- 1College of Agriculture and Biotechnology, Hunan University of Humanities, Science and Technology, Loudi, Hunan, China
- 2Key Laboratory of Characteristic Agricultural Resource Development and Quality Safety Control in Hunan Province, Hunan University of Humanities, Science and Technology, Loudi, Hunan, China
- 3Innovation and Entrepreneurship Education Center for Horticultural Production and Processing in Hunan Province, Hunan University of Humanities, Science and Technology, Loudi, Hunan, China
- 4Hunan University of Humanities, Science and Technology, Hunan Provincial Innovation and Entrepreneurship Demonstration Base, Loudi, Hunan, China
Introduction: This study reports for the first time the complete sequence characteristics of the mitochondrial genome of the tea plant cultivar Camellia sinensis cv. ‘Baihaozao’. It systematically unveils its multi-chromosomal structure, RNA editing patterns, and adaptive evolutionary mechanisms, providing critical theoretical insights into the structural complexity and evolutionary mechanisms of the tea plant mitochondrial genome.
Methods: The mitochondrial genome was fully analyzed using genome sequencing and annotation techniques. RNA editing sites were predicted to evaluate editing patterns. Codon usage bias analysis was conducted to identify high-frequency codons. Repeat sequence analysis was used to characterize dispersed and tandem repeats. Adaptive evolutionary analysis, based on Ka/Ks ratios, was performed to investigate gene selection pressures.
Results: The mitochondrial genome consists of 11 linear chromosomes, with a total length of 909,843 bp and a GC content of 45.62%. A total of 73 functional genes were annotated, among which 14 variable genes (e.g., ribosomal protein coding genes) retain intact functions without pseudogenization, which is rare among Theaceae plants. RNA editing site prediction revealed significant spatial heterogeneity, with the cox1 gene being a hotspot containing 19 editing sites. Approximately 58.49% of editing events were concentrated on the second base of codons, and 48.61% of the sites resulted in amino acid changes from hydrophilic to hydrophobic. Codon usage bias analysis showed significant enrichment of high-frequency codons, including UUU (phenylalanine), AUU (isoleucine), and UUC (phenylalanine). The genome’s repeat sequences were predominantly dispersed repeats (70.6%), with forward and palindromic repeats of 30–40 bp being dominant. Tandem repeats exhibited significant distribution heterogeneity among chromosomes. Adaptive evolution analysis showed that most PCGs (protein-coding genes) had Ka/Ks ratios below 1 (ranging from 0.07 to 0.78), with the atp9 gene showing the lowest ratio (0.07), while the mttB gene exhibited a significantly higher Ka/Ks ratio of 3.48. Additionally, 1.62% of the mitochondrial genome sequence was homologous to the chloroplast genome, carrying 26 complete functional genes, including 15 tRNA and 2 rRNA genes.
Discussion: Codon usage bias may be related to mutation pressure due to the high AT content of the genome or reflect adaptive selection pressures for translational efficiency. The Ka/Ks results align with the widespread purifying selection observed in mitochondrial genomes, while the high Ka/Ks ratio of the mttB gene suggests it might be under positive selection to adapt to environmental pressures. The evolutionary evidence of inter-organelle gene transfer highlights the homologous sequences between mitochondria and chloroplasts. Overall, these findings systematically elucidate the adaptive evolutionary mechanisms and functional regulation of the tea plant mitochondrial genome.
1 Introduction
The mitochondrial genome of plants, as a unique vector of maternal inheritance, holds significant scientific value in studies of phylogenetics and genetic diversity due to its dynamic recombination characteristics and adaptive evolutionary mechanisms (Lu et al., 2022; Li et al., 2023a). In recent years, breakthroughs in third-generation sequencing technologies have enabled research on the mitochondrial genomes of Camellia species, revealing key features such as gene recombination, dynamic equilibrium of repetitive sequences, and inter-organellar gene transfer (Lu et al., 2022; Li et al., 2023a). However, existing studies predominantly focus on cultivated varieties (e.g., C. sinensis var. assamica) and oilseed camellia (C. gigantocarpa) (Lu et al., 2022; Li et al., 2023a), while structural analyses of mitochondrial genomes in unique varieties of both economic and ecological significance—such as the early-germinating and stress-resistant tea cultivar Camellia sinensis cv. ‘Baihaozao’—remain unexplored. Traditional mitochondrial genome studies have largely been based on a single-chromosome model (Kozik et al., 2019). However, the recently revealed multichromosomal structures in species such as cucumber (Cucumis sativus) and poplar (Populus simonii) (Alverson et al., 2011; Bi et al., 2022) suggest that the mitochondrial genome of Camellia may exhibit greater structural diversity, which urgently requires investigation using advanced technological approaches.
Current research reveals that the mitochondrial genome of tea plants exhibits significant recombination polymorphism: the cultivated variety Duntsa (C. sinensis var. assamica cv. Duntsa) contains 18 homologous recombination regions, while different varieties (Camellia sinensis var. assamica OL, CSAOL and Camellia sinensis var. assamica OM, CSAOM) show a 44% difference in the distribution of repetitive sequences and the number of introns (Li et al., 2023a). Notably, the codon usage bias of the atp9 gene is driven by mutational pressure, whereas other genes are primarily influenced by natural selection (Li et al., 2023a). Additionally, the pseudogenization of the rps16 gene and mitochondrial-chloroplast gene transfer events (e.g., homologous sequences in the inverted repeat (IR) region accounting for over 90%) (Zhao et al., 2018) collectively outline the dynamic evolutionary landscape of the tea plant mitochondrial genome. However, these findings are based on the assumption of a single-chromosome model, overlooking critical issues such as the potential differences in allelic recombination frequency (Alverson et al., 2011) and replication autonomy (Bi et al., 2022) that may arise from multi-chromosomal structures. Of particular interest is the recent genome sequencing of Lingyun Baihao Tea, which reveals that wild germplasm differs significantly from cultivated varieties in morphological traits such as leaf area, serration density, and trichome coverage (He et al., 2023). This provides important insights into the relationship between mitochondrial genome structural variation and phenotypic plasticity.
As a unique early-maturing and high-quality germplasm native to China, Camellia sinensis cv. ‘Baihaozao’ serves as an ideal model for elucidating the adaptive evolution of mitochondrial genomes due to its distinctive stress-resistant metabolic pathways and rapid environmental response characteristics. This study employs an innovative hybrid sequencing strategy combining Illumina second-generation and third-generation sequencing technologies, overcoming the technical limitations of traditional circular genome assembly (Kozik et al., 2019), and successfully achieves the complete sequencing and assembly of a multi-chromosomal mitochondrial genome in Camellia. By integrating comparative genomics and adaptive evolution analyses, this study aims to elucidate: (1) the patterns of gene recombination under multi-chromosomal architectures and their impact on phenotypic plasticity; (2) the evolutionary relationship between rps16 gene pseudogenization and inter-organellar interactions; and (3) positive selection signals associated with stress-resistance traits and their molecular regulatory networks. These findings will fill the research gap in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ and provide a theoretical foundation for crop genetic improvement.
2 Materials and methods
2.1 Preparation of mitochondrial genome deoxyribonucleic acid
The experimental materials were obtained from tender shoots of tea plants (Camellia sinensis cv. ‘Baihaozao’) cultivated at Taohuayuan Modern Agriculture Development Co., Ltd., Xinhua County, Hunan Province, China (27°47′N, 110°51′E) (Figure 1). Total DNA was extracted using the TIANGEN Plant Genomic DNA Extraction Kit (DP305). Dual quality control was performed to ensure DNA integrity (fragment length >30 kb) and purity using a spectrophotometer (A260/A280 = 1.8–2.0, A260/A230 ≥ 2.0) and 1% agarose gel electrophoresis (0.8% agarose gel, 5 V/cm, 30 min) (Cui et al., 2024; Zhang et al., 2024a).
2.2 Multi-omics sequencing strategy
A hybrid sequencing library was constructed using Illumina NovaSeq 6000 (PE150 mode) and Oxford Nanopore PromethION (R10.4.1 flow cell). The construction of the second-generation library included standardized procedures such as ultrasonic fragmentation of 350 bp insert fragments, end repair using NEBNext Ultra II, A-tailing with Klenow polymerase, adapter ligation using T4 DNA ligase, and selection of target fragments with AMPure XP magnetic beads. The third-generation library was prepared using the Ligation Sequencing Kit (SQK-LSK114), followed by large-fragment selection using BluePippin (>30 kb selection window) (Shin et al., 2019; Cui et al., 2024).
Sequencing Data Preprocessing: Raw second-generation sequencing data were filtered using fastp (v0.20.0) with the following parameters: -q 5 -n 5 -l 50. Nanopore sequencing data underwent basecalling using Guppy (v6.4.2), removal of chimeric adapters using Porechop (v0.2.4), and error correction of long reads using LoRDEC (v0.6) (Shin et al., 2019; Yang and Duan, 2024). As a result, 20 Gb of second-generation clean data (Q30 > 90%) and third-generation data with 50× genome coverage (N50 > 30 kb) were obtained.
2.3 Multi-chromosomal assembly and validation
Using an adaptive k-mer weighting algorithm, de novo assembly of third-generation sequencing data was performed with Canu (v2.2) (parameters: genomeSize=600 k, corMhapSensitivity=high, corMinCoverage=50). Contigs were aligned to the NCBI reference genome (NC_037880.1) using Minimap2 (v2.24), and mitochondrial homologous sequences were identified via BlastN (e-value=1e-5) (Li, 2018; Yang and Duan, 2024).
Assembly Optimization Workflow: Iterative correction of the assembly was conducted using Pilon (v1.24) with second-generation data (3 rounds); Hybrid data polishing was performed using NextPolish (v1.3.1); linear structures were validated using Circlator (v1.5.5) and confirmed by reverse polymerase chain reaction (PCR) amplification (PrimeSTAR GXL DNA polymerase) and Sanger sequencing at the junction sites (Shin et al., 2019; Schiavone et al., 2022). The final assembly yielded a complete mitochondrial genome with a multi-chromosomal structure. Evaluation using QUAST (v5.2.0) demonstrated high assembly continuity (N50 >100 kb) and completeness (BUSCO > 95%), meeting reference genome standards (Schiavone et al., 2022; Yang and Duan, 2024).
2.4 Annotation of mitochondrial gene structure
The annotation of protein-coding genes and rRNA was performed using the BLAST algorithm (gapped BLAST and PSI-BLAST) through multiple sequence alignments with published plant mitochondrial reference genomes. Manual curation was conducted to ensure annotation accuracy. tRNA genes were systematically identified using tRNAscan-SE software, which employs a covariance model-based algorithm to efficiently detect secondary structural features of transfer RNAs, significantly reducing false-positive rates. Open reading frames were predicted using the national center for biotechnology information (NCBI) Open Reading Frame Finder with a minimum length threshold of 102 bp, and the results were manually corrected to exclude non-functional fragments (Tatusova et al., 2016). The final annotation results were visualized using the OGDRAW tool to generate a genomic circular map and submitted to the NCBI database in tbl format, adhering to standard mitochondrial genome annotation protocols (Tatusova et al., 2016; Li et al., 2023a).
2.5 Analysis of mitochondrial genome repeats
Dispersed repeat sequences were identified using vmatch (v2.3.0) in combination with Perl scripts. The parameters were set to a minimum length of 30 bp and a Hamming distance of 3, covering four types of repeats: direct, palindromic, reverse, and complementary. Simple sequence repeats (SSRs) were identified using the MISA software, which utilizes a statistical model based on repeat unit counts to systematically detect microsatellite loci, providing marker resources for subsequent population genetics studies (Li et al., 2023a).
2.6 Phylogenetic analysis
The mitochondrial genome sequences of 26 representative plant species from five different families were downloaded from the NCBI database. Phylogenetic trees were constructed using the maximum likelihood and neighbor-joining methods in MEGA 7.0 software. Bootstrap resampling was performed 1000 times to evaluate node support. The analysis was based on nucleotide substitution models (e.g., Kimura 2-parameter) to ensure the statistical reliability of the tree topology (Li et al., 2023a).
2.7 Ka/Ks analysis
Pairwise comparisons of homologous genes were conducted using MAFFT (v7.310) for multiple sequence alignment, followed by calculations of non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) using KaKs_Calculator v2.0. The MLWL algorithm was employed to optimize the evaluation of evolutionary pressure. The results were presented as box plots, revealing the functional divergence of different genes during adaptive evolution (Mao et al., 2005; Li et al., 2023a).
2.8 RNA editing sites and variation analysis of the mitochondrial genome
RNA editing sites were predicted using the PREPACT online platform, which employs an algorithm based on the conservation of plant mitochondrial RNA editing sites to identify C-to-U transition sites (Jühling et al., 2012). Nucleotide diversity (π value) was calculated using DNAsp v5 after global alignment with MAFFT (v7.427, –auto mode), reflecting the degree of sequence variation among species (Li et al., 2023a). Homologous regions between the chloroplast and mitochondrial genomes were identified using BLAST (similarity ≥70%, E-value ≤10-5) and visualized with Circos (v0.69-5) to depict genome-wide synteny relationships (Li et al., 2023a). Synteny analysis was performed using nucmer (v4.0.0beta2, –maxmatch parameter), and a dot-plot was generated to reveal genome rearrangement events (Li et al., 2023a).
3 Results
3.1 Multi-chromosomal structure of the mitochondrial genome in Camellia sinensis cv. ‘Baihaozao’
In this study, the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ was sequenced using the Illumina NovaSeq 6000 high-throughput sequencing platform. Quality analysis of the sequencing data showed that the Q20 (the proportion of bases with a quality score ≥ 20) and Q30 (the proportion of bases with a quality score ≥ 30) reached 97.00% and 92.27%, respectively, meeting the typical performance metrics of the platform, where Q20 generally exceeds 97% and Q30 surpasses 92% in transcriptome sequencing. After quality control of the sequencing data, a total of 16.14 Gb (16,140,581,400 bp) of clean data was obtained, which falls within the theoretical throughput range of a single run on the Illumina platform. Detailed data information is provided in Supplementary Table 1.
This study conducted deep sequencing and assembly of the mitochondrial genome of the tea cultivar Camellia sinensis cv. ‘Baihaozao’. The complete mitochondrial genome sequence has been submitted to GenBank (accession numbers: PV340641–PV340651). The results revealed that the mitochondrial genome is composed of 11 linear chromosomes (Figure 2; Supplementary Table 2), with a total length of 909,843 bp (individual chromosome lengths ranging from 1,718 to 205,065 bp) and a GC content of 46% (Supplementary Table 3). This multi-chromosomal structure is consistent with the characteristic mitochondrial genome organization commonly observed in Theaceae plants (Li et al., 2023a), but the total length is significantly shorter than that of C. sinensis var. assamica cv. ‘Duntsa’ (1.08 Mb), indicating notable intraspecific length polymorphism in tea mitochondrial genomes.
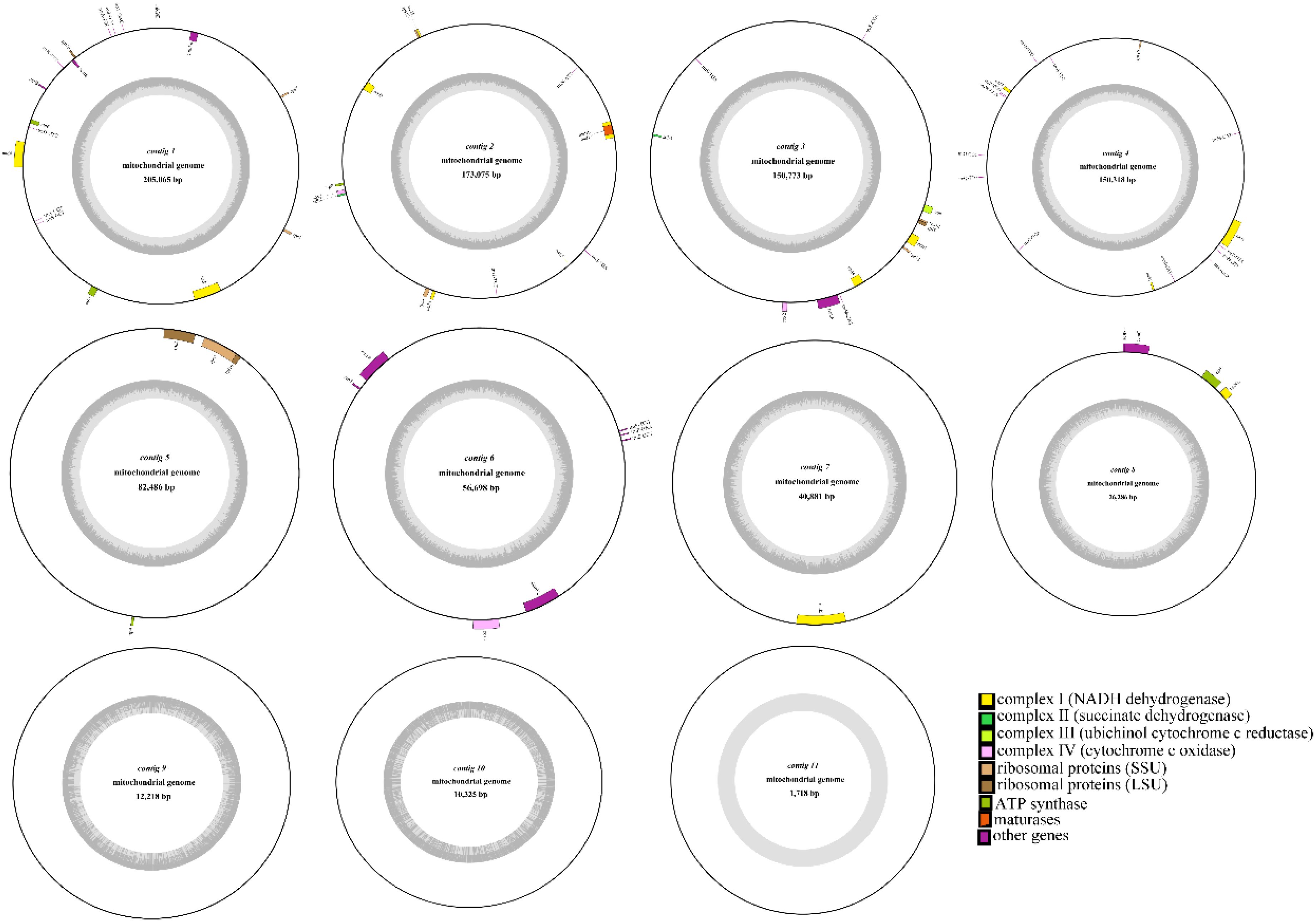
Figure 2. Mitochondrial genome map. The mitochondrial genome consists of 11 linear chromosomes. In the linear diagram, forward-strand genes are located on the outer side of the circle, while reverse-strand genes are on the inner side. The inner gray circle represents GC content.
Genome annotation identified a total of 73 functional genes, including 38 protein-coding genes, 30 tRNA genes, 3 rRNA genes, and 2 pseudogenes (Table 1; Supplementary Table 4). Among these, the GC content of protein-coding genes (43%) was significantly lower than that of tRNA genes (50%) and rRNA genes (52%), which may be attributed to differential selective pressures acting on functional elements (Li et al., 2023a). The core gene set includes 24 conserved genes, covering adenosine triphosphate (ATP) synthase subunits (atp1/4/6/8/9), nicotinamide adenine dinucleotide (NADH) dehydrogenase subunits (nad1-7/9), cytochrome c synthesis-related genes (ccmB/C/Fc/Fn), and cytochrome oxidase subunits (cox1-3). This gene composition aligns closely with the conserved features of mitochondrial genomes in Theaceae plants (Li et al., 2023a). Notably, the 14 variable genes include those encoding ribosomal large subunit proteins (rpl2/5/10/16) and small subunit proteins (rps1/3-4/7/12-14/19). These genes are frequently lost or pseudogenized in some Theaceae plants (Li et al., 2023a), yet they remain fully functional in Camellia sinensis cv. ‘Baihaozao’. This suggests that the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ exhibits a relatively high degree of evolutionary conservation.
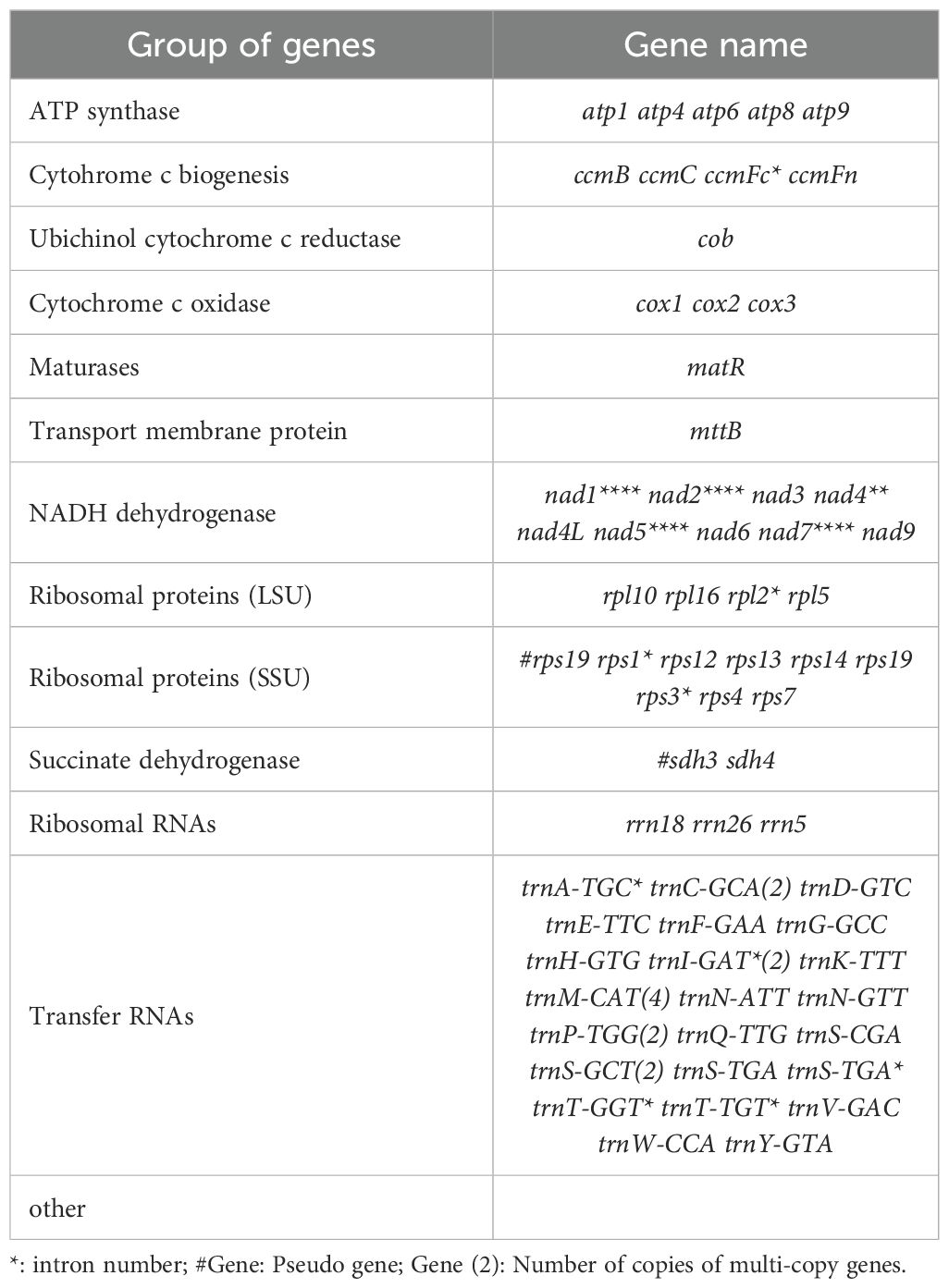
Table 1. Functional classification of genes in the Camellia sinensis cv. ‘Baihaozao’ mitochondrial genome.
Gene structure analysis revealed that trnM-CAT has four copies, while trnC-GCA and trnI-GAT are present in two copies. The distribution of introns showed significant heterogeneity: nad4 contains two introns, while nad1/2/5/7 each contain four introns, consistent with the intron expansion trend observed in the mitochondrial genomes of Theaceae plants (Li et al., 2023a). Notably, the sdh3 and rps19 genes each contain one pseudogene, which may be associated with gene fragmentation during mitochondrial genome recombination (Li et al., 2023a). Such structural variations might play important roles in the adaptive evolution of Camellia species. For example, the retention of the sdh4 gene may contribute to the assembly of the succinate dehydrogenase complex, potentially influencing energy metabolism pathways (Wei et al., 2017).
This study is the first to systematically reveal the multi-chromosomal structural characteristics of the mitochondrial genome in Camellia sinensis cv. ‘Baihaozao’. The gene composition shows significant differences compared to closely related species, such as C. sinensis var. assamica (Li et al., 2023a). A comparison with the published chloroplast genome data revealed that the GC content of the mitochondrial genome (46%) is significantly higher than that of the chloroplast genome (37%). This difference may be attributed to the varying selective pressures acting on the genomes of different organelles (Li et al., 2023a).
3.2 Prediction of RNA editing sites
RNA editing, a key regulatory mechanism in the expression of mitochondrial and chloroplast genes in eukaryotes, involves the specific modification of transcripts to maintain the functional integrity of organelles (Mower, 2005; Bi et al., 2020). In this study, the PREP-Mt algorithm was used to systematically analyze the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’, identifying 159 C-to-U RNA editing sites across 25 protein-coding genes (PCGs) (Figure 3). These findings align with previous reports on the mitochondrial genome of Phaseolus vulgaris (486 sites) and the chloroplast genome of Ginkgo biloba (255 sites), supporting the dominant role of C-to-U editing in plant organelles (He et al., 2016; Bi et al., 2020).
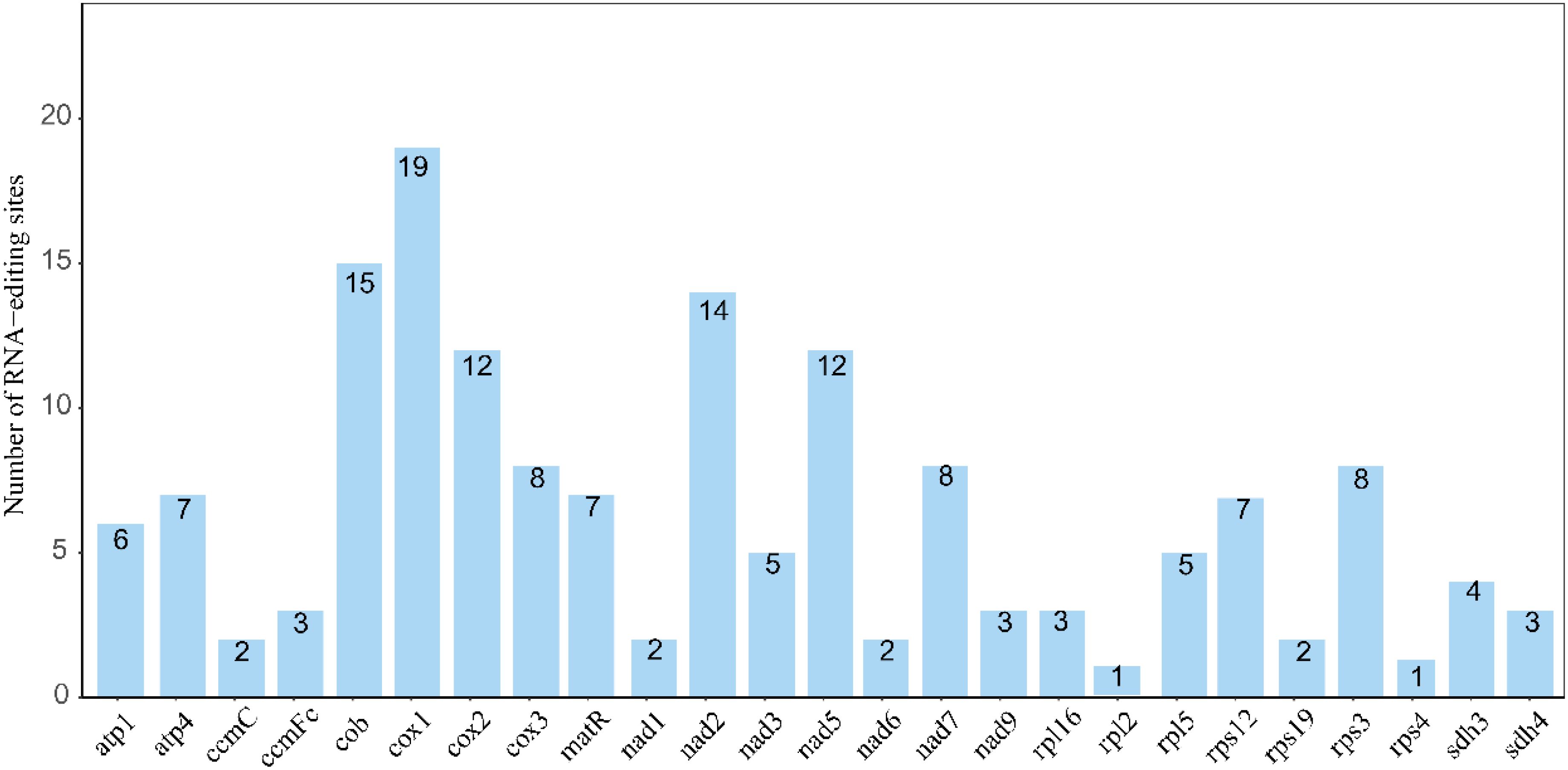
Figure 3. Statistics of RNA editing site numbers for each gene. The horizontal axis represents genes, and the vertical axis represents the number of RNA editing sites.
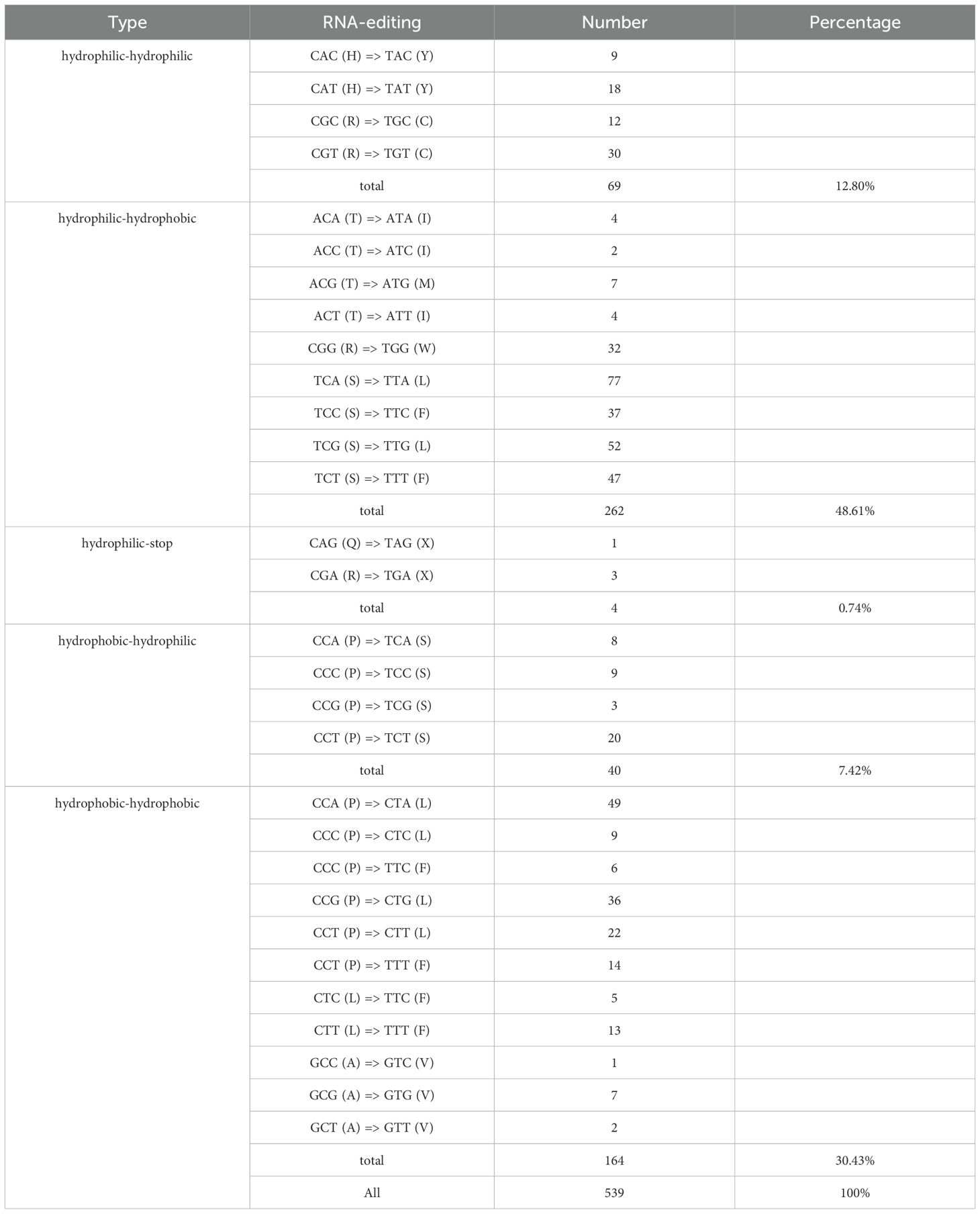
This study identified 19 RNA editing sites in the cox1 gene, significantly higher than the single editing site observed in the rpl2 and rps4 genes. This phenomenon is consistent with the general pattern of high editing frequencies observed in genes associated with mitochondrial complex I (Zhang et al., 2022b). Notably, a similar high number of editing sites in the cox1 gene has not been widely reported in other species, suggesting that this feature may be specific to mitochondrial genomes in the genus Camellia (Cuenca et al., 2010; Zhang et al., 2022b).Codon position analysis revealed that editing events were predominantly located at the first (32%) and second (58%) bases, with markedly fewer at the third position (9%). This contrasts sharply with the complete absence of third-position editing in Ilex metabaptista (Aquifoliaceae), suggesting that different plant lineages may have evolved distinct strategies for editing regulation (Yura et al., 2008).
From a molecular mechanism perspective, 96% of the editing events corresponded to C-to-T changes at the DNA level (i.e., C-to-U editing at the RNA level), consistent with observations in model plants such as Arabidopsis thaliana and rice (Cummings and Myers, 2004; Zheng et al., 2020). Notably, leucine (43%) emerged as the most frequently produced amino acid following editing, potentially reflecting its critical role in transmembrane protein domains (He et al., 2016). Among the amino acid changes induced by RNA editing, 49% involve a transition from hydrophilic to hydrophobic residues. This hydrophobicity-biased trend aligns with the hypothesis proposed in studies on Ginkgo chloroplasts that suggests “enhanced protein structural stability” as a potential outcome (Daley et al., 2002). Although functional optimization of membrane proteins may be an underlying driving factor, the specific mechanisms remain to be further validated. Additionally, 42% of the sites retain their original hydrophobicity, while only 7% undergo a reverse transition (Table 2), implying that RNA editing may contribute to the adaptive regulation of the mitochondrial energy metabolism system through multiple mechanisms. It is worth noting that the analysis of RNA editing sites in this study is based on computational prediction (in silico) and has yet to be experimentally validated using transcriptomic approaches such as RNA-Seq or RT-PCR. Recent studies have demonstrated that mitochondrial editing sites in tea plants can be experimentally confirmed through PCR amplification and sequencing techniques (e.g., Zhang et al. identified 139 mitochondrial editing sites) (Zhang et al., 2022b). Future research will integrate transcriptomic data and molecular experiments to further validate the biological functions and regulatory mechanisms of RNA editing sites.
3.3 Codon usage bias analysis
RSCU (Relative Synonymous Codon Usage) is a critical metric for assessing codon usage bias, and its calculation method and biological significance have been widely applied in genomic studies across multiple species (Qian et al., 2014; Yu et al., 2023). In this study, the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ contains a total of 10,674 codons across 38 protein-coding genes. Among these, the aliphatic amino acid leucine (Leu) is the most frequently used (1,093 occurrences, accounting for 10%), followed by serine (Ser, 999 occurrences, 9%) and arginine (Arg, 739 occurrences, 7%), while tryptophan (Trp) exhibits the lowest usage frequency (157 occurrences, 1%) (Figure 4; Supplementary Table 5). This distribution pattern is consistent with the amino acid usage preferences commonly observed in mitochondrial genomes, such as in the stonefly (Kamimuria wangi) and the land snail (Cernuella virgata), where Leu and Ser also show high expression trends (Qian et al., 2014; Lin et al., 2016).
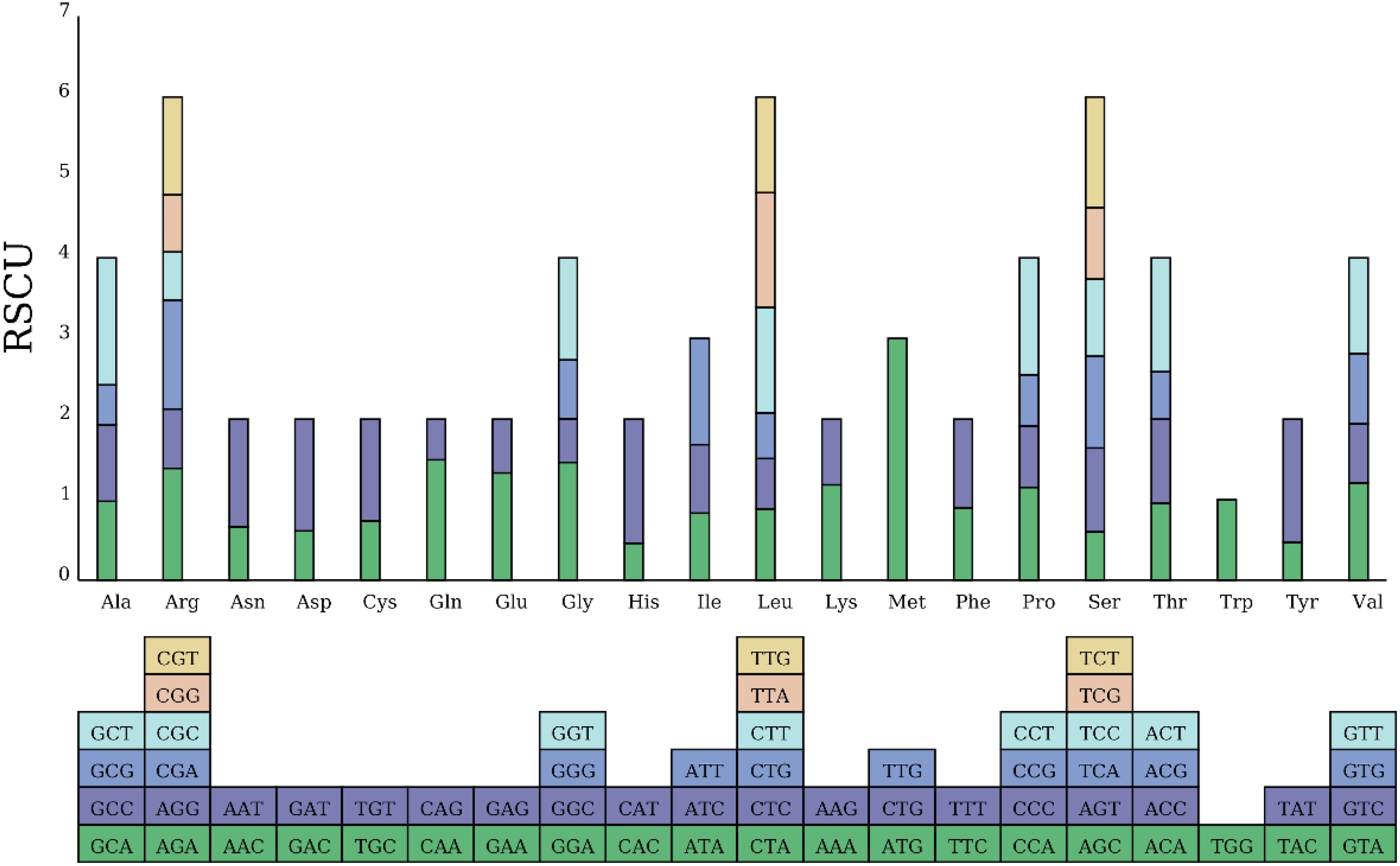
Figure 4. RSCU in the Camellia sinensis cv. ‘Baihaozao’ Mitochondrial Genome. The blocks below represent all codons encoding each amino acid, while the height of the bars above indicates the total RSCU values for all codons.
At the level of codon selection, there are 31 codons with RSCU > 1 in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’. Among them, the start codon AUG (methionine) exhibits the highest RSCU value (3.0), reflecting its conserved role as a translation initiation site and the strong selection pressure it undergoes (Sau et al., 2005; Yu et al., 2023). This study identified high-frequency codons UUU (371 occurrences, 3%), AUU (358 occurrences, 3%), and UUC (300 occurrences, 3%) in the mitochondrial genome. The preference for such codons may be associated with nucleotide compositional bias driven by the high AT content of the genome (Sun et al., 2015; Li et al., 2019), or it may reflect selective pressures related to translational efficiency or evolutionary processes (Bulmer, 1991; Hershberg and Petrov, 2008). Notably, the corresponding amino acids, such as phenylalanine (Phe) and isoleucine (Ile), are involved in mitochondrial energy metabolism pathways. However, the potential link between codon enrichment and metabolic function requires further experimental validation (Hershberg and Petrov, 2008; Sun et al., 2015). Notably, this preference for A/T-rich codons has also been reported in the mitochondrial genomes of adenoviruses (FAdV) and lepidopteran insects (Apostictopterus fuliginosus), suggesting that codon selection in mitochondrial genomes may be driven by both genomic adenine and thymine (AT) content and translation efficiency (Han et al., 2018; Niczyporuk, 2018).
Compared to other plant mitochondrial systems, the codon usage pattern of Camellia sinensis cv. ‘Baihaozao’ exhibits species-specific characteristics. For example, in creeping bentgrass (Agrostis stolonifera), the RSCU value of the alanine (Ala) codon GCU is significantly higher than other synonymous codons (Li et al., 2023b), whereas in Camellia sinensis cv. ‘Baihaozao’ the leucine codon UUA is the dominant type. This difference may reflect the varying adaptive functional requirements of mitochondrial genomes during the evolutionary process among different species (Wang et al., 2019).
3.4 Repetitive sequence characteristics of the mitochondrial genome in Camellia sinensis cv. ‘Baihaozao’
The systematic analysis of repetitive sequences in plant mitochondrial genomes is of great significance for understanding their structural dynamics and evolutionary mechanisms. In this study, a comprehensive analysis revealed that the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ contains three types of repetitive sequences: SSRs, tandem repeats, and dispersed repeats. This classification aligns with the classical definitions of repetitive sequences in plant organellar genomes. Specifically, SSRs, as a specialized type of tandem repeats, offer unique advantages in the development of genetic markers, while dispersed repeats significantly influence genome conformation by mediating homologous recombination (Zhu et al., 2024).
Quantitative analysis detected a total of 1,032 repetitive sequences in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’, of which SSRs (264 sequences) and tandem repeats (39 sequences) accounted for 26% and 4%, respectively, whereas dispersed repeats dominated with 729 sequences (71%) (Figure 5; Supplementary Table 6). This distribution pattern is similar to that of the mitochondrial genome in Prunus armeniaca (Rosaceae), where dispersed repeats account for 68%, suggesting that this type of repeat plays a widespread role in mitochondrial genome expansion in plants. Notably, the total length of dispersed repeats in Camellia sinensis cv. ‘Baihaozao’ reached 55,543 bp, comprising 6% of the genome, which is significantly lower than in Selenicereus monacanthus (85%, Solanaceae) (Lu et al., 2023), but higher than in common bean (Phaseolus vulgaris, 9%), reflecting substantial interspecies differences in the accumulation of repetitive sequences.
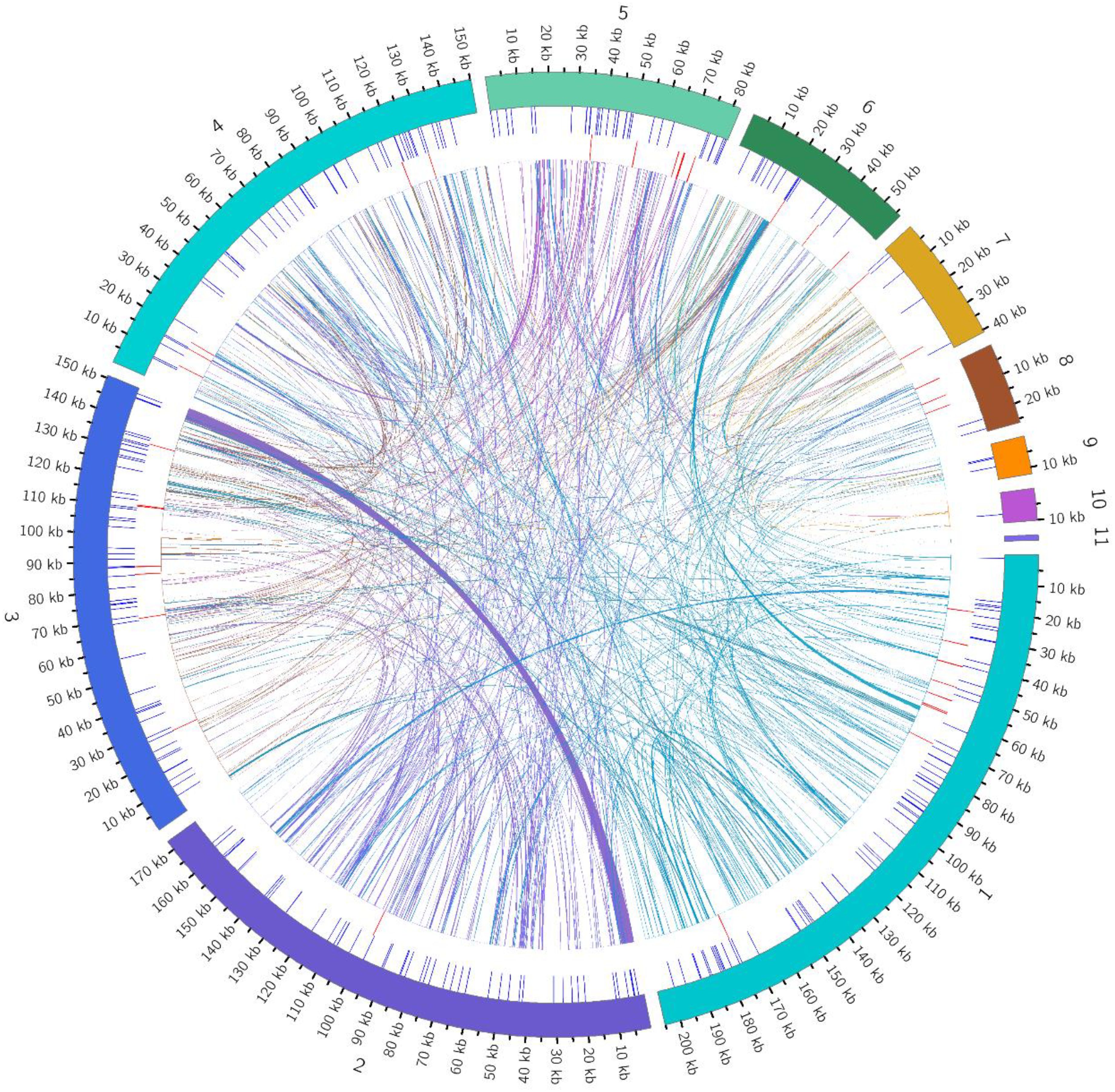
Figure 5. Distribution of repetitive sequences in the mitochondrial genome. The outermost circle represents the mitochondrial genome sequence, followed inward by simple sequence repeats, tandem repeats, and dispersed repeats.
Subtype analysis of dispersed repeats revealed that forward repeats (333 sequences, 46%) and palindromic repeats (396 sequences, 54%) are the predominant types (Supplementary Table 7). These results are consistent with the general trend observed in plant mitochondrial genomes, where forward and palindromic repeats dominate. The presence of the largest forward repeat (890 bp) and palindromic repeat (4,713 bp) suggests their potential roles in homologous recombination, contributing to the formation of subgenomic linear molecules. This phenomenon has been experimentally validated in the multichromosomal structure of potato (Solanum tuberosum, Solanaceae). Additionally, dispersed repeats in the 30–40 bp length range were the most abundant (55%–70%), consistent with the observed length threshold (≥28 bp) in this study, indicating its universality in the analysis of plant mitochondrial repetitive sequences.
The analysis of tandem repeats (satellite DNA) showed that the 39 tandem repeats in Camellia sinensis cv. ‘Baihaozao’ ranged in length from 12 to 54 bp, with a match percentage >70%. Their chromosomal distribution exhibited significant heterogeneity: chr1 and chr5 each contained nine tandem repeats, while chr9–11 were entirely devoid of such repeats (Supplementary Table 8). This uneven distribution pattern is consistent with the mitochondrial genome of Cymbidium ensifolium (Orchidaceae), where tandem repeats are enriched in chr2 and chr7 but absent in chr13 (Shen et al., 2024). A comparative analysis revealed that the number of tandem repeats in Camellia sinensis cv. ‘Baihaozao’ is significantly higher than in common bean (7 repeats) but lower than in hawthorn (Crataegus pinnatifida, 22 repeats) (Zhu et al., 2024), indicating high plasticity of this repeat type among species. Notably, tandem repeats located in rRNA coding regions (e.g., the rrnL gene in common bean) may influence transcriptional regulation. However, no tandem repeats were found in the coding regions of Camellia sinensis cv. ‘Baihaozao’, suggesting that their function may primarily be associated with maintaining the structural integrity of non-coding regions.
3.5 Phylogenetic inference
The phylogenetic tree constructed based on 38 mitochondrial protein-coding genes (Figure 6) revealed that the 26 representative species were clustered into five monophyletic groups. Brassicaceae species (Isatis tinctoria, Rorippa indica, and Lepidium apetalum) were used as the outgroup, while the remaining species were grouped into Actinidiaceae, Ebenaceae, Ericaceae, and Theaceae. Camellia sinensis cv. ‘Baihaozao’ was clearly clustered within the Camellia clade of Theaceae, forming a highly supported monophyletic group (bootstrap value >90%) with C. lanceoleosa, C. oleifera, and other species, confirming the taxonomic reliability of its classification.
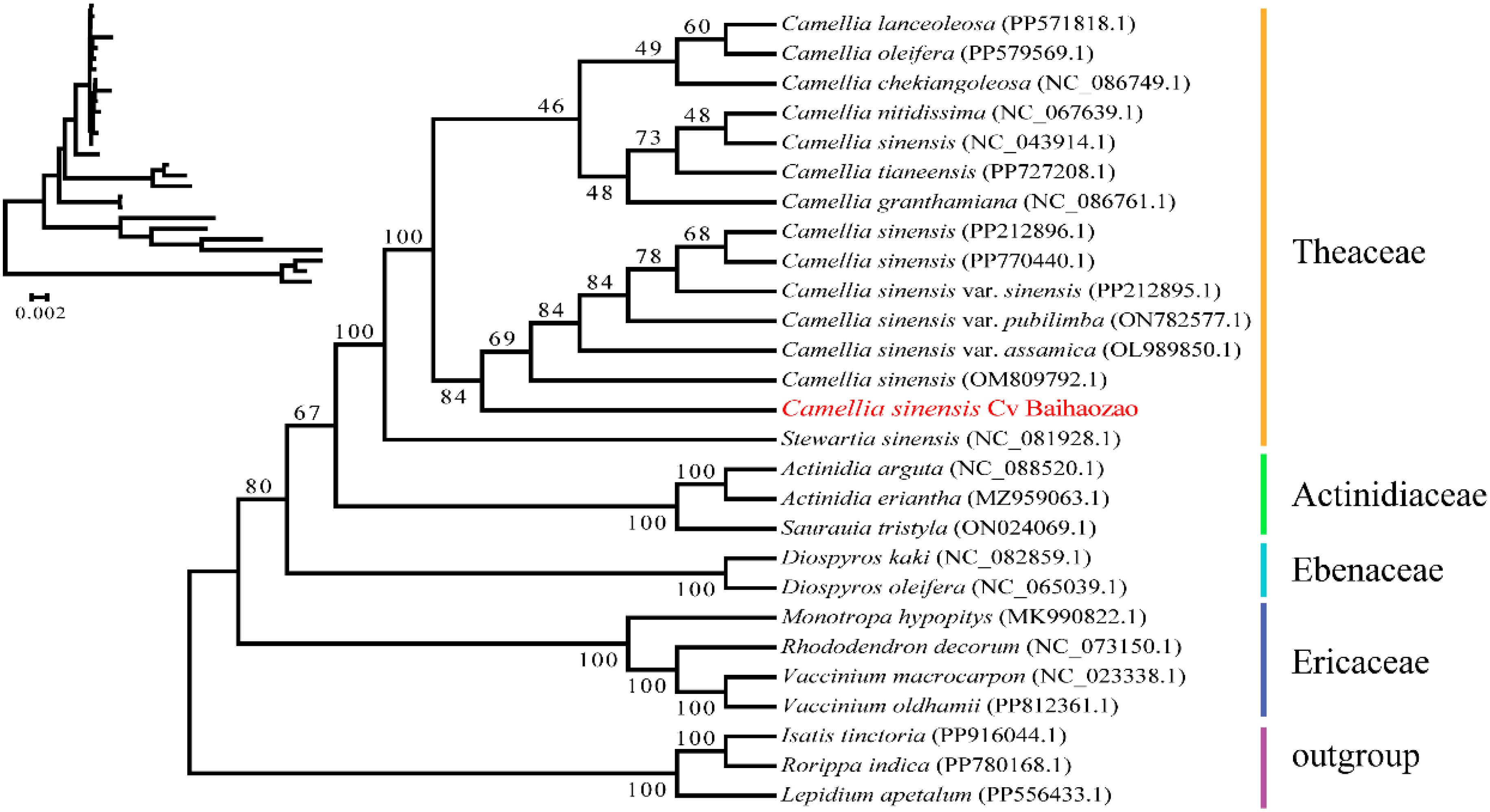
Figure 6. Phylogenetic relationships between Camellia sinensis cv. ‘Baihaozao’ and 26 other plant species.
Notably, Camellia sinensis cv. ‘Baihaozao’ exhibited the closest genetic distances to the two major cultivated tea tree varieties, Chinese small-leaf tea (C. sinensis var. sinensis) and Assam large-leaf tea (C. sinensis var. assamica), suggesting a possible origin from a shared ancestral lineage. This finding is consistent with the divergence time of CSS and CSA estimated from whole-genome sequencing, which ranges from 0.38 to 1.54 Mya (Wei et al., 2017; Li et al., 2024b).
At the family level, significant evolutionary divergence was observed within Theaceae. Although Camellia sinensis cv. ‘Baihaozao’ and Stewartia sinensis (NC_081928.1) shared the same primary branch, they displayed clear genetic isolation (genetic distance >0.12), which may be attributed to distinct adaptive radiation events experienced by species of the two genera. Of particular interest, Camellia and Polyspora formed a sister group, with their divergence predating the split between Camellia-Polyspora and Stewartia. This supports the topology of the Camellia-Polyspora clade observed in comparative studies of chloroplast genomes (Chen et al., 2023a; Li et al., 2021).
Molecular evolutionary analysis showed that genes encoding components of oxidative phosphorylation complexes (e.g., nad5 and cox1) in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ were under strong purifying selection (Ka/Ks < 0.5). In contrast, some genes involved in stress responses, such as ccmB, exhibited signals of positive selection (Ka/Ks > 1), suggesting specific functional optimization in response to environmental fluctuations (Chen et al., 2023b). This selection pattern is temporally and spatially associated with the whole-genome duplication events of secondary metabolic pathway genes in Camellia, such as those encoding theanine synthetase and flavonoid modification enzymes. These whole-genome duplication events have been shown to be key drivers in the evolution of tea quality traits (Wei et al., 2017; Li et al., 2024b).
3.6 Substitution rate analysis of Protein-Coding Genes
In molecular evolution studies, the ratio of Ka/Ks is widely used to assess the selective pressure acting on protein-coding genes. In this study, the nucleotide substitution model of Nei and Gojobori (1986) was applied in combination with multiple sequence alignments and Ka/Ks calculations using DnaSP v5 (Librado and Rozas, 2009). The evolutionary dynamics of 38 PCGs in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ were systematically compared with those of two species from the genus Camellia and six species from other genera.
The analysis indicates that among all PCGs, only 18 genes exhibit Ka/Ks values greater than 1, suggesting evidence of positive selection. Notably, the atp9 gene shows the lowest ratio (0.07), while the mttB gene displays a markedly higher value of 3.48, implying that it may have undergone strong adaptive evolution (Li et al., 2024b). These findings align with the general trend of purifying selection observed in mitochondrial genomes while also revealing the distinct evolutionary trajectories of specific genes (Li et al., 2024b) (Figure 7; Supplementary Table 9). This pattern indicates that the mitochondrial PCGs of Camellia species have been subject to strong purifying selection over long-term evolution, maintaining high functional conservation.

Figure 7. Boxplot of shared pairwise Ka/Ks values among mitochondrial genes in Camellia sinensis cv. ‘Baihaozao’.
It is worth noting that, although synonymous mutations have traditionally been considered neutral, recent studies suggest that synonymous sites may also be subject to selection due to their roles in splicing regulation, codon usage bias, or mRNA stability (Park et al., 2017; Savisaar and Hurst, 2024). For example, Savisaar and Hurst (2018) demonstrated that exon splicing regulation imposes strong selective pressure on synonymous sites (Savisaar and Hurst, 2024). However, the universally low Ka/Ks values observed in this study suggest that, even if non-neutral effects of synonymous substitutions exist, their overall impact on evolutionary rates is masked by the dominant effects of purifying selection (Moutinho and Walker, 2024).
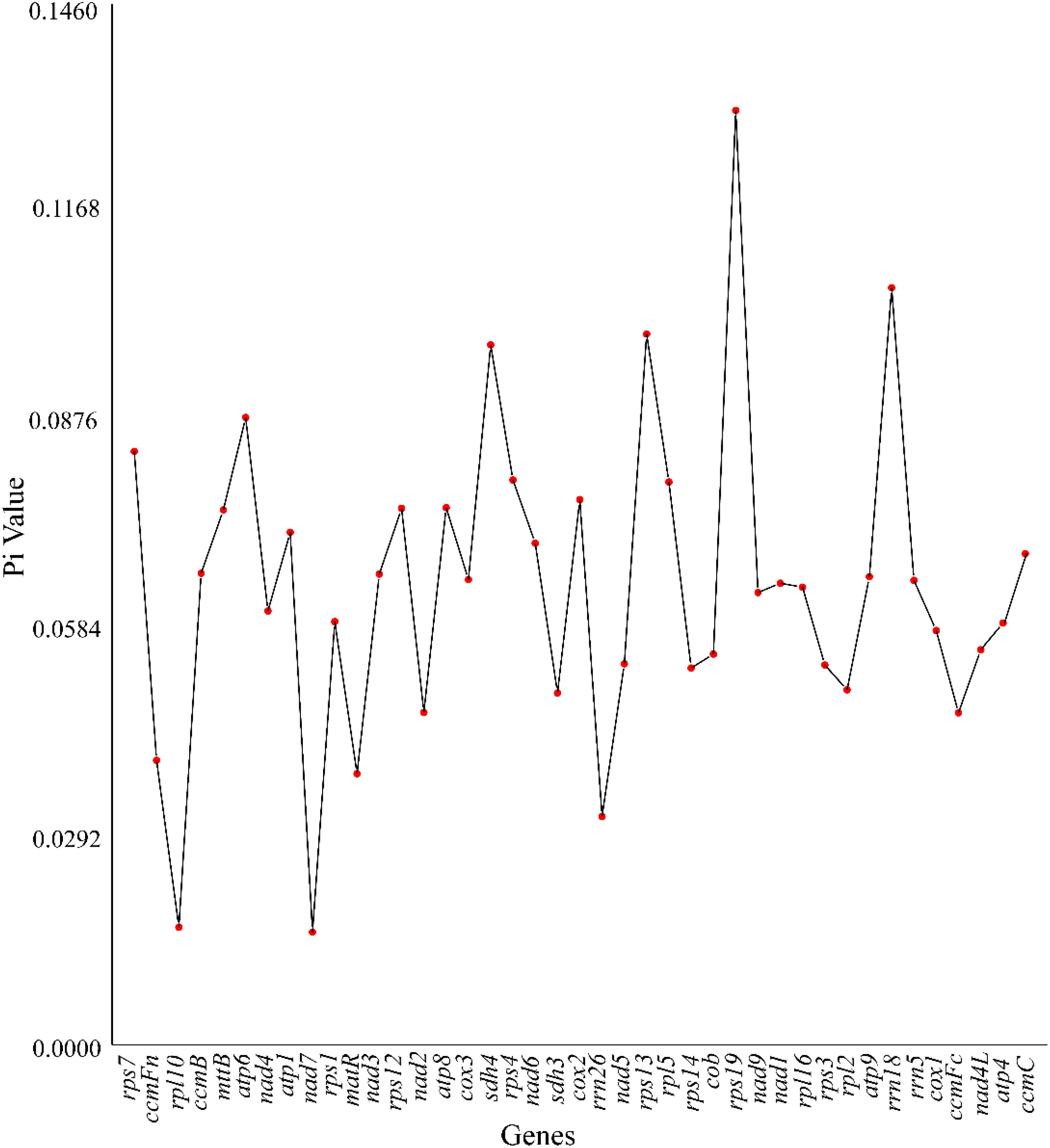
3.7 Nucleotide diversity analysis
This study analyzed the nucleotide diversity (Pi) of 38 PCGs and three rRNA genes in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ to reveal its genetic variation characteristics (Figure 8; Supplementary Table 10). The results showed that the Pi values of the 41 genes ranged from 0.02 to 0.13, with rps19 (Pi = 0.13) being the most variable PCG and nad7 (Pi = 0.02) the most conserved (Liang et al., 2024). Notably, nine PCGs, including rps13, sdh4, and atp6, had Pi values exceeding 0.08, suggesting that these genes may be under relatively weak selective pressure or could serve as key candidate genes in adaptive evolution (Liang et al., 2024). Similar to chloroplast genome studies, this study also observed that ribosomal subunit-encoding genes (e.g., the rps and rpl families) exhibited higher variability, indicating functional convergence between mitochondrion and chloroplasts in genetic marker screening.
Among the rRNA genes, rrn18 (Pi = 0.11) exhibited significantly higher variability than rrn26 (Pi = 0.03) and rrn5 (Pi = 0.07). This contrasts with the general pattern of high conservation observed in rRNA genes of chloroplast genomes in Camellia species (Yang et al., 2013; Li et al., 2021), possibly reflecting a distinct evolutionary trajectory of mitochondrial rRNA in species differentiation. For instance, the high variability of mitochondrial rrn18 may be associated with its role in regulating translation fidelity or adaptation to energy metabolism, a hypothesis that requires further functional validation (Yang et al., 2013; Chen et al., 2021).
Cross-species comparisons revealed significant differences in the Pi distribution of Camellia sinensis cv. ‘Baihaozao’ PCGs compared to closely related species such as Brassica rapa (NC_016125.1), particularly in energy metabolism-related genes like atp6 (Pi = 0.09) and sdh4 (Pi = 0.08). These differences may be related to the unique secondary metabolic demands of tea plants, such as the specific requirements of the tea polyphenol synthesis pathway for mitochondrial redox balance. Moreover, genome-wide association studies suggest that structural variations, such as long terminal repeat retrotransposon expansions, may influence Pi distribution by introducing frameshift mutations and nonsynonymous substitutions, consistent with the distribution patterns of highly variable genes observed in this study.
It is worth noting that the IR regions in chloroplast genomes are typically highly conserved (average Pi = 0.0008) (Liang et al., 2024), whereas the structural heterogeneity of mitochondrial genomes may result in different variation patterns across regions. A recent pan-genomic analysis of 625 tea germplasms revealed a geographic gradient in the genetic diversity of cultivated populations, highlighting the need for future studies to integrate the cultivation history of Camellia sinensis cv. ‘Baihaozao’ to explore the population differentiation mechanisms underlying its mitochondrial Pi values. Furthermore, the high variability of marker genes such as rps19 can be utilized in molecular-assisted breeding, and their potential as single nucleotide polymorphism markers requires experimental validation through population genetic studies.
3.8 Analysis of homologous fragments between mitochondrial and chloroplast genomes
This study conducted a systematic analysis of homologous fragments between the mitochondrial and chloroplast genomes of Camellia sinensis cv. ‘Baihaozao’. The mitochondrial genome (909,843 bp) was approximately 5.8 times larger than the chloroplast genome (157,052 bp; GC content: 37%), which exhibited the typical quadripartite structure of angiosperms (Large single-copy region: 86,586 bp; Small single-copy region: 18,227 bp; IRs: 52,162 bp) and encoded 133 functional genes. In this study, a total of 107 homologous sequence fragments were identified between the chloroplast and mitochondrial genomes, with their lengths ranging from 32 to 9,572 bp. Quantitative analysis revealed that the total length of homologous regions within the chloroplast genome was 27,578 bp, accounting for 17.56% of its total genome length. In contrast, the total length of homologous sequences within the mitochondrial genome was 14,733 bp, representing only 1.62% of its genome (Figure 9; Supplementary Tables 11, 12). This result indicates that the transfer of chloroplast genome sequences to the mitochondrial genome exhibits a distinct directional preference, consistent with the general pattern observed in multiple species, where mitochondrial genomes integrate chloroplast sequences (Hao et al., 2022; Xie et al., 2024).
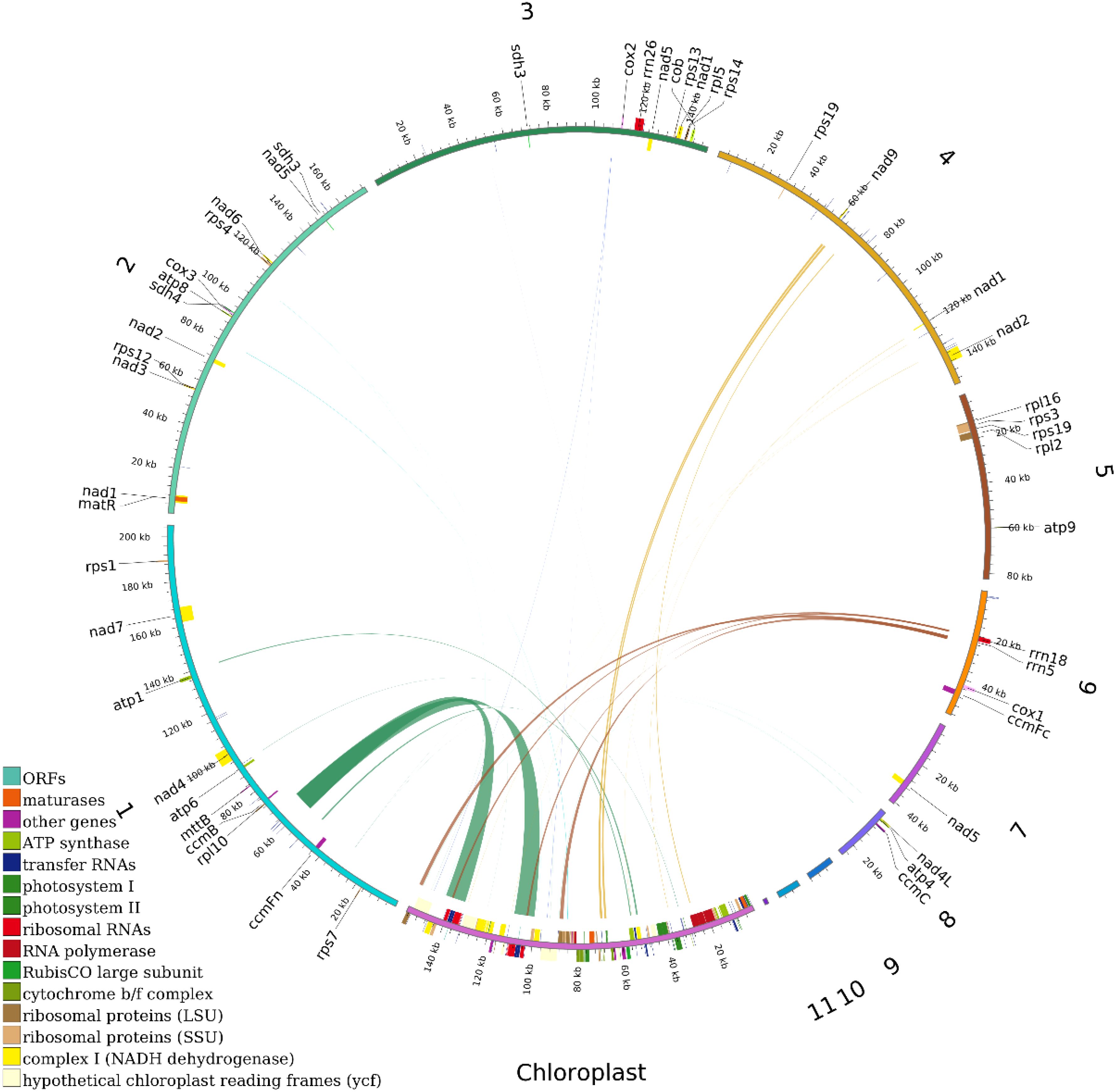
Figure 9. Distribution of homologous fragments between mitochondrial and chloroplast genomes in Camellia sinensis cv. ‘Baihaozao’. Chloroplast represents chloroplast sequences, while the others represent mitochondrial sequences. Genes belonging to the same complex are indicated by blocks of the same color. Blocks in the outer and inner rings represent genes on the forward and reverse strands, respectively. The connecting lines in the middle indicate homologous sequences.
Annotation analysis revealed significant sequence homology between the chloroplast and mitochondrial genomes in the tea plant Camellia sinensis cv. ‘Baihaozao’. Specifically, 29 complete homologous fragments were identified in the chloroplast genome, involving genes such as rrn23, rpl2, psbJ, and petL, as well as ribosomal RNA genes, photosynthesis-related genes, and tRNA genes (e.g., trnA-UGC, trnV-GAC). In the mitochondrial genome, 14 complete homologous fragments were identified, including tRNA genes such as trnV-GAC, trnA-TGC, and trnD-GTC (Supplementary Table 12). These homologous fragments are likely the result of DNA transfer events between organelles, potentially mediated by mechanisms such as recombination facilitated by repeat sequences or random insertions (Liu et al., 2013).
The mechanism of inter-organellar gene transfer may be associated with recombination events mediated by repetitive sequences. Multiple forward and palindromic repeats were detected within the transferred fragments (Figure 9), resembling the mechanism observed in Lithocarpus litseifolius, where mitochondrial recombination hotspots facilitated the integration of chloroplast DNA (Qiu et al., 2024). Although the functions of most transferred genes (e.g., rpl2, orf42) remain unclear, some tRNA genes (e.g., trnW-CCA) retain functional activity in mitochondrial genomes, such as in Stemona species, suggesting that such transfers may compensate for the loss of mitochondrial tRNAs and help maintain the stability of the translation system (Niu et al., 2022; Ou et al., 2024). These findings provide new insights into the co-evolution of plant organellar genomes. Future research should integrate transcriptomic and proteomic data to validate the functional expression patterns of transferred genes (Zhang et al., 2012; Ou et al., 2024).
3.9 Synteny and evolutionary dynamics analysis of the mitochondrial genome
To investigate the structural evolution and phylogenetic signals of the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’, a whole-genome alignment strategy was employed to analyze its synteny with the mtDNAs of other species in Theaceae and representative species from other families (Figure 10). The results showed that Camellia sinensis cv. ‘Baihaozao’ exhibited the highest degree of synteny with ‘Zhuyeqi’ mtDNA (62% of aligned blocks in the forward orientation and 25% in the reverse complementary orientation), followed by Camellia sinensis (58% forward alignment). This indicates significant structural conservation of mitochondrial genomes among these species. This phenomenon is highly consistent with the phylogenetic results of chloroplast genomes, suggesting that organellar genomes may have undergone coordinated evolution during the divergence of Camellia species.
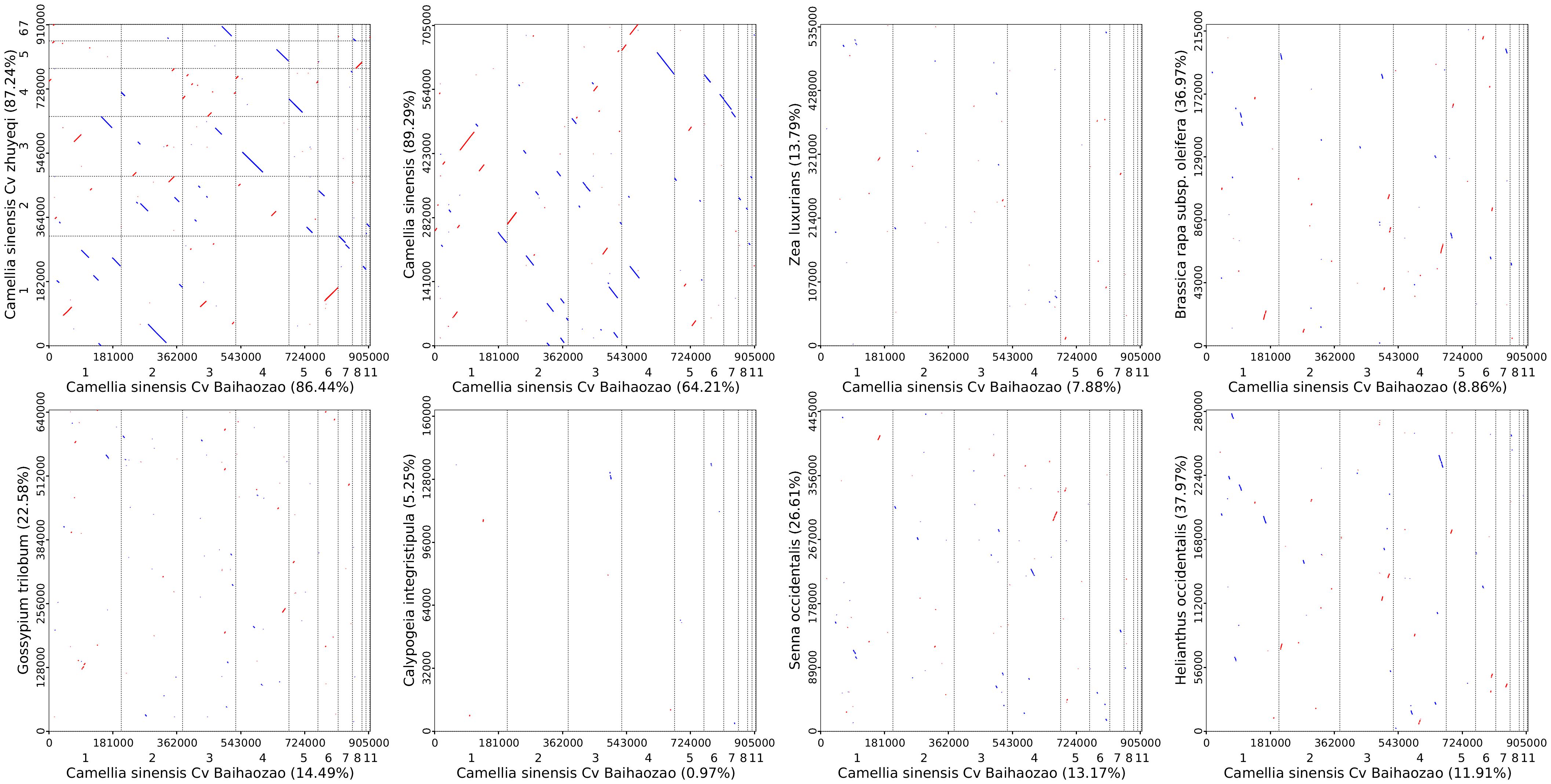
Figure 10. Homologous sequences among mitochondrial genomes of camellia species using Camellia sinensis cv. ‘Baihaozao’ as a Reference. The horizontal axis within each box represents the assembled sequence, while the vertical axis represents sequences from other species. The values in parentheses indicate the proportion of homologous sequences relative to the total genome size. Red lines within the boxes denote forward alignments, while blue lines represent reverse complementary alignments.
Further comparisons across families revealed that the number of homologous sequences among species within Theaceae (mean 1,428 ± 203) was significantly higher than that between Theaceae and species from other families, such as Rosaceae and Solanaceae (mean 327 ± 89) (Supplementary Table 13). This finding supports the “closely related species share a higher proportion of homologous sequences” hypothesis (Lai et al., 2022) and is consistent with the observed decline in synteny levels between genera in Solanaceae mitochondrial genome studies (Chen et al., 2024). Notably, although PCGs of mtDNA in Theaceae species exhibited high conservation (nucleotide similarity >99%), frequent structural rearrangements were observed in the non-coding regions, such as the dynamic expansion and contraction of IR boundaries (Liu et al., 2023). This feature resembles the evolutionary patterns of the mitochondrial genome in Rubus chingii (Rosaceae) (Shi et al., 2024).
4 Discussion
This study analyzed the multichromosomal structure, RNA editing characteristics, and adaptive evolutionary patterns of the mitochondrial genome in Camellia sinensis cv. ‘Baihaozao’ revealing its unique evolutionary dynamics and functional adaptations. These findings provide an important reference for mitochondrial genomics research in Theaceae. The 11 linear chromosomes of the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ (total length: 909,843 bp) challenge the traditional understanding of a single circular structure in angiosperm mitochondria (Sloan, 2013). Its multichromosomal differentiation may result from recombination and fragmentation mediated by repeat sequences, with dispersed repeats accounting for 71% of the genome (Wynn and Christensen, 2019). Notably, ribosomal protein-coding genes (rpl/rps), which are commonly pseudogenized in Theaceae (Kubo and Arimura, 2010), remain fully functional and unmethylated in Camellia sinensis cv. ‘Baihaozao’. This preservation may be linked to its multichromosomal architecture, which enhances recombination flexibility and facilitates the elimination of deleterious mutations as part of an adaptive mechanism (Marechal and Brisson, 2010; Zhou et al., 2023). The mechanism underlying this multichromosomal differentiation may be attributed to frequent homologous recombination events or horizontal gene transfer. The high proportion of dispersed repeats (71%; 55,543 bp) likely mediates chromosomal fragmentation and rearrangement through recombination (Kurtz et al., 2001). Notably, variable genes (e.g., ribosomal subunit genes rpl/rps) often exhibit pseudogenization trends in Theaceae (Han et al., 2024), while Camellia sinensis cv. ‘Baihaozao’ retains their full functionality, suggesting its adaptive requirements for nucleo-mitochondrial interactions or energy metabolism. The poly-chromosomal structure may accelerate the purging of deleterious mutations by enhancing recombination flexibility (Murat et al., 2010; Betran et al., 2012; Peer et al., 2017), while simultaneously maintaining the functional integrity of essential genes (e.g., rpl/rps) through inter-chromosomal functional complementation (Terasaki Hart and Wang, 2024). This mechanism aligns closely with the rapid environmental response characteristics of Camellia sinensis cv. ‘Baihaozao’ in stress-resistance metabolic pathways, suggesting that the poly-chromosomal architecture is a significant driving force of adaptive evolution (Terasaki Hart and Wang, 2024).
The spatial heterogeneity of RNA editing sites (e.g., 19 sites in cox1 vs. only 1 site in rpl2/rps4) reflects differences in functional constraints across genes. RNA editing predominantly occurred at the first and second codon positions (91%), which are closely associated with changes in amino acid properties. For instance, 49% of the editing sites resulted in hydrophilic-to-hydrophobic transitions, which may optimize protein conformation and enhance mitochondrial oxidative phosphorylation efficiency (Chen et al., 2012; Han et al., 2024). Additionally, the RNA editing site distribution pattern in Camellia sinensis cv. ‘Baihaozao’ is similar to that of other species such as Rehmannia chingii (Han et al., 2024). However, no reverse editing (U-to-C) was detected in Camellia sinensis cv. ‘Baihaozao’, indicating the conserved nature of the RNA editing mechanism in Theaceae (Zhang et al., 2020)). High-frequency editing genes (e.g., cox1) may dynamically regulate mitochondrial responses to environmental stress, consistent with findings of temperature-responsive RNA editing (e.g., dynamic editing in grapevine.
The high frequency of leucine (Leu, 10%) and serine (Ser, 9%) usage is closely associated with the functions of mitochondrial ATP synthase and the electron transport chain, and their codon preferences may be driven by translational efficiency or tRNA abundance (Lee et al., 2023). The highest RSCU value of the methionine initiation codon AUG (3.0) indicates its advantage in translation initiation efficiency. The preference for high-frequency codons such as UUU (Phe) and AUU (Ile) may be related to their critical roles in the hydrophobic cores of transmembrane proteins, further supporting the optimization of mitochondrial membrane system functionality (Han et al., 2024).
The dominance of dispersed repeats (71%) is consistent with the mitochondrial structural rearrangement patterns observed in species such as Fargesia qinlingensis (Wu et al., 2024), which likely facilitates genome plasticity through homologous recombination. The heterogeneous chromosomal distribution of tandem repeats (enrichment on chr1/chr5 vs. absence on chr9-11) may reflect local recombination hotspots or selective pressures in functional regions. For example, chr1 and chr5 harbor multiple core genes (e.g., atp and nad clusters), where tandem repeats might stabilize transcriptional regulation (Benson, 1999). Additionally, the high abundance of SSRs (264) could serve as molecular markers, providing valuable resources for genetic diversity analyses in Theaceae (Lee et al., 2023).
In the assessment of evolutionary selection pressures on mitochondrial PCGs, Ka/Ks ratio analysis reveals that the majority of genes (Ka/Ks < 1) are under purifying selection, consistent with typical molecular evolutionary patterns (Zhang et al., 2022a, 2024). Notably, the mttB gene exhibits a significant signal of positive selection (Ka/Ks = 3.48), with its exceptionally high ratio potentially reflecting adaptive evolution in specific metabolic pathways, such as metal ion transport (Zhang et al., 2024b). In contrast, the atp9 gene shows an extremely low ratio (0.07), indicating a highly conserved functional sequence. Pi analysis further reveals high variability in the ribosomal subunit gene rps19 (Pi = 0.13), which may be associated with evolutionary demands for regulating translational accuracy. Conversely, the strong conservation of the nad7 gene (Pi = 0.02) corresponds to its critical functional constraints in the assembly of mitochondrial complex I (Zhang et al., 2024b). This correlation between selection pressure and functional importance provides new insights into the evolutionary driving forces shaping mitochondrial genes.
The mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ contains 3% homologous fragments (29,491 bp) carrying 15 tRNA genes and 9 functional genes (e.g., psbE, petL), indicating frequent inter-organellar gene transfer events (Salmans et al., 2010; Han et al., 2024). These transferred fragments may enhance environmental adaptability by compensating for the loss of mitochondrial tRNAs or improving redox balance capabilities. Compared to the chloroplast gene transfer patterns observed in Oryza sativa (Notsu et al., 2002), the transferred genes in Camellia sinensis cv. ‘Baihaozao’ are more abundant, highlighting the unique role of Theaceae species in the co-evolution of organelles.
The multichromosomal architecture, dynamic RNA editing, and recombination mechanisms driven by repetitive sequences in the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’ collectively shaped its distinct evolutionary trajectory. These findings not only provide molecular markers for phylogenetic analyses of Theaceae but also offer new perspectives on the mechanisms underlying mitochondrial adaptive evolution. Future studies could integrate multi-omics data to further explore the dynamic regulation of RNA editing under environmental stress and the phenotypic associations of the multichromosomal structure.
5 Conclusions
This study presents the first complete sequencing and analysis of the mitochondrial genome of Camellia sinensis cv. ‘Baihaozao’. The genome consists of 11 linear chromosomes with a total length of 909,843 bp and a GC content of 46%, challenging the traditional view of a single circular mitochondrial structure. Among the 73 functional genes annotated in the genome, 24 core genes (from the atp, nad, ccm, and cox families) are highly conserved. Notably, ribosomal protein-coding genes (rpl/rps) have retained functional integrity despite the widespread pseudogenization observed in Theaceae plants. Their unmethylated state may be maintained by the recombination advantages conferred by the regulation of the poly-chromosomal structure. The genome displayed dynamic features, with dispersed repeats covering 71% (55,543 bp) of the sequence. Forward and palindromic repeats dominated and likely mediated frequent recombination events. RNA editing was unevenly distributed, with the cox1 gene identified as a hotspot (19 sites). Over 90% of editing events occurred at the first and second codon positions, and 49% of these caused transitions from hydrophilic to hydrophobic amino acids, suggesting precise regulation of mitochondrial energy metabolism.
Adaptive evolutionary analysis indicates that the vast majority of PCGs have Ka/Ks values less than 1 (ranging from 0.07 to 0.78), with the atp9 gene exhibiting the lowest ratio (0.07), consistent with the prevailing trend of purifying selection in mitochondrial genomes. Notably, the mttB gene displays a Ka/Ks value of 3.48, significantly higher than that of other genes, suggesting that it may have undergone positive selection to adapt to environmental pressures. Nucleotide diversity analysis revealed variability among functional genes: rps19 showed the highest diversity (Pi = 0.13), while nad7 was the most conserved (Pi = 0.02). Homologous fragments between the mitochondrial and chloroplast genomes (29,491 bp) carried 26 complete genes, 58% of which were tRNA genes, highlighting active inter-organellar gene transfer. Phylogenetic analysis clarified the evolutionary position of C. sinensis cv. ‘Baihaozao’ within Theaceae and provided insights into the plasticity of its mitochondrial genome, driven by its multichromosomal structure and dynamic repeats. This study establishes a genomic foundation for tea germplasm evaluation and molecular breeding. It also offers valuable insights into the evolutionary mechanisms of plant mitochondrial genomes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
No specific license was required for the collection of samples in this study. The study was conducted in compliance with relevant Chinese legislation.
Author contributions
ZC: Data curation, Funding acquisition, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. ZW: Data curation, Investigation, Methodology, Software, Validation, Writing – original draft. WZ: Data curation, Investigation, Methodology, Software, Validation, Writing – original draft. SL: Data curation, Investigation, Writing – original draft. YX: Writing – review & editing. YG: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Natural Science Foundation of Hunan Province (Zhiyin Chen) (grant numbers 2023JJ50465) and (Yihui Gong) (grant numbers 2024JJ7235), The science and technology innovation Program of Hunan Province (Zhiyin Chen) (grant numbers 2024RC8289), and the science and technology innovation Program of loudi city (Zhiyin Chen). (grant numbers 2023RC3501).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1604404/full#supplementary-material
Abbreviations
RNA, Ribonucleic acid; GC, Guanine-Cytosine; Ka, Non-synonymous substitution rate; Ks, Synonymous substitution rate; tRNA, Transfer ribonucleic acid; rRNA, Ribosomal ribonucleic acid; IR, Inverted repeat; DNA, Deoxyribonucleic acid; PCR, Polymerase chain reaction; NCBI, National Center for Biotechnology Information; SSR, Simple sequence repeat; bp, Base pair; ATP, Adenosine triphosphate; NADH, Nicotinamide adenine dinucleotide; PCG, Protein-coding genes; RSCU, Relative synonymous codon usage; AT, Adenine and thymine; Pi, Nucleotide diversity.
References
Alverson, A. J., Rice, D. W., Dickinson, S., Barry, K., and Palmer, J. D. (2011). Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23, 2499–2513. doi: 10.1105/tpc.111.087189
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Betran, E., Demuth, J. P., and Williford, A. (2012). Why chromosome palindromes? Int. J. Evol. Biol. 2012, 207958. doi: 10.1155/2012/207958
Bi, C. W., Lu, N., Xu, Y. Q., He, C. P., and Lu, Z. H. (2020). Characterization and analysis of the mitochondrial genome of common bean (Phaseolus vulgaris) by comparative genomic approaches. Int. J. Mol. Sci. 21, 3778. doi: 10.3390/ijms21113778
Bi, C. W., Qu, Y. S., Hou, J., Wu, K., Ye, N., and Yin, T. M. (2022). Deciphering the multi-chromosomal mitochondrial genome of populus simonii. Front. Plant Sci. 15, 914635. doi: 10.3389/fpls.2022.914635
Bulmer, M. (1991). The selection-mutation-drift theory of synonymous codon usage. Genetics 129, 897–907. doi: 10.1093/genetics/129.3.897
Chen, B. Y., Li, X., Tian, Y., Xing, D. D., Su, X. H., and Guan, C. P. (2024). Comparison and phylogenetic analysis of mitochondrial genome among lycium barbarum and other solanaceae plants. Genomics Appl. Biol. 43, 45–62. doi: 10.13417/j.gab.043.000045
Chen, C., Frankhouser, D., and Bundschuh, R. (2012). Comparison of insertional RNA editing in myxomycetes. PloS Comput. Biol. 8, e1002400. doi: 10.1371/journal.pcbi.1002400
Chen, X. X., Yuan, Z. W., Li, C., Dietrich, C. H., and Song, Y. H. (2021). Structural features and phylogenetic implications of two new mitogenomes of erythroneurine leafhopper (Hemiptera: cicadellidae: typhlocybinae). PloS One 16, e0251207. doi: 10.21203/rs.3.rs-74836/v1
Chen, Y. Q., Li, Y. F., Shen, C. W., and Xiao, L. Z. (2023a). Topics and trends in fresh tea (Camellia sinensis) leaf research A comprehensive bibliometric study. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1092511
Chen, Z. Y., Liu, Q., Xiao, Y., Zhou, G. H., Yu, P. H., Bai, J., et al. (2023b). Complete chloroplast genome sequence of camellia sinensis: genome structure, adaptive evolution, and phylogenetic relationships. J. Appl. Genet. 64, 419–429. doi: 10.1007/s13353-023-00767-7
Cuenca, A., Petersen, G., Seberg, O., Davis, J. I., and Stevenson, D. W. (2010). Are substitution rates and RNA editing correlated? BMC Evolutionary Biol. 10, 349. doi: 10.1186/1471-2148-10-349
Cui, Z. Y., Du, F., Yu, W. H., Wang, Z. X., Kong, F. Y., Xie, Z., et al. (2024). Alterations of mouse gut microbiome in alveolar echinococcosis. Heliyon 10, e32860. doi: 10.1016/j.heliyon.2024.e32860
Cummings, M. P. and Myers, D. S. (2004). Simple statistical models predict C-to-U edited sites in plant mitochondrial RNA. BMC Bioinf. 5, 132. doi: 10.1186/1471-2105-5-132
Daley, D. O., Clifton, R., and Whelan, J. (2002). Intracellular gene transfer: Reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. PNAS 99, 10510–10515. doi: 10.1073/pnas.122354399
Han, Y., Feng, Y. L., Wang, J., Zhu, S. S., Jin, X. J., Wu, Z. Q., et al. (2024). Comprehensive analysis of the complete mitochondrial genome of rehmannia chingii: an autotrophic species in the orobanchaceae family. Genes 15, 98. doi: 10.3390/genes15010098
Han, Y. K., Huang, Z. F., Tang, J., Chiba, H., and Fan, X. L. (2018). The complete mitochondrial genomes of two skipper genera (Lepidoptera: Hesperiidae) and their associated phylogenetic analysis. Sci. Rep. 8, 15762. doi: 10.1038/s41598-018-34107-1
Hao, J., Liang, Y. Y., Su, Y. J., and Wang, T. (2022). The complete mitochondrial genome of ophioglossum vulgatum L. Is with highly repetitive sequences: intergenomic fragment transfer and phylogenetic analysis. Genes (Basel) 13, 1287. doi: 10.3390/genes13071287
He, P., Huang, S., Xiao, G. H., Zhang, Y. Z., and Yu, J. N. (2016). Abundant RNA editing sites of chloroplast protein-coding genes in Ginkgo biloba and an evolutionary pattern analysis. BMC Plant Biol. 16, 257. doi: 10.1186/s12870-016-0944-8
He, S., Wang, X. H., Yin, M., Ye, J. P., Meng, J. Z., and Zhou, L. Q. (2023). Molecular identification of Lingyun Baihao wild and cultivated tea through genome-wide sequencing. Genet. Resour. Crop Evol. 70, 1407–1417. doi: 10.21203/rs.3.rs-1697952/v1
Hershberg, R. and Petrov, D. A. (2008). Selection on codon bias. Annu. Rev. Genet. 42, 287–299. doi: 10.1146/annurev.genet.42.110807.091442
Jühling, F., Pütz, J., Bernt, M., Donath, A., Middendorf, M., Florentz, C., et al. (2012). Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 40, 2833–2845. doi: 10.1093/nar/gkr1131
Kozik, A., Rowan, B. A., Lavelle, D., Berke, L., Schranz, M. E., Michelmore, R. W., et al. (2019). The alternative reality of plant mitochondrial DNA One ring does not rule them all. PloS Genet. 15, e1008373. doi: 10.1371/journal.pgen.1008373
Kubo, N. and Arimura, S.-I. (2010). Discovery of the rpl10 gene in diverse plant mitochondrial genomes and its probable replacement by the nuclear gene for chloroplast RPL10 in two lineages of angiosperms. DNA Res. 17, 1–9. doi: 10.1093/dnares/dsp024
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Lai, C. J., Wang, J., Kan, S. L., Zhang, S., Li, P., Reeve, W. G., et al. (2022). Comparative analysis of mitochondrial genomes of Broussonetia spp. (Moraceae) reveals heterogeneity in structure, synteny, intercellular gene transfer, and RNA editing. Front. Plant Sci., 13, 1052151. doi: 10.3389/fpls.2022.1052151
Lee, H. J., Lee, Y., Lee, S. C., Kim, C. K., Kang, J. N., Kwon, S. J., et al. (2023). Comparative analysis of mitochondrial genomes of Schisandra repanda and Kadsura japonica. Front. Plant Sci. 14, 1183406. doi: 10.3389/fpls.2023.1183406
Li, H. (2018). Minimap2 pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi: 10.1093/bioinformatics/bty191
Li, L., Hu, Y., He, M., Zhang, B., Wu, W., Cai, P., et al. (2021). Comparative chloroplast genomes: insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom. 22 (1), 138. doi: 10.1186/s12864-021-07427-2
Li, J. X., Chen, Y. L., Liu, Y. L., Wang, C., Li, L., and Chao, Y. H. (2023b). Complete mitochondrial genome of Agrostis stolonifera: insights into structure, Codon usage, repeats, and RNA editing. BMC Genomics 24, 466. doi: 10.1186/s12864-023-09573-1
Li, L., Li, X. R., Liu, Y., Li, J. D., Zhen, X. Y., Huang, Y., et al. (2024). Comparative analysis of the complete mitogenomes of Camellia sinensis var. sinensis and C. sinensis var. assamica provide insights into evolution and phylogeny relationship. Front. Plant Sci. 15, 1396389. doi: 10.3389/fpls.2024.1396389
Li, J., Tang, H., Luo, H., Tang, J., Zhong, N., and Xiao, L. Z. (2023a). Complete mitochondrial genome assembly and comparison of Camellia sinensis var. Assamica cv. Duntsa. Front. Plant Sci. 14, 1117002. doi: 10.3389/fpls.2023.1117002
Li, W., Zhang, C. P., Guo, X., Liu, Q. H., and Wang, K. L. (2019). Complete chloroplast genome of Camellia japonica genome structures, comparative and phylogenetic analysis. PloS One 14, e0216645–e0216684. doi: 10.1371/journal.pone.0216645
Liang, H., Qi, H. S., Wang, Y. D., Sun, X. X., Wang, C. M., and Zheng, D. J. (2024). Comparative chloroplast genome analysis of Camellia oleifera and C. meiocarpa: phylogenetic relationships, sequence variation and polymorphism markers. Trop. Plants. 3 (1), e023. doi: 10.48130/tp-0024-0022
Librado, P. and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, J. H., Zhou, W. C., Ding, H. L., Wang, P., and Ai, H. M. (2016). The mitochondrial genome of the land snail Cernuella virgata (Da Costa 1778): the first complete sequence in the family Hygromiidae (Pulmonata, Stylommatophora). ZooKeys 589, 55–69. doi: 10.3897/zookeys.589.7637
Liu, G. Z., Cao, D. D., Li, S. S., Su, A. G., Geng, J. N., Grover, C. E., et al. (2013). The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PloS One 8, e69476. doi: 10.1371/journal.pone.0069476
Liu, H. X., Qin, L., Chen, Y. L., Xu, S. Y., Zhou, X. W., Zhu, Y. L., et al. (2023). The complete mitochondrial genome of Camellia nitidissima Theaceae. Mitochondrial DNA Part B 8, 565–569. doi: 10.1080/23802359.2023.2209211
Lu, C., Gao, L. Z., and Zhang, Q. J. (2022). A high-quality genome assembly of the mitochondrial genome of the oil-tea tree camellia gigantocarpa (Theaceae). diversity 14, 850. doi: 10.3390/d14100850
Lu, G. L., Wang, W. H., Mao, J., Li, Q., and Que, Y. X. (2023). Complete mitogenome assembly of Selenicereus monacanthus revealed its molecular features, genome evolution, and phylogenetic implications. BMC Plant Biol. 23, 541. doi: 10.1186/s12870-023-04529-9
Mao, X. Z., Cai, T., Olyarchuk, J. G., and Wei, L. P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics (2005) 21 (19), 3787–3793. doi: 10.1093/bioinformatics/bti430
Marechal, A. and Brisson, N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186, 299–317. doi: 10.1111/j.1469-8137.2010.03195.x
Moutinho, A. F. and Walker, A. E. (2024). No evidence that selection on synonymous codon usage affects patterns of protein evolution in bacteria. Genome Biol. Evol. 16, evad232. doi: 10.1093/gbe/evad232
Mower, J. P. (2005). PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinf. 6, 96. doi: 10.1186/1471-2105-6-96
Murat, F., Xu, J. H., Tannier, E., Abrouk, M., Guilhot, N., Pont, C., et al. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20, 1545–1557. doi: 10.1101/gr.109744.110
Nei, M. and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3 (5), 418–426. doi: 10.1093/oxfordjournals.molbev.a040410
Niczyporuk, J. S. (2018). A complete analysis of Relative Synonymous Codon Usage in HVRs1–4 region in adenovirus genome. Polish J. Veterinary Sci. 21, 459–468. doi: 10.24425/122619
Niu, Y., Gao, C., and Liu, J. (2022). Complete mitochondrial genomes of three Mangifera species, their genomic structure and gene transfer from chloroplast genomes. BMC Genom. 23 (1), 147. doi: 10.1186/s12864-022-08383-1
Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., et al. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268, 434–445. doi: 10.1007/s00438-002-0767-1
Ou, T. Y., Wu, Z. N., Tian, C. Y., Yang, Y., and Li, Z. Y. (2024). Complete mitochondrial genome of Agropyron cristatum reveals gene transfer and RNA editing events. BMC Plant Biol. 24, 830. doi: 10.1186/s12870-024-05558-8
Park, S., Ruhlman, T. A., Weng, M. L., Hajrah, N. H., Sabir, J. S. M., and Jansen, R. K. (2017). Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of geranium. Genom. Biol. Evol. 9 (6), 1766–1780. doi: 10.1093/gbe/evx124
Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
Qian, Y. H., Wu, H. Y., Ji, X. Y., Yu, W. W., and Du, Y. Z. (2014). Mitochondrial genome of the stonefly kamimuriawangi (Plecoptera: perlidae) and phylogenetic position of plecoptera based on mitogenomes. PloS One 9, e86328. doi: 10.1371/journal.pone.0086328
Qiu, X. Y., Tian, Y. Q., Li, Z. Q., Wu, X. J., Xiang, Z. W., Wang, Y. Q., et al. (2024). Assembly and characterization analysis of the complete mitochondrial genome of lithocarpus litseifolius (Hance) chun. Genet. Resour. Crop Evol. 72, 295–313. doi: 10.21203/rs.3.rs-3230084/v1
Salmans, M. L., Chaw, S. M., Lin, C. P., Shih, A. C. C., Wu, Y. W., and Mulligan, R. M. (2010). Editing site analysis in a gymnosperm mitochondrial genome reveals similarities with angiosperm mitochondrial genomes. Curr. Genet. 56, 439–446. doi: 10.1007/s00294-010-0312-4
Sau, K., Sau, S., Mandal, S. C., and Ghost, T. C. (2005). Factors influencing the synonymous codon and amino acid usage bias in AT-rich pseudomonas aeruginosa phage phiKZ. Acta Biochim. Biophys. Sin. 37, 625–633. doi: 10.1111/j.1745-7270.2005.00089.x
Savisaar, R. and Hurst, L. D. (2024). Exonic splice regulation imposes strong selection at synonymous sites. Genome Res. 28, 1442–1454. doi: 10.1101/gr.233999.117
Savisaar, R. and Hurst, L. D. (2018). Exonic splice regulation imposes strong selection at synonymous sites. Genome Res. 28 (10), 1442–1454. doi: 10.1101/gr.233999.117
Schiavone, A., Pugliese, N., Samarelli, R., Cumbo, C., Minervin, C. F., Albano, F., et al. (2022). Factors affecting the quality of bacterial genomes assemblies by canu after nanopore sequencing. Appl. Sci. 12, 3110. doi: 10.3390/app12063110
Shen, B. M., Shen, A. R., Liu, L. N., Tan, Y., Li, S. N., and Tan, Z. M. (2024). Assembly and comparative analysis of the complete multichromosomal mitochondrial genome of Cymbidium ensifolium, an orchid of high economic and ornamental value. BMC Plant Biol. 24, 255. doi: 10.1186/s12870-024-04962-4
Shi, Y. J., Chen, Z., Jiang, J. Y., Wu, W. W., Xin, Y., and Zeng, W. (2024). Assembly and comparative analysis of the complete mitogenome of Rubus chingii var. suavissimus, an exceptional berry plant possessing sweet leaves. Front. Plant Sci. 15, 1504687. doi: 10.3389/fpls.2024.1504687
Shin, S. C., Kim, H., Lee, J. H., Kim, H. W., Park, J., Choi, B. S., et al. (2019). Nanopore sequencing reads improve assembly and gene annotation of the Parochlus steinenii genome. Sci. Rep. 9, 5095. doi: 10.1038/s41598-019-41549-8
Sloan, D. B. (2013). One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 200, 978–985. doi: 10.1111/nph.12395
Sun, G. L., Xu, W. J., Xing, T., Zhao, M. M., Yin, X. H., Xia, G. M., et al. (2015). Synonymous codon usage bias in plant mitochondrial genes is associated with intron number and mirrors species evolution. PloS One 10, e0131508. doi: 10.1371/journal.pone.0131508
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Terasaki Hart, D. E. and Wang, I. J. (2024). Genomic architecture controls multivariate adaptation to climate change. Global Change Biol. 30, e17179. doi: 10.1111/gcb.17179
Wang, X., Zhang, Y. H., Zhang, H. X., Qin, G., and Lin, Q. (2019). Complete mitochondrial genomes of eight seahorses and pipefishes (Syngnathiformes: Syngnathidae): insight into the adaptive radiation of syngnathid fishes. BMC Evolutionary Biol. 19, 119. doi: 10.1186/s12862-019-1430-3
Wei, C. L., Yang, H., Wang, S. B., Zhao, J., Liu, C., Gao, L. P., et al. (2017). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. PNAS 115, E4151–E4158. doi: 10.1073/pnas.1719622115
Wu, H., Li, X., Qu, K., Yang, L. L., Su, T., Yong, L. J., et al. (2024). Integration of illumina and pacBio hiFi sequencing reveals a three-linear-molecule mitogenome with RNA-editing sites and phylogeny in arrow bamboo (Fargesia qinlingensis). forests 15, 1267. doi: 10.3390/f15071267
Wynn, E. L. and Christensen, A. C. (2019). Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3 (Bethesda) 9, 549–559. doi: 10.1534/g3.118.200948
Xie, Y. N., Liu, W. Q., Guo, L. W., and Zhang, X. M. (2024). Mitochondrial genome complexity in Stemona sessilifolia: nanopore sequencing reveals chloroplast gene transfer and DNA rearrangements. Front. Genet. 15, 1395805. doi: 10.3389/fgene.2024.1395805
Yang, Y. L. and Duan, C. (2024). Mitochondrial genome features and systematic evolution of diospyros kaki thunb 'Taishuu'. BMC Genomics 25, 285. doi: 10.1186/s12864-024-10199-0
Yang, J. B., Yang, S. X., Li, H. T., Yang, J., and Li, D. Z. (2013). Comparative chloroplast genomes of camellia species. PloS One 8, e73053. doi: 10.1371/journal.pone.0073053
Yu, J. H., Chen, X., Liu, R. Y., Tang, Y. T., Nie, G. X., and Zhou, C. J. (2023). Mitochondrial genome of Acheilognathus barbatulus (Cypriniformes, Cyprinidae, Acheilognathinae): characterisation and phylogenetic analysis. Biodiversity Data J. 11, e93947. doi: 10.3897/BDJ.11.e93947
Yura, K., Miyata, Y., Arikawa, T., Higuchi, M., and Sugita, M. (2008). Characteristics and prediction of RNA editing sites in transcripts of the moss takakia lepidozioides chloroplast. DNA Res. 15, 309–321. doi: 10.1093/dnares/dsn016
Zhang, T. W., Fang, Y. J., Wang, X. M., Deng, X., Zhang, X. W., Hu, S. N., et al. (2012). The Complete Chloroplast and Mitochondrial Genome Sequences of Boea hygrometrica: Insights into the Evolution of Plant Organellar Genomes. PloS One 7, e30531. doi: 10.1371/journal.pone.0030531
Zhang, A. D., Jian, X. H., Zhang, F. P., Wang, T. F., and Zhang, X. J. (2020). Dynamic response of RNA editing to temperature in grape by RNA deep sequencing. Funct. Integr. Genomics 20, 421–432. doi: 10.1007/s10142-019-00727-7
Zhang, M. Y., Li, Z., Wang, Z. J., Xiao, Y., Bao, L., Wang, M., et al. (2022b). Exploring the RNA editing events and their potential regulatory roles in tea plant (Camellia sinensis L.). Int. J. Mol. Sci. 23, 13640. doi: 10.3390/ijms232113640
Zhang, T. X., Li, M. M., Zhu, X. Y., Li, S. X., Guo, M. Y., Guo, C. H., et al. (2024b). Comparative chloroplast genomes analysis provided adaptive evolution insights in medicago ruthenica. Int. J. Mol. Sci. 25, 8689. doi: 10.3390/ijms25168689
Zhang, H., Lu, C., Liu, Q., Zou, T. M., Qiao, G. X., and Huang, X. L. (2022a). Insights into the evolution of aphid mitogenome features from new data and comparative analysis. Anim. (Basel) 12, 1970. doi: 10.3390/ani12151970
Zhang, M., Zhang, X. H., Huang, Y. L., Chen, Z. X., and Chen, B. H. (2024a). Comparative mitochondrial genomics of Terniopsis yongtaiensis in Malpighiales structural, sequential, and phylogenetic perspectives. BMC Genomics 25, 853. doi: 10.1186/s12864-024-10765-6
Zhao, N., Wang, Y. M., and Hua, J. P. (2018). The roles of mitochondrion in intergenomic gene transfer in plants A source and a pool. Int. J. Mol. Sci. 19, 547. doi: 10.3390/ijms19020547
Zheng, P., Wang, D. X., Huang, Y. Q., Chen, H., Du, H., and Tu, J. M. (2020). Detection and analysis of C-to-U RNA editing in rice mitochondria-encoded ORFs. plants 9, 1277. doi: 10.3390/plants9101277
Zhou, S. X., Wei, N., Jost, M., Wanke, S., Rees, M., Liu, Y., et al. (2023). The Mitochondrial Genome of the Holoparasitic Plant Thonningia sanGuinea Provides Insights into the Evolution of the Multichromosomal Structure. Genome Biol. Evol. 15, evad155. doi: 10.1093/gbe/evad155
Keywords: Camellia sinensis cv. ‘Baihaozao’, mitochondrial genome, multichromosomal structure, adaptive evolution, phylogenetic relationships
Citation: Chen Z, Wang Z, Zhou W, Liu S, Xiao Y and Gong Y (2025) Complete sequencing of the mitochondrial genome of tea plant Camellia sinensis cv. ‘Baihaozao’: multichromosomal structure, phylogenetic relationships, and adaptive evolutionary analysis. Front. Plant Sci. 16:1604404. doi: 10.3389/fpls.2025.1604404
Received: 01 April 2025; Accepted: 27 May 2025;
Published: 13 June 2025.
Edited by:
Surendra Pratap Singh, Chhatrapati Shahu Ji Maharaj University, IndiaReviewed by:
Subodh Kumar Mishra, University at Albany, United StatesNoopur Singh, Indian Institute of Pulses Research (ICAR), India
Copyright © 2025 Chen, Wang, Zhou, Liu, Xiao and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyin Chen, Mjk0MEBodWhzdC5lZHUuY24=; Yihui Gong, Z3loemdoQDE2My5jb20=
 Zhiyin Chen
Zhiyin Chen Zixu Wang1,2,3,4
Zixu Wang1,2,3,4