- 1Guangzhou Key Laboratory of Crop Gene Editing, Guangdong Provincial Key Laboratory of Plant Adaptation and Molecular Design, Innovative Center of Molecular Genetics and Evolution, School of Life Sciences, Guangzhou University, Guangzhou, China
- 2Institute of Medical Plant Physiology and Ecology, School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
Cellulases are a crucial class of enzymes involved in cellulose synthesis and metabolism, significantly contributing to plant growth, development, and organ abscission. The role of Glycosyl hydrolase family 9 (GH9), a major gene family encoding cellulase, remains poorly elucidated in soybean. In this experiment, we identified 43 non-redundant GmGH9 genes in soybean through systematic genome-wide analysis. The physicochemical properties of GmGH9 proteins exhibit variability. Phylogenetic investigations revealed that class B constitutes the predominant evolutionary branch. The GmGH9B/C members display complex splicing patterns. GmGH9As contain typical transmembrane structural domains, while GmGH9Cs uniquely includes the carbohydrate-binding module 49 (CBM49) and signal peptide. Furthermore, we identified 13 distinct types of functional motifs, with light-responsive elements being predominant. Expression profiling of the GmGH9s in soybean revealed spatiotemporal and stress-regulated dynamics across organs, ethylene treatments, and photoperiodic conditions, especially for GmGH9A9 and GmGH9B19. Multi-species collinearity analysis of GH9 genes suggested that GmGH9A2 and GmGH9C4 exhibited greater conservation in pea, tomato, and soybean, which are distinguished by fruit abscission. Additional correlations between the haplotypes of GmGH9A2 and GmGH9C4 and yield-related traits indicated that soybean experienced selected pressure during domestication, resulting in a reduction in their genetic diversity.
1 Introduction
Plant organ abscission is a genetically organized process mediated by the coordinated activity of hydrolytic enzymes and phytohormones, which enable the specific destruction of cell wall constituents at defined abscission zone (AZ). This intricate process requires the concerted activity of various enzymes and signaling molecules, including cellulases, hemicellulases, pectinases (both polygalacturonases and pectinesterases), xylanases, expansins, peroxidases, and lipoxygenases. Notably, cellulases, which constitute one of the three biggest categories of industrial enzymes—catalyze the hydrolysis of β−1,4‐glycosidic bonds in cellulose, consequently compromising the mechanical integrity of the separation layer (Tranbarger et al., 2017).
Cellulase family comprises multiple members that can be categorized into three principal groups based on their mode of action: endo−β−1,4−glucanases (EGases), exo−β−1,4−glucanases, and β−glucosidases. The principal plant cellulases investigated to date are conventional β−1,4−glucanases, which belong to the glycosyl hydrolase family 9 (GH9). This multigene family is further divided into three distinct structural subclasses: A, B, and C. Subclass A encompasses membrane‐anchored isoforms (with or without a cytosolic domain), subclass B comprises secreted isoforms and subclass C includes secreted isoforms that contain a carbohydrate−binding module (CBM) (Urbanowicz et al., 2007). Sequence homology analyses have identified 25 GH9 gene family members in rice (Oryza sativa), Arabidopsis, and Populus (Libertini et al., 2004; Xie et al., 2013), and 24, 32, 42, and 74 GH9 genes in tomato, citrus, tobacco, and wheat, respectively (Du et al., 2015; Luo et al., 2022a; Deng et al., 2024).
The GH9 family is extensively documented for its essential functions in various biological processes, including plant organ abscission, fruit maturation, stress adaption, and developmental regulation (Robert et al., 2005; Flors et al., 2007; Lin et al., 2023; Meng et al., 2024). Members of this gene family generally display tissue-specific expression patterns influenced by environmental cues and hormone signaling. In tomato (Solanum lycopersicum), six members (SlCel1-6) of the GH9 family have been characterized, with SlCel5 exhibiting ethylene-responsive upregulation in AZ while maintaining constant expression in stem tissues (del Campillo and Bennett, 1996). Notably, the synergistic interaction between expansin SlExp1 and endoglucanase SlCel2 mediates cell wall disassembly during fruit softening (Su et al., 2024). Three members of the GH9A subfamily have been parsed in Spathiphyllum, SpGH9A1, SpGH9A2, and SpGH9A3, among which the differential expression of the SpGH9A3 gene in the leaves of Spathiphyllum may affect the cellulase activity, and consequently, the cellulose content of the leaves at different stages of expansion, which ultimately leads to the differences in leaf morphology (Yang et al., 2024a).
GH9s, express in plant pedicels, fruit stalks, and petioles, are regulated by a variety of hormones and light signals, and these regulatory mechanisms are key components of GH9 genes involved in plant organ abscission, as well as environmental adaptation. In litchi (Litchi chinensis), the transcription factor LcHB2 directly activates GH9 paralogs LcCEL2 and LcCEL8 within the fruit AZ, triggering sequential cellulase activation, cell wall degradation, and ultimately the organ abscission process (Li et al., 2019). Arabidopsis research reveals that AtCEL6 regulates silique dehiscence by coordinating cell differentiation timing in the valve and separation layer (He et al., 2018), while ATGH9A1/KORRIGAN1 participates in both cellulose biosynthesis and cellular elongation processes (Lane et al., 2001). The VaGH9A1 and VaGH1B were expressed in the AZ of blueberry fruit (Vaccinium ashei) and decreased as the fruit matured, whereas the expression of VaGH10B and VaGH10c continued to increase, which is thought to be a sign that the former is involved in the pre-formation stage of cell layer differentiation in the AZ and the latter is involved in the segregation stage where cell wall breakdown is active (Wang et al., 2023c). Additionally, phytohormonal regulation of cellulase activity has been demonstrated in bean (Phaseolus vulgaris), where ethylene and auxin signaling pathways converge to modulate BAC gene expression (Tucker et al., 2002). Recent evidence suggests that in tomato, SlIDL6 signaling may orchestrate pedicel abscission through transcriptional activation of cell wall hydrolases including TAPG1, TAPG4, and CEL2 (Li et al., 2021). SlGH9–15 has been identified as a key factor in the process of fruit cracking in tomatoes, which has been demonstrated to be activated by various hormones, such as ethylene and abscisic acid (ABA), as well as by abiotic stresses (Lin et al., 2023). Wheat GH9 gene family, which are rich in hormone-responsive and light-responsive elements, mediate the interaction between the jasmonic acid and ABA pathways to govern another development, with seven members identified as key regulators of cellulose levels via light and phytohormones, crucial for pollen fertility and anther dehiscence (Luo et al., 2022b).
Soybean (Glycine max) is an important source of vegetable fats and proteins. However, the higher flower and pod abscission rates limit soybean yields, and the process is strongly influenced by the environment, including temperature, light intensity, disease, water stress, and nutrient supply, among others (Estornell et al., 2013), while the GH9 family members are involved in those processes. Suppression of soybean GmCel7, a soybean homologue of AtGH9B2, showed increased resistance to soybean cyst nematode (Heterodera glycines) in soybean root (Woo et al., 2014). A hard-seed allele qHS1 from Glycine soja (G. soja) was identified as GmGH9B8 for increasing the amount of β-1,4-glucans in the outer layer of palisade cells of the seed (Jang et al., 2015). Using qHS1 loci for improvement of the modern cultivar “Tachinagaha” making its seed coat less permeable and more resistant to cracking. Similar to tomato, increased cellulase activity in the soybean AZ of pedicels and petioles may suggest that the GH9s family is also involved in the progress of organ abscission in soybean (Kemmerer and Tucker, 1994; Tucker et al., 2007). However, there are few reports about the roles of GH9s participating in the regulation of organic development in soybeans.
With the construction of the soybean pan-genome, the attainment of comprehensiveness, diversity, and in-depth functional resolution for soybean gene research is achieved (Liu et al., 2020). This study presents a comprehensive analysis of the chromosomal localization, phylogenetic relationships, gene structure, promoter regulatory motifs, and tissue expression profiles of the soybean cellulase gene family, characterized by the GH9 structural domain, at the genome-wide scale. We predicted the GmGH9s that are specifically expressed and responsive to ethylene hormone signaling in the soybean AZ to further explore the potential GH9 family members involved in soybean organ abscission and ultimately constructed the haplotypes of GmGH9A2 and GmGH9C4. Our study establishes a theoretical foundation for further understanding comprehension of GmGH9s’ function and offers a new perspective for investigating the mechanism of soybean organ abscission.
2 Materials and methods
2.1 GmGH9s identification and physicochemical properties analysis
Glycine max (Williams 82, Wm82) reference genome (Wm82 V6), along with its genomic sequences, protein annotations, and structural features in GFF3 format, was retrieved from SoyBase (Grant et al., 2010). Members of the GH9 family candidate were identified through a dual screening strategy. Homology-based screening: blastp alignments (E-value cutoff <1e-5) against Arabidopsis GH9 family members. Domain validation: Hidden Markov Model (HMM) profiling with the PF00759 seed alignment (Pfam v35.0) (Mistry et al., 2021) via HOMER v5.1 (Eddy, 2011), followed by SMART (Letunic et al., 2021) and CDD v3.21 (Wang et al., 2023b) database analyses to confirm GH9 specific catalytic domains (cd00254, glycosyl hydrolase family 9). Biochemical characteristics of GmGH9 proteins, such as molecular weight (MW), theoretical isoelectric point (pI), and grand average of hydropathicity (GRAVY), were computed using ExPASy ProtParam (Gasteiger et al., 2003) with default settings.
2.2 Chromosome localization and collinearity analysis of GmGH9s
Genomic coordinates of validated GmGH9 genes were extracted from the GFF3 file and visualized using TBtools-II v2.156 (Chen et al., 2023) with the “Advanced Chromosome Map” module. Whole-genome tandem duplication events were detected through MCScanX v1.0.0 (Wang et al., 2024) with default parameters (blastp E-value <1e-10, match score >50). Genomic and GFF3 files for pea, tomato, Arabidopsis, and maize were downloaded from the Ensembl plant database (release 60) (Bolser et al., 2016). Twelve soybean pangenomes (SoyW02, SoyW03, SoyL02, SoyL03, SoyC02, SoyL05, ZH13, SoyC11, SoyC12, SoyL09, and SoyC14) and their corresponding GFF3 files were downloaded from the SoyMD database (Yang et al., 2024b). Segmental duplication analysis through synteny blocks identified using JCVI v1.5.1 utilities (Tang et al., 2024). Ka/Ks ratios were calculated via KaKs_Calculator 3.0 (Zhang, 2022) to characterize evolutionary selection pressure.
2.3 Phylogenetic tree analysis
A multiple sequence alignment of soybean and other model plants (Arabidopsis [dicotyledonous] and maize [monocotyledonous]) was performed using the Clustalw algorithm in MEGA X (Kumar et al., 2018). The evolution analysis was performed using the maximum likelihood with default parameters, and a bootstrap of 1000 was applied. Finally, the evolutionary tree was visualized on the Evolview3 (Subramanian et al., 2019) webserver, with GH9s labeled accordingly.
2.4 Gene structure and conserved motif analysis
Exon-intron structures of GmGH9 genes were extracted from the GFF3 file, and gene structure schematics were visualized using TBtools-II v2.156. Conserved protein motifs were identified using the MEME Suite v5.5.2 (Bailey et al., 2006) with parameters set to a maximum of 4 motifs, E-value<1e−5, and motif widths ranging from 6 to 50 residues. The distribution of conserved motifs was mapped onto the phylogenetic tree using TBtools-II v2.156 to elucidate structural conservation across the GmGH9 family.
2.5 Promoter and cis-regulatory element analysis
Promoter sequences (2000 bp upstream of the transcription start site) of GmGH9 genes were retrieved from the soybean genome (Wm82.v6). Cis-regulatory elements were predicted using PlantCARE v2023 (Lescot et al., 2002) and categorized into functional groups (e.g., light-responsive, hormone-related, and stress-inducible elements). The distribution of these elements was visualized in R v4.3.1 using the ggplot2 package, with color-coded annotations highlighting regulatory diversity among subfamilies.
2.6 Haplotype analysis
SNP data of 4414 soybean accessions were obtained from the SoyMD database (Yang et al., 2024b). Yield trait data were obtained from the SoyOmics database (Liu et al., 2023). Haplotype blocks for GmGH9 genes were analyzed using geneHapR (Zhang et al., 2023), with haplotypes containing fewer than 10 individuals being excluded from the analysis. With a 20 kb window and 2 kb step, VCFtools v0.1.16 (Danecek et al., 2011) was used to calculate the nucleotide diversity (π) and fixation index (Fst) values. The top 10% of the π and Fst ratio (G. soja vs. landrace and landrace vs. cultivar) for the corresponding chromosome was used as the selective sweep threshold.
2.7 Material planting
Soybean variety Wm82 was selected and cultivated in a greenhouse. The growth conditions included a photoperiod of 16 hours (h) light/8 h dark, maintained at a temperature of 26°C. Fifteen days after flowering, soybean plants received a treatment of 100 mg/L ethephon, with water treatment serving as control. After 24 h, tissue samples were designated as ethylene-treated stem (ESt), ethylene-treated leaf (EL), ethylene-treated petiole (EYb), ethylene-treated flower (EF), ethylene-treated flower AZ (EFaz), ethylene-treated pod AZ (EPaz), ethylene-treated pod peel (EPp), ethylene-treated seed coat (ESc), and ethylene-treated embryo (ESe). Parallel samples were harvested from water-treated plants as control, designated as stem (St), leaf (L), petiole (Yb), flower (F), flower AZ (Faz), pod AZ (Paz), pod peel (Pp), seed coat (Sc), and embryo (Se). All samples were frozen in liquid nitrogen and stored at -80°C.
2.8 Analysis of the expression profile and RT-qPCR
Data on multi-organ expression (NCBI BioProject ID: PRJNA442256), AZ and NAZ in situ expression following ethylene treatment (Kim et al., 2016), and expression data from photoperiod conversion [plants were grown in the greenhouse under short-day (SD) (10 hours of light, 6:45–16:45) and long-day (LD) (16 hours of light, 6:45–22:45) conditions at 25°C and were sampled every four hours at six time points, T1-T6 (6:30, 10:30, 14:30, 18:30, 22:30 and 2:30). For a shift (Sh) experiment, plants were first grown under LD for three weeks and then transferred to SD for 5 days] (Wu et al., 2014) were downloaded from the publicly accessible transcriptome dataset in the NCBI database. Raw data was filtered using Trimmomatic software (Bolger et al., 2014), followed by alignment of clean data to the reference genome Wm82 v6 using STAR (Dobin et al., 2013). The counts of all genes were ultimately converted to gene expression values in transcripts per million (TPM).
RNA was isolated via the TRIzol method (Rio et al., 2010), and the concentration and integrity were evaluated with a NanoDrop spectrophotometer. RNA integrity was detected by 1.2% agarose gel electrophoresis. cDNA was synthesized using HiScript III RT SuperMix (Vazyme International LLC, Nanjing, China). Primers for real-time fluorescence quantitative PCR (RT-qPCR) were designed by Primer-BLAST online (Ye et al., 2012), and listed in Supplementary Table S6. RT-qPCR was performed using SYBR Green qPCR Mix (Thermo Fisher Scientific, Waltham, USA), with 40 cycles set. GmTublin was set as the internal reference gene, and gene expression level was calculated using the 2^(-ΔΔCt [cycle threshold]) method (Livak and Schmittgen, 2001).
2.9 Subcellular localization
GmGH-GFP (green fluorescent protein) fused expression cassettes were inserted into the pCAMBIA1302 vector with restriction sites (HindIII and KpnI), subsequently transformed into GV3101(pSoup-P19) Agrobacterium strain, and used to infect tobacco (Nicotiana benthamiana) leaves via injection. GFP fluorescence signals were detected after 48 h using a confocal laser scanning microscope (Zeiss LSM 880). AtPIP2A, connected to mCherry as a plasma membrane localization control (Li et al., 2024). The primer sequences used in this study are presented in Supplementary Table S6.
2.10 Data analysis
Heatmaps of expression data were generated in R v4.3.1 using the pheatmap package, with Z-score normalization. Statistical significance (p<0.05) was assessed via Student’s t-test to compare expression differences across tissues or treatments.
3 Results
3.1 Identification analysis of GmGH9s in Glycine max
Through systematic genome-wide analysis using HMMER3.0, we identified 43 non-redundant GmGH9 genes in soybeans, with strict elimination of redundant isoforms. Bioinformatic analysis revealed substantial variation in the physicochemical properties of encoded GH9 proteins. The polypeptides ranged from 126 amino acids (aa) (GmGH9B14) to 639 aa (GmGH9C4), corresponding to molecular weights between 14.62 kDa (GmGH9B14) and 70.76 kDa (GmGH9A16) (Supplementary Table S1). Theoretical isoelectric points (pI) exhibited broad diversity from 4.46 (GmGH9A6) to 9.52 (GmGH9B1), while the grand average of hydropathy (GRAVY) values spanned from -0.432 (GmGH9A16) to 0.291 (GmGH9A4). Notably, only two proteins (GmGH9A4 and GmGH9B18) displayed positive GRAVY scores, indicating that 95.3% of GmGH9 proteins are hydrophilic. These distinct physicochemical characteristics suggest functional diversification of GmGH9 proteins across cellular environments.
3.2 Phylogenetic analysis and classification of GmGH9s
To elucidate the evolutionary phylogenomics of GH9s glycosyl hydrolases, we performed maximum-likelihood phylogenetic reconstruction using MEGA X with 1000 bootstrap (BS) replicates, stratifying nodal support into three confidence tiers: low (BS ≤ 0.4), moderate (0.41<BS ≤ 0.8), and high (BS>0.8), with topological robustness quantified in strongly supported nodes (BS≥0.81) (Figure 1; Supplementary Table S1). The phylogram incorporated cross-kingdom homologs from Arabidopsis (Arabidopsis thaliana) and maize (Zea mays), revealing deep phylogenetic conservation of cellulose-metabolizing GH9 enzymes across embryophytes. According to the classification standards for Arabidopsis GH9s (Urbanowicz et al., 2007), the 43 GmGH9 predicted proteins segregated into three evolutionarily distinct classes: Class A (n=16, 37.21%), Class B (n=23, 53.49%), and Class C (n=4, 9.30%), demonstrating phyletic divergence consistent with subfunctionalization patterns. Class B emerged as the predominant clade, exhibiting functional diversification with varying nodal support (e.g., GmGH9B16/B7/B8) in contrast to an evolutionarily constrained cluster (e.g., GmGH9B11-B4/AT1G71380/AT1G22880/Zm00001eb423080). Intriguingly, phylogenetically cohesive subgroups of tandemly duplicated soybean paralogs (e.g., GmGH9A12/A2/A11/A3) formed species-specific expansions, potentially reflecting neofunctionalization events following whole-genome duplication.
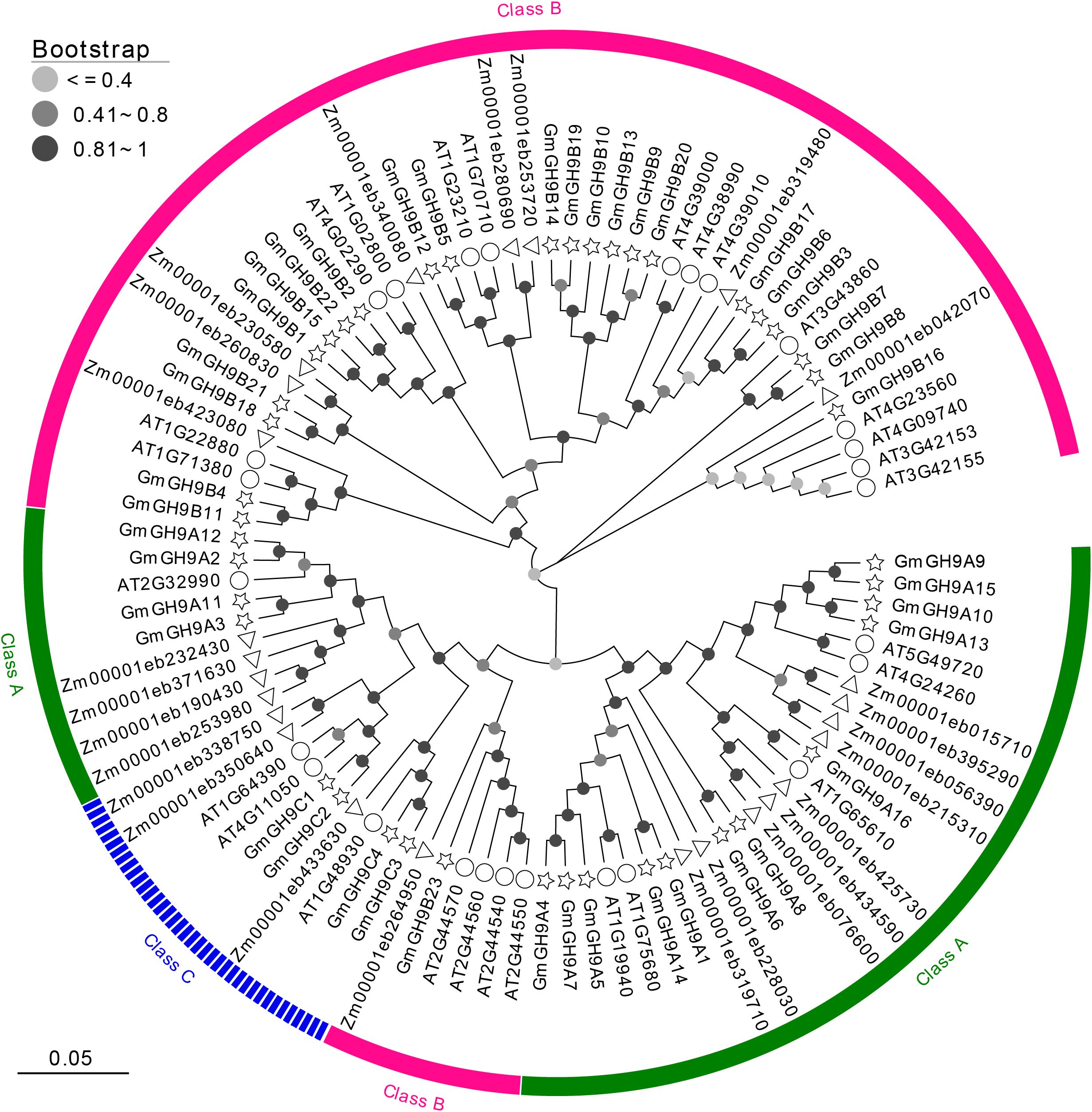
Figure 1. Phylogenetic tree of multi-species GH9s. Green, pink, and blue represent Class A, Class B, and Class C subgroups, respectively. Different leaf decorations represent different species. The color of the circle on the branch represents different bootstrap values.
3.3 Chromosomal location and collinearity analysis of GmGH9s
The 43 GmGH9s exhibited a non-random chromosomal distribution across 18 of the 20 soybean pseudomolecules, notably absent from Gm01 and Gm13 (Figure 2). Chromosomal allocation analysis revealed Gm06 as a genomic hotspot, harboring 23.3% (8/43) of GmGH9 members, followed by Gm02, Gm04, and Gm11, each with 4 genes, representing 9.3% per chromosome. Eight Class B members (GmGH9B7/B8, B9/B10, B13/B14, B19/B20) particularly formed tandem arrays within 200 kb intervals, predominantly localized in pericentromeric regions—a genomic architecture suggestive of non-reciprocal transpositions or unequal crossing-over events. A comparative analysis identified 28 segmental duplication pairs (Supplementary Figure S1), predominantly associated with ancient whole-genome duplication (WGD) events, as evidenced by synonymous substitution rates (Ks=0.09–2.61) and strong purifying selection (Ka/Ks=0.03–0.66; Supplementary Table S2). These evolutionary trajectories likely fine-tuned the spatiotemporal regulation of cell wall polysaccharide remodeling, facilitating adaptive responses to mechanical stress and pathogen threats in soybean ecotypes.
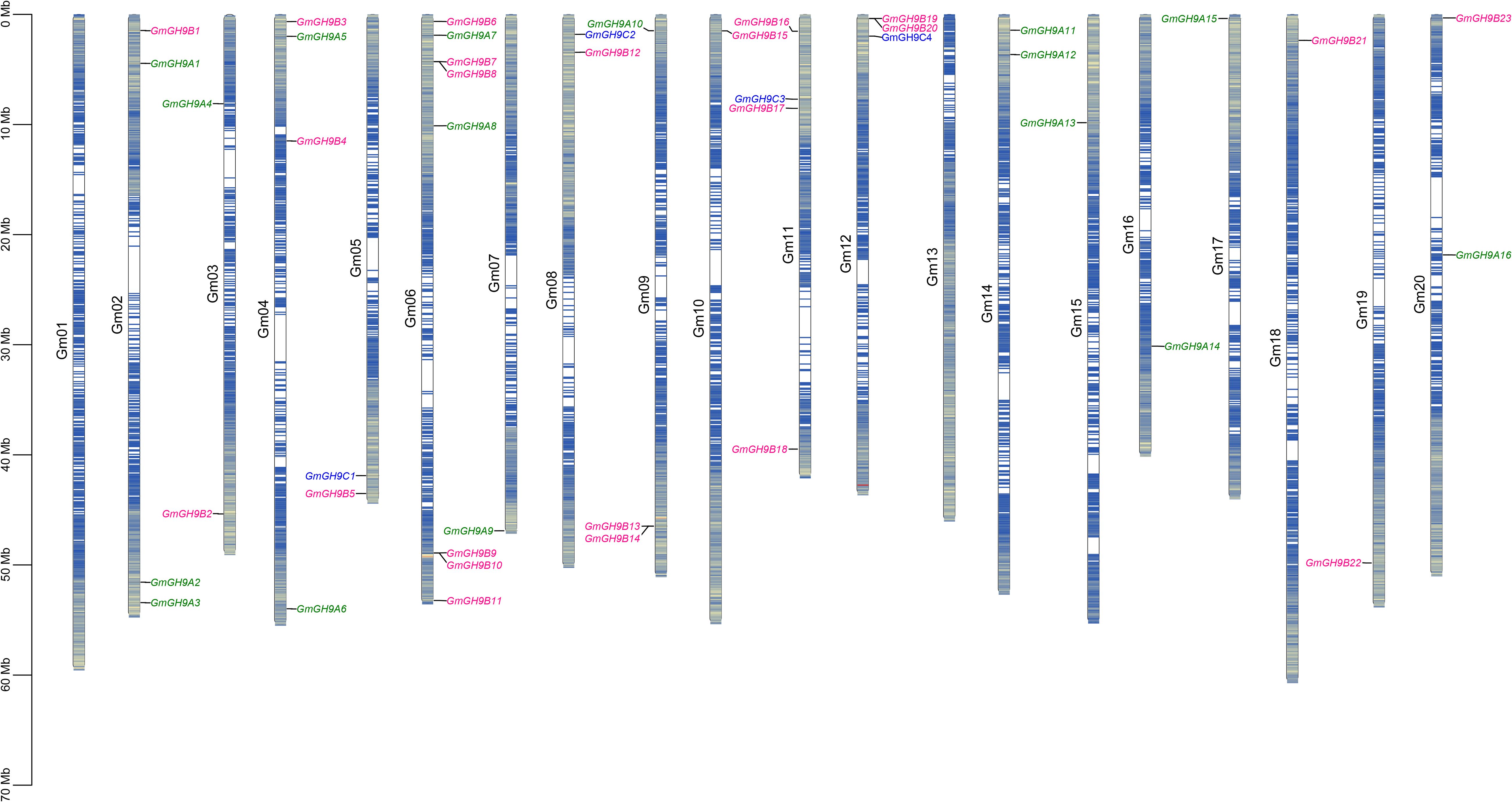
Figure 2. Chromosome distribution of GmGH9s. Green, pink, and blue represent Class A, Class B, and Class C subgroups, respectively.
3.4 Conserved motifs, conserved domains, and gene structures of the GH9 family in soybean
Comprehensive phylogeny-structural analysis of the soybean GmGH9 family unveiled conserved evolutionary modules and subfamily-specific innovations across GmGH9A/B/C clades. Exon-intron architectural complexity diverged significantly (Figure 3A), with GmGH9B/C members displaying elaborate splicing patterns (e.g., the 3’UTRs [three prime untranslated regions] of GmGH9B12 and GmGH9A13 contain introns) in contrast to the streamlined GmGH9A genes. These subfamily-differentiated UTR configurations, particularly intron-containing 3’UTRs, suggest cis-regulatory diversity via post-transcriptional regulation mediated by alternative splicing.
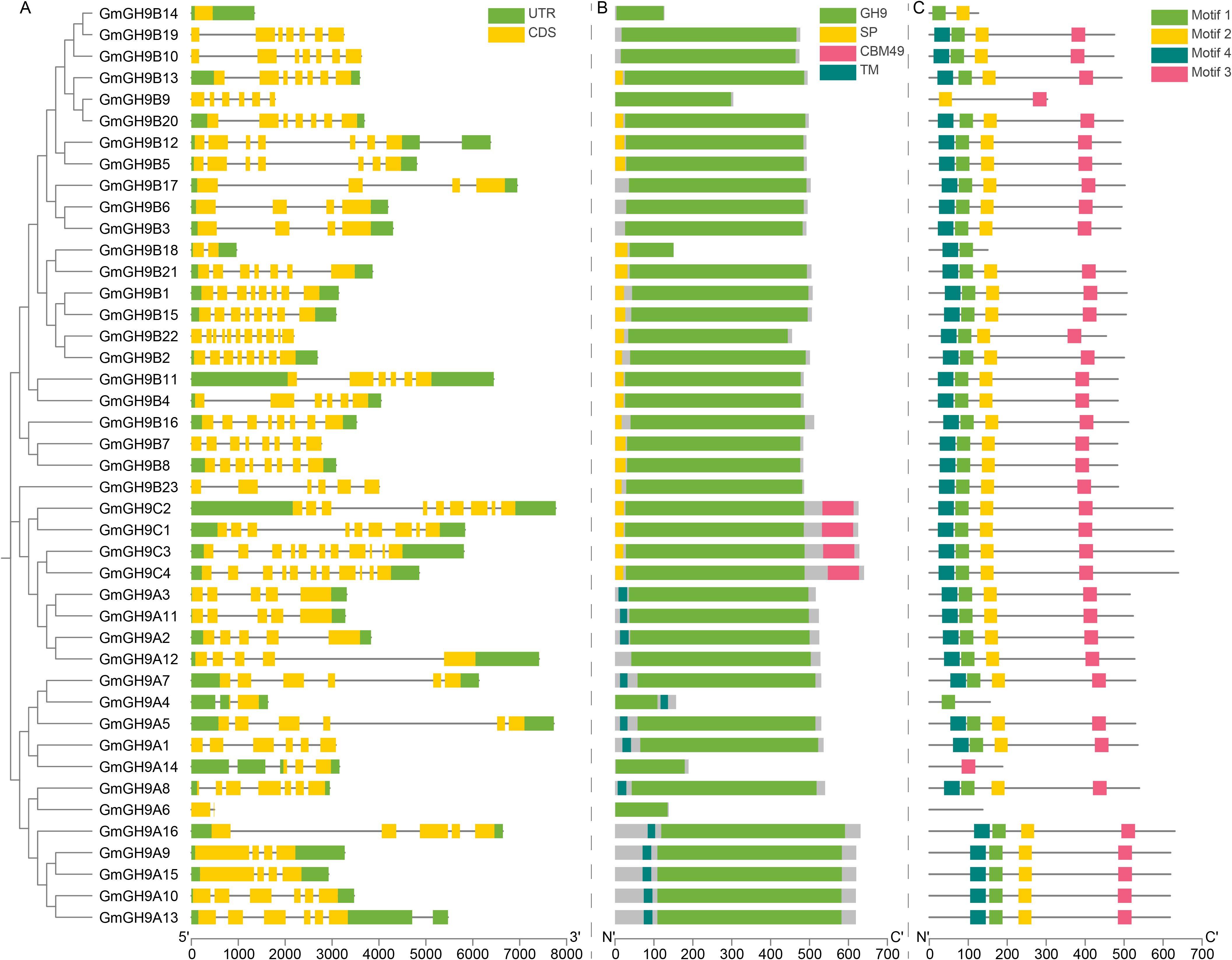
Figure 3. Molecular characteristics of GmGH9s. (A) GmGH9 structures; grey lines represent introns. (B) GmGH9 proteins conserved domain; the gray area is disordered region. (C) GmGH9 conserved motif; gray line is relatively disordered sequence.
Domain architecture (Figure 3B) highlighted the canonical GH9s catalytic domain, combined with distinctive characteristics peculiar to each subfamily: transmembrane (TM) domains were prominent in GmGH9As (except GmGH9A6/A12/A14), suggesting roles in membrane anchoring. GmGH9Cs uniquely harbored carbohydrate-binding module 49 (CBM49) and signal peptides (SP), implicating involvement in cellulose recognition and secretion mechanisms. These structural patterns align with phylogenetic divergence, indicating that the complexity and extension of the GmGH9Bs domain signify neofunctionalization in cellulose metabolism, whereas the streamlined architectures of GmGH9Cs imply conserved catalytic functions. Surprisingly, GmGH9A6 does not have a TM domain, and there is only a GH9 domain in the pan-gene (Supplementary Table S3). Phylogenetic and domain architecture may exhibit incongruence, potentially reflecting functional divergence through domain loss during lineage-specific evolution.
Motif decomposition revealed evolutionarily constrained modularity (Figure 3C; Supplementary Figure S2): four core motifs (1-4; 34–41 aa) showed positional conservation in 86.05% (n=37) of the members. Whereas features with functional loss, such as GmGH9B14/B9/B18, contain only two of the four motifs; GmGH9A4/A14 contains only one of the four motifs. GmGH9A6 inadvertently exhibited an absent pattern. The observed divergence among subfamilies may result from the tetraploidization and subsequent diploidization processes in soybean evolution, while lineage-specific adaptations may potentially underlie the cell wall remodeling strategies and ecological resilience mechanisms that govern soybean’s environmental responsiveness.
3.5 Analysis of cis-elements in the promoters and functional annotation of GmGH9s
Analysis of the promoters of the soybean GmGH9 gene family revealed a diverse array of cis-regulatory elements associated with biological processes and stress responses. A total of 13 types of functional motifs were identified (Figure 4), with all gene promoters containing light-responsive motifs, consistent with the diurnal regulation of cellulose metabolism (Wang et al., 2023a). Moreover, the distribution of light-responsive motifs is relatively dense, with GmGH9B5, GmGH9B21, and GmGH9B23 containing 27, 23, and 23 light signal motifs, respectively. Hormone-responsive elements are notably predominant, including ABA (39/43), gibberellin (GA, 24/43), methyl jasmonate (MeJA, 24/43), auxin (13/43), and salicylic acid (SA, 11/43) signaling pathways, suggesting tight hormonal regulation of GmGH9s expression. Stress-related motifs were highly enriched, particularly those linked to anaerobic conditions (37/43), drought (18/43), and temperature fluctuations (15/43), underscoring the family’s potential role in abiotic stress adaptation. Seed development-related motifs were also abundant, highlighting the family’s involvement in growth processes. GmGH9B5 exhibited a higher motif density for light and ABA metabolic regulation, whereas GmGH9B9 contained only 13 motifs. We performed GO (Supplementary Table S4) and KEGG (Supplementary Table S5) enrichment analysis to examine the function of GmGH9s. It was found that they were mainly engaged in the biological process of “cell wall modification”, subsequently followed by “response to cyclopentenone” and “β-glucan metabolic process” (related to cytokinin) (Supplementary Figure S3). Pathway enrichment analysis identified starch and sucrose metabolism as the most significantly enriched KEGG pathway. These findings collectively suggest that GmGH9 genes are regulated by a complex interplay of developmental, hormonal, and environmental cues, likely fine-tuning their roles in cell wall remodeling and stress resilience in soybeans.
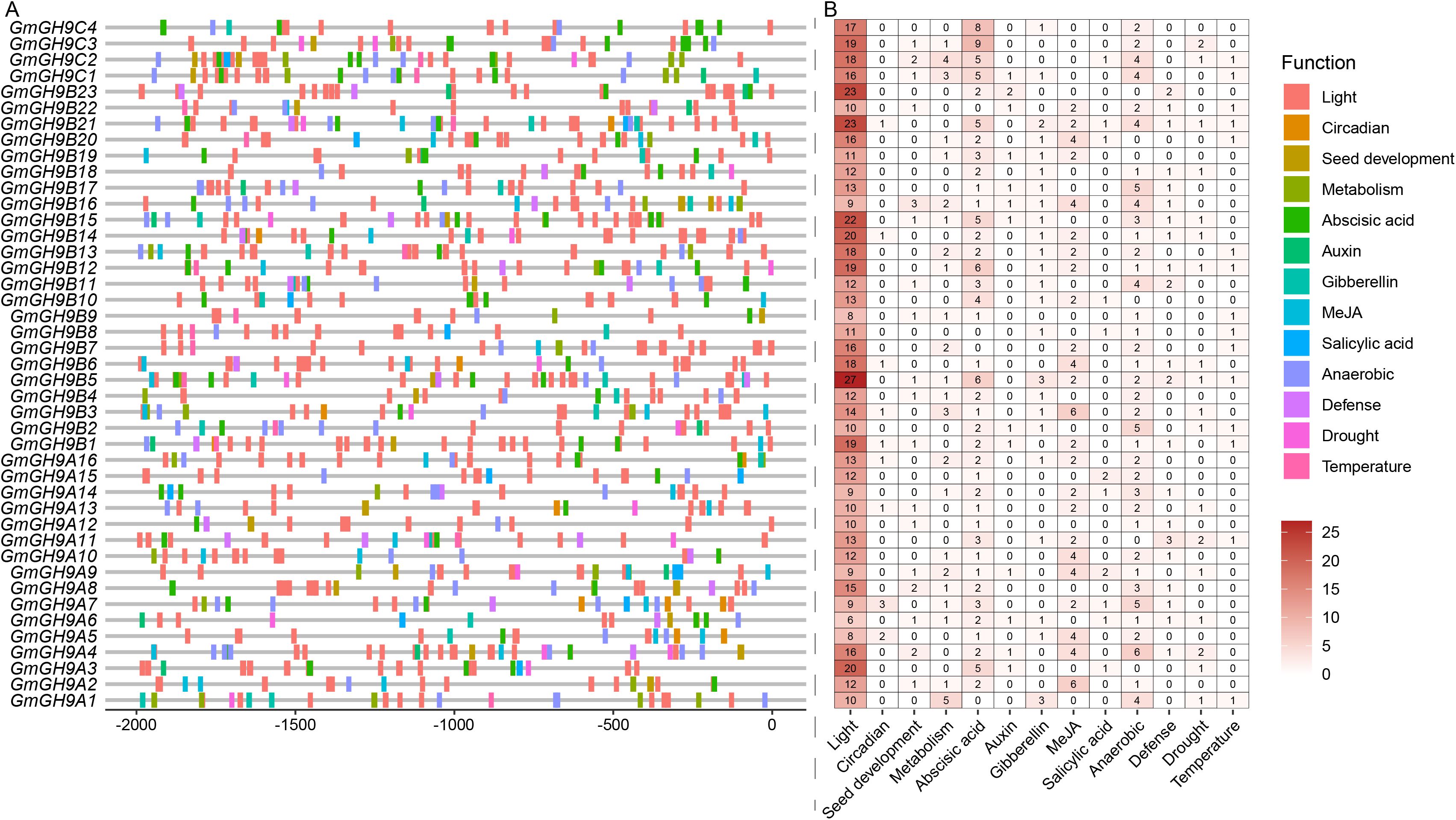
Figure 4. GmGH9s promoter motif distribution. (A) The distribution of different motifs in the promoter position. (B) The heatmap of motifs function classification statistics.
3.6 GmGH9s expression profiles under different conditions
The expression profiling of the GmGH9s family in soybean revealed spatiotemporal and stress-responsive regulatory dynamics across organs, ethylene treatment, and photoperiod conditions (Figures 5A–C). Organ-specific expression highlighted functional diversification (Figure 5A). GmGH9C4 was highly expressed in lateral roots, while GmGH9C1 and GmGH9C2 were specifically expressed at the apical regions (root and shoot). Seed-specific expression peaks of GmGH9A2 correlated with seed maturation. In addition, GmGH9A10/15/13/9 showed constitutive expression. Ethylene-responsive regulation (Figure 5B) revealed distinct expression patterns in leaf AZ (Laz) compared to non-AZ (Naz). GmGH9B15 was highly expressed in Laz-specific at 0 h after ethylene induction (AEI). GmGH9B19 was significantly induced by ethylene in Laz compared to Naz at 12 h AEI. Key genes (GmGH9B21/B11/A11/A3/B4/B16/B5) exhibited significant induction in Laz at 24–48 h AEI, consistent with ethylene’s role in cell wall loosening during abscission. In contrast, GmGH9A4 was downregulated in Laz but upregulated in Naz at 72 h AEI, indicating compartmentalized regulatory roles. Photoperiod-dependent expression (Figure 5C) demonstrated that LD conditions enhanced the expression of GmGH9B2/B22/A3/B1/B15/B5/A12/A2/C1/C2, while SD conditions suppressed it. SD significantly inhibited gene expression, except GmGH9A16 and GmGH9A8. Strikingly, GmGH9B12, GmGH9B11, and GmGH9B4 displayed rhythmic oscillations under LD, with peaks occurring at dawn (T5), and demonstrated elevated expression levels at T5 in Sh. Surprisingly, most genes showed a similar expression pattern in Clark and Wm82.
RT-qPCR was employed for examining the changes in gene expression of GmGH9 genes, which show different patterns (either tissue-specific high expression profiles across soybean organs, or induced or suppressed by ethylene) (Figure 5B). Stem, leaf, petiole, flower, Faz, Paz, pod peel, seed coat, and embryo were detected before and after ethylene treatment. As shown in Figures 6A, B, ethylene significantly enhanced the expression of GmGH9A9 and GmGH9B19 in leaves, whereas concurrently repressing their expression in flowers and the seed coat. GmGH9B12, GmGH9C1, and GmGH9B19 were found to be specifically highly expressed in the Paz. However, following ethylene treatment, their expression was dramatically decreased, showing a 15 to 35 fold reduction. In the Faz, both GmGH9B16 and GmGH9B21 exhibited increased expression levels in response to ethylene. Furthermore, GmGH9B16 was notable for its up-regulation in flower, whereas GmGH9B21 showed a significant rise in expression in the Paz, achieving a three-fold increase relative to control levels. The results indicate that GmGH9s may have multiple roles in abscission.
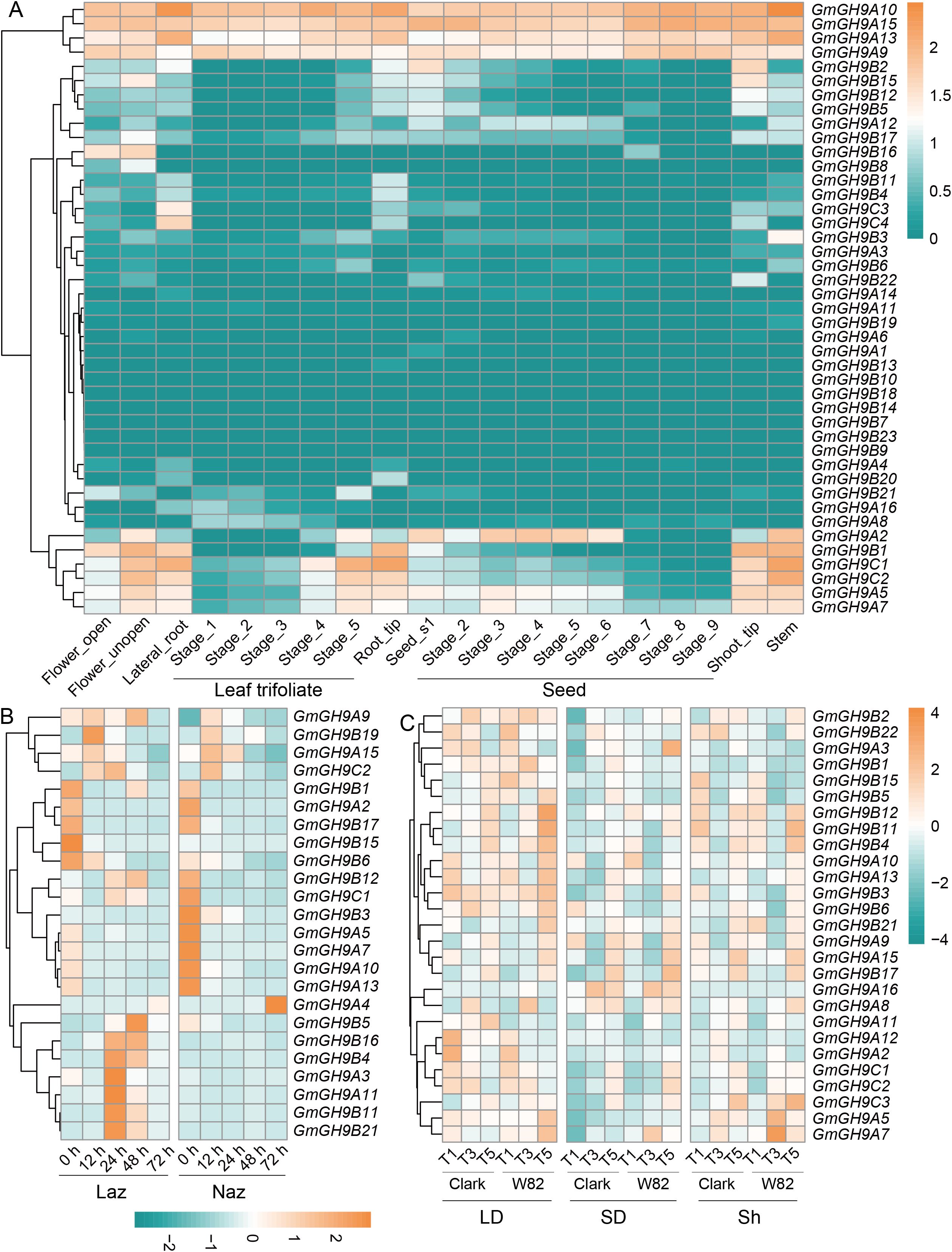
Figure 5. Spatio-temporal expression of GmGH9s. (A) The expression of GmGH9s in different organs and periods. (B) The expression of GmGH9s in leaf abscission zones (Laz) and non-leaf abscission zones (Naz) at 0 hour (h), 12 h, 24 h, 48 h and 72 h after ethylene induction. (C), GmGH9s in soybean varieties Clark and Williams 82 (Wm82) long-day (LD, 16 h of light, 6:45-22:45) and short-day (SD, 10 h of light, 6:45-16:45) period three weeks after germination. Expression patterns at T1 (6:30), T3 (14:30), and T5 (22:30) under LD, SD, and light conversion (Sh, first grown under LD for three weeks and then transferred to SD for 5 days) conditions.
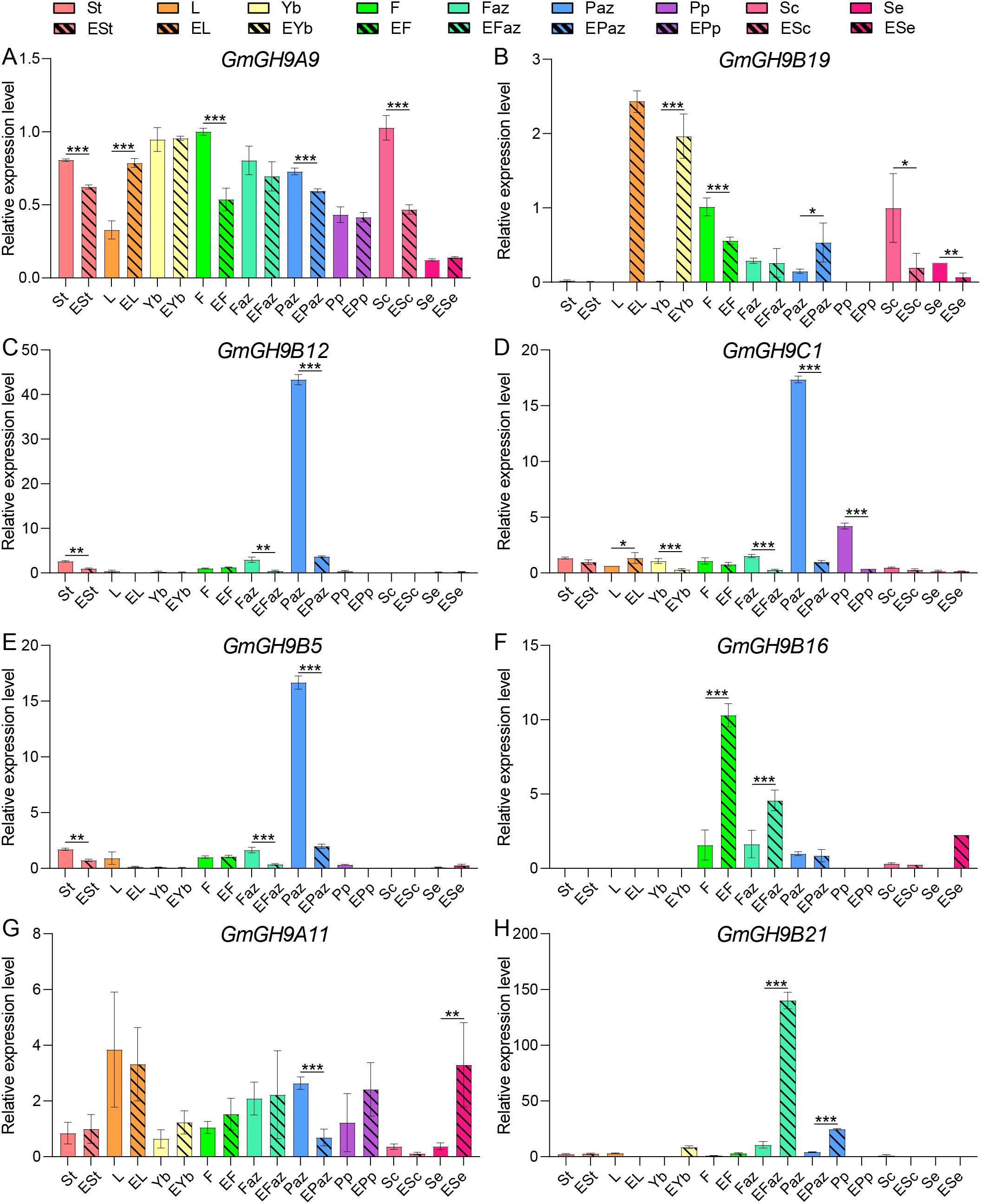
Figure 6. RT-qPCR analysis of the response of GmGH9s to ethylene in different tissues. (A–H) The relative expression levels of GmGH9A9, GmGH9B19, GmGH9B12, GmGH9C1, GmGH9B5, GmGH9B16, GmGH9A11, and GmGH9B21 respectively. ESt, EL, EYb, EF, EFaz, EPaz, EPp, ESc, and ESe denote ethylene-treated stem, leaf, petiole, flower, flower AZ, pod AZ, pod peel, seed coat, and embryo, respectively. Data are presented as means ± SEM. *, **, and *** indicate significant differences at p<0.05, p<0.01, and p<0.001, respectively.
3.7 Multi-species colinearity analysis of GH9s
The comparative synteny analysis of the GmGH9s family among soybean, pea (Pisum sativum), tomato (Solanum lycopersicum), Arabidopsis, and maize revealed both conserved and divergent evolutionary patterns. Colinearity analysis for the GH9s gene family revealed that pea, tomato, Arabidopsis, and maize exhibit 30, 25, 23, and 15 collinear events with soybean, respectively. The syntenic region shared among all species includes GmGHB3/B5/B13/B14/B17/B18. Moreover, seven GH9 syntenic regions—specifically GmGH9B1/A2/A4/A8/C2/C4/A15—are conserved in pea, tomato, and soybean, while they are absent in Arabidopsis and maize. These genes exhibit a high level of conservation throughout the soybean pan-genome (Figures 7B, C; Supplementary Figures S4–S8). Notably, GmGH9B1/A4/C2/A15 displayed differential expression in leaves between Laz and Naz after ethylene treatment (Figure 5B). GmGH9B1 exhibited specific expression in flowers following ethylene induction, with negligible levels detected in other organs (Figure 8A). GmGH9A2 showed a significant reduction in the AZ of flowers and pods after ethylene treatment (Figure 8B). Conversely, GmGH9A8 and GmGH9C4 exhibited significantly increased expression in the AZ of flowers and pods following ethylene treatment, suggesting their potential involvement in the regulation of flower and pod abscission (Figures 8C, F). In contrast, GmGH9C2 showed opposite ethylene responses in the AZ of flowers and pods, implying functional diversity (Figure 8E). Additionally, GmGH9A15 displayed differential expression in the Paz, whereas no such expression was observed in the Faz (Figure 8D). Fruit or pod abscission occurs during the growth of pea, tomato, and soybean. It is hypothesized that GmGH9A2/A8/A15/C2/C4 are linked to flower pod abscission, with GmGH9A8/C4 facilitating this process and GmGH9A2 serving as an inhibitor.
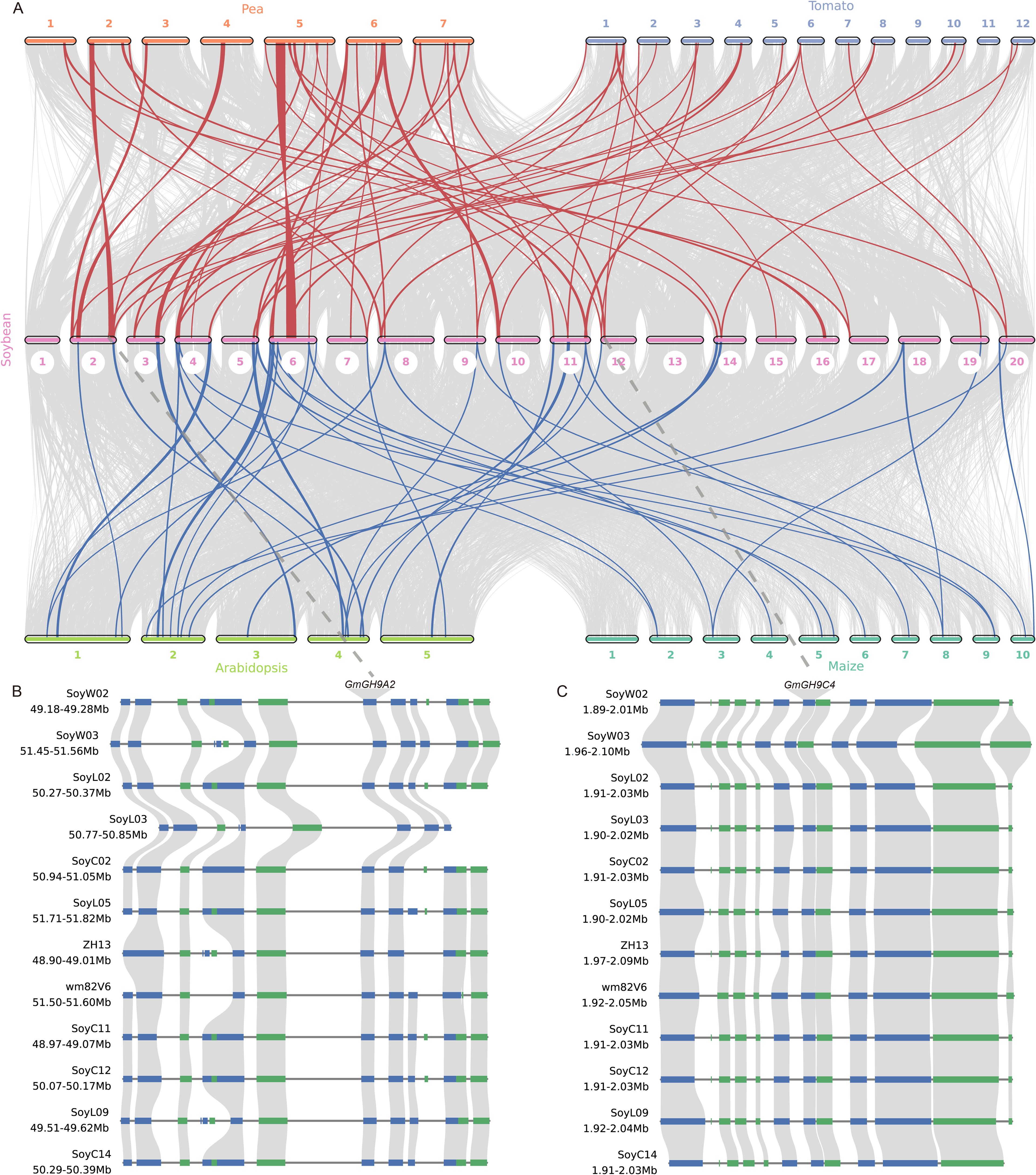
Figure 7. Co-linearity analysis of GH9s. (A) Multi-species genome-wide collinearity analysis. (B) Microsynteny analysis of the GmGH9A2 within soybean species. (C) Microsynteny analysis of the GmGH9C4 within soybean species. Green and blue represent genes on the negative and positive strands of chromosomes, respectively.
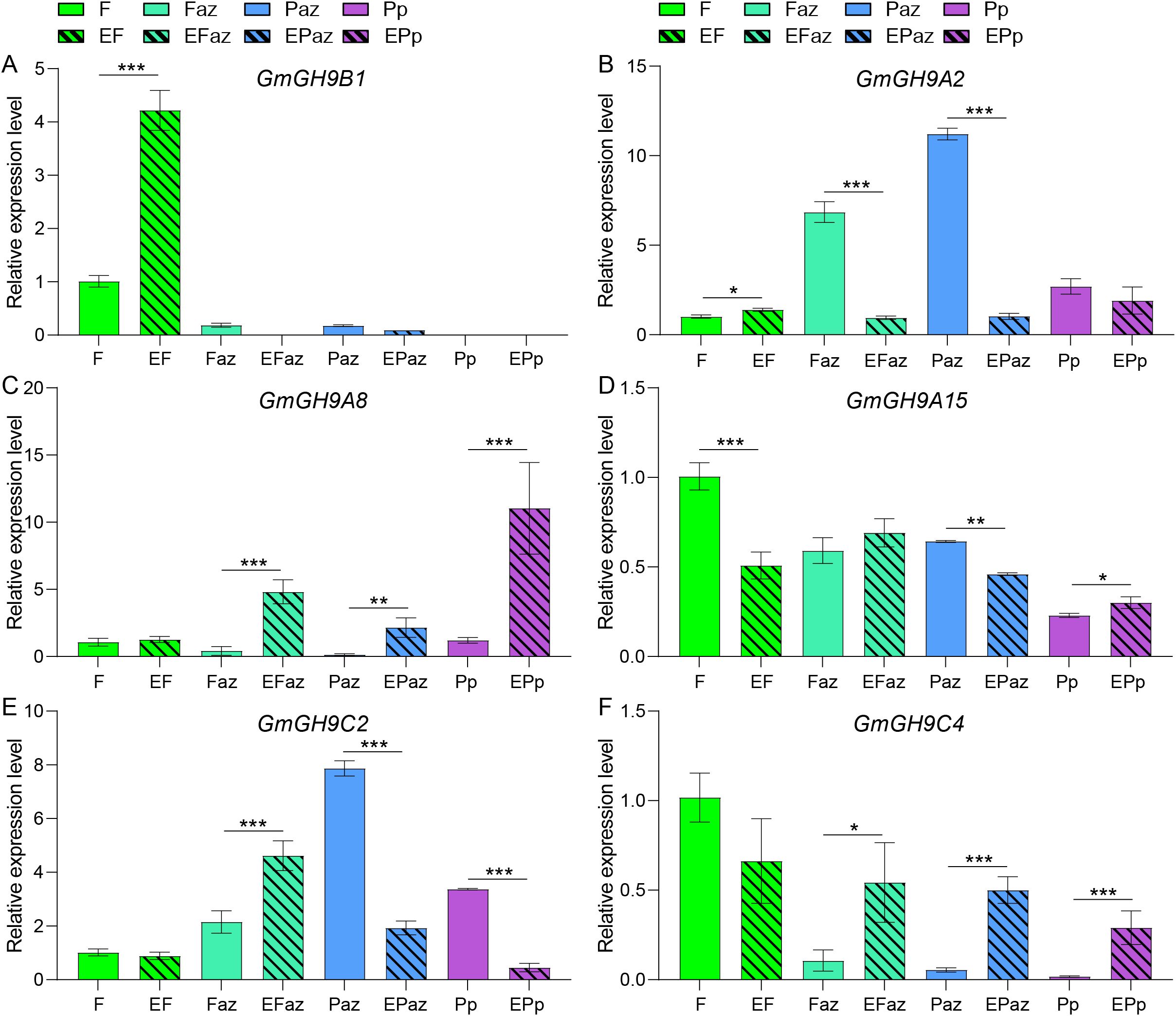
Figure 8. RT-qPCR analysis of the response of GmGH9s to ethylene in distinct floral and pod zones. (A–F) The relative expression levels of GmGH9B1, GmGH9A2, GmGH9A8, GmGH9A15, GmGH9C2, and GmGH9C4, respectively. Data are presented as means ± SEM. *, **, and *** indicate significant differences at p<0.05, p<0.01, and p<0.001, respectively.
3.8 Haplotype variations of GmGH9A2 and GmGH9C4
To clarify the effects of GmGH9s on soybean morphology, we examined the association between the haplotypes of GmGH9A2 and GmGH9C4 with yield-related traits, including 100‐seed weight (HSW), seed weight per plant (SWP), and total seed number per plant (TSN). GmGH9A2 and GmGH9C4 had twelve and five haplotypes, respectively (Figure 9A; Supplementary Figure S9A). Despite the absence of differences in TSN among the haplotypes of GmGH9A2, H001 exhibited a significantly higher HSW compared to others, resulting in a notable improvement in SWP for H001 (Figures 9B–D). An analysis of π and Fst in the 400 kb upstream and downstream regions of GmGH9A2 among G. soja, landrace, and cultivated soybeans revealed that the π ratio and Fst value for G. soja/landrace comparisons reached the top 10% threshold, while the Fst between landraces and cultivars was significantly lower than that observed between G. soja and landraces. The π ratio of landrace/cultivar, though not exceeding the threshold, was greater than 1. This suggests that H001 was selected for enhanced yield traits in soybean cultivation and indicates that intensive breeding practices have further eroded genetic variation in cultivated lines (Figure 9). In the case of GmGH9C4, although a slight decrease in the TSN of H003, the HSW was higher than that of H001 and H002, leading to a significant improvement in SWP (Supplementary Figures S9B–D). In contrast to GmGH9A2, the π ratio of landrace/cultivar approached 1, whereas the π ratio of G. soja/landrace exceeded that of landrace/cultivar. This suggests that selection pressure was exerted on G. soja during domestication, with only modest selection changes occurring during subsequent breeding improvement (Supplementary Figure S9E). These findings highlight the impact of domestication and breeding on the narrowing genetic diversity at functionally critical GmGH9A2 and GmGH9C4, potentially limiting adaptive potential in modern cultivars.
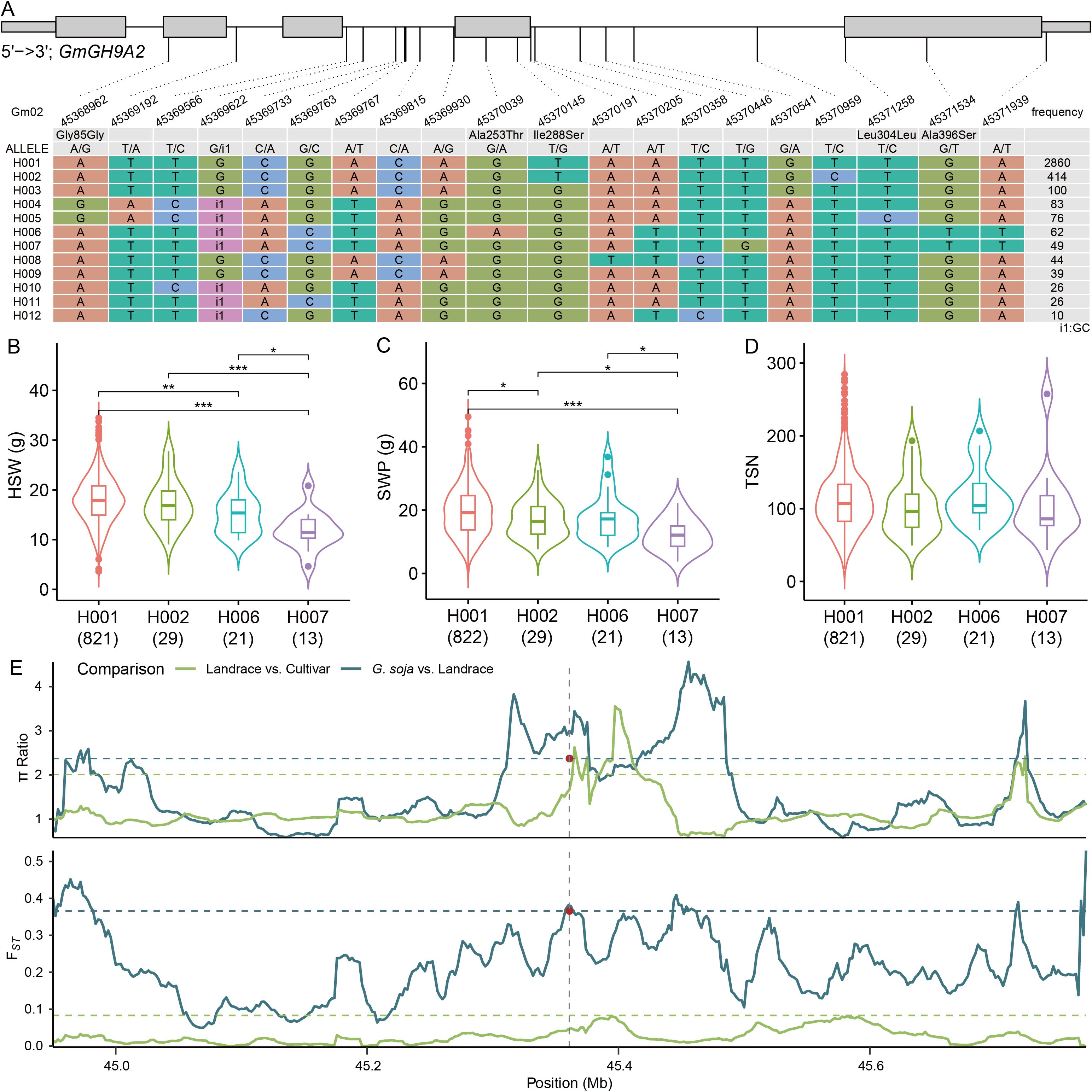
Figure 9. Analysis of genetic characteristics of GmGH9A2. (A) Haplotype of GmGH9A2. The wide and narrow gray boxes represent the exon and UTR regions, respectively. The gray line represents the intron. (B–D) The relationship between haplotypes with 100-seed weight (HSW), seed weight per plant (SWP), and total seed number (TSN). (E) The π ratio and Fst values of the flanking region at GmGH9A2 in (G) soja, landraces, and cultivars soybeans. The horizontal dashed lines indicate the genome-wide thresholds of (G) soja vs. landraces, and landraces vs. cultivars (top 10%). The middle position of GmGH9A2 is labeled by brown dot (Gm02: 45370242). *, **, and *** indicate significant differences at p<0.05, p<0.01, and p<0.001, respectively.
3.9 Subcellular localization of GmGH9s protein
To analyze the subcellular localization of GmGH9s, we generated recombinant constructs encoding 35S::GmGH9A2-GFP, 35S::GmGH9A8-GFP, and 35S::GmGH9C2-GFP. These were co-bombarded with the plasma membrane marker 35S::AtPIP2A-mCherry into Nicotiana benthamiana epidermal cells via transient expression. Confocal laser scanning microscopy revealed distinct localization patterns: while the 35S::GFP control exhibited diffuse cytoplasmic localization, all three GmGH9 fusion proteins showed pronounced extracellular compartmentalization (Figure 10). This extracellular localization profile aligns with their predicted roles in apoplastic processes involving cell wall remodeling.
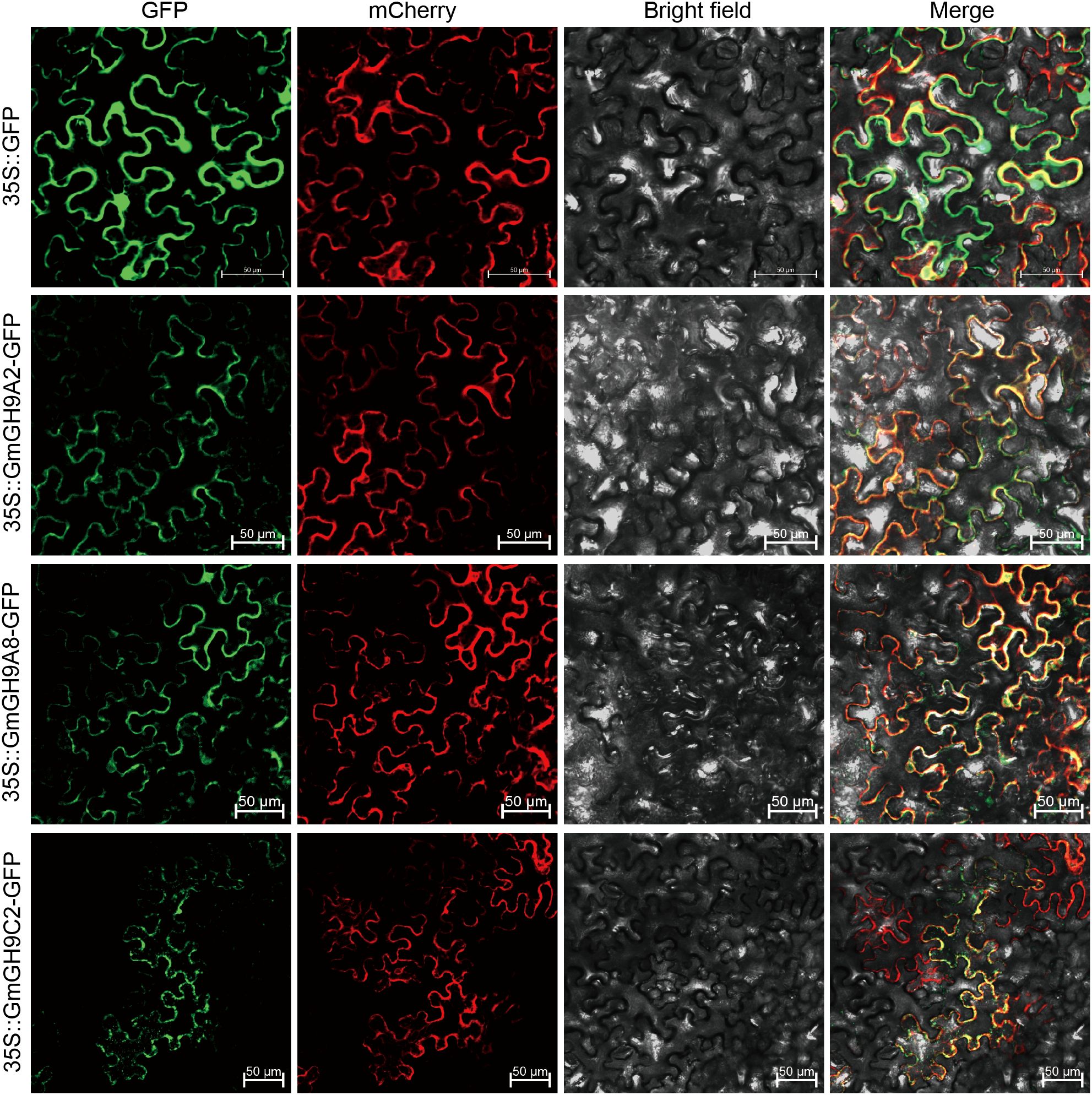
Figure 10. Subcellular localization of GmGH9s. GmGH9A2-GFP, GmGH9A8-GFP, and GmGH9C2-GFP fusion proteins, as well as GFP alone, were transiently expressed in Nicotiana benthamiana leave cells. An mCherry-based AtPIP2A labeling plasma membrane. Representative photos of the protein localization analysis conducted 48 h after infiltration are displayed. Individual images of GFP, mCherry, or bright field autofluorescence are shown. GFP merged with mCherry as well as bright field (Merge) images are also displayed. Scale bars=50 μm.
4 Discussion
Previous studies have documented the role of GH9s in the reassembly and degradation of the soybean cell wall (Nawaz et al., 2017). However, a comprehensive analysis of the expansion of the GH9 gene family and the subsequent functional differentiation within this family has not been conducted to date. This study performed a genome-wide identification and characterization of cellulase genes, facilitating the understanding of the genetic mechanisms underlying soybean organ abscission.
4.1 Gene family expansion and functional differentiation
GmGH9 family shows considerable expansion within the soybean genome, an ancient tetraploid specie, with a notable prevalence in the GmGH9B subfamily, comprising 23 out of 43 members, paralleling observations in other species (Luo et al., 2022b; Wang et al., 2023c). This expansion is likely related to the functional diversification of cellulose metabolism. The Class C family is quite conserved, particularly in the unique CBM49 structural domain (Figure 3B) and the transmembrane domains of GmGH9C. This suggests a distinct role in substrate recognition and membrane localization. All members present in the multispecies covariance belong to the Class C family (Figure 1). The GmGH9A subfamilies are more streamlined in structure and may retain conserved catalytic functions (Figures 3A, B). Intriguingly, while GmGH9A3/A11/A2/A12/B23 phylogenetically clusters near class C members, it notably lacks the characteristic CBM49 domain observed in this group. This structural divergence suggests a complex evolutionary relationship between subgroup A and B with C, though their precise phylogenetic origins require further molecular evidence to resolve.
Importantly, subfunctionalization or neofunctionalization after gene duplication can occur via structural domain reorganization, such as variations in motif arrangement (Sandve et al., 2018), thereby facilitating adaptation to the dynamic remodeling of the soybean cell wall, including pectin degradation and responses to environmental stressors. Tandem repeats and segmental repeat events on chromosomes may primarily driving this expansion, with a large number of tandem repeats in the soybean genome (Zhang et al., 2024). We identified 8 tandem repeats and 28 fragment repeats (Figure 2; Supplementary Figure S1). The structural hallmarks—tandem clusters in recombinationally active zones and retained paleoduplicates—imply compartmentalized functional diversification. This may occur through subfunctionalization that preserving ancestral cellulose synthase enhancement activity or neofunctionalization that introduces novel β-1,4-glucanase specificities, potentially driven by subgenome dominance during post-polyploid diploidization.
4.2 Adaptive significance of cis-regulatory elements
Homeostasis regulation is closely related to gene expression, with alteration in cis-regulatory sequences contributing to the diversification of gene functions in soybeans (Fang et al., 2023). Promoter analysis revealed a wide range of hormone- and adversity-responsive elements (e.g., ABA, MeJA, drought, and light-responsive elements) within the GmGH9s gene family, suggesting that its expression is regulated by complex environmental and developmental signals. The late-stage softening and ripening of fruit are closely related to light (Li et al., 2021; Lin et al., 2023; Wei et al., 2023; Su et al., 2024). The enrichment of light signals in the GmGH9s promoter suggests an important role for these genes in the late stage of soybean pod development (Figure 4). Enrichment of elements related to photoperiod and biological clock, such as GmGH9B5, GmGH9B21, and GmGH9B23, further supports a role for this family in circadian-regulated cellulose metabolism. Photoperiod-dependent circadian oscillations, such as the rhythmic expression of GmGH9A2 and GmGH9A12 under long day light (Figure 5B), may be associated with carbon allocation strategies. These observed patterns are highly compatible with cis-elements in promoters, indicating a synergistic optimization of regulatory and functional modules during evolution.
4.3 Spatio-temporal specificity of expression patterns
Expression profiling revealed that GmGH9 genes were highly specific in organ development, including root, flower, stem, and seed development, as well as in stress response such as ethylene-induced AZ formation. The high expression of GmGH9C1 in root tips may correlate with its involvement in root cell wall expansion, whereas the up-regulation of GmGH9A2 in late seed development (Figure 5A). Simultaneously, GmGH9A2 was also rapidly expressed in Laz and Naz after ethylene treatment; conversely, its expression was suppressed in the AZ of flowers and pods (Figures 5B, 8B). It reflects the functional diversity of GmGH9A2 in the petiole, flower and pod AZ. Low expression levels of certain genes were detected in different organs; however, these genes may have outstanding contributions at different times. Different organs of GmGH9A9 and GmGH9B19 have different sensitive responses to ethylene (Figures 6A, B), indicating that they have diverse functions. Despite GmGH9B4 displaying low expression levels in various organs, the expression of Laz was significantly higher than that of Naz following 24 h of ethylene treatment (Figure 5B). At the same time, GmGH9B4 also showed periodic expression, with high levels observed during the T5 stage of LD (Figure 5C). The up-regulation of GH9B3 (GmGH9B4 in this study) gene expression during homograft adhesion was consistently observed in soybeans, while it was not evident in interfamily grafts (Notaguchi et al., 2020). This space-time specific expression could enhance the practicality of soybeans.
4.4 Implications of covariance and evolutionary history
Syntenic blocks were identified between soybean chromosomes and homologous regions in other species, highlighting ancestral genomic conservation. Collinearity analyses showed that some members of the GmGH9 family, such as the GmGH9B subfamily, share conserved syntenic blocks (Figure 7A), which supports their derivation from a common ancestral core gene. This “conserved core-dynamic fringe” evolutionary model may have balanced functional stability with adaptive innovation. However, the absence of synteny in monocotyledons, such as Arabidopsis and maize, reflects lineage-specific adaptation. Abscission of pea, tomato and soybean fruits/pods occurs during the ripening stage (Ayeh et al., 2009; Kim et al., 2016; Lin et al., 2023). The presence of GmGH9B1/A2/A4/A8/C2/C4/A15 in pea, tomato and soybean suggests a potential relationship between these genes and fruit abscission. Overall, the synteny network underscores the dual evolutionary trajectory of the GmGH9 family: core functions are conserved through syntenic orthologs, while species-specific innovations driven by genomic plasticity.
4.5 Decay of genetic diversity and breeding challenges
The hydrolysis of plant cell walls leads to a series of reactions, such as HSW, a loss-of-function mutant allele of a glycosyl hydrolase gene, utilized to regulate seed weight during soybean domestication (Wei et al., 2023). qHS1, an endo-1,4-β-glucanase gene, harbors a SNP mutant that changes the permeability of soybean seed coat (Jang et al., 2015). Organ abscission can severely affect yield, with haplotypes of GmGH9A2 and GmGH9C4 showing a correlation to yield (Figure 8C; Supplementary Figure S9C). Genetic diversity analyses of GmGH9A2 revealed a significantly high π ratio and Fst between G. soja and landrace (Figure 9E), suggesting that artificial selection has led to a genetic bottleneck at this locus. This phenomenon may arise from directional selection for specific agronomic traits in breeding, such as cell wall strength or resistance to downy mildew. The attenuation of genetic diversity from G. soja soybean to cultivars serves as a caution that over-reliance on a limited genetic base may weaken the capacity of future varieties to adapt climate change (Tian et al., 2025). Therefore, the use of local germplasm or the restoration of key allelic variants through gene editing may provide new strategies to optimize GmGH9s function while balancing high yield and stress tolerance. Although GmGH9s are correlated with yield, further experimental evidence is required to determine whether abscission is the causative factor.
5 Conclusions
In this study, 43 GH9 genes were identified in soybean. These genes were classified into three groups. Notably, several genes exhibited significant responses to ethylene treatment and circadian variations, especially GmGH9A9 and GmGH9B19. Furthermore, multi-species synteny analysis indicated that GmGH9A2 and GmGH9C4 may be involved in organ abscission and significantly affect soybean yield, providing valuable insights for future soybean molecular breeding. Its evolutionary history and changes in genetic diversity offer important insights for crop improvement.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI BioProject, accession PRJNA442256 and PRJNA219510.
Author contributions
WZ: Formal analysis, Conceptualization, Data curation, Methodology, Validation, Writing – review & editing, Writing – original draft, Investigation. HH: Investigation, Writing – review & editing, Writing – original draft, Project administration, Data curation, Methodology. ZC: Data curation, Methodology, Writing – original draft, Investigation, Formal analysis. ZZ: Methodology, Software, Visualization, Data curation, Writing – original draft. XL: Data curation, Writing – original draft, Methodology, Investigation. YP: Data curation, Methodology, Investigation, Writing – original draft. ZD: Project administration, Validation, Resources, Writing – review & editing, Conceptualization. DQ: Conceptualization, Funding acquisition, Supervision, Resources, Project administration, Writing – review & editing. LJ: Resources, Writing – review & editing, Funding acquisition, Conceptualization, Project administration, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the China Postdoctoral Science Foundation (2022M720881), Guangdong Basic and Applied Basic Research Foundation (2023A1515110254), Graduate Student Innovation Ability Training Funding Program of Guangzhou University, and the Youth Talent Project of Guangdong Provincial Department of Education (2022KQNCX029).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AW declared a shared affiliation with the authors to the handling editor at the time of review.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1597668/full#supplementary-material
References
Ayeh, K. O., Lee, Y., Ambrose, M. J., and Hvoslef-Eide, A. K. (2009). Characterization and structural analysis of wild type and a non-abscission mutant at the development funiculus (def) locus in pisum sativum L. BMC Plant Biol. 9, 76. doi: 10.1186/1471-2229-9-76
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Method. Biochem. Anal. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolser, D., Staines, D. M., Pritchard, E., and Kersey, P. (2016). “Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data,” in Plant bioinformatics: methods and protocols. Ed. Edwards, D. (Springer, New York, NY), 115–140. doi: 10.1007/978-1-4939-3167-5_6
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Method. Biochem. Anal. 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
del Campillo, E. and Bennett, A. B. (1996). Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol. 111, 813–820. doi: 10.1104/pp.111.3.813
Deng, C., Guo, Y., Zhang, J., and Feng, G. (2024). Genome-wide investigation of glycoside hydrolase 9 (GH9) gene family unveils implications in orchestrating the mastication trait of Citrus sinensis fruits. BMC Genomics 25, 905. doi: 10.1186/s12864-024-10826-w
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Method. Biochem. Anal. 29, 15–21. doi: 10.1093/bioinformatics/bts635
Du, Q., Wang, L., Yang, X., Gong, C., and Zhang, D. (2015). Populus endo-β-1,4-glucanases gene family: genomic organization, phylogenetic analysis, expression profiles and association mapping. Planta 241, 1417–1434. doi: 10.1007/s00425-015-2271-y
Eddy, S. R. (2011). Accelerated profile HMM searches. PloS Comput. Biol. 7, e1002195. doi: 10.1371/journal.pcbi.1002195
Estornell, L. H., Agustí, J., Merelo, P., Talón, M., and Tadeo, F. R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 199–200, 48–60. doi: 10.1016/j.plantsci.2012.10.008
Fang, C., Yang, M., Tang, Y., Zhang, L., Zhao, H., Ni, H., et al. (2023). Dynamics of cis-regulatory sequences and transcriptional divergence of duplicated genes in soybean. Proc. Natl. Acad. Sci. 120, e2303836120. doi: 10.1073/pnas.2303836120
Flors, V., Leyva, M. D. L. O., Vicedo, B., Finiti, I., Real, M. D., García-Agustín, P., et al. (2007). Absence of the endo-β-1,4-glucanases Cel1 and Cel2 reduces susceptibility to Botrytis cinerea in tomato. Plant J. 52, 1027–1040. doi: 10.1111/j.1365-313X.2007.03299.x
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Grant, D., Nelson, R. T., Cannon, S. B., and Shoemaker, R. C. (2010). SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38, D843–D846. doi: 10.1093/nar/gkp798
He, H., Bai, M., Tong, P., Hu, Y., Yang, M., and Wu, H. (2018). CELLULASE6 and MANNANASE7 affect cell differentiation and silique dehiscence. Plant Physiol. 176, 2186–2201. doi: 10.1104/pp.17.01494
Jang, S.-J., Sato, M., Sato, K., Jitsuyama, Y., Fujino, K., Mori, H., et al. (2015). A single-nucleotide polymorphism in an endo-1,4-β-glucanase gene controls seed coat permeability in soybean. PloS One 10, e0128527. doi: 10.1371/journal.pone.0128527
Kemmerer, E. C. and Tucker, M. L. (1994). Comparative study of cellulases associated with adventitious root initiation, apical buds, and leaf, flower, and pod abscission zones in soybean. Plant Physiol. 104, 557–562. doi: 10.1104/pp.104.2.557
Kim, J., Yang, J., Yang, R., Sicher, R. C., Chang, C., and Tucker, M. L. (2016). Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Front. Plant Sci. 7, 125. doi: 10.3389/fpls.2016.00125
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lane, D. R., Wiedemeier, A., Peng, L., Höfte, H., Vernhettes, S., Desprez, T., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126, 278–288. doi: 10.1104/pp.126.1.278
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, X., Chen, Z., Li, H., Yue, L., Tan, C., Liu, H., et al. (2024). Dt1 inhibits SWEET-mediated sucrose transport to regulate photoperiod-dependent seed weight in soybean. Mol. Plant 17, 496–508. doi: 10.1016/j.molp.2024.02.007
Li, R., Shi, C.-L., Wang, X., Meng, Y., Cheng, L., Jiang, C.-Z., et al. (2021). Inflorescence abscission protein SlIDL6 promotes low light intensity-induced tomato flower abscission. Plant Physiol. 186, 1288–1301. doi: 10.1093/plphys/kiab121
Li, C., Zhao, M., Ma, X., Wen, Z., Ying, P., Peng, M., et al. (2019). The HD-Zip transcription factor LcHB2 regulates litchi fruit abscission through the activation of two cellulase genes. J. Exp. Bot. 70, 5189–5203. doi: 10.1093/jxb/erz276
Libertini, E., Li, Y., and McQueen-Mason, S. J. (2004). Phylogenetic analysis of the plant endo-β-1,4-glucanase gene family. J. Mol. Evol. 58, 506–515. doi: 10.1007/s00239-003-2571-x
Lin, H., Wu, Z., Zhou, R., Chen, B., Zhong, Z., and Jiang, F. (2023). SlGH9–15 regulates tomato fruit cracking with hormonal and abiotic stress responsiveness cis-elements. J. Integr. Agric. 22, 447–463. doi: 10.1016/j.jia.2022.09.013
Liu, Y., Du, H., Li, P., Shen, Y., Peng, H., Liu, S., et al. (2020). Pan-genome of wild and cultivated soybeans. Cell 182, 162–176.e13. doi: 10.1016/j.cell.2020.05.023
Liu, Y., Zhang, Y., Liu, X., Shen, Y., Tian, D., Yang, X., et al. (2023). SoyOmics: a deeply integrated database on soybean multi-omics. Mol. Plant 16, 794–797. doi: 10.1016/j.molp.2023.03.011
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, L., Bai, J., Yuan, S., Guo, L., Liu, Z., Guo, H., et al. (2022b). Genome wide identification and characterization of wheat GH9 genes reveals their roles in pollen development and anther dehiscence. Int. J. Mol. Sci. 23, 6324. doi: 10.3390/ijms23116324
Luo, G., Huang, X., Chen, J., Luo, J., Liu, Y., Tang, Y., et al. (2022a). Systematic analysis of the grafting-related glucanase-encoding GH9 family genes in pepper, tomato and tobacco. Plants 11, 2092. doi: 10.3390/plants11162092
Meng, F., Wang, P., Mo, F., Qi, H., Lv, R., Cheng, M., et al. (2024). Endocellulase SlGH9–21 significantly improves drought resistance and storage capacity of tomato. Scientia Hortic. 323, 112513. doi: 10.1016/j.scienta.2023.112513
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Nawaz, M. A., Rehman, H. M., Imtiaz, M., Baloch, F. S., Lee, J. D., Yang, S. H., et al. (2017). Systems identification and characterization of cell wall reassembly and degradation related genes in Glycine max (L.) merill, a bioenergy legume. Sci. Rep. 7, 10862. doi: 10.1038/s41598-017-11495-4
Notaguchi, M., Kurotani, K., Sato, Y., Tabata, R., Kawakatsu, Y., Okayasu, K., et al. (2020). Cell-cell adhesion in plant grafting is facilitated by β-1,4-glucanases. Science 369, 698–702. doi: 10.1126/science.abc3710
Rio, D. C., Ares, M., Hannon, G. J., and Nilsen, T. W. (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, pdb.prot5439. doi: 10.1101/pdb.prot5439
Robert, S., Bichet, A., Grandjean, O., Kierzkowski, D., Satiat-Jeunemaître, B., Pelletier, S., et al. (2005). An Arabidopsis endo-1,4-β-D-glucanase involved in cellulose synthesis undergoes regulated intracellular cycling. Plant Cell 17, 3378–3389. doi: 10.1105/tpc.105.036228
Sandve, S. R., Rohlfs, R. V., and Hvidsten, T. R. (2018). Subfunctionalization versus neofunctionalization after whole-genome duplication. Nat. Genet. 50, 908–909. doi: 10.1038/s41588-018-0162-4
Su, G., Lin, Y., Wang, C., Lu, J., Liu, Z., He, Z., et al. (2024). Expansin SlExp1 and endoglucanase SlCel2 synergistically promote fruit softening and cell wall disassembly in tomato. Plant Cell 36, 709–726. doi: 10.1093/plcell/koad291
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W.-H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Tang, H., Krishnakumar, V., Zeng, X., Xu, Z., Taranto, A., Lomas, J. S., et al. (2024). JCVI: a versatile toolkit for comparative genomics analysis. iMeta 3, e211. doi: 10.1002/imt2.211
Tian, Z., Nepomuceno, A. L., Song, Q., Stupar, R. M., Liu, B., Kong, F., et al. (2025). Soybean2035: a decadal vision for soybean functional genomics and breeding. Mol. Plant 18, 245–271. doi: 10.1016/j.molp.2025.01.004
Tranbarger, T. J., Tucker, M. L., Roberts, J. A., and Meir, S. (2017). Editorial: Plant organ abscission: from models to crops. Front. Plant Sci. 8, 196. doi: 10.3389/fpls.2017.00196
Tucker, M. L., Burke, A., Murphy, C. A., Thai, V. K., and Ehrenfried, M. L. (2007). Gene expression profiles for cell wall-modifying proteins associated with soybean cyst nematode infection, petiole abscission, root tips, flowers, apical buds, and leaves. J. Exp. Bot. 58, 3395–3406. doi: 10.1093/jxb/erm188
Tucker, M. L., Whitelaw, C. A., Lyssenko, N. N., and Nath, P. (2002). Functional analysis of regulatory elements in the gene promoter for an abscission-specific cellulase from bean and isolation, expression, and binding affinity of three TGA-type basic leucine zipper transcription factors. Plant Physiol. 130, 1487–1496. doi: 10.1104/pp.007971
Urbanowicz, B. R., Bennett, A. B., del Campillo, E., Catalá, C., Hayashi, T., Henrissat, B., et al. (2007). Structural organization and a standardized nomenclature for plant endo-1,4-β-glucanases (cellulases) of glycosyl hydrolase family 9. Plant Physiol. 144, 1693–1696. doi: 10.1104/pp.107.102574
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023b). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Wang, D., Hu, X., Ye, H., Wang, Y., Yang, Q., Liang, X., et al. (2023a). Cell-specific clock-controlled gene expression program regulates rhythmic fiber cell growth in cotton. Genome Biol. 24, 49. doi: 10.1186/s13059-023-02886-0
Wang, Y., Tang, H., Wang, X., Sun, Y., Joseph, P. V., and Paterson, A. H. (2024). Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 19, 2206–2229. doi: 10.1038/s41596-024-00968-2
Wang, Y., Xu, Y., Liao, F., Li, T., Li, X., Wu, B., et al. (2023c). Genome-wide identification of GH9 gene family and the assessment of its role during fruit abscission zone formation in Vaccinium ashei. Plant Cell Rep. 42, 1589–1609. doi: 10.1007/s00299-023-03049-y
Wei, S., Yong, B., Jiang, H., An, Z., Wang, Y., Li, B., et al. (2023). A loss-of-function mutant allele of a glycosyl hydrolase gene has been co-opted for seed weight control during soybean domestication. J. Integr. Plant Biol. 65, 2469–2489. doi: 10.1111/jipb.13559
Woo, M.-O., Beard, H., MacDonald, M. H., Brewer, E. P., Youssef, R. M., Kim, H., et al. (2014). Manipulation of two α-endo-β-1,4-glucanase genes, 6 and 7, reduces susceptibility to eterodera glycines in soybean roots. Mol. Plant Pathol. 15, 927–939. doi: 10.1111/mpp.12157
Wu, F., Price, B. W., Haider, W., Seufferheld, G., Nelson, R., and Hanzawa, Y. (2014). Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PloS One 9, e85754. doi: 10.1371/journal.pone.0085754
Xie, G., Yang, B., Xu, Z., Li, F., Guo, K., Zhang, M., et al. (2013). Global identification of multiple OsGH9 family members and their involvement in cellulose crystallinity modification in rice. PloS One 8, e50171. doi: 10.1371/journal.pone.0050171
Yang, S., Hu, M., Wu, R., Hou, Z., Zhang, H., He, W., et al. (2024a). Genetic evidence of SpGH9A3 in leaf morphology variation of Spathiphyllum ‘mojo.’. Genes 15, 1132. doi: 10.3390/genes15091132
Yang, Z., Luo, C., Pei, X., Wang, S., Huang, Y., Li, J., et al. (2024b). SoyMD: a platform combining multi-omics data with various tools for soybean research and breeding. Nucleic Acids Res. 52, D1639–D1650. doi: 10.1093/nar/gkad786
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. doi: 10.1186/1471-2105-13-134
Zhang, Z. (2022). KaKs_Calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genomics Proteomics Bioinf. 20, 536–540. doi: 10.1016/j.gpb.2021.12.002
Zhang, R., Jia, G., and Diao, X. (2023). geneHapR: an R package for gene haplotypic statistics and visualization. BMC Bioinf. 24, 199. doi: 10.1186/s12859-023-05318-9
Keywords: soybean, abscission cellulase, haplotype analysis, glycosyl hydrolase 9, ethylene response
Citation: Zhan W, Huang H, Cai Z, Zhao Z, Lin X, Peng Y, Dong Z, Qin D and Jiang L (2025) Genome-wide identification and characterization of soybean GH9 endo-1,4-β-glucanases. Front. Plant Sci. 16:1597668. doi: 10.3389/fpls.2025.1597668
Received: 31 March 2025; Accepted: 20 May 2025;
Published: 13 June 2025.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Aimin Wu, Guangzhou University, ChinaAshita Bisht, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, India
Longguo Jin, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2025 Zhan, Huang, Cai, Zhao, Lin, Peng, Dong, Qin and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Di Qin, cWluZGlAZ3podS5lZHUuY24=; Li Jiang, bGlqaWFuZ3Jlc0AxNjMuY29t
†These authors have contributed equally to this work
 Weimin Zhan
Weimin Zhan Huijuan Huang1†
Huijuan Huang1† Zhicheng Dong
Zhicheng Dong Li Jiang
Li Jiang