- 1School of Pharmacy, Shandong Second Medical University, Weifang, Shandong, China
- 2Chemical Drug Testing Laboratory, Weifang Inspection and Testing Center, Weifang, Shandong, China
- 3Dongying High level Talent Research Center, Dongying, Shandong, China
Triterpenoid saponins are a class of plant secondary metabolites with significant biological activities and are widely used in the pharmaceutical and nutritional supplement industries. However, the production of triterpenoid saponins is limited by their complicated biosynthetic pathways and the availability of glycosyl donors. UDP-glycosyltransferases (UGTs) play a key role in the glycosylation of triterpenoid saponins, significantly enhancing their structural diversity, solubility, pharmacological activity, and bioavailability. Therefore, the identification and modification of efficient, specific, and stable UGTs have attracted attention. This review focused on the advances in the glycosylation of triterpenoid saponins, with a particular emphasis on the application of multi-omics approaches in UGT mining. The combination of genomics, transcriptomics, and metabolomics has provided powerful tools for UGT screening, significantly improving the efficiency and accuracy of UGT identification. Additionally, the methods based on gene clusters, phylogenetic analysis, and the plant secondary product glycosyltransferase (PSPG) motif also offer new perspectives for UGT identification. Besides, the application of synthetic biology platforms has provided innovative approaches for high-throughput screening and functional validation of UGTs, laying a theoretical foundation for the functional modification of UGTs. We also discussed the latest research progress on UGT modification including directed evolution and rational design. These strategies, through amino acid mutations and structural optimization, are expected to enhance UGT catalytic activity, thermal stability, and broaden substrate specificity. Moreover, the diversity and availability of glycosyl donors directly influence the efficiency of glycosylation reactions and the diversity of the products. Thus, we discussed glycosyl donor synthesis, including in vitro and in vivo synthetic strategies. By optimizing metabolic pathways and introducing key enzyme genes, engineered microorganisms can efficiently synthesize various glycosyl donors, providing abundant substrates for glycosylation reactions. These studies offer new opportunities and challenges for the synthesis and application of triterpenoid saponins, promoting their industrial potential.
1 Introduction
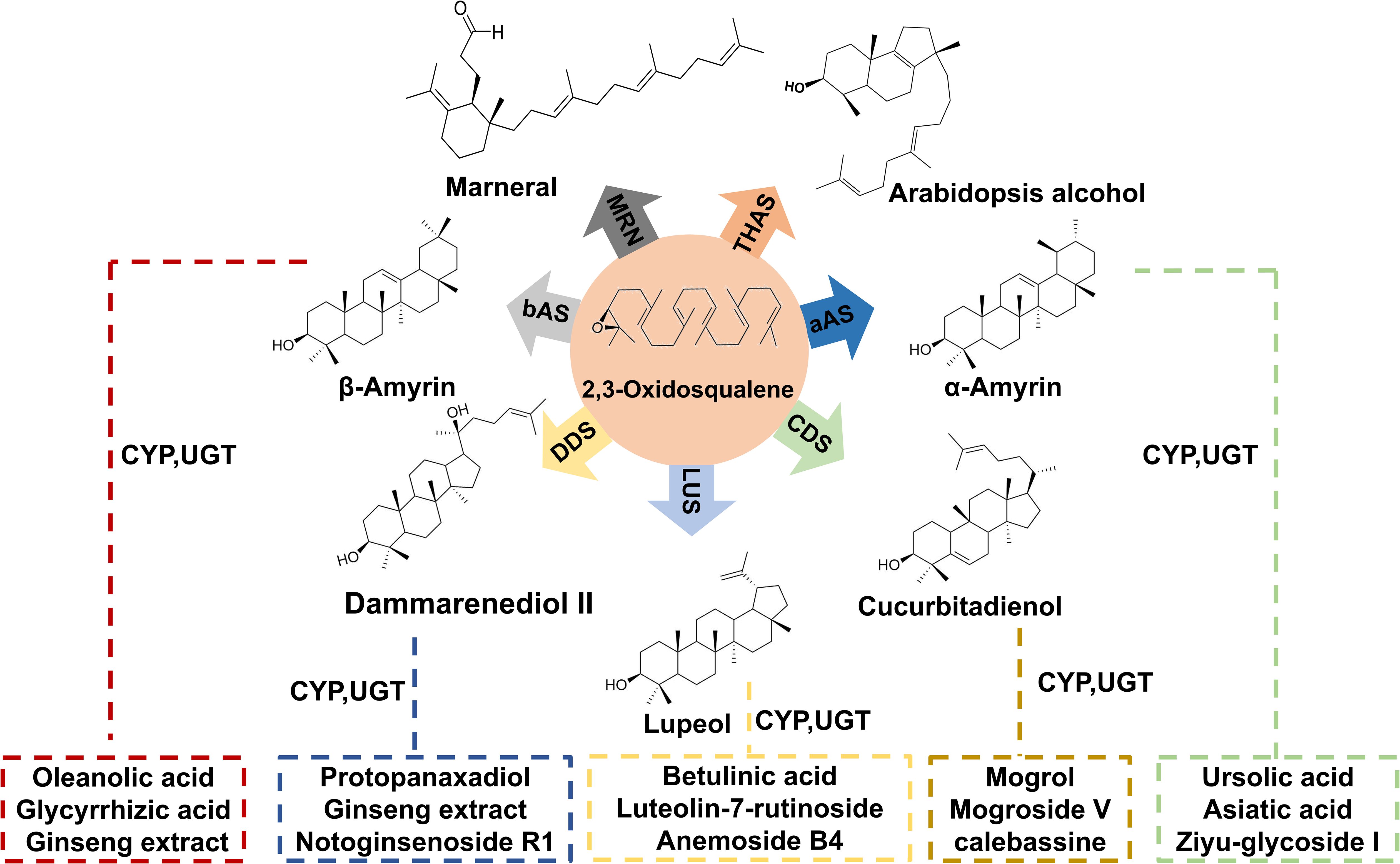
Triterpenoids are a class of terpenoid compounds with a basic skeleton formed by six isoprene units (C5H8). They can exist in free form or be conjugated with sugars to form glycosides or esters. Notably, triterpenoid saponins are widely distributed in nature and demonstrate significant pharmacological activities, including anti-inflammatory, antifungal, antibacterial, antiparasitic, and anticancer effects (Singh et al., 2024; Xu et al., 2024; Dong et al., 2023) (Figure 1). The biosynthesis of triterpenoids begins with the condensation of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), catalyzed by farnesyl pyrophosphate synthase (FPPS) to form the C15 molecule farnesyl pyrophosphate (FPP). Two molecules of FPP undergo a “head-to-head” condensation to form the linear C30 molecule squalene, which is then oxidized to yield the important precursor 2,3-oxidosqualene, possessing the triterpene skeleton. Squalene is oxidized to 2,3-oxidosqualene by the squalene epoxidase (SQLE) enzyme, which is a cytochrome P450 enzyme. This oxidation step is crucial as it introduces an epoxy group at the 2,3 positions of squalene, forming a reaction intermediate that serves as a branch point for different triterpene biosynthetic pathways. SQLE uses molecular oxygen and NADPH as cofactors to facilitate this oxidation process, which is the rate-limiting step in triterpene biosynthesis. 2,3-oxidosqualene enters various branched metabolic pathways under the action of different oxidosqualene cyclases (OSCs), leading to the formation of various cyclic precursors (Hazra et al., 2023) (Figure 2a). Next, the structural diversity of triterpenes arises from cyclization and oxidation steps, their pharmacological versatility is further amplified by glycosylation—a process mediated by UGTs (Li et al., 2023; Hucheng et al., 2023), which transfer activated sugar units from sugar nucleotide donors to a receptor, forming stable glycosidic bonds. According to the CAZy database, enzymes responsible for the glycosylation of small, lipophilic molecules in plants belong to the Glycoside Hydrolase Family 1 (GT1) and UDP-dependent glycosyltransferase (UGT) superfamily. Although more than 20,000 members have been identified in this family, less than 2% of them have been characterized (CAZy database statistics). UGTs play a key role in the structure and pharmacological properties of triterpenoid saponins (Gloster, 2014; Wilson and Tian, 2019).
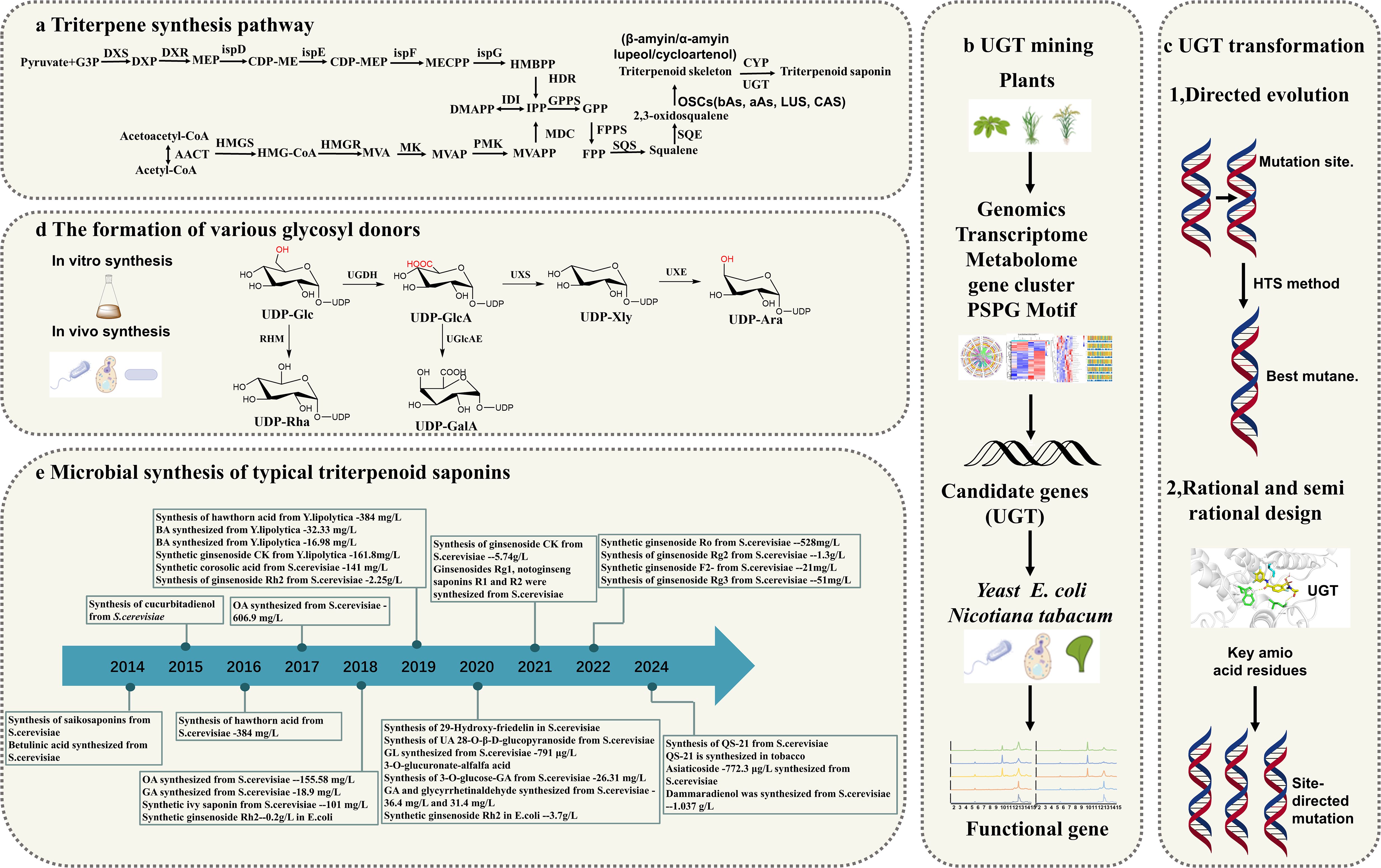
Figure 2. Overview of Triterpene Glycosyltransferase Research. (a) Triterpene Biosynthetic Pathways; (b) Discovery of UGTs (Uridine 5’-diphosphate Glucosyltransferases); (c) Modification of UGTs; (d) Glycosyl donor Biosynthetic Pathways; (e) Microbial Production of Triterpene Compounds.
Multi-omics methods including genomics, transcriptomics, and metabolomics, have become powerful tools mining UGT genes involved in triterpenoid saponins biosynthesis, enabling the systematic correlation of gene expression, metabolite profiles, and enzyme activity. Besides, genome clustering, phylogenetic analysis, and PSPG motif-based screening methods have offered new perspectives for identifying UGTs (Wang et al., 2023a). The PSPG motif is a conserved domain involved in glycosyl donor binding and is particularly important for identifying functional UGTs (Andong et al., 2022). Furthermore, the application of synthetic biology platforms has provided innovative methods for the high-throughput mining and functional validation of UGTs.
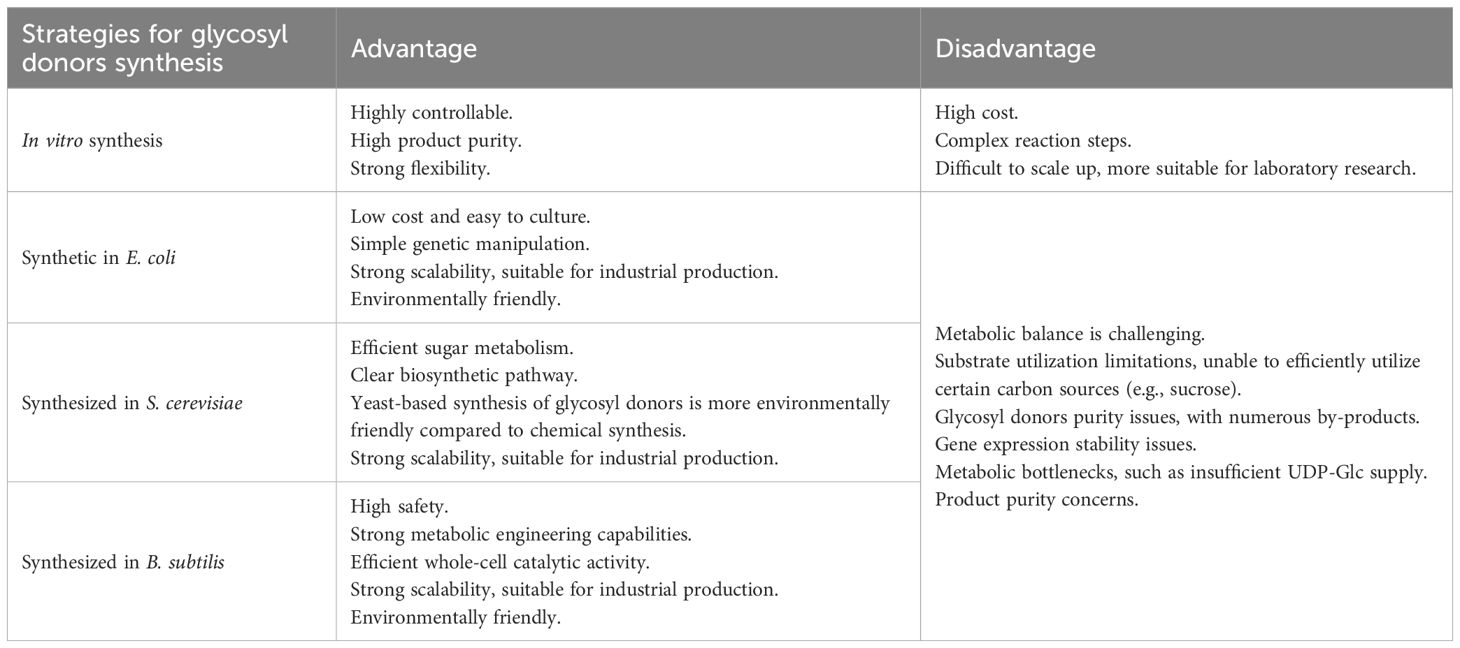
Glycosylation efficiency fundamentally depends on glycosyl donor availability. The diversity and accessibility of these donors directly influence reaction efficiency and product diversity. Key donor molecules include D-glucose (D-Glu), D-galactose (D-Gal), D-glucuronic acid (D-GlcA), L-rhamnose (L-Rha), D-xylose (D-Xyl), and L-arabinose (L-Ara), as established in current research (Kurze et al., 2022). This highlights the essential role of UDP-sugar biosynthesis in glycoside production. Existing manufacturing strategies divide into chemical synthesis and biological approaches. Due to the structure complexity of glycosyl donors, the steps for their chemical synthesis are costly and highly toxic, making it difficult to achieve their mass production., while engineered microbial systems (e.g., E. coli, yeast) have become main production platforms. Through metabolic engineering and enzyme optimization, these biological systems have demonstrated potential donor synthesis capabilities, overcoming traditional limitation of substrates (Cravens et al., 2019; Lu et al., 2023).
Recent studies have prioritized pathway engineering and heterologous production over basic UGT characterization, emphasizing integrated multi-omics and synthetic biology frameworks (Reed et al., 2023; Zhao et al., 2024; Xu et al., 2024). This review systematically examined advancements in triterpenoid glycosylation research, particularly highlighting multi-omics strategies for UGT discovery (Li et al., 2023; Wang et al., 2021a). We investigated innovative engineering tactics spanning genetic/protein modifications and evaluates both chemical and biological glycosyl donor production methods. These developments create new possibilities for optimizing triterpenoid saponin biosynthesis while presenting challenges for industrial applications (Figure 2).
2 Multi-strategy UGT mining for triterpene biosynthesis
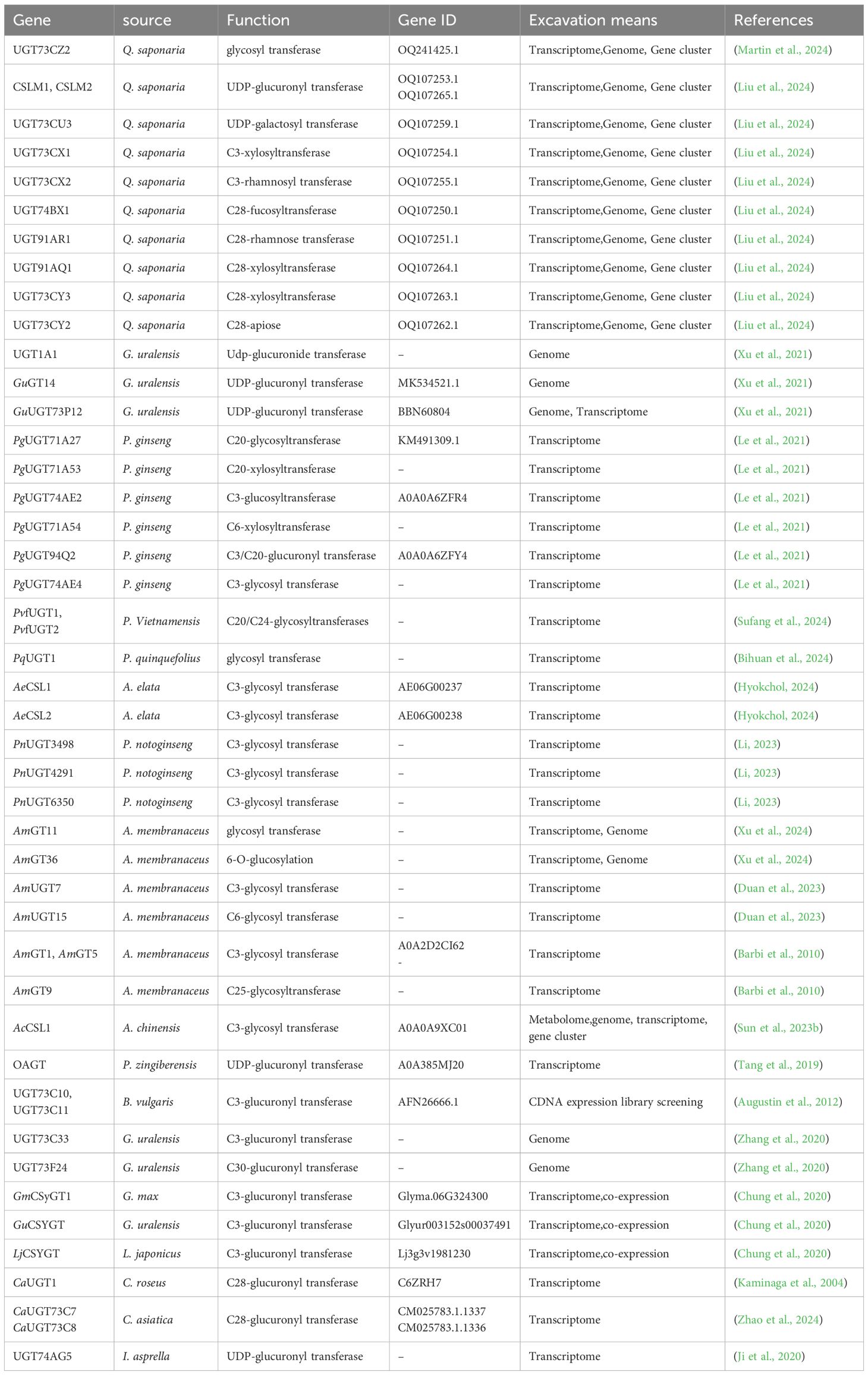
UGTs enzymes play critical roles in the post-modification of triterpenoid compounds, significantly contributing to their structural diversity. However, many UGT genes responsible for these modifications remain largely unexplored. Extensive studies have been performed on glucosyltransferases and glucuronic acid transferases, while far fewer investigations have focused on other UGTs, such as those responsible for the transfer of galactose, xylose, mannose, fucose, and arabinose (Li et al., 2014). In recent years, the omics technologies have provided new perspectives and tools for the identification and characterization of UGTs involved in triterpene glycosylation (Table 1). The combination of genomics, transcriptomics, and metabolomics allows researchers to systematically analyze the functions and mechanisms of UGTs from multiple dimensions (Dai and Shen, 2022). Additionally, strategies based on gene clusters, phylogenetic analysis and the presence of PSPG motifs have enhanced the efficiency and accuracy of UGT mining (Figure 2b). Furthermore, the application of synthetic biology platforms offers innovative approaches to explore and engineer these enzymes for improved catalytic efficiency and specificity. This review comprehensively explored the application of multi-omics approaches in the screening of UGTs related to triterpene glycosylation. It also analyzed strategies based on gene clusters, phylogenetic analysis and PSPG motifs, and discuss the advancements in synthetic biology platforms.
2.1 Multi-omics approaches for screening glycosyltransferases involved in triterpenoid glycosylation
Plant genomics and transcriptomics have provided extensive insights into structural genes, facilitating the identification of those involved in the biosynthesis of valuable secondary metabolites in plants (Jendoubi, 2021). Among various multi-omics approaches, genomics clarifies the structure, function, evolution, localization, and editing of genomes (You, 2023). Transcriptomics focuses on identifying and quantifying RNA, offering valuable insights into gene expression profiles under specific conditions and over time (Zhang et al., 2023). Metabolomics complements genomics and transcriptomics by uncovering the metabolic responses of organisms to external stimuli, environmental changes, or genetic modifications, thus providing a comprehensive view of cellular processes (Figure 2b). By integrating data from these three omics, researchers can systematically identify and screen UGT candidates involved in the biosynthesis of triterpenoids (Alami et al., 2023; Song et al., 2023).
2.1.1 Genome-based screening of glycosyltransferases involved in triterpenoid glycosylation
In plant genomics, gene annotation technology provides important clues to parse key DNA sequences. By tracking the common features of plant genomes, researchers can more clearly delineate evolutionary trajectories and taxonomic relationships. Notably, genome annotation data can also aid in the prediction of potential UGTs candidate genes (Song et al., 2023). Taking the G. uralensis genome study as an example, the Xu team accurately targeted UGT1A1 from 22 candidate UGTs and confirmed that the enzyme has efficient catalytic characteristics for the biosynthesis of glycyrrhizic acid GL and GAMG (Xu et al., 2021). The next results showed that GuGT14 and GuUGT73P12 could synergistically catalyze the directional connection of glucose and glucuronic acid at the C-3 position of the parent nucleus to form the characteristic glycyrrhizic acid (Xu et al., 2021). Genome analysis of A. chinensis Bunge discovered two gene groups for triterpene production: a 350 kb cluster containing multiple biosynthetic genes including oxygenase homologs and BAHD family members (Sun et al., 2023b). While identifying the gene clusters involved in triterpene biosynthesis by using triterpene pathway synthesis genes with known functions, such as OSCs and cleavage enzymes (CYPs), it also promotes the mining and identification of UGT on their synthetic pathways. Based on G. uralensis genome insights, researchers successfully identified UGT73C33, which specifically modified the 3-C position of triterpene cores (Zhang et al., 2020). This evidence confirmed that systematic examination of gene clusters associated with metabolic pathways can significantly improve the accuracy of UGT identification.
2.1.2 Transcriptome-based screening of glycosyltransferases involved in triterpenoid glycosylation
Compared with genomics, transcriptomics provides information into the temporal and spatial variations in gene expression. Thus, differentially expressed gene (DEG) and gene co-expression analyses are used to identify candidate genes (Rosati et al., 2024). In plants, triterpenoid biosynthesis usually occurs under biotic and abiotic stresses, and the genes related to triterpenoid synthesis get activated or upregulated due to increased triterpenoid accumulation after methyl jasmonate (MeJA) or dark treatment. Analyzing DEGs between treated and untreated plants has proven effective in identifying candidate genes related to triterpenoid synthesis. Han et al. utilized transcriptomic data from C. asiatica to screen 75 putative UGTs. Functional validation revealed that CaUGT1 specifically transfers glucose to the C-28 carboxyl group of asiatic acid and madecassic acid (De Costa et al., 2017). Based on a similar strategy, Zhou et al. identified 51 UGTs from the C. asiatica transcriptome and found that CaUGT73C7 and CaUGT73C8 could catalyze asiaticoside synthesis while capturing CaRRT that specifically transfer rhamnose. With these findings, the team achieved total synthesis of asiaticoside in S. cerevisiae with a yield of 772.3 μg/L (Zhao et al., 2024). The Sui team successfully identified two P450 enzymes and three UGTs as candidate genes for saikosaponin synthesis through the B. chinense transcriptome study (Sui et al., 2011).
The gene co-expression theory suggests that genes related to metabolic pathways have synergistic expression characteristics. Based on this principle, Chung et al. found a strong expression association between GmCSyGT1 and soybean saponin synthesis genes in the G. max co-function network database (Chung et al., 2020). Using GmyGTs as a probe, the researchers further identified GuCSyGT and LjCSyGT homologues in the transcriptome of G. uralensis and L. japonicus. Cross-species co-expression analysis confirmed the central role of CSyGTs in the synthesis of legume saponins, and the S. cerevisiae system verified that CsyGTs could catalyze the formation of glycyrrhetine-3-O-monoglucuronide (Chung et al., 2020). QS-21, a β-aromatic alcohol pentacyclic triterpene, contains three hydroxyl sites (glucuronic acid, galactose, xylose) and three carboxyl sites (fucose, rhamnose, xylose). The complexity of QS-21 molecular structure makes its chemical synthesis extremely challenging. Martin et al. analyzed the genes related to QS-21 synthesis based on the transcriptome data of saponins and achieved the compound heterologous synthesis in tobacco chassis through a 20-step catalytic reaction (Martin et al., 2024). Yao et al. performed RNA sequencing on different tissues of A. elata (Miq.), screened 64 triterpene skeleton synthesis related genes, 254 CYP450 and 122 UGTs, and identified 5 oleanolic acid 3-O-glucosyl transferase candidate genes through expression profiling (Cheng et al., 2020). The research team also identified key UGTs gene clusters involved in triterpene glycosylation, including PgUGT71A27, PvfUGT1, PqUGT1, etc., through transcriptome analysis of P. ginseng, P. Vietnamensis, P. quinquefolius and other species (Le et al., 2021; Sufang et al., 2024; Bihuan et al., 2024).
Transcriptome analysis offers a strategic approach for pinpointing UGT enzymes essential for triterpenoid modification. By tracking gene expression patterns under varying conditions, scientists can isolate environmentally responsive genes and validate their biochemical functions.
2.1.3 Multiple omics screening of glycosyltransferases for triterpenoid glycosylation
Multi-omics integration strategy has shown unique advantages in gene function analysis (Luo, 2015). Integrating different omics data significantly improves the efficiency and accuracy of UGTs identification (Liu et al., 2022).
Integrated genomic and transcriptomic strategies effectively pinpoint UGTs involved in triterpene glycosylation. Advanced sequencing technologies create gene maps that identify functional genes and track their evolutionary changes (Wang and Huo, 2022). Transcriptome analysis tracks UGT expression patterns under various conditions, identifying key metabolic pathway elements (Zhang et al., 2025). Notable advances included Zhou et al. discovered of 42 saponin-related SmUGTs distributed across 12 chromosomes in S. mukorossi (Zhou et al., 2024), and Wu et al. studied identifying 145 grapefruit UGTs through conserved motif analysis combined with developmental stage expression patterns (Wu et al., 2020). Jiang et al. analysed P. notoginseng data to isolate 27 UGTs, including PnUGT33 that efficiently extended sugar chains on ginsenosides, achieving 51mg/L Rg3 yields in lab cultures (Jiang et al., 2022).
Combining transcriptomics and metabolomics to link gene activity to metabolite production enables precise functional analysis of UGT enzymes (He, 2022). This approach clarified how UGTs operate within biochemical pathways (He et al., 2021). Taking the non-model medicinal plant A. flaccida as an example, Zhan et al. systematically identified the enzymes involved in the triterpene saponins synthesis pathway, including the key enzymes of the mevalonate pathway (MVA) and the methylerythritol phosphate pathway (MEP), through combined transcriptome and proteome analysis. In addition, 126 CYP450 enzymes and 32 UGTs were identified as triterpene modification candidate genes (Zhan et al., 2016). Wang et al. used the combined transcriptomics and metabolomics to predict genes related to triterpene and flavonoid synthesis pathways in B. rapa, and found that ERF transcription factors may play an important role in the synergistic synthesis of the two kinds of compounds (Wang et al., 2024). Rai group has successfully discovered candidate genes for C. officinalis triterpene synthesis by combining LC-QTOF-MS metabolomics and RNA-seq transcriptome data (Rai et al., 2020). Through multi-omics analysis of different tissues of B. chinense, He et al. revealed the difference of saikosaponin synthesis pathways in roots, stems, leaves, and flowers, and found that the regulation of P450 genes Bc95697 and Bc35434 may improve saponin production (He et al., 2021). Chen et al. identified 69 terpenoids in C. paliurus, of which triterpenoids accounted for more than 80%, and identified key genes in the triterpenoid synthesis pathway by transcriptome co-expression analysis (Chen et al., 2024). These reseach demonstrated that multi-omics integration technology provides an effective case for exploring the mechanism of secondary metabolism in non-model plants.
Integrated genomic, transcriptomic, and metabolomic approaches have proven particularly effective for studying exploration of plant systems, from evolutionary relationships to specialized metabolite production (Fu et al., 2021; Yang et al., 2023). Liu et al. developed a multi-stage screening method that involved in triterpene saponin biosynthesis in P. vulgaris, using multi-omics integration to identify key biosynthetic genes. Their enzyme validation protocol established an efficient framework for the discovery of key enzymes including UGTs (Liu et al., 2025). Feng et al. mapped tissue-specific metabolic profiles in I. hylonoma, combining multi-omics data to mine regulatory genes controlling triterpene production, including CYP450 enzymes and transcription factors (Feng et al., 2024).
Liu et al. integrated multi-omics data from P. vulgaris using phylogenetic analysis and co-expression networks, successfully identifying two OSCs, three CYP716s, and four UGT73s from hundreds of gene family members. Heterologous expression confirmed the function of these genes matched computational predictions (Liu et al., 2025). Lin et al. identified four CYP genes, one UGT, and associated transcription factors as key regulators of triterpene saponin biosynthesis in E. phaseoloides through multi-omics analysis (Lin et al., 2022). P. bretschneideri Rehd. genome data, Li et al. predicted 178 UGTs and linked 11 to arbutin glycosylation using transcriptomic and metabolomic correlation studies, which promoted the understanding of the glycosylation mechanism in pear plants (Li et al., 2022). Martin et al. identified the triterpene glycosyltransferase UGT73CZ2 (Martin et al., 2024) through multi-omics analysis of Q. saponaria, and Hassan et al. found that CSLM1 and CSLM2 in Q. saponaria (Hassan et al., 2024), Liu et al. further analyzed UGT73CU3, UGT73CX2 and other series of transferases in Q. saponaria (Liu et al., 2024). Xu et al. targeted AmGT11 and AmGT36 based on the multi-omics data of A. membranaceus (Xu et al., 2024), and Feng et al. revealed the regulatory network of triterpene saponins synthesis through the multi-omics study of I. hylonoma (Feng et al., 2024). These cases demonstrated that multi-omics integration has become a powerful tool to analyze the complex metabolic networks in plants.
2.2 Screening of glycosyltransferases for triterpenoid glycosylation based on gene clusters
The structural complexity of natural products frequently mirrors the evolutionary sophistication of their biosynthetic gene clusters (BGCs) (Jensen, 2016). Molecular phylogenetics helps track gene evolution and identify relationships between related sequences. Notably, clustering patterns of terpenoid-modifying enzyme genes create opportunities for targeted gene discovery.
Specialized databases now exist for gene cluster prediction, including BAGEL4 (prokaryotic ribosome synthesis analysis), antiSMASH (bacterial/fungal/plant secondary metabolism), plantiSMASH (plant-specific cluster detection), and PRISM (secondary metabolism prediction) (Blin et al., 2023; Kautsar et al., 2018; Skinnider et al., 2020). For triterpene glycosyltransferase identification, plantiSMASH analysis of Q. saponaria genomes revealed candidate genes Qs0321930 and Qs0321920 near CYP716A297 (Guo et al., 2018), while later studies detected co-expression patterns between CSL genes and QsbAS1 (Reed et al., 2023). Plant metabolic gene clusters have proven particularly valuable for research. The clustering tendency of terpenoid genes enables functional gene discovery through neighborhood analysis (Bharadwaj et al., 2021). In A. chinensis aescin synthesis, Sun et al. identified critical cluster components: AcCYP716A275 (C-14 hydroxylation), AcCYP716A278 (C-19 hydroxylation), AcCSL1 (C-3 glucuronidation), and AcBAHD3/6 (C-22 acetylation) (Sun et al., 2023b). These findings illuminate cluster component synergy and advance natural product biosynthesis understanding. Modern bioinformatics tools like BAGEL4 and antiSMASH now drive systematic BGC prediction and analysis (Jones et al., 2024; Chakraborty, 2022). These platforms accelerate both known gene validation and novel enzyme discovery, supporting metabolic pathway elucidation and pharmaceutical innovation.
2.3 Screening of Glycosyltransferases for triterpenoid glycosylation based on PSPG motif and phylogenetic analysis
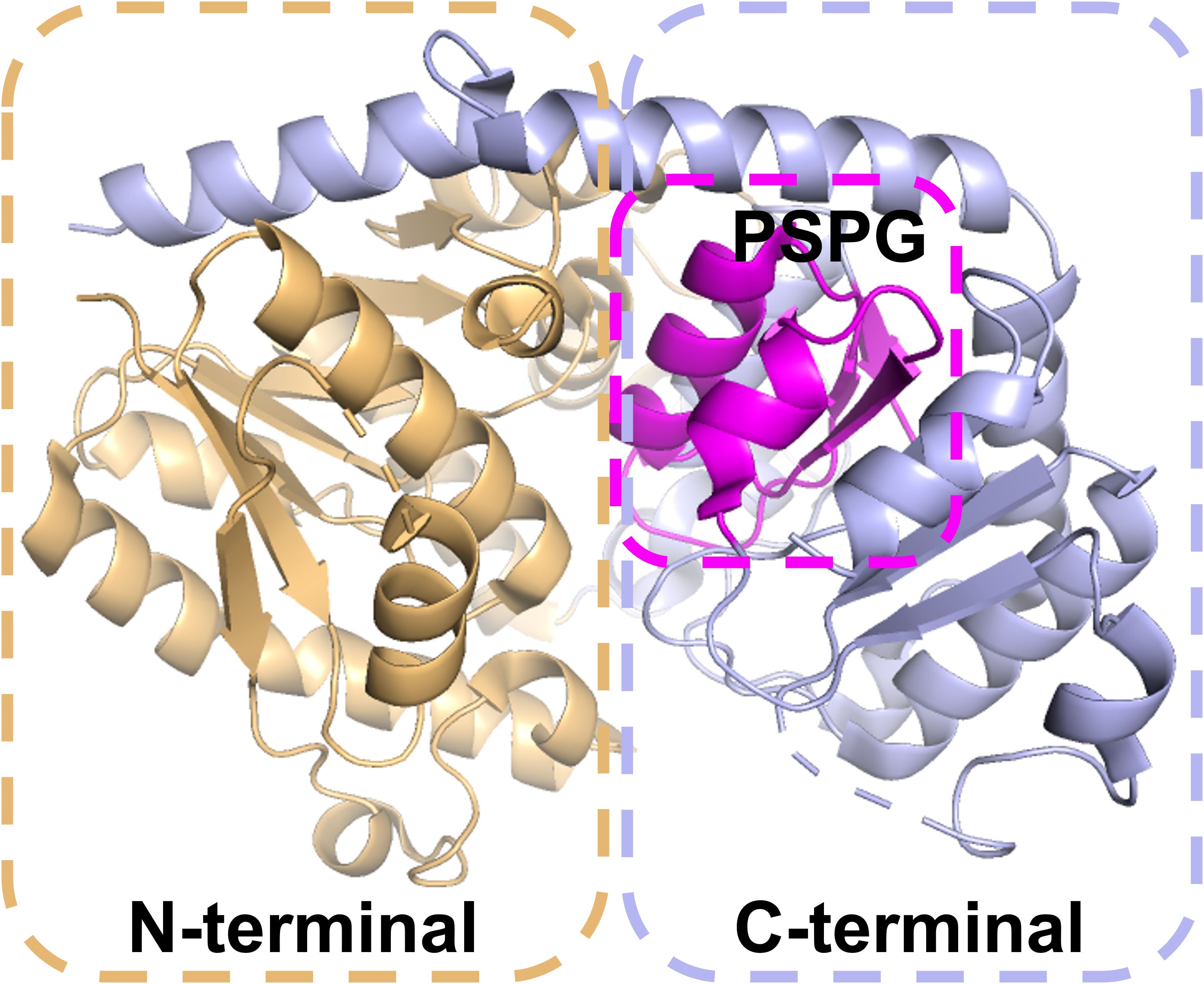
The characteristic cleft between the N-terminal and C-terminal domains of UGTs forms a binding cavity that accommodates the donor and acceptor. The C-terminal conserved PSPG motif is the signature structural feature of UGTs (Wang et al., 2023a) (Figure 3), and some of its 44 amino acid residues are highly conserved, which is directly related to glycosyl donor selection (Chen et al., 2022). The histidine (H) at the terminal of PSPG motif determines galactose/arabinose transfer activity, whereas glutamine (Q) dominates glucose transfer properties. Based on this property, researchers remodeled the enzyme function by site-directed mutagenesis. The glucosyltransferase was obtained by replacing H404 of AsAAT1 with proline (P154) by Louveau et al. (Louveau et al., 2018); Rahimi et al. converted the substrate specificity from UDP-Xly to UDP-Glc by the H to Q mutation of AtUGT78D3 (Rahimi et al., 2019).
Sequence variation in PSPG motifs can reflect functional differentiation. According to the N-terminal characteristic sequence, it can be divided into different subgroups: group A contains LPEGF, group D contains GW-PQ, group E contains WAPQ, group G contains WCPQ, group H contains RG-IV, and group L is characterized by WC-Q (Xueqing et al., 2022) (Figures 4, 5). Novel gene substrate preferences can be predicted by comparing the known functions of Arabidopsis UGTs with triterpene UGTs (Rahimi et al., 2019). The phylogenetic analysis in this review revealed that UGTs with similar PSPG motifs presented clustering characteristics on the evolutionary tree (Figure 4), indicating their functional relevance. This combined analysis strategy based on phylogenetic trees and PSPG motifs provides a new idea for the functional prediction and targeted modification of UGTs.
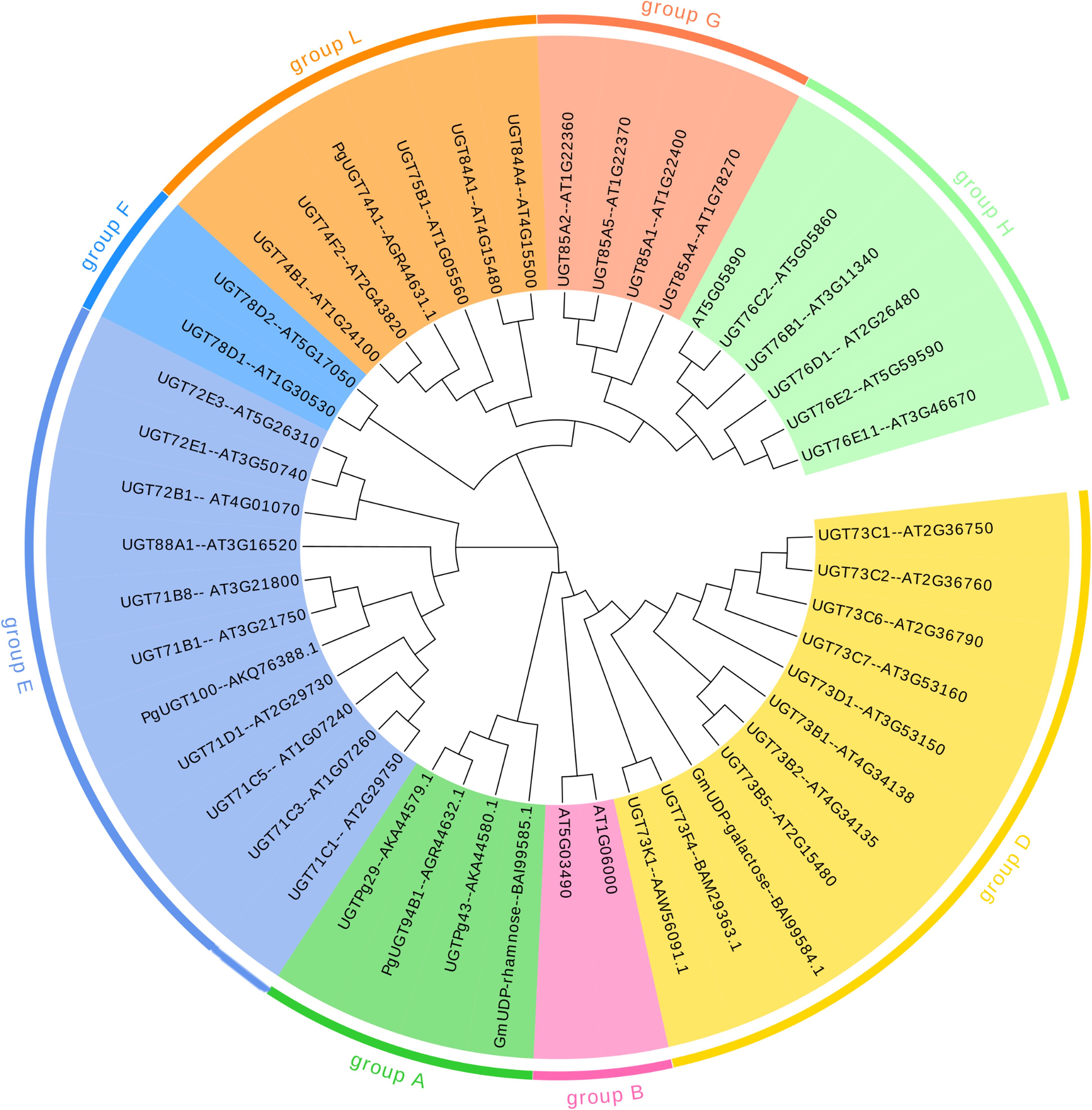
Figure 4. Phylogenetic tree analysis of UGTs. The tree is constructedbased on the PSPG motif sequences of UGTs involved in triterpenoidglycosylation. Key branches are annotated with PSPG groupings to highlight their substrate specificity.
2.4 Synthetic biology platform-based screening of glycosyltransferases for triterpenoid glycosylation
Wan et al. developed a modular yeast platform to dissect UGT-mediated glycosylation in plant natural products (PNPs), incorporating UDP-Glc and UDP-Xyl dependent enzymes (Wang et al., 2020). Their system employed a plug-and-play design to reconstitute PNP biosynthetic pathways in engineered yeast, enabling targeted investigation of glycosylation steps. Using this framework, they identified five triterpene glycosyltransferases from P. notoginseng, including the xylosyltransferase essential for the biosynthesis of Notoginsenoside R1.
Jian et al. elaborated the limited diversity of characterized UGT91H enzymes, a bottleneck in synthesizing trisaccharide-modified saponins. By analyzing evolutionary relationships among legume genomes which are rich in triterpenoid saponins, they selected 23 candidate genes using known UGT91H sequences as a seed. The results of functional identification confirmed conserved catalytic roles across 19 newly identified enzymes, demonstrating phylogenetic analysis as an efficient method for expanding this subfamily’s toolkit (Jian et al., 2024).
These parallel studies established complementary strategies: Wan et al. platform enables pathway reconstruction for functional validation, while Jian et al. phylogeny-driven approach accelerates enzyme discovery. Together, they advance both mechanistic understanding of plant glycosylation and practical tools for engineering bioactive compound production.
3 Catalytic mechanism of triterpenoid glycosyltransferases
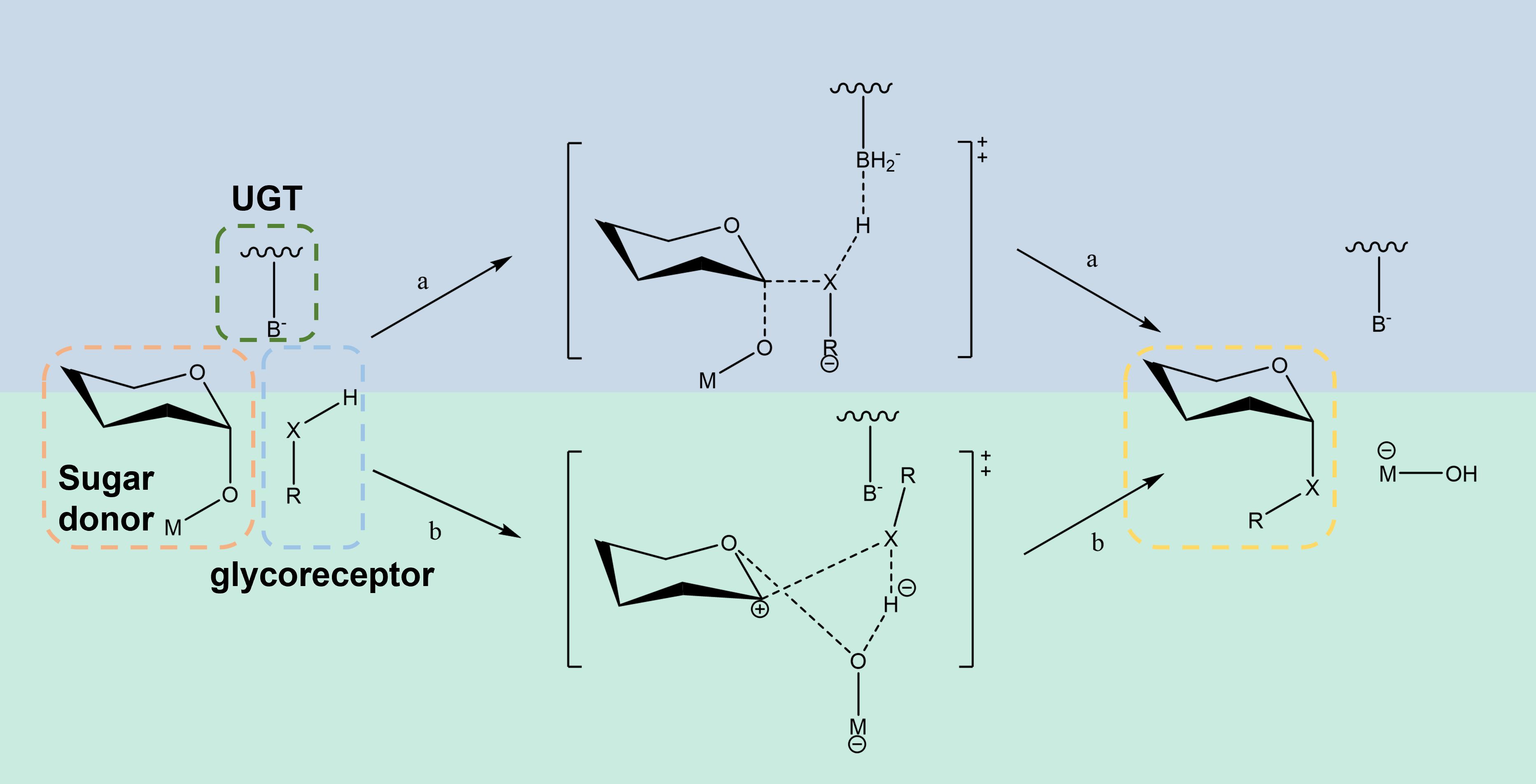
Triterpene saponins are consisted of glycosyl donors and a sugar acceptor, which are linked by glycosidylic linkages catalyzed by UGTs. Glycosylation can be classified into O-glycosidic (hydroxyl), C-glycosidic (carbon), N-glycosidic (amino) and S-glycosidic (sulfur) linkages according to the different linkage sites (Challinor and De Voss, 2013). Among them, O-glycosylation and C-glycosylation have attracted much attention because of their significant effects on the activity of compounds (Wang et al., 2023a). Glycosyltransferase catalysis generally follows the SN2 nucleophilic substitution mechanism (Figure 6), which consists of four key steps: substrate recognition, binding, glycosyltransfer, and product release. The active center of the enzyme binds to the substrate through hydrogen bonding, ion interaction and van der Waals force, thereby catalyzing the transfer of sugar groups from the donor to the acceptor (Xiao-chen et al., 2018).
The C-terminal domain of UGTs is primarily responsible for recognizing the UDP-glycosyldonor, whereas the N-terminus specifically binds receptor substrates (Osmani et al., 2009). Because the C-terminus of different UGTs needs to recognize the same or similar glycosyl donor, its conservation is higher than that of the N-terminal domain that recognizes heterologous receptors (Wang, 2009). An in-depth understanding of the catalytic mechanism of UGTs will not only help to reveal the law of natural product synthesis, but also provide a theoretical basis for enzyme engineering. Although natural UGTs play a central role in the synthesis of triterpene saponins, their catalytic efficiency, stability and substrate adaptability are often difficult to meet the needs of industrialization. Optimizing enzyme performance through protein engineering has become a current research hotspot.
3.1 Engineering of glycosyltransferases for enhanced catalytic properties
Synthetic biology constructs novel biosynthetic pathways through cross-species gene recombination. As the core components of the system, the natural catalytic properties of enzyme proteins often deviate from the industrial requirements, which restricts their wide application. Glycosylation modification of plant natural products is mainly mediated by UGTs (Siedhoff et al., 2020). However, the catalytic activity, stability, and substrate specificity of most natural UGTs are relatively low, which limits their application in the microbial biosynthesis of natural products. To address this challenge, enzyme modification technologies have been developed and can be broadly categorized based on their underlying principles: traditional enzyme engineering focuses on directed evolution, while rational and semi-rational redesign approaches leverage sequence and structural information to optimize natural enzymes (Guo et al., 2021) (Figure 2c).
3.1.2 Homology modeling and molecular docking in protein research
Zhang et al. investigated the substrate recognition mechanism of UGT73F24 from G. uralensis towards glycyrrhetinic acid and UDP-glucose using homology modeling and molecular docking analysis. Based on the identified recognition mechanism, they selected amino acids located near the C3-OH group of glycyrrhetinic acid and the glucose moiety of UDP-glucose, as well as residues in the substrate-binding pocket, as candidate sites for site-directed mutagenesis. They identified two key residues, I23 and L84, and the combination of mutations I23G/L84N resulted in a 4.1-fold increase in activity (Zhang et al., 2020). In a similar study, Chen et al. analyzed the hydrogen bond interactions between UGT76G1 from S. rebaudiana and UDP-glucose. They identified the conserved residues His17 and Asp359, and selected Asn358, located near the substrate channel, as a target for saturation mutagenesis. The N358F mutant, when incorporated into a multi-enzyme reaction system, led to a 60% increase in the yield of rebaudioside D (Chen et al., 2020). Bi et al. engineered the 1,6-glucosyltransferase CaUGT3 by substituting valine for residue T145, thereby converting the enzyme into a xylosyltransferase capable of catalyzing the conversion of cinnamyl alcohol monoglucoside (rosin) into rosavin E. Further enhancement of enzyme activity (2.9-fold increase) was achieved by introducing the N375Q mutation. The synthesis of rosavin E from glucose was successfully carried out, reaching a final concentration of 92.9 mg/L, by combining the CaUGT3T145V/N375Q variant with UDP-Xly synthase from S. meliloti 1021 (SmUXS) and the enzymes involved in rosin biosynthesis, all expressed in a phenylalanine-overproducing E.coli strain (Bi et al., 2022). Li et al. aimed to address the relatively low catalytic efficiency of triterpene-class UGTs. They selected UGT74AC1, a glycosyltransferase from S. grosvenorii, and analyzed its crystal structure. Using this structural insight as a foundation, they applied directed evolution and sequence/structure-based engineering to enhance its catalytic properties. Several resulting UGT variants demonstrated a remarkable 102- to 104-fold improvement in catalytic efficiency for triterpene glycosylation. One of these mutants has 4.17 × 104 times higher catalytic efficiency against mogrol and 1.53 × 104 times higher catalytic activity against UDP-glucose (Li et al., 2020). Chen et al. identified the AmGT1G146V/I mutants through sequence alignment, molecular docking, and site-directed mutagenesis, which specifically utilize UDP-Xyl while showing no activity toward UDP-Glc. By combining AmGT1/5/9 and AmGT1G146V/S with the previously reported AmGT8 and its mutant AmGT8A394F, they accomplished the combinatorial synthesis of 13 cycloartane-type saponins from A. membranaceus (Chen et al., 2023).
3.1.2 Directed evolution of enzymes for improved catalytic performance
Directed evolution mimics natural gene mutation and selection processes through repeated mutagenesis and screening (Sinha and Shukla, 2019). This approach identifies enzyme variants with increased activity, substrate specificity, or thermal stability using methods, such as random mutagenesis, site-directed mutagenesis, and DNA recombination (Figure 7A). Although powerful for engineering triterpene-producing enzymes, the technique faces challenges in managing vast mutation spaces during combinatorial mutagenesis, complicating optimization efforts (Thieker et al., 2022).
As for the biosynthesis of ginsenoside, low catalytic efficiency of UGTs often limits the yields. Researchers solved it by creating mutant libraries via error-prone PCR and screening variants through HPLC or fluorescent assays. This strategy was applied to PgUGT74AE2, resulting in mutants with higher catalytic efficiency and improved ginsenoside production (Ji et al., 2025). To expand the substrate range, glycyrrhizin pathway studies combined DNA recombination with rational design on GmUGAT, and engineered mutants capable of processing non-native substrates (Guo et al., 2021). Thermal stability improvements were achieved by evolving UGT variants through combinatorial mutagenesis, yielding heat-tolerant enzymes for industrial production (Bian et al., 2024).
The above cases demonstrated the capacity of directed evolution to increase enzyme activity by screening random mutations for desirable traits. The improved catalytic efficiency of PgUGT74AE2 reflected the improved yield, while the GmUGAT modification expanded the substrate range through optimization of the binding pocket. Despite the challenges in navigating extensive mutation field, technological advances promise to streamline mutant screening processes.
3.1.3 Rational and semi-rational design in enzyme engineering
Rational and semi-rational design have become fundamental approachfor enhancing enzymatic capabilities through structural and functional insights. These approaches commonly employ targeted amino acid substitutions or combinatorial mutation libraries, focusing on improving catalytic traits. Existing sequence databases combined with crystallographic evidence allow accurate identification of mutation hotspots, substantially streamlining experimental workflows while maintaining discovery efficiency. These strategies also illuminate fundamental structure-function relationships underlying enzymatic enhancement. Active site residues and adjacent regions are typically grouped into distinct layers based on their distance from the binding substrates. Structural alterations within active site residues occasionally yield unexpected stereochemical refinements, whereas mutations at sites farther from the active center often lead to measurable improvements in thermal tolerance and operational stability (Patsch et al., 2024) (Figure 7B). The expanding repository of high-resolution protein structures, augmented by computational modeling advancements, has enabled new frameworks for systematic enzyme engineering. Contemporary semi-rational platforms like 3DM and HotSpot Wizard integrate multidimensional structural data with evolutionary information, demonstrating particular promise for tailoring biocatalyst performance (Cho et al., 2021; Bendl et al., 2016). For instance, 3DM uses structure-based multiple sequence alignments to identify key residues affecting enzyme activity, drawing on structural-functional relationships from databases like PDB, GenBank, PubMed, and Swiss-Prot. These predictions are subsequently validated through multi-site saturation mutagenesis. Similarly, HotSpot Wizard integrates multiple databases and computational tools, both structural and evolutionary, to predict residues that influence enzyme stability, activity, and substrate specificity.
The practical application of these methods is evident in various case studies. Zhang et al. investigated A. membranaceus and discovered the first cycloartane-type triterpene glycosyltransferase, AmGT8, which catalyzes two consecutive glycosylation reactions at the 3-OH and 2-OH positions of cycloastragenol. Through targeted semi-rational engineering, researchers developed three AmGT8 variants (A394D, A394F, T131V) that showed specialized glycosylation patterns at distinct oxygen positions. These engineered catalysts enabled efficient biosynthesis of Astragalus-derived saponins through accurate glycosyl moiety attachment (Zhang et al., 2022). In parallel work, Xie et al. used a structurally directed mutagenesis technique to redesign the 4αGT from Synechocystis sp. PCC 6803. By analyzing residue spatial configurations through HotSpot Wizard, they created iincreased variants that improved enzymatic hydrolysis efficiency, subsequently scaled for commercial production (Jingwen et al., 2024). Comparative analysis of UGT91H_A1 and UGT91H8 revealed an important RTAS motif (R212/T213/A214/S215) governing substrate preference. Transplantation of this sequence into UGT91H8 resulted in the UGT91H8_6mu variant, achieving 15.9% catalytic conversion for compound 17 through optimized binding geometry. Computational modeling showed how this motif regulates substrate-channel interactions (Jian et al., 2024). Further reseach demonstrated that GuGT14 engineering yielded H47P and I182L mutants exhibiting 101.74% and 405.78% activity respectively versus wild-type. These optimized catalysts significantly improved Rh2 ginsenoside yields, establishing viable production routes for this pharmaceutically valuable compound (Yan et al., 2024).
Effective enzyme engineering requires a detailed understanding of structural-functional relationships governing catalytic behavior, which enables accurate alterations at key amino acid positions to achieve desired enzymatic improvements (Qian et al., 2021). The combination of rational and semi-rational design approaches has proved very effective in enzyme engineering, not only providing insights into the mechanisms of enzyme function.
4 Synthesis of glycosyl donors for glycosylation reactions
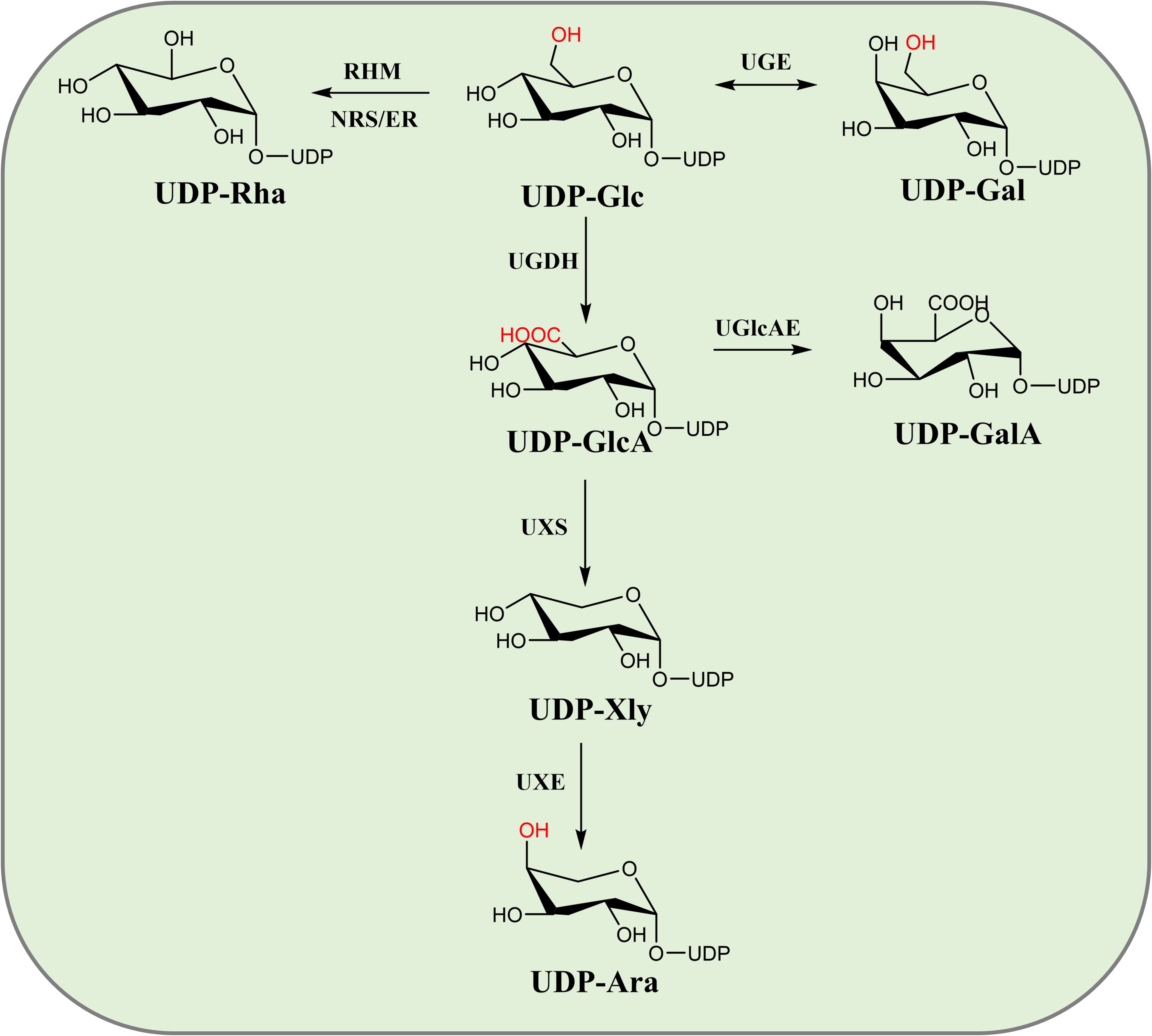
Triterpenoid saponins are composed of a glycoside moiety and an aglycone linked by glycosidic bonds, catalyzed by UGTs. The substrates for these enzymes include glycosyl acceptors and donors. However, the diversity of glycosyl donors in most host cells is limited, which may not meet the demands for synthesizing complex natural compounds. Various glycosyl donors can be interconverted (Figure 2d). Sucrose, under the catalysis of sucrose synthase (SUS), can produce UDP-Glc. The conversion of UDP-Glc to UDP-Ara involves three key metabolic enzymes in a three-step process: dehydrogenation, decarboxylation and epimerization. UDP-Glc is converted to UDP-GlcA by UDP-glucose dehydrogenase (UGDH). UDP-GlcA is then catalyzed by UDP-xylose synthase to form UDP-Xyl. UDP-Xyl is enzymatically transformed into UDP-Ara through UDP-xylose-4-epimerase (UXE)-mediated structural rearrangement. Meanwhile, rhamnose mutase (RHM) catalyzes the conversion of UDP-Glc to UDP-Rha, while UDP-arabinose 4-epimerase (UAXS) processes UDP-GlcA into UDP-Cel. The same UDP-GlcA precursor can alternatively yield UDP-galacturonic acid via UGlcAE activity (Sen and Jian-qiang, 2016) (Figure 8). These biochemical pathways have driven significant interest in synthesizing diverse glycosyl donors through exogenous gene integration, both in laboratory settings and biological systems.
4.1 In Vitro synthesis of glycosyl donors for glycosylation reactions
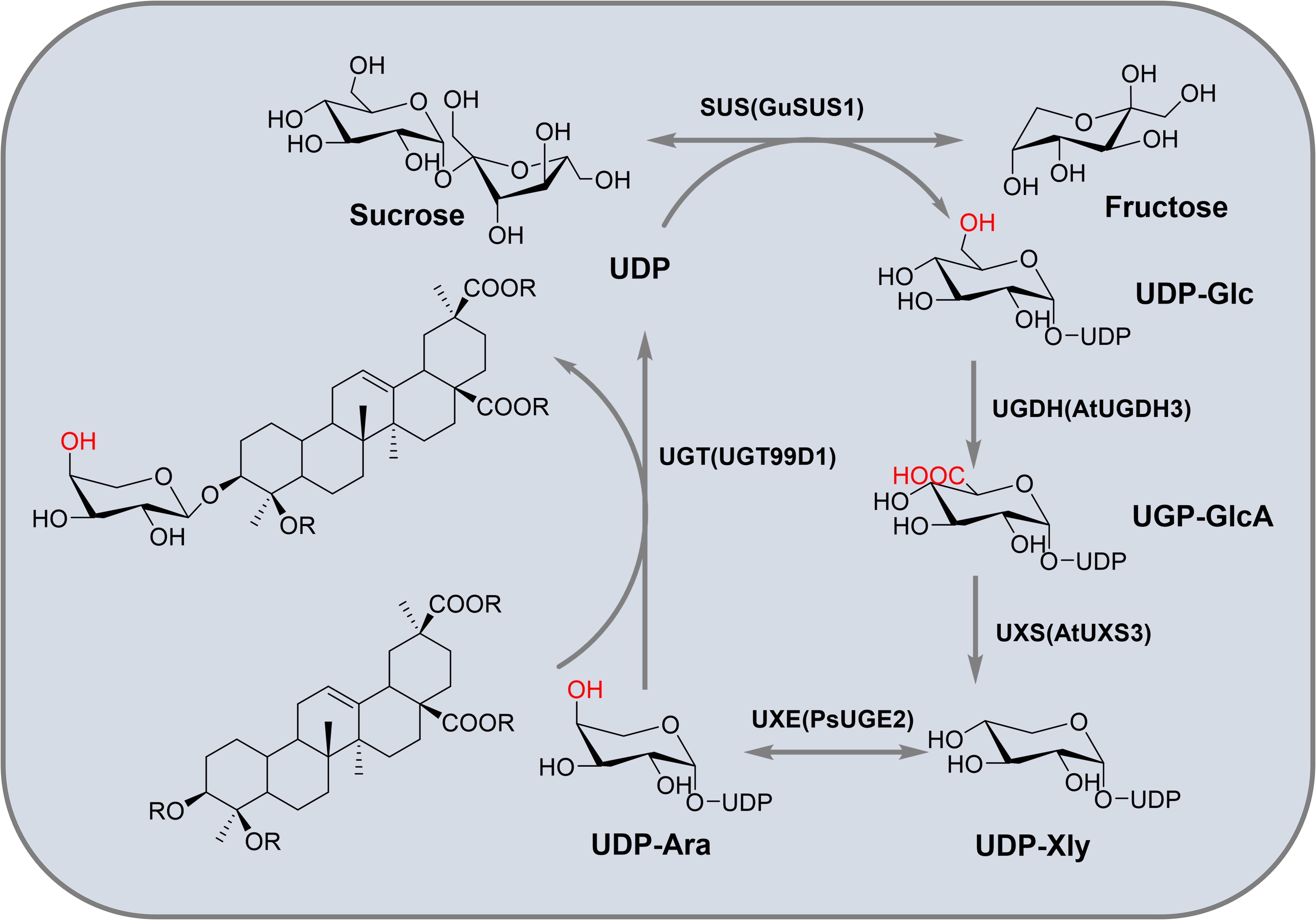
In vitro synthesis of glycosyl donors offers flexibility and cost-effectiveness. Sun et al. screened several key enzymes to facilitate the production of glycosyl donors in microbial systems. These included GuSUS1 from G. uralensis, AtUGDH3 and AtUXS3 from A. thaliana, and PsUGE2 from P. sativum, which collectively enable the conversion of sucrose into arabinose. The successful production of arabinose was confirmed by using the arabinose transferase UGT99D1 in A. sativaL (Sun et al., 2023a). This strategy not only facilitates the generation of glucose, glucuronic acid, xylose, and arabinose donors, but also addresses a significant challenge in the biosynthesis of triterpenoid saponins. Lay the foundation for the microbial synthesis of natural glycoside compounds. They successfully achieved the in vitro production of UDP-Ara and catalyzed the synthesis of Ara-BA in vitro (Figure 9).
UDP-glucose transferase can be coupled with sucrose synthase to construct synthetic pathways with different glycosyl donors, using inexpensive sucrose as the substrate for the glycosylation of natural products. However, the hydrolysis of sucrose leads to the accumulation of by-product fructose, which reduces the atomic economy of sucrose and inhibits in situ UDP recycling. To improve sucrose utilization, Wang et al. first demonstrated that a polyphosphate-dependent glucose kinase can convert fructose into fructose-6-phosphate without relying on expensive ATP. They then introduced the glucose kinase into the UDP-2E recycling system, constructing an improved three-enzyme UDP (UDP-3E) recycling system. This system enhances the glycosylation efficiency of triterpenoids by phosphorylating fructose, accelerates sucrose hydrolysis and UDP recycling, and improves the overall utilization of sucrose (Wang et al., 2023b).
Song et al. conducted a genomic analysis of the Pacific oyster and identified three genes responsible for catalyzing the conversion of UDP-Gal, UDP-GlcA, and UDP-Xyl. Enzymatic assays demonstrated that CGIUGE catalyzes the conversion of UDP-Glc to UDP-Gal, CGIUGD catalyzes the conversion of UDP-Glc to UDP-GlcA, and CGIUXS catalyzes the conversion of UDP-GlcA to UDP-Xyl. These findings enable the in vitro synthesis of UDP-Gal, UDP-GlcA, and UDP-Xyl using UDP-Glc as the starting substrate (Song, 2017).
Yin et al. cloned 17 key enzyme genes (OcUXS1-6, OcUAXS1/2, OcUGE1/2, OcUXE1/2, OcRHM1, OcNER1, OcGlcAE1/2/3) involved in the biosynthesis of diversified natural small molecule glycosides from O. caudatum, which catalyze the formation of key sugar nucleotide donors such as UDP-Xyl, UDP-Ara, UDP-Gal, UDP-GalA, and UDP-Rha. Through functional characterization of these enzymes, they successfully achieved the in vitro synthesis of UDP-Xyl, UDP-Ara, UDP-Gal, UDP-GalA, and UDP-Rha using UDP-Glc as a precursor (Yin, 2016). Jian et al. achieved de novo synthesis of UDP-Rha in vitro by coupling the GuSUS1-Δ9, AtRHM2, and AtNRS/ER genes. The UDP-Rha regeneration system, combined with UGT, holds great potential for the efficient production of glycosylated triterpenoid compounds (Jian et al., 2024). These studies suggest that in vitro synthesis holds great potential for the production of diverse glycosyl donors. They also highlight the potential of E. coli as a host for glycosyl donor synthesis.
4.2 The synthesis of glycosyl donors within microorganisms
The limitations of in vitro glycosyl donor synthesis highlight the advantages of microbial production systems that are more efficient, cost-effective, and scalable. Three microbial platforms including E. coli, S. cerevisiae, and B. subtilis dominate current research. The robust metabolism and rapid growth of E. coli enable efficient protein production, with shorter cultivation cycles making it ideal for industrial-scale operations (Rosano et al., 2019). S. cerevisiae stands out for its eukaryotic machinery, particularly the endoplasmic reticulum, which supports plant enzyme function, and a natural UDP-Glc reserve that can facilitate synthesis of glycosyl donor precursors (Long et al., 2024). B. subtilis is prominent in recombinant protein secretion and application in agricultural biotechnology (Pramastya et al., 2021).
Li et al. achieved de novo synthesis of UDP-Ara through coordinated expression of UGDH, XylS, and UXE genes, resulting in rosavin production. Bi et al. optimized UDP-Xly synthesis by overexpressing AtUXS3/SmUXS to produce rosavin E (Bi et al., 2022). Liu et al. established UDP-Rha/Gal biosynthesis pathway in E. coli to produce six different flavonoid glycosides (Liu et al., 2023). Zhao et al. demonstrated the complete madecassoside synthesis through VvRHM-NRS-mediated UDP-Rha conversion (Zhao et al., 2024).
S. cerevisiae platforms utilizes native UDP-Glc pools for donor synthesis. Oka and Jigami engineered yeast strains expressing plant AtUGD1/AtUXS3 to convert UDP-Glc to UDP-Xyl (Oka and Jigami, 2006). Zhang et al. achieved 2.53 g/L 2’-fucosyllactose production through integrated lactose/GDP-Fuc modules (Zhang et al., 2024b).
These microbial engineering strategies address critical challenges in triterpenoid saponin biosynthesis by diversifying glycosyl donor availability (Guihang et al., 2023). While each platform demonstrates unique strengths –the scalability of E. coli, the eukaryotic compatibility of yeast, and the secretion capacity of B. subtilis– future progress requires optimized chassis engineering (Xu et al., 2020; Shi and Jiang, 2020) and novel host exploration (Tianli et al., 2024). Strategic integration of synthetic biology tools will drive industrial-scale production of bioactive glycosides (Table 2).
5 Conclusion and future perspectives
In recent years, triterpene saponins have made important breakthroughs in the discovery and modification of UGTs and the synthesis of glycosyl donors (Li et al., 2023). These advances not only deepen the understanding of the biosynthetic mechanism of triterpene saponins, but also open up a new path for their industrial production. Using microbial platforms to synthesize triterpenoids has become an efficient and sustainable biomanufacturing strategy: Liu et al. achieved full synthesis of a complex vaccine adjuvant QS-21 in engineered yeast in 2024 (Liu et al., 2024); In the same year, Martin et al. completed the whole biosynthesis of QS-21 in tobacco by integrating a 20-step reaction (Martin et al., 2024). The Zhao team first synthesized asiaticoside in S. cerevisiae using oxidase and glycosyltransferase, and the yield reached 772.3 μg/L (Zhao et al., 2024). Wang et al. increased ginsenoside Rh2 production to 2.25 g/L by fed-feed fermentation (Wang et al., 2019). Microbial de novo synthesis of triterpenoid compounds such as ginsenosides has been achieved by many teams around the world through systematic metabolic engineering strategies such as heterologous pathway reconstruction and precursor supply optimization (Ren et al., 2022). The core of such platforms lies in pathway design, dynamic regulation and cross-scale optimization. With the development of synthetic biology tools such as CRISPR and AI enzyme design, the industrial production of more high-value triterpenoid compounds is expected to gradually replace the traditional plant extraction method (Li et al., 2023; Kim et al., 2017) (Figure 2e).
In terms of UGTs discovery and identification, the combination of genome-transcriptome-metabolome studies has significantly improved the UGTs screening efficiency (Chen et al., 2024). Gene cluster mapping, phylogenetic analysis, and application of PSPG motifs provide new insights into UGTs function (Zhan et al., 2022; Gharabli and Welner, 2024). In UGTs engineering, rational and semi-rational design strategies have effectively improved enzyme activity, substrate specificity, and stability (Chica et al., 2005). By using directed evolution, UGTs mutants with 2–5 times higher catalytic efficiency were obtained (Zhang et al., 2024a). In the field of glycan donor synthesis, the strategy of exogenous gene introduction has successfully overcome the limitation of the diversity of glycan donors in host cells: engineered microorganisms such as E. coli and yeast have been able to efficiently synthesize a variety of glycan nucleotides (Li et al., 2024; Oka and Jigami, 2006).
Despite progress, the field still faces many challenges. Functional annotation of UGTs requires a large number of experimental verification and the complexity of gene clusters and plant genome diversity increase the difficulty of comprehensive identification. The prediction of mutation effects in UGTs engineering is still challenging, and directed evolution is limited by the large mutation space and high-throughput screening requirements. The cost of in vitro synthesis of glycosyl donors is high, and the in vivo synthesis needs to optimize the metabolic network to improve the efficiency (Zhou et al., 2022).
In the future, it is necessary to deeply integrate multi-omics data to analyze the triterpene synthesis regulatory network (Chai et al., 2023) and develop a high-throughput screening platform to accelerate UGTs validation and glycodonor pathway optimization (Kwon et al., 2024). The application of machine learning and AI technology in enzyme function prediction and metabolic pathway design will accelerate the construction of efficient microbial factories (Jang et al., 2022). Developing a novel microbial host that can expand the synthesis pathway of glycans donors (Tianli et al., 2024) and in-depth understanding of the regulatory mechanism of triterpene synthesis will help to improve the yield of engineered strains (Yuanyuan et al., 2021). Continuous technological innovation will promote the industrial application of triterpene saponins in medicine, food and other fields.
Author contributions
LH: Validation, Writing – original draft, Writing – review & editing. YL: Data curation, Investigation, Writing – review & editing. GD: Data curation, Writing – review & editing. JL: Investigation, Writing – review & editing. KQ: Data curation, Investigation, Writing – review & editing. XL: Investigation, Writing – review & editing. YQ: Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82104031) and Science and Technology Support Plan for Youth Innovation of Colleges and Universities of Shandong Province of China 2022KJ264.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alami, M. M., Liu, S., Gong, D., Guo, S., Shaohua, S., Mei, Z., et al. (2023). Effects of excessive and deficient nitrogen fertilizers on triptolide, celastrol, and metabolite profile content in Tripterygium wilfordii Hook F. Industrial Crops Products 206(000), 14. doi: 10.1016/j.indcrop.2023.117577
Andong, Y., Lin, L., Ruicai, L., Junmei, K., Lin, C., Qingchuan, Y., et al. (2022). Function and application prospect of plant UDP-glycosyltransferase (UGT) (058-004). Plant Physiol. J. 58, 631–642. doi: 10.13592/j.cnki.ppj.2021.0128
Augustin, J. M., Drok, S., Shinoda, T., Sanmiya, K., Nielsen, J. K., Khakimov, B., et al. (2012). UDP-glycosyltransferases from the UGT73C subfamily in Barbarea vulgaris catalyze sapogenin 3-O-glucosylation in saponin-mediated insect resistance. Plant Physiol. 160, 1881–1895. doi: 10.1104/pp.112.202747
Barbi, M., Macabeo, A. P. G., Kreft, S., and Heilmann, J. R. (2010). Cycloastragenol glycosides from Astragalus illyricus. Biochem. Systematics Ecology. 38, 460–462. doi: 10.1016/J.BSE.2010.03.016
Bendl, J., Stourac, J., Sebestova, E., Vavra, O., Musil, M., Brezovsky, J., et al. (2016). HotSpot Wizard 2.0: automated design of site-specific mutations and smart libraries in protein engineering. Nucleic Acids Res. 44, W479–W487. doi: 10.1093/nar/gkw416
Bharadwaj, R., Kumar, S. R., Sharma, A., and Sathishkumar, R. (2021). Plant metabolic gene clusters: evolution, organization, and their applications in synthetic biology. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.697318
Bi, H., Qu, G., Wang, S., Zhuang, Y., Sun, Z., Liu, T., et al. (2022). Biosynthesis of a rosavin natural product in Escherichia coli by glycosyltransferase rational design and artificial pathway construction. Metab. Eng. 69, 15–25. doi: 10.1016/j.ymben.2021.10.010
Bian, J., Tan, P., Nie, T., Hong, L., and Yang, G. Y. (2024). Optimizing enzyme thermostability by combining multiple mutations using protein language model. mLife 3, 492–504. doi: 10.1002/mlf2.12151
Bihuan, C., Xiangyu, L., Cuixia, L., Yina, W., Sufang, P., Simei, H., et al. (2024). Prokaryotic expression and functional verification of rhamnose glycosyltransferase from panax quinquefolius. Mol. Plant Breed., 1–13. doi: 10.1038/s42003-022-03740-y
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–w50. doi: 10.1093/nar/gkad344
Chai, K., Chen, S., Wang, P., Kong, W., Ma, X., and Zhang, X. (2023). Multiomics analysis reveals the genetic basis of volatile terpenoid formation in oolong tea. J. Agric. Food Chem. 71, 19888–19899. doi: 10.1021/acs.jafc.3c06762
Chakraborty, P. (2022). Gene cluster from plant to microbes: Their role in genome architecture, organism’s development, specialized metabolism and drug discovery. Biochimie 193, 1–15. doi: 10.1016/j.biochi.2021.12.001
Challinor, V. L. and De Voss, J. J. (2013). Open-chain steroidal glycosides, a diverse class of plant saponins. Nat. Prod Rep. 30, 429–454. doi: 10.1039/c3np20105h
Chen, L., Cai, R., Weng, J., Li, Y., Jia, H., Chen, K., et al. (2020). Production of rebaudioside D from stevioside using a UGTSL2 Asn358Phe mutant in a multi-enzyme system. Microbial Biotechnol. 13, 974–983. doi: 10.1111/1751-7915.13539
Chen, Z., Ting, G., Jingjing, C., Tianjiao, C., and Jinling, Y. (2022). The glycosyltransferases involved in triterpenoid saponin biosynthesis: a review. Chinese J. Biotechnol. 38 (03), 1004–1024. doi: 10.13345/j.cjb.210587
Chen, D., Chen, X., Zheng, X., Zhu, J., and Xue, T. (2024). Combined metabolomic and transcriptomic analysis reveals the key genes for triterpenoid biosynthesis in Cyclocarya paliurus. BMC Genomics 25, 1197. doi: 10.1186/s12864-024-11125-0
Chen, K., Zhang, M., Gao, B., Hasan, A., Li, J., Bao, Y., et al. (2023). Characterization and protein engineering of glycosyltransferases for the biosynthesis of diverse hepatoprotective cycloartane-type saponins in Astragalus membranaceus. Plant Biotechnol. J. 21, 698–710. doi: 10.1111/pbi.13983
Cheng, Y., Liu, H., Tong, X., Liu, Z., Zhang, X., Li, D., et al. (2020). Identification and analysis of CYP450 and UGT supergene family members from the transcriptome of Aralia elata (Miq.) seem reveal candidate genes for triterpenoid saponin biosynthesis. BMC Plant Biol. 20, 214. doi: 10.1186/s12870-020-02411-6
Chica, R. A., Doucet, N., and Pelletier, J. N. (2005). Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 16, 378–384. doi: 10.1016/j.copbio.2005.06.004
Cho, E. S., Han, S., Lee, K. H., Kim, C. H., and Yoon, Y. G. (2021). 3DM: deep decomposition and deconvolution microscopy for rapid neural activity imaging. Optics express 29, 32700–32711. doi: 10.1364/oe.439619
Chung, S. Y., Seki, H., Fujisawa, Y., Shimoda, Y., Hiraga, S., Nomura, Y., et al. (2020). A cellulose synthase-derived enzyme catalyses 3-O-glucuronosylation in saponin biosynthesis. Nat. Commun. 11, 5664. doi: 10.1038/s41467-020-19399-0
Cravens, A., Payne, J., and Smolke, C. D. (2019). Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10, 2142. doi: 10.1038/s41467-019-09848-w
Dai, X. and Shen, L. (2022). Advances and trends in omics technology development. Front. Med. 9. doi: 10.3389/fmed.2022.911861
De Costa, F., Barber, C. J. S., Kim, Y. B., Reed, D. W., Zhang, H., Fett-Neto, A. G., et al. (2017). Molecular cloning of an ester-forming triterpenoid: UDP-glucose 28-O-glucosyltransferase involved in saponin biosynthesis from the medicinal plant Centella asiatica. Plant science: an Int. J. Exp. Plant Biol. 262, 9–17. doi: 10.1016/j.plantsci.2017.05.009
Dong, M., Li, J., Yang, D., Li, M., and Wei, J. (2023). Biosynthesis and pharmacological activities of flavonoids, triterpene saponins and polysaccharides derived from astragalus membranaceus. Molecules (Basel Switzerland) 28, (13). doi: 10.3390/molecules28135018
Duan, Y., Du, W., Song, Z., Chen, R., Xie, K., Liu, J., et al. (2023). Functional characterization of a cycloartenol synthase and four glycosyltransferases in the biosynthesis of cycloastragenol-type astragalosides from Astragalus membranaceus. Acta Pharm. Sin. B 13, 271–283. doi: 10.1016/j.apsb.2022.05.015
Feng, L., Yao, Y., Kang, M., Yang, W., Han, Y., Liu, W., et al. (2024). Integrated genomic, transcriptomic, and metabolomic analyses of Ilex hylonoma provide insights into the triterpenoid saponin biosynthesis. Plant journal: Cell Mol. Biol. 120, 1176–1189. doi: 10.1111/tpj.17046
Fu, A., Wang, Q., Mu, J., Ma, L., Wen, C., Zhao, X., et al. (2021). Combined genomic, transcriptomic, and metabolomic analyses provide insights into chayote (Sechium edule) evolution and fruit development. Horticulture Res. 8, 35. doi: 10.1038/s41438-021-00487-1
Gharabli, H. and Welner, D. H. (2024). The sugar donor specificity of plant family 1 glycosyltransferases. Front. bioengineering Biotechnol. 12, 697–711. doi: 10.3389/fbioe.2024.1396268
Gloster, T. M. (2014). Advances in understanding glycosyltransferases from a structural perspective. Curr. Opin. Struct. Biol. 28, 131–141. doi: 10.1016/j.sbi.2014.08.012
Guihang, C., Chun, L., and Xudong, F. (2023). Research progress of UDP-sugar biosynthesis. J. Biol. 40, 95–100. doi: 10.3969/j.issn.2095-1736.2023.02.095
Guo, L., Winzer, T., Yang, X., Li, Y., Ning, Z., He, Z., et al. (2018). The opium poppy genome and morphinan production. Science 362, 343–347. doi: 10.1126/science.aat4096
Guo, F., Zhang, L., X-d, F., and Li, C. (2021). Plant-derived UDP-glycosyltransferase and its molecular modification. China Biotechnol. 41, 78–91. doi: 10.13523/j.cb.2105001
Hassan, A. S., O’Donovan, L. A., Cowley, J. M., Akomeah, B., Phillips, R. J., Pettolino, F., et al. (2024). In planta ectopic expression of two sub-types of tomato cellulose synthase-like M (CslM) genes affects cell wall integrity and supports a role in arabinogalactan and/or rhamnogalacturonan-I biosynthesis. Plant Cell Physiol., 101–119. doi: 10.1093/pcp/pcae145
Hazra, A., Dutta, M., Dutta, R., Bhattacharya, E., Bose, R., and Biswas, S. M. (2023). Squalene synthase in plants – Functional intricacy and evolutionary divergence while retaining a core catalytic structure. Plant Gene, 100403. doi: 10.1016/j.plgene.2023.100403
He, Y. (2022). Study on synthetic pathway of saikosaponin by combining transcriptome and metabolome (Southwest University of Science and Technology). Master.
He, Y., Chen, H., Zhao, J., Yang, Y., Yang, B., Feng, L., et al. (2021). Transcriptome and metabolome analysis to reveal major genes of saikosaponin biosynthesis in Bupleurum chinense. BMC Genomics 22, 839. doi: 10.1186/s12864-021-08144-6
Hucheng, Y., Yini, X., Yuqi, R., Mohan, X., Ziyue, L., and Dexin, K. (2023). Progress on the classification, evolution and substrate prediction of natural product glycosyltransferase. Chin. Bull. Life Sci. 35, 1160–1168. doi: 10.13376/j.cbls/2023127
Hyokchol, R. (2024). Identification of the gene for the synthesis of pentacyclic triterpene saponins of Aralia and analysis of the catalytic mechanism of glycosyltransferase, doctorate. (Doctoral Electronic Journal Publishing Information). 10.27009/d.cnki.gdblu.2024.000134
Jang, W. D., Kim, G. B., Kim, Y., and Lee, S. Y. (2022). Applications of artificial intelligence to enzyme and pathway design for metabolic engineering. Curr. Opin. Biotechnol. 73, 101–107. doi: 10.1016/j.copbio.2021.07.024
Jendoubi, T. (2021). Approaches to integrating metabolomics and multi-omics data: A primer. Metabolites 11, (3). doi: 10.3390/metabo11030184
Jensen, P. R. (2016). Natural products and the gene cluster revolution. Trends Microbiol. 24, 968–977. doi: 10.1016/j.tim.2016.07.006
Ji, X., Lin, S., Chen, Y., Liu, J., Yun, X., Wang, T., et al. (2020). Identification of α-amyrin 28-carboxylase and glycosyltransferase from ilex asprella and production of ursolic acid 28-O-β-D-glucopyranoside in engineered yeast. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00612
Ji, Q., Liu, Y., Zhang, H., Gao, Y., Ding, Y., Ding, Y., et al. (2025). Structural insights into the substrate recognition of ginsenoside glycosyltransferase pq3-O-UGT2 (Baden-Wurttemberg, Germany: Weinheim). doi: 10.1002/advs.202413185
Jian, X., Sun, Q., Xu, W., Qu, H., Feng, X., and Li, C. (2024). Engineering the substrate specificity of UDP-glycosyltransferases for synthesizing triterpenoid glycosides with a linear trisaccharide as aided by ancestral sequence reconstruction. Angewandte Chemie (International ed English) 63, e202409867. doi: 10.1002/anie.202409867
Jiang, Z., Gao, H., Liu, R., Xia, M., Lu, Y., Wang, J., et al. (2022). Key glycosyltransferase genes of panax notoginseng: identification and engineering yeast construction of rare ginsenosides. ACS synthetic Biol. 11, 2394–2404. doi: 10.1021/acssynbio.2c00094
Jingwen, X., Chaohui, L., Meng, W., Wei, Z., and Jinxiang, Z. (2024). Semi-rational design to enhance the disproportionation activity of 4-α-glycosyltransferase and its application in industrial production. Food Fermentation Industries. 50, 45–52. doi: 10.13995/j.cnki.11-1802/ts.038159
Jones, J., Murphy, C. P., Sleator, R. D., and Culligan, E. P. (2024). An exploratory in silico analysis of bacteriocin gene clusters in the urobiome. Microbiome Res. Rep. 3, 24. doi: 10.20517/mrr.2023.78
Kaminaga, Y., Sahin, F. P., and Mizukami, H. (2004). Molecular cloning and characterization of a glucosyltransferase catalyzing glucosylation of curcumin in cultured Catharanthus roseus cells. FEBS Lett. 567, 197–202. doi: 10.1016/j.febslet.2004.04.056
Kautsar, S. A., Suarez Duran, H. G., and Medema, M. H. (2018). Genomic identification and analysis of specialized metabolite biosynthetic gene clusters in plants using plantiSMASH. Methods Mol. Biol. (Clifton NJ) 1795, 173–188. doi: 10.1007/978-1-4939-7874-8_15
Kim, S. K., Seong, W., Han, G. H., Lee, D. H., and Lee, S. (2017). CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb Cell Fact. 16, 188. doi: 10.1186/s12934-017-0802-x
Kurze, E., Wüst, M., Liao, J., McGraphery, K., Hoffmann, T., Song, C., et al. (2022). Structure-function relationship of terpenoid glycosyltransferases from plants. Natural product Rep. 39, 389–409. doi: 10.1039/d1np00038a
Kwon, K. K., Lee, J., Kim, H., Lee, D. H., and Lee, S. G. (2024). Advancing high-throughput screening systems for synthetic biology and biofoundry. Curr. Opin. Syst. Biol. 37, 1–8. doi: 10.1016/j.coisb.2023.100487
Le, D. D., Kim, W., Lim, S., Kim, S. C., and Choi, G. (2021). Identification of three groups of ginsenoside biosynthetic UDP-glycosyltransferases from Gynostemma pentaphyllum. Plant science: an Int. J. Exp. Plant Biol. 313, 111069. doi: 10.1016/j.plantsci.2021.111069
Li, Y. (2023). Functional study of panax notoginseng glycosyltransferase genes PnUGT6350, PnUGT3498 and PnUGT4291, Master. doi: 10.27009/d.cnki.gdblu.2023.000255
Li, Y., Li, P., Wang, Y., Dong, R., Yu, H., and Hou, B. (2014). Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 239, 1265–1279. doi: 10.1007/s00425-014-2050-1
Li, L., Liu, M., Bi, H., and Liu, T. (2024). High-level production of Rhodiola rosea characteristic component rosavin from D-glucose and L-arabinose in engineered Escherichia coli. Metab. Eng. 82, 274–285. doi: 10.1016/j.ymben.2024.02.017
Li, J., Liu, S., Cui, T., Ding, B., Zhou, Z., and Li, L. (2022). Identification and expression characteristics of UDP-glycosyltransferase genes in pear and their correlation with arbutin accumulation. Russian J. Plant Physiol. 69, 1–12. doi: 10.1134/S1021443722050120
Li, Y., Wang, J., Li, L., Song, W., Li, M., Hua, X., et al. (2023). Natural products of pentacyclic triterpenoids: from discovery to heterologous biosynthesis. Natural product Rep. 40, 1303–1353. doi: 10.1039/d2np00063f
Li, J., Yang, J., Mu, S., Shang, N., and Ma, Y. J. A. C. (2020). Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catalysis. 3629–3639. doi: 10.1021/acscatal.9b05232
Lin, M., Jian, J. B., Zhou, Z. Q., Chen, C. H., Wang, W., Xiong, H., et al. (2022). Chromosome-level genome of Entada phaseoloides provides insights into genome evolution and biosynthesis of triterpenoid saponins. Mol. Ecol. Resour. 22, 3049–3067. doi: 10.1111/1755-0998.13662
Liu, S., Li, D., Qin, Z., Zeng, W., and Zhou, J. (2023). Enhancing glycosylation of flavonoids by engineering the uridine diphosphate glucose supply in escherichia coli. J. Agric. Food Chem. 71, 17842–17851. doi: 10.1021/acs.jafc.3c05264
Liu, J., Li, W., Wang, L., Li, J., Li, E., and Luo, Y. (2022). Multi-omics technology and its applications to life sciences: a review. Sheng wu gong cheng xue bao = Chin. J. Biotechnol. 38, 3581–3593. doi: 10.13345/j.cjb.220724
Liu, S. J., Liu, Z., Shao, B. Y., Li, T., Zhu, X., Wang, R., et al. (2025). Deciphering the biosynthetic pathway of triterpene saponins in Prunella vulgaris. Plant journal: Cell Mol. Biol. 121, e17220. doi: 10.1111/tpj.17220
Liu, Y., Zhao, X., Gan, F., Chen, X., Deng, K., Crowe, S. A., et al. (2024). Complete biosynthesis of QS-21 in engineered yeast. Nature 629, 937–944. doi: 10.1038/s41586-024-07345-9
Long, Y., Han, X., Meng, X., Xu, P., and Tao, F. (2024). A robust yeast chassis: comprehensive characterization of a fast-growing Saccharomyces cerevisiae. mBio 15, e0319623. doi: 10.1128/mbio.03196-23
Louveau, T., Orme, A., Pfalzgraf, H., Stephenson, M. J., Melton, R., Saalbach, G., et al. (2018). Analysis of two new arabinosyltransferases belonging to the carbohydrate-active enzyme (CAZY) glycosyl transferase family1 provides insights into disease resistance and sugar donor specificity. Plant Cell 30, 3038–3057. doi: 10.1105/tpc.18.00641
Lu, X., Huang, L., Scheller, H. V., and Keasling, J. D. (2023). Medicinal terpenoid UDP-glycosyltransferases in plants: recent advances and research strategies. J. Exp. Bot. 74, 1343–1357. doi: 10.1093/jxb/erac505
Luo, J. (2015). Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 24, 31–38. doi: 10.1016/j.pbi.2015.01.006
Martin, L. B. B., Kikuchi, S., Rejzek, M., Owen, C., Reed, J., Orme, A., et al. (2024). Complete biosynthesis of the potent vaccine adjuvant QS-21. Nat. Chem. Biol. 20, 493–502. doi: 10.1038/s41589-023-01538-5
Oka, T. and Jigami, Y. (2006). Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J. 273, 2645–2657. doi: 10.1111/j.1742-4658.2006.05281.x
Osmani, S. A., Bak, S., and Møller, B. L. (2009). Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70, 325–347. doi: 10.1016/j.phytochem.2008.12.009
Patsch, D., Schwander, T., Voss, M., Schaub, D., Hüppi, S., Eichenberger, M., et al. (2024). Enriching productive mutational paths accelerates enzyme evolution. Nat. Chem. Biol. 20, 1662–1669. doi: 10.1038/s41589-024-01712-3
Pramastya, H., Song, Y., Elfahmi, E. Y., Sukrasno, S., and Quax, W. J. (2021). Positioning Bacillus subtilis as terpenoid cell factory. J. Appl. Microbiol. 130, 1839–1856. doi: 10.1111/jam.14904
Qian, W., Jie, B., and Hui-Feng, J. (2021). Research progress on technologies of enzyme engineering and design in synthetic biology. Chin. Bull. Life Sci. 33, 1493–1501. doi: 10.13376/j.cbls/2021168
Rahimi, S., Kim, J., Mijakovic, I., Jung, K. H., Choi, G., Kim, S. C., et al. (2019). Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 37, 107394. doi: 10.1016/j.bioteChadv.2019.04.016
Rai, A., Rai, M., Kamochi, H., Mori, T., Nakabayashi, R., Nakamura, M., et al. (2020). Multiomics-based characterization of specialized metabolites biosynthesis in Cornus Officinalis. DNA research: an Int. J. Rapid Publ. Rep. Genes Genomes 27, (2). doi: 10.1093/dnares/dsaa009
Reed, J., Orme, A., El-Demerdash, A., Owen, C., Martin, L. B. B., Misra, R. C., et al. (2023). Elucidation of the pathway for biosynthesis of saponin adjuvants from the soapbark tree. Sci. (New York NY) 379, 1252–1264. doi: 10.1126/science.adf3727
Ren, S., Sun, Q., Feng, X., and Li, C. (2022). Biosynthesis of pentacyclic triterpenoid saponins in microbial cell factories. Synthetic Biol. J. 3, 168–183. doi: 10.12211/2096-8280.2020-082
Rosano, G. L., Morales, E. S., and Ceccarelli, E. A. (2019). New tools for recombinant protein production in Escherichia coli: A 5-year update. Protein science: A Publ. Protein Soc. 28, 1412–1422. doi: 10.1002/pro.3668
Rosati, D., Palmieri, M., Brunelli, G., Morrione, A., Iannelli, F., Frullanti, E., et al. (2024). Differential gene expression analysis pipelines and bioinformatic tools for the identification of specific biomarkers: A review. Comput. Struct. Biotechnol. J. 23, 1154–1168. doi: 10.1016/j.csbj.2024.02.018
Sen, Y. and Jian-qiang, K. (2016). Biosynthetic pathway analysis of UDP-glycosyl donors. Chin. Medicinal Biotechnol. 11, 355–359. doi: 10.3969/j.issn.1673-713x2016.04.013
Shi, R. and Jiang, Z. (2020). Enzymatic synthesis of 2’-fucosyllactose: advances and perspectives. Synthetic Biol. J. 1, 14. doi: 10.12211/2096-8280.2020-033
Siedhoff, N. E., Schwaneberg, U., and Davari, M. D. (2020). Machine learning-assisted enzyme engineering. Methods enzymology 643, 281–315. doi: 10.1016/bs.mie.2020.05.005
Singh, S., Chhatwal, H., and Pandey, A. J. (2024). Deciphering the complexity of terpenoid biosynthesis and its multi-level regulatory mechanism in plants. J. Plant Growth Regulation 43 (10), 0721–7595. doi: 10.1007/s00344-024-11347-2
Sinha, R. and Shukla, P. (2019). Current trends in protein engineering: updates and progress. Curr. Protein Pept. Sci. 20, 398–407. doi: 10.2174/1389203720666181119120120
Skinnider, M. A., Johnston, C. W., Gunabalasingam, M., Merwin, N. J., Kieliszek, A. M., MacLellan, R. J., et al. (2020). Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 11, 6058. doi: 10.1038/s41467-020-19986-1
Song, H. (2017). Studies on enzymes related to the synthesis of UDP-galactose and UDP-xylose from Pacific oysters using UDP-glucose as substrate, doctorate. doi: 10.27244/d.cnki.gnjnu.2017.00026
Song, B., Ning, W., Wei, D., Jiang, M., Zhu, K., Wang, X., et al. (2023). Plant genome resequencing and population genomics: Current status and future prospects. Mol. Plant 16, 1252–1268. doi: 10.1016/j.molp.2023.07.009
Sufang, P., Xiaobo, L., Yuanyuan, W., Guisheng, X., Chunhua, M., Weiwei, J., et al. (2024). Prokaryotic Expression and Functional Verification of Glycosyltransferase from P. Vietnamensis var. fuscidiscus. Mol. Plant Breeding 1–14. Available at: https://link.cnki.net/urlid/46.1068.S.20241129.1007.005
Sui, C., Zhang, J., Wei, J., Chen, S., Li, Y., Xu, J., et al. (2011). Transcriptome analysis of Bupleurum chinense focusing on genes involved in the biosynthesis of saikosaponins. BMC Genomics 12, 539. doi: 10.1186/1471-2164-12-539
Sun, Q., Guo, F., Ren, S., Zhang, L., Liu, X., Li, C., et al. (2023a). Construction of a UDP-arabinose regeneration system for efficient arabinosylation of pentacyclic triterpenoids. ACS synthetic Biol. 12, 2463–2474. doi: 10.1021/acssynbio.3c00351
Sun, W., Yin, Q., Wan, H., Gao, R., Xiong, C., Xie, C., et al. (2023b). Characterization of the horse chestnut genome reveals the evolution of aescin and aesculin biosynthesis. Nat. Commun. 14, 6470. doi: 10.1038/s41467-023-42253-y
Tang, Q. Y., Chen, G., Song, W. L., Fan, W., Wei, K. H., He, S. M., et al. (2019). Transcriptome analysis of Panax zingiberensis identifies genes encoding oleanolic acid glucuronosyltransferase involved in the biosynthesis of oleanane-type ginsenosides. Planta 249, 393–406. doi: 10.1007/s00425-018-2995-6
Thieker, D. F., Maguire, J. B., Kudlacek, S. T., Leaver-Fay, A., Lyskov, S., and Kuhlman, B. (2022). Stabilizing proteins, simplified: A Rosetta-based webtool for predicting favorable mutations. Protein science: A Publ. Protein Soc. 31, e4428. doi: 10.1002/pro.4428
Tianli, S., Mingzhu, H., Bin, L., and Xuelan, C. (2024). Advances in microbial genome reduction and optimization based on synthetic biology. Sci. Technol. Food Industry. 46, 394. doi: 10.13386/j.issn1002-0306.2023120354
Wang, X. (2009). Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 583, 3303–3309. doi: 10.1016/j.febslet.2009.09.042
Wang, H., Han, T., Bai, A., Xu, H., Wang, J., Hou, X., et al. (2024). Potential regulatory networks and heterosis for flavonoid and terpenoid contents in pak choi: metabolomic and transcriptome analyses. Int. J. Mol. Sci. 25, (7). doi: 10.3390/ijms25073587
Wang, N. and Huo, Y. X. (2022). Using genome and transcriptome analysis to elucidate biosynthetic pathways. Curr. Opin. Biotechnol. 75, 102708. doi: 10.1016/j.copbio.2022.102708
Wang, M., Ji, Q., Lai, B., Liu, Y., and Mei, K. (2023a). Structure-function and engineering of plant UDP-glycosyltransferase. Comput. Struct. Biotechnol. J. 21, 5358–5371. doi: 10.1016/j.csbj.2023.10.046
Wang, Y., Sun, Q., Chi, Y., Liu, Z., Liu, H., Li, C., et al. (2023b). Constructing an intensified UDP recycling system for the glycosylation of natural products by phosphorylation of byproduct fructose. J. Agric. Food Chem. 71, 9441–9450. doi: 10.1021/acs.jafc.3c01077
Wang, D., Wang, J., Shi, Y., Li, R., Fan, F., Huang, Y., et al. (2020). Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab. Eng. 61, 131–140. doi: 10.1016/j.ymben.2020.05.007
Wang, P., Wang, J., Zhao, G., Yan, X., and Zhou, Z. (2021a). Systematic optimization of the yeast cell factory for sustainable and high efficiency production of bioactive ginsenoside compound K. Synthetic Syst. Biotechnol. 6, 69–76. doi: 10.1016/j.synbio.2021.03.002
Wang, P., Wei, W., Ye, W., Li, X., Zhao, W., Yang, C., et al. (2019). Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 5, 5. doi: 10.1038/s41421-018-0075-5
Wilson, A. E. and Tian, L. (2019). Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant journal: Cell Mol. Biol. 100, 1273–1288. doi: 10.1111/tpj.14514
Wu, B., Liu, X., Xu, K., and Zhang, B. (2020). Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 20, 459. doi: 10.1186/s12870-020-02655-2
Xiao-chen, L., Hu, L., Liang, Z., and Chun, L. (2018). Enzymatic glycosylation and its function in metabolic process of cells. China Biotechnol. 38, 69–77. doi: 10.13523/j.cb.20180109
Xu, B., Huang, J. P., Peng, G., Cao, W., Liu, Z., Chen, Y., et al. (2024). Total biosynthesis of the medicinal triterpenoid saponin astragalosides. Nat. Plants 10, 1826–1837. doi: 10.1038/s41477-024-01827-4
Xu, X., Liu, Y., Du, G., Ledesma-Amaro, R., and Liu, L. (2020). Microbial chassis development for natural product biosynthesis. Trends Biotechnol. 38, 779–796. doi: 10.1016/j.tibtech.2020.01.002
Xu, K., Zhao, Y. J., Ahmad, N., Wang, J. N., Lv, B., Wang, Y., et al. (2021). O-glycosyltransferases from Homo sapiens contributes to the biosynthesis of Glycyrrhetic Acid 3-O-mono-β-D-glucuronide and Glycyrrhizin in Saccharomyces cerevisiae. Synthetic Syst. Biotechnol. 6, 173–179. doi: 10.1016/j.synbio.2021.07.001
Xueqing, Z., Jing, Y., Yu, S., and Zhaohe, Y. (2022). Expression characteristics and recombinant expression analysis of PgUGT gene in pomegranate. Acta Botanica Boreali-Occidentalia Sin. 42, 390–397. Available at: https://kns.cnki.net/kcms2/article/abstract?v=TD_mLQSGK6tWbz3sEJ0IZaBf-d2Wm5Vsd0_U0IXG5zVxmTfMhVLejbEvm9lGc_h6cJVZblGMHuLJrVFdeHZpkT3KrmL6QEL1R4Pp02V28jruCm479Er-f4t93tGjRC2mmsw_neA6Ruars9zATD0M6Axb8C9zifkQs4XMm_gnVFTWmjDhA9TiOb0Qqz9G_uQfr16CHTM03cw=&uniplatform=NZKPT&language=CHS
Yan, L., Honghua, J., Jiajie, D., Huichang, H., and Kai, C. (2024). A glycosyltransferase mutant and its catalytic method for synthesizing ginsenoside rh2. National Intellectual Property Administration.
Yang, L., Yang, Y., Huang, L., Cui, X., and Liu, Y. (2023). From single- to multi-omics: future research trends in medicinal plants. Briefings Bioinf. 24, (1). doi: 10.1093/bib/bbac485
Yin, S. (2016). Cloning and functional study of key enzyme genes for glycoside biosynthesis of natural product of Ornithogalum caudatum, Master. (Medical and Health Science Technology Master's Electronic Journal Publishing Information).
You, F. M. (2023). Plant genomics-advancing our understanding of plants. Int. J. Mol. Sci. 24, (14). doi: 10.3390/ijms241411528
Yuanyuan, X. U., Chen, Z., Jia, L. M., and Weng, X. H. (2021). Advances in understanding of the biosynthetic pathway and regulatory mechanism of triterpenoid saponins in plants. SCIENTIA Sin. Vitae. 51, 525–555. doi: 10.1360/SSV-2020-0230
Zhan, C., Li, X., Zhao, Z., Yang, T., Wang, X., Luo, B., et al. (2016). Comprehensive analysis of the triterpenoid saponins biosynthetic pathway in anemone flaccida by transcriptome and proteome profiling. Front. Plant Sci. 7, 1094. doi: 10.3389/fpls.2016.01094
Zhan, C., Shen, S., Yang, C., Liu, Z., Fernie, A. R., Graham, I. A., et al. (2022). Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 27, 981–1001. doi: 10.1016/j.tplants.2022.03.002
Zhang, C., Cai, Y., Zhang, Z., Zheng, N., Zhou, H., Su, Y., et al. (2024a). Directed evolution of the UDP-glycosyltransferase UGT(BL)1 for highly regioselective and efficient biosynthesis of natural phenolic glycosides. J. Agric. Food Chem. 72, 1640–1650. doi: 10.1021/acs.jafc.3c07850
Zhang, F., Hao, X., Liu, J., Hou, H., Chen, S., and Wang, C. (2025). Herbal multiomics provide insights into gene discovery and bioproduction of triterpenoids by engineered microbes. J. Agric. Food Chem. 73, 47–65. doi: 10.1021/acs.jafc.4c08372
Zhang, X., Li, C., Hao, Z., and Liu, Y. (2023). Transcriptome analysis provides insights into coumarin biosynthesis in the medicinal plant Angelica dahurica cv. Yubaizhi. Gene 888, 147757. doi: 10.1016/j.gene.2023.147757
Zhang, L., Ren, S., Liu, X., Liu, X., Guo, F., Sun, W., et al. (2020). Mining of UDP-glucosyltrfansferases in licorice for controllable glycosylation of pentacyclic triterpenoids. Biotechnol. bioengineering 117, 3651–3663. doi: 10.1002/bit.27518
Zhang, Q., Xu, X., Zhang, W., Huang, Z., Wu, Y., Liu, Y., et al. (2024b). De novo 2’-fucosyllactose biosynthesis using glucose as the sole carbon source by multiple engineered Bacillus subtilis. Metab. Eng. 88, 85–93. doi: 10.1016/j.ymben.2024.12.004
Zhang, M., Yi, Y., Gao, B. H., Su, H. F., Bao, Y. O., Shi, X. M., et al. (2022). Functional characterization and protein engineering of a triterpene 3-/6-/2’-O-glycosyltransferase reveal a conserved residue critical for the regiospecificity. Angewandte Chemie (International ed English) 61, e202113587. doi: 10.1002/anie.202113587
Zhao, X., Wei, W., Li, S., Gao, S., Liu, S., Zeng, W., et al. (2024). Elucidation of the biosynthesis of asiaticoside and its reconstitution in yeast. ACS Sustain. Chem. Engineering. 12, 4028–4040. doi: 10.1021/acssuschemeng.3c07046
Zhou, M., Fan, J., Gao, Y., Zheng, C., Xu, Y., Jia, L., et al. (2024). Identification and analysis of UGT genes associated with triterpenoid saponin in soapberry (Sapindus mukorossi Gaertn.). BMC Plant Biol. 24, 588. doi: 10.1186/s12870-024-05281-4
Keywords: triterpenoid saponins, glycosylation, UDP-glycosyltransferases (UGTs), multi-omics approaches, enzyme engineering
Citation: Hao L, Liu Y, Dong G, Liu J, Qiu K, Li X and Qiao Y (2025) Multi-strategy ugt mining, modification and glycosyl donor synthesis facilitate the production of triterpenoid saponins. Front. Plant Sci. 16:1586295. doi: 10.3389/fpls.2025.1586295
Received: 13 March 2025; Accepted: 09 May 2025;
Published: 30 May 2025.
Edited by:
Richard Strasser, BOKU University, AustriaReviewed by:
Kunrong Mei, Tianjin University, ChinaGuozhi Xiao, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Hao, Liu, Dong, Liu, Qiu, Li and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanan Qiao, MTgyNTMxNjQ2NTRAMTYzLmNvbQ==
 Lin Hao1
Lin Hao1 Yanan Qiao
Yanan Qiao