- 1College of Life Science, Henan Normal University, Xinxiang, China
- 2The Observation and Research Field Station of Taihang Mountain Forest Ecosystems of Henan Province, Henan Normal University, Xinxiang, China
Long non-coding RNAs (lncRNAs) have been demonstrated to play key roles in plant response and adaptation to heavy metal stresses. However, the exact biological functions and potential regulatory mechanism, especially in wheat’s response to cadmium (Cd) stress, are still poorly understood. We have previously discovered a Cd stress-related lncRNA in wheat, namely TalncRNA18313. In this study, qRT-PCR analysis revealed that TalncRNA18313 was expressed extensively in wheat leaves, and its accumulation was highly induced by Cd stress. To further fully explore the function of lncRNA18313 in response to Cd stress, lncRNA18313 was cloned from wheat (Triticum aestivum L.), and was transformed into Arabidopsis. When TalncRNA18313 was heterologous expressed in Arabidopsis, the transgenic plants exhibited enhanced Cd tolerance characterized by lower malondialdehyde (MDA) levels and higher activities of key antioxidant enzymes, such as catalase (CAT), superoxide dismutase (SOD) and peroxidase (POD). Subsequently, RNA-sequencing (RNA-seq) analysis demonstrated that 370 genes were differentially expressed in lncRNA18313 overexpressing transgenic lines under Cd stress comparing to wild type plants. Among the genes regulated by lncRNA18313, the most significantly enriched were those involved in transcriptional regulation and antioxidative defense responses. These results suggest that TalncRNA18313 plays a crucial role in improving Cd tolerance in wheat by modulating key stress-related pathways, particularly those critical for coping with oxidative damage and regulating gene expression under Cd stress. This discovery contributes to the expanding understanding of knowledge about the involvement of lncRNAs in plant stress responses and offers promising potential for improving crop resilience to environmental stresses.
1 Introduction
Heavy metal pollution has emerged as one of the most severe and widespread environmental challenges in China, driven by the rapid expansion of modern industries, agriculture, and urbanization (Li et al., 2024). Among these heavy metals, cadmium (Cd) is particularly concerning due to its extremely toxic and highly mobile in natural environment. It readily accumulates in crop plants, posing a significant threat to both agricultural productivity and human health (Cao et al., 2023; Zhang et al., 2023). As a widely consumed crop globally, wheat (Triticum aestivum L.) has become a major and progressively growing source of Cd intake for humans, as it can enter the human body through the food chain, thereby posing considerable health risks (Zhu et al., 2023; Zhang et al., 2024). In addition to its harmful effects on human health, excessive Cd accumulation in wheat significantly disrupts plant growth, morphology, physiology, and metabolism, both directly and indirectly. This results in substantial losses in wheat yield and quality (Thind et al., 2020; Zhu et al., 2022; Rasheed et al., 2024). Therefore, it is imperative to dissect the regulatory mechanisms involved in Cd detoxification to develop effective strategies to reduce excessive Cd into wheat. Plants have evolved various defense mechanisms to detoxify Cd toxicity, including chelation, transportation and vacuolar sequestration, which help restricts Cd accumulation and minimize its harmful effects (Cao et al., 2023; Wang et al., 2025). Previous studies on stress gene regulation have primarily focused on identifying and analyzing of specific protein-encoding genes associated with Cd homeostasis and detoxification, such as key Cd absorption genes and various heavy metal transporters (Cao et al., 2023; Rasheed et al., 2024). However, the underlying molecular mechanism responsible for Cd stress in wheat is still not fully explored.
In recent years, long non-coding RNAs (lncRNAs) have become recognized as key regulators of plant growth, governing a variety of essential biological processes (Liu et al., 2012; Zhao et al., 2023). LncRNAs are transcripts longer than 200 nucleotides, typically lacking or having no capacity of protein-coding potential (Wierzbicki et al., 2021). A growing body of evidence indicates that lncRNAs play a crucial role in regulating gene expression in plants through a variety of complex mechanisms, such as interaction with hormones and transcription factors, as well as involvement in alternative splicing (Bardou et al., 2014; Liu et al., 2015).
Along with the advancement of next-generation RNA sequencing and recent improvements in bioinformatics tools, an increasing number of lncRNAs have been identified and confirmed to play a pivotal role in orchestrating plants’ responses to varying environmental stresses. These include drought stress in wheat (Li et al., 2022) and cassava (Ding et al., 2019), cold stress in wheat (Lu et al., 2020) and Brassica napus (Waseem et al., 2022), alkaline stress in wheat (Wei et al., 2022) and sugar beet (Zou et al., 2020), lead (Pb) stress in poplar (Chen et al., 2022), aluminum stress in Medicago truncatula (Gui et al., 2022), and Cd stress in rice (Chen et al., 2018) and Brassica napus (Feng et al., 2016). Moreover, several plant lncRNAs have been studied intensively. For instance, a salt-responsive lncRNA, lncRNA973, was identified in cotton, which was induced by salt stress. Transgenic Arabidopsis overexpressing lncRNA973 could facilitate salt tolerance by modulating genes involved in antioxidant defense, transcription factors and photosynthesis (Zhang et al., 2019). In Betula platyphylla, two Cd-responsive lncRNAs, lncRNA28068.1 and lncRNA30505.2, have been shown to increase Cd tolerance by modulating the expression of their target genes, heat shock protein (HSP18.1) and L-lactate dehydrogenase A (LDHA) (Wen et al., 2020). In our previous study, we identified 10,044 lncRNAs in the roots of wheat that responded to Cd stress using RNA-seq technology. Among these, 377 lncRNAs were differentially expressed in response to Cd stress. Furthermore, we found that overexpressing lncRNA37228 could confer Cd tolerance in Arabidopsis (Zhu et al., 2023). Although many Cd-responsive lncRNAs had been recognized or predicted in wheat, there may be a large number of lncRNAs whose biological functions have not yet been characterized, warranting further investigation.
The lncRNA showing the highest induction in response to Cd stress, namely lncRNA18313, in the wheat genome was identified in our previous research (Zhu et al., 2023). However, the biological function of lncRNA18313 in Cd stress response remained elusive. Here in this study, we cloned lncRNA18313 and examined its expression levels in wheat with and without Cd stress exposure. Subsequently, overexpression of lncRNA18313 in transgenic Arabidopsis plants demonstrated its positive role in enhancing Cd stress tolerance. To further explore the underlying mechanisms, transcriptome sequencing of the overexpressed lncRNA18313 plants and the control plants was conducted to investigate differentially expressed genes in response to Cd stress. These findings not only provide insights into the genetic and molecular mechanisms by which lncRNA18313 mediates Cd stress response in wheat but also lay a theoretical foundation for genetic improvement strategies aimed at enhancing Cd tolerance in plants.
2 Materials and methods
2.1 Plant materials and CdCl2 treatment
Wheat cultivar ‘Zhengmai 366” was kindly provided by Henan Academy of Agricultural Sciences. Seeds, surface-sterilized, were firstly germinated at 26°C for 2 days. The germinated seeds were subsequently transferred to Petri plates (16 cm in diameter) and cultivated in an illumination incubator with a day/night temperature of 25°C/16°C, 65%-75% relative humidity, and a 12-hour light/dark cycle (light intensity: 750 μmol m-² s-¹). When the seedlings were 10 days old (with two fully expanded leaves), uniform seedlings were transferred to a nutrient solution supplemented with 100 μM CdCl2·2.5H2O for 7 days. At 0, 1, 3, 5 and 7 days of CdCl2 stress, roots, stems and leaves were harvested separately, rapidly frozen in liquid nitrogen, and stored at -80°C.
Seeds of wild-type (WT) and transgenic plant were sown on plates containing half-strength Murashige and Skoog (1/2 MS) solid medium in the dark at 4°C for 2 days. Afterward, the plates were moved to a growth chamber with long-day conditions (16-hour photoperiod and light intensity of 120 μmol m-2 s-1) and maintained at 22°C for 8 days. Then, 8-day-old seedlings exhibiting uniform growth were transplanted into soil and placed back in a growth chamber under the same conditions. For Cd treatment, six-week-old WT and transgenic plants (with fully expanded rosette leaves) grown in soil were divided into two groups: control and Cd treatment. The plants were irrigated with 1/2 MS solution, with or without 100 μM CdCl2·2.5H2O, for 4 days. Each experimental group comprised at least three plants, and all treatments were conducted with three biological replicates. After 4 days of Cd treatment, rosette leaves were harvested, immediately frozen in liquid nitrogen, and stored at -80°C.
2.2 Vector construction and genetic transformation
A 414 bp full-length genomic sequence of lncRNA18313 was amplified by PCR from genomic DNA extracted from ten-day-old wheat seedlings using the primers 5′- TGCTCTAGA TCCGCTATTCCGATG -3′ and 5′- GGGGTACC GTTGGATAGGCT -3′. Sequencing confirmed PCR fragments were successfully inserted into pCAMBIA1300 expression vector driven by the cauliflower mosaic virus (CaMV) 35S promoter, using the restriction endonuclease XbaI and KpnI. These recombinant vectors were introduced into Agrobacterium EHA105 strain, and subsequently used to transform Arabidopsis (ecotype Columbia 0) through floral dip method as previously reported (Clough and Bent, 1998). Transgenic lines expressing lncRNA18313 were screened using 50 mg L-1 kanamycin and then further confirmed by PCR amplification. Subsequently, physiological and biochemical characterization, gene expression analysis, and RNA sequencing were performed on T3 homozygous lines unless otherwise specified.
2.3 RNA extraction and gene expression analysis
Total RNAs were extracted from wheat seedlings or Arabidopsis thaliana using Trizol reagent (Tiangen, Beijing, China) following the manufacturer’s instructions and M5 SuperFast plus qPCR RT kit (Tiangen, Beijing, China) was used for first-strand cDNA synthesis. Relative expression levels of lncRNA18313 and candidate genes were analyzed with qRT-PCR and elongation factor (TaEF-1α) as the reference gene was normalized gene expression using the 2-ΔΔCt method, as described by Livak and Schmittgen (2001). The primers used in the experiment are shown in Supplementary Table S1. Each sample was conducted with three biological replicates.
2.4 Transcriptome analysis
Rosette leaves from six-week-old wild-type and transgenic Arabidopsis thaliana (T3 generation) grown under Cd stress (100 μM CdCl2, 4 d) were collected and sent to Lianchuan Biological Technology Co. Ltd. (Hangzhou, China) for RNA sequencing (RNA-seq). Each sample included three biological replicates, with each replicate consisting of pooled leaves from six independent plants. For bioinformatics analysis, all raw reads that passed quality control in FastQC were trimmed to eliminate low quality sequences, and mapped to the Arabidopsis reference genome using HISAT2. Reads normalized expression level were quantified using RSEM (v1.2.8) to calculate FPKM values and differential expression analysis was evaluated using DESeq2 and edgeR package (version 1.20.0). Genes with p value ≤ 0.05 and | log2 (fold change) | ≥ 2 were defined as differentially expressed genes (DEGs). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs were conducted using GOseq R packages and KOBAS 2.0 software, respectively.
2.5 Determination of MDA content and antioxidant enzyme activity
Rosette leaves from six-week-old seedlings of WT and transgenic Arabidopsis grown under normal and Cd stress conditions (100 μM CdCl2) for 4 days were collected to quantify malondialdehyde (MDA) contents and antioxidant enzyme activity. MDA concentration was assessed using the thiobarbituric acid (TBA) method as described previously (Zhu et al., 2022). SOD activity was assessed by monitoring the inhibition of nitroblue tetrazolium at 560 nm, following the method described by Giannopolitis and Ries (1977). CAT activity was evaluated at 240 nm by observing a decrease in absorbance due to H2O2 degradation (Aebi, 1984). POD activity was assayed using guaiacol and hydrogen peroxide as substrates, with the reaction monitored at 470 nm (Zhang and Kirham, 1994).
2.6 Statistical analysis
The data was presented as mean ± SE based on three independent replications. One-way analysis of variance (ANOVA) was carried out to assess significance differences between the control and treatment groups, followed by Duncan’s multiple range test (p < 0.05).
3 Results
3.1 Characteristics and expression analysis of TalncRNA18313
In our previous study, 242 lncRNAs responsive to Cd stress were identified in wheat through RNA sequencing (RNA-Seq) (Zhu et al., 2023). Among dozens of identified Cd-responsive lncRNAs, lncRNA18313 was strongly induced by Cd stress in wheat. Therefore, TalncRNA18313 was chosen for further analysis. The sequence of TalncRNA18313 was cloned from wheat variety ‘Zhengmai No. 366’, with the sequence information provided in Supplementary Table S1. The length of TalncRNA18313 is 414 nucleotides, with the maximum open reading frames (ORFs) encoding 29 amino acids. However, it was predicted to lack protein-coding potential, as indicated by the Coding Potential Calculator (CPC) (Figure 1A). Given the low conservation of lncRNAs, the sequence of TalncRNA18313 was subjected to NCBI BLAST, but no significant homologous sequences were identified in Arabidopsis or other plants species.
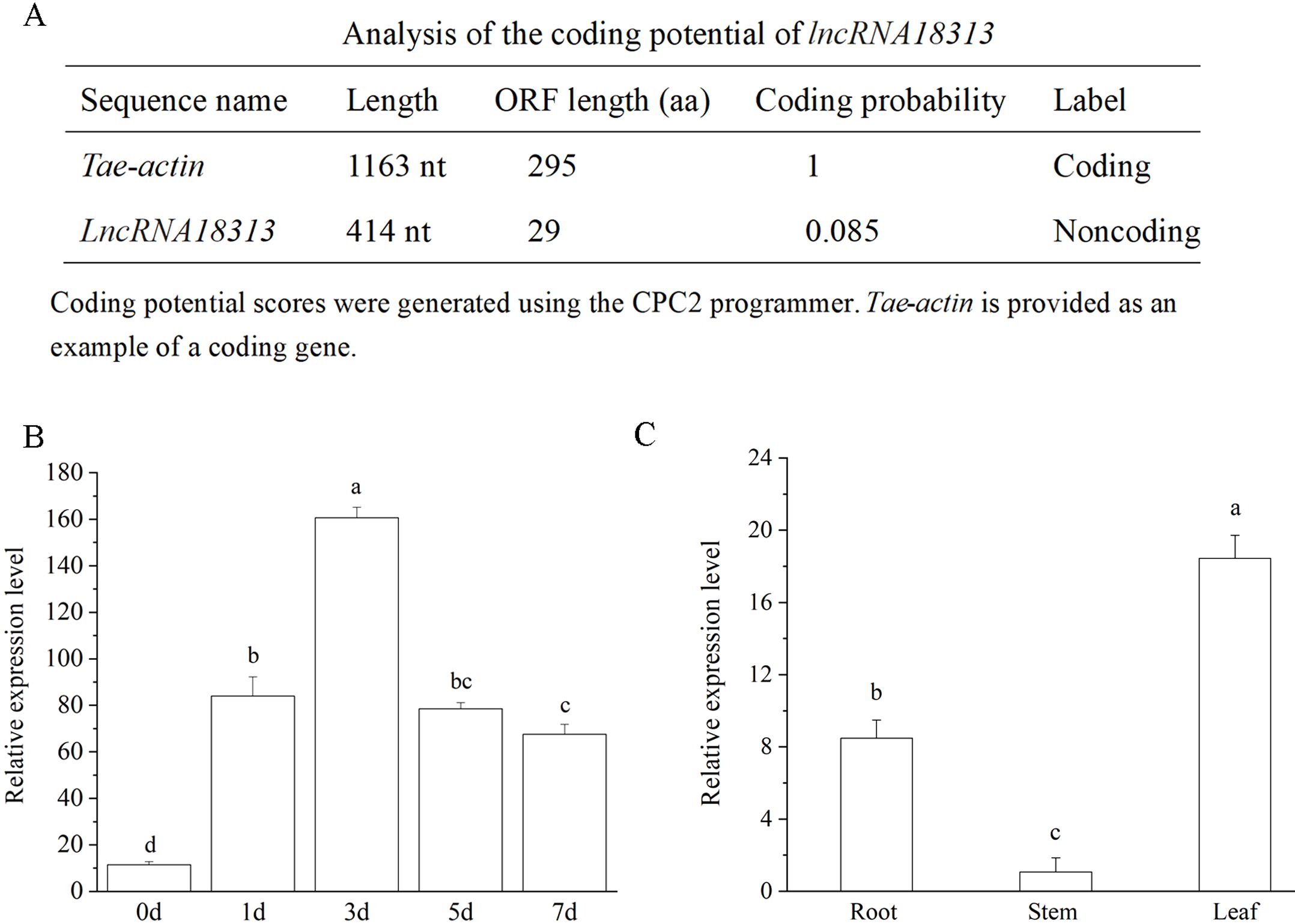
Figure 1. Characteristics and expression analysis of TalncRNA18313. (A) Analysis of the coding potential of lncRNA18313. (B) The expression levels of lncRNA18313 in wheat leaves treated with 100 μM CdCl2 at different time points. (C) The expression levels of lncRNA18313 in different tissues of wheat. Gene expression was measured by qRT-PCR, with the expression of lncRNA18313 normalized against the TaEF-1α gene. Each bar represents the mean ± standard error of six replicates. Columns labeled with different letters indicate a significant difference (p <0.05), as determined by Duncan’s multiple range test.
To elucidate the role of lncRNA18313 in Cd stress, we firstly used qRT-PCR to explore the expression of lncRNA18313 at different time points in wheat treated with 100 μM CdCl2 treatment. As shown in Figure 1B, the expression of lncRNA18313 in leaves was rapidly up‐regulated, reaching its peak at the 3th day after Cd treatment, then the expression of lncRNA18313 declined but remained at a higher expression level in contrast to that the seedlings under normal growth conditions (without Cd application). In agreement with our RNA-Seq data, the expression of lncRNA18313 was upregulated by 16.35-fold in wheat leaves after 3 days of Cd treatment. Additionally, no significant variation in lncRNA18313 expression was detected at different time points under normal growth conditions (Supplementary Figure S1). Those results suggested that lncRNA18313 was involved in wheat responsive to Cd stress.
We also performed qRT-PCR to assess the expression levels of lncRNA18313 in the leaves, stems and roots of wheat. Notably, a relatively high level of lncRNA18313 expression was found in the wheat leaves (Figure 1C), while its transcript level in the stem was very low, suggesting that lncRNA18313 exhibited highly tissue-specific expression patterns.
3.2 TalncRNA18313 overexpression positively regulates Cd tolerance in Arabidopsis
To directly characterize the role of TalncRNA18313 in plant response to Cd stress, transgenic Arabidopsis overexpressing TalncRNA18313 transcript were generated and two independent lines were randomly selected for further analysis. qRT-PCR analysis showed that the expression levels of lncRNA18313 in transgenic line 1 and 2 were approximately 20-fold higher than in the wild-type (WT) (Figure 2A), indicating that the transgenic lines overexpressing TalncRNA18313 had been successfully generated and could be used for further investigation.
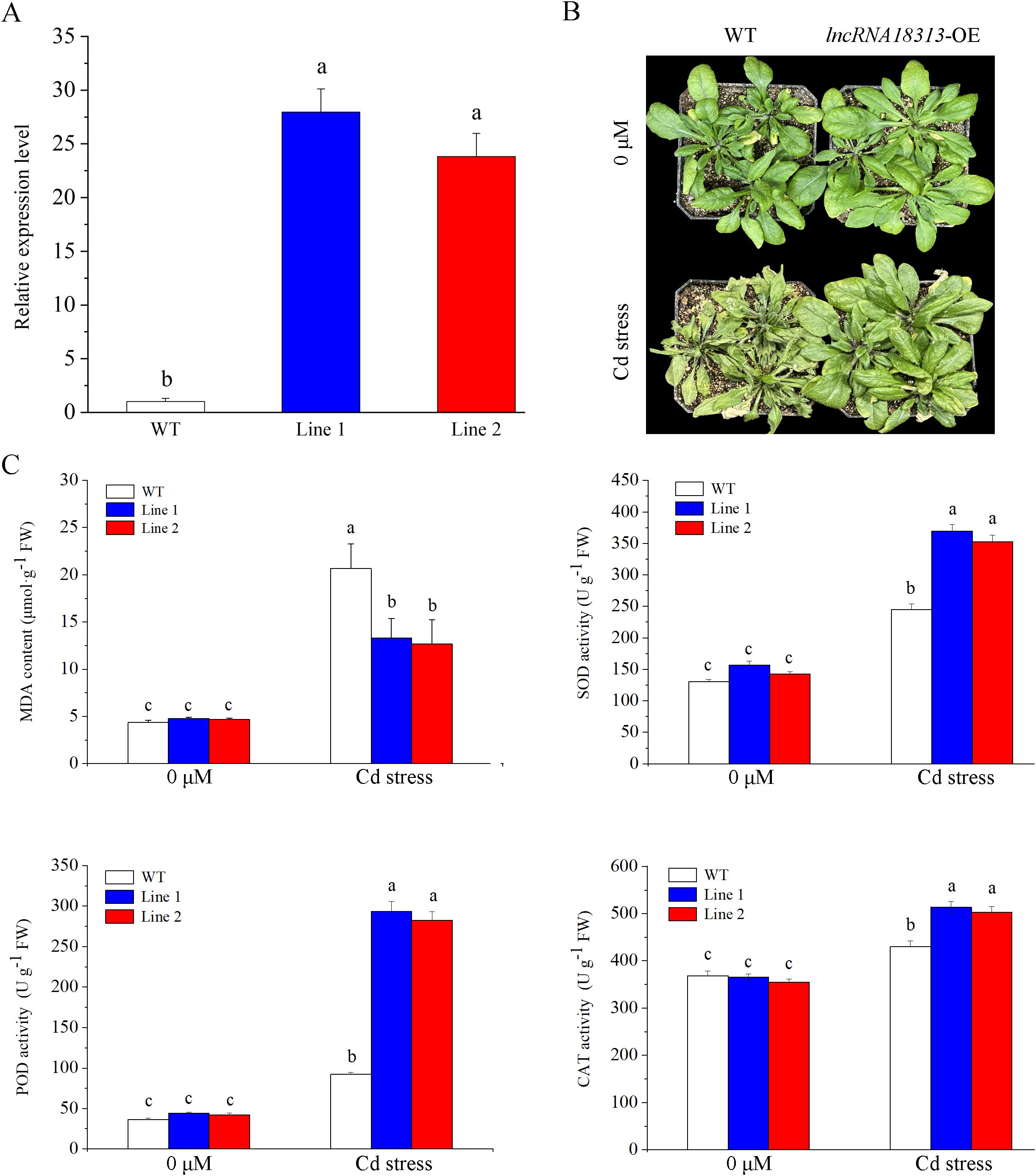
Figure 2. Overexpression of TalncRNA18313 confers Cd tolerance in Arabidopsis. (A) Expression levels of TalncRNA18313 in wild-type (WT) and lncRNA18313- overexpressing (OE) Arabidopsis lines. The expression of TalncRNA18313 was normalized to the AtActin gene. (B) Phenotypes of rosette leaves in 6-week-old WT and TalncRNA18313-OE Arabidopsis seedlings exposed to 0 μM and 100 μM CdCl2 for 4 days. (C) Determination of malondialdehyde (MDA) content and the activities of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) in WT and TalncRNA18313-OE lines under 100 μM CdCl2 treatment for 4 days. Each bar represents the mean ± SD for three biological replicates. Columns labeled with different letters indicate significant differences (p < 0.05), as determined by Duncan’s multiple range test.
To decipher the physiological functions of TalncRNA18313 in response to Cd stress, 6-week-old WT and lncRNA18313-OE Arabidopsis seedlings were treated with 100 μM CdCl2 in growth chambers. After 4 days of Cd treatment, WT plants exhibited severe damage, with most leaves completely withered and dry. In contrast, lncRNA18313-OE plants showed less damage, exhibiting fewer wilted leaves and maintaining a relatively healthy appearance compared with WT plants (Figure 2B). We thus quantified malondialdehyde (MDA) content as an indicator of membrane system damage. Consistent with the observation of leaf phenotypes, MDA contents in the leaves of lncRNA18313-OE lines were notably lower compared to those in WT plants (Figure 2C), suggesting that overexpression of TalncRNA18313 resulted in less oxidative damage under 100 μM CdCl2 stress. As previously reported, antioxidant enzymes are crucial for scavenging reactive oxygen species (ROS) during the initial stages of Cd stress. In an agreement, Cd stress led to a significant increase in the activities of SOD, CAT and POD in both WT and lncRNA18313-OE lines. However, after 4 days of Cd stress, the activities of these enzymes were notably higher in the lncRNA18313-OE lines compared to WT plants (Figure 2C), demonstrating an enhanced capacity for ROS scavenging in the transgenic plants. These findings indicate that TalncRNA18313 plays a vital role in mitigating Cd-induced oxidative damage, thereby improving plant tolerance to heavy metal stress.
3.3 Analysis of transcriptome sequencing data and identification of DEGs
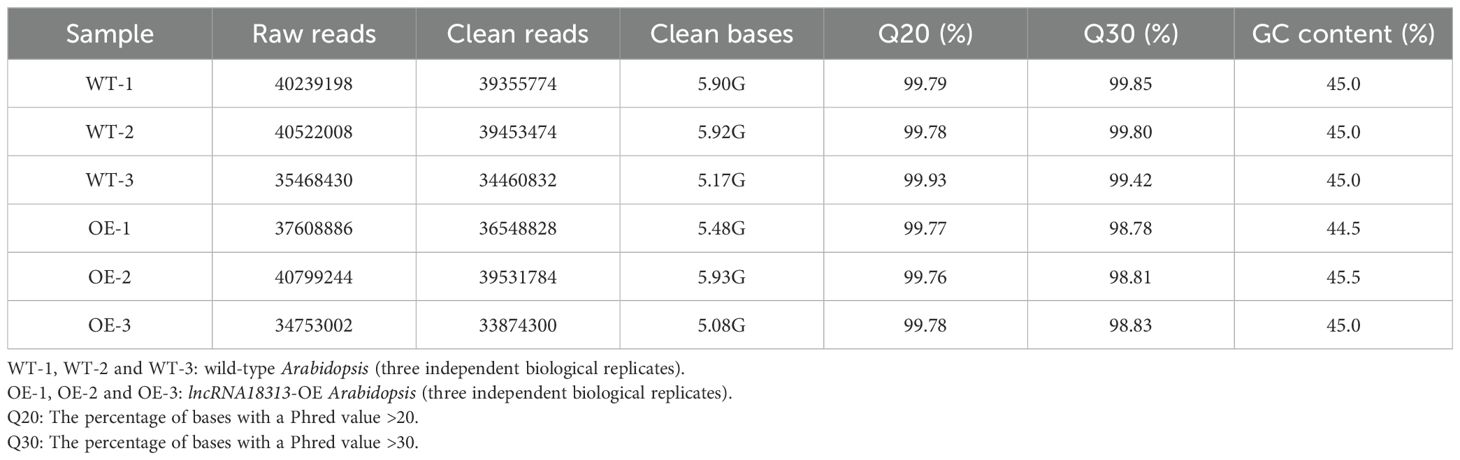
To further obtain potential genes regulated by lncRNA18313 in Cd tolerance, RNA sequencing (RNA‐seq) was performed on 6-week-old wild-type (WT-1, WT-2 and WT-3) and lncRNA18313-OE lines (OE-1, OE-2 and OE-3) in order to assess transcriptional changes under Cd stress conditions. Six libraries were constructed from the rosette leaves of WT and lncRNA18313-OE lines, with three biological replicates for each line and 5 plants per replicate. Each library contained over 5.08 Gb of clean bases, with a Q20 percentage exceeding 99.76%, a Q30 percentage over 98.78%, and a GC content ranging from 44.5 to 45.5% (Table 1). The clean data were subsequently aligned to the Arabidopsis reference genome, yielding the mapping ratio between 93.23% and 95.07%, indicating that RNA-seq data were of high quality and reliability. In order to gain a comprehensive view of transcript abundance in lncRNA18313-OE and WT plants under 100 μM CdCl2 conditions, a heatmap was constructed by clustering all DEGs, highlighting the changes in gene expression levels. In the present study, overexpression of lncRNA18313 in Arabidopsis was found to significantly alter transcript abundance under Cd stress conditions (Figure 3A). Compared with WT, lncRNA18313-OE can regulate 370 differentially expressed genes with more than a two-fold change in expression under Cd stress (Figure 3B). Of these, 203 differentially expressed genes (54.9%) were upregulated, while 167 genes (45.1%) were downregulated. Based on these DEGs, it is likely that TalncRNA18313 is positively associated with the expression of Cd-response genes.
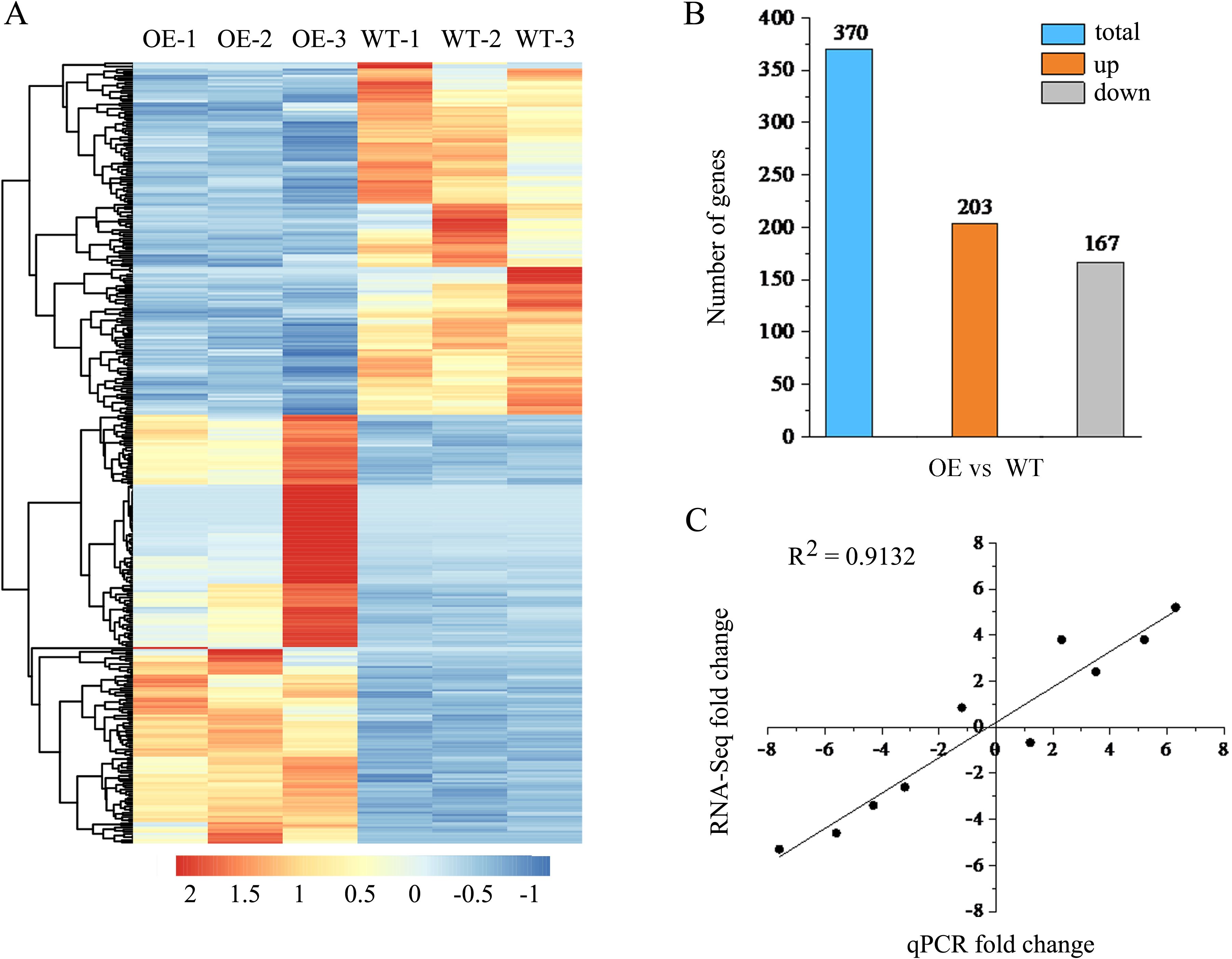
Figure 3. RNA-seq analysis of differentially expressed genes (DEGs) in lncRNA18313-OE and WT plants under 100 μM CdCl2 conditions for 4 days. (A) Hierarchical clustering of all DEGs between WT and lncRNA18313-OE under Cd stress, based on log10 RPKM (number of fragments per kilobase of transcript per million fragments mapped) values. The color scale (from blue to red) represents gene expression level from low to high. (B) Changes in DEGs between lncRNA18313-OE and WT plants under Cd stress. The number of up- and down-regulated genes between lncRNA18313-OE and WT is summarized. (C) Correlation between RNA-seq (y-axis) and qRT-PCR (x-axis) data, with the assay conducted for 10 randomly selected DEGs.
To verify the results of RNA-sequencing, a total of 10 DEGs involved in Cd tolerance regulated by lncRNA18313 were selected for qRT-PCR analysis. As shown in Figure 3C, the expression patterns of these 10 DEGs were in agreement with the RNA-seq results. The value of R square for the RNA-seq vs. qRT-PCR was 0.9132, further confirming the high reliability of identified DEGs in this study.
3.4 Potential functional analysis of differentially expressed genes regulated by lncRNA18313 under Cd stress
To infer the potential functions of DEGs regulated by lncRNA18313, GO term and KEGG pathway enrichment analyses were conducted on these significantly regulated genes. All DEGs were classified into 50 GO terms (Figure 4A), including 25 in “biological process” (BP), 10 in “molecular function” (MF), and 15 in “cellular component” (CC) (Figure 4A). GO terms associated with responses to various stresses, such as response to salt stress (GO: 0009651), response to water deprivation (GO: 0009414), response to oxidative stress (GO: 0006979) and response to wounding (GO: 0009611) were highly enriched in the biological process category. Molecular functions predominantly highlighted binding and catalytic activity. Additionally, KEGG pathway analysis showed that DEGs regulated by lncRNA18313 were significantly enriched in 52 KEGG pathways (Figure 4B). Notably, metabolic pathways (ath01100, 43), biosynthesis of secondary metabolites pathway (ath01110, 27), phenylpropanoid biosynthesis pathway (ath00940, 8) and glutathione metabolism pathway (ath00480, 5) were particularly enriched in lncRNA18313-OE plants under Cd stress based on KEGG pathways. Collectively, these findings suggest that the DEGs regulated by lncRNA18313 are implicated in a broad array of biological processes and regulatory networks, particularly in response to environmental stressors, such as Cd.
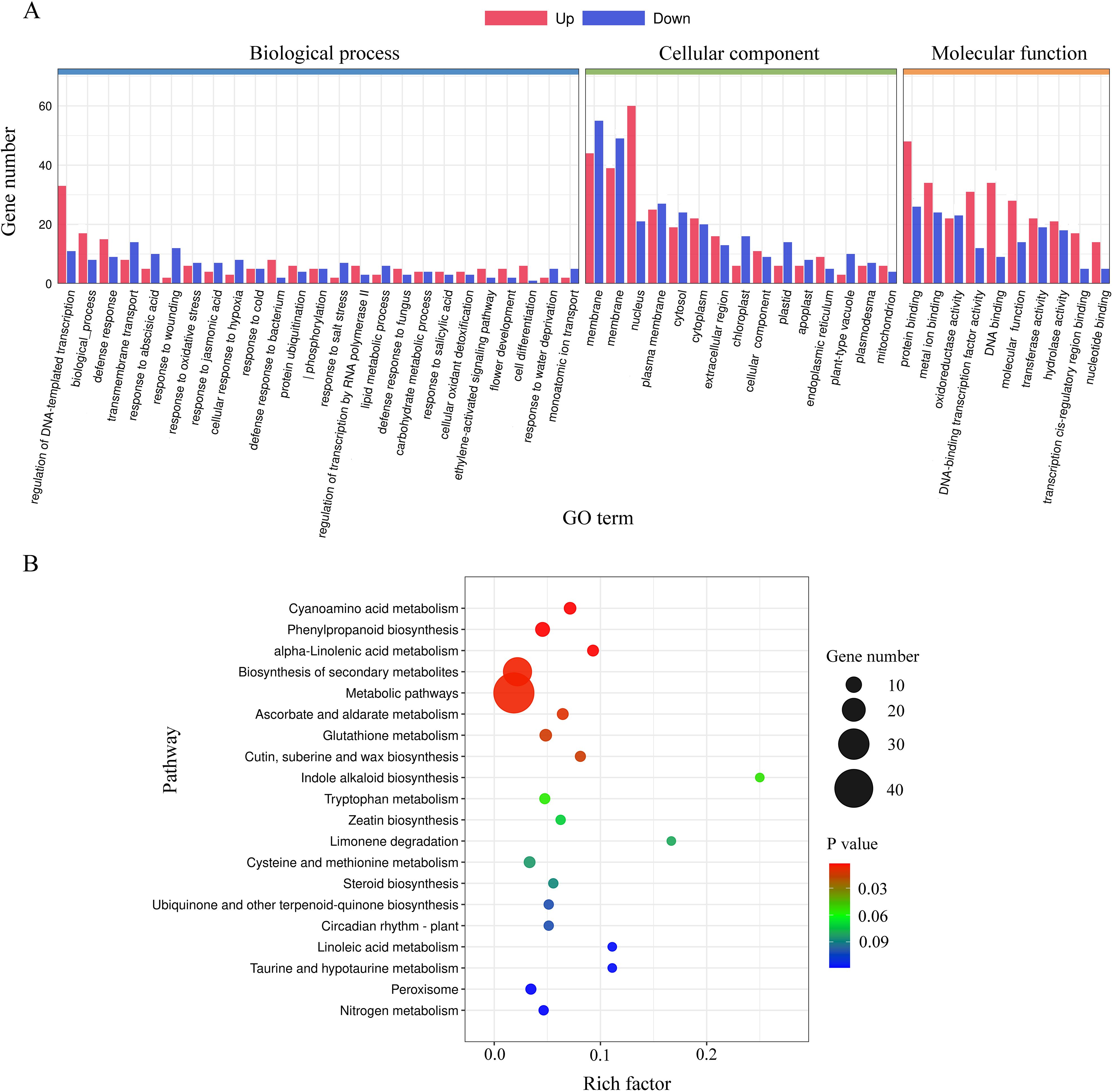
Figure 4. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes between lncRNA18313-OE and WT plants under 100 μM Cd stress. (A) GO enrichment analysis. The X-axis represents the GO term, while the Y-axis indicates the number of genes within each category. (B) KEGG enrichment analysis. The Y-axis indicates the KEGG pathway, the X-axis indicates rich factor, which is the ratio of differentially expressed gene numbers annotated in this pathway term to all gene numbers annotated in this pathway term. The color of the dot represents p-value, while the size of the dot represents the number of DEGs involved in the respective pathway.
3.5 LncRNA18313 regulates the expression of DEGs involved in antioxidative defense responses
RNA-Seq analysis indicated that genes associated with POD, SOD, and glutathione S-transferase (GST) were notably regulated in lncRNA18313-OE plants under Cd stress (Figure 5A). Compared to WT plants, 16 antioxidant enzyme genes were significantly upregulated in lncRNA18313-OE plants, including encoding three PODs (AT4G08770, AT5G58390, AT4G08780), two SODs (AT1G12520, AT1G08830), five catalases (CATs) (AT1G28480, AT3G02310, AT3G09940, AT3G28510, AT1G53860), and six ascorbate peroxidases (APXs) (AT5G24540, AT3G29240, AT4G21903, AT1G23300, AT3G25882, AT1G02450) (Figure 5A). Interestingly, three GST (AT2G29470, AT2G29420, AT2G29480) were significantly downregulated in lncRNA18313-OE plants compared to WT plants. To validate these findings, we further assessed the expression levels of POD (AT4G08770), APX (AT1G02450), SOD1 (AT1G08830), CAT (AT1G28480) by RT-qPCR in both WT and lncRNA18313-OE plants, with or without Cd treatment. Under normal condition, no dramatic difference were observed in the expression levels of the POD, APX, SOD1 and CAT between WT and lncRNA18313-OE plants. However, under Cd stress, the expression levels of POD, APX, SOD1 and CAT were significantly higher in the lncRNA18313-OE plants (Figure 5B). Furthermore, the expression trends of those four genes were also largely consistent with the RNA-Seq data, although minor variations in expression levels were observed.
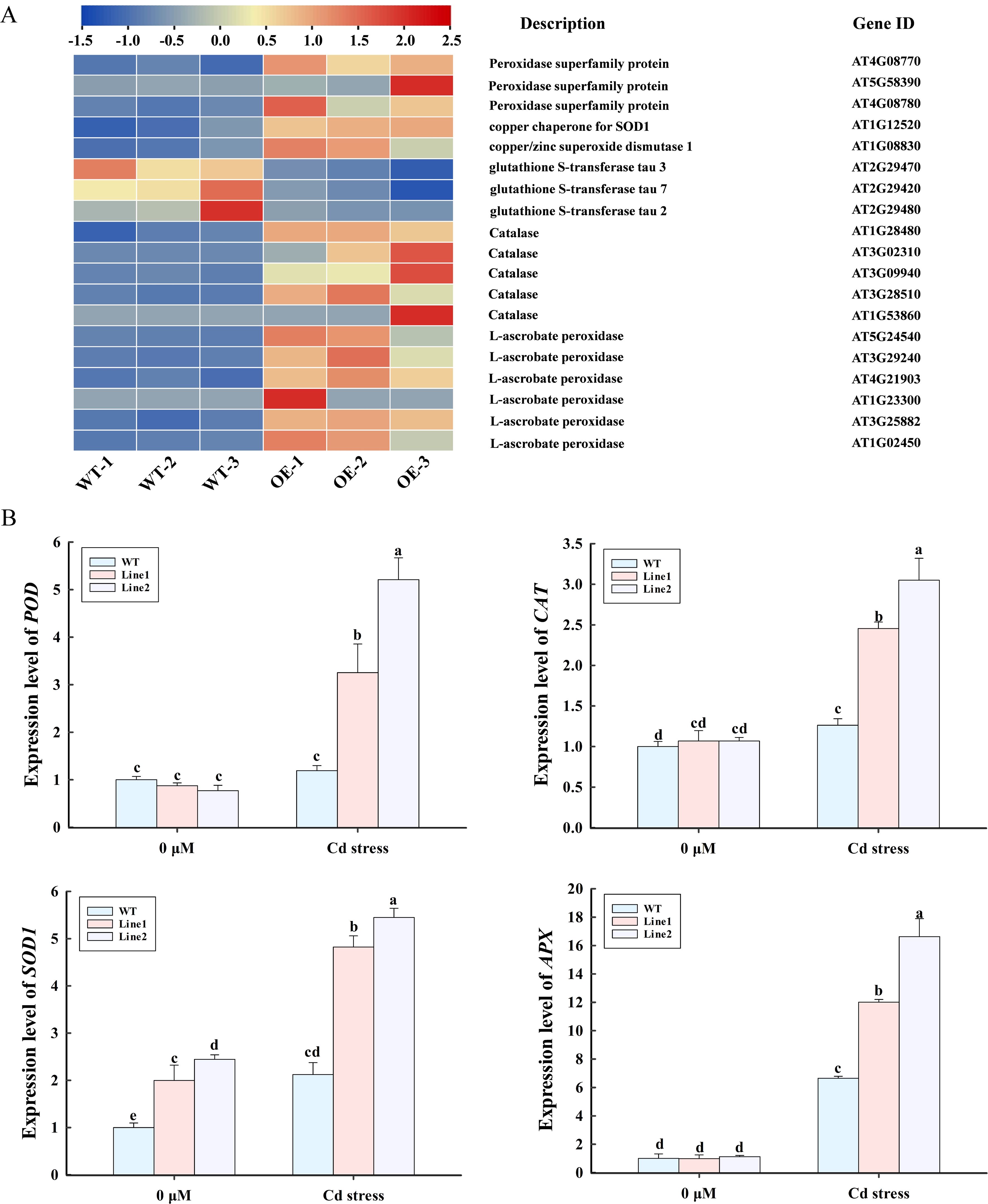
Figure 5. Effects of overexpressing lncRNA18313 on the expression of differentially expressed genes (DEGs) related to antioxidant enzymes in Arabidopsis under Cd stress. (A) Heatmap of DEG expressions. The bar represents the scale of the expression level for each gene as indicated by blue (low expression) and red rectangles (high expression). (B) RT-qPCR analysis of the expression profiles of four selected transcripts. 0 μM, no Cd treatment; Cd stress, 100 μM CdCl2. The bars (means ± SD, n = 3) labeled with different letters indicate significant differences (p < 0.05) between treatments as determined by one-way ANOVA with Duncan’s multiple range test.
3.6 LncRNA18313 regulates the expression of DEGs involved in transcriptional regulation
Transcriptome analysis in lncRNA18313-OE plants subjected to Cd stress revealed 43 differentially expressed transcription factors (TFs) from 14 TF families, with 35 upregulated and 8 downregulated (Supplementary Figure S2). Among them, 22 transcription factors encoding WRKY, MYB, bHLH and ERF were potentially involved in abiotic stress tolerance according to their functional annotation (Figure 6A). Compared to WT plants, 16 transcription factors genes, including encoding 7 WRKY (AT5G01900, AT5G22570, AT5G26170, AT2G40750, AT5G64810, AT3G56400, AT2G21900), 3 MYB (AT3G12820, AT1G57560, AT1G09540), 3 bHLH (AT4G00480, AT2G18300, AT4G36930), 3 ERF (AT5G61890, AT2G44840, AT4G32800) were significantly upregulated in lncRNA18313-OE plants under Cd stress (Figure 6A). In addition, 6 TF genes, including MYB (AT3G06490), bHLH (AT1G10585, AT1G10586, AT1G09530, AT1G43160), and ERF (AT5G64750), were significantly downregulated in the lncRNA18313-OE plants under Cd stress. Subsequently, the expression levels of 3 TFs, WRKY, MYB and ERF in lncRNA18313-OE plants under Cd stress were further validated by RT-qPCR. The results demonstrated a clear upregulation of the WRKY, MYB and ERF transcription factors in the lncRNA18313-OE plants under Cd stress when compared to the WT plants (Figure 6B). These findings suggest that these transcription factors, regulated by lncRNA18313, play a key role in mediating plant’s response to Cd-induced oxidative stress.
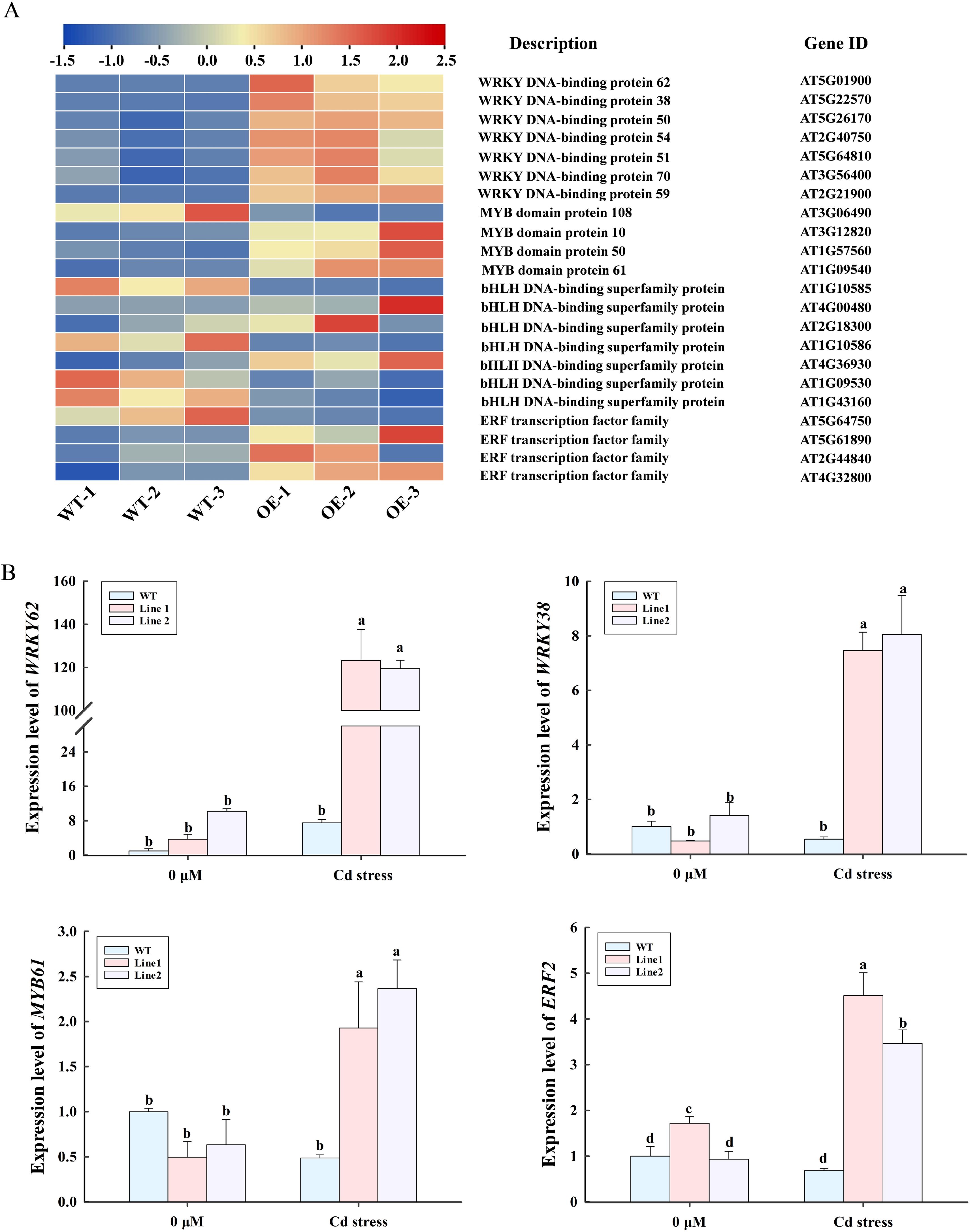
Figure 6. Effects of overexpressing lncRNA18313 on the expression of DEGs related to transcription factors in Arabidopsis under Cd stress. (A) Heatmap of DEG expression associated with transcription factors. The color scale represents gene expression levels, with blue indicating low expression and red indicating high expression. (B) RT-qPCR analysis of expression profiles in four selected transcripts under Cd stress. 0 μM, no Cd treatment; Cd stress, 100 μM CdCl2. Bars represent means ± SD (n = 3), with different letters indicating significant differences (p < 0.05) between treatments as determined by one-way ANOVA with Duncan’s multiple range test.
4 Discussion
4.1 LncRNA18313 enhances plant tolerance to Cd stress
With the rapid advancement of RNA-Seq technology, a growing number of lncRNAs have been emerging in various plant species, including key crops such as wheat and barley (Li et al., 2022; Bai et al., 2024). Some plant lncRNAs have been studied intensively, particularly those involved in responses to various stresses, such as cold stress in wheat (Lu et al., 2020) and cotton (Cao et al., 2021), drought stress in Arabidopsis (Qin et al., 2017), salt stress in Ginkgo biloba (Liu et al., 2025) and cotton (Zhang et al., 2021). However, the biological functions and molecular mechanisms of most lncRNAs still remain elusive, especially those involved in Cd stress in wheat. In our previous study, a Cd-responsive lncRNA, lncRNA18313 was identified in wheat through RNA sequencing (Zhu et al., 2023). The present study aims to functionally characterize TalncRNA18313 by overexpressing it in Arabidopsis, which lacks a homologous sequence. Because genetic transformation in wheat is still very difficult currently, we failed to generate transgenic wheat in this work. Therefore, we used Arabidopsis to overexpress TalncRNA18313. In order to explore whether lncRNA18313 is associated with plant tolerance to Cd stress, 6-week-old wild-type (WT) and TalncRNA18313-overexpressing Arabidopsis seedlings were incubated under 100 μM CdCl2 conditions in growth chambers. After 4 days of Cd stress, WT plants exhibited more severe damage compared to the lncRNA18313-OE Arabidopsis seedlings. Specifically, most of WT plant leaves were completely withered and dry, while the lncRNA18313-OE plants showed only slight damage, with fewer wilted leaves and a relatively healthy appearance compared to the WT. These results suggest that overexpression of TalncRNA18313 confers greater tolerance to Cd stress, similar to the function of other lncRNAs, such as lncRNA37228 in wheat (Zhu et al., 2023) and MSTRG.22608.1 in Populus tomentosa (Quan et al., 2021). To further characterize how TalncRNA18313 enhances Cd tolerance in Arabidopsis, the indicator of oxidative damage, malondialdehyde (MDA) was firstly measured. In the current study, MDA content increased significantly in WT plants under Cd stress. However, lncRNA18313-OE plants had slightly lower MDA content under Cd stress, indicating less oxidative damage to the membrane system. A similar reduction in MDA content was observed in DRIR (drought-induced lncRNA) overexpressing Arabidopsis plants under drought stress (Qin et al., 2017). Also, overexpression of wheat lncRNA37228 in Arabidopsis plants resulted in lower MDA content comparing to WT plants under 150 μM Cd treatment for 7 days (Zhu et al., 2023). Plants eliminate excessive reactive oxygen species (ROS) and mitigate oxidative damage by regulating the activities of key antioxidant enzymes, such as SOD, CAT and POD (Zhu et al., 2023; Wang et al., 2024). In this study, the activities of SOD, POD and CAT in the lncRNA18313-OE plant were significantly higher than those observed in WT plants under Cd stress. This result aligns with findings reported by Lu et al. (2020), who showed that overexpression of wheat lncRNA117 led to a significant increase in the activities of SOD and ascorbate peroxidase (APX) in transgenic Arabidopsis plants exposed to low-temperature stress. Similarly, Jia et al. (2024) demonstrated that overexpression of long non-coding RNA lncSIR1 from Betula platyphylla enhanced salt tolerance in transgenic Arabidopsis by boosting the activities of CAT, POD, and APX during salt stress for 21 days. Therefore, the increased activities of SOD, POD, and CAT in lncRNA18313-OE plants may play a crucial role in reducing oxidative damage, thereby contributing to improve Cd tolerance in Arabidopsis.
4.2 Regulation of transcription factors by lncRNA18313 confers Cd tolerance
GO enrichment analysis in the present study revealed that DEGs associated with response to oxidative stress were significantly enriched in lncRNA18313-OE plants under Cd stress compared to WT plants. Transcription factors (TFs) are critical in wheat response to various abiotic stresses, such as heavy metal (Zhu et al., 2022), salt (Wei et al., 2017) and drought (Xue et al., 2011), as demonstrated over the last decade. Many TFs from various families, such as ERF (ethylene responsive element binding protein), WRKY, bHLH (basic helix-loop-helix) and MYB (Myb-like DNA-binding domain) were considered as key players of transcriptional changes involved in plant stress adaptation response (Gao et al., 2016; Sheng et al., 2019). Li et al. (2021) studied the overexpression of CRIR1, a cold‐responsive intergenic lncRNA 1, in cassava and found that it positively modulated cold tolerance through upregulating MeNAC transcription factors. It was recently demonstrated that NAC transcription factors played a significant role in various abiotic stress responses (Shao et al., 2015). Overexpressing NAC in Arabdopsis has been found to confer salt stress in maize (Pan et al., 2024) and drought stress in wheat (Xue et al., 2011). Additionally, long non-coding RNA, LncY1 from birch improves salt tolerance by upregulating BpMYB96 transcription factors (Jia et al., 2023). Overexpression of MYB56 isolated from wheat, improved salt and freezing stress tolerance, accompanied by higher activities of POD, CAT and SOD in transgenic Arabidopsis (Zhang et al., 2012). In this study, sixteen transcription factors genes, including encoding seven WRKY, three MYB, three bHLH, three ERF were significantly upregulated in the lncRNA18313-OE plants exposed to Cd stress when compared to the WT plants. In birch (Betula platyphylla), overexpression of long noncoding RNA, BplncSIR1 in Arabidopsis enhanced salt tolerance by upregulating BpNAC2 to mediate ROS scavenging (Jia et al., 2024). In line with this, the RT-qPCR analysis further revealed a significant induction of WRKY, MYB and ERF transcripts in lncRNA18313-OE plants exposed to Cd stress. As reported previously, transgenic Arabidopsis overexpressing the TaMYB73, TaNAC29 and TaWRKY44 transcription factor genes from wheat, respectively significantly improved salt (He et al., 2012), drought (Huang et al., 2015) and cold (Wang et al., 2015) stress tolerance. Thus, the elevated transcriptional levels of NAC, WRKY and MYB transcription factor genes regulated by lncRNA18313 were likely to contribute to the increase tolerance of wheat seedlings to Cd stress. However, additional work is needed to test this possibility and to fully elucidate the function of lncRNA18313.
5 Conclusion
In this research, we functionally characterized the wheat long non-coding RNA, lncRNA18313, by overexpressing it in Arabidopsis. Through phenotypic, physiological, and transcriptomic analyses conducted in this work, we demonstrated that lncRNA18313 overexpression enhances Cd tolerance in the transgenic plants by modulating the expression of transcription factors to mediate ROS scavenging. However, our understanding of the complex regulatory role of lncRNA18313 is still in its infancy, and more attempts are still required to unravel its regulatory mechanism in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SZ: Formal Analysis, Methodology, Validation, Writing – original draft, Writing – review & editing. HB: Data curation, Methodology, Validation, Writing – original draft. ZF: Methodology, Validation, Writing – original draft. MZ: Formal Analysis, Supervision, Writing – review & editing. ZQ: Conceptualization, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Henan Province of China (242300420163) and Henan provincial key research projects for institutions of higher learning (25A180018).
Acknowledgments
Special thanks to reviewers for their valuable comments. In addition, the authors gratefully acknowledge every teacher, classmate, and friend who helped the authors with their experiment and writing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1583758/full#supplementary-material
References
Aebi, H. (1984). Catalase in vitro. Methods Enzymol. 105, 121–126. doi: 10.1016/S0076-6879(84)05016-3
Bai, Y., He, J., Yao, Y., An, L., Cui, Y., Li, X., et al. (2024). Identification and functional analysis of long non−coding RNA (lncRNA) and metabolites response to mowing in hulless barley (Hordeum vulgare L. var. nudum hook. f.). BMC Plant Biol. 24, 666. doi: 10.1186/s12870-024-05334-8
Bardou, F., Ariel, F., Simpson, C. G., Romero-Barrios, N., Laporte, P., Balzergue, S., et al. (2014). Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell. 30, 166–176. doi: 10.1016/j.devcel.2014.06.017
Cao, Y., Du, P., Zhang, J., Ji, J., Xu, J., and Liang, B. (2023). Dopamine alleviates cadmium stress in apple trees by recruiting beneficial microorganisms to enhance the physiological resilience revealed by high-throughput sequencing and soil metabolomics. Hortic. Res. 10, 112. doi: 10.1093/hr/uhad112
Cao, Z., Zhao, T., Wang, L., Zhao, T., Wang, L., Han, J., et al. (2021). The lincRNAXH123 is involved in cotton cold-stress regulation. Plant Mol. Biol. 106, 521–531. doi: 10.1007/s11103-021-01169-1
>Chen, L., Shi, S., Jiang, N., Khanzada, H., Wassan, G. M., Zhu, C., et al. (2018). Genome-wide analysis of long non-coding RNAs affecting roots development at an early stage in the rice response to cadmium stress. BMC Genomics 19, 460. doi: 10.1186/s12864-018-4807-6
>Chen, P., Song, Y., Liu, X., Xiao, L., Bu, C., Liu, P., et al. (2022). LncRNA PMAT-PtoMYB46 module represses PtoMATE and PtoARF2 promoting Pb2+ uptake and plant growth in poplar. J. Hazard Mater. 433, 128769. doi: 10.1016/j.jhazmat.2022.128769
Clough, S. J. and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Ding, Z., Wu, C., Tie, W., Yan, Y., He, G., and Wei, Hu. (2019). Strand-specific RNA-seq based identification and functional prediction of lncRNAs in response to melatonin and simulated drought stresses in cassava. Plant Physiol. Bioch. 140, 96–104. doi: 10.1016/j.plaphy.2019.05.008
Feng, S. J., Zhang, X. D., Liu, X. S., Tan, S. K., Chu, S. S., and Meng, J. G. (2016). Characterization of long non-coding RNAs involved in cadmium toxic response in Brassica napus. RSC Adv. 6, 82157. doi: 10.1039/C6RA05459E
Gao, Q., Li, X., Jia, J., Zhao, P., Byung-Hyun, L., Liu, G., et al. (2016). Overexpression of a novel cold-responsive transcript factor LcFIN1 from sheepgrass enhances tolerance to low temperature stress in transgenic plants. Plant Biotechnol. J. 14, 861–874. doi: 10.1111/pbi.12435
Giannopolitis, C. N. and Ries, S. K. (1977). Superoxide dismutase: I. occurrence in higher plants. Plant Physiol. 59, 309–314. doi: 10.1104/pp.59.2.309
Gui, Q., Yang, Z., Chen, C., Yang, F., Wang, S., and Dong, R. (2022). Identification and characterization of long noncoding RNAs involved in the aluminum stress response in Medicago truncatula via genome-wide analysis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1017869
He, Y., Li, W., Lv, J., Jia, Y., Wang, M., and Xia, G. (2012). Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 63, 1511–1522. doi: 10.1093/jxb/err389
Huang, Q., Wang, Y., Li, B., Chang, J. L., Chen, M. J., and Li, K. X. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15, 268. doi: 10.1186/s12870-015-0644-9
Jia, Y., Zhao, H., Niu, Y., and Wang, Y. (2023). Identification of birch lncRNAs and mRNAs responding to salt stress and characterization of functions of lncRNA. Hortic. Res. 10, uhac277. doi: 10.1093/hr/uhac277
Jia, Y., Zhao, H., Niu, Y., and Wang, Y. (2024). Long noncoding RNA from Betula platyphylla, BplncSIR1, confers salt tolerance by regulating BpNAC2 to mediate reactive oxygen species scavenging and stomatal movement. Plant Biotechnol. J. 22, 48–65. doi: 10.1111/pbi.14164
Li, S. X., Cheng, Z. H., Dong, S. M., Li, Z. B., Zou, L. P., Zhao, P. J., et al. (2021). Global identification of full-length cassava lncRNAs unveils the role of cold-responsive intergenic lncRNA 1 in cold stress response. Plant Cell Environ. 45, 412–426. doi: 10.1111/pce.14236
Li, C., Li, G., Wang, Y., Wang, J., Liu, H., Gao, W., et al. (2024). Supplementing two wheat genotypes with ZnSO4 and ZnO nanoparticles showed differential mitigation of Cd phytotoxicity by reducing Cd absorption, preserving root cellular ultrastructure, and regulating metal-transporter gene expression. Plant Physiol. Bioch. 206, 108199. doi: 10.1016/j.plaphy.2023.108199
Li, N., Liu, T., Guo, F., Yang, J., Shi, Y., Wang, S., et al. (2022). Identification of long non-coding RNA-microRNA-mRNA regulatory modules and their potential roles in drought stress response in wheat (Triticum aestivum L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1011064
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 24, 4333–4345. doi: 10.1105/tpc.112.102855
Liu, J., Wang, H., and Chua, N. H. (2015). Long noncoding RNA transcriptome of plants. Plant Biotech. J. 13, 319–328. doi: 10.1111/pbi.12336
Liu, S., Zhang, H., Meng, Z., Jia, Z., Fu, F., Jin, B., et al. (2025). The LncNAT11-MYB11-F3’H/FLS module mediates flavonol biosynthesis to regulate salt stress tolerance in Ginkgo biloba. J. Exp. Bot. 76, 1179–1201. doi: 10.1093/jxb/erae438
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real- time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, Q., Xu, Q., Guo, F., Lv, Y., Song, C., and Feng, M. (2020). Identification and characterization of long non-coding RNAs as competing endogenous RNAs in the cold stress response of Triticum aestivum. Plant Biol. 22, 635–645. doi: 10.1111/plb.13119
Pan, Y., Han, T., Yang, X., Wang, C., and Zhang, A. (2024). The transcription factor ZmNAC84 increases maize salt tolerance by regulating ZmCAT1 expression. Crop J. 12, 1344–1356. doi: 10.1016/j.cj.2024.09.005
Qin, T., Zhao, H., Cui, P., Albesher, N., and Xiong, L. M. (2017). A nucleus localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 75, 1321–1336. doi: 10.1104/pp.17.00574
Quan, M., Liu, X., Xiao, L., Chen, P., Song, F., Lu, W., et al. (2021). Transcriptome analysis and association mapping reveal the genetic regulatory network response to cadmium stress in Populus tomentosa. J. Exp. Bot. 72, 576–591. doi: 10.1093/jxb/eraa434
Rasheed, A., AL-Huqail, A. A., Ali, B., Alghanem, S. M., Shah, A. A., Azeem, F., et al. (2024). Molecular characterization of genes involved in tolerance of cadmium in Triticum aestivum (L.) under Cd stress. J. Hazard. Mater. 464, 132955. doi: 10.1016/j.jhazmat.2023.132955
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00902
Sheng, Y., Yan, X., Huang, Y., Han, Y., Zhang, C., Ren, Y., et al. (2019). The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 42, 891–903. doi: 10.1111/pce.13457
Thind, S., Hussain, I., Ali, S., Hussain, S., Rasheed, R., Ali, B., et al. (2020). Physiological and biochemical bases of foliar silicon-induced alleviation of cadmium toxicity in wheat. J. Soil Sci. Plant Nutt. 4, 2714–2730. doi: 10.1007/s42729-020-00337-4
Wang, L., Shi, Y. P., Tang, Y. N., Xian, B. S., Ren, X. T., Ren, M. Y., et al. (2025). The tartary buckwheat FtMYB46-FtNRAMP3 module enhances plant lead and cadmium tolerance. Plant Cell Environ. doi: 10.1111/pce.15518
Wang, Y., Sun, S., Feng, X., Li, N., and Song, X. (2024). Two lncRNAs of Chinese cabbage confer Arabidopsis with heat and drought tolerance. Vegetable Res. 4, e029. doi: 10.48130/vegres-0024-0029
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., and Sun, T. (2015). Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00615
Waseem, M., Yang, X., Aslam, M. M., Li, M., Zhu, L., Chen, S., et al. (2022). Genome-wide identification of long non-coding RNAs in two contrasting rapeseed (Brassica napus L.) genotypes subjected to cold stress. Environ. Exp. Bot. 201, 104969. doi: 10.1186/s12870-020-2286-9
Wei, Q., Luo, Q., Wang, R., Zhang, F., He, Y., Zhang, Y., et al. (2017). A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01374
Wei, L., Zhang, R., Zhang, M., Xia, G., and Liu, S. (2022). Functional analysis of long non-coding RNAs involved in alkaline stress responses in wheat. J. Exp. Bot. 73, 5698–5714. doi: 10.1093/jxb/erac211
Wen, X., Ding, Y., Tan, Z., Wang, J., Zhang, D., and Wang, Y. (2020). Identification and characterization of cadmium stress-related lncRNAs from Betula platyphylla. Plant Sci. 299, 110601. doi: 10.1016/j.plantsci.2020.110601
Wierzbicki, A. T., Blevins, T., and Swiezewski, S. (2021). Long non-coding RNAs in plants. Annu. Rev. Plant Biol. 72, 245–271. doi: 10.1146/annurev-arplant-093020-035446
Xue, G. P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., and McIntyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi: 10.1093/mp/ssr013
Zhang, X., Dong, J., Deng, F., Wang, W., Cheng, Y., Song, L., et al. (2019). The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 19, 459. doi: 10.1186/s12870-019-2088-0
Zhang, J. X. and Kirham, M. B. (1994). Drought stress-induced changes in activities of superoxide dismutase, catalase and peroxidase in wheat species. Plant Cell Physiol. 35, 785–791. doi: 10.1016/S0168-9452(98)00246-5
Zhang, D., Liu, J., Zhang, Y., Wang, H., Wei, S., Xu, Z., et al. (2023). Morphophysiological, proteomic and metabolomic analyses reveal cadmium tolerance mechanism in common wheat (Triticum aestivum L.). J. Hazard Mater. 445, 130499. doi: 10.1016/j.jhazmat.2022.130499
Zhang, X., Shen, J., Xu, Q., Dong, J., Song, L., Wang, W., et al. (2021). Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton. Plant Cell Environ. 44, 3302–3321. doi: 10.1111/pce.14133
Zhang, D., Wang, H., Zhang, Y., Su, Z., Hu, T., Liu, J., et al. (2024). Methyl jasmonate enhances the safe production ability of Cd-stressed wheat by regulating the antioxidant capacity, Cd absorption, and distribution in wheat. Plant Physiol. Bioch. 212, 108788. doi: 10.1016/j.plaphy.2024.108788
Zhang, L. C., Zhao, G. Y., Xia, C., Jia, J. Z., Liu, X., and Kong, X. Y. (2012). Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic Arabidopsis. Gene 505, 100–107. doi: 10.1016/j.gene.2012.05.033
Zhao, M., Tian, R., Sun, X., and Zhang, W. H. (2023). lncRNA MtCIR2 positively regulates plant-freezing tolerance by modulating CBF/DREB1 gene clusters. Plant Cell Environ. 46, 2450–2469. doi: 10.1111/pce.14611
Zhu, M., Duan, X., Zeng, Q., Liu, Y., and Qiu, Z. (2022). He-Ne laser irradiation ameliorates cadmium toxicity in wheat by modulating cadmium accumulation, nutrient uptake and antioxidant defense system. Ecotoxi. Environ. Safe. 236, 113477. doi: 10.1016/j.ecoenv.2022.113477
Zhu, M., Liu, Y., Bai, H., Zhang, W., Liu, H., and Qiu, Z. (2023). Integrated physio-biochemical and RNA sequencing analysis revealed mechanisms of long non-coding RNA-mediated response to cadmium toxicity in wheat. Plant Physiol. Bioch. 203, 108028. doi: 10.1016/j.plaphy.2023.108028
Keywords: long non-coding RNA, wheat, Cd stress, RNA-Seq, heavy metal
Citation: Zhao S, Bai H, Fan Z, Zhu M and Qiu Z (2025) A long non-coding RNA lncRNA18313 regulates resistance against cadmium stress in wheat. Front. Plant Sci. 16:1583758. doi: 10.3389/fpls.2025.1583758
Received: 26 February 2025; Accepted: 19 May 2025;
Published: 02 June 2025.
Edited by:
Xinyang Wu, China Jiliang University, ChinaCopyright © 2025 Zhao, Bai, Fan, Zhu and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongbo Qiu, cWl1em9uZ2JvQDEyNi5jb20=
 Sujing Zhao
Sujing Zhao Hongxia Bai
Hongxia Bai Ziyi Fan1
Ziyi Fan1 Mo Zhu
Mo Zhu Zongbo Qiu
Zongbo Qiu