- State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, China
Plant disease poses a great threat to crop production. The mechanisms underlying plant-pathogen interactions are critical research topics worldwide. In recent years, significant breakthrough studies have been reported, broadening our understanding of plant immunity. Based on these findings, many strategies have been developed to improve plant defense against various diseases. Here, we summarize these strategies and their applications in studies aimed at promoting crop resistance. Besides domain swapping, gene shuffling, and random mutation, three additional strategies have been developed in the last decade. The first strategy is gene editing of host susceptibility (S) genes to prevent pathogen infection. Editing of Mlo and DMR6 gene in many species are good examples of this approach. The second strategy is editing the promoters of host S genes or resistance (R) genes. This strategy is widely used to counteract Xanthomonas, such as modifying the promoters of LOB1 and SWEET genes in several crops to enhance resistance. The third strategy is designing R gene products, especially nucleotide-binding and leucine-rich repeat (NLR) receptors. This approach is based on the growing knowledge of the structural features and mechanisms of NLRs, which have seen significant advances recently. To date, all NLR-engineering attempts have focused on rice paired NLRs, such as Pikp-1/Pikp-2 (allelic to Pikm-1/Pikm-2) and RGA4/RGA5. The bioengineering of these NLRs provides a promising method to combat diverse pathogens. Detailed studies in many crops are also discussed in this review, organized around these strategies. In summary, with progresses in understanding plant immune mechanism, many innovative molecular strategies are available to mitigate the threat of plant pathogens in the future.
An overview of plant pathology development
Plants are very important to human society. Besides releasing oxygen, they provide food, fruits and even clothing for our survival. In the world today, the population is continually growing and the demand for more crops is also increasing every day, which brings more pressure on crop production. In agriculture, crop yield is threatened by many biotic and abiotic stresses. Various pathogens constitute the main biotic stresses in nature, which have caused significant yield losses throughout history.
To solve these problems, the first step is to understand whether plants can develop resistance to pathogens. In the 1950s, the gene-for-gene hypothesis was proposed by Flor, based on his studies on flax rust. In this model, the resistance occurs when the plant’s resistance (R) gene and the pathogen’s avirulence (Avr) gene are both present. This hypothesis demonstrates that plants can genetically resist pathogens and laid the foundation for crop disease resistance breeding and molecular plant pathology. Since the 1990s, hundreds of Avr and R genes have been cloned from different pathogens and plants, benefitting the improvement of crop breeding.
Based on the accumulation of knowledge about Avr and R genes, researchers have begun to realize that plants have developed many strategies to overcome the invasion of pathogens during evolution. Unlike vertebrates, plants lack an adaptive immune system and rely entirely on innate immunity to defend against pathogens. This type of immunity is highly evolved and comprises two tiers of response. The first layer of defense takes place on or outside the plant cell membrane, which can respond to pathogen invasion immediately. Usually, pathogens possess small conserved molecules, called pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs), on their surface. Plants utilize membrane-anchored receptors, known as pattern recognition receptors (PRRs), to recognize PAMPs and trigger PAMP-triggered immunity (PTI). The PTI process is effective against a broad range of pathogens. The most well-known PRRs are FLS2 and CERK1, which are receptor-like kinases (RLKs) for bacterial flagellin and fungal chitin respectively (Gomez-Gomez and Boller, 2000; Miya et al., 2007). However, this immunity is relatively weak, and many pathogens can release effectors or other molecules to suppress it. Meanwhile, pathogens can also inject other effectors into plant cells to manipulate host metabolism and gene expressions for their survival and reproduction. This process is called effector-triggered susceptibility (ETS). To counteract ETS, plants evolved a second layer of defense, known as effector-triggered immunity (ETI), which involves the recognition of pathogen effectors by different kinds of resistance (R) gene products. This model is commonly referred to as the ‘zig-zag’ model (Jones and Dangl, 2006). Pathogen effectors are highly diverse, even among strains of the same species, resulting in ETI specificity. The ETI process usually induces a strong immune response, often accompanied by hypersensitive response (HR) that limits pathogen spread in a very short time. Nonetheless, immutable boundaries do not exist between PTI and ETI, and they are indispensable and function together in nature sometimes (Yuan et al., 2021).
Plant R genes function in diverse ways to defend against pathogens
Among all the cloned R genes, NLR-encoding genes constitute the largest group and are present in almost all land plants (Gao et al., 2018). Typical NLRs share similar protein structures except for their N-termini, which are categorized into coiled-coil (CC) motif, Toll/Interleukin-1 receptor (TIR) domain and resistance to powdery mildew 8 (RPW8) domain. Based on this, typical NLRs are classified as CC-NLRs (CNLs), TIR-NLRs (TNLs) and RPW8-NLRs (RNLs) (Maruta et al., 2022). Recently, significant progresses have been achieved in NLR protein structure and mechanism studies. It has been revealed that CNLs and TNLs can interact with pathogen effectors directly or indirectly, while RNLs primarily mediate downstream signaling of CNLs and TNLs (Zhang et al., 2022b). All three NLRs classes can be assembled into resistosomes to trigger resistance, inducing Ca2+ influx and producing small bioactive molecules (Figure 1) (Wang et al., 2019a; Ma et al., 2020; Jacob et al., 2021). These breakthroughs advance our understanding of plant innate immunity and enable applications in breeding practices, such as wheat Sr35 (Zhao et al., 2022).
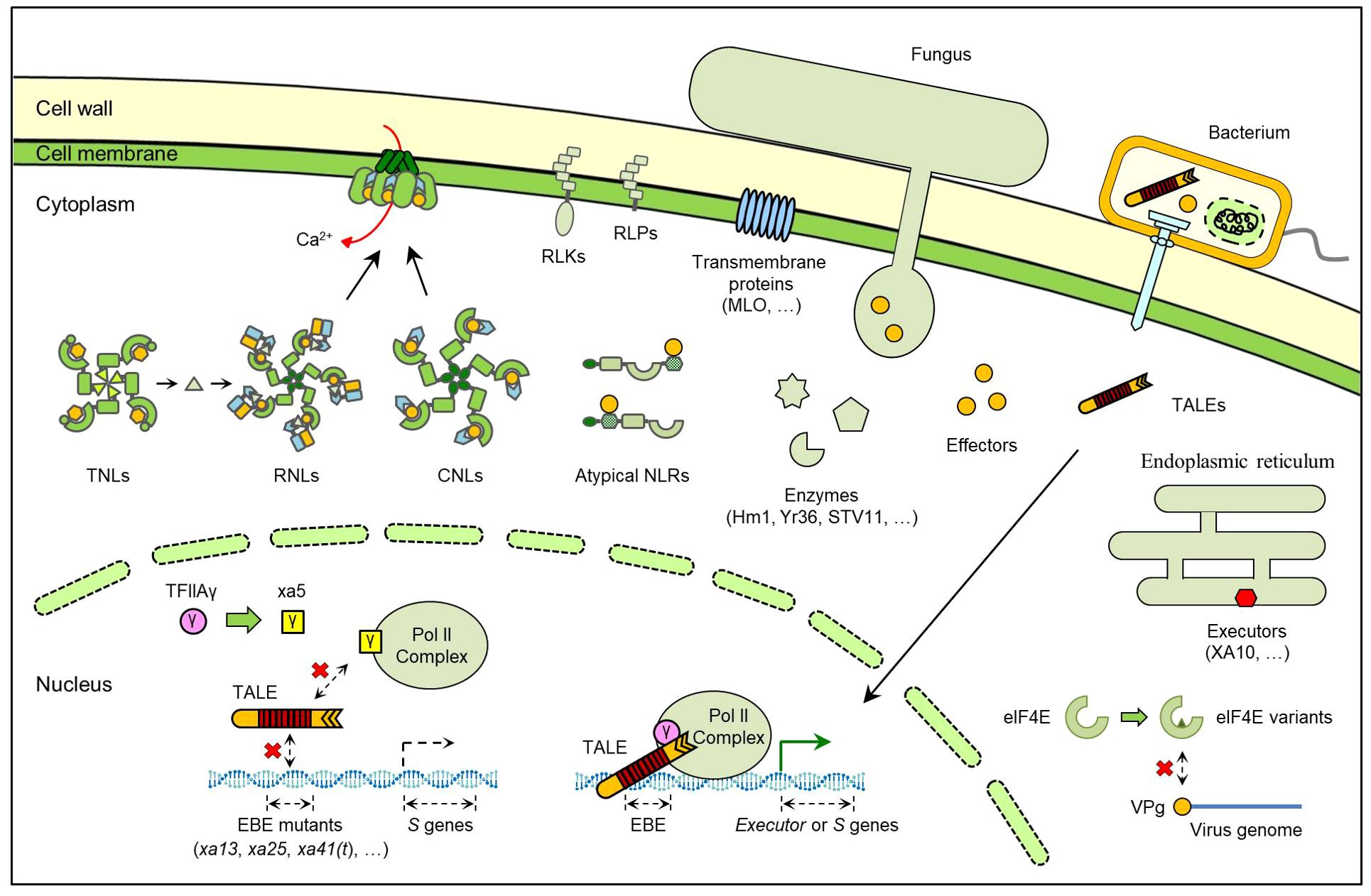
Figure 1. Model of different R gene products in plant-pathogen interactions. The products encoded by cloned R genes primarily include NLRs, RLKs/RLPs, executors and enzymes. Recessive R genes are usually mutations of S genes, including protein variants and promoter mutations.
Meanwhile, many crop NLRs are atypical, which contain additional integrated domains (IDs), including WRKY, kinase, heavy metal-associated (HMA), and zinc-finger BEAF and DREF (zf-BED) domains (Zhang et al., 2022b). In several atypical NLRs, the IDs act as ‘integrated decoys’ to bind pathogen effectors (Figure 1) (Jones and Dangl, 2006). However, not all the IDs serve as decoys; for example, the zf-BED domains of rice XA1 and XA14 could not directly interact with pathogen effectors (Zhang et al., 2020; Yoshihisa et al., 2022). Additionally, many atypical NLRs require partner NLRs, usually known as ‘helper NLRs’, to form NLR pairs (also called paired NLRs) and confer resistance (Jones et al., 2016). In rice, many cloned blast R genes encode NLR pairs, such as RGA4/RGA5, Pikp-1/Pikp-2 and its allelic genes (Zhang et al., 2022b).
RLKs and receptor-like proteins (RLPs) represent another major class of R gene products. Although classified as PRRs in many reports, their discoveries stemmed from the gene-for-gene hypothesis. The examples include Xa21 and Cf-9 (Figure 1). Xa21 is the first cloned R gene in rice and confers resistance to bacterial blight (Song et al., 1995). Cf-9 is the first cloned tomato R gene for resistance to the fungus Cladosporium fulvum (Jones et al., 1994). Xa21 protein is an RLK containing an extracellular leucine-rich repeat (LRR) domain and an intracellular kinase domain, while Cf-9 is an LRR-RLP lacking the kinase domain (Jones et al., 1994; Song et al., 1995). There are also some other RLKs that contain different kinds of extracellular domains, such as the XA4 and Pi-d2. XA4 is a cell wall-associated kinase (WAK) conferring bacterial blight resistance in rice, and Pi-d2 contains an extracellular B-lectin domain and mediates rice blast resistance (Chen et al., 2006; Hu et al., 2017).
Besides NLRs and RLKs, another interesting class of R gene products is the executors. The executor R genes harbor special effector binding elements (EBEs) in their promoters and can be induced by transcription activator-like effectors (TALEs) secreted by Xanthomonas, which includes many species that can infect large numbers of crops worldwide (Figure 1) (Timilsina et al., 2020; Zhang et al., 2022a). The common feature of these executors is that they trigger significant defense responses, even cell death (Zhang et al., 2015). To date, all the reported executor R genes originate from pepper and rice. Among them, Bs3 and Bs3-E encode flavin-dependent monooxygenase (FMO) homologs. The others, including Bs4C-R, Xa7, Xa10, Xa23 and Xa27, all encode small special proteins with low similarity to known proteins (Zhang et al., 2015, 2022).
Notably, there are many special recessive R genes. These genes can be considered as mutations of susceptibility (S) genes targeted by pathogen effectors (Figure 1) (Timilsina et al., 2020; Zhang et al., 2022a). The well-known examples of this class include barley mildew resistance locus O (mlo) and rice xa13 genes. Mlo encodes a seven-transmembrane (7-TM) protein that negatively regulates cell death and plant defense (Buschges et al., 1997; Piffanelli et al., 2002). Homozygous mlo mutants can confer resistance to powdery mildew fungus in barley (Buschges et al., 1997). The recessive xa13 results from a mutation in the EBE of OsSWEET11, which is targeted by Xanthomonas oryzae pv. oryzae (Xoo) effector PthXo1 (Chu et al., 2006; Yang et al., 2006; Moscou and Bogdanove, 2009). Xoo TALEs can also bind to the EBEs of other SWEET genes in rice, and mutations in these EBEs have generated other recessive R genes, such as xa25 and xa41(t) (Liu et al., 2011; Hutin et al., 2015).
Some recessive R genes are alternative forms of the corresponding S genes, because they are essential components for gene expression or translation (Figure 1). The rice xa5 gene encodes a protein harboring a V39E amino acid substitution in the gamma subunit of the basal transcription factor IIA 5 (TFIIAγ5) (Iyer and McCouch, 2004; Jiang et al., 2006). In Xa5-carrying rice, Xoo TALEs hijacks TFIIAγ5 for inducing host gene expression. In contrast, the mutated TFIIAγ5V39E blocks TALE binding and leads to passive resistance (Zhang and Wang, 2013; Yuan et al., 2016). Similar results have been found in cloning of R genes against viruses. The potyviruses use a genome-linked viral protein, VPg, to recruit host eukaryotic translation initiation factor 4E (eIF4E) or its isoform eIF(iso)4E for viral replication (Wang and Krishnaswamy, 2012). Coincidentally, many R genes mediating resistance to different potyviruse species were finally found to be variants of eIF4E or eIF(iso)4E (Wang and Krishnaswamy, 2012).
Other R gene products also exhibit great functional diversity (Figure 1). The first cloned plant R gene, Hm1 in maize, encodes an enzyme that detoxifies the Helminthosporium carbonum (HC) toxin from Cochliobolus carbonum (Johal and Briggs, 1992). Wheat Yr36 encodes a kinase-START domain protein that phosphorylates other proteins to increase reactive oxygen species (ROS) and suppresses photosynthesis during stripe rust infection (Fu et al., 2009; Wang et al., 2019b). The rice STV11 gene encodes a sulfotransferase that converts salicylic acid (SA) into sulphonated SA (SSA) during the resistance to rice stripe virus (RSV) (Wang et al., 2014a). Due to space limitations, other cloned plant R gene products are not discussed here. Collectively, all these R genes provide a foundation for developing new breeding strategies to enhance crop resistance.
Strategies for improving crop resistance
Strategy 1. Domain swapping and gene shuffling of R genes
Domain swapping is a technique to construct chimeric genes or proteins formed by exchanging functional domains. Gene shuffling, in contrast, involves generating constructs through PCR amplification using homologous R genes as templates. Both methods have long served as the strategic approaches in plant immunity research to identify critical sequences or residues in cloned R gene products (Figure 2). The earliest application of these methods can be traced back to the studies on flax L and tomato Cf4/9 loci (Ellis et al., 1999; Wulff et al., 2001). The L locus harbors multiple allelic NLR genes that confer strain-specific resistance against the flax rust fungus Melampsora lini via recognition of distinct effector proteins. Intragenic exchanges among these genes had produced chimeric genes, which revealed the TIR and LRR domains as key determinants of resistance spectrum (Ellis et al., 1999).
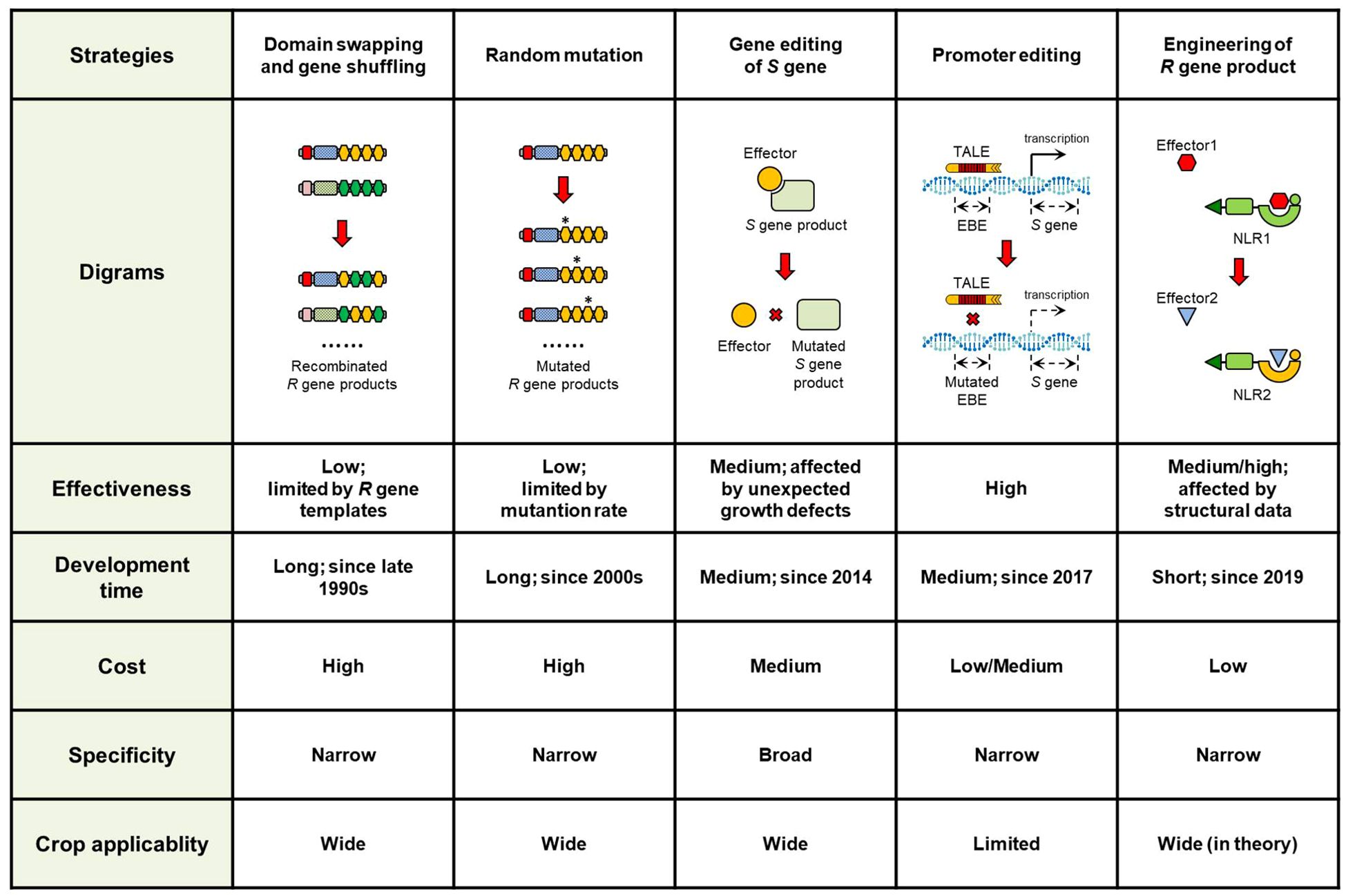
Figure 2. Comparison of different strategies to improve plant disease resistance. The five strategies were compared based on effectiveness, development time, cost, specificity, and crop applicability.
These methods have also been used to generate novel R genes with enhanced specificity. Using the L locus again as an example, L5 and L6 are two alleles that confer resistance to different variants of the fungal effector AvrL567. After domain swapping, a chimera protein exhibited recognition of a previously non-recognized AvrL567 variant when expressed in tobacco (Ravensdale et al., 2012).
Practically speaking, the value of these methods lies on discovering the critical functional residues in R gene products, rather than exploiting for novel recognition profile. Both approaches require homologous R genes as initial templates, making them particularly suitable for NLR- and RLK-encoding genes, as recombination within LRR domains would expands combinatorial possibilities (Figure 2). However, the templates would also be the limits of the resistance spectrum potential in most cases, and the new recognition profiles are often limited within the same pathogen species (Zhao et al., 2009; Ravensdale et al., 2012).
Strategy 2. Random mutation of R genes
Mutation is the engine of evolution. As a result, random mutagenesis has emerged as a common strategy to expand R gene recognition profiles (Figure 2). For instance, Rx encodes an NLR that confers resistance to potato virus X (PVX) by recognizing its coat protein, CP-TK. Error-prone PCR was used to mutate the LRR domain of Rx. After screening more than two thousand clones, mutants with broader recognition spectra for CP-KR variants and poplar mosaic virus (PopMV) were identified (Farnham and Baulcombe, 2006). In another mutant library targeting the remaining regions of Rx, several mutants with increased resistance to PopMV were also obtained (Harris et al., 2013). The potato NLR R3a detects Phytophthora infestans effector AVR3a but shows limited response to the AVR3aEM variant. By constructing a random mutant library of R3a, researchers successfully isolated multiple variants with enhanced sensitivity to AVR3aEM, thereby broadening its resistance spectrum (Segretin et al., 2014).
Similar studies have been reported in other pathosystems to generate artificially evolved R genes with expanded recognition specificity by using random mutagenesis (Sueldo et al., 2015). The efficiency varies across different R genes. The critical requirement is generating sufficient mutants to ensure adequate coverage of the whole variations (Figure 2) (Farnham and Baulcombe, 2006; Segretin et al., 2014; Sueldo et al., 2015). Therefore, the balance between outcome and cost should be considered before performance.
Strategy 3. Gene editing of S genes
In the last decade, gene editing has emerged as one of the most attractive technologies in research. It has developed spans from zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) to clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) system (Wang and Doudna, 2023). These technologies enable precise modification of target genes/sequences for specific purposes. Consequently, their applications have rapidly expanded from functional genomic study to crop improvement (Zhang et al., 2022a; Wang and Doudna, 2023). For improving disease resistance, editing crop S genes represents a straightforward strategy in practice (Figure 2; Table 1).
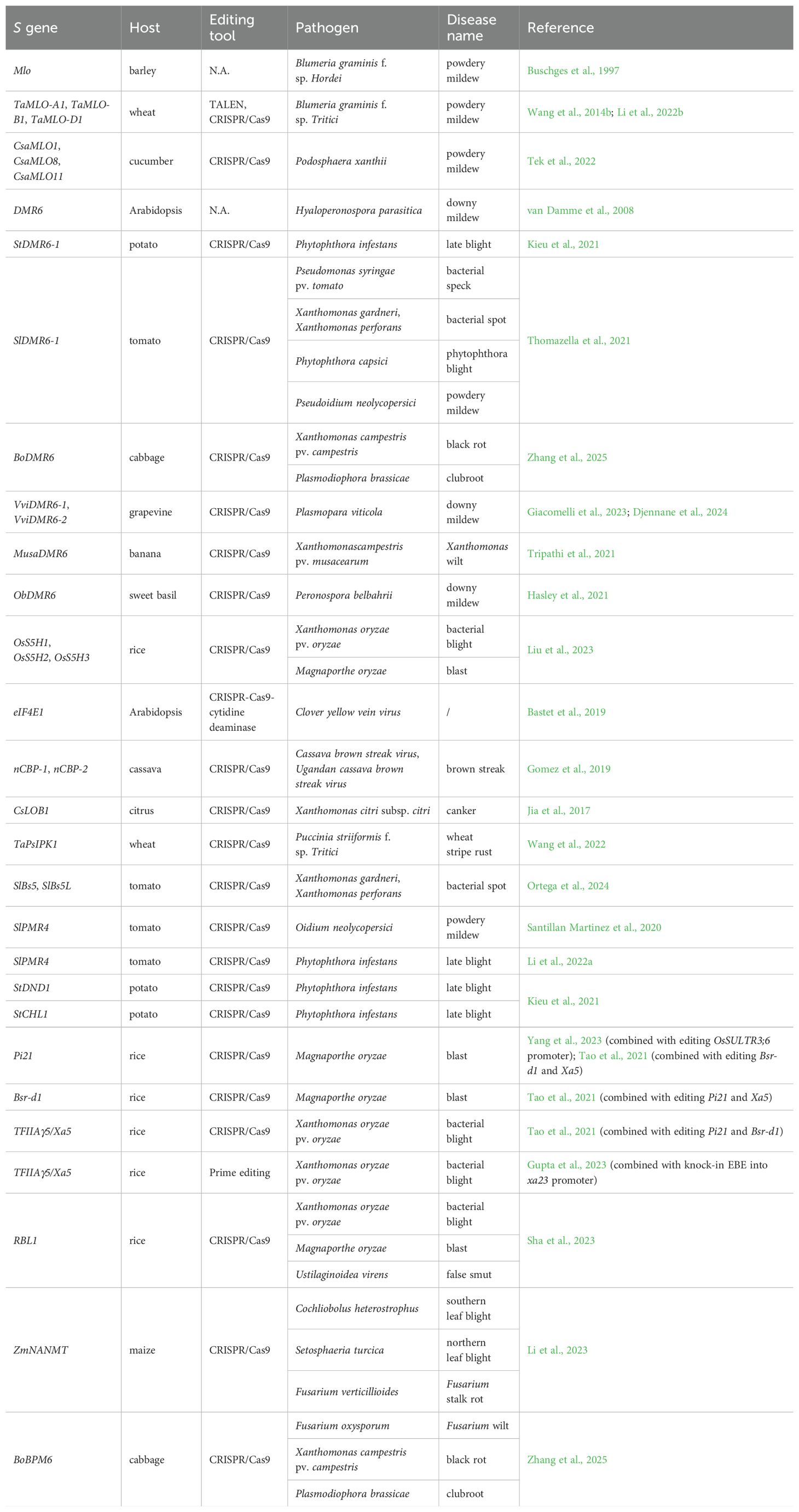
The best-characterized example is loss-of-function mutations in the Mlo gene. The barley mlo mutants have provided durable broad-spectrum resistance against almost all Blumeria graminis f.sp. hordei (Bgh) isolates in agriculture since the late 1970s (Kusch and Panstruga, 2017). The mlo gene was isolated via map-based cloning almost three decades ago (Buschges et al., 1997). It encodes a seven-transmembrane protein that is critical for mediating powdery mildew susceptibility. As a calcium-regulated calmodulin-binding protein, MLO protein suppresses host defense reactions including cell wall reinforcement and hypersensitive responses, facilitating pathogen invasion (Piffanelli et al., 2002). Subsequent studies demonstrated that loss function of Mlo confers resistance in over 10 plant species, including Arabidopsis, wheat, and grapevine (Kusch and Panstruga, 2017). Using different gene editing tools, Mlo-edited plants have been generated in wheat and cucumber (Table 1). Unlike barley, bread wheat is hexaploid, and only complete loss of all the three Mlo homologs, TaMLO-A1, TaMLO-B1 and TaMLO-D1, had led to the resistance to Blumeria graminis f. sp. tritici (Bgt) (Wang et al., 2014b; Li et al., 2022b). Although cucumber is a diploid, it has many Mlo homologs. Triple mutants of CsaMLO1, CsaMLO8 and CsaMLO11 were generated by CRISPR/Cas system and exhibited complete resistance to Podosphaera xanthii (Tek et al., 2022).
Downy Mildew Resistant 6 (DMR6) is an S gene first identified in Arabidopsis and then applied in many crops (Table 1). It encodes a salicylic acid 5-hydroxylase (S5H) that converts salicylic acid (SA) into 2,5-Dihydroxybenzoic acid (2,5-DHBA) and plays a negative role in plant immunity (Zhang et al., 2017). The Arabidopsis dmr6 mutants displayed resistance to Hyaloperonospora parasitica, the pathogen of downy mildew (DM) (van Damme et al., 2008). The potato genome contains two DMR6 homologs. CRISPR/Cas9-generated deletion mutants of StDMR6–1 showed increased resistance to the oomycete pathogen Phytophthora infestans, which is the causal agent of late blight (Kieu et al., 2021). Inactivation of SlDMR6–1 by CRISPR/Cas9 made tomato resistant to a broad spectrum of pathogen, including bacteria, oomycetes, and fungi (Thomazella et al., 2021). In grapevine, CRISPR/Cas9 editing of VviDMR6–1 and VviDMR6–2 could reduce the susceptibility to downy mildew (Giacomelli et al., 2023; Djennane et al., 2024). Knocking-out of BoDMR6 in cabbage resulted in lower disease index of black rot and clubroot (Zhang et al., 2025). Editing MusaDMR6 in banana and ObDMR6 in sweet basil enhanced their resistance to Xanthomonas campestris pv. musacearum (Xcm) and Peronospora belbahrii respectively (Hasley et al., 2021; Tripathi et al., 2021). In rice, the homologs of DMR6, also called OsS5H1, OsS5H2, and OsS5H3, were simultaneously edited by CRISPR/Cas9. The triple mutant exhibited stronger resistance to Xoo and Magnaporthe oryzae (M. oryzae) than any single mutants (Liu et al., 2023).
Other attempts at S gene editing have also been carried out (Table 1). To combat potyviruses, eIF4E1, nCBP-1 and nCBP-2 were modified in Arabidopsis and cassava (Bastet et al., 2019; Gomez et al., 2019). To overcome citrus canker, the CsLOB1 gene was mutated using CRISPR/Cas9 technology (Jia et al., 2017). The wheat kinase gene TaPsIPK1 was recently discovered as an S gene, and its inactivation by CRISPR/Cas9 conferred resistance to wheat stripe rust (Wang et al., 2022). In tomato, CRISPR/Cas9-mediated mutagenesis of SlBs5, SlBs5L and SlPMR4 increased the resistance to bacterial spot, powdery mildew and late blight (Santillan Martinez et al., 2020; Li et al., 2022a; Ortega et al., 2024). Knocking out StDND1 and StCHL1 made potato plants resistant to late blight (Kieu et al., 2021). Editing Pi21, Bsr-d1 and TFIIAγ5/Xa5 genes mimicked the resistance of pi21, bsr-d1 and xa5 in rice (Tao et al., 2021; Gupta et al., 2023; Yang et al., 2023). Meanwhile, gene editing of rice RESISTANCE TO BLAST1 (RBL1), maize ZmNANMT and cabbage BoBPM6 could even result in broad-spectrum resistance (Li et al., 2023; Sha et al., 2023; Zhang et al., 2025).
S gene inactivation has been attracting increasing attention in studies because of its high effectiveness. However, there are still some considerations to be noted. The most important consideration is that knockout of many S genes could lead to unexpected developmental impairments (Figure 2). This phenomenon has been observed in multiple crops. To address this, one option is to select alternative S genes for editing, while another is to screen large-scale mutant populations for ideal phenotypes. The later approach has proven feasible in wheat and rice. Knockout of Mlo homologs in wheat caused reduced plant height and grain yield. Through screening additional mlo mutants, the Tamlo-R32 mutant without growth penalties was recently identified (Li et al., 2022b). The rbl1 mutant exhibited many growth defects in rice, whereas the rblΔ12 mutant, generated via multiplex gene editing, achieved a balance between growth and resistance (Sha et al., 2023).
The success in S gene inactivation-mediated resistance has extended its application to studies on resistance negative regulators. Some negative regulator genes even function similarly to S genes in plant-microbe interactions. For example, the GbWAKL14 gene negatively regulated defense response by modulating reactive oxygen species (ROS) levels, and its CRISPR/Cas9-mediated inactivation enhanced the wilt disease resistance in cotton (Zhao et al., 2024). Although their mechanistic contributions to pathogen invasion may vary, these genes provide valuable resources for improving crop resistance. Given the limited availability of cloned R and S genes in certain crops, modification of negative regulators offers a viable short-term alternative option.
Strategy 4. Promoter editing for resistance
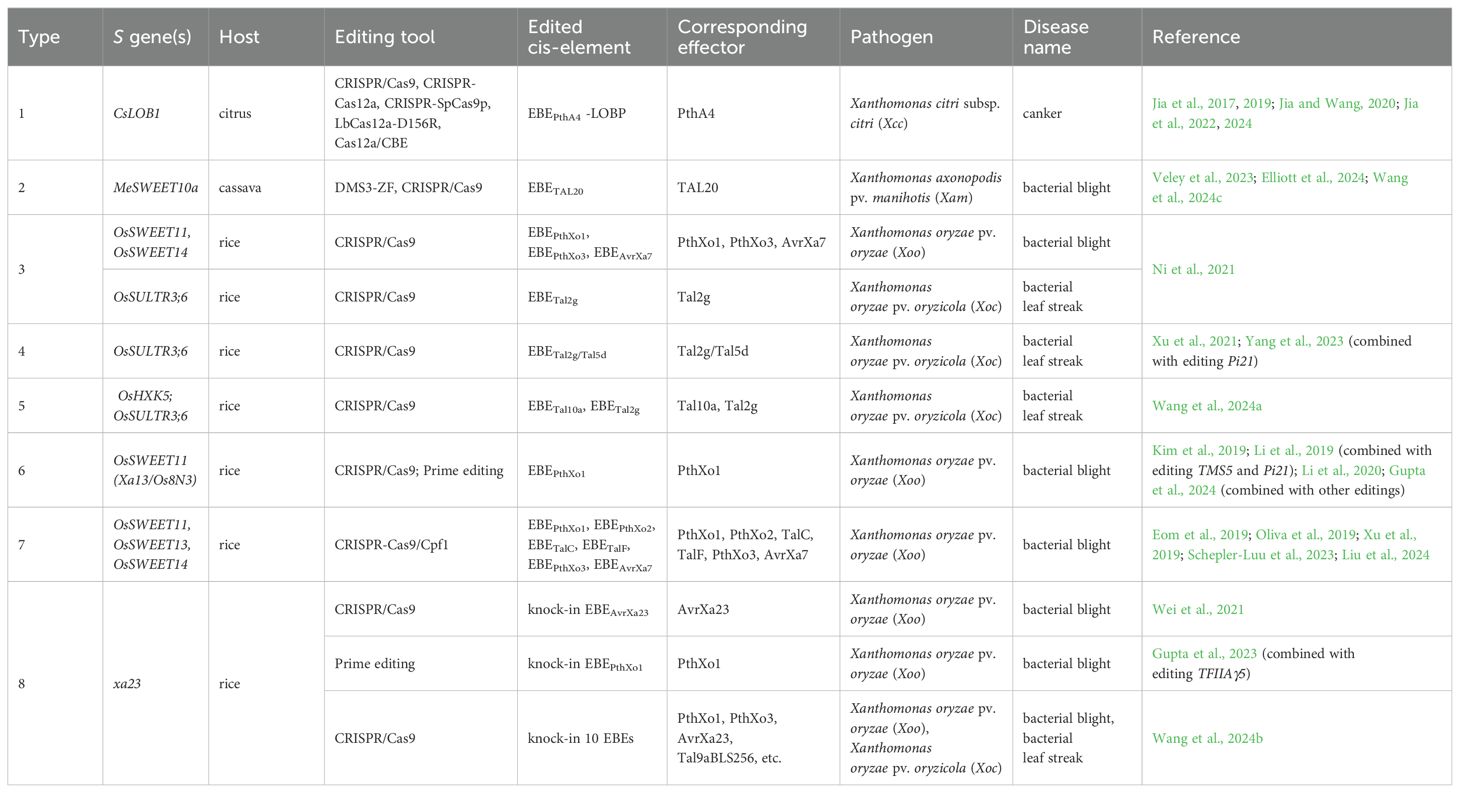
Besides editing the coding sequences of crop S genes, promoter modification is another strategy for improving resistance, especially when combating Xanthomonas. Many Xanthomonas species exploit TALEs to induce the host S genes to acquire nutrients and induce disease symptoms. As a result, artificially editing EBEs in S gene promoters will attenuate their expression activation, mimicking the function of some recessive R genes (Figure 2; Table 2). In citrus, the CsLOB1 gene is induced by Xanthomonas citri subsp. citri (Xcc) effector PthA4. It has been reported in many studies that editing the PthA4 EBE in CsLOB1 promoter significantly reduced or even eliminated the canker symptoms (Jia et al., 2017, 2019; Jia and Wang, 2020; Jia et al., 2022, 2024). Xanthomonas axonopodis pv. manihotis (Xam), also called Xanthomonas phaseoli pv. manihotis (Xpm), causes bacterial blight in cassava. Its major virulence effector, TAL20, upregulates the expression of MeSWEET10a during infection. Editing the TAL20 EBE in MeSWEET10a promoter resulted in bacterial blight resistance in cassava (Elliott et al., 2024; Wang et al., 2024c). Moreover, using a DMS3-ZF system, which fused the artificial zinc-fingers (ZFs) to a DNA methylation-related protein, the methylation within and around the EBETAL20 was increased. This modification prevented the transcription activation of MeSWEET10a and decreased the disease symptoms in cassava (Veley et al., 2023). Bacterial blight and bacterial leaf streak disease in rice are both caused by Xanthomonas, and several rice S genes targeted by TALEs have been identified until now. OsSWEET11 (also called Xa13 or Os8N3), OsSWEET13, and OsHXK5 are activated by Xoo effectors PthXo1, PthXo2, and Tal10a, respectively; OsSWEET14 is induced by four Xoo TALEs, PthXo3, AvrXa7, TalC, and TalF, which bind distinct EBEs; The OsSULTR3;6 gene is targeted by Xanthomonas oryzae pv. oryzicola (Xoc) effectors Tal2g and Tal5d (Zhang et al., 2022a). As a result, editing the EBEs in these genes led to resistance in rice, and different combinations of these edits had shown broader resistance spectra in multiple studies (Eom et al., 2019; Kim et al., 2019; Li et al., 2019; Oliva et al., 2019; Xu et al., 2019; Li et al., 2020; Ni et al., 2021; Xu et al., 2021; Schepler-Luu et al., 2023; Yang et al., 2023; Gupta et al., 2024; Liu et al., 2024; Wang et al., 2024a).
After long-term evolution, some crops have developed ‘imitative’ EBEs for TALEs in the promoters of executor-encoding genes to trigger defense responses (Zhang et al., 2022a). Using gene editing tools, promoters of the recessive alleles of executor-encoding genes could be modified to respond to pathogen invasion. Such attempts are mainly focused on rice xa23, which encodes the same executor as Xa23 but lacks the EBEAvrXa23 in its promoter (Table 2). Using the CRISPR/Cas9 system, EBEAvrXa23 could be knocked into the xa23 promoter with very low probability, rendering rice plants resistant to Xoo (Wei et al., 2021). In another study, EBEPthXo1 was knocked into the xa23 promoter to broaden the resistance spectrum of TFIIAγ5-edited plants (Gupta et al., 2023). Even an artificial EBE array, which consisted of different EBEs for TALEs of Xoo and Xoc, could be knocked into the promoter of xa23, conferring broad-spectrum resistance in rice (Wang et al., 2024b).
To date, promoter modification of non-R or non-S genes for resistance enhancement remains unreported. This strategy is also theoretically feasible for genes triggering cell death and defense responses. However, the cis-elements specially responding to pathogens are not always clear in many cases, which would limit the applicability of this approach (Figure 2).
Strategy 5. Engineering of R gene products
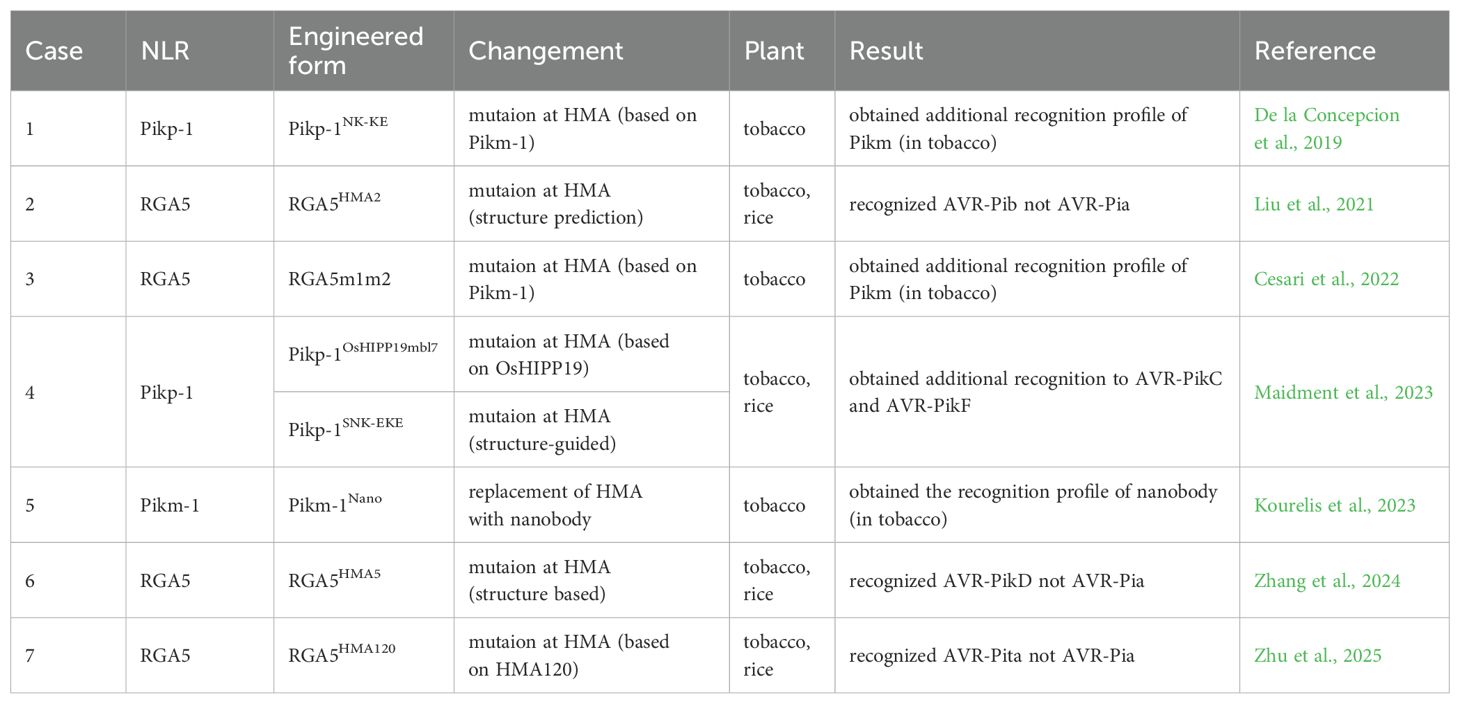
In recent years, rapid developments have taken place in structural biology. These advancements has provided accumulating details on interactions between R gene products and pathogen effectors, enabling the engineering of novel R gene products. Such pioneering attempts and studies mainly focused on rice blast resistance (Figure 2; Table 3). The allelic R genes Pikp and Pikm are each composed of two tandem NLR genes. Both Pikp-1 and Pikm-1 contain a Heavy Metal Associated (HMA) domain that interacts with effectors from M. oryzae. Based on structural insights into protein interactions, specific residues from the Pikm-1 HMA domain were swapped into Pikp-1. And one mutant, Pikp-1NK-KE, exhibited altered binding to AVR-PikA and AVR-PikE, effectors recognized by Pikm but not Pikp (De la Concepcion et al., 2019). Similar engineering was applied to RGA5, another NLR containing an HMA domain at the carboxyl terminus. Key residues in RGA5 were replaced with Pikp-1-derived AVR-PikD-interacting residues. The engineered NLR, RGA5m1m2, retained the recognition of AVR-Pia and AVR1-CO39 while gaining additional AVR-PikD specificity (Cesari et al., 2022). However, these engineered NLRs conferred resistance only in tobacco. Transgenic rice plants expressing these constructs failed to resist blast disease, possibly due to insufficient structural data on full-length NLR proteins at the time (De la Concepcion et al., 2019; Cesari et al., 2022).
With advancements in structural analysis and prediction techniques, engineering NLRs has become feasible in practice (Table 3). In a study on RGA5, mutations were introduced into its HMA domain based on structure predictions to interact with a non-cognate effector, AvrPib. The engineered NLR, RGA5HMA2, successfully shifted recognition from AvrPia to AvrPib in both tobacco and rice (Liu et al., 2021). A similar outcome was observed with another designed NLR, RGA5HMA5, which altered resistance specificity from AVR-Pia to AVR-PikD in rice (Zhang et al., 2024). Through large-scale screening of rice HMA domains, HMA120 was identified as an AVR-Pita-interacting domain. After integrating HMA120 into RGA5, the chimeric RGAHMA120 could confer resistance to AVR-Pita-carrying pathogen in rice (Zhu et al., 2025). Likewise, the HMA domain of OsHIPP19 was able to interact with AVR-PikC and AVR-PikF with high affinity. Using structural modeling, two engineered NLRs, Pikp-1OsHIPP19-mbl7 and Pikp-1SNK-EKE, were generated. Both of them mediated additional resistance to M. oryzae strains expressing AVR-PikC and AVR-PikF in rice (Maidment et al., 2023).
The most groundbreaking advance in R gene product engineering is the fusion of NLRs with nanobodies (Table 3). The nanobodies are small fragments of camelid antibodies and are widely used in biotechnology. Replacing the HMA domain with nanobodies specific to fluorescent proteins (FPs), Pikm-1Nano and Pikm-2 formed a ‘Pikobody’ that conferred resistance against Potato virus X (PVX) variants expressing FPs in transgenic tobacco (Kourelis et al., 2023). The Pikobody established an artificial adaptive immunity-like system, enabling plants to combat diverse pathogens, including emerging strains, with potential applications in future crop production. However, it should be noticed that no effector-targeted Pikobody has been engineered to date. As a consequence, more practice is needed to further optimize the system to enhance its practicality.
Future perspectives
Over three decades of researches have significantly advanced our understanding of plant defense mechanisms against pathogens. Although there are still many unknowns, current insights already provide actionable guidance for crop resistance breeding. Pathogen invasion relies on multiple steps, many of which involve hijacking and exploiting proteins or other molecules in host cells for paying the minimum cost. Conversely, perturbations in pathogen invasion processes can disrupt pathogen viability and lead to infection failure. Such dynamics underpin the evolutionary emergence of recessive resistance genes in plants. As a result, targeted intervention in critical molecular nodes of pathogen-host interplay will still be a common strategy in the future. These nodes include both S genes and resistance-negative regulator-encoding genes. More studies on the editing of both the coding regions and promoters of these genes will appear, especially in combination with editing of other functional genes.
With rapid developments in synthetic biology, attempts to engineer specialized NLRs have already appeared. Although these attempts have not reached the point where it can be applied in crop production now, the prospect is broad and attractive. The ‘Pikobody’ system has shown us the possibility that artificial immune receptor proteins could be a general solution in dealing with diverse pathogen threats. Nonetheless, more work should be done to optimize it to meet the needs of different crops. Additionally, expanding the spectrum of engineered receptors will be a key direction in the near future. This could be achieved by targeting PAMPs or other common features of distinct pathogens, as well as by combining recognition regions from multiple receptors. Moreover, synthetic promoters will be another focus. With the help of pathogen-responsive cis-elements, many genes could be used to improve crop disease resistance. Currently, such attempts have been made, such as the ‘promoter trap’ in several plants to resist Xanthomonas (Zhang et al., 2022a). In the future, more sophisticated promoters will be engineered to combat multiple pathogens.
Artificial intelligence (AI) has emerged as a transformative force in life sciences, redefining many research paradigms. Last year, the Nobel Prize was awarded to three scientists for their contribution to computational protein design and protein structure prediction. Both techniques can be applied in research on plant-pathogen interaction and crop disease resistance. To date, many direct interactions between plant R gene products and pathogen effectors remain unclear. And this challenge will soon be addressed with the aid of platforms like AlphaFold. Not only can AI assist in prediction, but structure-based de novo design of plant R gene products will also be possible in the near future. Recently, artificial antibodies have been successfully designed through a fully de nove method and have bound specific epitopes as expected (Bennett et al., 2025). This approach will facilitate the engineering of plant immune receptors. In addition, machine learning will play a key role in identifying essential genes in the response to pathogens based on multi-omics data and in optimizing CRISPR targets to increase editing efficiency.
Ecological factors and impacts should be considered in disease resistance breeding. Plants are colonized by communities of microbes in nature, which include not only pathogens but also beneficial microbes. Some of them can protect plants from being infected by various pathogens. If the plants are engineered to facilitate their colonization, the disease resistance will be enhanced (Ge and Wang, 2025). Meanwhile, plants and pathogens are co-evolving with each other. Therefore, the variation and evolution of pathogens should be considered when engineered R genes are applied in breeding. Monitoring the effector diversity among different strains from various locations would provide important information for adapting engineering strategies.
The strategies discussed here largely rely on transgenic process. The public’s attitude towards genetically modified organisms (GMOs) is worth noting. Most of the concerns about GMOs stem from misunderstandings. Communication and transparent policies would be useful to change public perception. Scientists should make more efforts to demonstrate to the public that using gene editing tools can now minimize changes to crop genomes; policymakers should let the public know that all the GMOs are regulated by a series of policies and the GMOs are optional, not mandatory, in daily life. Open-access databases documenting the safety and efficacy of edited traits will also be helpful for the public. In China, the acceptance of GMOs has improved compared with ten years ago. With continuous efforts, disease resistance breeding paradigms will be reshaped in the future.
Author contributions
DW: Writing – original draft. RY: Writing – original draft. ML: Writing – original draft. HlL: Investigation, Writing – original draft. HL: Supervision, Funding acquisition, Visualization, Writing – review & editing. WY: Funding acquisition, Writing – review & editing. HZ: Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from the Natural Science Foundation of Hubei Province (2023AFB650) and National Natural Science Foundation of China (31872811 and 32172028).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bastet, A., Zafirov, D., Giovinazzo, N., Guyon-Debast, A., Nogue, F., Robaglia, C., et al. (2019). Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 17, 1736–1750. doi: 10.1111/pbi.13096
Bennett, N. R., Watson, J. L., Ragotte, R. J., Borst, A. J., See, D. L., Weidle, C., et al. (2025). Atomically accurate de novo design of antibodies with RFdiffusion. bioRxiv 2024, 3.14.585103. doi: 10.1101/2024.03.14.585103
Buschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. doi: 10.1016/s0092-8674(00)81912-1
Cesari, S., Xi, Y., Declerck, N., Chalvon, V., Mammri, L., Pugniere, M., et al. (2022). New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 13, 1524. doi: 10.1038/s41467-022-29196-6
Chen, X., Shang, J., Chen, D., Lei, C., Zou, Y., Zhai, W., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. doi: 10.1111/j.1365-313X.2006.02739.x
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
De la Concepcion, J. C., Franceschetti, M., MacLean, D., Terauchi, R., Kamoun, S., and Banfield, M. J. (2019). Protein engineering expands the effector recognition profile of a rice NLR immune receptor. eLife 8, e47713. doi: 10.7554/eLife.47713
Djennane, S., Gersch, S., Le-Bohec, F., Piron, M. C., Baltenweck, R., Lemaire, O., et al. (2024). CRISPR/Cas9 editing of Downy mildew resistant 6 (DMR6-1) in grapevine leads to reduced susceptibility to Plasmopara viticola. J. Exp. Bot. 75, 2100–2112. doi: 10.1093/jxb/erad487
Elliott, K., Veley, K. M., Jensen, G., Gilbert, K. B., Norton, J., Kambic, L., et al. (2024). CRISPR/Cas9-generated mutations in a sugar transporter gene reduce cassava susceptibility to bacterial blight. Plant Physiol. 195, 2566–2578. doi: 10.1093/plphys/kiae243
Ellis, J. G., Lawrence, G. J., Luck, J. E., and Dodds, P. N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. doi: 10.1105/tpc.11.3.495
Eom, J. S., Luo, D., Atienza-Grande, G., Yang, J., Ji, C., Thi Luu, V., et al. (2019). Diagnostic kit for rice blight resistance. Nat. Biotechnol. 37, 1372–1379. doi: 10.1038/s41587-019-0268-y
Farnham, G. and Baulcombe, D. C. (2006). Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. U.S.A. 103, 18828–18833. doi: 10.1073/pnas.0605777103
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Gao, Y., Wang, W., Zhang, T., Gong, Z., Zhao, H., and Han, G. Z. (2018). Out of water: The origin and early diversification of plant R-genes. Plant Physiol. 177, 82–89. doi: 10.1104/pp.18.00185
Ge, A. H. and Wang, E. (2025). Exploring the plant microbiome: A pathway to climate-smart crops. Cell 188, 1469–1485. doi: 10.1016/j.cell.2025.01.035
Giacomelli, L., Zeilmaker, T., Giovannini, O., Salvagnin, U., Masuero, D., Franceschi, P., et al. (2023). Simultaneous editing of two DMR6 genes in grapevine results in reduced susceptibility to downy mildew. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1242240
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Gomez-Gomez, L. and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
Gupta, A., Liu, B., Chen, Q. J., and Yang, B. (2023). High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice. Plant Biotechnol. J. 21, 1454–1464. doi: 10.1111/pbi.14049
Gupta, A., Liu, B., Raza, S., Chen, Q. J., and Yang, B. (2024). Modularly assembled multiplex prime editors for simultaneous editing of agronomically important genes in rice. Plant Commun. 5, 100741. doi: 10.1016/j.xplc.2023.100741
Harris, C. J., Slootweg, E. J., Goverse, A., and Baulcombe, D. C. (2013). Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. U.S.A. 110, 21189–21194. doi: 10.1073/pnas.1311134110
Hasley, J. A. R., Navet, N., and Tian, M. (2021). CRISPR/Cas9-mediated mutagenesis of sweet basil candidate susceptibility gene ObDMR6 enhances downy mildew resistance. PloS One 16, e0253245. doi: 10.1371/journal.pone.0253245
Hu, K., Cao, J., Zhang, J., Xia, F., Ke, Y., Zhang, H., et al. (2017). Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009. doi: 10.1038/nplants.2017.9
Hutin, M., Sabot, F., Ghesquiere, A., Koebnik, R., and Szurek, B. (2015). A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042
Iyer, A. S. and McCouch, S. R. (2004). The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17, 1348–1354. doi: 10.1094/MPMI.2004.17.12.1348
Jacob, P., Kim, N. H., Wu, F., El-Kasmi, F., Chi, Y., Walton, W. G., et al. (2021). Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373, 420–425. doi: 10.1126/science.abg7917
Jia, H., Omar, A. A., Xu, J., Dalmendray, J., Wang, Y., Feng, Y., et al. (2024). Generation of transgene-free canker-resistant Citrus sinensis cv. Hamlin in the T0 generation through Cas12a/CBE co-editing. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1385768
Jia, H., Orbovic, V., and Wang, N. (2019). CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 17, 1928–1937. doi: 10.1111/pbi.13109
Jia, H. and Wang, N. (2020). Generation of homozygous canker-resistant citrus in the T0 generation using CRISPR-SpCas9p. Plant Biotechnol. J. 18, 1990–1992. doi: 10.1111/pbi.13375
Jia, H., Wang, Y., Su, H., Huang, X., and Wang, N. (2022). LbCas12a-D156R efficiently edits LOB1 effector binding elements to generate canker-resistant citrus plants. Cells 11, 315. doi: 10.3390/cells11030315
Jia, H., Zhang, Y., Orbovic, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jiang, G. H., Xia, Z. H., Zhou, Y. L., Wan, J., Li, D. Y., Chen, R. S., et al. (2006). Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAgamma1. Mol. Genet. Genomics 275, 354–366. doi: 10.1007/s00438-005-0091-7
Johal, G. S. and Briggs, S. P. (1992). Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258, 985–987. doi: 10.1126/science.1359642
Jones, J. D. and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, D. A., Thomas, C. M., Hammond-Kosack, K. E., Balint-Kurti, P. J., and Jones, J. D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. doi: 10.1126/science.7973631
Jones, J. D., Vance, R. E., and Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395. doi: 10.1126/science.aaf6395
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, B. L., and Andreasson, E. (2021). Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11, 4487. doi: 10.1038/s41598-021-83972-w
Kim, Y. A., Moon, H., and Park, C. J. (2019). CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice (N. Y.) 12, 67. doi: 10.1186/s12284-019-0325-7
Kourelis, J., Marchal, C., Posbeyikian, A., Harant, A., and Kamoun, S. (2023). NLR immune receptor-nanobody fusions confer plant disease resistance. Science 379, 934–939. doi: 10.1126/science.abn4116
Kusch, S. and Panstruga, R. (2017). mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30, 179–189. doi: 10.1094/MPMI-12-16-0255-CR
Li, Y. J., Gu, J. M., Ma, S., Xu, Y., Liu, M., Zhang, C., et al. (2023). Genome editing of the susceptibility gene ZmNANMT confers multiple disease resistance without agronomic penalty in maize. Plant Biotechnol. J. 21, 1525–1527. doi: 10.1111/pbi.14078
Li, C., Li, W., Zhou, Z., Chen, H., Xie, C., and Lin, Y. (2020). A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 18, 313–315. doi: 10.1111/pbi.13217
Li, S., Lin, D., Zhang, Y., Deng, M., Chen, Y., Lv, B., et al. (2022b). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460. doi: 10.1038/s41586-022-04395-9
Li, R., Maioli, A., Yan, Z., Bai, Y., Valentino, D., Milani, A. M., et al. (2022a). CRISPR/Cas9-based knock-out of the PMR4 gene reduces susceptibility to late blight in two tomato cultivars. Int. J. Mol. Sci. 23, 14542. doi: 10.3390/ijms232314542
Li, S., Shen, L., Hu, P., Liu, Q., Zhu, X., Qian, Q., et al. (2019). Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 61, 1201–1205. doi: 10.1111/jipb.12774
Liu, L., Li, Y., Wang, Q., Xu, X., Yan, J., Wang, Y., et al. (2024). Constructed rice tracers identify the major virulent transcription activator-like effectors of the bacterial leaf blight pathogen. Rice (N. Y.) 17, 30. doi: 10.1186/s12284-024-00704-0
Liu, X., Yu, Y., Yao, W., Yin, Z., Wang, Y., Huang, Z., et al. (2023). CRISPR/Cas9-mediated simultaneous mutation of three salicylic acid 5-hydroxylase (OsS5H) genes confers broad-spectrum disease resistance in rice. Plant Biotechnol. J. 21, 1873–1886. doi: 10.1111/pbi.14099
Liu, Q., Yuan, M., Zhou, Y., Li, X., Xiao, J., and Wang, S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x
Liu, Y., Zhang, X., Yuan, G., Wang, D., Zheng, Y., Ma, M., et al. (2021). A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors. Proc. Natl. Acad. Sci. U.S.A. 118, e2110751118. doi: 10.1073/pnas.2110751118
Ma, S., Lapin, D., Liu, L., Sun, Y., Song, W., Zhang, X., et al. (2020). Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370, eabe3069. doi: 10.1126/science.abe3069
Maidment, J. H. R., Shimizu, M., Bentham, A. R., Vera, S., Franceschetti, M., Longya, A., et al. (2023). Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor. eLife 12, e81123. doi: 10.7554/eLife.81123
Maruta, N., Burdett, H., Lim, B. Y. J., Hu, X., Desa, S., Manik, M. K., et al. (2022). Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 74, 5–26. doi: 10.1007/s00251-021-01242-5
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Moscou, M. J. and Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. doi: 10.1126/science.1178817
Ni, Z., Cao, Y., Jin, X., Fu, Z., Li, J., Mo, X., et al. (2021). Engineering resistance to bacterial blight and bacterial leaf streak in rice. Rice (N. Y.) 14, 38. doi: 10.1186/s12284-021-00482-z
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Ortega, A., Seong, K., Schultink, A., de Toledo Thomazella, D. P., Seo, E., Zhang, E., et al. (2024). CRISPR/Cas9-mediated editing of Bs5 and Bs5L in tomato leads to resistance against Xanthomonas. Plant Biotechnol. J. 22, 2785–2787. doi: 10.1111/pbi.14404
Piffanelli, P., Zhou, F., Casais, C., Orme, J., Jarosch, B., Schaffrath, U., et al. (2002). The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. doi: 10.1104/pp.010954
Ravensdale, M., Bernoux, M., Ve, T., Kobe, B., Thrall, P. H., Ellis, J. G., et al. (2012). Intramolecular interaction influences binding of the Flax L5 and L6 resistance proteins to their AvrL567 ligands. PloS Pathog. 8, e1003004. doi: 10.1371/journal.ppat.1003004
Santillan Martinez, M. I., Bracuto, V., Koseoglou, E., Appiano, M., Jacobsen, E., Visser, R. G. F., et al. (2020). CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 20, 284. doi: 10.1186/s12870-020-02497-y
Schepler-Luu, V., Sciallano, C., Stiebner, M., Ji, C., Boulard, G., Diallo, A., et al. (2023). Genome editing of an African elite rice variety confers resistance against endemic and emerging Xanthomonas oryzae pv. oryzae strains. eLife 12, e84864. doi: 10.7554/eLife.84864
Segretin, M. E., Pais, M., Franceschetti, M., Chaparro-Garcia, A., Bos, J. I., Banfield, M. J., et al. (2014). Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol. Plant Microbe Interact. 27, 624–637. doi: 10.1094/MPMI-02-14-0040-R
Sha, G., Sun, P., Kong, X., Han, X., Sun, Q., Fouillen, L., et al. (2023). Genome editing of a rice CDP-DAG synthase confers multipathogen resistance. Nature 618, 1017–1023. doi: 10.1038/s41586-023-06205-2
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Sueldo, D. J., Shimels, M., Spiridon, L. N., Caldararu, O., Petrescu, A. J., Joosten, M. H., et al. (2015). Random mutagenesis of the nucleotide-binding domain of NRC1 (NB-LRR Required for Hypersensitive Response-Associated Cell Death-1), a downstream signalling nucleotide-binding, leucine-rich repeat (NB-LRR) protein, identifies gain-of-function mutations in the nucleotide-binding pocket. New Phytol. 208, 210–223. doi: 10.1111/nph.13459
Tao, H., Shi, X., He, F., Wang, D., Xiao, N., Fang, H., et al. (2021). Engineering broad-spectrum disease-resistant rice by editing multiple susceptibility genes. J. Integr. Plant Biol. 63, 1639–1648. doi: 10.1111/jipb.13145
Tek, M. I., Calis, O., Fidan, H., Shah, M. D., Celik, S., and Wani, S. H. (2022). CRISPR/Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1081506
Thomazella, D. P. T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 118, e2026152118. doi: 10.1073/pnas.2026152118
Timilsina, S., Potnis, N., Newberry, E. A., Liyanapathiranage, P., Iruegas-Bocardo, F., White, F. F., et al. (2020). Xanthomonas diversity, virulence and plant-pathogen interactions. Nat. Rev. Microbiol. 18, 415–427. doi: 10.1038/s41579-020-0361-8
Tripathi, J. N., Ntui, V. O., Shah, T., and Tripathi, L. (2021). CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol. J. 19, 1291–1293. doi: 10.1111/pbi.13614
van Damme, M., Huibers, R. P., Elberse, J., and Van den Ackerveken, G. (2008). Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 54, 785–793. doi: 10.1111/j.1365-313X.2008.03427.x
Veley, K. M., Elliott, K., Jensen, G., Zhong, Z., Feng, S., Yoder, M., et al. (2023). Improving cassava bacterial blight resistance by editing the epigenome. Nat. Commun. 14, 85. doi: 10.1038/s41467-022-35675-7
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014b). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, J. Y. and Doudna, J. A. (2023). CRISPR technology: A decade of genome editing is only the beginning. Science 379, eadd8643. doi: 10.1126/science.add8643
Wang, Y., Geng, M., Pan, R., Zhang, T., Lu, X., Zhen, X., et al. (2024c). Editing of the MeSWEET10a promoter yields bacterial blight resistance in cassava cultivar SC8. Mol. Plant Pathol. 25, e70010. doi: 10.1111/mpp.70010
Wang, J., Hu, M., Qi, J., Han, Z., Wang, G., Qi, Y., et al. (2019a). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870. doi: 10.1126/science.aav5870
Wang, A. and Krishnaswamy, S. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. doi: 10.1111/j.1364-3703.2012.00791.x
Wang, M., Li, S., Li, H., Song, C., Xie, W., Zuo, S., et al. (2024b). Genome editing of a dominant resistance gene for broad-spectrum resistance to bacterial diseases in rice without growth penalty. Plant Biotechnol. J. 22, 529–531. doi: 10.1111/pbi.14233
Wang, S., Li, Q. P., Wang, J., Yan, Y., Zhang, G. L., Yan, Y., et al. (2019b). YR36/WKS1-mediated phosphorylation of PsbO, an extrinsic member of photosystem II, inhibits photosynthesis and confers stripe rust resistance in wheat. Mol. Plant 12, 1639–1650. doi: 10.1016/j.molp.2019.10.005
Wang, J., Liao, Z., Jin, X., Liao, L., Zhang, Y., Zhang, R., et al. (2024a). Xanthomonas oryzae pv. oryzicola effector Tal10a directly activates rice OsHXK5 expression to facilitate pathogenesis. Plant J. 119, 2423–2436. doi: 10.1111/tpj.16929
Wang, Q., Liu, Y., He, J., Zheng, X., Hu, J., Liu, Y., et al. (2014a). STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 5, 4768. doi: 10.1038/ncomms5768
Wang, N., Tang, C., Fan, X., He, M., Gan, P., Zhang, S., et al. (2022). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961–2974.e2919. doi: 10.1016/j.cell.2022.06.027
Wei, Z., Abdelrahman, M., Gao, Y., Ji, Z., Mishra, R., Sun, H., et al. (2021). Engineering broad-spectrum resistance to bacterial blight by CRISPR-Cas9-mediated precise homology directed repair in rice. Mol. Plant 14, 1215–1218. doi: 10.1016/j.molp.2021.05.012
Wulff, B. B., Thomas, C. M., Smoker, M., Grant, M., and Jones, J. D. (2001). Domain swapping and gene shuffling identify sequences required for induction of an Avr-dependent hypersensitive response by the tomato Cf-4 and Cf-9 proteins. Plant Cell 13, 255–272. doi: 10.1105/tpc.13.2.255
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446. doi: 10.1016/j.molp.2019.08.006
Xu, X., Xu, Z., Li, Z., Zakria, M., Zou, L., and Chen, G. (2021). Increasing resistance to bacterial leaf streak in rice by editing the promoter of susceptibility gene OsSULRT3;6. Plant Biotechnol. J. 19, 1101–1103. doi: 10.1111/pbi.13602
Yang, J., Fang, Y., Wu, H., Zhao, N., Guo, X., Mackon, E., et al. (2023). Improvement of resistance to rice blast and bacterial leaf streak by CRISPR/Cas9-mediated mutagenesis of Pi21 and OsSULTR3;6 in rice (Oryza sativa L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1209384
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yoshihisa, A., Yoshimura, S., Shimizu, M., Sato, S., Matsuno, S., Mine, A., et al. (2022). The rice OsERF101 transcription factor regulates the NLR Xa1-mediated immunity induced by perception of TAL effectors. New Phytol. 236, 1441–1454. doi: 10.1111/nph.18439
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Yuan, M., Ke, Y., Huang, R., Ma, L., Yang, Z., Chu, Z., et al. (2016). A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife 5, e19605. doi: 10.7554/eLife.19605
Zhang, B., Han, X., Yuan, W., and Zhang, H. (2022a). TALEs as double-edged swords in plant-pathogen interactions: Progress, challenges, and perspectives. Plant Commun. 3, 100318. doi: 10.1016/j.xplc.2022.100318
Zhang, Y., Liu, J., Li, Y., Ma, H., Ji, J., Wang, Y., et al. (2025). Generation of novel bpm6 and dmr6 mutants with broad-spectrum resistance using a modified CRISPR/Cas9 system in Brassica oleracea. J. Integr. Plant Biol. doi: 10.1111/jipb.13842. online ahead of print.
Zhang, B., Liu, M., Wang, Y., Yuan, W., and Zhang, H. (2022b). Plant NLRs: Evolving with pathogen effectors and engineerable to improve resistance. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1018504
Zhang, X., Liu, Y., Yuan, G., Wang, S., Wang, D., Zhu, T., et al. (2024). The synthetic NLR RGA5HMA5 requires multiple interfaces within and outside the integrated domain for effector recognition. Nat. Commun. 15, 1104. doi: 10.1038/s41467-024-45380-2
Zhang, H. and Wang, S. (2013). Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr. Opin. Plant Biol. 16, 188–195. doi: 10.1016/j.pbi.2013.02.008
Zhang, J., Yin, Z., and White, F. (2015). TAL effectors and the executor R genes. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00641
Zhang, B., Zhang, H., Li, F., Ouyang, Y., Yuan, M., Li, X., et al. (2020). Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to Xanthomonas oryzae pv. oryzae. Plant Commun. 1, 100088. doi: 10.1016/j.xplc.2020.100088
Zhang, Y., Zhao, L., Zhao, J., Li, Y., Wang, J., Guo, R., et al. (2017). S5H/DMR6 encodes a salicylic acid 5-hydroxylase that fine-tunes salicylic acid homeostasis. Plant Physiol. 175, 1082–1093. doi: 10.1104/pp.17.00695
Zhao, J., Fu, J., Li, X., Xu, C., and Wang, S. (2009). Dissection of the factors affecting development-controlled and race-specific disease resistance conferred by leucine-rich repeat receptor kinase-type R genes in rice. Theor. Appl. Genet. 119, 231–239. doi: 10.1007/s00122-009-1032-3
Zhao, N., Guo, A., Wang, W., Li, B., Wang, M., Zhou, Z., et al. (2024). GbPP2C80 interacts with GbWAKL14 to negatively co-regulate resistance to Fusarium and Verticillium wilt via MPK3 and ROS signaling in sea island cotton. Adv. Sci. (Weinh) 11, e2309785. doi: 10.1002/advs.202309785
Zhao, Y. B., Liu, M. X., Chen, T. T., Ma, X., Li, Z. K., Zheng, Z., et al. (2022). Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism. Sci. Adv. 8, eabq5108. doi: 10.1126/sciadv.abq5108
Keywords: plant-pathogen interaction, plant immunity, susceptibility gene, resistance gene, NLR, pathogen effector, gene editing, engineering
Citation: Wang D, Yang R, Liu M, Li H, Li H, Yuan W and Zhang H (2025) Recent advances in innovative strategies for plant disease resistance breeding. Front. Plant Sci. 16:1586375. doi: 10.3389/fpls.2025.1586375
Received: 02 March 2025; Accepted: 28 April 2025;
Published: 21 May 2025.
Edited by:
Zhaohui Chu, Wuhan University, ChinaReviewed by:
Zhiyuan Ji, Chinese Academy of Agricultural Sciences (CAAS), ChinaJiehua Qiu, Chinese Academy of Agricultural Sciences, China
Tao Wu, Yangzhou University, China
Copyright © 2025 Wang, Yang, Liu, Li, Li, Yuan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Zhang, emh0QGh1YnUuZWR1LmNu; Wenya Yuan, d3l5dWFuQGh1YnUuZWR1LmNu; Haitao Li, bGh0QGh1YnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Dekun Wang
Dekun Wang Ruojiao Yang
Ruojiao Yang Mengting Liu
Mengting Liu Haolong Li
Haolong Li Haitao Li
Haitao Li Wenya Yuan
Wenya Yuan Haitao Zhang
Haitao Zhang