- 1College of Life Sciences, Sichuan Agricultural University, Ya’an, China
- 2College of Modern Technology, Mianyang City College, Mianyang, China
Introduction: Tartary buckwheat (Fagopyrum tataricum Gaertn.), classified as a food and herbal medicinal crop, offers substantial nutritional benefits but suffers from poor yields and quality. Studies indicate that Serendipita indica positively impacts Tartary buckwheat's yield and quality, yet the underlying processes remain largely unexplored.
Methods: This study aimed to examine the genetic transcript of Tartary buckwheat in both colonized and uncolonized S. indica.
Results: It was discovered that the pathway for producing phenylpropanoids in Tartary buckwheat, both in colonized and uncolonized S. indica, both in colonized and uncolonized S. indica, was found to be enriched in KEGG (Kyoto Encyclopedia of Genes and Genomes). Genetic expression analysis of lignin and flavonoid biosynthesis pathways in colonized S. indica showed a comparison between lignin biosynthesis pathway genes in colonized S. indica and those in uncolonized S. indica in Tartary buckwheat. Research revealed a decrease in certain genes linked to lignin synthesis and an increase in others associated with flavonoid biosynthesis in both colonized and uncolonized S. indica Tartary buckwheat. Furthermore, research revealed a reduction in lignin levels in Tartary buckwheat stems and seeds both colonized and uncolonized by S. indica, in contrast to an increase in flavonoid levels in leaves and seeds of Tartary buckwheat colonized and uncolonized by the same fungi.
Discussion: Findings indicate that the process of synthesizing lignin and flavonoids could offer valuable insights into how S. indica enhances Tartary buckwheat's yield and quality.
1 Introduction
Tartary buckwheat is an annual herb of the Polygonaceae buckwheat. Tartary buckwheat, recognized as a cash crop, boasts significant nutritional content and resembles authentic grains (Giménez-Bastida and Zieliński, 2015). Its composition includes flavonoids (Kim et al., 2007; Kalinova and Vrchotova, 2009), starch (Giménez-Bastida and Zieliński, 2015), raw protein (Janovská et al., 2021) and trace elements (Aubert et al., 2021; Kasar et al., 2021) along with various other healthful compounds. Consequently, Tartary buckwheat has gained favor in the global market and is considered a strategic crop that can provide human nutrition (Chettry and Chrungoo, 2021). Nonetheless, the production and quality of Tartary buckwheat face distinct challenges. The cultivation of Tartary buckwheat often occurs in regions with elevated temperatures, arid conditions, and barren soils, which affect its growth and lead to reduced crop yields (Liu et al., 2025). Concurrently, the challenge of dehulling Tartary buckwheat in the industry lies in its complexity. While steam dehulling aids in this process, its drawbacks include being time-intensive, expensive, and diminishing the quality of Tartary buckwheat, which fails to meet the substantial market demand (Wang and Campbell, 2007). Consequently, enhancing the production and quality of Tartary buckwheat has become a significant challenge that must be addressed. Presently, certain endophytic fungi are known to enhance plant nutrient absorption and growth by improving nutrient absorption efficiency, acclimatizing the plant to its growing conditions and maintaining plant health, thereby fostering growth and increasing yield (Almario et al., 2017).
The endophytic fungus Serendipita indica, extracted from the soil of the Indian desert, establishes itself in plant roots (Verma et al., 1998; Varma et al., 1999). The fungus Serendipita indica is a member of the Sebacinaceae family, classified under the Basidiomycota division (Weiss et al., 2004; Li et al., 2023a). S. indica offers numerous advantages in facilitating nutrient absorption by plants, resisting diseases, enduring stress, and enhancing growth, and is commonly used in agricultural and ecological fields (Li et al., 2023a). It has been shown that S. indica promotes the growth of Cerasus humilis plants and enhances their antioxidant enzymes by increasing the indole acetic acid (IAA) content of the plants, which in turn improves the quality of Cerasus humilis (Yin et al., 2024). Extensive research has demonstrated that symbiotic colonization by S. indica enhances abiotic stress resilience in diverse plant species, including Hordeum vulgare L (Waller et al., 2005), Centella asiatica (Jisha et al., 2018), Passiflora edulis (Yan et al., 2021) and Tartary buckwheat (Zheng et al., 2023). This mutualistic association not only improves plant growth parameters but also elevates secondary metabolite production, while concurrently strengthening both abiotic stress toleranceand biotic stress resistance against phytopathogens. Tartary buckwheat roots can be colonized by S. indica in abiotic stress conditions, and via phytohormone metabolism in Tartary buckwheat, it enhances both its growth and that of the plant. The build-up of soluble sugars, proteins, flavonoids, and phenolics has been shown to increase Tartary buckwheat’s abiotic stress (Zheng et al., 2023). Nevertheless, it remains uncertain how S. indica enhances Tartary buckwheat production and bolsters its ability to withstand stress.
Here, we hypothesized that S. indica has a promoting effect on Tartary buckwheat yield and quality. To explore this possibility, we investigated the effect of S. indica on Tartary buckwheat in the absence of stress through a transcriptomic approach. This study deepens S. indica’s understanding of the regulatory mechanisms of lignin and flavonoid biosynthesis in Tartary buckwheat, and provides theoretical guidance for improving Tartary buckwheat yield and quality.
2 Materials and methods
2.1 Plant materials and fungi preparation
The seeds were immersed in water at a temperature of 42°C for 2 hours. Full-sized samples were sterilized using a 0.1% mercuric chloride solution for 10 minutes before use (Sasidharan and Jayachitra, 2017). S. indica was inoculated in solid complete medium (CM) and maintained at 28°C for two weeks. Additionally, S. indica, measuring 7.5 mm, was introduced into 200 mL of CM liquid medium and kept at 28°C and 120 rpm for 20 days. Subsequently, 2 g of mycelium were gathered and combined with 100 mL of sterilized double-distilled water to achieve a 2% (w/v) (± 1.5 × 107 spores/mL) S. indica suspension (Abdelaziz et al., 2019). The soils used for potting trials were obtained from Ya’an, China(29°58’N, 102°59’E), where one batch of soil was inoculated with S. indica liquid culture at 100 mL/kg named P group, while a separate batch received an identical volume of distilled water for control purposes named CK group. Each containing 8 replicate pots (12 cm diameter × 10cm height) with four Tartary buckwheat plants per pot. Cultivation occurred under natural photoperiod conditions (11/13 h light/dark cycle) with average daytime temperature maintained at 20 ± 5°C and watered every two days. Roots, stems, leaves and seeds of Tartary buckwheat were collected after the 40th day of cultivation.
2.2 Colonization of S. indica in roots and measurement of plant growth indices
After 15 days of co-cultivation, collection of roots of colonized and uncolonized Tartary buckwheat plants, soaked in 10% KOH for 30 min, then in 1% HCl for 20 min, then stained with 0.02% Trypan Blue solution for 2 h, and finally decolorized with 50% lactophenol solution for 2 h (Rai et al., 2001). The colonization of S. indica in Tartary buckwheat roots was observed with an Olympus fluorescence microscope, BX53 (Olympus, Shanghai, China). S. indica colonization in buckwheat roots was observed under an Olympus fluorescence microscope BX53 (Olympus, Shanghai, China). Next, colonized and uncolonized Tartary buckwheat plants (three samples per group) were selected and weighed using an MTQ300 electronic scale (Mellen, Shenzhen, China) for weighing the fresh weight of the above-ground and below-ground parts of Tartary buckwheat. The plant samples were then dried in an oven at 60°C until constant weight, and the dry weight of the plants was weighed using an MTQ300 electronic scale (Meilun, Shenzhen, China).
2.3 Determination of total lignin and flavonoid content
After about 40 days of co-cultivation, 4 plants were taken from each pot to determine the lignin and the flavonoid content. To determine the lignin content of S. indica colonized and uncolonized Tartary buckwheat, the samples were dried in an oven until constant weight. After passing through a 35-mesh sieve, 3 mg of the sample was weighed, and the lignin content of Tartary buckwheat was determined using a lignin content detection kit (Beijing Box Biotechnology Co., Ltd., Beijing, China) and an enzyme labeling instrument, Multiskan SkyHigh 500C (ThermoFisher Scientific, Shanghai, China). Each replicate was performed three times. To determine the flavonoid content of S. indica colonized and uncolonized Tartary buckwheat, the samples were dried in an oven at 60°C until constant weight. After passing through a 35-mesh sieve, 0.1 g of the sample was weighed and extracted by ultrasonic extraction at 300 w, 60°C for 30 min. The supernatant was collected by centrifugation at 12000 rpm, 25°C for 10 min. The flavonoid content of Tartary buckwheat was determined using a Plant Flavonoid Content Assay Kit (Micro method) (Beijing Solabao Technology Co., Ltd., Beijing, China) and an enzyme labeling instrument, Multiskan SkyHigh 500C (ThermoFisher Scientific, Shanghai, China). Each replicate was performed three times.
2.4 Lignin staining
The paraffin sections were routinely dewaxed and hydrated, then stained with Senna (1% prepared in ultrapure water) for 1 to 2 h; washed slightly under running water; and then stained with solid green (1% prepared in 95% ethanol) for 30–60 s. The sections were then stained with anhydrous ethanol I for 2 min, anhydrous ethanol II for 2 min and anhydrous ethanol II for 2 min. Then they were sequentially dehydrated by anhydrous ethanol I for 2 min and anhydrous ethanol II for 2 min, then by xylene I for 1 min and xylene II for 2 min, and finally sealed with neutral gum and examined microscopically (Brandizzi, 2000).
2.5 RNA isolation, library construction and sequencing
Roots of colonized and uncolonized Tartary buckwheat were collected and a total of 18 samples were collected. Three biological replicates were made for each treatment group. Total RNA was extracted from the plants, and the integrity and total amount of RNA were accurately detected using an Agilent 2100 bioanalyzer. After RNA extraction, purification and library construction, the libraries were subjected to paired-end (PE) sequencing using Next-Generation Sequencing (NGS) based on the Illumina sequencing platform. The transcriptome sequencing was performed by Suzhou PANOMIX Biomedical Tech Co. DEGs were identified based on a fold change (FC) >1 and a false discovery rate (FDR) value below 0.05.
2.6 Quantitative real-time PCR analysis
To validate the lignin-flavonoid-related pathway, key enzyme genes were selected and primers were designed for further analysis (Supplementary Table S1). Total RNA was extracted in fresh plant samples using a total RNA isolation kit (Vazyme, Nanjing, China). Total RNA (1 µg) was reverse-transcribed into first-strand cDNA using the HiScript III RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme, Nanjing, China).Genes were amplified using the ChamQ Universal SYBR qPCR Master Mix kit (Vazyme, Nanjing, China) using gene-specific primers according to the manufacturer’s instructions. QPCR amplification was performed under the following thermal cycling conditions: initial denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 10 s and annealing/extension at 60°C for 30 s. Gene expression was calculated by using the 2-ΔΔCT method. To normalize the gene transcript levels, the FtH3 gene was co-amplified as a reference gene. Three biological replicates were done for each sample.
2.7 Data analysis
The graphs in the paper were plotted and statistically calculated using origin 2021 software. Student’s t-test was performed using IBM SPSS Statistics 27 program to test the significance of the data obtained, a significance threshold of P < 0.05. Data are means and standard deviations of three independent biological replicates.
3 Results
3.1 Differences in phenotypes of Tartary buckwheat colonized by S. indica
Tartary buckwheat in colonized S. indica had a significant change from that of uncolonized S. indica, with a tendency for larger leaves and thicker rhizomes in colonized S. indica (Figure 1A; Supplementary Figures S1, S2). S. indica led to a significant increase in significant increase in aboveground part and root biomass of Tartary buckwheat, where the aboveground part and root fresh weight increased by 1.68 and 2.20 times, and the aboveground part and root dry weight increased by 1.91 and 2.21 times (Figure 1B). The results showed that there was a significant difference between Tartary buckwheat colonized with S. indica and uncolonized with S. indica, and S. indica significantly increased the biomass of Tartary buckwheat.
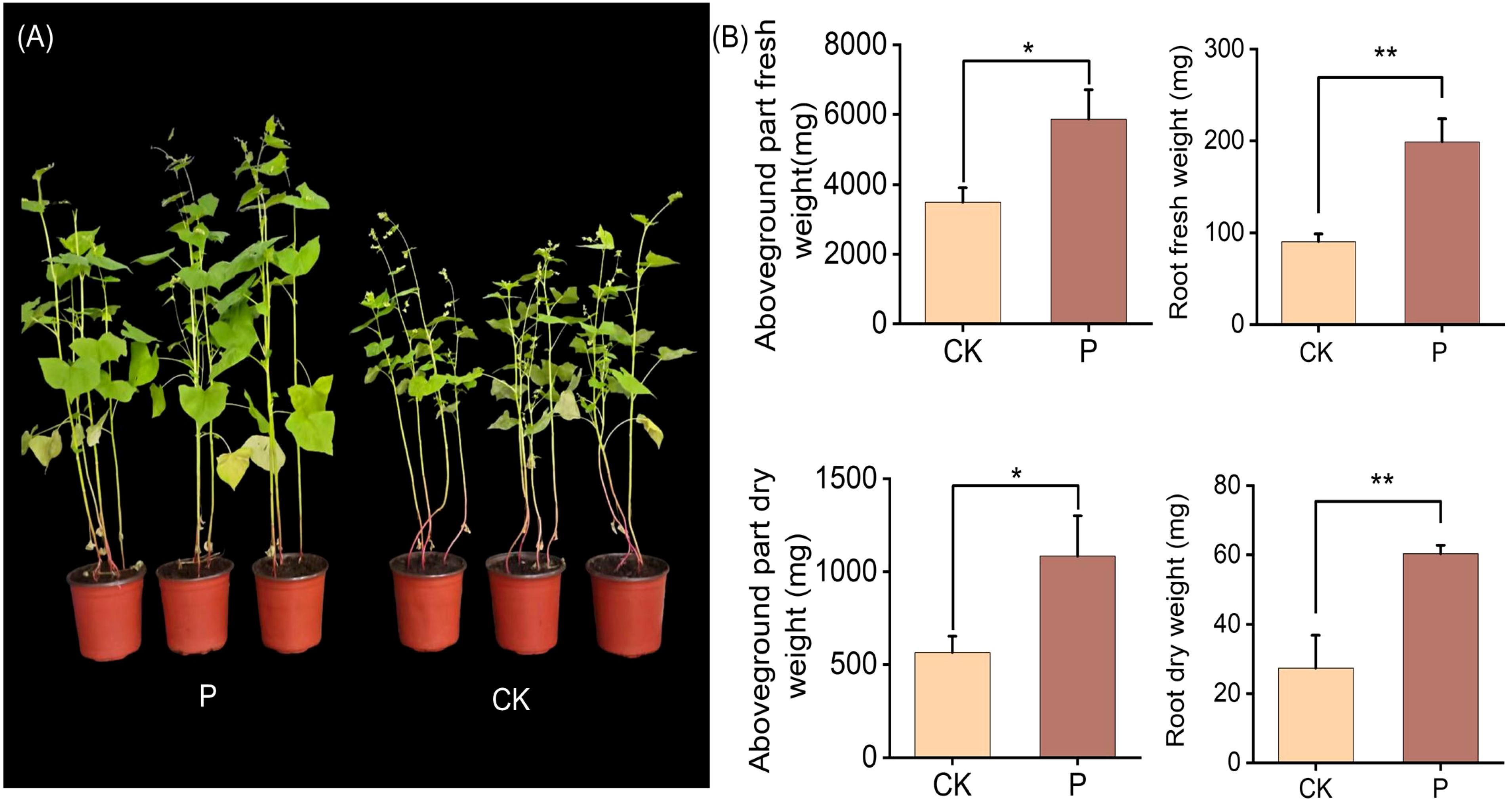
Figure 1. Analyzing plant phenotypes of Tartary buckwheat after inoculation with S. indica. (A) Control (CK) and S. indica colonized (P) Tartary buckwheat plants; (B) Fresh and dry weights of Aboveground parts and Root of CK and P Tartary buckwheat plants. Values are expressed as mean ± SD (standard deviation) of three replicates. *p < 0.05; **p < 0.01.
3.2 Overall RNA-seq results and quality control in Tartary buckwheat colonized by S. indica
In order to explore the regulatory mechanisms of S. indica on Tartary buckwheat in terms of growth promotion and resistance, the transcriptomes of the roots of Tartary buckwheat with significant phenotypic differences (divided into two groups, each group was repeated three times) were selected for sequencing. Total RNA was extracted for transcriptome sequencing and gene expression profiling. The statistics of RNA sequencing (RNA-seq) reads for each sample are shown in Supplementary Table S2. These six libraries yielded a total of 39,306,277,876 raw reads from Illumina sequencing, with an average of 6,551,046,313 reads per sample. After quality control filtering, these six libraries yielded a total of 38,691, 425,229 clean reads, with an average of 6,448,570,872 clean reads per sample. the average quality scores of clean reads, Q20 and Q30, were 98.72% and 96.27%, respectively, indicating that the clean reads obtained were of high quality. The differentially expressed genes (DEGs) statistics of Tartary buckwheat are presented in Figure 2A. It is evident that 403 genes were significantly up-regulated and 471 genes were significantly down-regulated (log2 (fc)) by the addition of S. indica treatment. The volcano plot (Figure 2B) depicted the gene expression pattern by revealing the number of genes that were up-regulated, down-regulated and those that did not undergo significant changes. Further, the reliability of the data was determined by correlation analysis between the samples, and as shown in Figure 3A, the correlation coefficients (R2) between the groups were above 98%, indicating high correlation between the groups and reliable data. In addition, hierarchical clustering of DEG patterns was represented by heatmaps to present the clustering results (Figure 3B). Genes with similar expression patterns may have common functions or participate in common metabolic and signaling pathways.
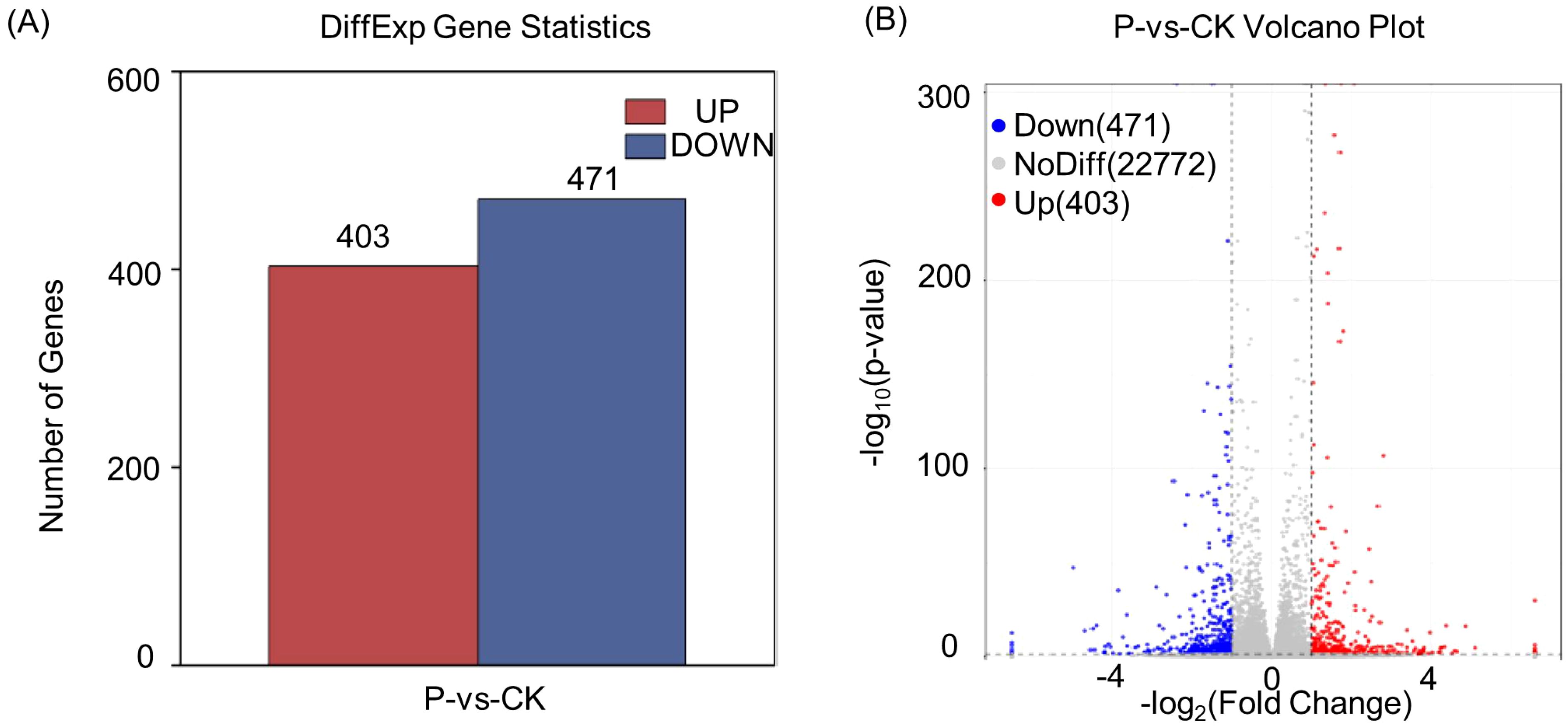
Figure 2. Differentially expressed gene statistics (A) and the volcano plot of P vs CK (B). P: S. indica colonized Tartary buckwheat plants and CK: S. indica uncolonized Tartary buckwheat plants.
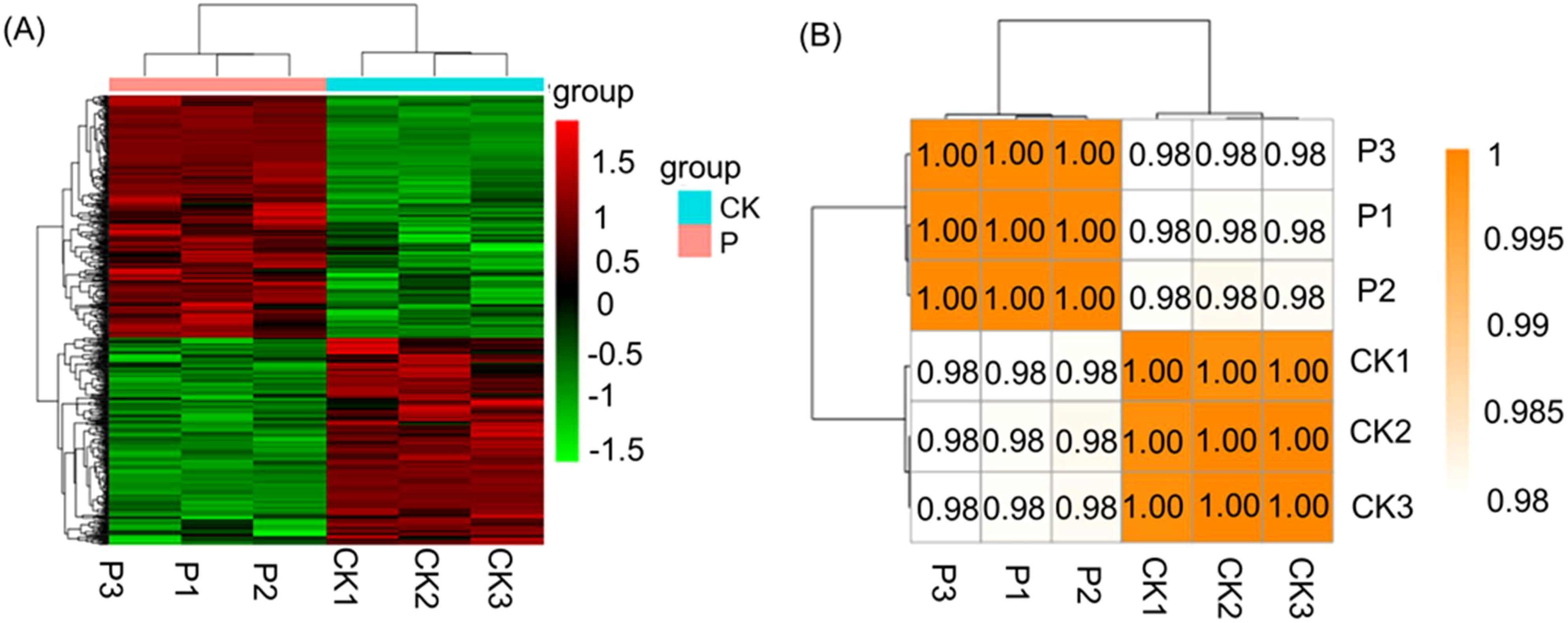
Figure 3. Sample correlation heatmap (A) and Comparison group of P vs CK heatmap (B). (A) The red color shows the number of up regulated genes, and the green color shows the number of down regulated genes. P: S. indica colonized Tartary buckwheat plants and CK: S. indica uncolonized Tartary buckwheat plants.
3.3 Functional enrichment analysis of DEGs
Functional enrichment of DEGs in Tartary buckwheat roots of colonized and uncolonized S. indica was analyzed using GO and KEGG databases. To research the function of shared DEGs between the two groups, we annotated the DEGs and performed enrichment analysis using the Gene Ontology (GO) database. In Figure 4A, the top 10 GO terms with the smallest p-values that were significantly enriched in the biological process (BP), cellular component (CC), and molecular function (MF) categories are shown. The GO term with the smallest p-value in the BP category was oxidation-reduction process (GO:0055114), which had DEG counts of 88. the next 14 DEGs were enriched in drug catabolic process (GO:0042737). The items with the highest DEG numbers in MF were catalytic activity (GO:0003824), oxidoreductase activity (GO:0016491), and cofactor binding (GO:0048037), with 288, 99, and 56 DEGs, respectively. In contrast, the highest number of differential genes in CC was for membrane (GO: 0016020), with 75 DEGs. Subsequently, we compared the annotations of up-regulated and down-regulated DEGs in the GO database, and found that up-regulated DEGs were mainly enriched in the cofactor binding (GO:0048037), tetrapyrrole binding (GO:0046906) and heme binding (GO:0020037), and down-regulated genes were mainly enriched in catalytic activity (GO:0003824), oxidoreductase activity (GO:0016491), membrane (GO:0016020) (Figures 4, 5A).

Figure 4. Histogram of the GO enrichment. Analyses with the top 10 entries with the smallest p-value selected for each Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The left histogram is the -log10 (p-value) and the right histogram is the DEGs number of the corresponding.
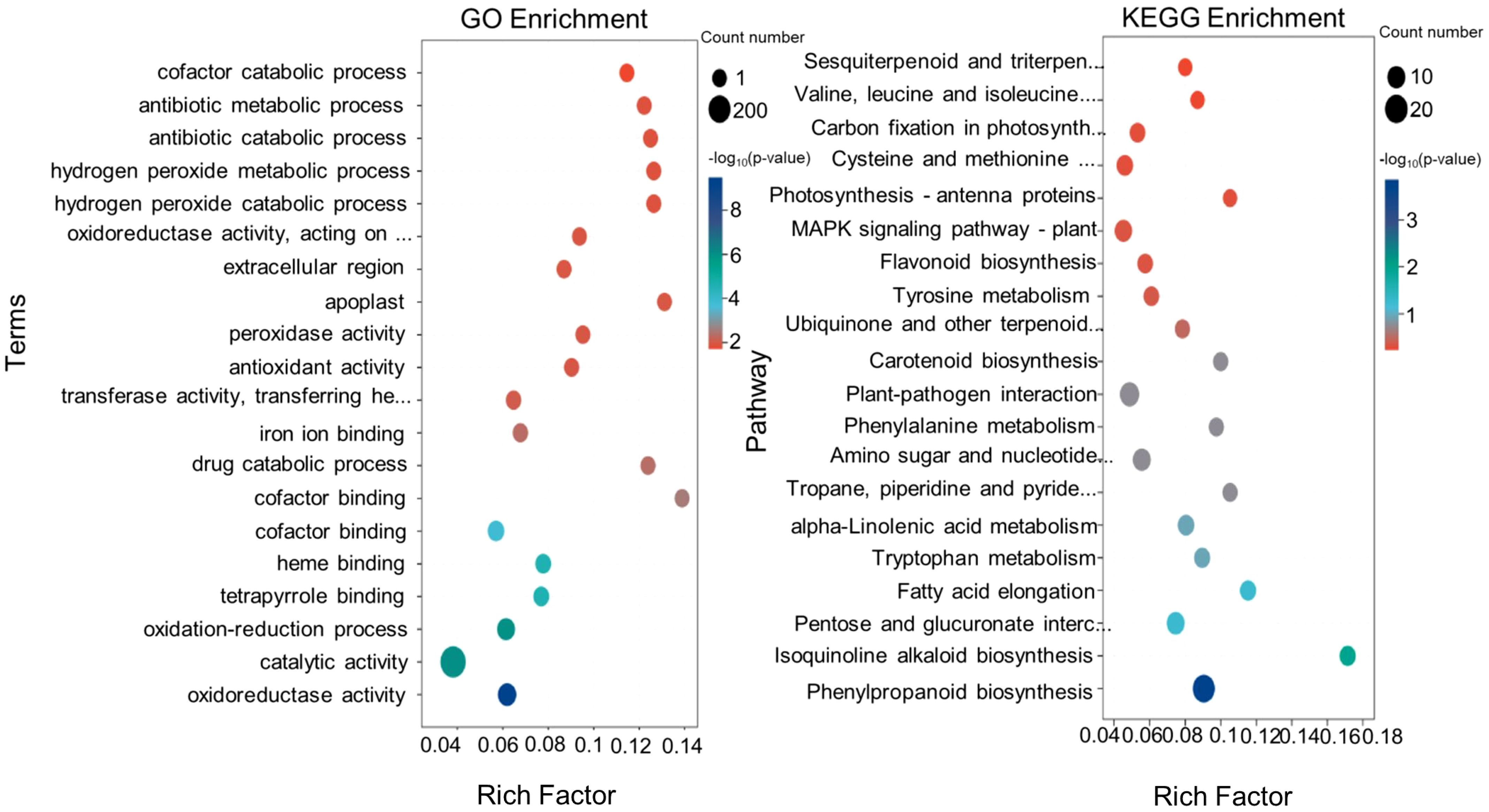
Figure 5. The GO and KEGG enrichment analyses of DEGs. (A) Bubble diagram for GO enrichment analysis of differential genes. (B) Bubble diagram for KEGG enrichment analysis of differential genes.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that KEGG enrichment analysis identified the top 20 pathways with the lowest p-values (Figure 5B). Among them, there were 12 pathways with p-value <0.5 (Supplementary Table S3), which were Phenylpropanoid biosynthesis (KO00940), Isoquinoline alkaloid biosynthesis (KO00950), Fatty acid elongation (KO00062), Pentose and glucuronate interconversions (KO00040), Alpha-Linolenic acid metabolism (KO00592), Tyrosine Metabolism (KO00350), Piperidine and pyridine alkaloid biosynthesis (KO00960), Amino sugar and nucleotide sugar metabolism (KO00520), Carotenoid biosynthesis (KO00906), Plant-pathogen interaction (KO04626), Phenylalanine metabolism (KO00360), Ubiquinone and other terpenoid-quinone biosynthesis (KO00130). The results show that most DEGs were significantly enriched in phenylalanine biosynthesis (KO00940), Pentose and glucuronate interconversions (KO00130), Amino sugar and nucleotide sugar metabolism (KO00520) and other In the pathway. Among them, there was a significant enrichment of DEGs with up-regulated expression in the Phenylpropanoid biosynthesis pathway, Amino sugar and nucleotide sugar metabolism was the most significantly down-regulated.
3.4 Effect of S. indica on phenylpropanoid biosynthesis in Tartary buckwheat
The content of lignin in the stems and seeds of Tartary buckwheat was determined, and the results showed that the content of lignin in the stems and seeds of Tartary buckwheat was reduced after the addition of S. indica (Figure 6A). Key enzymes related to lignin synthesis were selected for further analysis, and the results of quantitative gene analysis showed that Tartary buckwheat seeds inoculated with S. indica showed reduced expression of the FtHCT gene, significantly lower expression of the FtCCOAOM and FtC3H1 genes (p < 0.01), and a highly significant reduction in the expression of the FtCAD6 gene (p < 0.001) compared with the control (Figure 6B). In S. indica colonized Tartary buckwheat stems, the expression of the FtCCOAOM gene was reduced, the expression of the FtHCT gene was significantly reduced (p < 0.05), and the expression of the FtCAD6 gene was significantly reduced (p < 0.01) compared with the control (Figure 6C). It was further proved that the change of lignin gene expression level was basically consistent with the trend of content change. Next, the lignin staining of Tartary buckwheat stems was carried out with Safranin-fixed green dyeing, and the results showed that the color of lignin in the stems of Tartary buckwheat colonized with S. indica was significantly lighter than that in the stems of Tartary buckwheat uncolonized with S. indica, which indicated that the lignin content of Tartary buckwheat colonized with S. indica was reduced (Figure 6D).

Figure 6. Lignin content and expression levels of genes of S. indica colonized (P) and non-colonized (CK) Tartary buckwheat. (A) Lignin content in stems and seeds of P and CK Tartary buckwheat. (B) Expression levels of genes related to the lignin biosynthesis pathway in P and CK Tartary buckwheat of seeds. (C) Expression levels of genes related to the lignin biosynthesis pathway in P and CK Tartary buckwheat of stems. (D) Histochemical analysis of stem cross-section lignin with Safranin-fixed green dyeing. Scale bar in (D) = 50 μm. Values are presented as mean ± SD (standard deviation) of three replicates. *p < 0.05; **p < 0.01; ***p < 0.001; ns p > 0.05.
3.5 Effect of S. indica on flavonoid biosynthesis in Tartary buckwheat
Further, the content of flavonoids in leaves and seeds of Tartary buckwheat was determined by S. indica, and the results showed that the content of flavonoids in leaves and seeds of Tartary buckwheat was significantly elevated by the addition of S. indica (Figure 7A). The key enzymes related to flavonoid synthesis were selected for further analysis, and the results of quantitative gene analysis showed that Tartary buckwheat seeds inoculated with S. indica showed significantly higher (p<0.01) expression of FtCHI, FtANS and FtCHS1 genes, and highly significantly higher (p<0.001) expression levels of FtF3H, FtC4H and FtF3’5’H genes, as compared with the control (Figure 7B). The results indicated that the alteration of flavonoid gene expression levels in Tartary buckwheat colonized with S. indica was consistent with the trend of altered flavonoid content.
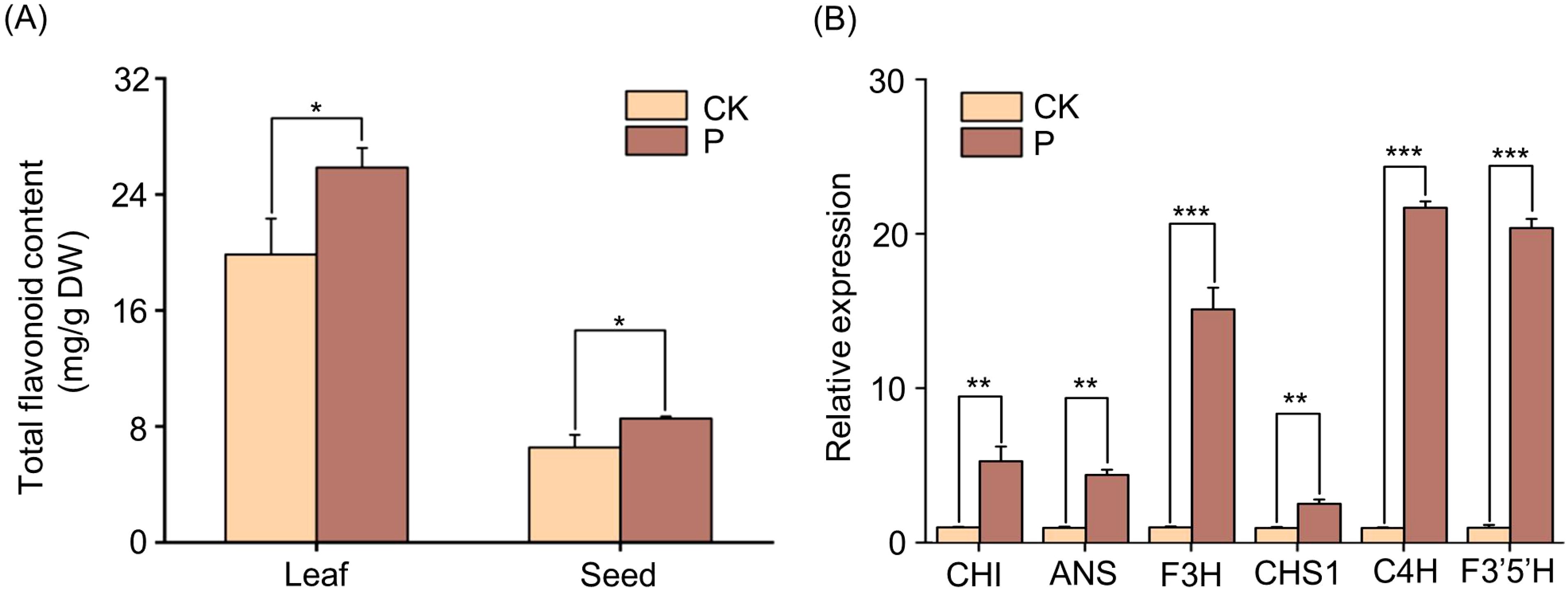
Figure 7. Flavonoids content and expression levels of genes of S. indica colonized (P) and non-colonized (CK) Tartary buckwheat. (A) Total flavonoids in leaves and seeds of P and CK Tartary buckwheat. (B) Expression levels of genes related to the flavonoid biosynthesis pathway in P and CK Tartary buckwheat seeds. Values are presented as mean ± SD (standard deviation) of three replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
4 Discussion
Several researches have shown that S. indica can extensively colonize conductive plants (Mensah et al., 2020) and establish a symbiotic relationship with the plants, and it can significantly improve the yield and quality of the plants (Chen et al., 2022; Kundu et al., 2022; Solanki et al., 2023). It has been reported that S. indica significantly increased the yield of Tartary buckwheat under both drought and normal conditions (Zheng et al., 2023), which is consistent with this paper where S. indica which is consistent with this paper that S. indica increased the yield of Tartary buckwheat (Figure 1). However, current research on the effect of S. indica on Tartary buckwheat is mainly focused on phenotypes and other aspects. Research on the ways in which S. indica improves the seed quality of Tartary buckwheat is very limited, which makes the nutritional and economic value of S. indica on Tartary buckwheat overlooked.
Lignin usually plays an important role in plant secondary walls (Geng et al., 2020). Its synthesis is mainly divided into three parts, the first of which is the deamination of phenylalanine to produce cinnamic acid, the second of which is the formation of lignin monomers through a series of hydroxylation, O-methylation, and reduction reactions, and the last part of which polymerizes the lignin monomers to form lignin (Liu et al., 2011). In the present research, we have further focused on the transcriptome analysis of the Phenylpropanoid biosynthesis pathway (Figure 5B). And found that cinnamyl-alcohol dehydrogenase (CAD), the key enzyme for the synthesis of lignin precursors such as p-Coumaryl alcohol, Caffeyl alcohol, Coniferyl alcohol, and Sinapyl alcohol, was basically down-regulated in roots (Figure 8). According to research, CAD is usually involved in the last step of lignin monomer synthesis, and the expression level of CAD gene is positively correlated with the total lignin content (Reddy et al., 2005; Tao et al., 2009; Shi et al., 2022). Caffeoyl-CoA O-methyltransferase (CoAOMT), CAD, and shikimate O-hydroxycinnamoyl transferase (HCT) are key enzymes for lignin biosynthesis. In this experiment, the relationship between lignin content and the expression of key enzyme genes FtCoAOMT, FtCAD and FtHCT was determined, and it was found that the lignin content was basically consistent with the trend of gene expression (Figures 6A, B). Therefore, it is hypothesized that S. indica may have reduced the lignin content of Tartary buckwheat seeds, and further experiments on functional expression are needed to show the causal relationship between changes in gene expression and phenotypic results. Tartary buckwheat is a food and herbal medicinal crop, due to its thick, hard and completely closed shells, the increase in lignin content increases the hardness of the shells of Tartary buckwheat, thus down-regulating the expression of genes in the lignin synthesis pathway and reducing lignin synthesis, which may lead to the thinning of the shell thickness and the decrease in lignin content. may lead to thinner hull thickness, thereby promoting fruit dehiscence (Li et al., 2020). Tartary buckwheat faces production and processing challenges of difficult hull removal, and S. indica makes the lignin content in the seeds of Tartary buckwheat decrease, which is hypothesized to facilitate the dehiscence of Tartary buckwheat. However, the thickness of hulls is closely related to the ratio of lignin to cellulose, and the species and proportion of lignin (Song et al., 2019; Yang et al., 2024). It is necessary to pay further attention to the content of cellulose in the hulls of Tartary buckwheat and the expression of genes, and the proportion of different lignin species in the hulls, to determine the effect of S. indica on the hulling of Tartary buckwheat.

Figure 8. Lignin biosynthetic pathway involved in Tartary buckwheat the response to S.indica. Red and green indicate upregulation and downregulation, respectively. Intensity of the colors is proportional to the log2FC.
Phenylpropanoid biosynthesis pathway includes lignin biosynthesis pathway and flavonoid biosynthesis pathway. In this research, we found that the lignin content in the phenylpropanoid biosynthesis pathway was reduced in the Tartary buckwheat of S. indica colonized compared with that of S. indica uncolonized. However, p-Coumaroyl-CoA, p-Coumaric acid p-Coumaric acid and other substances to rise. These substances are precursors for the synthesis of lignin and flavonoids, and it is hypothesized that these substances may flow into the flavonoid biosynthesis pathway, leading to an increase in the flavonoid content of Tartary buckwheat colonized with S. indica (Figure 9) (Song et al., 2019). Therefore, in this experiment, we determined the flavonoid content of Tartary buckwheat content with the expression of the corresponding key enzyme genes such as FtCHI, FtANS, FtCHS1, FtCHI, FtANS, FtCHS1, and it was found that the content was up-regulated in a consistent relationship with the expression of the genes (Figure 7), which led to a significant increase in the flavonoid content of the leaves and seeds of Tartary buckwheat colonized with S. indica. While, flavonoid compounds include isoflavones, flavonols and anthocyanins, flavonoids etc. Flavonoids affect the growth and development of plants and also act as antioxidants or signaling molecules to help plants to cope with adverse environments (Wang et al., 2017; Andrade et al., 2018; Liu et al., 2022; Wu et al., 2023; Yu et al., 2024). In addition to this, flavonoids have beneficial effects on human health and help to prevent diseases such as cancer, cardiovascular diseases and diabetes (Havsteen, 2002; Zhang et al., 2012; Sebastian et al., 2017; Ragheb et al., 2020). Therefore, S. indica has a significant impact on the enhancement of nutritional value of Tartary buckwheat and also provides new ways to improve the quality of Tartary buckwheat.

Figure 9. Summary diagram for the promotive effects of S. indica on tartarybuckwheat growth. Yellow and green arrows indicate the flavonoid biosynthesis pathway and lignin biosynthesis pathway, respectively. Red and green boxes indicate up and down regulation of gene expression.
The study revealed a notable enrichment of the phenylpropanoid biosynthetic route within the transcriptome. The creation of phenylpropanoids stands as a crucial secondary synthesis route, beginning with phenylalanine and evolving into ρ-coumaroyl-coenzyme A through the activity of essential enzymes like phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate–CoA ligase (4CL), followed by the transformation of ρ-coumaroyl-coenzyme A, the initial substrate, into lignin, flavonoids, and various other biosynthesis routes (Garibay-Hernández et al., 2021; Tohge et al., 2017). Transcriptome analysis revealed that the Phenylpropanoid biosynthesis pathway in Tartary buckwheat colonized with S. indica was significantly enriched in the KEGG database, leading to a series of alterations in lignin and flavonoid biosynthesis. A similar phenomenon has been found in several studies, where S. indica colonization in different plants increases the activity of genes within the Phenylpropanoid biosynthesis, leading to the up-regulation of lignin-related genes in some plants. This is different from the phenomenon observed in this study of lignin gene expression and content in Tartary buckwheat buckwheat (Bajaj et al., 2018; Li et al., 2023b). Whereas a large number of studies have also shown that S. indica increases the flavonoid content of the plant under abiotic stress, which is also different from the results of the present study, suggesting that S. indica may be able to increase the flavonoid content of Tartary buckwheat both under abiotic stress and under normal conditions. Furthermore, it has been shown that changes in key enzymes involved in the synthesis of lignin and flavonoids in the Phenylpropanoid biosynthesis pathway directly or indirectly lead to changes in the metabolic equilibrium between the lignin and flavonoid biosynthesis pathways (Nabavi et al., 2020; Wang et al., 2024). Further suggesting that the colonization of Tartary buckwheat by S. indica leads to a change in the control of lignin and flavonoids, resulting in Tartary buckwheat sacrificing part of the structural defense against lignin synthesis and prioritizing the chemical defense against flavonoid synthesis and nutrient growth. In addition, our data represent only one period of time during which S. indica altered lignin and flavonoid metabolism in Tartary buckwheat, and more sampling sites may be needed to clarify whether changes in these substances are species-specific or time-related. In conclusion, further attention needs to be paid to the mechanism of flavonoid accumulation in order to explain the enhancement of the nutritional value of Tartary buckwheat by woad from a more comprehensive point of view.
5 Conclusion
In summary, our results showed that S. indica colonization on Tartary buckwheat significantly increased the yield of aboveground parts of Tartary buckwheat along with roots. Further transcriptome analysis further revealed that phenylpropanoid biosynthetic was highly enriched in the KEGG database. Studies have focused on lignin and flavonoid synthesis pathways downstream of phenylacetone biosynthesis. S. indica was found to promote flavonoid synthesis in buckwheat seeds in addition to inhibiting lignin content in Tartary buckwheat seeds, and the effect of S. indica on key enzymes of lignin and flavonoid synthesis in Tartary buckwheat was further explored in terms of molecular mechanisms. The results showed that S. indica could not only reduce the lignin content but also increase the flavonoid content of Tartary buckwheat seeds, which provided new clues and potential methods to improve the nutritional and economic value of Tartary buckwheat seeds.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
WW: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. SZ: Methodology, Writing – original draft. WT: Validation, Writing – review & editing. XZ: Validation, Writing – review & editing. SL: Validation, Writing – review & editing. BD: Writing – review & editing. TW: Writing – review & editing. TB: Supervision, Writing – review & editing. ZT: Investigation, Writing – review & editing. QL: Project administration, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by Natural Science Foundation of Sichuan Province (grant no.2024ZYD0191).
Acknowledgments
Thank you to Shaoshuai Liu (Beijing Academy of Agricultural and Forestry Sciences, Beijing, China) for donating the Serendipita indica strain and Chenglei Li (Sichuan Agricultural University, Sichuan, China) for providing the Tartary buckwheat seeds.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1595781/full#supplementary-material
References
Abdelaziz, M. E., Abdelsattar, M., Abdeldaym, E. A., Atia, M. A. M., Mahmoud, A. W. M., Saad, M. M., et al. (2019). Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress. Scientia Hortic. 256, 108532. doi: 10.1016/j.scienta.2019.05.059
Almario, J., Jeena, G., Wunder, J., Langen, G., Zuccaro, A., Coupland, G., et al. (2017). Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl. Acad. Sci. 114, E9403–E9412. doi: 10.1073/pnas.1710455114
Andrade, A. W. L., MaChado, K. da C., MaChado, K. da C., Figueiredo, D. D. R., David, J. M., Islam, M. T., et al. (2018). In vitro antioxidant properties of the biflavonoid agathisflavone. Chem. Cent. J. 12, 75. doi: 10.1186/s13065-018-0443-0
Aubert, L., Decamps, C., Jacquemin, G., and Quinet, M. (2021). Comparison of Plant Morphology, Yield and Nutritional Quality of Fagopyrum esculentum and Fagopyrum tataricum Grown under Field Conditions in Belgium. Plants 10, 258. doi: 10.3390/plants10020258
Bajaj, R., Huang, Y., Gebrechristos, S., Mikolajczyk, B., Brown, H., Prasad, R., et al. (2018). Transcriptional responses of soybean roots to colonization with the root endophytic fungus reveals altered phenylpropanoid and secondary metabolism. Sci. Rep. 8, 10227. doi: 10.1038/s41598-018-26809-3
Brandizzi, F. (2000). Ruzin SE. 1999. Plant microtechnique and microscopy. 322 pp. Oxford, New York: Oxford University Press. £32.50 (softback). Ann. Bot. 86, 708. doi: 10.1006/anbo.2000.1231
Chen, W., Lin, F., Lin, K.-H., Chen, C., Xia, C., Liao, Q., et al. (2022). Growth Promotion and Salt-Tolerance Improvement of Gerbera jamesonii by Root Colonization of Piriformospora indica. J. Plant Growth Regul. 41, 1219–1228. doi: 10.1007/s00344-021-10385-4
Chettry, U. and Chrungoo, N. K. (2021). Beyond the cereal box: breeding buckwheat as a strategic crop for human nutrition. Plant Foods Hum. Nutr. 76, 399–409. doi: 10.1007/s11130-021-00930-7
Garibay-Hernández, A., Kessler, N., Józefowicz, A. M., Türksoy, G. M., Lohwasser, U., and Mock, H.-P. (2021). Untargeted metabotyping to study phenylpropanoid diversity in crop plants. Physiol. Plant. 173, 680–697. doi: 10.1111/ppl.13458
Geng, P., Zhang, S., Liu, J., Zhao, C., Wu, J., Cao, Y., et al. (2020). MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 182, 1272–1283. doi: 10.1104/pp.19.01070
Giménez-Bastida, J. A. and Zieliński, H. (2015). Buckwheat as a functional food and its effects on health. J. Agric. Food Chem. 63, 7896–7913. doi: 10.1021/acs.jafc.5b02498
Havsteen, B. H. (2002). The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 96, 67–202. doi: 10.1016/S0163-7258(02)00298-X
Janovská, D., Jágr, M., Svoboda, P., Dvořáček, V., Meglič, V., and Hlásná Čepková, P. (2021). Breeding buckwheat for nutritional quality in the Czech Republic. Plants 10, 1262. doi: 10.3390/plants10071262
Jisha, S., Gouri, P. R., Anith, K. N., and Sabu, K. K. (2018). Piriformospora indica cell wall extract as the best elicitor for asiaticoside production in Centella asiatica (L.) Urban, evidenced by morphological, physiological and molecular analyses. Plant Physiol. Biochem. 125, 106–115. doi: 10.1016/j.plaphy.2018.01.021
Kalinova, J. and Vrchotova, N. (2009). Level of Catechin, Myricetin, Quercetin and Isoquercitrin in Buckwheat (Fagopyrum esculentum Moench), Changes of Their Levels during Vegetation and Their Effect on The Growth of Selected Weeds. J. Agric. Food Chem. 57, 2719–2725. doi: 10.1021/jf803633f
Kasar, C., Thanushree, M. P., Gupta, S., and Inamdar, A. A. (2021). Milled fractions of common buckwheat (Fagopyrum esculentum) from the Himalayan regions: grain characteristics, functional properties and nutrient composition. J. Food Sci. Technol. 58, 3871–3881. doi: 10.1007/s13197-020-04848-x
Kim, S.-J., Zaidul, I. S. M., Maeda, T., Suzuki, T., Hashimoto, N., Takigawa, S., et al. (2007). A time-course study of flavonoids in the sprouts of tartary (Fagopyrum tataricum Gaertn.) buckwheats. Scientia Hortic. 115, 13–18. doi: 10.1016/j.scienta.2007.07.018
Kundu, A., Mishra, S., Kundu, P., Jogawat, A., and Vadassery, J. (2022). Piriformospora indica recruits host-derived putrescine for growth promotion in plants. Plant Physiol. 188, 2289–2307. doi: 10.1093/plphys/kiab536
Li, L., Feng, Y., Qi, F., and Hao, R. (2023a). Research progress of Piriformospora indica in improving plant growth and stress resistance to plant. J. Fungi 9, 965. doi: 10.3390/jof9100965
Li, L., Hao, R., Yang, X., Feng, Y., and Bi, Z. (2023b). Piriformospora indica Increases Resistance to Fusarium pseudograminearum in Wheat by Inducing Phenylpropanoid Pathway. Int. J. Mol. Sci. 24, 8797. doi: 10.3390/ijms24108797
Li, H.-Y., Wu, C.-X., Lv, Q.-Y., Shi, T.-X., Chen, Q.-J., and Chen, Q.-F. (2020). Comparative cellular, physiological and transcriptome analyses reveal the potential easy dehulling mechanism of rice-tartary buckwheat (Fagopyrum Tararicum). BMC Plant Biol. 20, 505. doi: 10.1186/s12870-020-02715-7
Liu, L., Li, L., Feng, Y., Wang, T., Li, C., Wu, H., et al. (2025). Impact of heat stress on the development, physiological and biochemical characteristics of Tartary buckwheat flowers, and its transcriptomic analysis. Plant Physiol. Biochem. 220, 109535. doi: 10.1016/j.plaphy.2025.109535
Liu, H., Liu, S., Wang, H., Chen, K., and Zhang, P. (2022). The flavonoid 3′-hydroxylase gene from the Antarctic moss Pohlia nutans is involved in regulating oxidative and salt stress tolerance. Biotechnol. Appl. Biochem. 69, 676–686. doi: 10.1002/bab.2143
Liu, C.-J., Miao, Y.-C., and Zhang, K.-W. (2011). Sequestration and transport of lignin monomeric precursors. Molecules 16, 710–727. doi: 10.3390/molecules16010710
Mensah, R. A., Li, D., Liu, F., Tian, N., Sun, X., Hao, X., et al. (2020). Versatile Piriformospora indica and its potential applications in horticultural crops. Hortic. Plant J. 6, 111–121. doi: 10.1016/j.hpj.2020.01.002
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., et al. (2020). Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 38, 107316. doi: 10.1016/j.bioteChadv.2018.11.005
Ragheb, S. R., El Wakeel, L. M., Nasr, M. S., and Sabri, N. A. (2020). Impact of Rutin and Vitamin C combination on oxidative stress and glycemic control in patients with type 2 diabetes. Clin. Nutr. ESPEN 35, 128–135. doi: 10.1016/j.clnesp.2019.10.015
Rai, M., Acharya, D., Singh, A., and Varma, A. (2001). Positive growth responses of the medicinal plants Spilanthes calva and Withania somnifera to inoculation by Piriformospora indica in a field trial. Mycorrhiza 11, 123–128. doi: 10.1007/s005720100115
Reddy, M. S. S., Chen, F., Shadle, G., Jackson, L., Aljoe, H., and Dixon, R. A. (2005). Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. 102, 16573–16578. doi: 10.1073/pnas.0505749102
Sasidharan, P. and Jayachitra, A. (2017). Direct shoot bud regeneration from shoot tip explants of Enicostema axillare: an important medicinal plant. Agroforest Syst. 91, 471–477. doi: 10.1007/s10457-016-9943-x
Sebastian, R. S., Wilkinson Enns, C., Goldman, J. D., and Moshfegh, A. J. (2017). Dietary flavonoid intake is inversely associated with cardiovascular disease risk as assessed by body mass index and waist circumference among adults in the United States. Nutrients 9, 827. doi: 10.3390/nu9080827
Shi, J., Yan, X., Sun, T., Shen, Y., Shi, Q., Wang, W., et al. (2022). Homeostatic regulation of flavonoid and lignin biosynthesis in phenylpropanoid pathway of transgenic tobacco. Gene 809, 146017. doi: 10.1016/j.gene.2021.146017
Solanki, S., Gupta, S., Kapoor, R., and Varma, A. (2023). Chemically synthesized AgNPs and Piriformospora indica synergistically augment nutritional quality in black rice. J. Fungi 9, 611. doi: 10.3390/jof9060611
Song, C., Ma, C., and Xiang, D. (2019). Variations in accumulation of lignin and cellulose and metabolic changes in seed hull provide insight into dehulling characteristic of tartary buckwheat seeds. Int. J. Mol. Sci. 20, 524. doi: 10.3390/ijms20030524
Tao, S., Khanizadeh, S., Zhang, H., and Zhang, S. (2009). Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci. 176, 413–419. doi: 10.1016/j.plantsci.2008.12.011
Tohge, T., de Souza, L. P., and Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. doi: 10.1093/jxb/erx177
Varma, A., Savita Verma, Sudha, Sahay, N., Bütehorn, B., and Franken, P. (1999). Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 65, 2741–2744. doi: 10.1128/AEM.65.6.2741-2744.1999
Verma, S., Varma, A., Rexer, K.-H., Hassel, A., Kost, G., Sarbhoy, A., et al. (1998). Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia 90, 896–903. doi: 10.1080/00275514.1998.12026983
Waller, F., Achatz, B., Baltruschat, H., Fodor, J., Becker, K., Fischer, M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. 102, 13386–13391. doi: 10.1073/pnas.0504423102
Wang, Y. and Campbell, C. G. (2007). Tartary buckwheat breeding (Fagopyrum tataricum L. Gaertn.) through hybridization with its Rice-Tartary type. Euphytica 156, 399–405. doi: 10.1007/s10681-007-9389-3
Wang, W., Gao, T., Yang, H., Sun, Y., Yang, J., Zhou, J., et al. (2024). The balance between lignin and flavonoid metabolism has a central role in the changes of quality in young shoots of the tea plant (Camellia sinensis). Scientia Hortic. 338, 113788. doi: 10.1016/j.scienta.2024.113
Wang, X., Wang, M., Cao, J., Wu, Y., Xiao, J., and Wang, Q. (2017). Analysis of flavonoids and antioxidants in extracts of ferns from Tianmu Mountain in Zhejiang Province (China). Ind. Crops Prod. 97, 137–145. doi: 10.1016/j.indcrop.2016.12.013
Weiss, M., Selosse, M.-A., Rexer, K.-H., Urban, A., and Oberwinkler, F. (2004). Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential*. Mycol. Res. 108, 1003–1010. doi: 10.1017/S0953756204000772
Wu, J., Lv, S., Zhao, L., Gao, T., Yu, C., Hu, J., et al. (2023). Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses. Planta 257, 108. doi: 10.1007/s00425-023-04136-w
Yan, C., Muhammad Rizwan, H., Liang, D., Reichelt, M., Mithöfer, A., Scholz, S. S., et al. (2021). The effect of the root-colonizing Piriformospora indica on passion fruit (Passiflora edulis) development: Initial defense shifts to fitness benefits and higher fruit quality. Food Chem. 359, 129671. doi: 10.1016/j.foodchem.2021.129671
Yang, W., Duan, H., Yu, K., Hou, S., Kang, Y., Wang, X., et al. (2024). Integrative dissection of lignin composition in tartary buckwheat seed hulls for enhanced dehulling efficiency. Adv. Sci. 11, 2400916. doi: 10.1002/advs.202400916
Yin, L., Qu, P., Wang, D., Yan, S., Gong, Q., Yang, R., et al. (2024). The Influence of Piriformospora indica Colonization on the Root Development and Growth of Cerasus humilis Cuttings. Plants 13, 1482. doi: 10.3390/plants13111482
Yu, Q., Liu, C., Sun, J., Ding, M., Ding, Y., Xu, Y., et al. (2024). McWRKY43 Confers Cold Stress Tolerance in Michelia crassipes via Regulation of Flavonoid Biosynthesis. Int. J. Mol. Sci. 25, 9843. doi: 10.3390/ijms25189843
Zhang, Z.-L., Zhou, M.-L., Tang, Y., Li, F.-L., Tang, Y.-X., Shao, J.-R., et al. (2012). Bioactive compounds in functional buckwheat food. Food Res. Int. 49, 389–395. doi: 10.1016/j.foodres.2012.07.035
Keywords: Tartary buckwheat, Serendipita indica, RNA-seq, lignin biosynthesis, flavonoid biosynthesis, phenylpropanoid biosynthesis
Citation: Wang W, Zhong S, Tang W, Zhou X, Li S, Ding B, Wang T, Bu T, Tang Z and Li Q (2025) Transcriptome analysis reveals changes in lignin and flavonoid biosynthesis in Serendipita indica colonized Tartary buckwheat. Front. Plant Sci. 16:1595781. doi: 10.3389/fpls.2025.1595781
Received: 18 March 2025; Accepted: 08 May 2025;
Published: 05 June 2025.
Edited by:
Ricardo Aroca, Spanish National Research Council (CSIC), SpainCopyright © 2025 Wang, Zhong, Tang, Zhou, Li, Ding, Wang, Bu, Tang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingfeng Li, cWluZ2ZlbmcubGlAc2ljYXUuZWR1LmNu; Zizhong Tang, MTQxMjZAc2ljYXUuZWR1LmNu
 Wenjing Wang1
Wenjing Wang1 Zizhong Tang
Zizhong Tang Qingfeng Li
Qingfeng Li