyi. Catalysts 14, 504. doi: 10.3390/catal14080504
Guo, C., Qin, L., Ma, Y., and Qin, J. (2022). Integrated metabolomic and transcriptomic analyses of the parasitic plant Cuscuta japonica Choisy on host and non-host plants. BMC Plant Biol. 22, 393. doi: 10.1186/s12870-022-03773-9
Han, S., Xu, X., Yuan, H., Li, S., Lin, T., Liu, Y., et al. (2023). Integrated transcriptome and metabolome analysis reveals the molecular mechanism of rust resistance in resistant (Youkang) and susceptive (Tengjiao) zanthoxylum armatum cultivars. Int. J. Mol. Sci. 24, 1476. doi: 10.3390/ijms241914761
Hao, J., Lou, P., Han, Y., Zheng, L., Lu, J., Chen, Z., et al. (2022). Ultraviolet-B irradiation increases antioxidant capacity of pakchoi (Brassica rapa L.) by inducing flavonoid biosynthesis. Plants 11, 766. doi: 10.3390/plants11060766
Hao, S., Lu, Y., Peng, Z., Wang, E., Chao, L., Zhong, S., et al. (2021). McMYB4 improves temperature adaptation by regulating phenylpropanoid metabolism and hormone signaling in apple. Horticult. Res. 8, 182. doi: 10.1038/s41438-021-00620-0
Haskett, T. L., Cooke, L., Green, P., and Poole, P. S. (2025). Regulation of rhizobial nodulation genes by flavonoid-independent nodD supports nitrogen-fixing symbioses with legumes. Environ. Microbiol. 27, e70014. doi: 10.1111/1462-2920.70014
Hassan, S. and Mathesius, U. (2012). The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions. J. Exp. Bot. 63, 3429–3444. doi: 10.1093/jxb/err430
Hernández-Elvira, M. and Sunnerhagen, P. (2022). Post-transcriptional regulation during stress. FEMS Yeast Res. 22, foac025. doi: 10.1093/femsyr/foac025
Hérouart, D., Baudouin, E., Frendo, P., Harrison, J., Santos, R., Jamet, A., et al. (2002). Reactive oxygen species, nitric oxide and glutathione: a key role in the establishment of the legume–Rhizobium symbiosis? Plant Physiol. Biochem. 40, 619–624. doi: 10.1016/S0981-9428(02)01415-8
Huang, Z., Chen, S., He, K., Yu, T., Fu, J., Gao, S., et al. (2024). Exploring salt tolerance mechanisms using machine learning for transcriptomic insights: case study in Spartina alterniflora. Horticult. Res. 11, uhae082. doi: 10.1093/hr/uhae082
Huang, X., Chu, G., Wang, J., Luo, H., Yang, Z., Sun, L., et al. (2023). Integrated metabolomic and transcriptomic analysis of specialized metabolites and isoflavonoid biosynthesis in Sophora alopecuroides L. under different degrees of drought stress. Indust. Crops Prod. 197, 116595. doi: 10.1016/j.indcrop.2023.116595
Huang, Q., Lei, Z., Xiang, L., Zhang, W., Zhang, L., and Gao, Y. (2022). Transcriptomic Analysis of Sunflower (Helianthus annuus) Roots Resistance to Orobanche cumana at the Seedling Stage. Horticulturae 8, 701. doi: 10.3390/horticulturae8080701
Ilyas, N., Gull, R., Mazhar, R., Saeed, M., Kanwal, S., Shabir, S., et al. (2017). Influence of salicylic acid and jasmonic acid on wheat under drought stress. Commun. Soil Sci. Plant Anal. 48, 2715–2723. doi: 10.1080/00103624.2017.1418370
Iqbal, S., Wang, X., Mubeen, I., Kamran, M., Kanwal, I., Díaz, G. A., et al. (2021). Phytohormones trigger drought tolerance in crop plants: outlook and future perspectives. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.799318
Jardim-Messeder, D., de Souza-Vieira, Y., and Sachetto-Martins, G. (2025). Dressed up to the nines: the interplay of phytohormones signaling and redox metabolism during plant response to drought. Plants 14, 208. doi: 10.3390/plants14020208
Ji, J., Li, X., Guo, X., Liu, X., Ma, X., Wang, M., et al. (2025). X-ray irradiation delayed the browning development of fresh-cut bulbs of Lanzhou Lily (Lilium davidii) during room temperature storage. Radiat. Phys. Chem. 226, 112168. doi: 10.1016/j.radphyschem.2024.112168
Jia, C., Guo, B., Wang, B., Li, X., Yang, T., Li, N., et al. (2022). Integrated metabolomic and transcriptomic analysis reveals the role of phenylpropanoid biosynthesis pathway in tomato roots during salt stress. Front. Plant Sci. 13, 1023696. doi: 10.3389/fpls.2022.1023696
Jia, L., Li, Y., Liu, G., and He, J. (2023). UV-C delays senescence in ‘Lingwu long’jujube fruit by regulating ROS and phenylpropanoid metabolism. Plant Physiol. Biochem. 194, 383–393. doi: 10.1016/j.plaphy.2022.11.030
Jiang, S., Jin-Long, C., and Li, X.-k. (2021). MicroRNA-mediated gene regulation of secondary metabolism in plants. Crit. Rev. Plant Sci. 40, 459–478. doi: 10.1080/07352689.2022.2031674
Jiang, L., Xiao, M., Huang, R., and Wang, J. (2025). The regulation of ROS and phytohormones in balancing crop yield and salt tolerance. Antioxidants 14, 63. doi: 10.3390/antiox14010063
Jie, H., He, P., Zhao, L., Ma, Y., and Jie, Y. (2023). Molecular mechanisms regulating phenylpropanoid metabolism in exogenously-sprayed ethylene forage ramie based on transcriptomic and metabolomic analyses. Plants 12, 3899. doi: 10.3390/plants12223899
Jin, R., Yan, M., Li, G., Liu, M., Zhao, P., Zhang, Z., et al. (2024). Comparative physiological and transcriptome analysis between potassium-deficiency tolerant and sensitive sweetpotato genotypes in response to potassium-deficiency stress. BMC Genomics 25, 61. doi: 10.1186/s12864-023-09939-5
Karamian, R., Ghasemlou, F., and Amiri, H. (2020). Physiological evaluation of drought stress tolerance and recovery in Verbascum sinuatum plants treated with methyl jasmonate, salicylic acid and titanium dioxide nanoparticles. Plant Biosystems-An Int. J. Deal. all Aspects Plant Biol. 154, 277–287. doi: 10.1080/11263504.2019.1591535
Keylor, M. H., Matsuura, B. S., and Stephenson, C. R. (2015). Chemistry and biology of resveratrol-derived natural products. Chem. Rev. 115, 8976–9027. doi: 10.1021/cr500689b
Kim, J. I., Zhang, X., Pascuzzi, P. E., Liu, C. J., and Chapple, C. (2020). Glucosinolate and phenylpropanoid biosynthesis are linked by proteasome-dependent degradation of PAL. New Phytol. 225, 154–168. doi: 10.1111/nph.16108
Knollenberg, B. J., Liu, J., Yu, S., Lin, H., and Tian, L. (2018). Cloning and functional characterization of a p-coumaroyl quinate/shikimate 3′-hydroxylase from potato (Solanum tuberosum). Biochem. Biophys. Res. Commun. 496, 462–467. doi: 10.1016/j.bbrc.2018.01.075
Koeslin-Findeklee, F., Rizi, V. S., Becker, M. A., Parra-Londono, S., Arif, M., Balazadeh, S., et al. (2015). Transcriptomic analysis of nitrogen starvation-and cultivar-specific leaf senescence in winter oilseed rape (Brassica napus L.). Plant Sci. 233, 174–185. doi: 10.1016/j.plantsci.2014.11.018
Kumar, G. M. and Knowles, N. R. (2003). Wound-induced superoxide production and PAL activity decline with potato tuber age and wound healing ability. Physiol. Plant. 117, 108–117. doi: 10.1034/j.1399-3054.2003.1170114.x
Kundu, P., Shinde, S., Grover, S., Sattler, S. E., and Louis, J. (2025). Caffeic acid O-methyltransferase-dependent flavonoid defenses promote sorghum resistance to fall armyworm infestation. Plant Physiol. 197, kiaf071. doi: 10.1093/plphys/kiaf071
La, V. H., Tran, D. H., Han, V. C., Nguyen, T. D., Duong, V. C., Nguyen, V. H., et al. (2023). Drought stress-responsive abscisic acid and salicylic acid crosstalk with the phenylpropanoid pathway in soybean seeds. Physiol. Plant. 175, e14050. doi: 10.1111/ppl.v175.5
Lee, S. K., Liao, P. Z., Lin, C. Y., Chen, H. W., Hsieh, M. S., Lin, Y. P., et al. (2024). Wild mungbean resistance to the nematode Meloidogyne enterolobii involves the induction of phenylpropanoid metabolism and lignification. Physiol. Plant. 176, e14533. doi: 10.1111/ppl.v176.5
Lehari, K. and Kumar, D. (2024). “Chapter 10 - Metabolic engineering and production of secondary metabolites,” in Secondary Metabolites and Biotherapeutics. Eds. Kumar, A. and Kumar, S. (India: Academic Press), 215–244. doi: 10.1016/B978-0-443-16158-2.00004-5
Li, C., Aluko, O. O., Shi, S., Mo, Z., Nong, T., Shi, C., et al. (2023). Determination of optimal NH4+/K+ concentration and corresponding ratio critical for growth of tobacco seedlings in a hydroponic system. Front. Plant Sci. 14, 1152817. doi: 10.3389/fpls.2023.1152817
Li, N., Chen, W., Wang, B., Zhang, C., Wang, Y., Li, R., et al. (2024). Arbuscular mycorrhizal fungi improve the disease resistance of Lycium barbarum to root rot by activating phenylpropane metabolism. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1459651
Li, Z., Jiang, H., Jiang, X., Zhang, L., and Qin, Y. (2023). Integrated physiological, transcriptomic, and metabolomic analyses reveal that low-nitrogen conditions improve the accumulation of flavonoids in snow chrysanthemum. Indust. Crops Prod. 197, 116574. doi: 10.1016/j.indcrop.2023.116574
Li, C., Jiang, Y., Xu, C., and Mei, X. (2024). Editorial: Contribution of phenylpropanoid metabolism to plant development and stress responses. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1456913
Li, J., Li, A., Wang, Y., Wu, Z.-H., Li, X., Meng, J., et al. (2025). An extensin-like gene OsPEX1 plays key roles in temperature stress response in rice. Plant Cell Tissue Organ Cult. (PCTOC) 161, 4. doi: 10.1007/s11240-025-03019-0
Li, W., Lin, Y.-C. J., Chen, Y.-L., Zhou, C., Li, S., De Ridder, N., et al. (2024). Woody plant cell walls: Fundamentals and utilization. Mol. Plant 17, 112–140. doi: 10.1016/j.molp.2023.12.008
Li, F., Liu, J., Dewer, Y., Ahsan, M. H., and Wu, C. (2025). Quercetin, a natural flavonoid induced by the spider mite Tetranychus urticae or alamethicin, is involved in the defense of lima bean against spider mites. Pest Manage. Sci. doi: 10.1002/ps.8681
Li, J., Liu, X., Xu, L., Li, W., Yao, Q., Yin, X., et al. (2023). Low nitrogen stress-induced transcriptome changes revealed the molecular response and tolerance characteristics in maintaining the C/N balance of sugar beet (Beta vulgaris L.). Front. Plant Sci. 14, 1164151. doi: 10.3389/fpls.2023.1164151
Li, Y., Liu, S., Zhang, D., Liu, A., Zhu, W., Zhang, J., et al. (2023a). Integrative Omic Analysis Reveals the Dynamic Change in Phenylpropanoid Metabolism in Morus alba under Different Stress. Plants 12, 3265. doi: 10.3390/plants12183265
Li, Y., Shen, Y., Shi, R., Yang, Z., Chen, Y., Luo, W., et al. (2024). The synthesis and secretion of key substances in the flavonoid metabolic pathway responding to different nitrogen sources during early growth stages in Robinia pseudoacacia. Plant Soil 494, 373–393. doi: 10.1007/s11104-023-06286-y
Li, Y., Tian, Q., Wang, Z., Li, J., Liu, S., Chang, R., et al. (2023b). Integrated analysis of transcriptomics and metabolomics of peach under cold stress. Front. Plant Sci. 14, 1153902. doi: 10.3389/fpls.2023.1153902
Li, S.-H. and Wang, P. (2025). Pumpkin (Cucurbita pepo L.) CpVQ20 increases resistance to powdery mildew of tobacco through flavonoid and lignin biosynthesis pathways. Physiol. Mol. Plant Pathol. 136, 102562. doi: 10.1016/j.pmpp.2025.102562
Li, Y., Zhang, R., Sun, L., and Cao, C. (2025). Resistance of Populus davidiana × P. bolleana overexpressing cinnamoyl-CoA reductase gene to Lymantria dispar larvae. Transgenic Res. 34, 10. doi: 10.1007/s11248-024-00426-5
Li, J.-W., Zhou, P., Hu, Z.-H., Xiong, A.-S., Li, X.-H., Chen, X., et al. (2025). The transcription factor CsPAT1 from tea plant (Camellia sinensis) is involved in drought tolerance by modulating phenylpropanoid biosynthesis. J. Plant Physiol. 308, 154474. doi: 10.1016/j.jplph.2025.154474
Liao, Q., Xiao, H., Chen, L., Shen, C., Wang, B., Zhou, W., et al. (2025). Potassium alters cadmium accumulation and translocation by regulating root cell wall biosynthesis in Brassica napus. Sci. Hortic. 339, 113833. doi: 10.1016/j.scienta.2024.113833
Ling, Y., Wang, D., Peng, Y., Peng, D., and Li, Z. (2025). Cross-stressful adaptation to drought and high salinity is related to variable antioxidant defense, proline metabolism, and dehydrin b expression in white clover. Agronomy 15, 126. doi: 10.3390/agronomy15010126
Liu, S., An, X., Xu, C., Guo, B., Li, X., Chen, C., et al. (2024). Exploring the dynamic adaptive responses of Epimedium pubescens to phosphorus deficiency by Integrated transcriptome and miRNA analysis. BMC Plant Biol. 24, 480. doi: 10.1186/s12870-024-05063-y
Liu, W., Feng, Y., Yu, S., Fan, Z., Li, X., Li, J., et al. (2021). The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 22, 12824. doi: 10.3390/ijms222312824
Liu, Q., Hou, S., Zhang, Y., Zhou, D., Guo, L., Zhao, S., et al. (2024). Dielectric barrier discharge cold plasma improves storage stability in paddy rice by activating the phenylpropanoid biosynthesis pathway. J. Agric. Food Chem. 72, 25066–25077. doi: 10.1021/acs.jafc.4c04316
Liu, H., Jiang, W., Bi, Y., and Luo, Y. (2005). Postharvest BTH treatment induces resistance of peach (Prunus persica L. cv. Jiubao) fruit to infection by Penicillium expansum and enhances activity of fruit defense mechanisms. Postharvest Biol. Technol. 35, 263–269. doi: 10.1016/j.postharvbio.2004.08.006
Liu, M., Li, X., Liu, Y., and Cao, B. (2013). Regulation of flavanone 3-hydroxylase gene involved in the flavonoid biosynthesis pathway in response to UV-B radiation and drought stress in the desert plant, Reaumuria soongorica. Plant Physiol. Biochem. 73, 161–167. doi: 10.1016/j.plaphy.2013.09.016
Liu, Y., Liao, L., Yin, F., Song, M., Shang, F., Shuai, L., et al. (2022). Integration of metabolome and transcriptome profiling reveals the effect of 6-Benzylaminopurine on the browning of fresh-cut lettuce during storage. Postharvest Biol. Technol. 192, 112015. doi: 10.1016/j.postharvbio.2022.112015
Liu, H., Liu, Y., Xu, N., Sun, Y., Li, Q., Yue, L., et al. (2022). Chrysanthemum× grandiflora leaf and root transcript profiling in response to salinity stress. BMC Plant Biol. 22, 240. doi: 10.1186/s12870-022-03612-x
Liu, X., Xie, Z., Xin, J., Yuan, S., Liu, S., Sun, Y., et al. (2024). OsbZIP18 is a positive regulator of phenylpropanoid and flavonoid biosynthesis under UV-B radiation in rice. Plants 13, 498. doi: 10.3390/plants13040498
Liu, H., Xu, M., Guo, Y., Dan, Z., Liu, X., Zhang, J., et al. (2024). Combined transcriptome and metabolome analysis of alfalfa responses to aphid infestation. Agriculture 14, 480. doi: 10.3390/agriculture14091545
Liu, S., Zainuddin, I. M., Vanderschuren, H., Doughty, J., and Beeching, J. R. (2017). RNAi inhibition of feruloyl CoA 6′-hydroxylase reduces scopoletin biosynthesis and post-harvest physiological deterioration in cassava (Manihot esculenta Crantz) storage roots. Plant Mol. Biol. 94, 185–195. doi: 10.1007/s11103-017-0602-z
Lohmaneeratana, K., Leetanasaksakul, K., and Thamchaipenet, A. (2024). Transcriptomic profiling of sugarcane white leaf (SCWL) canes during maturation phase. Plants (Basel) 13, 1551. doi: 10.3390/plants13111551
Lu, Y.-l., Song, G.-P., Wang, Y.-H., Wang, L.-B., Xu, M.-Z., Zhou, L.-P., et al. (2023). Combining nitrogen effects and metabolomics to reveal the response mechanisms to nitrogen stress and the potential for nitrogen reduction in maize. J. Integr. Agric. 22, 2660–2672. doi: 10.1016/j.jia.2023.03.002
Lu, L., Zhang, Y., Li, L., Yi, N., Liu, Y., Qaseem, M. F., et al. (2021). Physiological and transcriptomic responses to nitrogen deficiency in Neolamarckia cadamba. Front. Plant Sci. 12, 747121. doi: 10.3389/fpls.2021.747121
Lunz, K. and Stappen, I. (2021). Back to the roots—An overview of the chemical composition and bioactivity of selected root-essential oils. Molecules 26, 3155. doi: 10.3390/molecules26113155
Luo, F., Yan, P., Xie, L., Li, S., Zhu, T., Han, S., et al. (2022). Molecular Mechanisms of Phenylpropane-Synthesis-Related Genes Regulating the Shoot Blight Resistance of Bambusa pervariabilis x Dendrocalamopsis grandis. Int. J. Mol. Sci. 23, 6760. doi: 10.3390/ijms23126760
Ma, D., Hu, H., Feng, J., Xu, B., Du, C., Yang, Y., et al. (2025). Metabolomics and transcriptomics analyses revealed overexpression of TaMGD enhances wheat plant heat stress resistance through multiple responses. Ecotoxicol. Environ. Saf. 290, 117738. doi: 10.1016/j.ecoenv.2025.117738
Ma, H., Su, L., Zhang, W., Sun, Y., Li, D., Li, S., et al. (2025). Epigenetic regulation of lignin biosynthesis in wood formation. New Phytol. 245, 1589–1607. doi: 10.1111/nph.20328
Ma, Q., Xu, J., Feng, Y., Wu, X., Lu, X., and Zhang, P. (2022). Knockdown of p-coumaroyl shikimate/quinate 3′-hydroxylase delays the occurrence of post-harvest physiological deterioration in cassava storage roots. Int. J. Mol. Sci. 23, 9231. doi: 10.3390/ijms23169231
Ma, Y., Zhao, S., Ma, X., Dong, G., Liu, C., Ding, Y., et al. (2025). A high temperature responsive UDP-glucosyltransferase gene OsUGT72F1 enhances heat tolerance in rice and Arabidopsis. Plant Cell Rep. 44, 48. doi: 10.1007/s00299-025-03438-5
Maeda, H. and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Mao, P., Xu, Y., Qin, H., Sun, Q., Ma, C., Xu, Z., et al. (2024). Transcriptome and metabolome analyses reveal the regulation effect of ultraviolet-B irradiation on secondary metabolites in pakchoi. Postharvest Biol. Technol. 209, 112702. doi: 10.1016/j.postharvbio.2023.112702
Marcela, V.-H., Gerardo, V.-M., Agustín, A.-R. C., Diego, C.-P., and Cruz-Hernández, A. (2019). Secondary metabolites production. Plant Physiol. Aspects Phenol. Comp. 51, 33–51. doi: 10.5772/intechopen.83804
Martínez-Silvestre, K. E., Santiz-Gómez, J. A., Luján-Hidalgo, M. C., Ruiz-Lau, N., Sánchez-Roque, Y., and Gutiérrez-Miceli, F. A. (2022). Effect of UV-B radiation on flavonoids and phenols accumulation in tempisque (Sideroxylon capiri Pittier) callus. Plants 11, 473. doi: 10.3390/plants11040473
Maslennikova, D., Ivanov, S., Petrova, S., Burkhanova, G., Maksimov, I., and Lastochkina, O. (2023). Components of the phenylpropanoid pathway in the implementation of the protective effect of sodium nitroprusside on wheat under salinity. Plants 12, 2123. doi: 10.3390/plants12112123
Miao, C., Hu, Z., Liu, X., Ye, H., Jiang, H., Tan, J., et al. (2025). Transcriptome analysis of nitrate enhanced tobacco resistance to aphid infestation. Plant Physiol. Biochem. 220, 109514. doi: 10.1016/j.plaphy.2025.109514
Mohi-Ud-Din, M., Talukder, D., Rohman, M., Ahmed, J. U., Jagadish, S. K., Islam, T., et al. (2021). Exogenous application of methyl jasmonate and salicylic acid mitigates drought-induced oxidative damages in french bean (Phaseolus vulgaris L.). Plants 10, 2066. doi: 10.3390/plants10102066
Morse, A. M., Whetten, R. W., Dubos, C., and Campbell, M. M. (2009). Post-translational modification of an R2R3-MYB transcription factor by a MAP Kinase during xylem development. New Phytol. 183, 1001–1013. doi: 10.1111/j.1469-8137.2009.02900.x
Mukami, A., Juma, B. S., Mweu, C., Oduor, R., and Mbinda, W. (2024). CRISPR-Cas9-induced targeted mutagenesis of feruloyl CoA 6′-hydroxylase gene reduces postharvest physiological deterioration in cassava roots. Postharvest Biol. Technol. 208, 112649. doi: 10.1016/j.postharvbio.2023.112649
Naikoo, M. I., Dar, M. I., Raghib, F., Jaleel, H., Ahmad, B., Raina, A., et al. (2019). Role and regulation of plants phenolics in abiotic stress tolerance: An overview. Plant Signaling mol., 157–168. doi: 10.1016/B978-0-12-816451-8.00009-5
Nayak, S. P., Prasad, P., Fakhrah, S., Pattanaik, D., Bag, S. K., and Mohanty, C. S. (2025). Differential miRNA expression and regulatory mechanisms in pigmentation and fiber development of white and brown cotton (Gossypium hirsutum). Funct. Integr. Genomics 25, 61. doi: 10.1007/s10142-025-01568-3
Nejad, S. M., Özgüneş, H., and Başaran, N. (2017). Pharmacological and toxicological properties of eugenol. Turkish J. pharma. Sci. 14, 201. doi: 10.4274/tjps.62207
Ninkuu, V., Han, T., and Dakora, F. D. (2025). Development of the soybean industry in Africa: safeguarding food security in Africa and China—A perspective. Engineering. doi: 10.1016/j.eng.2025.03.008
Ninkuu, V., Liu, Z., and Sun, X. (2023a). Genetic regulation of nitrogen use efficiency in Gossypium spp. Plant Cell Environ. 46, 1749–1773. doi: 10.1111/pce.14586
Ninkuu, V., Yan, J., Fu, Z., Yang, T., Zhang, L., Ren, J., et al. (2023b). Genome-wide identification, phylogenomics, and expression analysis of benzoxazinoids gene family in rice (Oryza sativa). Plant Stress 10, 100214. doi: 10.1016/j.stress.2023.100214
Ninkuu, V., Yan, J., Fu, Z., Yang, T., Ziemah, J., Ullrich, M. S., et al. (2023c). Lignin and its pathway-associated phytoalexins modulate plant defense against fungi. J. Fungi 9 (1), 52. doi: 10.3390/jof9010052
Ninkuu, V., Yan, J., Zhang, L., Fu, Z., Yang, T., Li, S., et al. (2022). Hrip1 mediates rice cell wall fortification and phytoalexins elicitation to confer immunity against Magnaporthe oryzae. Front. Plant Sci. 13, 980821. doi: 10.3389/fpls.2022.980821
Ninkuu, V., Zhang, L., Yan, J., Fu, Z., Yang, T., and Zeng, H. (2021). Biochemistry of terpenes and recent advances in plant protection. Int. J. Mol. Sci. 22 (11), 5710. doi: 10.3390/ijms22115710
Ninkuu, V., Zhou, Y., Liu, H., Sun, S., Liu, Z., Liu, Y., et al. (2024). Regulation of nitrogen metabolism by COE2 under low sulfur stress in Arabidopsis. Plant Sci. 346, 112137. doi: 10.1016/j.plantsci.2024.112137
Olson, D. R. and Ruhland, C. T. (2024). Ultraviolet-B Stress Increases Epidermal UV-Screening Effectiveness and Alters Growth and Cell-Wall Constituents of the Brown Midrib bmr6 and bmr12 Mutants of Sorghum bicolor. J. Agron. Crop Sci. 210, e12723. doi: 10.1111/jac.v210.4
Oro, C. E. D., Wancura, J. H., Santos, M. S., Venquiaruto, L. D., Dallago, R. M., and Tres, M. V. (2025). High-pressure extraction techniques for efficient recovery of flavonoids and coumarins from flower seeds. Processes 13, 300. doi: 10.3390/pr13020300
Park, Y. J., Kwon, D. Y., Koo, S. Y., Truong, T. Q., Hong, S.-C., Choi, J., et al. (2023). Identification of drought-responsive phenolic compounds and their biosynthetic regulation under drought stress in Ligularia fischeri. Front. Plant Sci. 14, 1140509. doi: 10.3389/fpls.2023.1140509
Park, S. Y., Yang, D., Ha, S. H., and Lee, S. Y. (2022). Production of phenylpropanoids and flavonolignans from glycerol by metabolically engineered Escherichia coli. Biotechnol. Bioengineering 119, 946–962. doi: 10.1002/bit.28008
Peña Barrena, L. E., Mats, L., Earl, H. J., and Bozzo, G. G. (2024). Phenylpropanoid Metabolism in Phaseolus vulgaris during Growth under Severe Drought. Metabolites 14, 319. doi: 10.3390/metabo14060319
Peng, M.-Y., Ren, Q.-Q., Lai, Y.-H., Zhang, J., Chen, H.-H., Guo, J., et al. (2023). Integration of physiology, metabolome, and transcriptome for the understanding of the adaptive strategies to long-term nitrogen deficiency in Citrus sinensis leaves. Sci. Hortic. 317, 112079. doi: 10.1016/j.scienta.2023.112079
Peng, Z., Tian, J., Luo, R., Kang, Y., Lu, Y., Hu, Y., et al. (2020). MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ. 43, 1148–1159. doi: 10.1111/pce.13697
Pinar, O. and Rodríguez-Couto, S. (2025). Biologically active secondary. Organic Chem. Editor’s Pick 2024. doi: 10.3389/fchem.2024.1363354
Qin, X., Yin, Y., Zhao, J., An, W., Fan, Y., Liang, X., et al. (2022). Metabolomic and transcriptomic analysis of Lycium chinese and L. ruthenicum under salinity stress. BMC Plant Biol. 22, 1–18. doi: 10.1186/s12870-021-03375-x
Rabeh, K., Hnini, M., and Oubohssaine, M. (2025). A comprehensive review of transcription factor-mediated regulation of secondary metabolites in plants under environmental stress. Stress Biol. 5, 15. doi: 10.1007/s44154-024-00201-w
Rao, M. J. and Zheng, B. (2025). The role of polyphenols in abiotic stress tolerance and their antioxidant properties to scavenge reactive oxygen species and free radicals. Antioxidants 14, 74. doi: 10.3390/antiox14010074
Razavi, B. M., Ghasemzadeh Rahbardar, M., and Hosseinzadeh, H. (2021). A review of therapeutic potentials of turmeric (Curcuma longa) and its active constituent, curcumin, on inflammatory disorders, pain, and their related patents. Phytother. Res. 35, 6489–6513. doi: 10.1002/ptr.v35.12
Reid, D., Nadzieja, M., Novák, O., Heckmann, A. B., Sandal, N., and Stougaard, J. (2017). Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 175, 361–375. doi: 10.1104/pp.17.00832
Ren, R. C., Kong, L. G., Zheng, G. M., Zhao, Y. J., Jiang, X., Wu, J. W., et al. (2024). Maize requires arogenate dehydratase 2 for resistance to Ustilago maydis and plant development. Plant Physiol. 195, 1642–1659. doi: 10.1093/plphys/kiae115
Rezaie, R., Abdollahi Mandoulakani, B., and Fattahi, M. (2020). Cold stress changes antioxidant defense system, phenylpropanoid contents and expression of genes involved in their biosynthesis in Ocimum basilicum L. Sci. Rep. 10, 5290. doi: 10.1038/s41598-020-62090-z
Ri, I., Pak, S., Pak, U., Yun, C., and Tang, Z. (2024). How does UV-B radiation influence the photosynthesis and secondary metabolism of Schisandra chinensis leaves? Indust. Crops Prod. 208, 117832. doi: 10.1016/j.indcrop.2023.117832
Rizi, M. R., Azizi, A., Sayyari, M., Mirzaie-Asl, A., and Conti, L. (2021). Increased phenylpropanoid production in UV-B irradiated Salvia verticillata as a consequence of altered genes expression in young leaves. Plant Physiol. Biochem. 167, 174–184. doi: 10.1016/j.plaphy.2021.07.037
Rodrigues, R., Oliveira, M. B. P. P., and Alves, R. C. (2023). Chlorogenic acids and caffeine from coffee by-products: A review on skincare applications. Cosmetics 10, 12. doi: 10.3390/cosmetics10010012
Rosatti, S., Rojas, A. M., Moro, B., Suarez, I. P., Bologna, N. G., Chorostecki, U., et al. (2024). Principles of miRNA/miRNA* function in plant MIRNA processing. Nucleic Acids Res. 52, 8356–8369. doi: 10.1093/nar/gkae458
Ryu, H., Cho, H., Choi, D., and Hwang, I. (2012). Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol. Cells 34, 117–126. doi: 10.1007/s10059-012-0131-1
Safdar, H., Amin, A., Shafiq, Y., Ali, A., Yasin, R., Shoukat, A., et al. (2019). A review: Impact of salinity on plant growth. Nat. Sci. 17, 34–40. doi: 10.7537/marsnsj170119.06
Samanta, S. and Roychoudhury, A. (2025). Molecular crosstalk of jasmonate with major phytohormones and plant growth regulators during diverse stress responses. J. Plant Growth Regul. 44, 62–88. doi: 10.1007/s00344-024-11412-w
Samarina, L., Malyukova, L., Wang, S., Li, Y., Doroshkov, A., Bobrovskikh, A., et al. (2024). Nitrogen deficiency differentially affects lignin biosynthesis genes and flavanols accumulation in tolerant and susceptible tea genotypes (Camellia sinensis (L.) Kuntze). Plant Stress 14, 100581. doi: 10.1016/j.stress.2024.100581
Shanker, M. A. and Rana, S. S. (2025). Prospects of cold plasma in enhancing food phenolics: analyzing nutritional potential and process optimization through RSM and AI techniques. Front. Nutr. 11, 1504958. doi: 10.3389/fnut.2024.1504958
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., and Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24 (13), 2452. doi: 10.3390/molecules24132452
Shi, J., Wang, Y., Fan, X., Li, R., Yu, C., Peng, Z., et al. (2024). A novel plant growth regulator B2 mediates drought resistance by regulating reactive oxygen species, phytohormone signaling, phenylpropanoid biosynthesis, and starch metabolism pathways in Carex breviculmis. Plant Physiol. Biochem. 213, 108860. doi: 10.1016/j.plaphy.2024.108860
Shomali, A., Das, S., Arif, N., Sarraf, M., Zahra, N., Yadav, V., et al. (2022). Diverse physiological roles of flavonoids in plant environmental stress responses and tolerance. Plants 11, 3158. doi: 10.3390/plants11223158
Siebeneichler, T. J., Crizel, R. L., Rombaldi, C. V., and Galli, V. (2024). Regulation of phenylpropanoid biosynthesis in strawberry ripening: molecular and hormonal mechanisms. Phytochem. Rev. 23, 923–941. doi: 10.1007/s11101-023-09907-7
Silva, L. F., Barreto, K. F. M., Silva, H. C., de Souza, I. D., Meneses, C. H. S. G., Uchôa, A. F., et al. (2025). Insights of cellular and molecular changes in sugarcane response to oxidative signaling. BMC Plant Biol. 25, 54. doi: 10.1186/s12870-024-06036-x
Singh, N., Maurya, V., Gupta, K., Sharma, I., Sharma, A., and Kumar, R. (2025). Salt stress and its eco-friendly management using biostimulants in grain legumes: a review. Discov. Agric. 3, 1–29. doi: 10.1007/s44279-024-00150-y
Singh, P., Singh, A., and Choudhary, K. K. (2023). Revisiting the role of phenylpropanoids in plant defense against UV-B stress. Plant Stress 7, 100143. doi: 10.1016/j.stress.2023.100143
Song, Y., Ma, B., Guo, Q., Zhou, L., Lv, C., Liu, X., et al. (2022). UV-B induces the expression of flavonoid biosynthetic pathways in blueberry (Vaccinium corymbosum) calli. Front. Plant Sci. 13, 1079087. doi: 10.3389/fpls.2022.1079087
Song, X., Zhu, Y., and Bao, Y. (2025). Identification and characteristics of differentially expressed genes under UV-B stress in Gossypium hirsutum. Front. Plant Sci. 15, 1529912. doi: 10.3389/fpls.2024.1529912
Sulis, D. B. and Wang, J. P. (2020). Regulation of lignin biosynthesis by post-translational protein modifications. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00914
Sun, J., Wang, Y., Zhang, X., Cheng, Z., Song, Y., Li, H., et al. (2024). Transcriptomic and metabolomic analyses reveal the role of phenylalanine metabolism in the maize response to stalk rot caused by fusarium proliferatum. Int. J. Mol. Sci. 25 (3), 1492. doi: 10.3390/ijms25031492
Sun, T., Yang, W., Zhang, W., Liu, Y., Li, L., Cheng, S., et al. (2025). Phenylpropanoid pathway mediated the defense response of ‘Korla’ fragrant pear against Alternaria alternata infection. Postharvest Biol. Technol. 220, 113318. doi: 10.1016/j.postharvbio.2024.113318
Sun, T., Zhang, J., Zhang, Q., Li, X., Li, M., Yang, Y., et al. (2023). Transcriptional and metabolic responses of apple to different potassium environments. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1131708
Tiwari, A., Yadav, V. K., Singh, S. K., Shukla, A. K., and Kumar, M. (2025). “Marine-derived Natural Flavonoids and Their Biological Activities,” in Nutraceuticals in Cardiac Health Management (New York: Apple Academic Press), 311–325.
Tzin, V. and Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3, 956–972. doi: 10.1093/mp/ssq048
Uddin, N., Li, X., Ullah, M. W., Sethupathy, S., Ma, K., Elboughdiri, N., et al. (2024). Lignin developmental patterns and Casparian strip as apoplastic barriers: A review. Int. J. Biol. Macromol. 260, 129595. doi: 10.1016/j.ijbiomac.2024.129595
Vanderschuren, H., Nyaboga, E., Poon, J. S., Baerenfaller, K., Grossmann, J., Hirsch-Hoffmann, M., et al. (2014). Large-scale proteomics of the cassava storage root and identification of a target gene to reduce postharvest deterioration. Plant Cell 26, 1913–1924. doi: 10.1105/tpc.114.123927
Vargas-Tah, A. and Gosset, G. (2015). Production of cinnamic and p-hydroxycinnamic acids in engineered microbes. Front. Bioengineering Biotechnol. 3. doi: 10.3389/fbioe.2015.00116
Wagay, N. A., Rafiq, S., Khan, A., Kaloo, Z. A., Malik, A. R., and Pulate, P. (2023). “). Impact of phenolics on drought stress and expression of phenylpropanoid pathway gen,” in Plant Phenolics in Abiotic Stress Management (Singapore: Springer), 265–285.
Wahengbam, E. D., Devi, C. P., Sharma, S. K., Roy, S. S., Maibam, A., Dasgupta, M., et al. (2023). Reactive oxygen species turnover, phenolics metabolism, and some key gene expressions modulate postharvest physiological deterioration in cassava tubers. Front. Microbiol. 14, 1148464. doi: 10.3389/fmicb.2023.1148464
Wang, Y., Chu, J., Zhang, H., Ju, H., Xie, Q., and Jiang, X. (2024a). Integrated transcriptomics and metabolomics analyses provide new insights into cassava in response to nitrogen deficiency. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1488281
Wang, Y., Chu, J., Zhang, H., Ju, H., Xie, Q., and Jiang, X. (2025a). Integrated transcriptomics and metabolomics analyses provide new insights into cassava in response to nitrogen deficiency. Front. Plant Sci. 15, 1488281. doi: 10.3389/fpls.2024.1488281
Wang, J. P., Chuang, L., Loziuk, P. L., Chen, H., Lin, Y. C., Shi, R., et al. (2015). Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc. Natl. Acad. Sci. U S A 112, 8481–8486. doi: 10.1073/pnas.1510473112
Wang, B., Jiang, H., Bi, Y., He, X., Wang, Y., Li, Y., et al. (2019). Preharvest multiple sprays with sodium nitroprusside promote wound healing of harvested muskmelons by activation of phenylpropanoid metabolism. Postharvest Biol. Technol. 158, 110988. doi: 10.1016/j.postharvbio.2019.110988
Wang, Y., Jiang, W., Li, C., Wang, Z., Lu, C., Cheng, J., et al. (2024b). Integrated transcriptomic and metabolomic analyses elucidate the mechanism of flavonoid biosynthesis in the regulation of mulberry seed germination under salt stress. BMC Plant Biol. 24, 132. doi: 10.1186/s12870-024-04804-3
Wang, J., Li, F., Zhang, X., Sun, W., Ali, M., Li, X., et al. (2023). Combined transcriptomic and targeted metabolomic analysis reveals the mechanism of flesh browning in cold stored ‘Fuji’ apple fruit. Sci. Hortic. 320, 112195. doi: 10.1016/j.scienta.2023.112195
Wang, Y., Qin, J., Wei, M., Liao, X., Shang, W., Chen, J., et al. (2025b). Verticillium dahliae elicitor vdSP8 enhances disease resistance through increasing lignin biosynthesis in cotton. Plant Cell Environ. 48, 728–745. doi: 10.1111/pce.15170
Wang, Y., Wang, X., Fang, J., Yin, W., Yan, X., Tu, M., et al. (2023). VqWRKY56 interacts with VqbZIPC22 in grapevine to promote proanthocyanidin biosynthesis and increase resistance to powdery mildew. New Phytol. 237, 1856–1875. doi: 10.1111/nph.18688
Wang, M., Wang, Y., Li, X., Zhang, Y., Chen, X., Liu, J., et al. (2024). Integration of metabolomics and transcriptomics reveals the regulation mechanism of the phenylpropanoid biosynthesis pathway in insect resistance traits in Solanum habrochaites. Horticult. Res. 11, uhad277. doi: 10.1093/hr/uhad277
Wang, C., Wu, J., Tang, Y., Min, Y., Wang, D., Ma, X., et al. (2023). Understanding the changes of phenylpropanoid metabolism and lignin accumulation in wounded cassava root during postharvest storage. Sci. Hortic. 310, 111765. doi: 10.1016/j.scienta.2022.111765
Wang, Y., Yang, Q., Jiang, H., Wang, B., Bi, Y., Li, Y., et al. (2020). Reactive oxygen species-mediated the accumulation of suberin polyphenolics and lignin at wound sites on muskmelons elicited by benzo (1, 2, 3)-thiadiazole-7-carbothioic acid S-methyl ester. Postharvest Biol. Technol. 170, 111325. doi: 10.1016/j.postharvbio.2020.111325
Wang, J., Yuan, B., and Huang, B. (2019). Differential heat-induced changes in phenolic acids associated with genotypic variations in heat tolerance for hard fescue. Crop Sci. 59, 667–674. doi: 10.2135/cropsci2018.01.0063
Wang, H., Zhang, Y., Jiang, H., Ding, Q., Wang, Y., Wang, M., et al. (2025). Transcriptomic and metabolomic analysis reveals the molecular mechanism of exogenous melatonin improves salt tolerance in eggplants. Front. Plant Sci. 15, 1523582. doi: 10.3389/fpls.2024.1523582
Wang, W., Zhou, X., Hu, Q., Wang, Q., Zhou, Y., Yu, J., et al. (2025). Lignin Metabolism Is Crucial in the Plant Responses to Tambocerus elongatus (Shen) in Camellia sinensis L. Plants 14. doi: 10.3390/plants14020260
Wei, H., Dhanaraj, A. L., Arora, R., Rowland, L. J., Fu, Y., and Sun, L. (2006). Identification of cold acclimation-responsive Rhododendron genes for lipid metabolism, membrane transport and lignin biosynthesis: importance of moderately abundant ESTs in genomic studies. Plant Cell Environ. 29, 558–570. doi: 10.1111/j.1365-3040.2005.01432.x
Wei, X., Du, Y., Zhang, W., Zhao, Y., Yang, S., Su, H., et al. (2024). Comparative Metabolome and Transcriptome Analysis Reveals the Defense Mechanism of Chinese Cabbage (Brassica rapa L. ssp. pekinensis) against Plasmodiophora brassicae Infection. Int. J. Mol. Sci. 25 (19), 10440. doi: 10.3390/ijms251910440
Wei, J., Xu, L., Shi, Y., Cheng, T., Tan, W., Zhao, Y., et al. (2023). Transcriptome profile analysis of Indian mustard (Brassica juncea L.) during seed germination reveals the drought stress-induced genes associated with energy, hormone, and phenylpropanoid pathways. Plant Physiol. Biochem. 200, 107750. doi: 10.1016/j.plaphy.2023.107750
Wu, Q., Yang, L., Liang, H., Yin, L., Chen, D., and Shen, P. (2022). Integrated analyses reveal the response of peanut to phosphorus deficiency on phenotype, transcriptome and metabolome. BMC Plant Biol. 22, 524. doi: 10.1186/s12870-022-03867-4
Wu, Y., Zhu, K., Wang, C., Li, Y., Li, M., and Sun, Y. (2024). Comparative metabolome and transcriptome analyses reveal molecular mechanisms involved in the responses of two carex rigescens varieties to salt stress. Plants 13, 2984. doi: 10.3390/plants13212984
Xiao, J., Cao, B., Tang, W., Sui, X., Tang, Y., Lai, Y., et al. (2025). The CaCAD1-CaPOA1 module positively regulates pepper resistance to cold stress by increasing lignin accumulation. Int. J. Biol. Macromol. 290, 139979. doi: 10.1016/j.ijbiomac.2025.139979
Xiao, S., Li, D., Tang, Z., Wei, H., Zhang, Y., Yang, J., et al. (2023). Supplementary UV-B radiation effects on photosynthetic characteristics and important secondary metabolites in eucommia ulmoides leaves. Int. J. Mol. Sci. 24, 8168. doi: 10.3390/ijms24098168
Xin, W., Zhang, L., Zhang, W., Gao, J., Yi, J., Zhen, X., et al. (2019). An integrated analysis of the rice transcriptome and metabolome reveals differential regulation of carbon and nitrogen metabolism in response to nitrogen availability. Int. J. Mol. Sci. 20, 2349. doi: 10.3390/ijms20092349
Xu, C., Shen, Y., He, F., Fu, X., Yu, H., Lu, W., et al. (2019). Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 222, 752–767. doi: 10.1111/nph.15658
Xu, J., Zhang, Z., Li, X., Wei, J., and Wu, B. (2019). Effect of nitrous oxide against Botrytis cinerea and phenylpropanoid pathway metabolism in table grapes. Sci. Hortic. 254, 99–105. doi: 10.1016/j.scienta.2019.04.061
Xue, Z., Wang, B., Qu, C., Tao, M., Wang, Z., Zhang, G., et al. (2023). Response of salt stress resistance in highland barley (Hordeum vulgare L. var. nudum) through phenylpropane metabolic pathway. PloS One 18, e0286957. doi: 10.1371/journal.pone.0286957
Yan, Q., Chen, W., Zhang, H., Liu, P., and Zhang, Y. (2025). PbrMYB14 enhances pear resistance to alternaria alternata by regulating genes in lignin and salicylic acid biosynthesis pathways. Int. J. Mol. Sci. 26 (3), 972. doi: 10.3390/ijms26030972
Yan, J., Ninkuu, V., Fu, Z., Yang, T., Ren, J., Li, G., et al. (2023). OsOLP1 contributes to drought tolerance in rice by regulating ABA biosynthesis and lignin accumulation. Front. Plant Sci. 14, 1163939. doi: 10.3389/fpls.2023.1163939
Yang, M., Chen, S., Huang, Z., Gao, S., Yu, T., Du, T., et al. (2023). Deep learning-enabled discovery and characterization of genes in Spartina alterniflora. Plant J. 116, 690–705. doi: 10.1111/tpj.16397
Yang, X., Gil, M. I., Yang, Q., and Tomás-Barberán, F. A. (2022). Bioactive compounds in lettuce: Highlighting the benefits to human health and impacts of preharvest and postharvest practices. Compr. Rev. Food Sci. Food Saf. 21, 4–45. doi: 10.1111/1541-4337.12877
Yang, K., Zhou, G., Chen, C., Liu, X., Wei, L., Zhu, F., et al. (2024). Joint metabolomic and transcriptomic analysis identify unique phenolic acid and flavonoid compounds associated with resistance to fusarium wilt in cucumber (Cucumis sativus L.). Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1447860
Yasir, M., Bhasin, A., Maibam, B. D., and Sharma, M. (2024). Review on finger millet (Eleusine coracana L.): Nutritional composition, modification, its effect on physicochemical, structural, functional properties, and its applications. J. Food Composit. Anal. 135, 106623. doi: 10.1016/j.jfca.2024.106623
Yu, W., Gong, F., Xu, H., and Zhou, X. (2024). Molecular mechanism of exogenous ABA to enhance UV-B resistance in rhododendron chrysanthum pall. by modulating flavonoid accumulation. Int. J. Mol. Sci. 25, 5248. doi: 10.3390/ijms25105248
Yuan, H., Cheng, M., Wang, R., Wang, Z., Fan, F., Wang, W., et al. (2024). miR396b/GRF6 module contributes to salt tolerance in rice. Plant Biotechnol. J. 22, 2079–2092. doi: 10.1111/pbi.14326
Yuan, Y., Ma, X., Li, C., Zhong, X., Li, Y., Zhao, J., et al. (2025). Integration of transcriptome and metabolome reveals key regulatory defense pathways associated with high temperature stress in cucumber (Cucumis sativus L.). BMC Plant Biol. 25, 6. doi: 10.1186/s12870-024-05876-x
Zeng, J., He, X., Quan, X., Cai, S., Han, Y., Nadira, U. A., et al. (2015). Identification of the proteins associated with low potassium tolerance in cultivated and Tibetan wild barley. J. Proteomics 126, 1–11. doi: 10.1016/j.jprot.2015.05.025
Zeng, J., Quan, X., He, X., Cai, S., Ye, Z., Chen, G., et al. (2018). Root and leaf metabolite profiles analysis reveals the adaptive strategies to low potassium stress in barley. BMC Plant Biol. 18, 1–13. doi: 10.1186/s12870-018-1404-4
Zhan, X., Qi, J., Shen, Q., He, B., and Mao, B. (2022). Regulation of phenylpropanoid metabolism during moderate freezing and post-freezing recovery in Dendrobium officinale. J. Plant Interact. 17, 290–300. doi: 10.1080/17429145.2022.2036377
Zhang, S., Dai, B., Wang, Z., Qaseem, M. F., Li, H., and Wu, A.-M. (2023a). The key physiological and molecular responses of Neolamarckia cadamba to phosphorus deficiency stress by hydroponics. Indust. Crops Prod. 202, 117065. doi: 10.1016/j.indcrop.2023.117065
Zhang, X., Gou, M., and Liu, C.-J. (2013). Arabidopsis kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25, 4994–5010. doi: 10.1105/tpc.113.119644
Zhang, S., He, C., Wei, L., Jian, S., and Liu, N. (2023b). Transcriptome and metabolome analysis reveals key genes and secondary metabolites of Casuarina equisetifolia ssp. incana in response to drought stress. BMC Plant Biol. 23, 200. doi: 10.1186/s12870-023-04206-x
Zhang, M., Ma, Y., Wang, Y., Gao, H., Zhao, S., Yu, Y., et al. (2024). MAPK and phenylpropanoid metabolism pathways involved in regulating the resistance of upland cotton plants to Verticillium dahliae. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1451985
Zhang, L., Sun, Y., Ji, J., Zhao, W., Guo, W., Li, J., et al. (2023). Flavonol synthase gene MsFLS13 regulates saline-alkali stress tolerance in alfalfa. Crop J. 11, 1218–1229. doi: 10.1016/j.cj.2023.05.003
Zhang, J., Wang, Y., Li, J., Zhu, Y., Wang, L., Li, Z., et al. (2024). Overexpression of chalcone isomerase-like genes, gmCHI4A and gmCHI4B, enhances salt tolerance of cotyledon hairy roots and composite plant in soybean (Glycine max (L.) merr.). Agronomy 14, 731. doi: 10.3390/agronomy14040731
Zhang, Q., Wang, S., Qin, B., Sun, H., Yuan, X., Wang, Q., et al. (2023). Analysis of the transcriptome and metabolome reveals phenylpropanoid mechanism in common bean (Phaseolus vulgaris) responding to salt stress at sprout stage. Food Energy Secur. 12, e481. doi: 10.1002/fes3.481
Zhang, M., Xing, Y., Ma, J., Zhang, Y., Yu, J., Wang, X., et al. (2023). Investigation of the response of Platycodongrandiflorus (Jacq.) A. DC to salt stress using combined transcriptomics and metabolomics. BMC Plant Biol. 23, 589.
Zhang, L., Yan, J., Fu, Z., Shi, W., Ninkuu, V., Li, G., et al. (2021). FoEG1, a secreted glycoside hydrolase family 12 protein from Fusarium oxysporum, triggers cell death and modulates plant immunity. Mol. Plant Pathol. 22, 522–538. doi: 10.1111/mpp.13041
Zhang, T., Zhang, C., Wang, W., Hu, S., Tian, Q., Li, Y., et al. (2025). Effects of drought stress on the secondary metabolism of Scutellaria baicalensis Georgi and the function of SbWRKY34 in drought resistance. Plant Physiol. Biochem. 219, 109362. doi: 10.1016/j.plaphy.2024.109362
Zhang, Y., Zhang, H., Zhang, Y., Wang, D., Meng, X., and Chen, J. (2025). Utilizing physiologies, transcriptomics, and metabolomics to unravel key genes and metabolites of Salvia miltiorrhiza Bge. seedlings in response to drought stress. Front. Plant Sci. 15, 1484688. doi: 10.3389/fpls.2024.1484688
Zhang, Q., Zheng, G., Wang, Q., Zhu, J., Zhou, Z., Zhou, W., et al. (2022). Molecular mechanisms of flavonoid accumulation in germinating common bean (Phaseolus vulgaris) under salt stress. Front. Nutr. 9, 928805. doi: 10.3389/fnut.2022.928805
Zhang, X., Zhu, X., Chen, L., Fan, H., Liu, X., Yang, N., et al. (2024). MiR398b Targets Superoxide Dismutase Genes in Soybean in Defense Against Heterodera glycines via Modulating Reactive Oxygen Species Homeostasis. Phytopathology 114, 1950–1962. doi: 10.1094/phyto-09-23-0343-r
Zhao, X., Jiang, X., Li, Z., Song, Q., Xu, C., and Luo, K. (2023). Jasmonic acid regulates lignin deposition in poplar through JAZ5-MYB/NAC interaction [Original Research. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1232880
Zhao, S., Ma, Y., Ding, Y., Dong, G., Liu, C., Ma, X., et al. (2025). Rice glycosyltransferase UGT706F1 functions in heat tolerance through glycosylating flavonoids under the regulation of transcription factor MYB61. Plant J. 121, e17252. doi: 10.1111/tpj.17252
Zhao, B., Wang, L., Pang, S., Jia, Z., Wang, L., Li, W., et al. (2020). UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Indust. Crops Prod. 151, 112483. doi: 10.1016/j.indcrop.2020.112483
Zheng, L., Chen, Y., Ding, D., Zhou, Y., Ding, L., Wei, J., et al. (2019). Endoplasmic reticulum–localized UBC34 interaction with lignin repressors MYB221 and MYB156 regulates the transactivity of the transcription factors in Populus tomentosa. BMC Plant Biol. 19, 1–17. doi: 10.1186/s12870-018-1600-2
Zheng, T., Lv, J., Sadeghnezhad, E., Cheng, J., and Jia, H. (2022). Transcriptomic and metabolomic profiling of strawberry during postharvest cooling and heat storage. Front. Plant Sci. 13, 1009747. doi: 10.3389/fpls.2022.1009747
Zheng, X., Zhang, X., and Zeng, F. (2025). Biological functions and health benefits of flavonoids in fruits and vegetables: A contemporary review. Foods 14, 155. doi: 10.3390/foods14020155
Zhou, Y., Bai, Y. H., Han, F. X., Chen, X., Wu, F. S., Liu, Q., et al. (2024). Transcriptome sequencing and metabolome analysis reveal the molecular mechanism of Salvia miltiorrhiza in response to drought stress. BMC Plant Biol. 24, 446. doi: 10.1186/s12870-024-05006-7
Zhou, L., Wang, E., Yang, Y., Yang, P., Xu, L., and Ming, J. (2024). Antioxidant Enzyme, Transcriptomic, and Metabolomic Changes in Lily (Lilium spp.) Leaves Induced by Aphis gossypii Glover. Genes (Basel) 15. doi: 10.3390/genes15091124
Keywords: phenylpropanoids, plant interactions, post-transcription, post-translation, epigenetics modifications, plant development
Citation: Ninkuu V, Aluko OO, Yan J, Zeng H, Liu G, Zhao J, Li H, Chen S and Dakora FD (2025) Phenylpropanoids metabolism: recent insight into stress tolerance and plant development cues. Front. Plant Sci. 16:1571825. doi: 10.3389/fpls.2025.1571825
Received: 06 February 2025; Accepted: 14 April 2025;
Published: 26 June 2025.
Edited by:
Igor Cesarino, University of São Paulo, BrazilReviewed by:
Masood Jan, University of Florida, United StatesShouchuang Wang, Hainan University, China
Navin Kumar, Agricultural Research Organization (ARO), Israel
Copyright © 2025 Ninkuu, Aluko, Yan, Zeng, Liu, Zhao, Li, Chen and Dakora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vincent Ninkuu, bmlua3V1LnZpbmNlbnRAeWFob28uY29t; Songbi Chen, c29uZ2JpY2hlbkBob3RtYWlsLmNvbQ==; Felix Dapare Dakora, ZGFrb3JhZmRAdHV0LmFjLnph
†These authors have contributed equally to this work
 Vincent Ninkuu
Vincent Ninkuu Oluwaseun Olayemi Aluko
Oluwaseun Olayemi Aluko Jianpei Yan
Jianpei Yan Hongmei Zeng
Hongmei Zeng Guodao Liu3
Guodao Liu3 Huihui Li
Huihui Li Songbi Chen
Songbi Chen Felix Dapare Dakora
Felix Dapare Dakora