- Institute of Fruits and Vegetables, Xinjiang Academy of Agricultural Sciences, Key Laboratory of Genome Research and Genetic Improvement of Xinjiang Characteristic Fruits and Vegetables, Scientific Observation and Experimental Station for Pomology of Xinjiang, Ministry of Agriculture and Rural Affairs, Urumqi, Xinjiang, China
As calcium responders, calcium dependent protein kinases (CDPKs) play an important role in plant growth and development and in response to biotic and abiotic stresses. However, information on CDPKs in jujube (Ziziphus jujuba Mill.) (ZjCDPKs) is limited. In the current study, a total of 21 ZjCDPKs were identified, which are located on eight chromosomes. Gene structure and conserved motif analysis showed that all ZjCDPKs have similar gene structures and conserved motifs, except for ZjCDPK9 and ZjCDPK21. The CDPKs from Arabidopsis, rice, tomato, alfalfa, and jujube were divided into eight subgroups, and the members of ZjCDPKs were unevenly distributed across these subgroups. Colinear analysis revealed that 12 homozygous CDPKs were detected between jujube and Arabidopsis, and 14 pairs were found between jujube and tomato. Additionally, four types of cis-acting elements were identified in the promoters of the ZjCDPKs, including hormone, stress, development, and light response elements. The expression profiles of ZjCDPKs at different fruit growth stages, in response to phytoplasma infection, cold, and salt stresses revealed that most ZjCDPKs were either up- or down-regulated. Finally, varying numbers of transcription factors were observed to interact with the promoter region of ZjCDPK4/6/7/8/10/14/16 and showed opposite expression patterns in response to cold or salt stress. The systematic analysis of ZjCDPKs provides important information for further functional characterization of CDPKs in jujube in response to multiple biological processes.
1 Introduction
In nature, plants have evolved sophisticated mechanisms to respond and adapt to a wide variety of biotic and abiotic stresses (Wang et al., 2020b). Calcium (Ca2+), as an important second messenger, plays a crucial role in signal transduction during stress adaptation and plant growth and development (Wang et al., 2023a; Tuteja and Mahajan, 2007; Wang et al., 2020b). Three calcium sensors have been identified in plants, including calmodulin (CaM)/calmodulin-like proteins (CML), calcineurin B-like protein (CBL), and calcium dependent protein kinase (CDPK) (Wang et al., 2023b). Among them, the signaling system composed of CBLs and their interacting protein kinases participates in the regulation of plant response to low-temperature stress through the phosphorylation induction and decoding of Ca2+ signals (Ma et al., 2020). CMLs can directly bind with Ca2+, which is involved in the responses to numerous stresses (Wang et al., 2023b). Moreover, previous research has reported the activation of CDPKs by Ca2+ to perform phosphorylation, indicating the key role of CDPKs as Ca2+ responders (Dekomah et al., 2022a).
CDPKs are a class of serine/threonine type protein kinases with four characteristic domains, including a variable N-terminal domain (VNTD), a catalytic Ser/thr protein kinase domain (PKD) (which can bind the ATP phosphate donor and phosphorylates the serine and threonine residues of its substrates), an autoinhibitory domain (AIR), and a carboxyl-terminal calmodulin-like domain (CLD) (which has one to four conserved EF-hand motifs for calcium-binding) (Cheng et al., 2001; Hrabak et al., 2003; Liese and Romeis, 2013). Apart from the functions of PKD and CLD, the VNTD domain typically contains N-myristoylation sites or N-myristoylation and S-palmitoylation sites, and plays an important role in subcellular location and substrate recognition. The AIR domain, also denoted as the junction domain function, acts as a pseudo substrate to maintain CDPK inactive in the absence of Ca2+ stimulation (Hrabak et al., 2003; Hamel et al., 2014). When Ca2+ increases, the EF-hand can bind motifs, leading to conformation changes in CLD and the subsequent activation of the CDPK kinase domain, which can recognize and phosphorylate downstream targets (Dekomah et al., 2022a).
Similar to CaM/CML and CBLs, CDPKs play an important role in the signal transduction pathway, including secondary metabolites, hormone regulation, and stress tolerance (Dekomah et al., 2022a). For example, in maize, the overexpression of ZmCDPK7 can enhance thermotolerance by decreasing the accumulation of hydrogen peroxide (H2O2) and malondialdehyde (MDA) (Zhao et al., 2021). In tomato, virus-induced gene silencing stimulates the silencing of ShCDPK6, while ShCDPK26 shows less resistance to Botrytis cinerea, cold, and drought stress (Li et al., 2022). In potato, StCDPK21/22 and StCDPK3 can regulate the content of MDA and proline to facilitate drought tolerance (Dekomah et al., 2022b). During peach storage, PpCDPK7 has been reported to interact with PpRBOH, which further mediates the Ca2+ and reactive oxygen species (ROS) signal cascades to enhance chilling tolerance (Zhao et al., 2022). In wheat, TaCDPK25-U is significantly induced by drought stress and can be positively regulated by TaDREB3 to improve drought tolerance (Linghu et al., 2023). In Arabidopsis, AtCPK28 can be activated by Ca2+ and phosphorylated downstream NLP7 to improve cold tolerance (Ding et al., 2022). Moreover, resveratrol levels can increase following the overexpression of VaCPK20 or VaCPK29 in Vitis amurensis cells (Aleynova-Shumakova et al., 2014). AtCPK6 acts as a positive regulator in stomatal movement or closure by ABA and methyl jasmonate, respectively, in a signaling-dependent manner (Xu et al., 2010; Ye et al., 2013). With the development of plant genome sequencing and the important biological function of CDPKs, the CDPKs members in different plant species have been widely identified, including 34 members in Arabidopsis thaliana (Hrabak et al., 2003), 31 in rice (Kong et al., 2013), 29 in tomato (Wang et al., 2016), 40 in maize (Kong et al., 2013), 30 in pear (Liu et al., 2022), and 17 in peach (Zhao et al., 2022). However, the identification of CDPKs in jujube and their biological function has not yet been reported.
Jujube (Ziziphus jujuba Mill.) belongs to the Rhamnaceae family and has important economic and ecological value in China (Liu et al., 2020). Jujube is widely planted in sandy alkali arid areas, which has facilitated its advanced tolerance to salt, drought, and cold tolerance (Wang et al., 2020a; Gao et al., 2021; Wang et al., 2025). With the whole genome sequencing completed in jujube, it has become an ideal fruit tree for research on abiotic stress mechanisms. Thus, in the current study, the identification, phylogenetic analysis, gene structure, and conserved motifs of CDPKs in jujube were conducted. The RNA-seq data was then used to analyze the expression levels of CDPKs in response to various biotic and abiotic stresses and fruit growth development. The results provide a theoretical basis for clarifying the biological function of CDPKs in jujube.
2 Materials and methods
2.1 Identification and analysis of the physicochemical properties of the CDPKs in jujube
To screen the CDPKs members in jujube, the complete genome and annotated information files of jujube were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov), and the CDPK Hidden Markov model (PF00069 and PF13499) was downloaded from the InterPRO database (https://www.ebi.ac.uk/interpro/). HMMER with an E value of 1e-3 parameter was employed to search for the CDPK protein sequences in the jujube genome database and obtain the initial members of potential CDPKs. Following this, 34 CDPKs in Arabidopsis were retrieved from the TAIR database (https://www.arabidopsis.org) to perform BLASTP analysis against the jujube genome. The Conserved Domain Database (CDD) with a P-score cutoff 0.03 (http://www.ncbi.nlm.nih.gov/cdd/) was then employed to determine the conservative domains, remove the redundant sequence (Cao et al., 2024b), and finally obtain the CDPK members in jujube (ZjCDPKs).
The online ExPASy tool (http://web.expasy.org/protparam/) was used to determine the molecular weight (kDa), isoelectric points (pI), and other physicochemical properties of the ZjCDPKs (Wilkins et al., 1999).
2.2 Chromosomal location prediction of ZjCDPKs
The CDS, protein sequences (Supplementary Table 1), and chromosome position information of the ZjCDPKs were obtained from the NCBI database.
2.3 Gene structure and conserved motif analysis
TBtools was employed to extract the CDS and related genome formation of the ZjCDPKs for the visualization analysis of the gene structure (Chen et al., 2020). The MEME online tool (http://meme-suite.org/) was used to analyze the conserved motifs of the ZjCDPKs, with a maximum motif number of 10 and an optimal motif width for the 6–50 amino acid residues (Bailey et al., 2009). The conserved motifs were then visualized in TBtools.
2.4 Phylogenetic analysis of the CDPKs in jujube and four other species
The CDPK protein sequences of Arabidopsis, rice, tomato, alfalfa, and jujube were retrieved from the NCBI database. MEGA 5.0 was used to construct the phylogenetic tree using the neighbor joining (NJ) method, with a bootstrap value of 1,000 (Tamura et al., 2011). The visualization of the phylogenetic tree was improved with the iTOL online tool (https://itol.embl.de).
2.5 Collinearity and cis-acting element analysis
The genome data of Z. jujuba Mill., Arabidopsis thaliana, and Solanum lycopersicum were used to determine duplication events and perform collinearity analysis of the CDPKs using MCScanX with Ka/Ks value (Qi et al., 2024). The results were visualized in TBtools.
For the cis-acting elements analysis, the 2 kb sequences upstream of each ZjCDPK were extracted as the promoter region and submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) to search for cis-acting elements (Higo et al., 1998). TBtools was used to visualize the results and create the heat maps.
2.6 Expression profile analysis of ZjCDPKs during jujube fruit development and in response to biotic and abiotic stresses
To understand the biological functions of ZjCDPKs, the expression profiles of ZjCDPKs were mined from the transcriptome data in response to low-temperature and salt stresses, phytoplasma infection materials, and different jujube fruit growth stages of ‘Jinsixiaozao’ and ‘Jinkuiwang’, respectively. The transcriptome assembly process could be referred to Cao et al. (2024a, 2025) and Jiang et al. (2025). Briefly, for cold treatment, samples of Z. jujuba Mill. ‘Dongzao’ and its autotetraploid were treated under a cold temperature (4°C) for 0, 6 and 24 h (Gao et al., 2021). For salt stress, 400 mM NaCl was treated on sour jujube seedlings for 0, 6, 24, and 192 h (Zhu et al., 2023). For phytoplasma infection analysis, different growth stages (S1, S2, and S3) of Z. jujuba Mill. ‘Pozao’ (‘PZ’) and Z. jujuba Mill. ‘T13’ were selected for RNA-seq analysis, where PZ_D and T13_D denote samples infected by phytoplasma, respectively, and PZ_H and T13_H denote healthy samples (Wang et al., 2022). For the jujube fruit growth analysis of ‘Jinsixiaozao’ (JS) and ‘Jinkuiwang’ (JKW), fruit from nine growth stages, namely, the early stage of young fruit (F1), the middle stage of young fruit (F2), the early stage of stone formation (F3), the stone formation stage (F4), the white mature stage (F5), the late white mature stage (F6), the quarter coloring stage (F7), the half red stage (F8), and the full red stage (F9), were selected for RNA-seq analysis (Zhao et al., 2023). The expression levels of ZjCDPKs were presented on a heat map in TBtools.
2.7 Mining and expression analysis of related transcription factor of ZjCDPKs
The PlantRegMap database (http://plantregmap.gao-lab.org/network.php) was employed to identify the potential transcription factors (TFs) upstream of ZjCDPK4/6/7/8/10/14/16 and their expression profiles in response to cold and salt stresses were then evaluated (Tian et al., 2020).
3 Results
3.1 Identification, physicochemical characteristics, and chromosome locations of ZjCDPKs
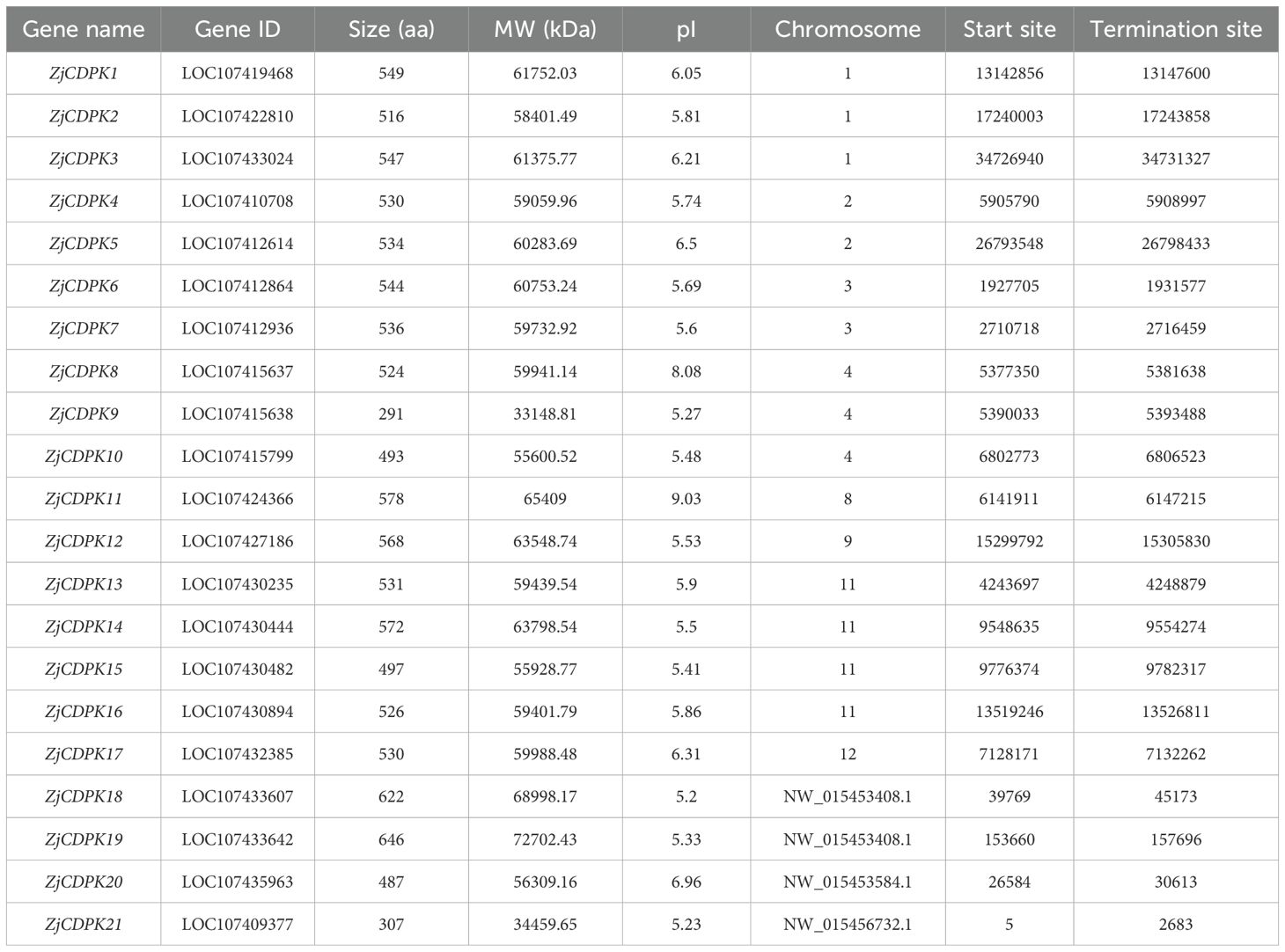
Through the systematic and genome-wide identification of ZjCDPKs, a total of 21 ZjCDPK members were screened and denoted as ZjCDPK1–ZjCDPK21 based on their chromosome positions. As shown in Table 1, the physicochemical characteristics (e.g., amino acid length and theoretical isoelectric point (pI) values) of the ZjCDPKs varied. The number of amino acids ranged from 291aa (ZjCDPK9) to 646 aa (ZjCDPK19). The molecular weight (mw) ranged from 33.15 kD (ZjCDPK9) to 72.70 kD (ZjCDPK19) and the pI values ranged from 5.20 (ZjCDPK18) to 9.03 (ZjCDPK11). In addition, the ZjCDPKs were located on eight chromosomes (Chr). Among them, three members of the ZjCDPKs (ZjCDPK1–ZjCDPK3) were located on Chr1, while ZjCDPK8– ZjCDPK10 was located on Chr4. Two and one ZjCDPKs were identified on Chr2/3 and Chr8/9/12, respectively. Four members of the ZjCDPKs were located on Chr 11, while ZjCDPK18–ZjCDPK21 did not match with any Chrs.
3.2 Conserved motifs and gene structure of ZjCDPKs
CDPKs have four characteristic domains, namely, VNTD, PKD, AIR, and CLD. Through MEME analysis, 10 conserved motifs were identified in the ZjCDPKs (Supplementary Table 2). As shown in Figure 1A, all the ZjCDPKs contained nine or ten conserved motifs, except for ZjCDPK9 and ZjCDPK21, which only contained five and six, respectively. The upstream of motif1 was found to be the kinase domain and the auto-inhibitory junction region was observed in motif4. Moreover, the EF-hand domains were identified in the motifs. The ZjCDPKs exhibited a similar number and spatial distribution of motifs in the same subgroup, indicating that the ZjCDPK proteins have evolutionary conservatism.
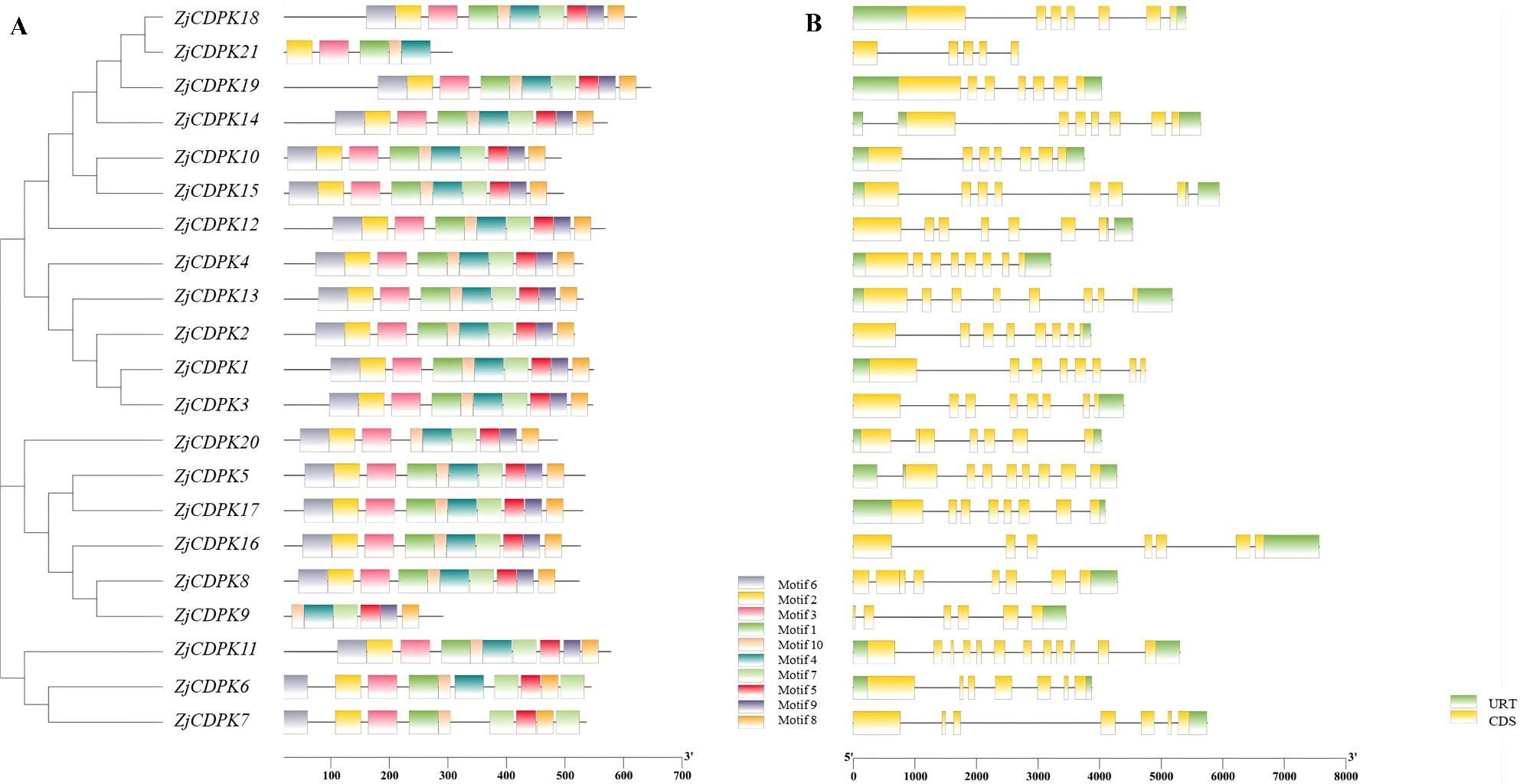
Figure 1. Conservative motifs and gene structures of ZjCDPKs. (A) Schematic diagram of the conserved motifs of ZjCDPKs. Ten motifs are represented by different colored boxes. (B) Gene structures of ZjCDPKs. Exons, introns, and UTR are represented by yellow boxes, black lines, and green boxes, respectively.
Gene structure analysis showed that all ZjCDPKs had similar gene structures and were interrupted by 5–11 introns. In particular, nine ZjCDPKs had seven introns and seven ZjCDPKs had six introns (Figure 1B). ZjCDPK9, ZjCDPK20, and ZjCDPK21 had fewer introns (five, five, and four, respectively), while ZjCDPK11 had the largest number of introns (eleven). The similarity and diversity of the ZjCDPKs gene structures may indicate their similar and distinct biological functions.
3.3 Phylogenetic analysis of the CDPKs
To study the evolutionary relationship of the CDPKs in jujube, Arabidopsis, rice, tomato, and alfalfa, we constructed an NJ phylogenetic tree using CDPK protein sequences from the above species. As shown in Figure 2, the CDPKs were divided into eight subgroups. The 21 ZjCDPKs members were distributed into different subgroups, including four in subgroup A, two in subgroup B, one in subgroup C, two in subgroup D, and three in subgroups E, F, G, and L, accounting for 18.18%, 13.33%, 8.30%, 12.50%, 12.50%, 23.07%, 15.00%, and 27.27% respectively. In Arabidopsis, three (13.63%), three (20.00%), three (25%), three (18.75%), ten (41.67%), three (23.07%), five (25.00%), and three (27.27%) CDPKs were categorized in subgroups A to H, respectively. The same was observed for rice, tomato, and alfalfa. In addition, the proportion of ZjCDPKs in different subgroups was similar to that of Arabidopsis, rice, tomato, and alfalfa, indicating that the evolution of CDPK in different plant species was conservative.
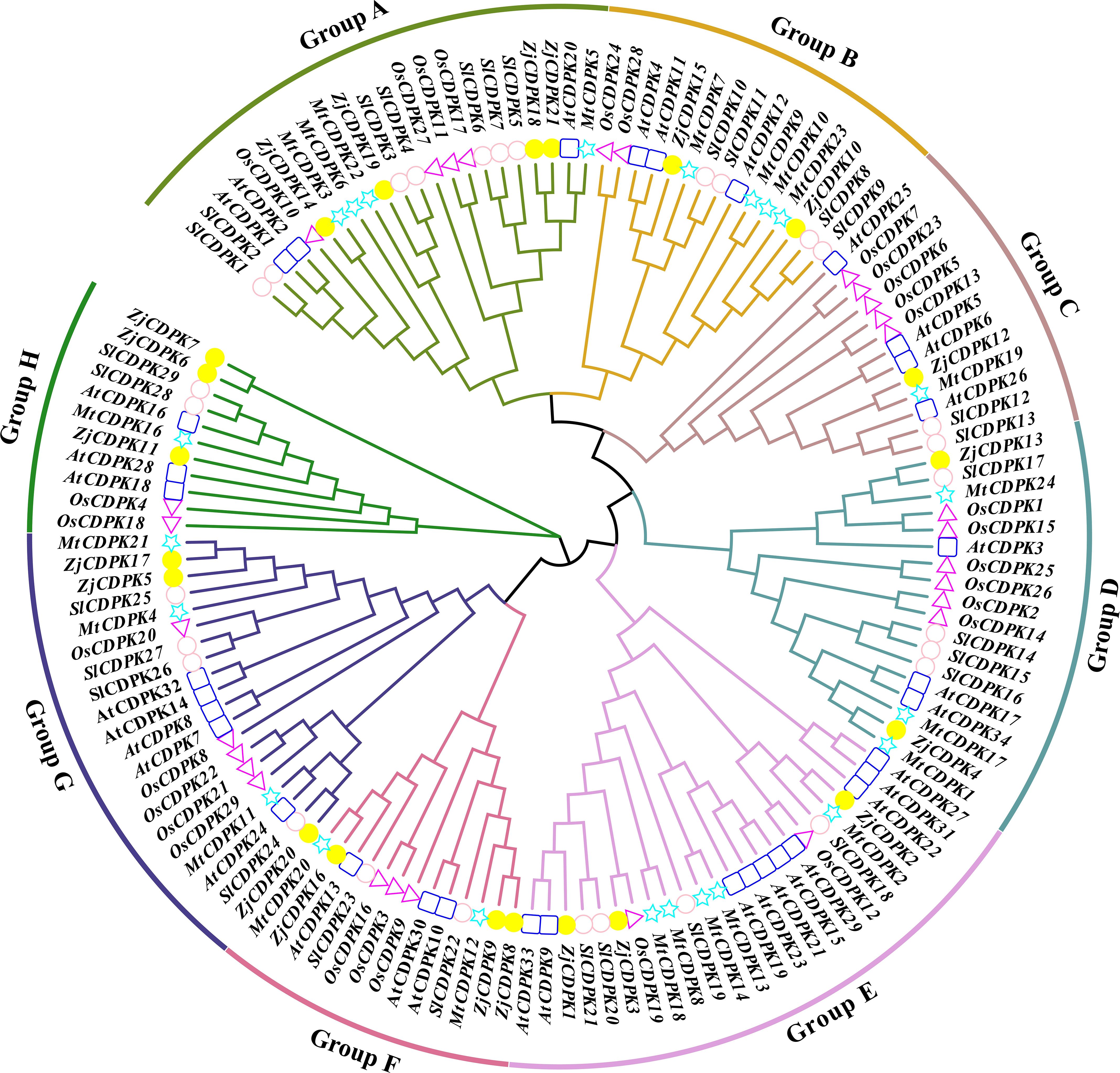
Figure 2. Phylogenetic tree analysis of the CDPKs in jujube, Arabidopsis, rice, tomato, and alfalfa, represented by yellow circles, blue squares, purple triangles, light purple circles, and green triangles, respectively. Different subgroups are represented by different line colors.
3.4 Colinear analysis of ZjCDPKs
The tandem and segmental duplication functions are important in gene evolution analysis. Here, through collinearity analysis in the jujube genome, two pairs of ZjCDPKs (ZjCDPK7/9 and ZjCDPK14/15) exhibited tandem duplication (Figure 3). In addition, one pair of collinearity genes (ZjCDPK6/7) was identified, indicating that segmental duplication occurred on the same chromosome.

Figure 3. Colinear analysis of the ZjCDPKs. The outermost circle represents 12 chromosomes. The gray and colored connecting genes represent collinear blocks and segmental duplication events. The color lines in the middle and inner layers of the circle represent the gene density of the chromosome.
To explore the evolutionary relationship of the CDPKs in different species, collinearity analysis of the CDPKs from jujube, Arabidopsis, and Solanum lycopersicum was performed. The collinearity plot identified 12 pairs of homozygous genes between jujube and Arabidopsis, while two ZjCDPK genes (ZjCDPK1 and ZjCDPK11) simultaneously formed homozygous gene pairs with three Arabidopsis CDPKs (Figure 4). In addition, a total of 14 homologous gene pairs were detected in jujube and tomato, and ZjCDPK1 and ZjCDPK4 also formed homologous gene pairs with three tomato CDPKs. This suggests that these genes may play an important role in the phylogeny of the CDPKs.
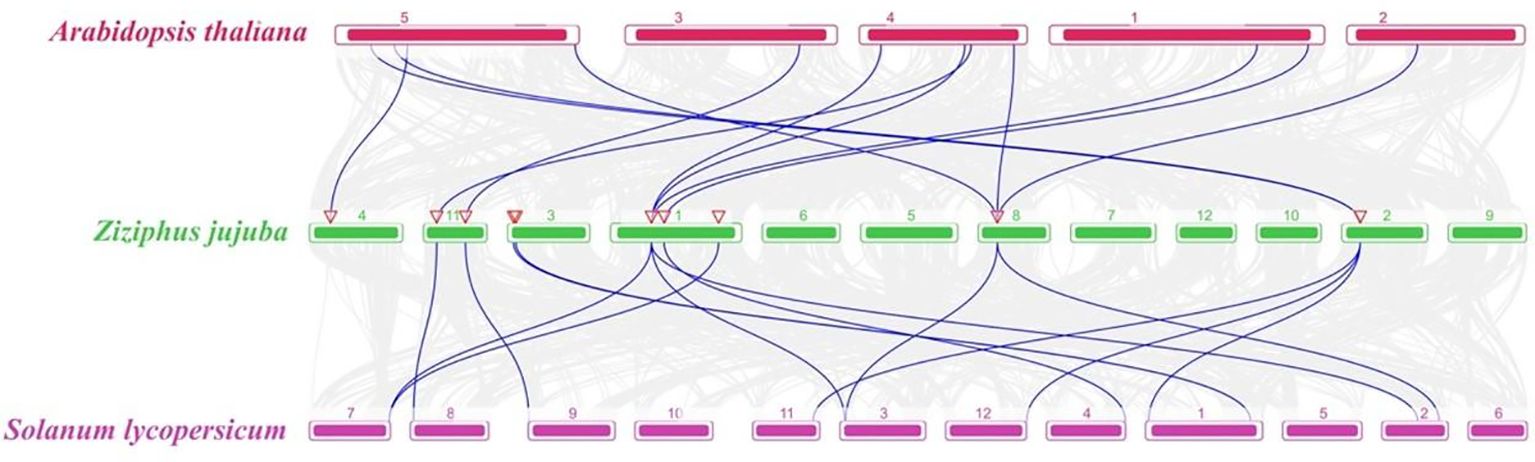
Figure 4. Colinear analysis of the CDPKs from jujube, Arabidopsis, and Solanum lycopersicum. The gray line represents the collinear blocks among jujube, Arabidopsis, and Solanum lycopersicum genomes, while the blue line represents the collinearity of CDPK gene pairs among these three species.
3.5 Cis-acting element analysis of ZjCDPKs
To investigate the possible transcriptional regulation network of the ZjCDPKs, TBtools was used to extract the 2 kb sequences upstream of the ZjCDPKs. These sequences were uploaded to PlantCARE to identify the cis-acting elements (Supplementary Table 3). As shown in Figure 5, four types of cis-acting elements were identified in the promoters of ZjCDPKs, including hormone, stress, development, and light response elements. Hormone-related cis-acting elements mainly included the abscisic acid response element (ABRE), the auxin response element (TGA element), the gibberellin response element (GARE motif/P-box/TATC box), the methyl jasmonate response element (CGTCA motif/TGACG motif), and the salicylic acid response element. The methyl jasmonate response element was most abundant in the ZjCDPKs promoters (21 occurrences), followed by the salicylic acid response element (10 occurrences). The auxin response element was the least abundant (4 occurrences), and was identified in the ZjCDPK3, ZjCDPK10, ZjCDPK15, and ZjCDPK18 promoter regions, respectively.
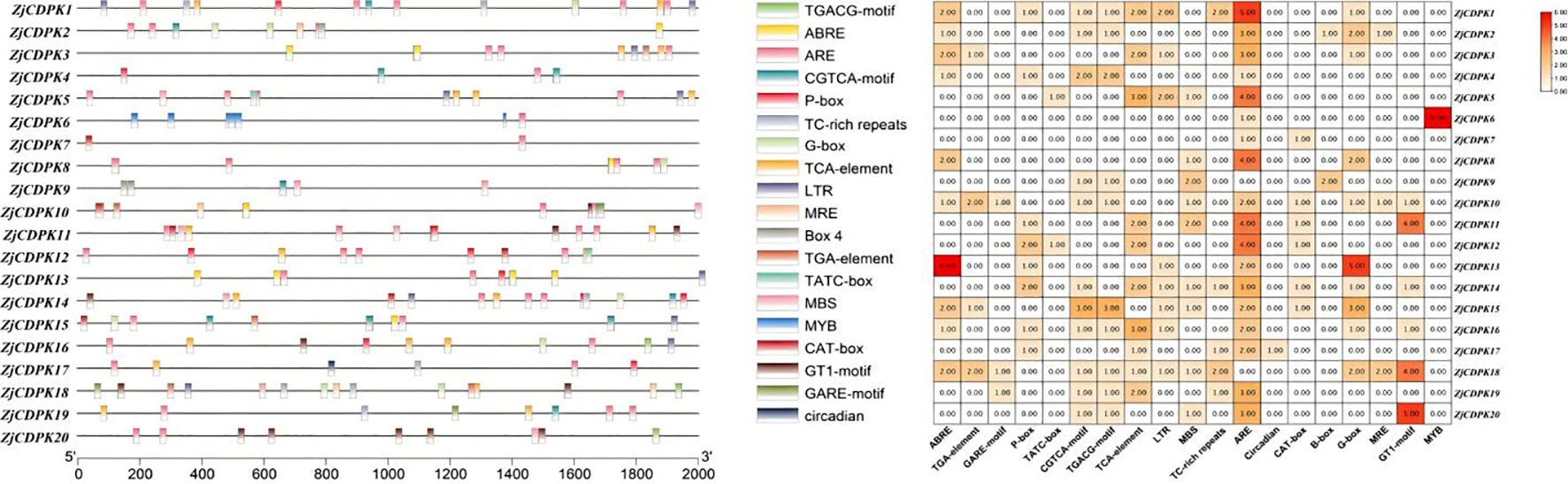
Figure 5. Identification of the cis-acting elements of the ZjCDPK promoters. The different types of cis-acting elements in the promoter region of the ZjCDPKs are represented by different colors. The different colors in the grid represent cis-acting elements.
We identified three stress-related cis-acting elements, namely, MBS, TC rich repeats, and LTR. Among them, MBS was observed the most (9 times), followed by LTR (8 times) and TC rich repeats (5 times).
Developmental response cis-acting elements, including circadian, CAT box, and ARE, and photoreactive elements such as G-BOX, MRE, and GT1 motifs, were also identified. All ZjCDPK promoters contained ARE, except for ZjCDPK9 and ZjCDPK18. ZjCDPK18 promoter contained a relatively large number of cis-acting elements, suggesting that it may participate in more biological and stress-related processes than other ZjCDPK members.
3.6 Expression profiles of the ZjCDPKs during fruit growth
Plant CDPKs play important roles in plant growth and development. Thus, to understand how ZjCDPKs are involved in fruit growth development, the expression levels of the ZjCDPKs in fruit nine growth stages (F1–F9) of ‘Jinkuiwang’ (large fruit size) and ‘Jinsixiaozao’ (small fruit size) were analyzed. The different expression patterns of 17 ZjCDPKs were identified (Figure 6). The majority of ZjCDPKs (e.g., ZjCDPK1/2/4/18) showed a down-regulation pattern from F1 to F9 in both cultivars, while other ZjCDPKs (e.g., ZjCDPK 3/5/14/15) did not exhibit any significant expressing patterns and had high expression levels. Most of the ZjCDPKs showed higher expression levels from F1 to F4 and were subsequently down-regulated in the other stages. The expression levels of ZjCDPK11/14/17/19 increased from F1 to F2 in ‘Jinkuiwang’, but decreased in ‘Jinsixiaozao’. This may indicate the important functions of these ZjCDPKs in the different fruit size formation between ‘Jinkkuiwang’ and ‘Jinsixiaozao’.
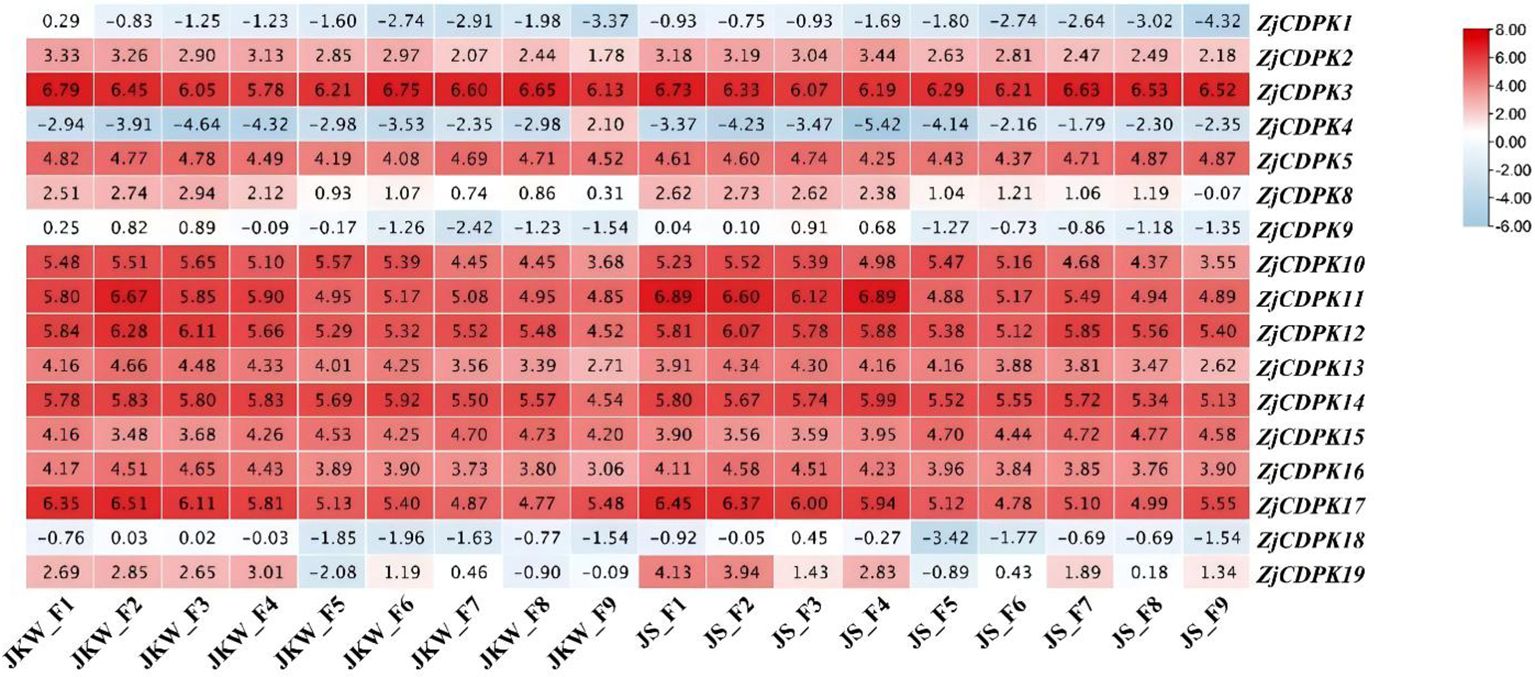
Figure 6. Expression profiles of ZjCDPKs during the fruit growth of ‘Jinkuiwang’ (JWK) and ‘Jinsixiaozao’ (JS). F1 to F9 are defined in Section 2.6.
3.7 Expression profiles of ZjCDPKs in response to phytoplasma infection
CDPKs are involved in the regulating mechanism of biotic stress. Thus, we analyzed the expression profiles of the ZjCDPKs in response to phytoplasma infection on the susceptible cultivar ‘Pozao’ and resistant cultivar ‘T13’. As shown in Figure 7, compared to the healthy control at stage S1, most of the ZjCDPKs exhibited down-regulation in PZ and only ZjCDPK8/9/11/17/19 was upregulated, while ZjCDPK3/8/9/11/12/14/15/17/19 showed an upregulating pattern in ‘T13’. As phytoplasma infection growth progressed (S2 to S3) in ‘PZ’, more ZjCDPKs were upregulated (e.g., ZjCDPK1/2/3/5/8/9/10/11/12/13/14/16/17/19/21) compared to the healthy control. Moreover, most of the ZjCDPK expression levels in ‘T13’ did not exhibit any significant changes at S2 and were significantly down-regulated in S3. These results may demonstrate the varying expression patterns of the ZjCDPKs involved in the different resistant levels to phytoplasma infection between ‘Pozao’ and ‘T13’.

Figure 7. Expression profiles of ZjCDPKs in response to phytoplasma infection in ‘Pozao’ (PZ) and ‘T13’ from the S1 to S3 growth stages. H, healthy; D, diseased.
3.8 Expression profiles of ZjCDPKs in response to salt stress
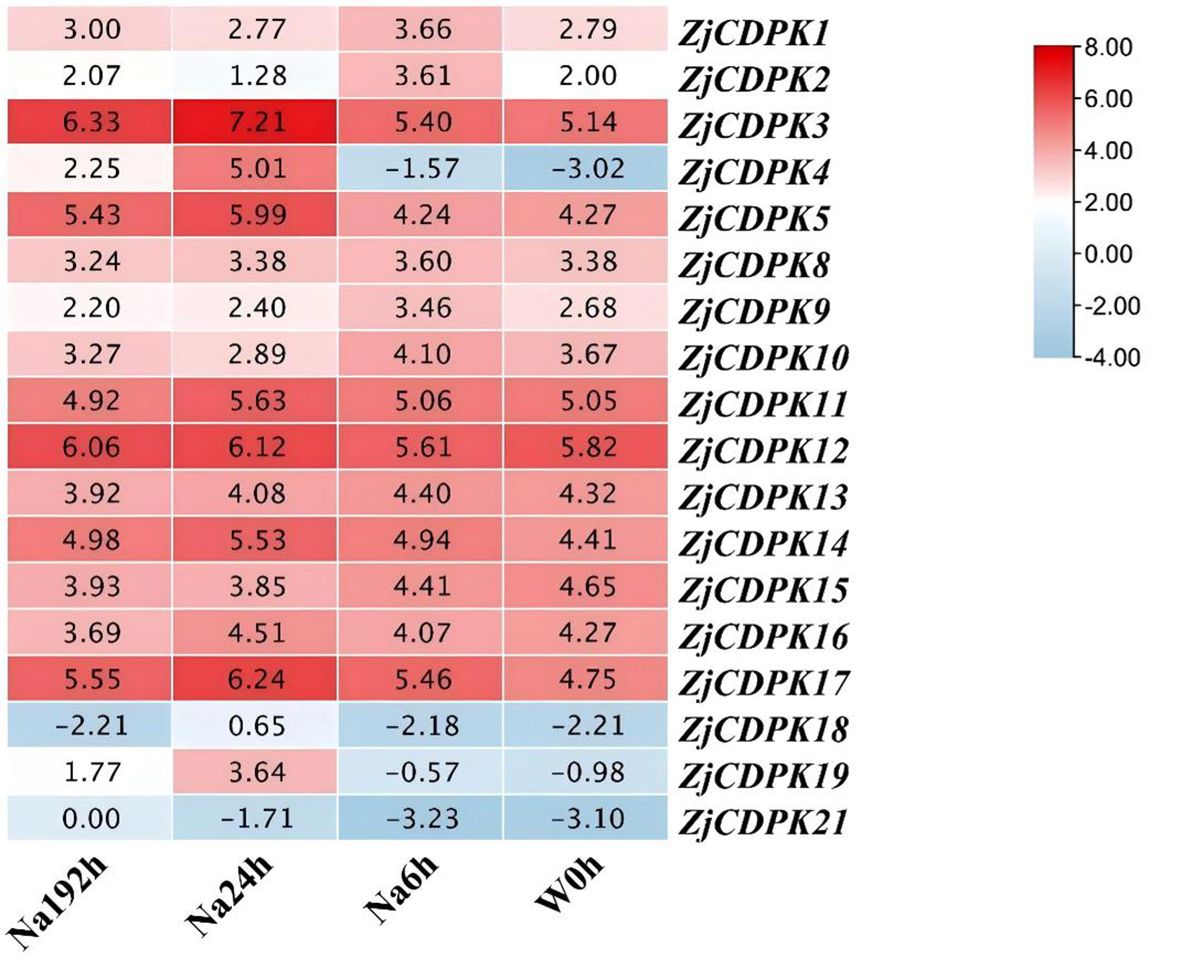
CDPKs also play important roles in response to abiotic stress. Thus, the expression levels of ZjCDPKs in response to salt stress were investigated. In response to salt stress, most of the ZjCDPKs were upregulated from 6 h to 24 h, while others initially exhibited a down-regulation pattern and were then regulated at 192 h (Figure 8). Among them, ZjCDPK15 maintained a down-regulation trend from 0 h to 192 h, while the expression levels of ZjCDPK3/5/11/12/17 increased from 0 h to 192 h and stayed high at 192 h, indicating that these ZjCDPKs are positively involved in salt stress.
3.9 Expression profiles of ZjCDPKs in response to cold stress
ZjCDPKs may have important functions in salt stress, and thus the expression profiles of ZjCDPKs in response to cold stress were further analyzed. As shown in Figure 9, the expression pattern of ZjCDPKs exhibited down- and upregulation patterns in ‘Dongzao’ and its autotetraploid. Among them, most of the ZjCDPKs, (e.g., ZjCDPK1/8/9/10/13/14/15/16) exhibited a down-regulation trend, while the expression levels of ZjCDPK3/11/12/17 were upregulated from 0 h to 24 h. In addition, the expression level of ZjCDPK19 was significantly induced in the ‘Dongzao’ autotetraploid compared to ‘Dongzao’, while the expression level of ZjCDPK16 was significantly upregulated in ‘Dongzao’ and down-regulated in its autotetraploid. This may indicate the important gene functions of this gene in cold differential resistance between ‘Dongzao’ and its autotetraploid.
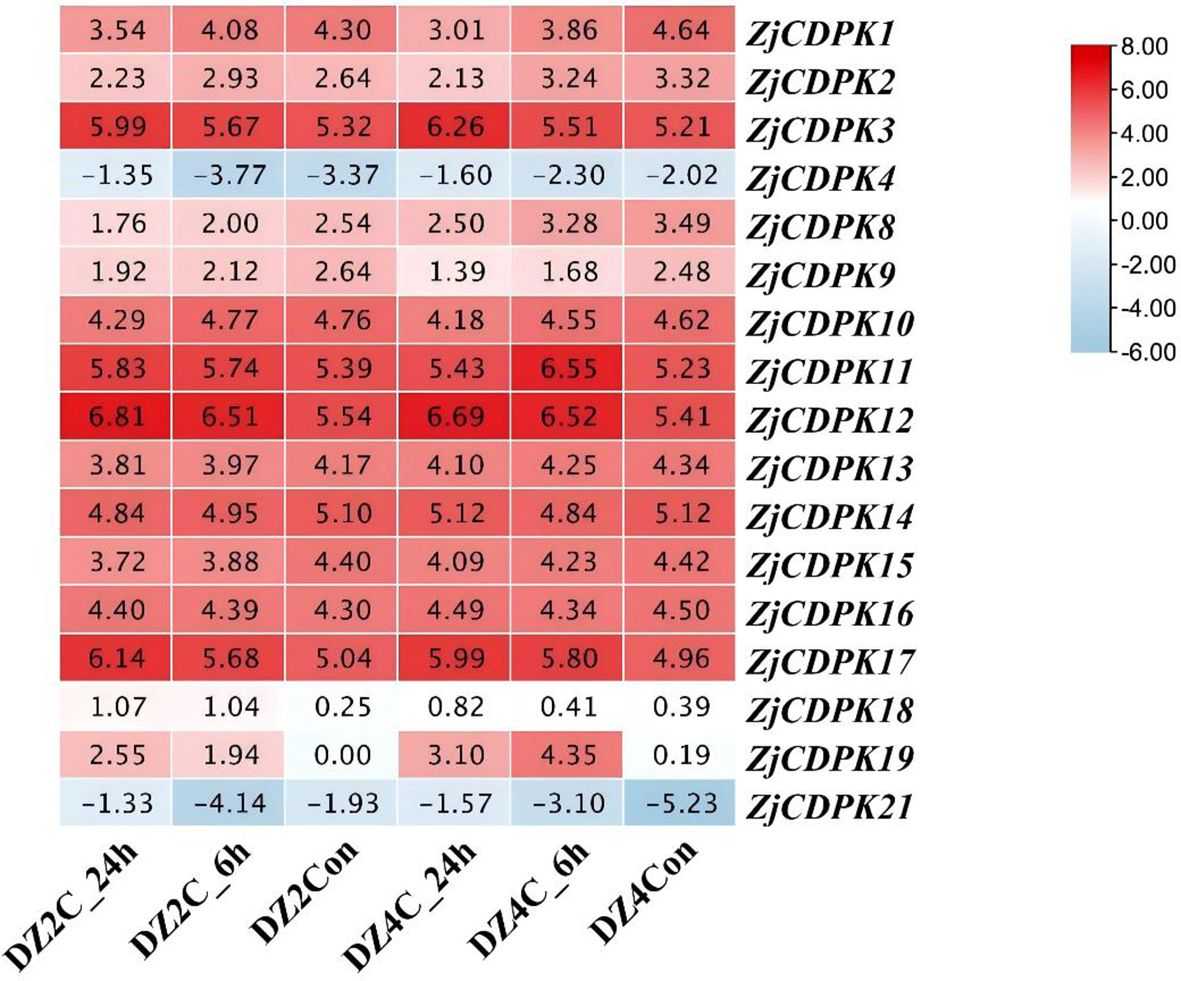
Figure 9. Expression profiles of ZjCDPKs in response to cold stress for ‘Dongzao’ and its autotetraploid at 0, 6, and 24 h respectively.
3.10 Prediction of ZjCDPK-related TFs and their expression analysis in response to cold and salt stress
To explore the regulatory network of ZjCDPKs under cold or salt stress, the TFs that could modulate the expression pattern of ZjCDPKs were predicted. As shown in Figure 10A, ZjCDPK4/6/7/8/10/14/16 interacted with six, one, one, two, two, one, and three TFs, respectively. The TFs mainly belonged to the ERF, DOF, MADS, and BBM families. Moreover, the expression profiles of these TFs in response to cold and salt stress (Figures 10B, C) were analyzed. In response to cold stress, the expression patterns of most TFs showed a down-regulation pattern. However, TFs such as LOC107405089 of ZjCDPK10 and LOC101706029 of ZjCDPK16 were significantly upregulated, indicating that these two TFs may play important regulatory roles in cold stress. In addition, under salt stress, the expression patterns of most related TFs showed opposite expressing patterns to that under cold stress. Most TFs were upregulated, while LOC107408788 and LOC107409268 of ZjCDPK4 were significantly inhibited under salt stress.
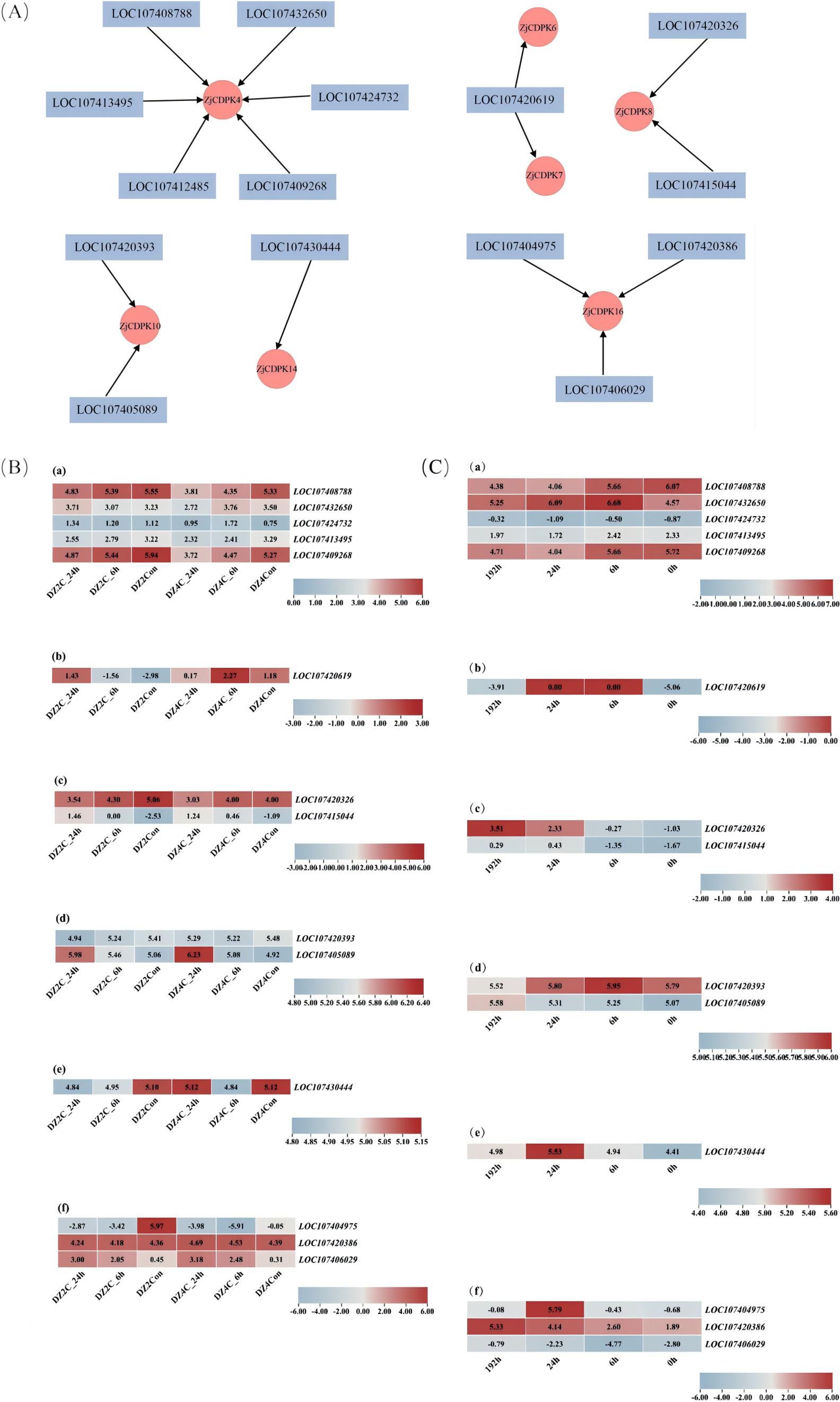
Figure 10. Prediction analysis of ZjCDPK-related transcription factors (A) and their expression patterns in response to cold (B) and salt stress (C).
4 Discussion
CDPKs, as serine/threonine type protein kinases widely found in plants, which function as calcium sensor and responder. Plant CDPKs and their mediated signaling cascades regulate plant growth and development, participate in hormone signal transduction, and play a role in abiotic stress response. The CDPKs family has undergone a long evolutionary process, which can be traced back to the earliest terrestrial plants, such as pteridophytes and bryophytes (Dekomah et al., 2022a). In the current study, 21 CDPKs were identified in the jujube genome using bioinformatics, which is fewer than the number identified in other species such as Arabidopsis, poplar, banana, and tomato. The number of gene families may be related to the extensive genomic diversity and tandem and segment duplications that took place in the history of plant evolution (Shi and Zhu, 2022). Thus, the smaller number of ZjCDPKs may be attributed to the small number of segment and tandem gene duplications that occurred in the jujube genome.
CDPK has four typical conserved domains, and the conserved kinase domain is a typical feature of Ser/Thr protein kinase. Calcium sensors mainly rely on the binding of Ca2+ to the EF-hand motif, which is a unique and conserved helical-ring-helical structure (Dekomah et al., 2022a). Our study showed that ZjCDPKs contain two highly conserved domains such as kinase and EF-hand domains. Among them, ZjCDPKs have 2–4 conserved EF-hand motifs, which is consistent with those in Arabidopsis (Yip Delormel and Boudsocq, 2019). In addition, gene structure analysis showed that the ZjCDPKs have multiple introns. The existence of more introns may increase the functional diversity of ZjCDPKs through alternative splicing and exon shuffling (Zhu et al., 2016). Moreover, cis-acting element analysis revealed that there was numerous hormone, developmental, and stress response elements in the promoters of ZjCDPKs. We also found that the methyl jasmonate hormone response element appeared frequently in the ZjCDPKs. Methyl jasmonate has many physiological functions, and CDPKs that were associated with plant hormones were also involved in the plant defense and development process. For example, AtCDPK32 can bind and phosphorylate the ABA response transcription factor ABF4, while overexpressing AtCDPK32 exhibits ABA sensitivity phenotype (Choi et al., 2005). Furthermore, exogenous ABA treatment increases the expression of BrrCDPK38/42 and FaCDPK4/11 in brassica and strawberry, respectively (Wang et al., 2017; Crizel et al., 2020). However, studies on the relationship between methyl jasmonate and CDPKs are limited, and methyl jasmonate may play an important role in the CDPK-mediated biological process of plant development and defense against various stresses.
Although calcium functions are important in plant growth and development, the function of calcium sensors such as CDPKs during fruit development has rarely been reported. In our study, we found that most of the ZjCDPKs in ‘Jinkuiwang’ and ‘Jinsixiaozao’ showed higher expressing levels during stages F1 to F4 and were then down-regulated during the subsequent stages. Among them, the expression levels of ZjCDPK11/14/17/19 increased from F1 to F2 in ‘Jinkuiwang’, but decreased in ‘Jinsixiaozao’. ‘Jinkuiwang’ and ‘Jinsixiaozao’ are big- and small-sized fruit cultivars of jujube, respectively. During fruit development, stages F1 to F3 belong to the rapid growth period, and stages F4 to F9 belong to the slow and pre-mature growth period (Zhao et al., 2022). Thus, the different expression levels of ZjCDPK11/14/17/19 between ‘Jinkuiwang’ and ‘Jinsixiaozao’ at F1 to F2 may determine the fruit size differences. However, further functional verification experiments should be conducted to confirm this hypothesis.
Calcium signaling plays a key role in response to biotic and abiotic stresses. For example, Ca2+-permeable channels can be regulated by phytoplasma infection, which can further affect Ca2+ signaling in sieve elements (Musetti et al., 2013). In our study, we found that more ZjCDPKs were induced with the severe jujube witches’ broom (‘Zaofeng’) disease symptoms in ‘Pozao’ at S3 after phytoplasma infection (Wang et al., 2022). In contrast, when the witches’ broom symptoms recovered in ‘T13’ at S2, the expression levels of most ZjCDPKs were maintained constant compared with healthy plants. These results may indicate that the early calcium signal compared with the most expression of ZjCDPKs conferred the phytoplasma resistance in ‘T13’. Moreover, CDPKs play an important role in cold stress response. For example, PbCDPK2, PbCDPK7, PbCDPK10, and PbCDPK13 have been associated with post-harvest low-temperature stress in peach. In particular, PbCDPK7 can interact with PbRBOH4 on the cell membrane, which can induce Ca2+-ROS signaling and maintain intracellular ROS homeostasis to reduce chilling injury in peach (Zhao et al., 2022). In rice, OsCDPK13 (Komatsu et al., 2007), OsCDPK17 (Almadanim et al., 2017), and OsCDPK24 (Liu et al., 2018) participate in cold stress responses, while AtCDPK28 in Arabidopsis (Ding et al., 2022) functions as a positive regulatory factor for cold resistance. In our study, we found that the expression level of ZjCDPK16 was significantly upregulated in ‘Dongzao’ and down-regulated in its autotetraploid. Previous research reported that ‘Dongzao’ is more cold-tolerant than its autotetraploid (Gao et al., 2021). Therefore, ZjCDPK16 may have an important function in cold differential resistance between ‘Dongzao’ and its autotetraploid. Moreover, CDPKs play a role in salt stress resistance. In Arabidopsis, the overexpression of AtCDPK6 can increase the accumulation of proline and reduce the content of MDA, thereby improving salt stress resistance (Xu et al., 2010). In rice, OsCDPK7 and OsCDPK12 play important roles in response to salt stress (Saijo et al., 2001; Asano et al., 2012). VpCDPK9 is involved in salt stress regulation in grapevine (Zhang et al., 2015). Here, we found that the expression level of ZjCDPK3/5/11/12/17 increased from 0 h to 192 h and maintained a high level at 192 h in response to salt stress. The transcription factor analysis revealed that most TFs belonging to the MADS and ERF families were significantly upregulated in response to salt stress, demonstrating that TFs could regulate related ZjCDPKs to facilitate salt tolerance in sour jujube. The systematic analysis of ZjCDPKs can provide important information for further functional analysis of the CDPKs in jujube during fruit development and in response to biotic and abiotic stresses. However, the relationship between TFs and ZjCDPKs, and their biological functions should be solidified by molecular experiments in the future.
5 Conclusion
In this study, a total of 21 ZjCDPKs were identified, which are located on eight chromosomes. Gene structure and conserved motif analysis showed that all ZjCDPKs have similar gene structures and conserved motifs, except for ZjCDPK9 and ZjCDPK21. All the CDPKs from the Arabidopsis, rice, tomato, alfalfa, and jujube were divided into eight subgroups and the members of the ZjCDPKs were unevenly distributed across these subgroups. Colinear analysis showed that 12 homozygous CDPKs were detected between jujube and Arabidopsis, and 14 pairs were found between jujube and tomato. Additionally, four types of cis-acting elements were identified in the promoters of ZjCDPKs, including hormone, stress, development and light response elements were identified. The expression profiles of ZjCDPKs in response to phytoplasma infection and cold and salt stresses, during different fruit growth stages indicated that most ZjCDPKs were up or down-regulated. Finally, a varying number of TFs could interact with the promoter regions of ZjCDPK4/6/7/8/10/14/16 and exhibited opposite expression patterns in response to cold and salt stress.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: [https://www.ncbi.nlm.nih.gov/, accession number PRJNA798569].
Author contributions
LL: Formal analysis, Visualization, Data curation, Writing – original draft, Writing – review & editing. YY: Writing – original draft, Software, Data curation, Visualization, Formal analysis. BS: Software, Formal analysis, Resources, Writing – original draft, Data curation. CC: Validation, Project administration, Writing – original draft, Methodology, Supervision. LY: Validation, Resources, Writing – original draft, Project administration, Visualization. JJ: Formal analysis, Data curation, Methodology, Writing – original draft, Supervision. DF: Visualization, Resources, Funding acquisition, Formal analysis, Writing – review & editing. QH: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Science and Technology Innovation Team Project (2022TSYCTD0010).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1593446/full#supplementary-material
Supplementary Data Sheet 1 | Amino acid sequences of ZjCDPKs
Supplementary Data Sheet 2 | 10 motifs were identified from ZjCDPKs
Supplementary Data Sheet 3 | The cis-acting elements identification of ZjCIPKs
References
Aleynova-Shumakova, O., Dubrovina, A., Manyakhin, A., Karetin, Y., and Kiselev, K. (2014). VaCPK20 gene overexpression significantly increased resveratrol content and expression of stilbene synthase genes in cell cultures of Vitis amurensis Rupr. Appl. Microbiol. Biotechnol. 98, 5541–5549. doi: 10.1007/s00253-014-5625-7
Almadanim, M. C., Alexandre, B. M., Rosa, M. T., Sapeta, H., Leitão, A. E., Ramalho, J. C., et al. (2017). Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell environment. 40, 1197–1213. doi: 10.1111/pce.12916
Asano, T., Hayashi, N., Kobayashi, M., Aoki, N., Miyao, A., Mitsuhara, I., et al. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69, 26–36. doi: 10.1111/j.1365-313X.2011.04766.x
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Cao, Y. P., Feng, X. F., Ding, B. P., Huo, H. Q., Abdullah, M., Hong, J. Y., et al. (2025). Gap-free genome assemblies of two Pyrus bretschneideri cultivars and GWAS analyses identify a CCCH zinc finger protein as a key regulator of stone cell formation in pear fruit. Plant Communications. 6, 101238. doi: 10.1016/j.xplc.2024.101238
Cao, Y. P., Hong, J. Y., Zhao, Y., Li, X. X., Feng, X. F., Wang, H., et al. (2024a). De novo gene integration into regulatory networks via interaction with conserved genes in peach. Horticulture Res. 11, uhae252. doi: 10.1093/hr/uhae252
Cao, Y. P., Mo, W. H., Li, Y. L., Xiong, Y., Wang, H., Zhang, Y. J., et al. (2024b). Functional characterization of NBS−LRR genes reveals an NBS−LRR gene that mediates resistance against Fusarium wilt. BMC Biol. 22, 45. doi: 10.1186/s12915-024-01836-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, S. H., Sheen, J., Gerrish, C., and Bolwell, G. P. (2001). Molecular identification of phenylalanine ammonia-lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS letters. 503, 185–188. doi: 10.1016/s0014-5793(01)02732-6
Choi, H. I., Park, H. J., Park, J. H., Kim, S., Im, M. Y., Seo, H. H., et al. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139, 1750–1761. doi: 10.1104/pp.105.069757
Crizel, R. L., Perin, E. C., Vighi, I. L., Woloski, R., Seixas, A., da Silva Pinto, L., et al. (2020). Genome-wide identification, and characterization of the CDPK gene family reveal their involvement in abiotic stress response in Fragaria x ananassa. Sci. reports. 10, 11040. doi: 10.1038/s41598-020-67957-9
Dekomah, S. D., Bi, Z., Dormatey, R., Wang, Y., Haider, F. U., Sun, C., et al. (2022a). The role of CDPKs in plant development, nutrient and stress signaling. Front. Genet. 13. doi: 10.3389/fgene.2022.996203
Dekomah, S. D., Wang, Y., Qin, T., Xu, D., Sun, C., Yao, P., et al. (2022b). Identification and expression analysis of calcium-dependent protein kinases gene family in potato under drought stress. Front. Genet. 13. doi: 10.3389/fgene.2022.874397
Ding, Y., Yang, H., Wu, S., Fu, D., Li, M., Gong, Z., et al. (2022). CPK28-NLP7 module integrates cold-induced Ca2+ signal and transcriptional reprogramming in Arabidopsis. Sci. Adv. 8, ebn7901. doi: 10.1126/sciadv.abn7901
Gao, M., Wang, L., Li, M., Sun, P., Sadeghnezhad, E., Shi, H., et al. (2021). Physiological and transcriptome analysis accentuates microtubules and calcium signaling in Ziziphus jujuba Mill ‘Dongzao’autotetraploids with sensitive cold tolerance. Scientia Horticulturae. 285, 110183. doi: 10.1016/j.scienta.2021.110183
Hamel, L. P., Sheen, J., and Séguin, A. (2014). Ancient signals: comparative genomics of green plant CDPKs. Trends Plant science. 19, 79–89. doi: 10.1016/j.tplants.2013.10.009
Higo, K., Ugawa, Y., Iwamoto, M., and Higo, H. (1998). PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 26, 358–359. doi: 10.1093/nar/26.1.358
Hrabak, E. M., Chan, C. W., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Jiang, L., Li, X. X., Lyu, K., Wang, H., Li, Z. Y., Qi, W., et al. (2025). Rosaceae phylogenomic studies provide insights into the evolution of new genes. Hortic. Plant J. 11, 389–405. doi: 10.1016/j.hpj.2024.02.002
Komatsu, S., Yang, G., Khan, M., Onodera, H., Toki, S., and Yamaguchi, M. (2007). Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277, 713–723. doi: 10.1007/s00438-007-0220-6
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14, 1–15. doi: 10.1186/1471-2164-14-433
Li, Y., Zhang, H., Liang, S., Chen, X., Liu, J., Zhang, Y., et al. (2022). Identification of CDPK gene family in solanum habrochaites and its function analysis under stress. Int. J. Mol. Sci. 23, 4227. doi: 10.3390/ijms23084227
Liese, A. and Romeis, T. (2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta (BBA)-Molecular Cell Res. 1833, 1582–1589. doi: 10.1016/j.bbamcr.2012.10.024
Linghu, B., Xu, Z., Chu, Y., Yan, Y., Nie, X., and Weining, S. (2023). Genome-wide analysis of calcium-dependent protein kinase (CDPK) family and functional characterization of TaCDPK25-U in response to drought stress in wheat. Environ. Exp. Botany. 209, 105277. doi: 10.1016/j.envexpbot.2023.105277
Liu, S., Li, J., Li, N., Zhou, P., and Li, L. (2022). Genome-wide identification of calcium-dependent protein kinases (CDPKs) in pear (Pyrus bretschneideri Rehd) and characterization of their responses to Venturia nashicola infection. Horticulture Environment Biotechnol. 63, 903–915. doi: 10.1007/s13580-022-00444-4
Liu, M., Wang, J., Wang, L., Liu, P., Zhao, J., Zhao, Z., et al. (2020). The historical and current research progress on jujube-a superfruit for the future. Horticulture Res. 7, 119. doi: 10.1038/s41438-020-00346-5
Liu, Y., Xu, C., Zhu, Y., Zhang, L., Chen, T., Zhou, F., et al. (2018). The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 60, 173–188. doi: 10.1111/jipb.12614
Ma, X., Li, Q.-H., Yu, Y.-N., Qiao, Y.-M., Haq, S. U., and Gong, Z.-H. (2020). The CBL–CIPK pathway in plant response to stress signals. Int. J. Mol. Sci. 21, 5668. doi: 10.3390/ijms21165668
Musetti, R., Buxa, S. V., De Marco, F., Loschi, A., Polizzotto, R., Kogel, K.-H., et al. (2013). Phytoplasma-triggered Ca2+ influx is involved in sieve-tube blockage. Mol. Plant-Microbe Interactions. 26, 379–386. doi: 10.1094/MPMI-08-12-0207-R
Qi, C. F., Wang, Q. F., Niu, Y. H., Zhang, Y., Liu, M. J., Liu, Z. G., et al. (2024). Characteristics of ZjCIPKs and ZjbHLH74-ZjCIPK5 regulated cold tolerance in jujube. Int. J. Biol. Macromolecules. 264, 130429. doi: 10.1016/j.ijbiomac.2024.130429
Saijo, Y., Kinoshita, N., Ishiyama, K., Hata, S., Kyozuka, J., Hayakawa, T., et al. (2001). A Ca2+-dependent protein kinase that endows rice plants with cold-and salt-stress tolerance functions in vascular bundles. Plant Cell Physiol. 42, 1228–1233. doi: 10.1093/pcp/pce158
Shi, G. and Zhu, X. (2022). Genome-wide identification and functional characterization of CDPK gene family reveal their involvement in response to drought stress in Gossypium barbadense. PeerJ. 10, e12883. doi: 10.7717/peerj.12883
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., and Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Tuteja, N. and Mahajan, S. (2007). Calcium signaling network in plants: an overview. Plant Signal Behav. 2, 79–85. doi: 10.4161/psb.2.2.4176
Wang, L., Li, M., Liu, Z., Dai, L., Zhang, M., Wang, L., et al. (2020a). Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genomics 21, 1–16. doi: 10.1186/s12864-020-6601-5
Wang, L., Liu, S., Gao, M., Wang, L., Wang, L., Wang, Y., et al. (2022). The crosstalk of the salicylic acid and jasmonic acid signaling pathways contributed to different resistance to phytoplasma infection between the two genotypes in Chinese jujube. Front. Microbiology. 13. doi: 10.3389/fmicb.2022.800762
Wang, L., Liu, Z., Han, S., Liu, P., Sadeghnezhad, E., and Liu, M. (2023b). Growth or survival: What is the role of calmodulin-like proteins in plant? Int. J. Biol. Macromolecules 242, 124733. doi: 10.1016/j.ijbiomac.2023.124733
Wang, Q., Qi, C., Wang, L., Li, M., Niu, Y., Muhammad, N., et al. (2025). ZjMAPKK4 interacted with ZjNAC78 regulates cold tolerance response in jujube. Plant Cell Environ. 48, 3691–3707. doi: 10.1111/pce.15381
Wang, L., Sadeghnezhad, E., and Nick, P. (2020b). Upstream of gene expression: what is the role of microtubules in cold signalling? J. Exp. Bot. 71, 36–48. doi: 10.1093/jxb/erz419
Wang, L., Wang, L. X., Zhang, M., Qu, Y., Yuan, Y., Ehsan, S., et al. (2023a). A cyclic effect of cAMP and calcium signaling contributes to jujube growth and development. J. Integr. Agriculture. 22, 2094–2110. doi: 10.1016/j.jia.2023.04.039
Wang, J.-P., Xu, Y.-P., Munyampundu, J.-P., Liu, T.-Y., and Cai, X.-Z. (2016). Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genomics 291, 661–676. doi: 10.1007/s00438-015-1137-0
Wang, Q., Yin, X., Chen, Q., Xiang, N., Sun, X., Yang, Y., et al. (2017). Genome-wide survey indicates diverse physiological roles of the turnip (Brassica rapa var. rapa) calcium-dependent protein kinase genes. Sci. Reports. 7, 15803. doi: 10.1038/s41598-017-16102-0
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xu, J., Tian, Y.-S., Peng, R.-H., Xiong, A.-S., Zhu, B., Jin, X.-F., et al. (2010). AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta. 231, 1251–1260. doi: 10.1007/s00425-010-1122-0
Ye, W., Muroyama, D., Munemasa, S., Nakamura, Y., Mori, I. C., and Murata, Y. (2013). Calcium-dependent protein kinase CPK6 positively functions in induction by yeast elicitor of stomatal closure and inhibition by yeast elicitor of light-induced stomatal opening in Arabidopsis. Plant Physiol. 163, 591–599. doi: 10.1104/pp.113.224055
Yip Delormel, T. and Boudsocq, M. (2019). Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytologist. 224, 585–604. doi: 10.1111/nph.16088
Zhang, K., Han, Y. T., Zhao, F. L., Hu, Y., Gao, Y. R., Ma, Y. F., et al. (2015). Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 15, 1–19. doi: 10.1186/s12870-015-0552-z
Zhao, Y., Du, H., Wang, Y., Wang, H., Yang, S., Li, C., et al. (2021). The calcium-dependent protein kinase ZmCDPK7 functions in heat-stress tolerance in maize. J. Integr. Plant Biol. 63, 510–527. doi: 10.1111/jipb.13056
Zhao, L., Xie, B., Hou, Y., Zhao, Y., Zheng, Y., and Jin, P. (2022). Genome-wide identification of the CDPK gene family reveals the CDPK-RBOH pathway potential involved in improving chilling tolerance in peach fruit. Plant Physiol. Biochem. 191, 10–19. doi: 10.1016/j.plaphy.2022.09.015
Zhao, X., Zhao, Z. X., Cheng, S. S., Wang, L. H., Luo, Z., Ai, C. F., et al. (2023). ZjWRKY23 and zjWRKY40 promote fruit size enlargement by targeting and downregulating cytokinin oxidase/dehydrogenase 5 expression in chinese jujube. J. Agric. Food Chem. 71, 18046–18058. doi: 10.1021/acs.jafc.3c04377
Zhu, W., Cao, H., Wang, Q., Niu, Y., Sadeghnezhad, E., Han, S., et al. (2023). Transcriptome analysis revealed MAPK and hormone pathway involving in exogenous melatonin-regulated salt tolerance in sour jujube. Fruit Res. 3, 19. doi: 10.48130/FruRes-2023-0019
Keywords: CDPKs, jujube, phylogenetic analysis, expression profiles, transcription factor
Citation: Li L, Yuan Y, Shen B, Chen C, Yang L, Jin J, Fan D and Hao Q (2025) Genome-wide identification of CDPKs in jujube and expression profile analysis in response to multiple biological processes. Front. Plant Sci. 16:1593446. doi: 10.3389/fpls.2025.1593446
Received: 14 March 2025; Accepted: 11 April 2025;
Published: 27 May 2025.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Noor Muhammad, Hebei Agricultural University, ChinaNisar Uddin, Hazara University, Pakistan
Copyright © 2025 Li, Yuan, Shen, Chen, Yang, Jin, Fan and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dingyu Fan, ZmFuZGluZ3l1MjQxM0AxNjMuY29t; Qing Hao, aGFvcWluZ3hqQHNvaHUuY29t
 Lili Li
Lili Li Ye Yuan
Ye Yuan Bingqi Shen
Bingqi Shen Chong Chen
Chong Chen Juan Jin
Juan Jin Dingyu Fan
Dingyu Fan Qing Hao
Qing Hao