- 1Institute of Cash Crops, Hebei Academy of Agriculture and Forestry Sciences, Shijiazhuang, China
- 2Institute of Coastal Agriculture, Hebei Academy of Agriculture and Forestry Sciences, Tangshan, China
- 3Department of Horticulture, University of Arkansas, Fayetteville, AR, United States
Introduction: Excessive nitrate accumulation in leafy vegetables poses health risks and indicates inefficient nitrogen use in conventional agriculture. While nitrogen metabolism has been extensively studied, the molecular mechanisms linking nutrient deprivation to nitrate reduction in vegetables remain unclear.
Methods: In this study, we investigated these mechanisms in lettuce (Lactuca sativa var. ramosa Hort.) by analyzing nitrate content and gene expression in leaves and roots under nutrient deprivation.
Results: After five days of treatment, nitrate content decreased by 57.49% in leaves and 50.77% in roots. Transcriptome sequencing identified 323 and 3,494 differentially expressed genes (DEGs) in leaves and roots, respectively, with 78 shared DEGs. KEGG enrichment analysis revealed involvement of DEGs in pyrimidine metabolism, base excision repair, hormone signaling, terpenoid biosynthesis, and triglyceride metabolism, indicating cross-talk between nitrate regulation and stress responses. Nitrate transporter genes NRT2.4 and NRT2.5 were upregulated in roots, while NRT1 was induced in leaves, suggesting enhanced nitrate redistribution. Additionally, antioxidant genes such as POD, LOX, and cytochrome P450 were upregulated in roots, whereas SODC was downregulated in both tissues. These results suggest that lettuce responds to nutrient deprivation by activating nitrate transport and antioxidant pathways to reduce nitrate levels and enhance nitrogen use efficiency.
Discussion: This study provides a foundation for optimizing pre-harvest strategies to improve lettuce quality and identifies candidate genes (e.g., NRT2.5, LOX) for breeding low-nitrate varieties suited for nitrogen-limited environments.
Introduction
Nitrogen is indispensable for the synthesis of amino acids, nucleotides, and chlorophyll (Tatiana et al., 2011; Meng et al., 2015; Xiong et al., 2018; Reddy et al., 2020). Nitrate, the primary form of nitrogen absorbed by plants, is taken up through roots and assimilated into organic compounds (Vandna et al., 2024). The absorption and metabolism of nitrate are influenced by multiple factors, including nutrient availability, environmental conditions, and the physiological state of the plant (Yuan et al., 2021; Rui et al., 2023; Vandna et al., 2024). Nutrient availability in the soil is not only affected by nitrogen fertilizer application strategies but also by factors such as soil pH, ion competition, and microbial activity. Environmental conditions, such as photoperiod, can further regulate nutrient uptake through hormone signaling pathways (Noorden et al., 2016; Dai et al., 2024). From a physiological perspective, nitrate demand varies across developmental stages—being higher during vegetative growth—and is further influenced by abiotic stresses such as drought or nitrogen deficiency, which can activate nitrogen utilization pathways to maintain homeostasis, for example, through glutathione biosynthesis or reduced amino acid synthesis (Li et al., 2023). However, nitrogen regulation and utilization are complex, and plant responses to nitrogen deficiency can vary across species and conditions (Yin et al., 2019). These complex regulatory networks form the molecular basis for precision agriculture practices aimed at improving nitrogen use efficiency. Future research must therefore integrate multi-omics technologies to elucidate the synergistic interactions among environmental signals, gene expression, and metabolism.
Lettuce (Lactuca sativa var. ramosa) is a widely cultivated leafy vegetable with high nutritional value (Kim et al., 2016). However, its classification as a high-nitrogen-demand crop presents a critical challenge: while sufficient nitrogen promotes biomass accumulation, excessive nitrate accumulation in edible tissues poses food safety risks. Epidemiological data estimate that vegetables contribute approximately 80% of total dietary nitrate intake in humans (Liu et al., 2011a; Li et al., 2013), making nitrate management in crops like lettuce a priority for sustainable agriculture. Previous research indicates that nutrient deprivation can trigger nitrate remobilization through autophagy-mediated protein degradation and upregulation of vacuolar nitrate efflux transporters (Yuan et al., 2007; Rengel et al., 2022). However, tissue-specific responses—especially in leafy vegetables where leaf nitrate content directly impacts food safety—remain poorly understood. Lettuce is highly responsive to nutrient availability, and nutrient deprivation not only affects its growth and productivity but also significantly influences nitrate content, with direct implications for human health (Zhao et al., 2024). Thus, studying nutrient deprivation in lettuce offers a unique opportunity to deepen our understanding of nitrate metabolism and provide foundational knowledge for high-yield, high-quality hydroponic cultivation. Despite its importance, the mechanisms by which nutrient deprivation influences nitrate metabolism in lettuce are still insufficiently explored.
Transcriptome analysis offers powerful insights into gene expression changes under specific physiological conditions. Transcription factors (TFs) play crucial roles in modulating gene expression and regulating metabolic pathways (Fan et al., 2023). Advances in transcriptomics have enabled the identification of key genes and TFs involved in metabolite biosynthesis and stress responses (Huang et al., 2023). For example, Dof1.7 and NIGT1 TFs mediate complex regulation of NRT2 gene expression under nitrogen deficiency (Zhuo et al., 2024), while transcriptomic studies in wheat have revealed calcium-mediated signaling pathways involved in nitrogen stress tolerance (Yan et al., 2021). These findings highlight the potential of tissue-specific transcriptomics to uncover both conserved and species-specific regulatory mechanisms. Nonetheless, current knowledge is largely derived from model species and cereals, with limited data available for fast-growing leafy vegetables, in which nitrate accumulation has direct consequences for consumer health and marketability.
Optimizing nutrient management can effectively reduce nitrate accumulation in crops. For instance, Song et al. (2023) demonstrated that nutrient deprivation five days prior to harvest significantly lowered nitrate content in Brassica rapa ssp. chinensis. This study provides a useful reference for our research. Here, we examine the impact of nutrient deprivation on nitrate content in lettuce under standard and reduced nutrient conditions. Using transcriptome sequencing, we analyze gene expression patterns in lettuce roots and leaves. This study establishes a theoretical framework for understanding the molecular response of lettuce to nutrient deprivation and provides practical guidance for optimizing nitrogen management in lettuce production systems.
Materials and methods
Plant materials and experimental conditions
The lettuce variety Lactuca sativa (Dasusheng, Beijing Fengming Yashi Technology Development Co., Ltd., Beijing, China) was selected for this study. Seeds were sown in cavity trays filled with a substrate mixture of cottonseed husk and vermiculite (2:1 ratio).
Lettuce was grown under hydroponic conditions, following the methodology outlined in our previous study (Song et al., 2023). After 25 days of growth in a complete nutrient solution, seedlings exhibiting consistent growth were selected and transferred to two identical hydroponic racks for cultivation. One rack was subjected to nutrient deprivation, where the nutrient solution was completely replaced with water, thereby completely eliminating the supply of essential nutrients to the plants. The other hydroponic rack maintained unchanged culture conditions and served as the control group (CK). A harvest of the control group plants was performed just before the onset of nutrient deprivation, corresponding to day 0 (D0) of the nutrient deprivation experiment.
Determination of biomass and nitrate content
On the day of nutrient deprivation application (D0), control plants were harvested in three replicates, each consisting of four plants, to ensure an adequate sample size for subsequent analysis. Samples were collected daily thereafter. The fresh weight of each tissue sample was recorded, and each sample was then divided into two homogeneous batches. One batch was stored at -80°C for molecular analysis, while the other was used for nitrate content determination. Nitrate content was measured using salicylic acid-sulfuric acid colorimetry, as described by Cao et al. (2007). Data were analyzed using one-way ANOVA followed by Tukey’s test (SPSS v27.0).
RNA extraction and Illumina sequencing
On the 5th day, leaves (CL, TL) and roots (CR, TR) from both the control group and the nutrient deprivation treatment were immediately frozen in liquid nitrogen and stored at -80°C. The tissue samples were then sent to BMK for transcriptome sequencing.
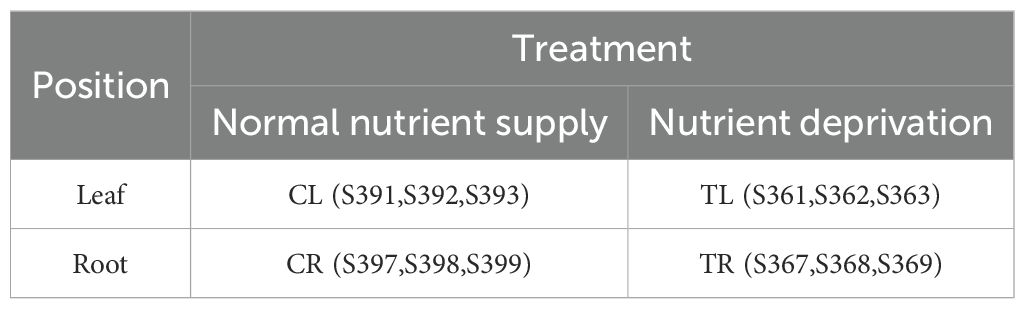
Total RNA was extracted from the leaves and roots of lettuce under both conditions (CL/CR for normal nutrient supply and TL/TR for nutrient deprivation) using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE). Twelve libraries representing four sample groups (three replicates per group) were constructed for transcriptome sequencing. RNA quality, quantification, and sequencing were conducted with the assistance of BMKCloud (www.biocloud.net). Following cluster generation, the library preparations were sequenced on an Illumina platform, and paired-end reads were produced. The transcriptomic samples and their corresponding experimental groupings are detailed in Table 1.
Screening and functional analysis of DEGs
The sequencing data (clean data) were aligned using HISAT2 (v2.2.1) software, and the reads were spliced and assembled with StringTie to obtain reference sequences for subsequent analysis. High-quality reads were mapped to the spliced transcriptome to calculate gene expression levels. DEGs were identified using a fold change ≥ 1.5 and a P-value ≤ 0.01 as the screening criteria. Gene Ontology (GO) and KEGG enrichment analyses were then performed on the identified DEGs to explore their functional roles.
Real-time PCR
Quantitative reverse transcription PCR (qRT-PCR) was performed to validate the RNA-seq results. RNA samples were reverse-transcribed using the Evo M-MLV RT Mix Kit (Accurate Biology). Primers for the analysis were designed with Primer 5.0 software (PREMIER Biosoft) (Table 2). The expression of the Lactuca sativa actin gene (LSAT_V11C800436410) was used as internal control. SG Green qPCR Mix (Bysbio) was used for real-time PCR reactions. The Ct values of DEGs and the internal reference gene were measured, and each gene was tested in triplicate. The 2-ΔΔCt method was employed to calculate the relative expression levels of the genes.
Results
Nutrient deprivation triggers nitrate content reduction
The biomass of lettuce remained unaffected during the first five days of nutrient deprivation (Figure 1). While the nitrate content in both the leaves and roots of lettuce exhibited a rapid decline. The most substantial reduction in nitrate content occurred on day 3 in the leaves and day 2 in the roots. Overall, nitrate levels decreased by 57.49% in the leaves and 50.77% in the roots. By the end of the treatment, nitrate concentrations remained within the prescribed limit for green, pollution-free vegetables (less than 3000 mg/kg) (Figure 2). It shows that short-term nutrient deprivation can be used as an effective nitrate reduction strategy.
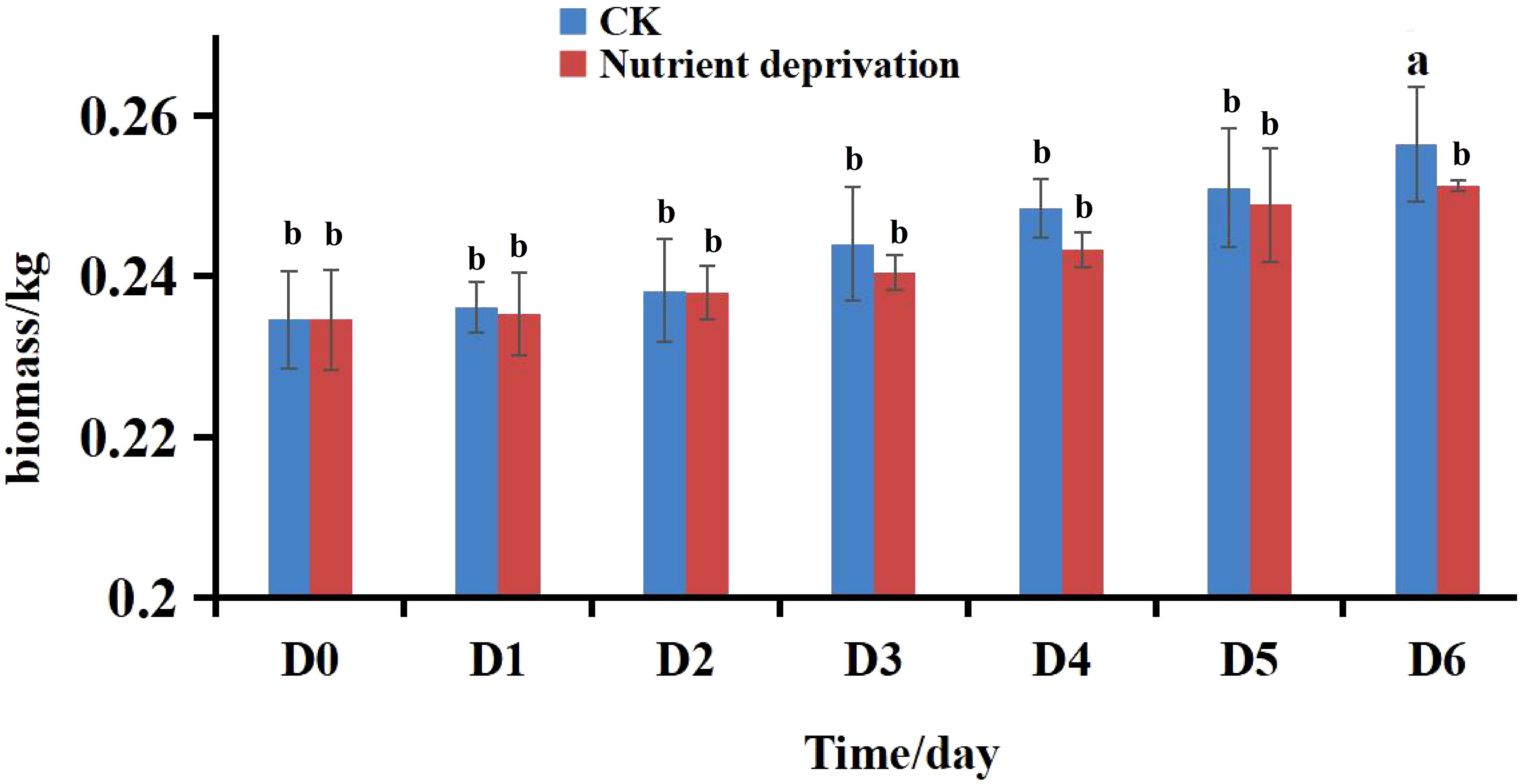
Figure 1. Biomass of lettuce under different conditions. Different lowercase letters in the same day indicate significant differences among different treatments at 0.05 level.

Figure 2. Nitrate content of lettuce (A) leaf; (B) root. Data represent means ± standard deviation, and asterisks indicate significant differences (*p<0.05, ***p<0.001).
Transcriptome sequencing and alignment
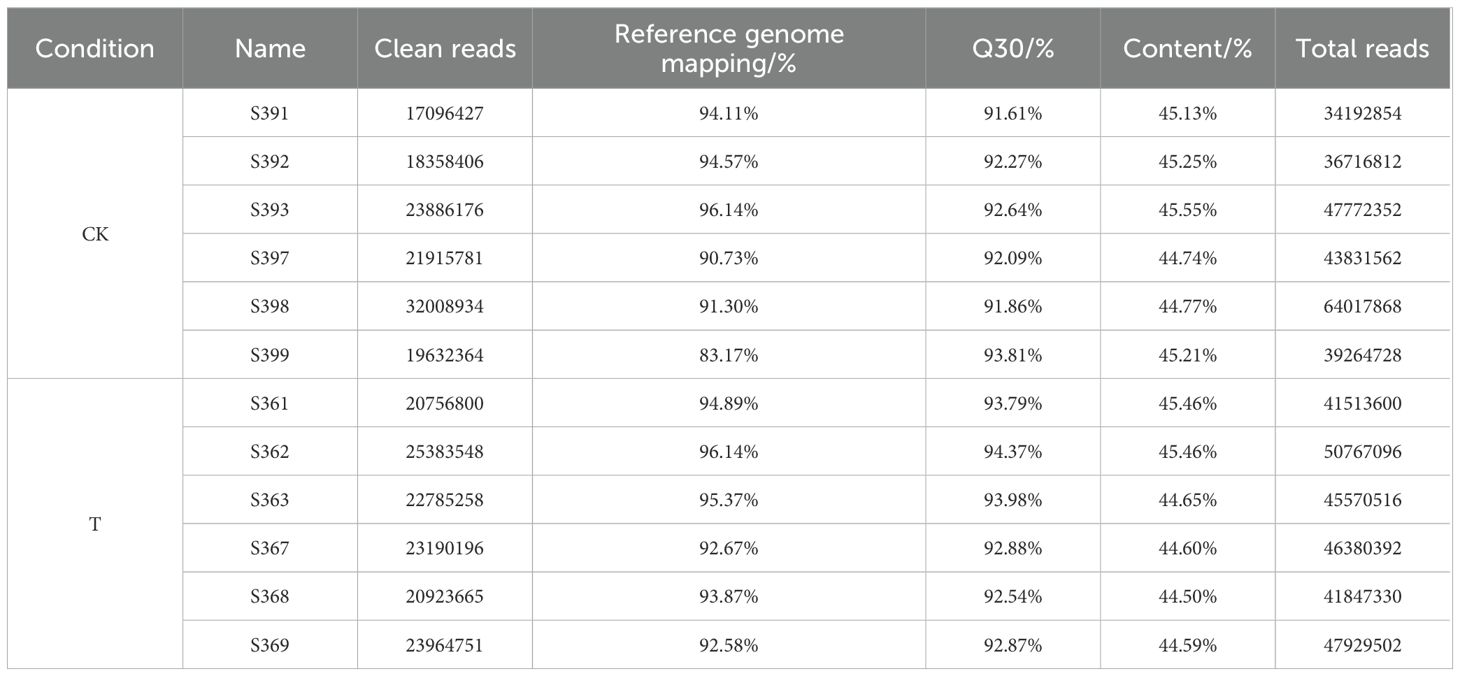
Transcriptome sequencing of lettuce roots and leaves generated a total of 80.86 Gb of clean data after quality control (Table 3). The sequencing quality was high, with the percentage of Q30 bases (sequencing error rate ≤ 0.1%) in all samples exceeding 91.61%. The GC content of the samples ranged from 44.50% to 45.55%. The comparison efficiency between the reads of each sample and the reference genome ranged from 83.17% to 96.14%, indicating strong alignment and providing reliable data for further analysis.
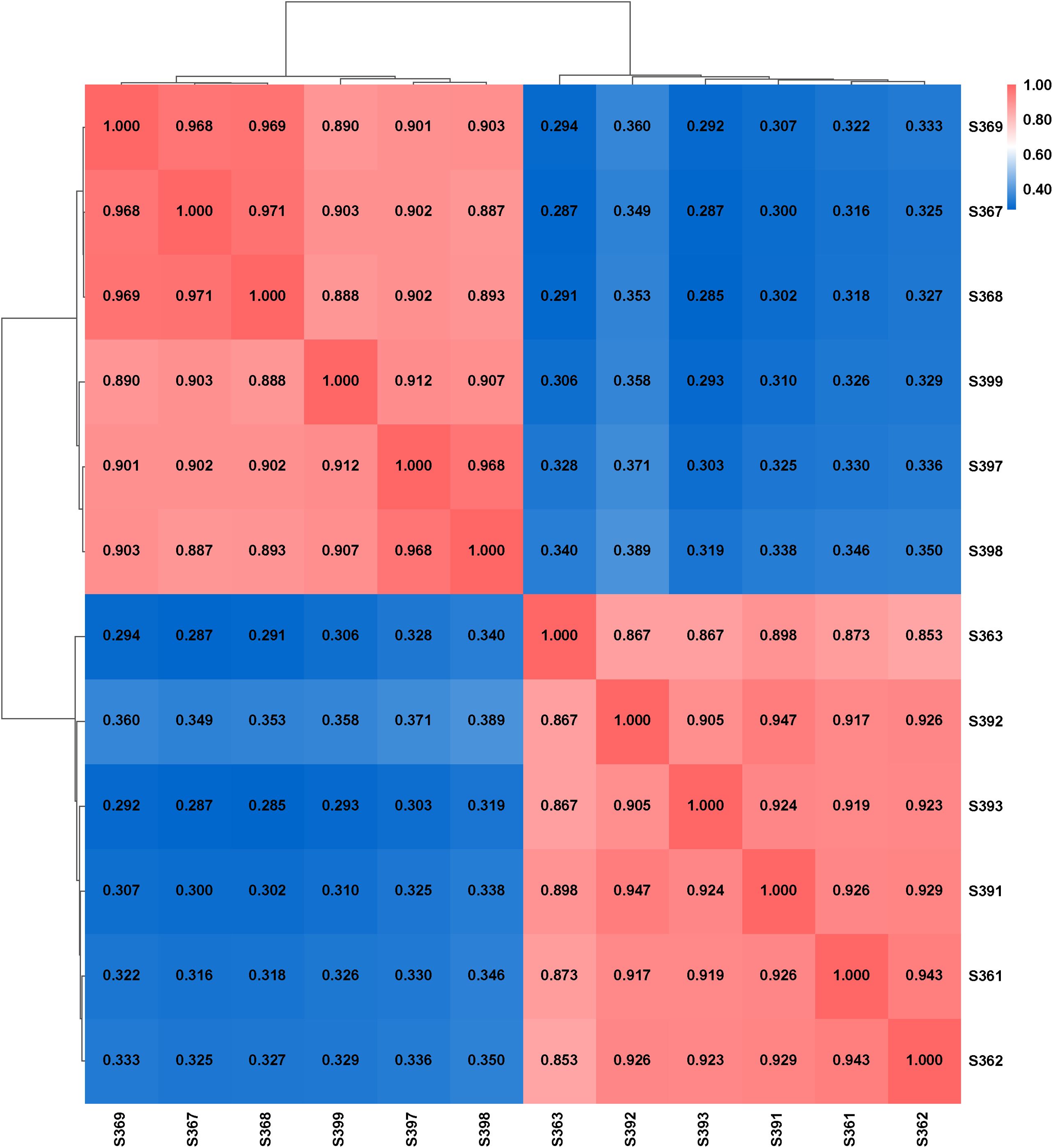
Spearman’s correlation coefficient (r) was used to assess the correlation between biological replicates based on the quantitative FPKM (Fragments Per Kilobase of exon model per Million mapped reads) results. The closer the correlation coefficient (r²) is to 1, the stronger the correlation between the two replicate samples. A heat map (Figure 3) was generated to visualize gene expression correlations across the 12 samples. As shown in the figure, the correlation between the three biological replicates for both the leaf and root samples remained consistently above 0.853. This indicates a strong correlation among the replicates, which supports the reliability of the experimental results and the appropriateness of the sample selection.
DEGs in leaves and roots of lettuce under nutrient deprivation
DEGs between nutrient deprivation and control samples were screened and identified using the DESeq2_edgeR. DEGs were selected based on a fold change (FC) ≥ 1.5 and a significant difference (P-value ≤ 0.01). To identify genes associated with nitrate accumulation and explore the relationship between DEGs in lettuce roots and leaves under nutrient deprivation conditions, expression levels were analyzed and compared with the control group. Differential expression analysis revealed 323 DEGs in the leaves, consisting of 212 upregulated genes and 111 downregulated genes. In the roots, 3494 DEGs were identified, with 1894 upregulated genes and 1600 downregulated genes (Figure 4A). Notably, 78 DEGs were found in both the leaves and roots (Figure 4B). The number of DEGs in roots was greater than those in leaves, indicating that the roots undergo more significant differential expression, which might be because roots were the first to respond to stress stimuli.

Figure 4. DEGs analysis of leaves and roots under nutrient deprivation conditions. (A) Volcano plot of DEGs of different positions (roots, leaves) under nutrient deprivation. (B) Venn diagram displaying the DEGs in leaf (CL_vs_TL) and root (CR_vs_TR) under nutrient deprivation.
191 DEGs in the leaves and 1955 DEGs in the roots were enriched in three main GO categories: biological process, cellular component, and molecular function (Figure 5), which suggest that the DEGs play roles in essential cellular processes, highlighting their potential involvement in nitrate accumulation and the plant’s response to nutrient deprivation.

Figure 5. GO analysis of DEGs (red) biological process; (green) cell component; (blue) molecular function.
Low nitrogen stress impacts secondary metabolic pathways in plants, aiding in stress mitigation and damage repair. These pathways include phenylpropanoid metabolism, with sub-pathways like lignin and flavonoid biosynthesis. Studies have shown that regulatory factors influence nitrogen metabolism by modulating target genes at the transcriptional level (Liu et al., 2024b). To further explore the metabolic responses of lettuce to nutrient deprivation, the top 20 most significantly enriched pathways in DEGs from both leaves and roots were analyzed (Figure 6).
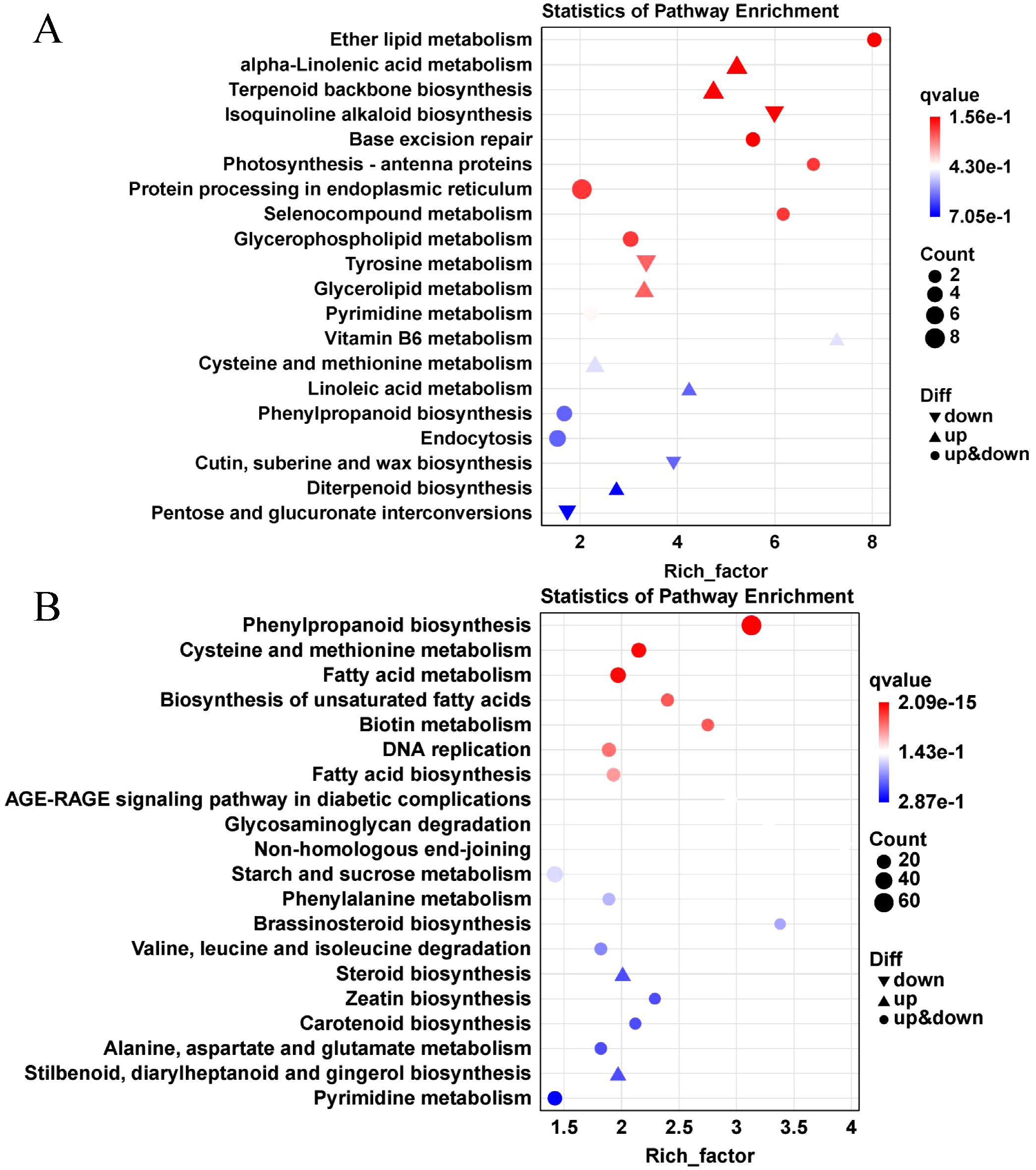
Figure 6. The top 20 KEGG pathway enrichment analysis of DEGs after nutrient deprivation. The picture shows the top 20 KEGG pathway enrichment bubble of DEGs in leaves (A) and roots (B) after nutrient deprivation; The X-axis represents the enrichment factor value (Rich Factor). The larger the data, the more obvious the enrichment results. The Y-axis represents the path name. The size of the dots represents the number of DEGs. The depth of the dots indicates the Q-value, the smaller the value, the more significant the enrichment results.
In leaves, DEGs were primarily enriched in pathways such as protein processing in the endoplasmic reticulum (duress-based repair), and plant hormone signal transduction (growth regulation). In roots, DEGs were enriched in phenylpropanoid biosynthesis (antioxidant), starch and sucrose metabolism (carbon-nitrogen balance), and amino acid biosynthesis (nitrogen reuse). These pathways are involved in antioxidant defense and growth regulation, indicating the plant’s adaptive mechanisms to nutrient deprivation stress.
Functional enrichment analysis of upregulated and downregulated DEGs in lettuce leaves and roots after nutrient deprivation was conducted using the KEGG database. The results (Figure 7) revealed the following:
● Leaves: The upregulated DEGs were significantly enriched in α-linolenic acid metabolism, terpenoid backbone biosynthesis, cysteine and methionine metabolism, endocytosis, glycerolipid metabolism, and glycerophospholipid metabolism. The downregulated DEGs were concentrated in protein processing in the endoplasmic reticulum, starch and sucrose metabolism, isoquinoline alkaloid biosynthesis, and tyrosine metabolism.
● Roots: The upregulated DEGs were mainly enriched in phenylpropanoid biosynthesis, starch and sucrose metabolism, ribosome function, fatty acid metabolism, plant-pathogen interaction, amino acid biosynthesis, carbon metabolism, cysteine and methionine metabolism, and DNA replication. The downregulated DEGs were primarily involved in plant hormone signal transduction, purine metabolism, protein processing in the endoplasmic reticulum, spliceosome function, α-linolenic acid metabolism, and glutathione metabolism.
These findings highlight significant differences in the nitrogen response genes between the leaves and roots of lettuce, with distinct enrichment in pathways related to carbon metabolism (such as starch and sucrose metabolism) and nitrogen metabolism (such as amino acid metabolism) under nutrient deprivation conditions.
Analysis of DEGs associated with nutrient deprivation
To explore the molecular mechanisms underlying lettuce’s response to nutrient deprivation stress, we further screened key genes among the DEGs. A total of 130 TFs, classified into 23 families, were identified in the root transcripts (TR). The AR2/ERF family was the most abundant, with 35 genes (26.92%), followed by WRKY (25 genes, 19.23%), bHLH (16 genes, 12.31%), MYB (15 genes, 11.54%), and NAC (5 genes, 3.85%). In leaf tissues, 11 TFs were identified in response to nutrient deprivation stress, classified into 5 TF families, including ERF, bHLH, and MYB. Notably, 3 MYB family TFs (MYB3, MYB108, APL) were upregulated. A partial list of TFs involved in NO3- response is provided in Table 4. These TFs likely play a critical role in lettuce’s adaptation to nutrient deprivation stress.
Essential DEGs in nitrate transport and assimilation
To understand the effects of nutrient deprivation on nitrate transport and assimilation, we examined the nitrate accumulation mechanism in the roots and leaves of lettuce. The results revealed that one nitrate transporter protein, NRT1, and three SPX structural domain DEGs were upregulated in the leaves. In the roots, 16 nitrate transporters were detected, with high-affinity transporters NRT2.4 and NRT2.5 being upregulated, indicating that plants adapted to nutrient deprivation by enhancing nitrate assimilation. While, the remaining low-affinity transporters (NRT1) were downregulated, suggested that the root absorption capacity is inhibited. Additionally, 10 SPX domains were almost all downregulated. One assimilation enzyme, glutamine synthetase (GS) was upregulated, which can promote NH4+ assimilation. While two MADS-box family TFs were downregulated.
Interestingly, under low nitrogen stress, plants typically improve nitrogen absorption efficiency by inhibiting the expression of bHLH130 (Wang et al., 2022b). However, in this study, bHLH130 expression was upregulated after nutrient deprivation. These findings suggest that lettuce is assimilated by a unique nitrate redistribution strategy. And a partial list of TFs involved, or potentially involved, in NO3- response is provided in Table 4.
Therefore, the synergistic regulation of core transcription factors NRT2, NRT1 and bHLH family member bHLH130. These findings systematically clarify the molecular mechanism of nitrogen metabolism reprogramming of lettuce under nutrient deprivation, and provide target genes for the improvement of low nitrate varieties.
Effects of nutrient deprivation on the antioxidant systems of lettuce
The reactive oxygen species (ROS) scavenging system plays a crucial role in plant defense against abiotic stresses. Under stress conditions, plants enhance their protective mechanisms and survival competitiveness by increasing the production of secondary metabolites (Wang et al., 2022a). Following nutrient deprivation, an imbalance in free radical metabolism within plant cells leads to toxicity and damage, resulting in the accumulation of hydrogen peroxide and superoxide anion (O2-).
DEGs related to ROS-metabolizing enzymes were identified in lettuce roots. Most peroxidase (POD) DEGs were upregulated. Lipoxygenase (LOX), which generates ROS during the catalytic process of unsaturated fatty acids (Zhang et al., 2024c), showed upregulation in 4 out of 5 DEGs. Additionally, two copper/zinc superoxide dismutase (SODC) DEGs were downregulated, while most cytochrome P450 DEGs were upregulated. In the leaves, one LOX gene was upregulated, two SODC genes were downregulated, and most cytochrome P450 DEGs were upregulated. These results suggest that DEGs related to POD, LOX, and cytochrome P450 were predominantly upregulated in the roots under nutrient deprivation stress, while cytochrome P450 and LOX were widely upregulated in the leaves, and SODC-related DEGs were downregulated in both tissues.
Thus, we infer that the LOX activity in leaves is positively correlated with the POD activity in roots, indicating that roots and leaves adapt resistance through metabolic division of labor: roots are responsible for ROS clearance, and leaves preferentially activate lipid peroxidation signals.
qRT-PCR validation
To validate the RNA-seq data, 18 DEGs were randomly selected, and their expression profiles were confirmed via qRT-PCR (Figure 8). The results demonstrated that the expression trends observed through qRT-PCR were consistent with those from RNA-seq, confirming the reliability of the transcriptome data. This consistency supports the use of the RNA-seq data for further analysis with confidence.
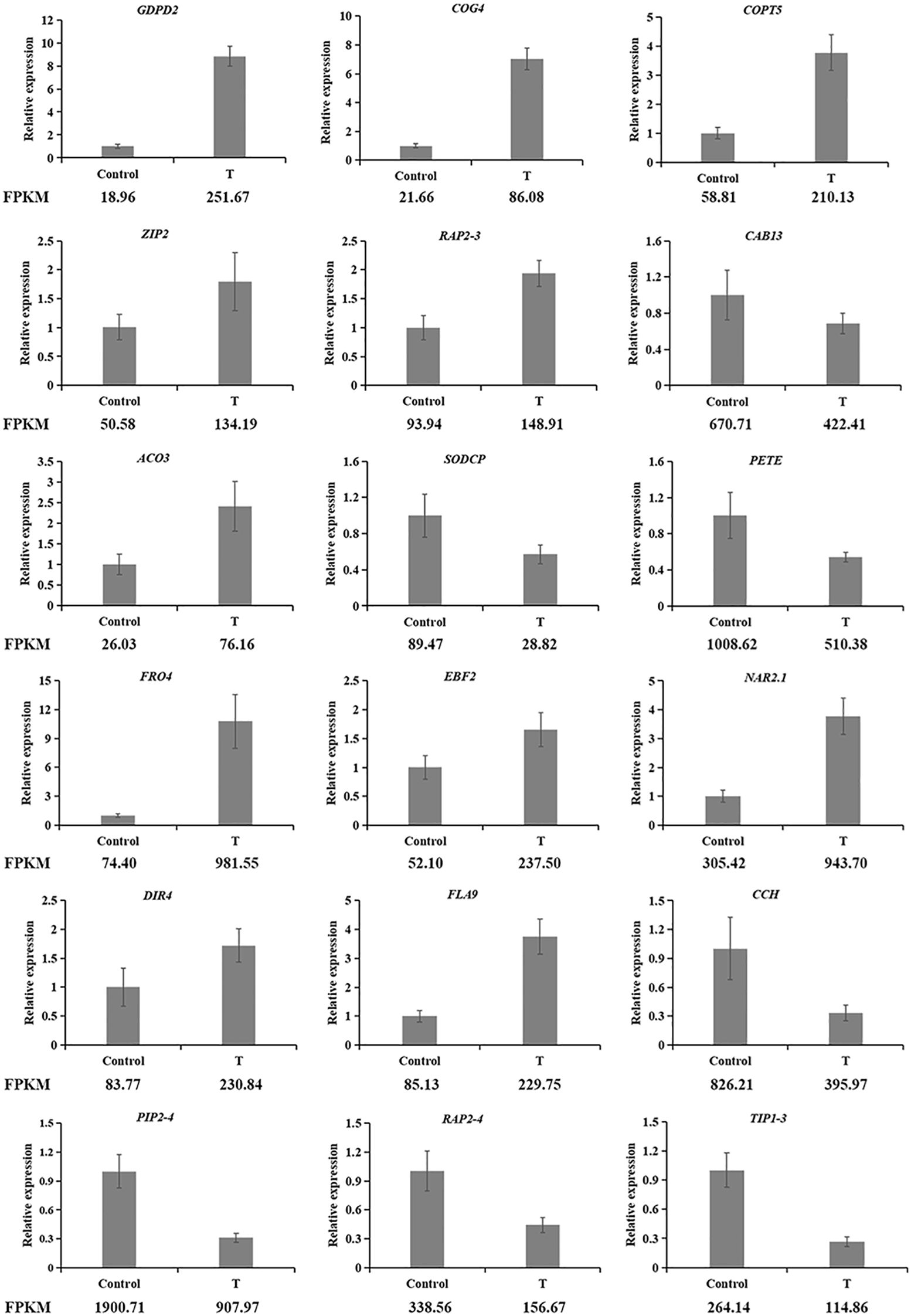
Figure 8. qRT-PCR vertification of DEGs identified by transcriptome analysis (LsACT was used as an internal control. GDPD2, COG4, COPT5, ZIP2, RAP2-3, CAB13, ACO3, SODCP and PETE are derived from the leaf; FRO4, EBF2, NAR2.1, DIR4, FLA9, CCH, PIP2-4, RAP2–4 and TIP1–3 are derived from the root.).
Discussion
Nitrogen is a vital element for plant growth and development, with nitrate being the primary form of nitrogen metabolism in vegetables, significantly influencing plant growth and development (Li et al., 2004; Liu et al., 2024a). While, nitrate acted as both a vital nutrient and a potential quality risk factor when accumulated excessively (Okushima et al., 2011). Our study corroborates that nutrient deprivation triggers a transcriptional reprogramming in lettuce, characterized by downregulation of nitrate transporters (e.g., NRT1.1) and activation of ammonium assimilation genes (e.g., GS). In flue-cured tobacco, midribs with threefold higher nitrate content than lamina exhibited suppressed expression of NRT2.4 and NRT2.5, mirroring our observation that lettuce roots upregulate these high-affinity transporters under nitrogen limitation to enhance nitrate scavenging efficiency. This parallel suggests NRT2.4/2.5 could serve as universal targets for nitrate reduction across leafy vegetables (Feng et al., 2022; Prasanna et al., 2023).
It has been observed that the nitrate content in vegetables decreases as plant age increases, and appropriate late harvesting can help reduce the nitrate content in vegetables (Liu et al., 2011b; Liu, 2014; Yi et al., 2015). In this study, the nitrate content was determined within 5 days after nutrient deprivation (Figure 2), confirming this trend. Additionally, transcriptome analysis of lettuce leaves and roots under normal and nutrient deprivation conditions revealed that upregulated DEGs after nutrient deprivation were primarily involved in cysteine and methionine metabolism, terpenoid backbone biosynthesis, endocytosis, and α-linolenic acid metabolism. The downregulated DEGs were mainly associated with phenylpropanoid biosynthesis, plant hormone signal transduction, carbon metabolism, isoquinoline alkaloid biosynthesis, and purine metabolism (Figure 6). These findings align with previous analyses conducted in tobacco and maize (Feng, 2024; Ji, 2024), emphasizing the synergistic regulation of nitrogen metabolism and phenylpropanoid biosynthesis under nutrient deprivation conditions.
Nitrogen deficiency enhances the uptake activity of ammonium and nitrate in plants (Iqbal, 2020). Transcript analysis also revealed significant changes in the expression of genes related to nitrate metabolism in lettuce after nutrient deprivation. These changes likely involve the regulation of nitrate transport, assimilation, and metabolic pathways. For example, the NRT2 family of high-affinity nitrate transporters functions effectively under low nitrate concentrations (Zhang et al., 2024a). Studies in wild soybean have shown that NRT2.4 and NRT2.5 are upregulated under low nitrogen stress (Sun et al., 2021; Yu, 2021), and in this study, we found that NRT2.4 and NRT2.5 in lettuce roots were similarly upregulated after nutrient deprivation, suggesting these genes play a crucial role in nitrogen absorption and utilization in a low nitrate environment. In the leaves, we found that 1 nitrate transporter protein, NRT1, was upregulated after nutrient deprivation. These expression changes may impact the response and metabolism of lettuce under nutrient deprivation stress. SPX domains are significantly responsive to low-nitrogen stress. Subcellular localization studies have shown that SPX proteins function in the nucleus and are involved in regulating nitrogen and phosphorus metabolism, as well as plant stress responses (Zhang et al., 2024b). In this study, 3 DEGs with SPX domains in leaves were all upregulated. Additionally, the CHLORIDE CHANNEL (CLC) family mediates nitrate absorption from the soil and its translocation and redistribution processes. We detected 1 DEG in both leaves and roots, which further emphasizes the role of CLCs in nitrate transport and distribution in lettuce (Wang et al., 2012, 2018).
In addition, TFs play a crucial role in the regulation of nitrate metabolism. For example, many TFs from the AR2/ERF family are upregulated in lettuce roots and leaves, and WRKY and bZIP family TFs are also involved in plant responses to nitrate stress (Liu et al., 2015; Chen et al., 2017; Jiang et al., 2017; Zhang and Yang, 2017; Pan et al., 2022). In this study, WRKY family TFs (WRKY18, WRKY60, WRKY70, etc.) and bZIP family TFs (bZIP30, bZIP34) were upregulated in lettuce roots (Table 4). The MADS-box family of genes influences growth hormone transport by regulating PIN gene expression, thus controlling root growth under varying NO3- concentrations (Maurya et al., 2020). In this study, we observed that the expression of two MADS-box family TFs was downregulated after nutrient deprivation. Additionally, the bHLH family gene bHLH130, which is significantly downregulated under low nitrogen conditions and negatively correlated with flavonoid pathway genes (Wang et al., 2022b), was upregulated after nutrient deprivation in this experiment. This upregulation of bHLH130 may suggest a synergistic effect with other TFs to enhance plant tolerance to stress.
Nitrogen deficiency affects plant quality by altering photosynthetic rates, osmotic regulation, and the accumulation of reactive oxygen species, resulting in significant inhibition of growth, development, and appearance (Guo et al., 2023). Wang et al. sequenced the transcriptomes of wheat seedlings treated with low nitrogen and found that the stress response involves signaling, carbon/nitrogen metabolism, and antioxidant activity (Wang et al., 2019). In this study, most of the DEGs related to POD, LOX, and cytochrome P450 were upregulated in lettuce roots under nutrient deprivation, with most of the cytochrome P450 and LOX genes also upregulated in leaves, while SODC-related DEGs were downregulated in both leaves and roots. These findings suggest that the antioxidant system’s response to nutrient deprivation stress differs between leaves and roots. Additionally, nitrogen deficiency leads to changes in protein expression and enzyme activity, which subsequently alters plant metabolism. For instance, nitrogen deficiency reduces the activity of ribulose bisphosphate carboxylase/oxygenase (Rubisco) (Yang, 2022). In this study, the expression of Rubisco accumulation factor 2 decreased in lettuce roots after nutrient deprivation, indicating potential effects on photosynthesis, respiration, stress tolerance, and secondary metabolite synthesis in lettuce, ultimately altering the plant’s metabolic state.
Conclusion
This study investigated the effects of nutrient deprivation on nitrate metabolism in lettuce, revealing its impact on growth, development, and metabolism. Nutrient deprivation significantly reduced nitrate content in both lettuce leaves and roots. Transcriptome sequencing analysis indicated that DEGs related to nitrate metabolism were predominantly enriched in pyrimidine metabolism, plant hormone signal transduction, and phenylpropanoid biosynthesis pathways. These findings suggest that lettuce may regulate nitrate metabolism by modulating gene expression in these pathways.
Key nitrate transport and assimilation genes, such as NRT2.4 and NRT2.5, were upregulated in roots, while NRT1 was upregulated in leaves, indicating an enhanced nitrate transport mechanism under nutrient deprivation stress. Additionally, DEGs related to peroxidase (POD), lipoxygenase (LOX), and cytochrome P450 were predominantly upregulated in roots, while cytochrome P450 and LOX were primarily upregulated in leaves. Conversely, DEGs related to superoxide dismutase copper (SODC) were downregulated in both tissues.
Nitrogen deficiency also induced changes in protein expression and enzyme activity, affecting plant metabolism. For example, the reduced expression of Rubisco accumulation factor 2 in roots suggests potential impacts on photosynthesis and overall plant metabolism.
In summary, applying nutrient deprivation 5 days before harvest can effectively reduce nitrate content, enhance nitrogen metabolism, and improve lettuce quality without compromising yield. These results provide valuable insights into the molecular mechanisms underlying lettuce responses to low-nitrogen stress and offer practical guidance for optimizing lettuce cultivation practices.
Future outlook
This study offers valuable insights into the molecular response of lettuce to nutrient deprivation stress and provides potential molecular strategies for reducing nitrate levels in vegetables. However, additional research is needed to fully elucidate the genetic and molecular mechanisms underlying these responses. Future studies could focus on the functional validation of key TFs identified in this work, as well as investigating the signal transduction pathways and regulatory networks involved in metabolic processes such as nitrate assimilation and oxidative stress response. Furthermore, exploring the role of other potential regulatory molecules and their interactions with environmental factors would be crucial for improving nutrient management efficiency in vegetable production. These efforts will contribute to optimizing strategies for enhancing vegetable quality and reducing harmful nitrate accumulation, ultimately benefiting plant health and consumer safety.
Data availability statement
The RNA-seq raw data used in this study have been deposited in Sequence Read Archive (SRA) database in NCBI under accession number PRJNA1224991 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1224991).
Author contributions
ZY: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. XL: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. SL: Investigation, Methodology, Writing – review & editing. MW: Investigation, Validation, Writing – review & editing. XH: Data curation, Investigation, Writing – review & editing. AS: Conceptualization, Writing – review & editing. JG: Data curation, Funding acquisition, Resources, Validation, Writing – review & editing. LD: Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the Technical system of vegetable industry in Hebei province, Southern Hebei high quality vegetable technology promotion post (HBCT2023100205).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be perceived as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cao, J. K., Jiang, W. B., and Zhao, Y. M. (2007). Experiment guidance of postharvest physiology and biochemistry of fruits and vegetables (Beijing: China: Light Industry Press).
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Dai, M. D., Tan, X. F., Ye, Z. R., Chen, X. T., Zhang, Y., Ruan, Y. J., et al. (2024). Analysis of lettuce transcriptome reveals the mechanism of different light/dark cycle in promoting the growth and quality. Front. Plant Sci. 15, 1394434. doi: 10.3389/fpls.2024.1394434
Fan, J. S., Jia, Y. P., He, S. Y., Tan, Z. L., Li, A. Q., Li, J., et al. (2023). GlnR activated transcription of nitrogen metabolic pathway genes facilitates biofilm formation by mycobacterium abscessus. Int. J. Antimicrob. Ag. 63, 107025–107025. doi: 10.1016/J.IJANTIMICAG.2023.107025
Feng, Y. Q. (2024). Study on the mechanism of nitrate accumulation in flue-cured and burley tobacco and regulation technologies of nitrate. [dissertation]. Zhengzhou, China: Henan Agricultural University.
Feng, Y. Q., Zhao, Y. Y., Li, Y. F., Zhou, J., Li, Y. J., and Shi, H. Z. (2022). Physiological and transcriptome analysis reveals the differences in nitrate content between lamina and midrib of flue-cured tobacco. Sci. Rep. 12, 2932–2932. doi: 10.1038/s41598-022-07011-y
Guo, T. L., Yang, Z. H., Bao, R., Fu, X. M., Wang, N., Liu, C. H., et al. (2023). The m6A reader MhYTP2 regulates the stability of its target mRNAs contributing to low nitrogen tolerance in apple (Malus domestica). Hortic. Res. 10, uhad094–uhad094. doi: 10.1093/hr/uhad094
Huang, Y. C., Ren, D. L., He, B., Zhao, Y. M., Gong, X. J., Chen, J. X., et al. (2023). Research progress in transcriptomics and metabolomics in plant abiotic stress. Jiangsu Agric. Sci. 51, 1–7. doi: 10.15889/j.issn.1002-1302.2023.22.001
Iqbal, A. (2020). The physiological and molecular mechanisms of nitrogen use efficiency in cotton [dissertation]. Beijing, China: Chinese Academy of Agricultural Sciences.
Ji, M. G. (2024). Multi-omics analysis of maize root response to low nitrogen and identification of genes associated with nitrogen efficiency. [master's thesis]. Yangzhou, China: Yangzhou University.
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., and Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Kim, M. J., Moon, Y., Tou, J. C., Mou, B., and Waterland, N. L. (2016). Nutritional value, bioactive compounds and health benefits of lettuce ( Lactuca sativa L.). J. Food Compos. Anal. 49, 19–34. doi: 10.1016/j.jfca.2016.03.004
Li, J. J., Liu, X. Y., Xu, L. Q., Li, W. S., Yao, Q., Yin, X. L., et al. (2023). Low nitrogen stress-induced transcriptome changes revealed the molecular response and tolerance characteristics in maintaining the C/N balance of sugar beet (Beta vulgaris L.). Front. Plant Sci. 14, 1164151. doi: 10.3389/fpls.2023.1164151
Li, J. Y., Lu, L. M., Zhang, R. M., Shen, H. B., Li, Z. Z., and Zhu, E. (2013). Study onpakchoi’s nitrate content and major influencing factors. Acta Agric. Shanghai 29, 116–118. doi: 10.3969/j.issn.1000-3924.2013.05.027
Li, H. H., Wang, Z. Y., and Li, B. Z. (2004). Relationship between vegetable nutrition and nitrate content. Chin. J. Appl. Ecol. 09), 1667–1672. doi: 10.3321/j.issn:1001-9332.2004.09.035
Liu, S. Y. (2014). Leaf vegetables N nutrition absorption and accumulation law. Harbin, China: Northeast Agricultural University.
Liu, Y. J., Ji, X. Y., Nie, X. G., Qu, M., Zheng, L., Tan, Z. L., et al. (2015). Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 207, 692–709. doi: 10.1111/nph.2015.207.issue-3
Liu, X. Y., Wang, H., Li, J. J., Xu, L. Q., Xing, W., Liu, D. L., et al. (2024b). Research progress of microRNAs related to low nitrogen stress. Plant Physiol. 60, 1–8. doi: 10.13592/j.cnki.ppj.300204
Liu, H., Wu, Z. Z., Zhang, W. Z., Wang, L. S., Li, Z. M., and Liu, H. (2024a). Synergistic effect between green light and nitrogen concentration on nitrate primary metabolism in lettuce (Lactuca sativa L.). Sci. Hortic. 328, 112848. doi: 10.1016/j.scienta.2024.112848
Liu, W. K., Yang, Q. C., and Du, L. F. (2011a). Short-term treatment with hydroponic solutions containing osmotic ions and ammonium molybdate decreased nitrate concentration in lettuce. Acta Agric. Scandinavica Section B - Soil Plant Sci. 61, 573–576. doi: 10.1080/09064710.2010.521762
Liu, W. K., Yang, Q. C., and Qiu, Z. P. (2011b). Effects of nitrogen-free hydroponic solutions on nitrate and vitamin C content in lettuce grown hydroponically. Acta Agric. Boreali-Sinica 26, 114–116. doi: 10.7668/hbnxb.2011.S1.024
Maurya, J., Bandyopadhyay, T., and Prasad, M. (2020). Transcriptional regulators of nitrate metabolism: key players in improving nitrogen use in crops. J. Biotech. 324, 121–133. doi: 10.1016/j.jbiotec.2020.10.001
Meng, S., Peng, J. S., He, Y. N., Zhang, G. B., Yi, H. Y., Fu, Y. L., et al. (2015). Arabidopsis NRT1.5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol. Plant 9, 461–470. doi: 10.1016/j.molp.2015.12.015
Noorden, G., Verbeek, R., Dinh, Q. D., Jin, J., Green, A., Ng, J. L. P., et al. (2016). Molecular signals controlling the inhibition of nodulation by nitrate in medicago truncatula. Int. J. Mol. Sci. 17, 1060. doi: 10.3390/ijms17071060
Okushima, Y., Inamoto, H., and Umeda, M. (2011). A high concentration of nitrate causes temporal inhibition of lateral root growth by suppressing cell proliferation. Plant Biotechnol. 28, 413–416. doi: 10.5511/plantbiotechnology.11.0722a
Pan, L. Y., Ma, J. J., Li, J. M., Yin, B. B., and Fu, C. (2022). Advances of salt stress-responsive transcription factors in plants. Chin. J. Biotechnol. 38, 50–65. doi: 10.13345/j.cjb.210135
Prasanna, J. A., Mandal, V. K., Kumar, D., Chakraborty, N., and Raghuram, N. (2023). Nitrate-responsive transcriptome analysis of rice RGA1 mutant reveals the role of G-protein alpha subunit in negative regulation of nitrogen-sensitivity and use efficiency. Plant Cell Rep. 42, 1987–2010. doi: 10.1007/s00299-023-03078-7
Reddy, K., Mallesham, B., Aakanksha, W., and Jagadis, G. K. (2020). An overview of important enzymes involved in nitrogen assimilation of plants. Methods Mol. Biol. 2057, 1–13. doi: 10.1007/978-1-4939-9790-9_1
Rengel, Z., Cakmak, I., and White, P. (2022). Marschner’s mineral nutrition of plants (Amsterdam: Academic Press RM).
Rui, H., Yue, L., Congcong, S., Gu, F., and Jie, S. (2023). Osmotic regulation beyond nitrate nutrients in plant resistance to stress: a review. Plant Growth Regul. 103, 1–8. doi: 10.1007/S10725-023-01093-Y
Song, D. H., Jiao, Y. G., Shi, L. Q., Yang, Y. B., Guo, J. H., and Dong, L. D. (2023). Transcriptomics analysis of the influence of nutrient-breaking on nitrate content in Brassica rapa L. ssp. chinensis L. Acta Agric. Boreali-Sinica 38, 74–83. doi: 10.7668/hbnxb.20193217
Sun, Q., Lu, H. R., Zhang, Q., Wang, D., Chen, J., Xiao, J. L., et al. (2021). Transcriptome sequencing of wild soybean revealed gene expression dynamics under low nitrogen stress. J. Appl. Genet. 62, 1–16. doi: 10.1007/s13353-021-00628-1
Tatiana, K., Gras, D. E., Gutiérrez, A. G., Bernardo, G., and Gutiérrez, R. A. (2011). A holistic view of nitrogen acquisition in plants. J. Exp. Bot. 62, 1455–1466. doi: 10.1093/jxb/erq425
Vandna, Sharma, V., Usha, K., Singh, D., Gupta, R., Gupta, V. K., et al. (2024). Nitrogen nutrition-induced changes in macronutrient content and their indirect effect on N-metabolism via an impact on key N-assimilating enzymes in bread wheat (Triticum aestivum L.). Nat. Environ. Pollut. 23, 1471–1482. doi: 10.46488/NEPT.2024.V23I03.017
Wang, X. N., Chai, X. F., Gao, B. B., Deng, C., Günther, C. S., Wu, T., et al. (2022b). Multiple-omics reveal the role of transcription factor bHLH130 during low nitrogen in apple rootstock. Plant Physiol. 191, 1305–1323. doi: 10.1093/PLPHYS/KIAC519
Wang, Y. Y., Cheng, Y. H., Chen, K. E., and Tsay, Y. F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, annurev–arplant-042817-040056. doi: 10.1146/annurev-arplant-042817-040056
Wang, Y. Y., Hsu, P. K., and Tsay, Y. F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 458–467. doi: 10.1016/j.tplants.2012.04.006
Wang, J., Song, K., Sun, L., Qin, Q., Sun, Y., Pan, J., et al. (2019). Morphological and transcriptome analysis of wheat seedlings response to low nitrogen stress. Plants 8, 98. doi: 10.3390/plants8040098
Wang, B., Zhang, T. X., Liu, C. Q., Zu, Y. Y., Li, Y. F., and Meng, X. C. (2022a). Research progress in the effects of abiotic stress on reactive oxygen species metabolism in medicinal plants. Res. Pract. Chin. Med. 36, 94–98. doi: 10.13728/j.1673-6427.2022.03.018
Xiong, X., Chang, L., Khalid, M., Zhang, J., and Huang, D. (2018). Alleviation of drought stress by nitrogen application in Brassica campestris ssp. Chinensis L. Agron. J. 8, 66–66. doi: 10.3390/agronomy8050066
Yan, H. S., Shi, H. W., Hu, C. M., Luo, M. Z., Xu, C. J., Wang, S. G., et al. (2021). Transcriptome differences in response mechanisms to low-nitrogen stress in two wheat varieties. Int. J. Mol. Sci. 22, 12278–12278. doi: 10.3390/ijms222212278
Yang, X. L. (2022). Transcriptome analysis physiological mechanism of Quinoa in response to different nitrogen supply. [master's thesis]. Taiyuan, China: Shanxi Normal University.
Yi, Y., Chang, Y. Q., and Ke, L. W. (2015). Quality improvement efficiency of hydroponic lettuce after replacement two nitrogen level nutrient solutions by nitrogen-free solution. Acta Agric. Boreali-Sinica 30, 446–448. doi: 10.7668/hbnxb.2015.S1.081
Yin, L., Xu, H., Dong, S., Chu, J., Dai, X., and He, M. (2019). Optimised nitrogen allocation favours improvement in canopy photosynthetic nitrogen-use efficiency: evidence from late-sown winter wheat. Environ. Exp. Bot. 159, 75–86. doi: 10.1016/j.envexpbot.2018.12.013
Yu, D. C. (2021). Transcriptome sequencing and expression analysis of nitrate assimilation-associated genes in pyropia yezoensis under N-sufficient conditions. [master's thesis]. Suzhou, China: Soochow University.
Yuan, Y. J., Hao, T. W., Miao, Z., Yang, Z. Q., Xin, D. W., Xin, Z. Y., et al. (2021). STOP1 activates NRT1.1-mediated nitrate uptake to create a favorable rhizospheric pH for plant adaptation to acidity. Plant Cell 33, 3658–3674. doi: 10.1093/PLCELL/KOAB226
Yuan, L. X., Loqué, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19, 2636–2652. doi: 10.1105/tpc.107.052134
Zhang, B. H., Liu, J. J., Tian, X., Tian, X. Z., Dong, K., Wu, Y. J., et al. (2024b). Cloning, expression, and functional analysis of wheat (Triticum aestivum L.) TaSPX1 gene in low nitrogen stress tolerance. Acta Agronomica Sin. 50, 576–589. doi: 10.3724/SP.J.1006.2024.31025
Zhang, Y. J., Sang, H. T., Wang, H. Q., Shi, Z. Z., Li, L., Wang, X., et al. (2024c). Research progress of plant signaling in systemic responses to abiotic stresses. Chin. Bull. Entomol. 59, 122–133. doi: 10.11983/CBB23063
Zhang, B., Xu, Y. X., Zhang, L. W., Yu, S. Y., Zhu, Y. Y., Liu, C. J., et al. (2024a). Root endodermal suberization induced by nitrate stress regulate apoplastic pathway rather than nitrate uptake in tobacco (Nicotiana tabacum L.). Plant Physiol. Biochem. 216, 109166–109166. doi: 10.1016/j.plaphy.2024.109166
Zhang, R. J. and Yang, Z. R. (2017). Molecular regulatory network of nitrate absorption and transportation. J. Shanxi Agric. Sci. 45, 861–866. doi: 10.3969/j.issn.1002-2481.2017.05.46
Zhao, Q. S., Dong, J. J., Li, S. B., Lei, W. X., and Liu, A. K. (2024). Effects of micro/nano-ozone bubble nutrient solutions on growth promotion and rhizosphere microbial community diversity in soilless cultivated lettuces. Front. Plant Sci. 15, 1393905. doi: 10.3389/fpls.2024.1393905
Keywords: lettuce, nutrient deprivation, nitrate, N metabolism, transcriptome analysis
Citation: Zhang Y, Liu X, La S, Wang M, Hu X, Shi A, Guo J and Dong L (2025) Transcriptome analysis reveals nutrient deprivation reduces nitrate content in lettuce (Lactuca sativa var. ramosa Hort.) and enhances nitrogen metabolism. Front. Plant Sci. 16:1585955. doi: 10.3389/fpls.2025.1585955
Received: 01 March 2025; Accepted: 22 April 2025;
Published: 19 May 2025.
Edited by:
Guoxiang Jiang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2025 Zhang, Liu, La, Wang, Hu, Shi, Guo and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinghua Guo, MTU4MzExMzg5MThAMTYzLmNvbQ==; Lingdi Dong, ZG9uZ2xpbmdkaUAxNjMuY29t
 Yingying Zhang
Yingying Zhang Xuena Liu1
Xuena Liu1 Ainong Shi
Ainong Shi