- 1Sanya Nanfan Research Institute, Hainan University, Sanya, Hainan, China
- 2School of Tropical Agriculture and Forestry, Hainan University, Haikou, Hainan, China
- 3Rice Innovation Team, Yazhouwan National Laboratory, Sanya, Hainan, China
- 4Hainan Seed Innovation Research Institute, Sanya, Hainan, China
Introduction: The RWP-RK transcription factor family plays a pivotal role in nitrogen response and gametophyte development. Despite its biological importance, the evolutionary relationships and functional characteristics of RWP-RK genes in rice remain incompletely understood. This study aimed to investigate the structure, classification, expression patterns, and potential regulatory roles of RWP-RK transcription factors in rice (Oryza sativa), with a focus on their involvement in nitrogen signaling and reproductive development.
Methods: A comprehensive genome-wide analysis was conducted to identify RWP-RK genes in rice. A total of 13 genes encoding 14 proteins, including two alternative splicing variants of OsNLP5, were identified and mapped across 8 of the 12 rice chromosomes. Phylogenetic analysis was used to classify the proteins into subfamilies, and gene/protein structure characteristics were examined, including coding sequence length, exon number, and domain composition. Collinearity analysis was performed to explore evolutionary relationships between rice and Arabidopsis. Promoter regions of the RWP-RK genes were analyzed for cis-regulatory elements, and tissue-specific as well as nitrogen-responsive gene expression patterns were evaluated using expression profiling.
Results: Phylogenetic analysis grouped the 14 RWP-RK proteins into four clades: groups 1–3 were assigned to the NLP subfamily and group 4 to the RKD subfamily. NLP members contained both RWP-RK and PB1 domains, while RKD proteins possessed only the RWP-RK domain. Structural analysis revealed that NLP genes generally have longer CDS, more exons, and larger proteins than RKD genes. Collinearity analysis suggested that rice and Arabidopsis RWP-RK genes share a common ancestor, with evidence of gene recombination and species-specific divergence. Promoter analysis revealed numerous hormone- and stress-responsive cis-elements. Expression profiling showed that OsNLP genes are broadly expressed in all tissues, whereas OsRKD genes are predominantly active in reproductive organs. Upon nitrogen resupply after nitrogen starvation, expression levels of all OsNLP genes and three OsRKDs (OsRKD1, OsRKD3, OsRKD5) showed dynamic changes.
Discussion: The findings provide new insights into the classification, structure, and expression dynamics of the RWP-RK transcription factor family in rice. The distinct domain architectures and expression patterns between the NLP and RKD subfamilies suggest functional divergence, with NLP genes potentially playing broader roles in general nitrogen regulation, while RKD genes may be more specialized for reproductive development. The nitrogen-responsive expression changes highlight the potential regulatory role of these transcription factors in nutrient signaling. Overall, this study lays a valuable foundation for future functional investigations into the OsRWP-RK family's roles in nitrogen response and gametophyte development in rice.
1 Introduction
Nitrogen is an essential plant macronutrient, constituting a fundamental component of proteins, nucleic acids, and chlorophyll. Soil nitrogen mainly originates from microbial nitrogen fixation and anthropogenic input (Li et al., 2024). Its deficiency severely impairs plant growth and development. In rice, nitrogen shortage reduces biomass, tiller number, and ultimately affects yield and quality (Hong et al., 2022). However, plant roots absorb only 30% of the available soil nitrogen, with the remaining 70% lost through leaching or gaseous emissions, causing environmental degradation, such as eutrophication and the greenhouse effect (Zhang et al., 2024). Therefore, developing crop varieties that efficiently acquire and utilize nitrogen, while maintaining high yields under reduced nitrogen input is a sustainable solution (Tegeder and Masclaux-Daubresse, 2018). Transcription factors containing the RWP-RK domain play a crucial role in regulating genes involved in nitrogen signaling and metabolism, which are vital for plant growth and development (Yan and Nambara, 2023).
RWP-RK transcription factors contain a conserved 60-amino acid RWP-RK motif, an ancient motif predating the emergence of Viridiplantae (green algae and land plants) (Sakuraba et al., 2022). In the process of evolution, they are classified into two subfamilies—NIN-like proteins (NLPs) and RWP-RK domain proteins (RKDs)—based on protein sequence (Chardin et al., 2014). NLPs feature three characteristic domains: an RWP-RK domain that binds cis-acting elements in NUE-related gene promoters (Konishi and Yanagisawa, 2013), a PB1 domain for protein-protein interactions (Konishi and Yanagisawa, 2019), and an N-terminal GAF-like domain (widely present in animals, bacteria and fungi, but is absent in plants) potentially involved in signal transduction (Chardin et al., 2014; Liu et al., 2017). RKDs, on the other hand, contain a conserved RWP-RK domain, regulating genes involved in gametogenesis and embryogenesis (Koszegi et al., 2011).
NLPs are involved in the transcriptional activation of key genes related to nitrogen uptake, assimilation, and nitrate signaling, ensuring efficient nitrogen for plants under nutrient-limited conditions (Yan and Nambara, 2023). A mutant of seven Arabidopsis NLP transcription factors (nlp2, 4, 5, 6, 7, 8, 9) impairs primary nitrate responses and developmental processes of seedlings (Liu et al., 2022). The function of AtNLP7 has been extensively studied, it acts as both a transcription activator and an intracellular nitrate sensor, regulated by nitrate through a nuclear retention mechanism. AtNLP7 binds and modulates most known nitrate signaling and assimilation genes (Marchive et al., 2013; Liu et al., 2022; Durand et al., 2023). AtNLP2, AtNLP6, and AtNLP8 also play vital roles in nitrate signaling, with AtNLP2 regulating early nitrate responses and linking nitrate assimilation to energy and carbon supply (Durand et al., 2023), AtNLP6 activating nitrate signaling and supporting root meristem growth (Guan et al., 2017), and AtNLP8 promoting nitrate-dependent seed germination (Yan et al., 2016). In rice, OsNLP3, the homolog of AtNLP7, together with OsNLP1 and OsNLP4, directly binds to the promoters of nitrate-responsive genes (Hu et al., 2019; Alfatih et al., 2020; Wang et al., 2021; Wu et al., 2021; Zhang et al., 2022), playing overlapping yet distinct roles in nitrogen acquisition, assimilation, and nitrogen use efficiency (NUE), thereby enhancing grain yield (Yan and Nambara, 2023). OsNLP3 and OsNLP4 primarily govern nitrate metabolism, with mutants showing significant growth inhibition and decreased NUE under nitrate conditions, but no notable differences under ammonium conditions (Wang et al., 2021; Wu et al., 2021; Zhang et al., 2022). In contrast, OsNLP1 regulates both nitrate and ammonium metabolism, coordinating the expression of relevant genes and conferring broad adaptability to these nitrogen sources (Alfatih et al., 2020). OsNLP3 and OsNLP1 transcripts are both induced by nitrogen starvation, with OsNLP1 protein localizing in the nucleus, while OsNLP3 nucleo-cytosolic shuttling is specifically regulated by nitrate (Alfatih et al., 2020; Zhang et al., 2022). OsNLP2 localizes to the nucleus and negatively regulates ferroptotic cell death and immune responses during Magnaporthe oryzae infection [20]. Additionally, NLP2-NR-associated NO enhances salt stress tolerance by promoting ABA catabolism during seed germination (Yi et al., 2022).
The RKD (RWP-RK domain-containing) transcription factors possess a highly conserved RWP-RK domain (Koszegi et al., 2011). Recent studies have demonstrated that RKD transcription factors are crucial in germ cell differentiation and embryo development in land plants. In Arabidopsis, AtRKD1 and AtRKD2 are predominantly expressed in egg cells; their ectopic expression promotes cell proliferation and activates an egg cell marker (Koszegi et al., 2011). AtRKD3, an ovule development-specific gene, regulates germ cell differentiation and embryo development through transcriptional activation or repression of target genes (Tedeschi et al., 2017). AtRKD4 is preferentially expressed in early embryos and activates early embryo-specific genes (Waki et al., 2011). Moreover, in rice, OsRKD3 induces somatic embryo formation in the normally somatic embryogenesis-resistant Indonesian black rice landrace (Cempo Ireng) (Purwestri et al., 2023).
The RWP-RK transcription factor involves various physiological processes, such as nitrogen response, gametophyte development, and abiotic stress regulation (Chardin et al., 2014). In many plants, including wheat (Kumar et al., 2018), maize (Ge et al., 2018), brassica (Liu et al., 2018), and soybean (Amin et al., 2023), genome-wide identification and characterization of the RWP-RK family have been reported. RWP-RK transcription factors are crucial for rice nitrogen utilization and embryo development, but genome-wide analyses of rice RWP-RKs are scarce. Leif Schauser’s study on OsNLP1, OsNLP2, and OsNLP3 revealed that NLPs play a key role in rice nitrogen signaling (Schauser et al., 2005). In 2020, B. Jagadhesan investigated OsNLP1-6 in four rice lines (APO, IR83929-B-B-291-3-1-1, Nerica-L-42, and Pusa Basmati 1) and identified OsNLP1 and OsNLP3 as major contributors to nitrogen use efficiency (Jagadhesan et al., 2020). However, these studies focus mainly on individual genes or genotypes, lacking comprehensive genome-wide identification and expression analyses. In this study, 13 OsRWP-RK genes (14 proteins, including two proteins of OsNLP5) were identified, their conserved motifs, gene structure, protein structures, and promoter elements were analyzed. The expression patterns of OsRWP-RK genes were examined using transcriptome data. Additionally, expression changes in roots were assessed after transferring nitrogen-deprived rice seedlings to nitrogen-sufficient conditions. This study provides a deeper understanding of RWP-RK genes in rice. It lays a theoretical foundation for further research on the nitrogen signaling pathway and gametophyte development.
2 Materials and methods
2.1 Identification of RWP-RK transcription factors family members in rice
To identify RWP-RK domain-containing transcription factors in rice (Oryza sativa), the rice protein database was obtained from JGI (https://phytozome-next.jgi.doe.gov; accessed on 5 Jan 2025; Osativa 323 v7.0) (Goodstein et al., 2012). Raw Hidden Markov Model (HMM) data of the conserved RWP-RK (PF02042) domain were downloaded from the Pfam database (http://pfam.xfam.org; accessed on 5 Jan 2025) and used to search for orthologues in the rice genome (Mistry et al., 2021). The RWP-RK domain in candidate proteins was verified using the NCBI Conserved Domains search tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi; accessed on 5 Jan 2025) (Marchler-Bauer et al., 2017; Wang et al., 2023). The physicochemical properties of OsRWP-RK proteins were predicted using ProtParam (https://web.expasy.org/protparam/; accessed on 5 Jan 2025) (Wilkins et al., 1999), and their subcellular localizations were predicted using Cell-PLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/; accessed on 5 Jan 2025) (Chou and Shen, 2010).
2.2 Phylogenetic tree, chromosomal distribution, and synteny analysis
Arabidopsis RWP-RK protein sequences were obtained from PlantTFDB v5.0 (https://planttfdb.gao-lab.org; accessed on 3 Jan 2025) (Jin et al., 2017). Sequences of 28 RWP-RK proteins from Arabidopsis thaliana and Oryza sativa were aligned using Mafft, and a phylogenetic tree was constructed with the maximum likelihood (ML) method in Fasttree, including bootstrap analysis with 500 replicates. Chromosomal localization and synteny analysis were performed using TBtools (version 2.194) (Chen et al., 2023).
2.3 Gene structures, conserved structural domains, conserved motifs
Gene structure data were obtained from the genome annotation file on JGI (https://phytozome-next.jgi.doe.gov; accessed on 5 Jan 2025; Osativa 323 v7.0) and analyzed using TBtools (version 2.194) (Goodstein et al., 2012; Chen et al., 2023). Conserved motifs of OsRWP-RK proteins were identified with MEME (http://meme-suite.org/tools/meme; accessed on 3 Jan 2025) (Bailey et al., 2015), and the top three motifs were extracted. Conserved domains were predicted using NCBI-CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; accessed on 3 Jan 2025) and visualized in TBtools (version 2.194) (Marchler-Bauer et al., 2017; Chen et al., 2023; Wang et al., 2023).
2.4 Protein 3D structure prediction
The 3D structure of the OsRWP-RK proteins was predicted using the AlphaFold Protein Structure Database (https://alphafold.com/; accessed on 14 Jan 2025) (Jumper et al., 2021; Varadi et al., 2023).
2.5 OsRWP-RK gene promoter analysis
To analyze the promoter sequences of OsRWP-RK transcription factors, 2000 bp upstream of each gene’s initiation codon was extracted using TBtools (version 2.194) (Chen et al., 2023). Cis-elements were predicted with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html; accessed on 6 Jan 2025) and visualized using TBtools (version 2.194) (Lescot et al., 2002; Chen et al., 2023).
2.6 Transcription profile of OsRWP-RK genes
Expression data for five rice tissues (leaf, root, stem, inflorescence, and seed) were obtained from the Rice Gene Annotation Project (https://rice.uga.edu/expression.shtml; accessed on 7 Jan 2025). Heatmaps and histograms were generated using GraphPad Prism (version 9.5).
2.7 Nitrogen induces treatments, RNA extraction, and qRT-PCR analysis
Rice seedlings of the Japonica variety ZH11 were grown for 15 days in a modified Kimura B nutrient solution (1 mM NO3- + 1 mM NH4+) and then transferred to a nitrogen-free solution for 5 days. After nitrogen starvation, plants were returned to the modified solution, and roots were collected at 0, 10, 30, 60, and 120 minutes to obtain RNA. RNA was extracted using RNA isolator Total RNA Extraction Reagent (R401-01, Vazyme) and reverse transcribed with HiScript III All-in-one RT SuperMix (R333, Vazyme). Quantitative PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme) on a LightCycler® 480 Instrument II. The primers used for qRT-PCR are listed in Supplementary Data Sheet S1. The experiment was conducted with three biological replicates.
3 Result
3.1 Identification and classification of OsRWP-RK genes
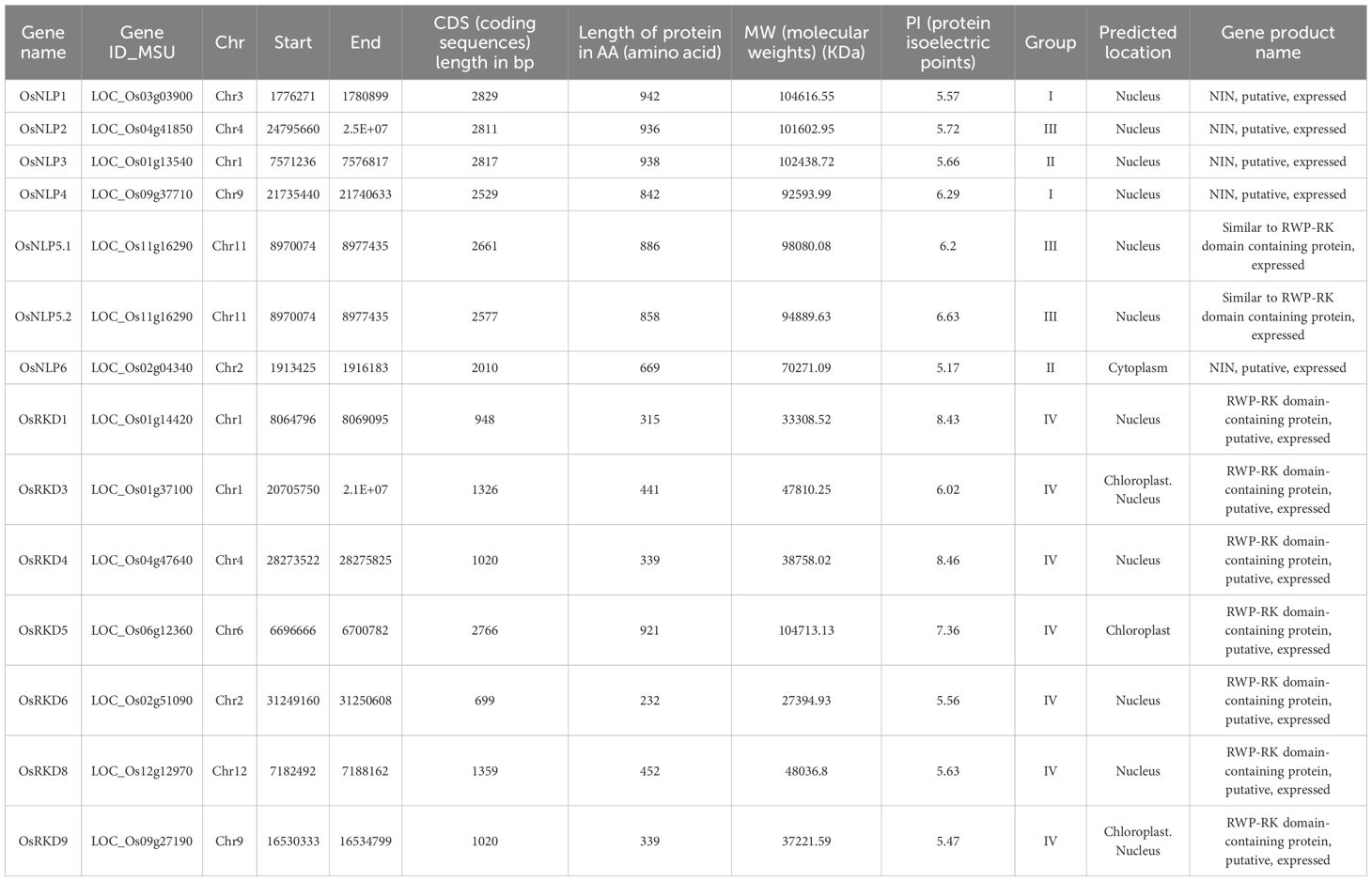
The Oryza sativa genome contains 13 RWP-RK genes encoding 14 protein isoforms, including two variants of OsNLP5 (Table 1), all harboring the conserved RWP-RK domain (Figure 1). The coding sequence (CDS) lengths of the OsRWP-RK family range from 699 bp (OsRKD6) to 2829 bp (OsNLP1), with corresponding protein lengths spanning 232 (OsRKD6) to 942 amino acids (OsNLP1). Their molecular weights vary from 27.4 kDa (OsRKD6) to 104.7 kDa (OsRKD5), and predicted protein isoelectric points range from 5.17 (OsNLP6) to 8.46 (OsRKD4) (Table 1). Proteins in the NLP subfamily generally possess longer CDS and higher molecular weights compared to those in the RKD subfamily (Table 1, Supplementary Figure 1), whereas OsRKD5 exhibits similar features to the NLP subfamily (Table 1).
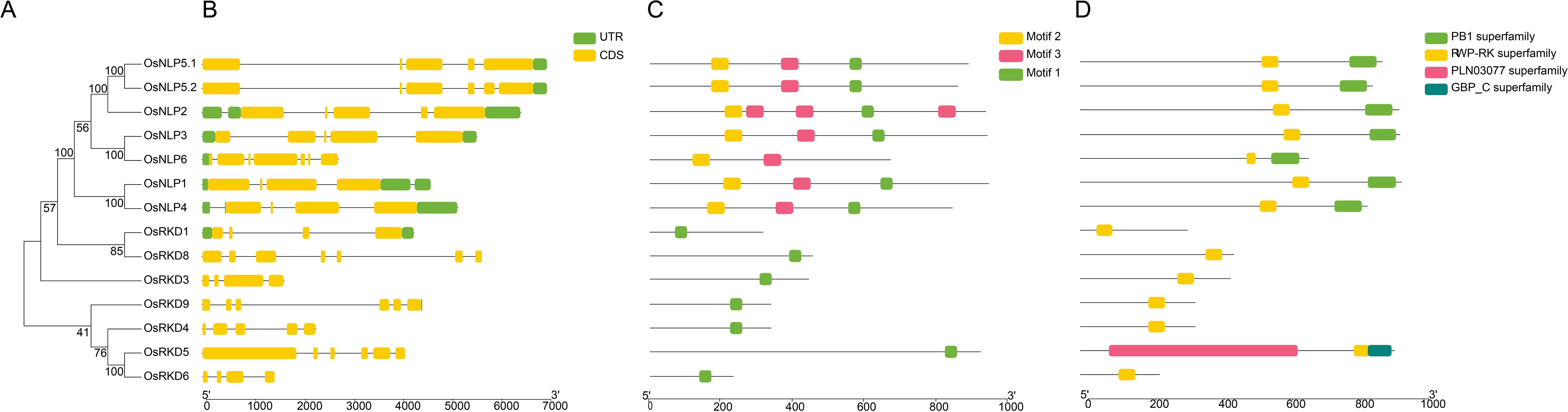
Figure 1. Phylogenetic analysis, gene structure, and conserved motifs and domain of OsRWP-RK proteins in Oryza sativa. (A) Phylogenetic tree of 14 OsRWP-RK proteins. (B) Gene structure showing exons (yellow bars), introns (black lines), and upstream/downstream regions (green bars). (C) Conserved motif distribution, with each motif represented by a distinct color. (D) Conserved domain organization, with different domains highlighted in colored boxes.
Subcellular localization prediction indicated that the majority (10/14) localized exclusively to the nucleus, consistent with their predicted roles as transcription factors (Table 1). Notably, two exceptions were identified: OsNLP6 and OsRKD5 exhibited cytoplasmic localization (Table 1), suggesting potential non-nuclear functions such as post-translational regulation or signaling. Additionally, OsRKD3 and OsRKD9 showed dual cytoplasm-nucleus distribution (Table 1), implying shuttling dynamics that may regulate their activity.
These 13 OsRWP-RK genes are unevenly distributed across 8 of the 12 chromosomes, with chromosome 1 harboring three genes, chromosomes 2, 4, and 9 each containing two, and chromosomes 3, 6, 11, and 12 each with one gene (Figure 2A). Notably, OsNLP1, OsNLP4, and OsNLP6 are positioned near chromosome ends, suggesting potential telomeric regulatory roles (Figure 2A).
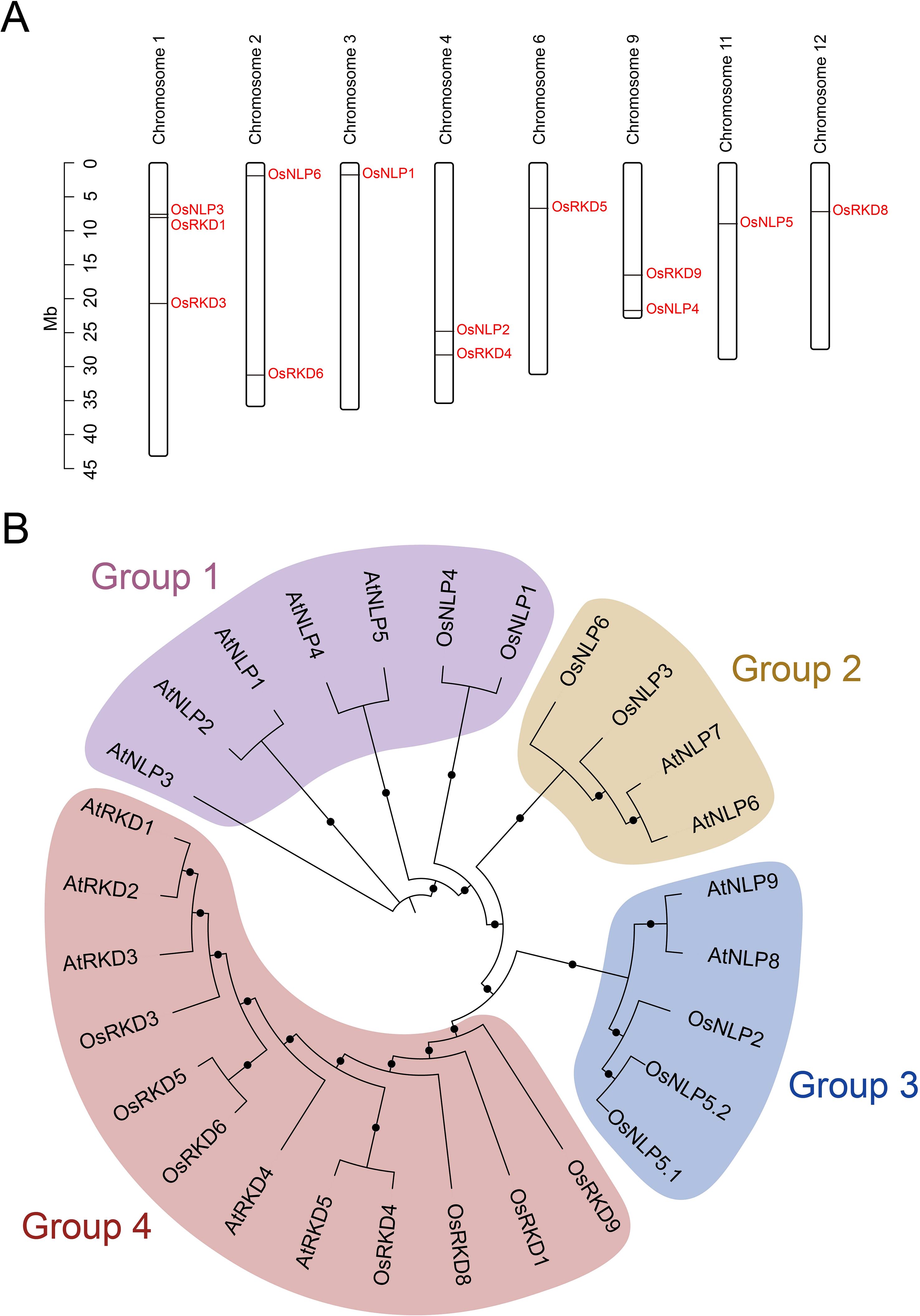
Figure 2. OsRWP-RK transcriptional factor family. (A) Chromosomal distribution of 13 OsRWP-RK genes (6 OsNLPs, 7 OsRKDs) across eight chromosomes. (B) Phylogenetic tree of 14 OsRWP-RK proteins, classified into four groups. Color bars denote phylogenetic groups: Group 1 (Purple), Group 2 (Brown), Group 3 (Blue), Group 4 (Red). .
A phylogenetic tree of RWP-RK proteins from Oryza sativa and Arabidopsis thaliana was constructed to investigate their evolutionary relationships (Figure 2B). The analysis revealed that RWP-RK proteins are classified into four major groups, each containing specific genes from both species. Group 1 contains AtNLP1-5 and OsNLP1, OsNLP4. Group 2 includes AtNLP6-7, OsNLP3, and OsNLP6. Group 3 clusters AtNLP8-9, OsNLP2, OsNLP5.1, and OsNLP5.2. Group 4 contains all OsRKDs and AtRKDs. The NLP subfamily is distributed across Groups 1-3, while Group 4 represents the RKD subfamily, with clear differentiation between the two subfamilies, indicating functional divergence during evolution. The presence of multiple members in each group suggests that gene duplication events contributed to the expansion of the gene family. Furthermore, the clustering of genes from both Arabidopsis and rice indicates a shared evolutionary origin, suggesting that the RWP-RK gene family is ancient and conserved across species.
3.2 Gene structures, conserved motifs, and protein domains
To elucidate structural diversity among OsRWP-RK genes, we analyzed their exon-intron organization, conserved motifs, and protein domain (Figures 1A, B). The genes consist of 3-6 introns and 4-7 exons, exhibiting variation in exon/intron length and arrangement. Additionally, structural differences between the two subfamilies may result in distinct regulatory patterns or expression profiles, potentially influencing their biological functions.
Motifs are small, conserved sequence fragments with functional significance in proteins. Motif analysis helps determine protein function. Using the MEME tool, three conserved motifs were identified in OsRWP-RK proteins (Figure 1C). Each protein contained 1-5 conserved motifs. The RKD subfamily retained the same functional motifs, while the NLP subfamily acquired two additional motifs, reflecting functional differences between the subfamilies.
Protein conserved domain analysis reveals core functional regions, evolutionary history, and biological functions of proteins. In this study, conserved domains were identified using the NCBI CDD database (Figure 1D). The results demonstrated that OsRKD subfamily members contain a single RWP-RK domain. In contrast, OsNLP proteins possess both the RWP-RK and PB1 domains (Figure 1D). This suggests that NLP subfamily members may adapt to specific functions by acquiring new domains, while RKD members retain their original domains to maintain basic functions. Additionally, the PB1 domain in the OsNLP subfamily is predominantly located at the C-terminus (Figure 1D), and the conserved spatial relationship between the RWP-RK and PB1 domains suggests a shared origin. Notably, OsRKD5 contains both a PLN03077 and a GBP_C domain (Figure 1D), indicating that the gene acquired new functions during evolution, enhancing rice’s adaptation to its environment.
3.3 Protein structure and evolutionary features
The 3D structure of a protein dictates its function, with features such as the catalytic site and ligand binding site determined by its spatial arrangement. We predicted the 3D structures of OsRWP-RK family proteins, including OsNLP and OsRKD subfamily members (Figure 3; Supplementary Figure 2). The models reveal differences in domain composition and overall folding. The OsNLP subfamily exhibits a relatively conserved 3D structure. Two large conserved domains (green and orange) are consistently located at the N-terminus (Figure 3), forming the structural core and occupying a substantial volume. Two smaller regions, located near the C-terminus (with OsNLP4 containing only one), are spatially arranged around the large regions (Figure 3). These domains are connected by flexible loops, suggesting potential cooperative function. Based on Figure 1D, the two small C-terminal domains are likely the RWP-RK domain (purple) and the PB1 domain (pink). Their spatial positioning suggests favorable orientation for DNA binding and nitrate signal perception (Konishi and Yanagisawa, 2013, 2019). In contrast, OsRKD proteins display greater structural variability, indicating potential functional diversity. Most OsRKD members exhibit a simplified architecture, often retaining only a single RWP-RK domain. This minimal structure may be sufficient for specific transcriptional regulation, such as the activation of genes involved in reproduction (Purwestri et al., 2023). Notably, OsRKD5 possesses a more complex structure, potentially due to the presence of additional domains—PLN03077 and GBP_C—as shown in Figure 1D. These structural characteristics provide valuable insights into the functional specialization and regulatory mechanisms of the OsRWP-RK protein family.
Figure 3. Predicted 3D structures of OsRWP-RK proteins. Helices are represented as spirals, β-pleated sheets as broad strips, and coils as thin loops. Each protein contains up to four domains, labeled from the N-terminus to the C-terminus in green, orange, purple, and pink, respectively.
Synteny analysis of gene families reveals their evolutionary history, functional conservation, and inter-species relationships, and provides insights into adaptation, environmental response, and functional evolution. In this study, synteny analysis of the OsRWP-RK family highlights the conservation of the RWP-RK genes in rice and Arabidopsis, shedding light on their evolutionary patterns (Figure 4). The analysis shows significant collinearity within the rice genome, such as between OsRKD6 (Chromosome 6) and OsRKD5 (Chromosome 2) (Figure 4A). Additionally, OsNLP2 (Chromosome 4) exhibits strong collinearity with a region on chromosome 2 (Figure 4A), suggesting gene duplication and evolutionary divergence, leading to the distribution and functional diversification of the OsRWP-RK family in rice. Furthermore, synteny between the rice and Arabidopsis RWP-RK gene families reveals four collinear gene pairs (Figure 4B), indicating that these families have retained conservation through evolution, likely originating from a common ancestor and undergoing gene recombination and species-specific divergence over time.
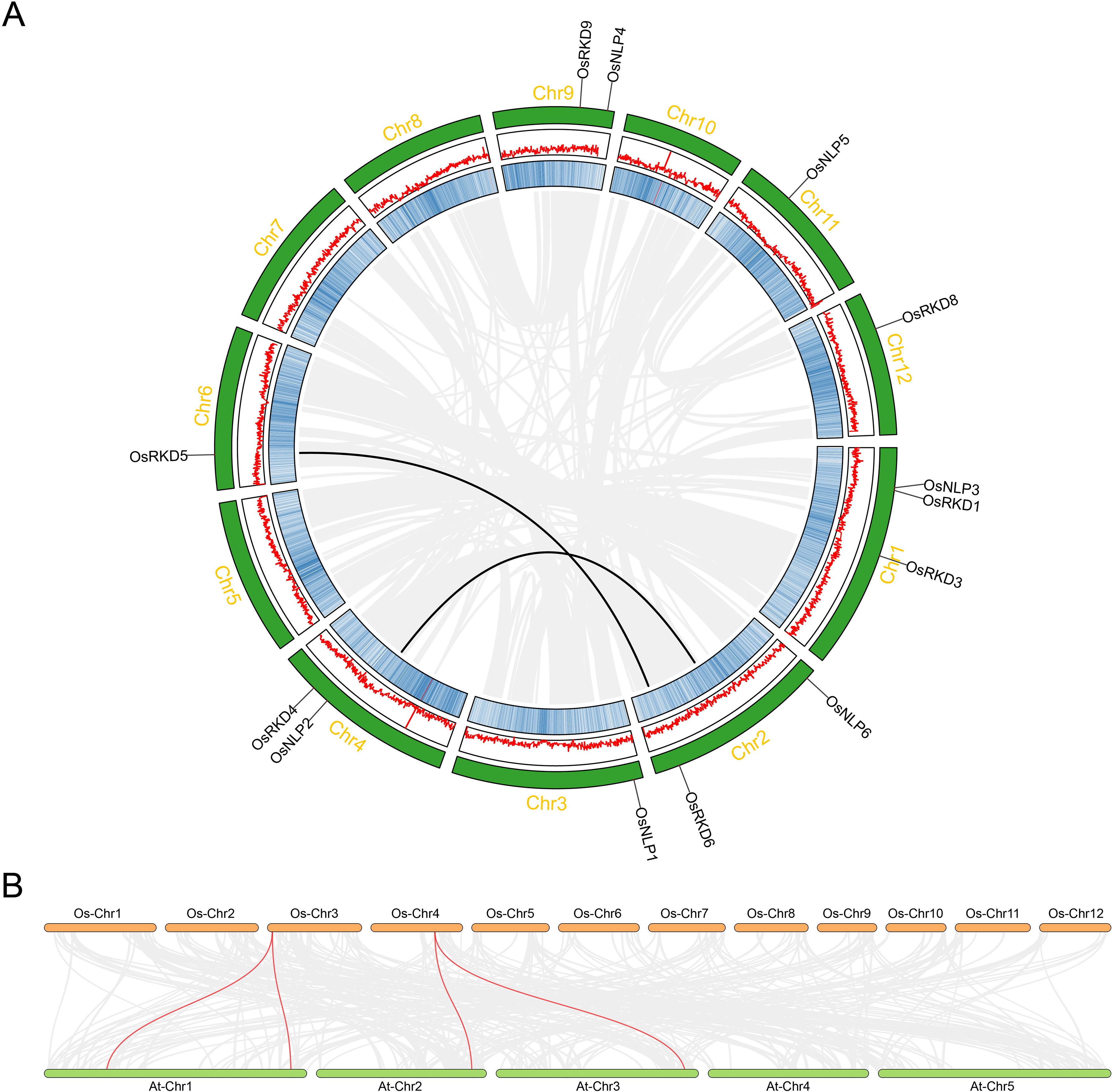
Figure 4. Collinearity and synteny analysis of OsRWP-RK genes. (A) Collinearity analysis of OsRWP-RK genes in Oryza sativa. Gray lines inside the circle represent all collinear gene pairs within the rice genome, while red lines highlight segmental duplication events within the OsRWP-RK family. Chromosome numbers are labeled outside the green boxes. Gene density is depicted using a blue heatmap in the inner boxes and red lines in the middle boxes. The chromosomal locations of OsRWP-RK genes are marked by black lines. (B) Synteny analysis between Oryza sativa and Arabidopsis thaliana. Gray lines indicate all collinear gene pairs between the two species, whereas red lines highlight collinear OsRWP-RK gene pairs.
3.4 Analysis of OsRWP-RK gene promoters
Cis-acting elements are crucial for gene regulation and can predict gene expression patterns under different tissue, organ, or environmental conditions. In this study, promoter analysis revealed diverse cis-regulatory elements in the 2 kb upstream regions of OsRWP-RK genes. Among the 80 detected elements, the CAAT-box and TATA-box are the most critical elements (Figure 5B; Supplementary Data Sheet S2). The CAAT-box enhances basal transcription by binding to CAAT-binding proteins, while the TATA-box facilitates the assembly of transcription initiation complexes, aiding RNA polymerase II in locating the transcription start site. Abundant cis-elements associated with hormone and abiotic stress responses were also identified (Figure 5A). Notable hormone-related cis-elements include ABRE (abscisic acid response), TGACG-motif and CGTCA-motif (jasmonic acid response), and as-1 (salicylic acid response). Abiotic stress-related elements include MYB and G-box (light response), MYC and MBS (drought response), MYC and LTR (low-temperature response), ARE (anaerobic response), and STRE (stress-responsive) (Figures 5A, B; Supplementary Data Sheet S2). All in all, the promoter region of the OsRWP-RK family contains various cis-acting elements that regulate gene expression and are involved in hormonal and stress responses.
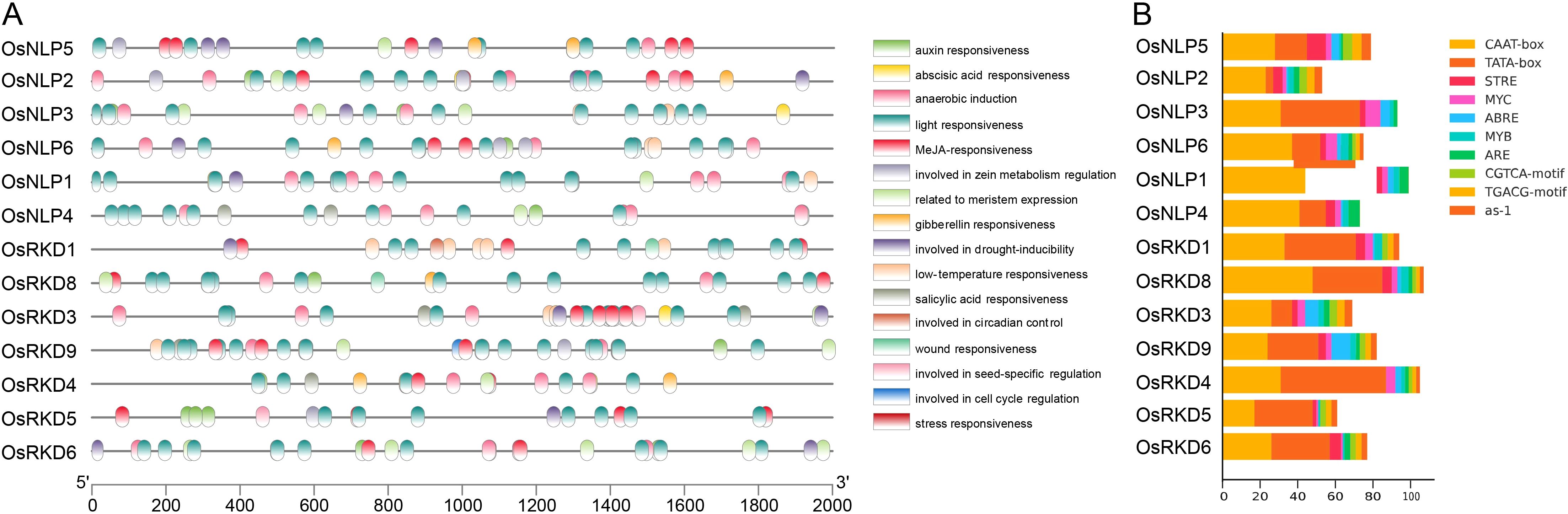
Figure 5. Cis-element analysis in OsRWP-RK gene promoters. (A) Distribution of cis-elements in OsRWP-RK gene promoters, with each element represented by a distinct color. (B) Top 10 cis-element count for each OsRWP-RK gene.
3.5 Transcription profiles of OsRWP-RK genes in different tissues
To investigate the transcription patterns of OsRWP-RK genes, we obtained a gene expression matrix from the Rice Gene Annotation Project (https://rice.uga.edu/expression.shtml; accessed on 7 Jan 2025) (Figure 6). Most NLPs are expressed across all tissues, suggesting their role in growth regulation in rice. (Figures 6A, B). OsNLP1-3, OsNLP5.1, and OsNLP6 are expressed at significantly higher levels in roots (TPM>10; Figures 6A, B), suggesting shared transcriptional regulation under variational nutrient conditions. Furthermore, the OsRKD subfamily is generally expressed at low levels across tissues (TPM<10; Figure 6A), while OsRKD1, OsRKD4, OsRKD6, OsRKD8, and OsRKD9 show relatively higher expression in inflorescences, exceeding levels in other tissues by at least two-fold (Figures 6A, B), further supporting subfamily-specific functional roles.
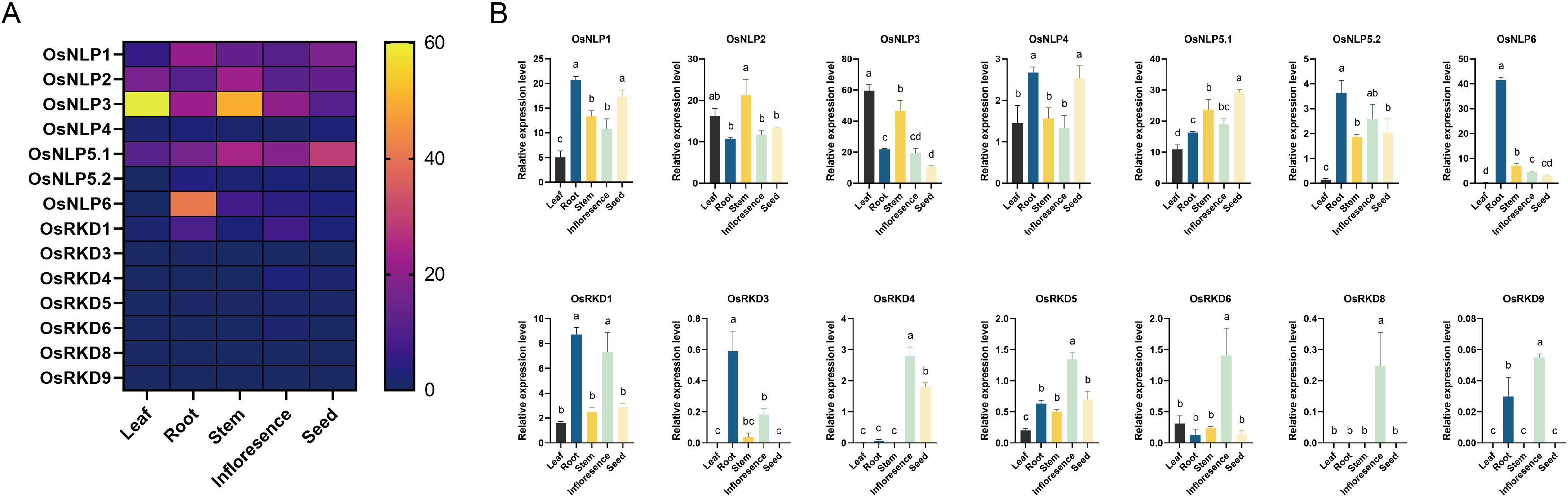
Figure 6. Expression patterns of OsRWP-RK genes in Oryza sativa. (A) Heatmap showing the expression levels of all OsRWP-RK genes across different tissues. (B) Bar graph depicting the expression levels of individual OsRWP-RK genes. Different letters indicate statistically significant differences between tissues (p < 0.05).
3.6 Nitrogen-induced expression dynamics of OsRWP-RK genes
To further explore the diversity of transcriptional responses among OsRWP-RK genes upon nitrogen re-supply, we analyzed temporal expression profiles of 10 representative genes using qRT-PCR across five time points (0, 10, 30, 60, and 120 minutes; Figure 7). Most genes responded rapidly to nitrogen, with peak expression occurring at 10 minutes. For example, OsNLP1 and OsRKD3 showed strong and transient upregulation, followed by a sharp decline to near-baseline levels by 30 minutes. These early-transient responders may act as immediate nitrogen sensors or signal initiators. OsNLP2, OsNLP4, and OsNLP5.1 were also initially induced at 10 minutes, but their transcript levels subsequently declined to below or near baseline. This pattern suggests a transient and tightly controlled regulatory role, potentially functioning in early signaling checkpoints. OsNLP5.2 and OsRKD1 maintained elevated expression levels throughout the time course, indicating prolonged involvement in transcriptional regulation during the nitrogen response. OsNLP6 exhibited a unique two-phase expression pattern, with an initial increase at 10 minutes, a transient decrease at 30 minutes, and a second rise at 60 minutes. This oscillatory behavior may reflect feedback regulation or involvement in multiple signaling phases. OsNLP3 displayed a gradual increase in expression, reaching peak levels at 60 minutes. This delayed response suggests a role downstream of initial nitrogen signaling, possibly in the regulation of secondary targets (Zhang et al., 2022).
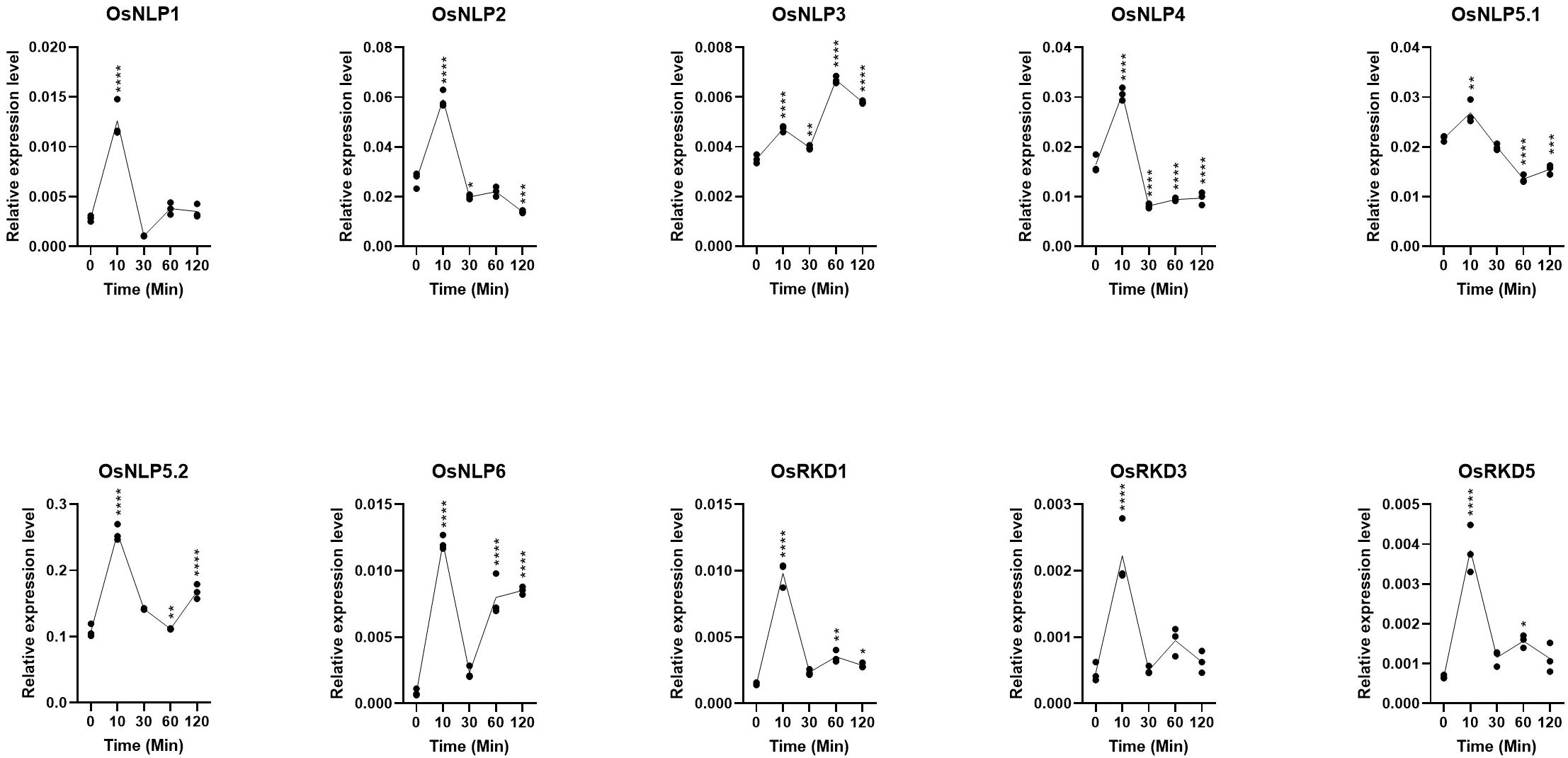
Figure 7. Fluctuations in OsRWP-RK gene expression after nitrogen-starved seedlings were reintroduced to a nitrogen-contain environment. (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001, ****p ≤ 0.0001).
4 Discussion
The discovery and functional analysis of RWP-RK proteins revolutionized our understanding of nitrogen signaling and gametophyte development across plants (Marchive et al., 2013; Chardin et al., 2014; Tedeschi et al., 2017). Rice, as a major cereal crop, represents an exemplary model system for dissecting these processes. Although previous studies reported 15 RWP-RK genes in rice (Zheng et al., 2024), our analysis revealed that the proteins of OsRKD7 (LOC_Os08g19820) and OsRKD10 (LOC_Os02g20530) in the Nipponbare cultivar lack RWP-RK domains. Thus, 13 OsRWP-RK genes were identified in this study (Table 1) and unevenly distributed across eight of the 12 chromosomes (Figure 2A). Additionally, 37 RWP-RK genes were identified in wheat (Kumar et al., 2018) and 12 ZmNLP genes in maize (Ge et al., 2018). Compared to wheat and maize, the RWP-RK gene family in Oryza sativa underwent a conservative expansion. Moreover, NLP subfamily proteins generally have longer CDS and higher molecular weights (Table 1; Figure 1), similar to those in wheat and maize (Ge et al., 2018; Kumar et al., 2018).
The RWP-RK transcription factor family is divided into two subfamilies: RKDs and NLPs. RKDs contain a conserved RWP-RK domain (Yin et al., 2020). NLPs, in contrast, possess both RWP-RK and PB1 domains, along with a GAF-like structure domain (Schauser et al., 2005). All OsNLPs possess PB1 domains—facilitating protein–protein interactions—and RWP-RK domains, indicating roles in transcriptional complexes responsive to nitrogen (Figure 1D). In contrast, RKDs retain only the RWP-RK domain, possibly reflecting specialized roles in gametophyte development (Figure 1D). OsRKD5 uniquely contains PLN03077 and GBP_C domains, suggesting an evolutionary innovation that may broaden its regulatory scope (Figure 1D).
Gene duplication is a key driver of gene functional diversification and species evolution (Bowers et al., 2003). In eukaryotes, duplication events occur primarily through tandem or segmental duplication (Cannon et al., 2004). Studies suggest that the NLP subfamily in Arabidopsis evolved via segmental rather than tandem duplication (Schauser et al., 2005). In this study, we identified two segmentally duplicated gene pairs (Figure 4A), suggesting that segmental duplication also contributed to the expansion of OsNLP genes. Synteny analysis of the rice and Arabidopsis RWP-RK gene families reveals four collinear gene pairs (Figure 4B), suggesting they share a common ancestor and have conserved through evolution.
Cis-acting elements are crucial for gene regulation and can predict gene expression patterns under different tissue, organ, or environmental conditions. We identified 80 types of cis-acting elements in OsRWP-RK promoters (Supplementary Data Sheet S2), which may contribute to the differential expression of RWP-RK genes across tissues. However, some genes with diverse cis-acting elements exhibited consistently low expression in all tested tissues (Figures 5, 6), suggesting that gene expression is influenced by additional factors beyond promoter elements. Epigenetic modifications, such as DNA methylation, may contribute to this low expression (Zhang et al., 2018). Furthermore, an analysis combining gene expression patterns and promoter cis-element composition revealed that most genes showed higher expression in leaves and roots (Figures 5, 6A), potentially due to the abundance of light-responsive elements and stress-responsive OsRWP-RK promoters (Figure 5). Additionally, the presence of numerous hormone-responsive elements (Figure 5) raises the intriguing possibility that these elements mediate crosstalk between the RWP-RK gene family and hormone signaling pathways, warranting further investigation.
In Oryza sativa, 13 OsRWP-RK genes (14 transcripts) were identified across leaves, roots, stems, inflorescences, and seeds. OsNLP genes exhibited broad expression, particularly in roots, stems, and leaves (Figure 6), supporting a broad regulatory role in various physiological processes, such as nitrogen uptake, transport, and redistribution to regulate plant growth and development (Liu et al., 2018). In contrast, most OsRKD members are expressed higher in inflorescences than elsewhere, exceeding other tissues by at least twofold. Previous research has found that OsRKD3 in Indonesian black rice (Cempo Ireng), which is resistant to somatic embryogenesis, can induce somatic embryo formation (Purwestri et al., 2023). Similarly, the RKD-type RWP-RK transcription factor Shohai1 is essential for embryo and endosperm development in maize (Mimura et al., 2018). This indicated that RKDs contribute to gametophyte development.
Furthermore, the expression levels of all OsNLP genes and OsRKD1, OsRKD3, and OsRKD5 fluctuated upon transferring nitrogen-starved seedlings to a nitrogen-sufficient environment. This suggests that OsRWP-RK genes play pivotal roles in nitrate response and nitrogen starvation signaling. Given the evolutionary expansion and sub-functionalization of the OsRWP-RK subfamily, these genes likely employ diverse molecular mechanisms to respond to external nitrogen fluctuations.
In summary, RWP-RK genes in Oryza sativa show diverse expression patterns, with OsNLP genes primarily expressed in nutritive organs and OsRKD genes involved in gametogenesis and embryogenesis. This study offers new insights into the role of RWP-RKs in seed development and nitrogen response.
5 Conclusion
In this study, we performed a genome-wide identification of the RWP-RK family in Oryza sativa, analyzed their expression across five tissues, and examined their transcriptional responses to nitrogen availability. A total of 13 RWP-RK genes were identified, including OsNLP5, which encodes two proteins containing RWP-RK domains. These genes are classified into two subfamilies—OsNLP (six genes) and OsRKD (seven genes)—distributed across eight of the 12 rice chromosomes.
Significant structural differences exist between the two subfamilies: OsNLP genes are longer in gene and coding sequence (CDS) length, protein size, and molecular weight, with more complex 3D structures than OsRKD genes. While OsRKD members retain a single RWP-RK domain, OsNLP members possess both RWP-RK and PB1 domains, suggesting functional specialization. Promoter analysis revealed diverse cis-acting elements associated with hormone and stress responses.
Expression analysis indicated that OsNLP genes exhibited broad expression, supporting a broad regulatory role in various physiological processes. In contrast, OsRKD genes generally showed low expression but were highly expressed in inflorescences, implying involvement in gametophyte development. Additionally, all NLP genes, along with OsRKD1, OsRKD3, and OsRKD5, exhibited similar overall expression fluctuations with subtle differences upon nitrogen reintroduction, suggesting diverse molecular responses to nitrate availability.
In summary, this genome-wide characterization of RWP-RK transcription factors in Oryza sativa provides a theoretical framework for future research on the regulatory mechanisms of germ cell differentiation and embryo development. Moreover, this study could contribute to the development of rice varieties with improved nitrogen use efficiency, which is crucial for sustainable agriculture.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. ZG: Software, Writing – original draft. SG: Writing – review & editing. LX: Software, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Science Foundation of China (Grant No. 32130095).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1597029/full#supplementary-material
Supplementary Figure 1 | The differences in gene and protein structures between the OsNLP and OsRKD subfamilies. The OsNLP subfamily is larger than the OsRKD subfamily in CDS length (A), protein length (B), protein molecular weight (C), and protein domain number (D).
Supplementary Figure 2 | Predicted 3D structures of OsRWP-RK proteins. Helices are represented as spirals, β-pleated sheets as broad strips, and coils as thin loops. AlphaFold produces a per-residue model confidence score (pLDDT) between 0 and 100. Some regions below 50 pLDDT may be unstructured in isolation (dark blue, pLDDT > 90, light blue, 90 > pLDDT > 70, yellow, 70 > pLDDT > 50, orange, pLDDT < 50).
References
Alfatih, A., Wu, J., Zhang, Z. S., Xia, J. Q., Jan, S. U., Yu, L. H., et al. (2020). Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 71, 6032–6042. doi: 10.1093/jxb/eraa292
Amin, N., Ahmad, N., Khalifa, M. A. S., Du, Y. Y., Mandozai, A., Khattak, A. N., et al. (2023). Identification and molecular characterization of RWP-RK transcription factors in soybean. Genes 14. doi: 10.3390/genes14020369
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chardin, C., Girin, T., Roudier, F., Meyer, C., and Krapp, A. (2014). The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65, 5577–5587. doi: 10.1093/jxb/eru261
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chou, K. C. and Shen, H. B. (2010). Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5. doi: 10.1371/journal.pone.0011335
Durand, M., Brehaut, V., Clement, G., Kelemen, Z., Mace, J., Feil, R., et al. (2023). The Arabidopsis transcription factor NLP2 regulates early nitrate responses and integrates nitrate assimilation with energy and carbon skeleton supply. Plant Cell 35, 1429–1454. doi: 10.1093/plcell/koad025
Ge, M., Liu, Y., Jiang, L., Wang, Y., Lv, Y., Zhou, L., et al. (2018). Genome-wide analysis of maize NLP transcription factor family revealed the roles in nitrogen response. Plant Growth Regul. 84, 95–105. doi: 10.1007/s10725-017-0324-x
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guan, P. Z., Ripoll, J. J., Wang, R. H., Vuong, L., Bailey-Steinitz, L. J., Ye, D. N., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. United States America 114, 2419–2424. doi: 10.1038/s41467-017-01187-x
Hong, W. Y., Chen, Y. J., Huang, S. H., Li, Y. Z., Wang, Z. M., Tang, X. R., et al. (2022). Optimization of nitrogen-silicon (N-Si) fertilization for grain yield and lodging resistance of early-season fragrant rice under different planting methods. Eur. J. Agron. 136. doi: 10.1016/j.eja.2022.126508
Hu, B., Jiang, Z. M., Wang, W., Qiu, Y. H., Zhang, Z. H., Liu, Y. Q., et al. (2019). Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Jagadhesan, B., Sathee, L., Meena, H. S., Jha, S. K., Chinnusamy, V., Kumar, A., et al. (2020). Genome wide analysis of NLP transcription factors reveals their role in nitrogen stress tolerance of rice. Sci. Rep. 10. doi: 10.3389/fpls.2020.574176
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1105/tpc.16.00500
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Konishi, M. and Yanagisawa, S. (2013). An NLP-binding site in the 3’ flanking region of the nitrate reductase gene confers nitrate-inducible expression in Arabidopsis thaliana (L.) Heynh. Soil Sci. Plant Nutr. 59, 612–620. doi: 10.1080/00380768.2013.809602
Konishi, M. and Yanagisawa, S. (2019). The role of protein-protein interactions mediated by the PB1 domain of NLP transcription factors in nitrate-inducible gene expression. BMC Plant Biol. 19. doi: 10.1186/s12870-019-1692-3
Koszegi, D., Johnston, A. J., Rutten, T., Czihal, A., Altschmied, L., Kumlehn, J., et al. (2011). Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 67, 280–291. doi: 10.1104/pp.111.176867
Kumar, A., Batra, R., Gahlaut, V., Gautam, T., Kumar, S., Sharma, M., et al. (2018). Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PloS One 13. doi: 10.1371/journal.pone.0208409
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, P., Tian, Y., Yang, K., Tian, M., Zhu, Y., Chen, X., et al. (2024). Mechanism of microbial action of the inoculated nitrogen-fixing bacterium for growth promotion and yield enhancement in rice (Oryza sativa L.). Advanced Biotechnol. 2, 32. doi: 10.1007/s44307-024-00038-4
Liu, M., Chang, W., Fan, Y. H., Sun, W., Qu, C. M., Zhang, K., et al. (2018). Genome-wide identification and characterization of NODULE-INCEPTION-like protein (NLP) family genes in brassica napus. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19082270
Liu, K. H., Liu, M. H., Lin, Z. W., Wang, Z. F., Chen, B. Q., Liu, C., et al. (2022). NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 377, 1419–1425. doi: 10.1126/science.add1104
Liu, K. H., Niu, Y. J., Konishi, M., Wu, Y., Du, H., Chung, H. S., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316. doi: 10.1038/nature22077
Liu, T., Ren, T., White, P. J., Cong, R. H., and Lu, J. W. (2018). Storage nitrogen co-ordinates leaf expansion and photosynthetic capacity in winter oilseed rape. J. Exp. Bot. 69, 2995–3007. doi: 10.1093/jxb/ery134
Marchive, C., Roudier, F., Castaings, L., Bréhaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4. doi: 10.1038/ncomms2650
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Mimura, M., Kudo, T., Wu, S., McCarty, D. R., and Suzuki, M. (2018). Autonomous and non-autonomous functions of the maize Shohai1 gene, encoding a RWP-RK putative transcription factor, in regulation of embryo and endosperm development. Plant J. 95, 892–908. doi: 10.1111/tpj.13996
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Purwestri, Y. A., Lee, Y. S., Meehan, C., Mose, W., Susanto, F. A., Wijayanti, P., et al. (2023). RWP-RK Domain 3 (OsRKD3) induces somatic embryogenesis in black rice. BMC Plant Biol. 23. doi: 10.1186/s12870-023-04220-z
Sakuraba, Y., Zhuo, M. N., and Yanagisawa, S. (2022). RWP-RK domain-containing transcription factors in the Viridiplantae: biology and phylogenetic relationships. J. Exp. Bot. 73, 4323–4337. doi: 10.1093/jxb/erac229
Schauser, L., Wieloch, W., and Stougaard, J. (2005). Evolution of NIN-Like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60, 229–237. doi: 10.1007/s00239-004-0144-2
Tedeschi, F., Rizzo, P., Rutten, T., Altschmied, L., and Bäumlein, H. (2017). RWP-RK domain-containing transcription factors control cell differentiation during female gametophyte development in Arabidopsis. New Phytol. 213, 1909–1924. doi: 10.1111/nph.14293
Tegeder, M. and Masclaux-Daubresse, C. (2018). Source and sink mechanisms of nitrogen transport and use. New Phytol. 217, 35–53. doi: 10.1111/nph.14876
Varadi, M., Bertoni, D., Magana, P., Paramval, U., Pidruchna, I., Radhakrishnan, M., et al. (2023). AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375. doi: 10.1093/nar/gkac1031
Waki, T., Hiki, T., Watanabe, R., Hashimoto, T., and Nakajima, K. (2011). The arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr. Biol. 21, 1277–1281. doi: 10.1016/j.cub.2011.07.001
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Wang, M. Y., Hasegawa, T., Beier, M., Hayashi, M., Ohmori, Y., Yano, K., et al. (2021). Growth and nitrate reductase activity are impaired in rice osNLP4 mutants supplied with nitrate. Plant Cell Physiol. 62, 1156–1167. doi: 10.1093/pcp/pcab035
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. 2-D Proteome Anal. Protoc. 112, 531–552.
Wu, J., Zhang, Z. S., Xia, J. Q., Alfatih, A., Song, Y., Huang, Y. J., et al. (2021). Rice NIN-LIKE PROTEIN 4 plays a pivotal role in nitrogen use efficiency. Plant Biotechnol. J. 19, 448–461. doi: 10.1111/pbi.13475
Yan, D. W., Easwaran, V., Chau, V., Okamoto, M., Ierullo, M., Kimura, M., et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7. doi: 10.1038/ncomms13179
Yan, D. W. and Nambara, E. (2023). Conserved and unique functions of NIN-like proteins in nitrate sensing and signaling. Plant Sci. 336. doi: 10.1016/j.plantsci.2023.111842
Yi, Y. K., Peng, Y. Q., Song, T., Lu, S. Q., Teng, Z. N., Zheng, Q., et al. (2022). NLP2-NR module associated NO is involved in regulating seed germination in rice under salt stress. Plants-Basel 11. doi: 10.3390/plants11060795
Yin, M., Zhang, Z., Xuan, M., Feng, H., Ye, W., Zheng, X., et al. (2020). Conserved subgroups of the plant-specific RWP-RK transcription factor family are present in oomycete pathogens. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01724
Zhang, Q., Liang, Z., Cui, X., Ji, C., Li, Y., Zhang, P., et al. (2018). N6-methyladenine DNA methylation in japonica and indica rice genomes and its association with gene expression, plant development, and stress responses. Mol. Plant 11, 1492–1508. doi: 10.1016/j.molp.2018.11.005
Zhang, Z. S., Xia, J. Q., Alfatih, A., Song, Y., Huang, Y. J., Sun, L. Q., et al. (2022). Rice NIN-LIKE PROTEIN 3 modulates nitrogen use efficiency and grain yield under nitrate-sufficient conditions. Plant Cell Environ. 45, 1520–1536. doi: 10.1111/pce.14294
Zhang, D., Yu, H., Yu, X., Yang, Y., Wang, C., Wu, K., et al. (2024). Mechanisms underlying the interactions and adaptability of nitrogen removal microorganisms in freshwater sediments. Advanced Biotechnol. 2, 21. doi: 10.1007/s44307-024-00028-6
Keywords: RWP-RK, NLP proteins, RKD proteins, transcription factors, nitrogen starvation, rice
Citation: Du M, Guo Z, Gong S and Xu L (2025) Genome-wide analysis of RWP-RK transcription factor family reveals its roles in nitrogen response in rice (Oryza sativa). Front. Plant Sci. 16:1597029. doi: 10.3389/fpls.2025.1597029
Received: 20 March 2025; Accepted: 20 August 2025;
Published: 08 September 2025.
Edited by:
Yogeshwar Vikram Dhar, Ruhr University Bochum, GermanyReviewed by:
Yang Zhiquan, Guangzhou University, ChinaMo-Xian Chen, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Du, Guo, Gong and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Du, ZHVtZWkxN0BtYWlscy51Y2FzLmVkdS5jbg==
 Mei Du
Mei Du Ziwen Guo3
Ziwen Guo3