- 1School of Plant, Environmental and Soil Sciences, Louisiana State University Agricultural Center, Baton Rouge, LA, United States
- 2Southern Regional Research Center, USDA-ARS, New Orleans, LA, United States
Both biotic and abiotic stresses pose serious threats to the growth and productivity of crop plants, including maize worldwide. Identifying genes and associated networks underlying stress resistance responses in maize is paramount. A meta-transcriptome approach was undertaken to interrogate 39,756 genes differentially expressed in response to biotic and abiotic stresses in maize were interrogated for prioritization through seven machine learning (ML) models, such as support vector machine (SVM), partial least squares discriminant analysis (PLSDA), k-nearest neighbors (KNN), gradient boosting machine (GBM), random forest (RF), naïve bayes (NB), and decision tree (DT) to predict top-most significant genes for stress conditions. Improved performances of the algorithms via feature selection from the raw gene features identified 235 unique genes as top candidate genes across all models for all stresses. Three genes such as Zm00001eb176680, Zm00001eb176940, and Zm00001eb179190 expressed as bZIP transcription factor 68, glycine-rich cell wall structural protein 2, and aldehyde dehydrogenase 11 (ALDH11), respectively were commonly predicted as top-most candidates between abiotic stress and combined stresses and were identified from a weighted gene co-expression network as the hub genes in the brown module. However, only one gene Zm00001eb038720 encoding RNA-binding protein AU-1/Ribonuclease E/G, predicted by the PLSDA algorithm, was found commonly expressed under both biotic and abiotic stress. Genes involved in hormone signaling and nucleotide binding were significantly differentially regulated under stress conditions. These genes had an abundance of antioxidant responsive elements and abscisic acid responsive elements in their promoter region, suggesting their role in stress response. The top-ranked genes predicted to be key players in multiple stress resistance in maize need to be functional validated to ascertain their roles and further utilization in developing stress-resistant maize varieties.
Introduction
Plants are constantly subjected to various biotic and abiotic stresses that have negative effects on the growth, development, and productivity of economically important crops including maize (Zea mays L.) (Ramegowda and Senthil-Kumar, 2015). Maize is a main grain, forage, and energy crop as well as a genetic model plant (Farooqi et al., 2022). It is one of the most important cereal crops cultivated worldwide, mainly in Africa and South America (Kimotho et al., 2019). After wheat and rice, maize is the most frequent cereal food in Mexico (Nazari et al., 2023). United States remains the largest producer of maize. According to USDA, corn production for the 2024/25 marketing year is projected to be around 377.63 million metric tons, a 1% reduction than last year, which is attributed to extreme drought and heat during the 2024 crop year (https://www.fas.usda.gov/data/production/commodity/0440000).
Abiotic stresses such as drought, cold, submergence, salinity, waterlogging, heavy metal contamination, or nutrient deficiency can reduce crop yield by more than >50% (Mallikarjuna et al., 2020). Similarly, biotic stresses, caused by living organisms such as bacteria, viruses, fungi, or nematodes, negatively affect the productivity of maize by approximately 10% (Nazari et al., 2023). Plants undergo genome-wide transcriptome reprogramming in response to external stressors, which leads to induction and/or repression of genes associated with various mechanisms at whole plant, organellar, cellular, and molecular levels. Recent advances in various omics technologies have accelerated the identification of genes and biological processes controlling stress responses in plants. Analysis of transcriptomes has made it possible to identify genes overexpressed/repressed under specific stress (Sharma et al., 2013). However, in field conditions, plants are repeatedly exposed to multiple stresses simultaneously, which requires the plants to exercise efficient molecular mechanisms to recognize a host of signals to effectively respond to more than one stress (Sharma et al., 2013). Both biotic and abiotic stress factors and their various combinations in natural conditions elicit modified stress responses in plants. In addition to several genes, transcription factors (TFs) are known to be significantly involved in stress response in plants (Zhu, 2002). Many TFs from AP2/IREP, bZIP, MYB, NAC, and WRKY families have been found to improve stress resistance by regulating the expression of other stress-responsive genes in plants (Qin et al., 2011). Thus, the identification and characterization of key genes that are co-expressed in plants’ response to both abiotic and biotic stresses would provide targets for genetic strategies to improve stress tolerance.
Over the years, several stress-regulated genes have been identified in maize using both microarray and RNA-seq approaches (Hayford et al., 2024). However, deciphering unique genes responding to specific or multiple stresses requires significant computational maneuvers. Meta-analysis is recognized as a reasonable yet statistically powerful approach where the results from multiple independent studies can be combined to eliminate the challenges due to variations between individual studies (Ramasamy et al., 2008; Keel and Lindholm-Perry, 2022). Meta-analysis has been used successfully on transcriptomic data of several crops including maize to identify potentially top candidate genes that are regulated in plants to cope with stress (Baisakh et al., 2023; Hayford et al., 2024; Wang et al., 2022; Nazari et al., 2024).
The multidimensionality of RNA-seq and microarray data owing to the high number of variables and genes with minimal sample size necessitates a gene selection technique to find the most informative, expressed genes and remove the redundancy in the original space (Mahendran et al., 2020). Machine Learning (ML), which is a subset of artificial intelligence, focuses on training the algorithms based on available datasets thus enabling models to learn to make decisions on their own from data without explicit programming. ML uses feature extraction and selection as a reduction technique for classification performance to make decisions on the top features (Kira and Rendell, 1992). In this study, we conducted a meta-analysis for integrating RNA-seq data across independent studies on biotic and abiotic stresses in maize to predictably identify the top-most significantly differentially expressed genes using ML tools. Furthermore, we compared the results from multiple ML models and integrated gene co-expression network analysis to identify the most useful stress-responsive genes.
Materials and methods
RNA-seq data collection and feature counts
All RNA-seq datasets related to both biotic and abiotic stresses in maize (Supplementary Table 1) were searched online using the publicly available sequence repository database i.e., NCBI Gene Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra). Raw sequence reads were subjected to quality control and filtering following the method described earlier (Bedre et al., 2015). Clean sequence reads were mapped against the B73 reference genome NAM 5.0 using HISAT2 (Kim et al., 2019) with default parameters. Mapped reads were then counted for genomic features such as genes, chromosomal locations, etc. using the FeatureCounts program (Liao et al., 2014).
Read counts preprocessing and merging
Gene expression data are often associated with high inconsistency due to noise and pieces attributed to the differences in sample numbers, labels, experimental conditions, etc. To correct these biases caused by non-biological conditions, data were normalized using “normalize.quantiles” in R package preprocessCore version 1.56.0 following Schadt et al. (2001). However, normalization procedures do not adjust the sample data for batch effects (BE) when merging batches of data from multiple experiments that contain large batch-to-batch variation. Therefore, BE correction was performed using the ‘ComBat’ function within the SVA package in R as described earlier (Zhang et al., 2022), which uses empirical Bayes method that estimates the LS parameters (mean and variance) for each gene and merges information from multiple genes with similar expression attributes in each batch.
Machine learning gene selection approaches
Gene expression data in the form of classified attributes with feature counts of several thousands of genes with expression patterns from multiple stress samples were analyzed to identify the most important genes, which required extraction and selection of features with discriminating ability. For ML, 80% of the data was used as a training set and the remaining 20% as the test/validation set. The classification of maize samples for the gene expression under control and (a)biotic stress was used to select the ML models that can best identify the top stress-responsive genes. We used seven ML algorithms (i.e., SVM, support vector machine; PLSDA, partial least squares discriminant analysis, KNN: K-nearest neighbors, GBM, gradient boosting machine; RF, random forest; NB, naïve bayes; and DT, decision tree) for the identification of most informative genes from the differentially expressed genes (DEGs). Variable importance (VarImp) evaluation functions were grouped with and without the model information. A model-based approach is more closely tied to its performance, which can integrate the correlation structure between the predictors (genes in this case) into the importance calculation. Each gene is assigned a separate variable importance for each class in classification models where all importance measures are scaled at minimum and maximum values of 0 and 100, respectively. The area under the receiver operating characteristic (ROC) curve (AUC) values for each gene was obtained using the “filterVarImp” function.
The SVM algorithm was used to identify top genes with R package e1071 version 1.7–6 using the function of SVM-radial and default code: RFE (x = data, y = as.numeric (as.factor (group)), sizes = c (seq (2, 40, by = 2)), RFE-Control = rfeControl (functions = caretFuncs, method = “cv”), methods = “svmRadial”). Another model, Random Forest (version 4.6–14), was implemented using the RF algorithm with the following parameters: ntree = 100–500 and mtry = 1 – 8 (Kim et al., 2022). The relevance score and ranking of the genes in RF and SVM were determined following the recursive feature elimination (REF) method as per the program manual.
The PLS-DA method was implemented in R package PLSDA with PLS regression where Y is a set of binary response variables describing the categories (control or stress) of a categorical variable on X, where X is the gene expression matrix using the equation of Pérez-Enciso and Tenenhaus (2003) that used the algorithm of Wold et al. (1983) to allow for missing values. We identified the top genes using variable importance in projection (VIP) for each gene (Eriksson et al., 1999). GBM, an ensemble method (Hastie et al., 2009) is also used for regression and classification methods with reduced variance and bias in simple prediction models (Hastie et al., 2009). The caret package in R was used with the GBM function for selecting the top genes. The caret package was also used for KNN model with 10-fold cross-validation and default parameter tune-Length =10 to select the top-ranked genes for different stresses. Similarly, the DT model used the R package Rpart to select the top genes through mtree function. Another method, Naïve Bayes version 0.9.7 was adopted for the NB algorithm with 10-fold cross-validation for identifying the most important genes. The performance of each ML model was determined by classification metrics such as accuracy, precision, specificity, sensitivity (recall), F1-score, Mathews correlation coefficient (MCC), and ROC derived from the confusion matrix following the equations described in Sabanci et al. (2022). The confusion matrix was prepared with the maize gene under control labeled as 1 and stress as -1.
Gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) is used in systems biology to make clusters (modules) of highly correlated genes based on the module eigengene (ME) and to identify an intramodular hub gene. The expression values of genes after normalization and batch effect correction were used in the R package WGCNA version 1.66 (Langfelder and Horvath, 2008) for weighted co-expression network construction where the similar matrix between each pair of genes across all samples was evaluated based on the Pearson’s correlation values. Modules were identified by the blockwiseModules function of the WGCNA package with default parameters and a tree cut height of 0.4. The modules were defined at a cut height of 0.98 and a size at 30. Similar modules were merged when dissimilarity of module eigengenes was <0.25. The function signedKME was used to calculate module eigengene values (KME) based on the correlation of the module eigengene (ME) with the corresponding gene. Modules with correlation value (r) >0.8 between genes and P-value <0.01 were considered as significant modules (de Silva et al., 2022). Gene significance was calculated based on the p value of the linear regression between the gene expression profile with multiple stress conditions. The hub genes in a module were identified based on gene significance value (GS) >0.5, module eigengene (KME) >0.8, and with maximum connections with other genes (Baisakh et al., 2023). Gene co-expression network was visualized using Cytoscape (version 3.10.3) software in R (Shannon et al., 2003; Baisakh et al., 2023).
Gene ontology and promoter analysis
The ontology of the genes was assigned by the singular enrichment analysis within AgriGO version 2 (https://systemsbiology.cau.edu.cn/agriGOv2/) and gene ontology enrichment was performed using Fisher’s t-test (P<0.05) and FDR correction by the Hochberg method. Metabolic pathway enrichment analysis was performed using the DAVID tool version 6.7 (https://davidbioinformatics.nih.gov/tools.jsp). The top genes common between two or more models were used to extract 2000 bp upstream flanking region of sequence using the Ensembl Plants database (http://plants.ensembl.org). Promoter prediction and identification of cis-regulatory elements (CREs) within a promoter were performed using PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Results and discussion
DEGs under biotic and abiotic stress
Raw sequence reads of a total of 3,052 samples, which included 976 from biotic (bacteria, fungus, insect, nematode, and weed) and 2,076 from abiotic (drought, heat, cold, waterlogging, salt, nutrient, and mechanical wounding) stresses, were obtained from 52 RNA-seq studies conducted with 12 stress conditions of which five were biotic and seven were abiotic (Supplementary Table 1). Finally, 39,756 differentially expressed genes from 1,452 samples (451 from biotic and 1001 from abiotic) and 22 of 52 studies after normalization and batch effect correction, respectively were used in ML models sets to identify the topmost significant genes.
Identification of top stress-responsive genes by ML models
Identifying top stress-responsive DEGs was conducted for biotic or abiotic stress individually and in combination to identify top genes unique to a specific stress category and common between the stress categories.
Prediction of top genes under biotic stress
Based on the confusion matrix (Supplementary Table 2) of the seven ML models, SVM performed the best with the highest average accuracy of 76.6% followed by RF (73.0%) and GBM (71.8%) whereas NB, KNN, DT, and PLSDA performed very poorly with an average accuracy of 57% (Table 1A). SVM also had the highest specificity (0.81) and MCC (0.54) with precision (0.81) behind KNN (0.84) and sensitivity (0.73) behind NB (0.83), RF (0.80), and GBM (0.75). Interestingly, SVM had the lowest F1-score (0.56). However, MCC, which considers all four parameters in the confusion matrix, is considered a better performance matrix as compared to F1-score, which only considers precision/recall. DT had the worst performance in terms of accuracy (53.5%), sensitivity (0.58), and MCC (0.03), although it had a slightly better specificity over PLSDA and NB, higher precision than NB, and higher F1-score than SVM. DT was the least sensitive algorithm correctly detecting only 0.58 for positive samples.
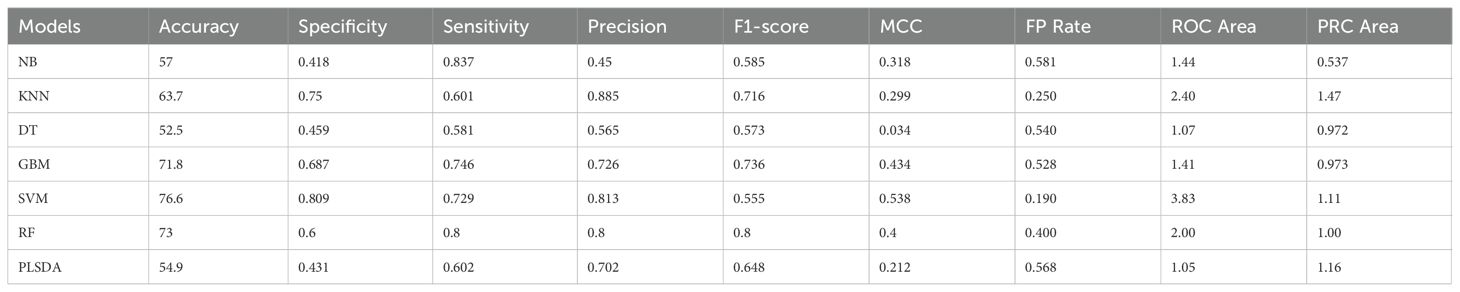
Table 1A. Confusion matrices of machine learning algorithms used with gene expression values under biotic stress in maize.
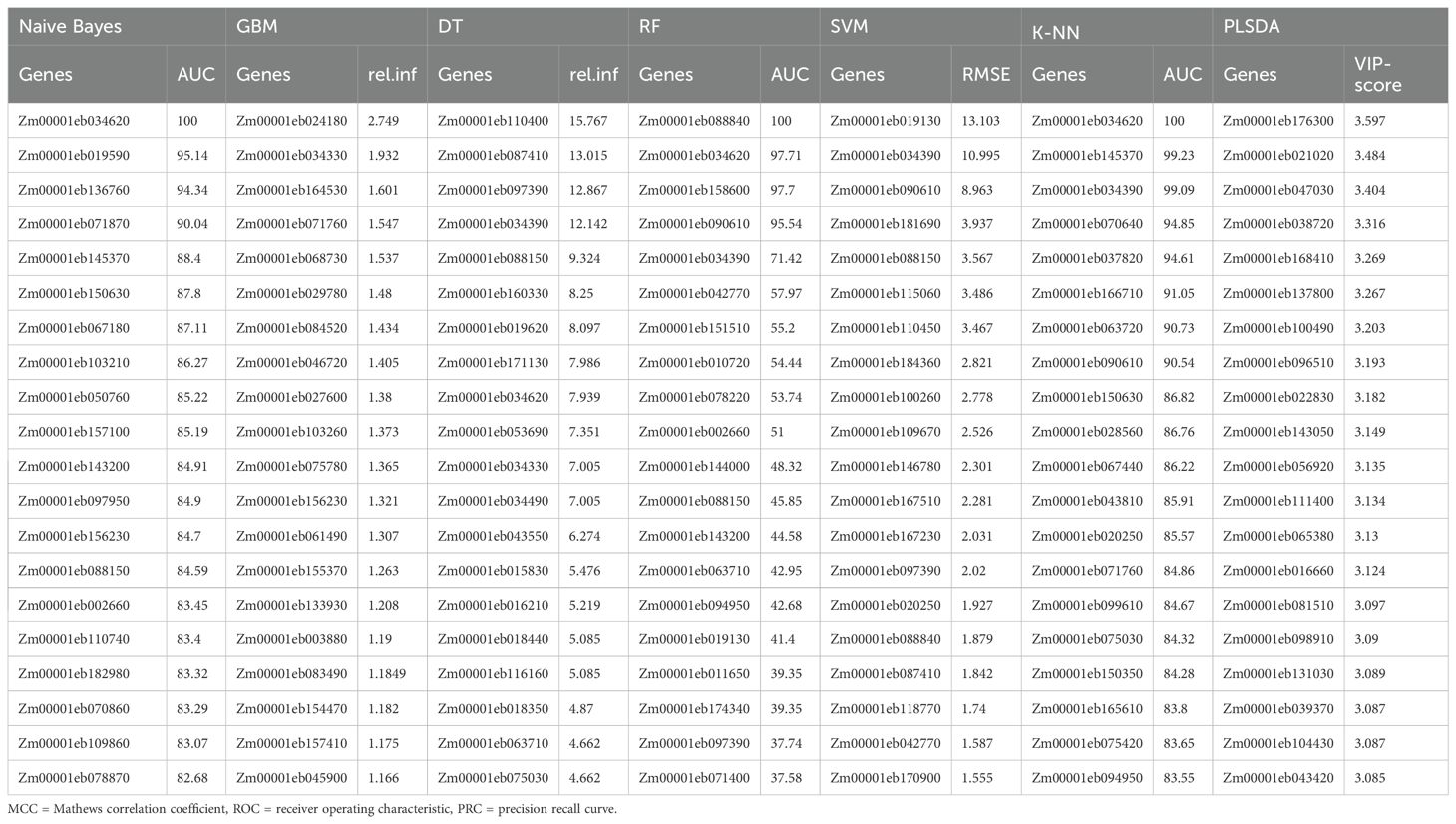
The top 20 genes predicted by each model resulted in a total of 111 unique top significantly differentially regulated genes by all seven models (Table 1B). Of these, 16 genes were predicted by at least two models. Four genes, Zm00001eb034390, Zm00001eb088150, Zm00001eb042770, and Zm00001eb097390 predicted as top genes by two or more models including the two high-performing models, SVM and RF, were considered the most significantly differentially expressed genes under biotic stress.
Prediction of top genes under abiotic stress
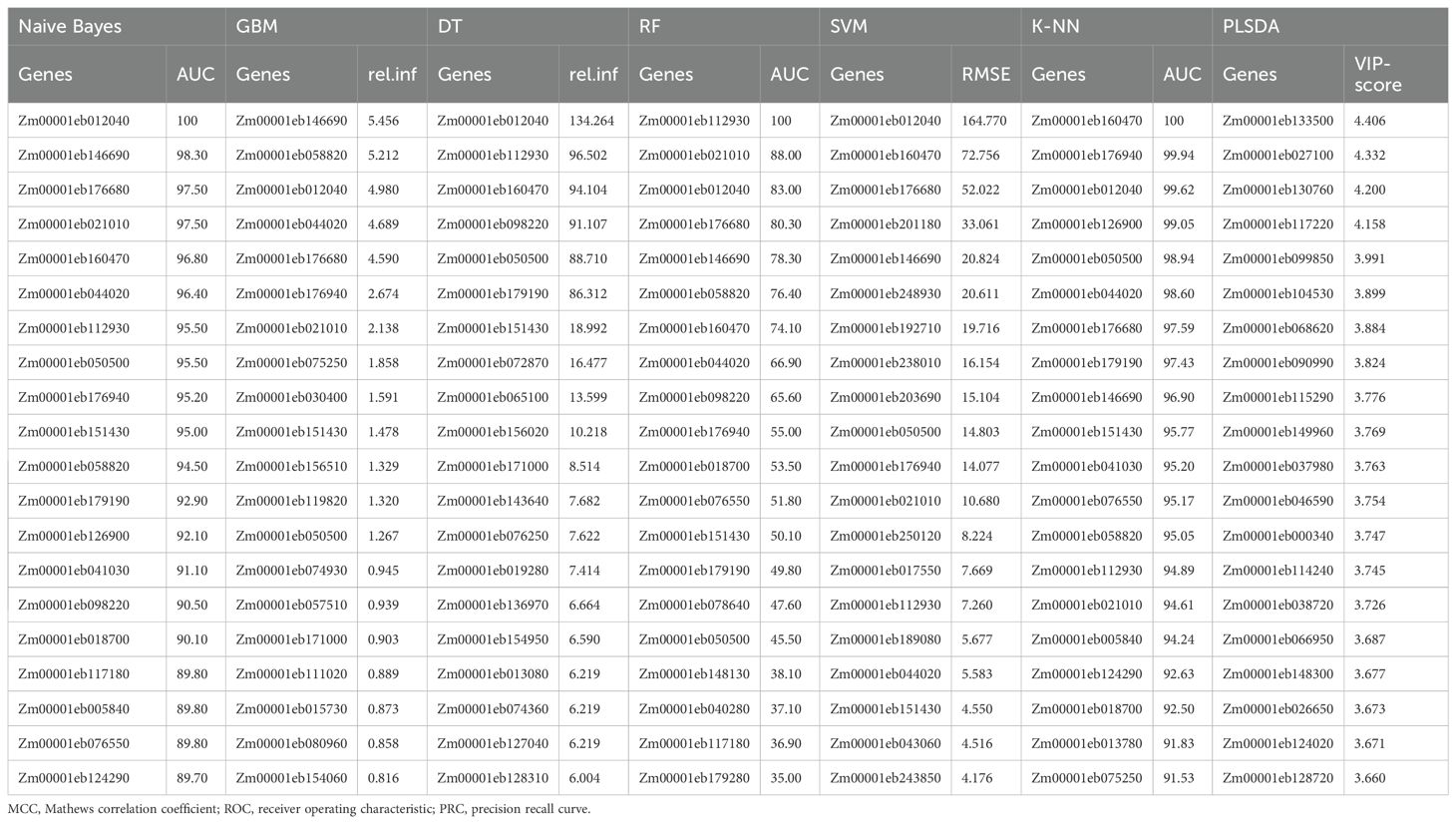
The confusion matrix (Supplementary Table 2) for the abiotic stress responsive genes revealed that the highest average accuracy of 88.6% was obtained by GBM, closely followed by SVM (87.5%) and RF (87.0%) algorithms (Table 2A). RF had the highest MCC value (0.72) followed by SVM at 0.71 and GBM (0.6). SVM had the highest specificity (0.94) followed by RF at 0.89 and precision (0.86). Altogether, 68 unique genes were identified as most informative by all seven models. Twenty-one genes were consistently predicted as top abiotic stress-related genes by at least two models of which 12 were common in four or more models, which included GBM, SVM, RF and KNN models that had higher accuracy and/or model performance matrices compared to other models (Table 2B). Interestingly, only one gene Zm00001eb038720, predicted by PLSDA, was consistent between the biotic and abiotic stress conditions.
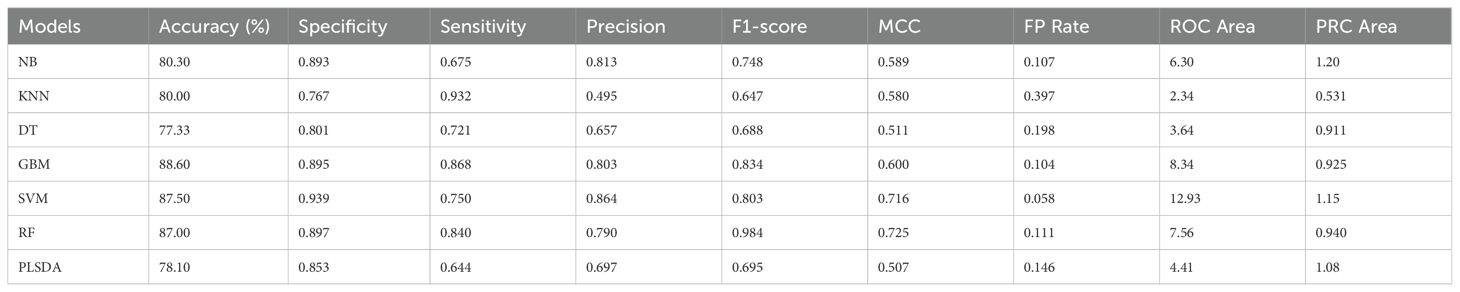
Table 2A. Confusion matrices of machine learning algorithms used with gene expression values under abiotic stress in maize.
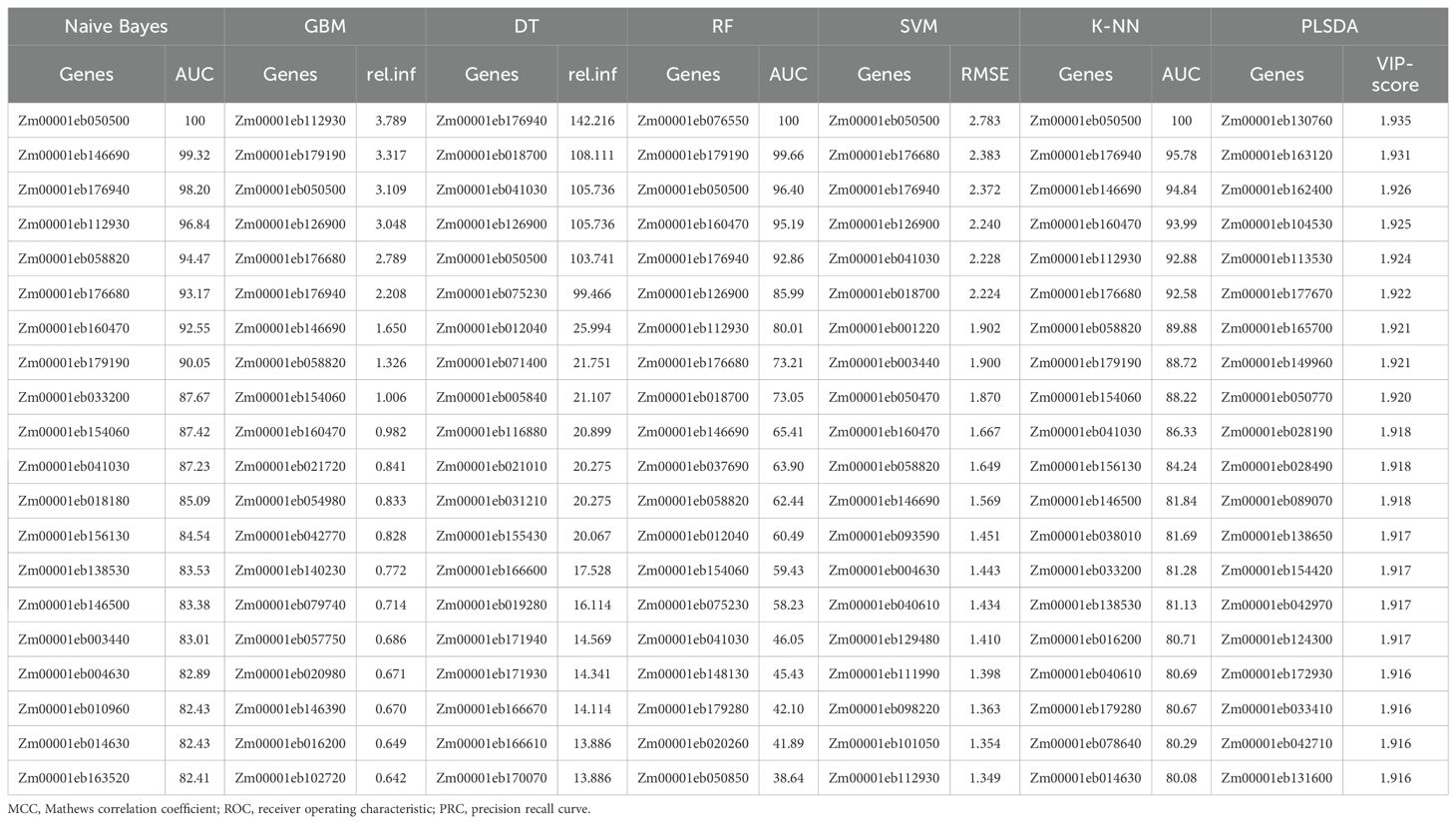
Prediction of top genes under combined stress conditions
When the genes responsive to biotic and/or abiotic stress conditions based on the datasets in the literature were combinedly used for prediction by seven models, RF outperformed others with the highest accuracy (81.7%) and other performance matrices except for sensitivity (0.75), which was behind SVM (0.80) and GBM (0.77) (Table 3A; Supplementary Table 2). SVM predicted the top 20 genes with the highest sensitivity, an accuracy of 81.0%, F1-score (0.77) and MCC (0.61) second to only RF. GBM also performed good with 79.0% accuracy, nearly equal specificity and same sensitivity as SVM. NB was the worst predictor algorithm with the lowest values recorded for accuracy as well as other parameters. A total of 83 unique top significant genes were reported for combined stress by the seven models of which 11 genes were commonly predicted by four or more models. Among biotic and combined stress conditions, 23 genes were found common. On the other hand, only two genes were found within biotic and combined stress conditions (Table 3B).
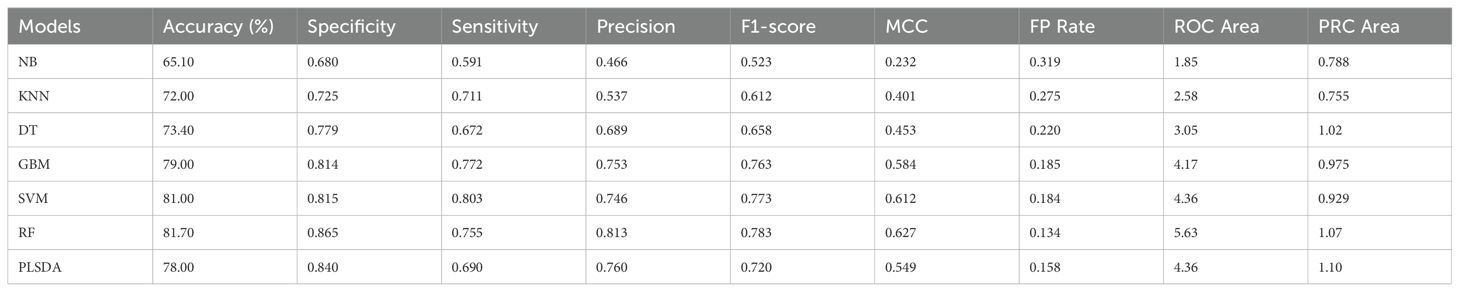
Table 3A. Confusion matrices of machine learning algorithms used with gene expression values under combined stress conditions in maize.
Taken together, SVM, RF, and GBM were identified as the best models in predicting top significant (a)biotic stress responsive genes with high accuracy in our study. However, Nazari et al. (2023) found KNN (82.0%) and Ensemble (85.7%) to be more accurate for predicting biotic stress tolerance genes while modeling gene expression data from microarray studies in maize. Interestingly, none of the significant genes identified by these authors matched the top genes selected by the seven models used in our study.
Significant modules and potential hub genes
The weighted gene co-expression network analysis based on a height cut-off at 0.25 to merge the modules, detected four modules of which Turquoise and Brown modules with 438 and 14 genes, respectively were significant with r >0.8 and GS (P-value¾0.01) (Supplementary Table 3; Figure 1a). Three significant hub genes, Zm00001eb176680, Zm00001eb176940, and Zm00001eb179190 with KME>0.8, were identified in the brown module (Figure 1b) for abiotic stress and one gene Zm00001eb150630 (turquoise model) for biotic stress (Figure 1c) that were predicted by two or more models.
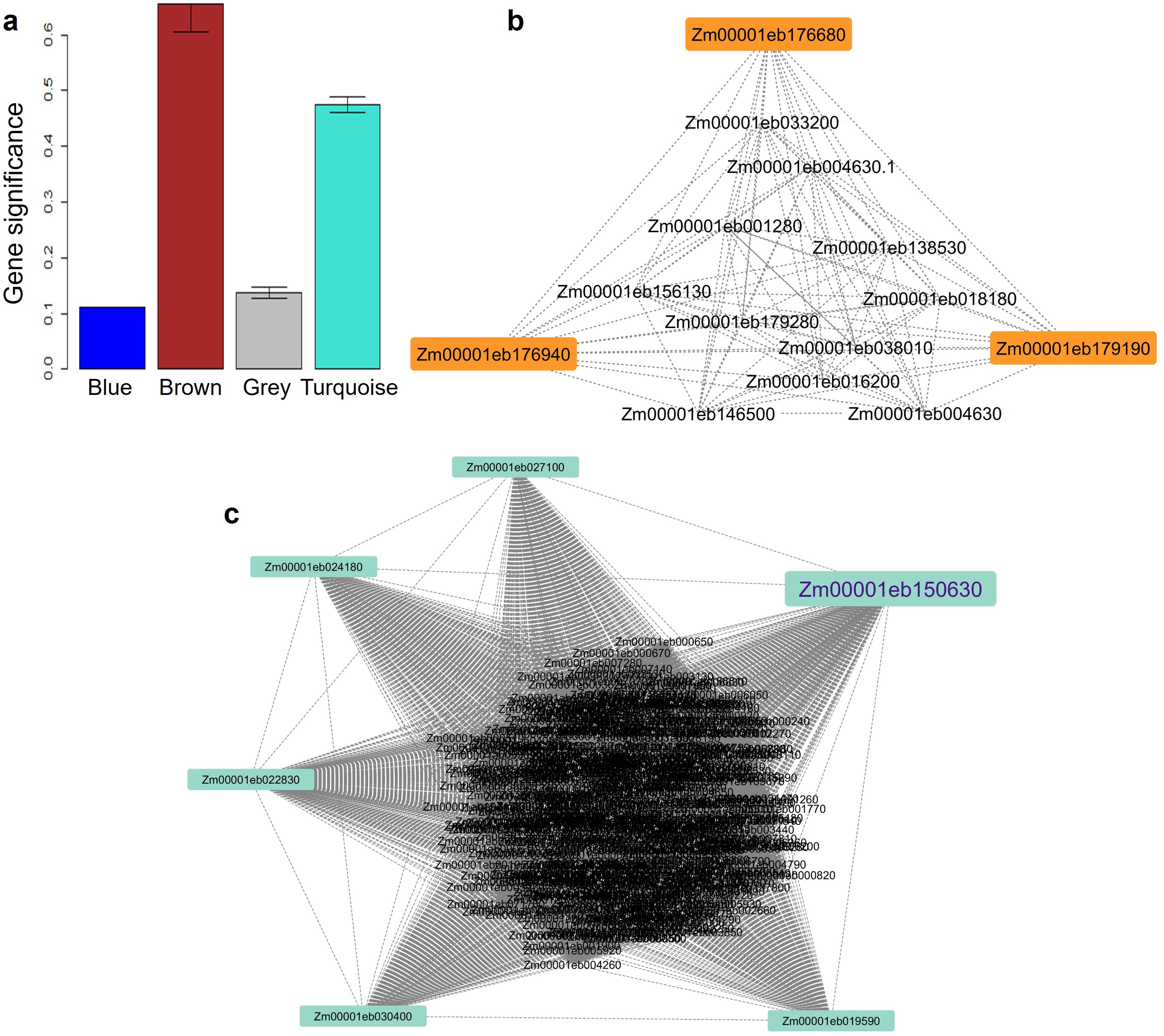
Figure 1. The gene significance values (P-value=8.9e-124) across co-expression network modules (a). Brown (b) and Turquoise (c) significant modules showing three and six potential hub genes, respectively.
Functional involvement of the top highly significant genes
GO analysis of top genes representing conserved up and down-regulation under stress conditions identified 47 and 3 significant GO terms (p-value<0.001; FDR <0.05) for abiotic and biotic stress, respectively whereas 17 significant GO terms were identified for combined stress (Supplementary Table 4). The most significantly enriched molecular function with the highest number of genes was associated with nucleotide binding for biotic stress whereas binding followed by response to stimulus were the most enriched processes under abiotic stress. On the other hand, cell communication, cellular response to alcohol, lipid, external stimuli, and abscisic acid, and hormone signaling were the two most enriched biological processes under combined stress conditions. The biological pathways involving the top-most significant genes involved are presented in Supplementary Table 5. In biotic stress, genes involved in calcium ion binding were the most enriched whereas genes in abscisic acid activated signaling pathway, cation binding and myosin phosphate activity were the most significant in abiotic stress. Genes associated with seed nutrient storage activity, cation binding, and plant hormone signal transduction were significant under combined, biotic and abiotic stresses.
Of the four top-most significant genes associated with biotic stress response and predicted by the two high-performing models SVM and RF, Zm00001eb034390 (GO:0005509) coding for EF-hand 1 calcium binding protein (1.23-fold), Zm00001eb088150 (GO:0098754) for UDP-glucuronosyl/UDP-glucosyltransferase (0.74-fold), Zm00001eb042770 (GO:0005634) for ribonuclease 3-like protein 3 (0.23) were significantly upregulated, whereas gene Zm00001eb097390 (GO:0006457) encoding GrpE nucleotide exchange factor was downregulated (-0.14) under fungal infection (Supplementary Table 6) (Shu et al., 2017; Kebede et al., 2018; Shi et al., 2018; Han et al., 2020; Lambarey et al., 2020; Musungu et al., 2020; He et al., 2021; Liu et al., 2021; Schurack et al., 2021). Zm00001eb088150 has been shown to be involved in the detoxification of several exogenous and endogenous compounds, and it plays multiple roles in plant responses to biotic as well as abiotic stresses (Gharabli et al., 2023) providing protection against mycotoxins (Wetterhorn, 2018), pathogens, drought, heat, cold, and salinity (Van Aken, 2008; Meale et al., 2015). UDP-glucuronosyl/UDP-glucosyltransferase was found to be one of the 26 genes commonly regulated between maize, peanut, and cotton in response to Aspergillus flavus, and it was upregulated in both pericarp and seed tissues of cotton (Mehanathan et al., 2018).
Among the 12 most-significant genes predicted by SVM, RF, and GBM for abiotic stress, eight were upregulated under drought (Kakumanu et al., 2012; Song et al., 2017; Li et al., 2017; Thirunavukkarasu et al., 2017; SkZ et al., 2018; Yang et al., 2019; Danilevskaya et al., 2019; Zenda et al., 2019; Jia et al., 2020; Li et al., 2021; Kim et al., 2021; Maheswari et al., 2021; Bai et al., 2022), cold (Sobkowiak et al., 2014; Mao et al., 2017; Goering, 2017; Li et al., 2017; Avila et al., 2018; Waititu et al., 2021; Li et al., 2021), heat (Li et al., 2014; Shi et al., 2017; Li et al., 2017 and Zhao et al., 2019), and salt (Zhang et al., 2015; Du et al., 2017; Li et al., 2017; Chen et al., 2020; Zhang et al., 2021; 2022) stress of which four were downregulated under waterlogging condition (Supplementary Table 6) (Arora et al., 2017; Yao, 2021). Also, eight of the genes were commonly predicted between abiotic and combined stresses. Of the four genes specifically uniquely predicted for abiotic stress, Zm00001eb012040 (GO:0006470) is expressed as a PPM-type phosphatase protein and was upregulated in all but waterlogging conditions whereas Zm00001eb160470, which also codes for a protein phosphatase, was upregulated under drought, heat and salt but downregulated under cold and waterlogging conditions (Supplementary Table 6). These genes are known to dephosphorylate serine/threonine in stress responsive genes in maize, thus impacting its response to multiple stresses (drought, salt) via hormone signal transductions (He et al., 2019). On the other hand, Zm00001eb021010 (cysteine-rich and transmembrane domain-containing protein WIH1) was downregulated in all abiotic stresses except slight upregulation under waterlogging conditions. Several members of the cysteine-rich peptide family have been shown to respond extensively to various abiotic stresses in different plants including maize (Xu et al., 2018). Zm00001eb050500 (GO:0009653), a top gene under abiotic as well as combined stress, encodes expansin, which plays an important role in stress relaxation of (arabino) xylan-cellulose networks within the cell wall (Yennawar et al., 2006). bZip-transcription factors (e.g., Zm00001eb058820) have been extensively studied for their critical roles in regulating plants response to abiotic stresses through morphological adaptations (Guo et al., 2024). In maize, a member of bZIP family positive regulated stress resistance through ABA-dependent signaling (He et al., 2024). Zm00001eb044020 (a P-loop containing nucleoside triphosphate hydrolase), which was moderately upregulated under cold and waterlogging conditions, but highly upregulated in response to drought, heat and salt, was also identified to be induced by proline under low water potential (Verslues et al., 2014) and was located within the genomic region associated with heat stress (Bashir et al., 2025). Only one gene Zm00001eb038720, expressed as Ribonuclease E/G, was found commonly predicted between biotic and abiotic stress, although it was not identified as one of the top-most significant genes by the two or more high-performing accurate models. Ribonuclease E/G are chloroplastic endoribonuclease that play crucial roles in plant’s response to biotic and abiotic stresses because of their involvement in cleavage-mediated RNA homeostasis (Schein et al., 2008). The RNase E/G enzymes influence RNA modifications in plant adaptation to stresses by controlling mRNA stability and subsequent translation of genes (Cai et al., 2025).
Three genes that were commonly predicted as top-most candidates between abiotic stress and combined stresses as well as identified as the hub genes in the brown module, coded for a bZIP transcription factor 68 (Zm00001eb176680), glycine-rich cell wall structural protein 2 (Zm00001eb176940), and aldehyde dehydrogenase 11 (ALDH11; Zm00001eb179190). bZIP transcription factor 68 showed a negative response to cold stress in transgenic maize plants (Zeng et al., 2021). bZIP68 interacts with mitogen-activated protein kinase 8, which is also a negative regulator of the cold-stress response. A 358-bp indel in the bZIP68 promoter region increased its expression resulting in decreased cold tolerance in maize (Li et al., 2022). ZmbZIP4 was also shown to be differentially stimulated by high salinity, drought, heat, cold, and abscisic acid treatment in maize seedlings (Ma et al., 2018). Glycine-rich cell wall structural proteins (GRCWSPs) are known to be involved in plant’s response to abiotic stress, especially under osmotic stress because of their roles in maintaining cell wall integrity during dehydration by connecting the lignin rings to strengthen the cell wall (Ryser et al., 2004). The GRCWSPs interact with cell-wall associated kinases to initiate recognition of environmental stimuli and subsequent signal transduction (Park et al., 2001). ALDH11 showed upregulation under drought, cold, heat and salt and negative regulation under waterlogging conditions. Several ALDH genes have been reported to contribute to improving salt and drought tolerance in plants (Wang et al., 2024).
Promoter motifs in the top significant genes
Promoter regions corresponding to the top-most significantly predicted genes for different stresses showed 19 different CREs. As expected of the promoters, CAAT-box, TATA-box, and Unnamed_4 motifs existed in all promoters in high numbers (Supplementary Table 7). While there was no overall pattern in the distribution of the CREs among the promoters of the genes, the top biotic stress responsive genes, except Zm00001eb042770, had less overall total CREs. For most of the abiotic and combined stress responsive genes, the number of antioxidant responsive element (ARE) and ABRE (abscisic acid responsive element) were higher than the biotic stress responsive genes. The number of CREs were higher for two genes Zm00001eb176940 (168) and Zm00001eb179190 (169) predicted common between abiotic and combined stresses than the other genes. The ABRE and G-Box were found in more abundance in genes Zm00001eb012040, Zm00001eb058820, Zm00001eb112930, Zm00001eb018700, Zm00001eb126900, Zm00001eb018700, and Zm00001eb126900 that were identified as top-most significant genes for abiotic and combined stresses.
Conclusion
The integration of a large volume of gene expression data from several RNA-seq studies with machine learning methods increased the generalizability and statistical power and allowed us to analyze the stress responses of the genes to identify a set of top-most genes with significant associations with (a)biotic stress in maize. The GO and KEGG pathway enrichment analysis of top genes provided clues to the mechanisms underlying maize’s response to both biotic and abiotic stress conditions. Furthermore, the randomization procedure used in WGCNA method led to the identification of the hub genes validating their gene connectivity and possible interaction with other genes. Some of the genes identified in this study through ML were also found related to Aspergillus flavus resistance in maize in previous studies. Further functional validation of the roles of the 19 unique top-ranked genes including hub genes, predicted by the high-performing models, will lead to their utilization in developing multiple stress-resistant maize varieties.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Author contributions
AP: Data curation, Formal Analysis, Software, Visualization, Writing – original draft. PG: Data curation, Formal Analysis, Methodology, Writing – review & editing. KR: Funding acquisition, Visualization, Writing – review & editing. NB: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research was funded by the USDA-ARS (NACA Agreement # 58-6054-0-010) and USDA-NIFA Hatch grant (Accession # 7006617). The manuscript was accepted for publication by the Louisiana Agricultural Experiment Station as MS# 2025-306-40333.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor GS declared a past co-authorship with the author AP.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1611784/full#supplementary-material
Abbreviations
ML, machine learning; AI, artificial intelligence; SVM, support vector machine; PLSDA, partial least squares discriminant analysis; KNN, k-nearest neighbors; GBM, gradient boosting machine; RF, random forest; NB, naïve bayes; DT, decision tree; BE, batch effect; DEGs, differentially expressed genes; VarImp, Variable importance; REF, recursive feature elimination; ROC, receiver operating characteristic; AUC, area under the curve; MCC, Mathews correlation coefficient; VIP, variable importance in projection; WGCNA, weighted gene co-expression network analysis; ME, module eigengene; GS, gene significance; CREs, cis-regulatory elements.
References
Arora, K., Panda, K. K., Mittal, S., Mallikarjuna, M. G., Rao, A. R., Dash, P. K., et al. (2017). RNAseq revealed the important gene pathways controlling adaptive mechanisms under waterlogged stress in maize. Sci. Rep. 7, 10950. doi: 10.1038/s41598-017-10561-1
Avila, L. M., Obeidat, W., Earl, H., Niu, X., Hargreaves, W., and Lukens, L. (2018). Shared and genetically distinct Zea mays transcriptome responses to ongoing and past low temperature exposure. BMC Genomics 19, 761. doi: 10.1186/s12864-018-5134-7
Bai, M., Zeng, W., Chen, F., Ji, X., Zhuang, Z., Jin, B., et al. (2022). Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize. Biotechnol. Letters. 44, 367–386. doi: 10.1007/s10529-022-03221-6
Baisakh, N., Da Silva, E. A., Pradhan, A. K., and Rajasekaran, K. (2023). Comprehensive meta-analysis of QTL and gene expression studies identify candidate genes associated with Aspergillus flavus resistance in maize. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1214907
Bashir, L., Budhlakoti, N., Pradhan, A.K., Mehmood, A., Haque, M., Jacob, S. R, et al. (2025). Unraveling the genetic basis of heat tolerance and yield in bread wheat: QTN discovery and its KASP-assisted validation. BMC Plant Biol. 25 (268). doi: 10.1186/s12870-025-06285-4
Bedre, R., Rajasekaran, K., Mangu, V. R., Sanchez Timm, L. E., Bhatnagar, D., and Baisakh, N. (2015). Genome-wide transcriptome analysis of cotton (Gossypium hirsutum l.) identifies candidate gene signatures in response to aflatoxin producing fungus Aspergillus flavus. PloS One 10, e0138025. doi: 10.1371/journal.pone.0138025
Cai, J., Shen, L., Kang, H., and Xu, T. (2025). RNA modifications in plant adaptation to abiotic stresses. Plant Communications, 6(2). doi: 10.1016/j.xplc.2024.101229
Chen, F., Fang, P., Zeng, W., Ding, Y., Zhuang, Z., and Peng, Y. (2020). Comparing transcriptome expression profiles to reveal the mechanisms of salt tolerance and exogenous glycine betaine mitigation in maize seedlings. PloS One 15, e0233616. doi: 10.1371/journal.pone.0233616
Danilevskaya, O. N., Yu, G., Meng, X., Xu, J., Stephenson, E., Estrada, S., et al. (2019). Developmental and transcriptional responses of maize to drought stress under field conditions. Plant Direct. 3, e00129. doi: 10.1002/pld3.129
de Silva, K.K., Dunwell, J.M., and Wickramasuriya, A.M. (2022). Weighted gene correlation network Aaalysis (WGCNA) of arabidopsis somatic embryogenesis (SE) and identification of key gene modules to uncover SE‐Associated hub genes. Int. J. Genom. 2022 (1), 7471063. doi: 10.1155/2022/7471063
Du, X., Wang, G., Ji, J., Shi, L., Guan, C., and Jin, C. (2017). Comparative transcriptome analysis of transcription factors in different maize varieties under salt stress conditions. Plant Growth Regulation. 81, 183–195. doi: 10.1007/s10725-016-0192-9
Eriksson, L., Johansson, E., Kettaneh-Wold, N., Trygg, J., Wikström, C., and Wold, S. (1999). Multi- and Megavariate Data Analysis: Principles and Applications. Umetrics Academy, Umea, Sweden, 533.
Farooqi, M. Q.U., Nawaz, G., Wani, S. H., Choudhary, J. R., Rana, M., Sah, R. P, et al. (2022). Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.). Frontiers in Plant Science, 13, 965878. doi: 10.3389/fpls.2022.965878
Gharabli, H., Della Gala, V., and Welner, D. H. (2023). The function of UDP-glycosyltransferases in plants and their possible use in crop protection. Biotechnol. Advances. 67, 108182. doi: 10.1016/j.bioteChadv.2023.108182
Goering, R. N. (2017). Uncovering candidate cold tolerance genes in maize (Zea mays). Departmental Honors Projects. 54, 22–23. Available online at: https://digitalcommons.hamline.edu/dhp/54.
Guo, Z., Dzinyela, R., Yang, L., and Hwarari, D. (2024). bZIP transcription factors: Structure, modification, abiotic stress responses, and application in plant improvement. Plants, 13(15), 2058. doi: 10.3390/plants13152058
Han, G., Li, C., Xiang, F., Zhao, Q., Zhao, Y., Cai, R., et al. (2020). Genome-wide association study leads to novel genetic insights into resistance to Aspergillus flavus in maize kernels. BMC Plant Biol. 20, 1–11. doi: 10.1186/s12870-020-02404-5
Hastie, T., Tibshirani, R., and Friedman, J. (2009). The elements of statistical learning: data mining, inference, and prediction (2nd ed.). (New York: Springer). doi: 10.1007/978-0-387-84858-7
Hayford, R. K., Haley, O. C., Cannon, E. K., Portwood, II, J. L., Gardiner, J. M., Andorf, C. M., and Woodhouse, M. R. (2024). Functional annotation and meta-analysis of maize transcriptomes reveal genes involved in biotic and abiotic stress. BMC Genomics, 25, 533. doi: 10.1186/s12864-024-10443-7
He, Z., Wu, J., Sun, X., and Dai, M. (2019). The maize clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int. J. Mol. Sci. 20, 3573. doi: 10.3390/ijms20143573
He, S., Li, Y., Wang, Y., and Zhang, X. (2024). Molecular characterization of a stress-response bZIP transcription factor in banana. Plant Cell, Tissue and Organ Culture, 113(2), 173–187. doi: 10.1007/s11240-013-0395-2
He, W., Zhu, Y., Leng, Y., Yang, L., Zhang, B., Yang, J., et al. (2021). Transcriptomic analysis reveals candidate genes responding maize gray leaf spot caused by Cercospora zeina. Plants 10, 2257. doi: 10.3390/plants10112257
Jia, S., Li, H., Jiang, Y., Tang, Y., Zhao, G., Zhang, Y., et al. (2020). Transcriptomic analysis of female panicles reveals gene expression responses to drought stress in maize (Zea mays L.). Agronomy 10, 313. doi: 10.3390/agronomy10020313
Kakumanu, A., Ambavaram, M. M., Klumas, C., Krishnan, A., Batlang, U., Myers, E., et al. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 160, 846–867. doi: 10.1104/pp.112.200444
Kebede, A. Z., Johnston, A., Schneiderman, D., Bosnich, W., and Harris, L. J. (2018). Transcriptome profiling of two maize inbreds with distinct responses to Gibberella ear rot disease to identify candidate resistance genes. BMC Genomics 19, 1–12. doi: 10.1186/s12864-018-4513-4
Keel, B. N. and Lindholm-Perry, A. K. (2022). Recent developments and future directions in meta-analysis of differential gene expression in livestock RNA-Seq. Front. Genet. 13. doi: 10.3389/fgene.2022.983043
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology, 37(8), 907–915. doi: 10.1038/s41587-019-0201-4
Kim, H., Lee, S., and Park, J. (2022). Identification of key genes in stress response using machine learning models. Journal of Computational Biology, 29(4), 345–358. doi: 10.1089/cmb.2022.0123
Kim, K. H., Song, K., Park, J. M., Kim, J. Y., and Lee, B. M. (2021). RNA-Seq analysis of gene expression changes related to delay of flowering time under drought stress in tropical maize. Appl. Sci. 11, 4273. doi: 10.3390/app11094273
Kimotho, R. N., Baillo, E. H., and Zhang, Z. (2019). Transcription factors involved in abiotic stress responses in Maize (Zea mays L.) and their roles in enhanced productivity in the post genomics era. PeerJ 7, e7211. doi: 10.7717/peerj.7211
Kira, K. and Rendell, A. L. (1992). “The feature selection problem: Traditional methods and a new algorithm,” in Proceedings of the tenth national conference on Artificial intelligence (AAAI'92). (San Jose, CA: AAAI Press) 129–134.
Lambarey, H., Moola, N., Veenstra, A., Murray, S., and Suhail Rafudeen, M. (2020). Transcriptomic analysis of a susceptible African maize line to Fusarium verticillioides infection. Plants 9, 1112. doi: 10.3390/plants9091112
Langfelder, P. and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 1–13. doi: 10.1186/1471-2105-9-559
Li, P., Cao, W., Fang, H., Xu, S., Yin, S., Zhang, Y., et al. (2017). Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00290
Li, Y., Hu, J., Liu, J., Suo, H., Yu, Y., and Han, F. (2014). Genome‐wide analysis of gene expression profiles during early ear development of sweet corn under heat stress. Plant Breeding, 134(1), 17–27. doi: 10.1111/pbr.12235
Li, Z., Fu, D., Wang, X., Zeng, R., Zhang, X., Tian, J., et al. (2022). The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell. 34, 2833–2851. doi: 10.1093/plcell/koac137
Li, H., Yue, H., Xie, J., Bu, J., Li, L., Xin, X., et al. (2021). Transcriptomic profiling of the high-vigour maize (Zea mays L.) hybrid variety response to cold and drought stresses during seed germination. Sci. Rep. 11, 19345. doi: 10.1038/s41598-021-98907-8
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: An efficient general-purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. doi: 10.1093/bioinformatics/btt656
Liu, H., Wu, H., Wang, Y., Wang, H., Chen, S., and Yin, Z. (2021). Comparative transcriptome profiling and co-expression network analysis uncover the key genes associated with early-stage resistance to Aspergillus flavus in maize. BMC Plant Biol. 21, p.216. doi: 10.1186/s12870-021-02983-x
Ma, H., Liu, C., Li, Z., Ran, Q., Xie, G., Wang, B., et al. (2018). ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 178, 753–770. doi: 10.1104/pp.18.00436
Mahendran, N., Durai Raj Vincent, P. M., Srinivasan, K., and Chang, C. Y. (2020). Machine learning based computational gene selection models: a survey, performance evaluation, open issues, and future research directions. Front. Genet. 11. doi: 10.3389/fgene.2020.603808
Maheswari, M., Varalaxmi, Y., Sarkar, B., Ravikumar, N., Vanaja, M., Yadav, S. K., et al. (2021). Tolerance mechanisms in maize identified through phenotyping and transcriptome analysis in response to water deficit stress. Physiol. Mol. Biol. Plants. 27, 1377–1394. doi: 10.1007/s12298-021-01003-4
Mallikarjuna, M. G., Thirunavukkarasu, N., Sharma, R., Shiriga, K., Hossain, F., Bhat, J. S., et al. (2020). Comparative transcriptome analysis of iron and zinc deficiency in maize (Zea mays L.). Plants 9, 1812. doi: 10.3390/plants9121812
Mao, J., Yu, Y., Yang, J., Li, G., Li, C., Qi, X., et al. (2017). Comparative transcriptome analysis of sweet corn seedlings under low-temperature stress. Crop J. 5, 396–406. doi: 10.1016/j.cj.2017.03.005
Meale, A. D., Blomstedt, C. K. M., Hamill, J. D., Gaff, D. F., Griffiths, C., Islam, S., et al. (2015). “Method for improving crop productivity,” in International Application Published Under The Patent Cooperation Treaty (PCT).
Mehanathan, M., Bedre, R., Mangu, V., Rajasekaran, K., Bhatnagar, D., and and Baisakh, N. (2018). Identification of candidate resistance genes of cotton against Aspergillus flavus infection using a comparative transcriptomics approach. Physiology and Molecular Biology of Plants, 24(3), 513–519. doi: 10.1007/s12298-018-0522-7
Musungu, B., Bhatnagar, D., Quiniou, S., Brown, R. L., Payne, G. A., O’Brian, G., et al. (2020). Use of dual RNA-seq for systems biology analysis of Zea mays and Aspergillus flavus interaction. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00853
Nazari, L., Aslan, M. F., Sabanci, K., and Ropelewska, E. (2023). Integrated transcriptomic meta-analysis and comparative artificial intelligence models in maize under biotic stress. Sci. Rep. 13, 15899. doi: 10.1038/s41598-023-42984-4
Nazari, L., Zinati, Z., and Bagnaresi, P. (2024). Identification of biomarker genes from multiple studies for abiotic stress in maize through machine learning. Journal of Biosciences, 49(1), 1. doi: 10.1007/s12038-023-00392-w
Park, A., Cho, R., S., Yun, K., U., Jin, J., M., Lee, Y., S., Sachetto-Martins, H., Park, G., O., K., et al. (2001). Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. Journal of Biological Chemistry, 276(28), 26688–26693. doi: 10.1074/jbc.M101283200
Pérez-Enciso, M. and Tenenhaus, M. (2003). Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Hum. Genet. 112, 581–592. doi: 10.1007/s00439-003-0921-9
Qin, F., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52, 1569–1582. doi: 10.1093/pcp/pcr106
Ramasamy, A., Mondry, A., Holmes, C. C., and Altman, D. G. (2008). Key issues in conducting a meta-analysis of gene expression microarray datasets. PloS Med. 5, e184. doi: 10.1371/journal.pmed.0050184
Ramegowda, V. and Senthil-Kumar, M. (2015). The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 176, 47–54. doi: 10.1016/j.jplph.2014.11.008
Ryser, U., Schorderet, M., Guyot, R., and Keller, B. (2004). A new structural element containing glycine-rich proteins and rhamnogalacturonan I in the protoxylem of seed plants. Journal of Cell Science, 117(7), 1179–1190. doi: 10.1242/jcs.00966
Sabanci, K., Aslan, M. F., Ropelewska, E., Unlersen, M. F., and Durdu, A. (2022). A novel convolutional-recurrent hybrid network for sunn pest–damaged wheat grain detection. Food Analytical Methods, 15, 1748–1760. doi: 10.1007/s12161-022-02251-0
Schadt, E. E., Li, C., Ellis, B., and Wong, W.H. (2001). Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. Journal of Cellular Biochemistry, 80(2), 192–202. doi: 10.1002/jcb.10073
Schurack, S., Depotter, J. R., Gupta, D., Thines, M., and Doehlemann, G. (2021). Comparative transcriptome profiling identifies maize line specificity of fungal effectors in the maize–Ustilago maydis interaction. Plant J. 106, 733–752. doi: 10.1111/tpj.15195
Schein, A., Sheffy-Levin, S., Glaser, F., and Schuster, G. (2008). The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA, 14(6), 1057–1068. doi: 10.1261/rna.907608
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, R., De Vleesschauwer, D., Sharma, M. K., and Ronald, P. C. (2013). Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant 6, 250–260. doi: 10.1093/mp/sss147
Shi, J., Yan, B., Lou, X., Ma, H., and Ruan, S. (2017). Comparative transcriptome analysis reveals the transcriptional alterations in heat-resistant and heat-sensitive sweet maize (Zea mays L.) varieties under heat stress. BMC Plant Biol. 17, 1–10. doi: 10.1186/s12870-017-0973-y
Shi, F., Zhang, Y., Wang, K., Meng, Q., Liu, X., Ma, L., et al. (2018). Expression profile analysis of maize in response to Setosphaeria turcica. Gene 659, 100–108. doi: 10.1016/j.gene.2018.03.030
Shu, X., Livingston, D. P., III, Woloshuk, C. P., and Payne, G. A. (2017). Comparative histological and transcriptional analysis of maize kernels infected with Aspergillus flavus and Fusarium verticillioides. Front. Plant Science. 8. doi: 10.3389/fpls.2017.02075
SkZ, A., Vardharajula, S., and Vurukonda, S. S. K. P. (2018). Transcriptomic profiling of maize (Zea mays L.) seedlings in response to Pseudomonas putida stain FBKV2 inoculation under drought stress. Ann. Microbiol. 68, 331–349. doi: 10.1007/s13213-018-1341-3
Sobkowiak, A., Jończyk, M., Jarochowska, E., Biecek, P., Trzcinska-Danielewicz, J., Leipner, J., et al. (2014). Genome-wide transcriptomic analysis of response to low temperature reveals candidate genes determining divergent cold-sensitivity of maize inbred lines. Plant Mol. Biol. 85, 317–331. doi: 10.1007/s11103-014-0187-8
Song, K., Kim, H. C., Shin, S., Kim, K. H., Moon, J. C., Kim, J. Y., et al. (2017). Transcriptome analysis of flowering time genes under drought stress in maize leaves. Front. Plant Science. 8. doi: 10.3389/fpls.2017.00267
Thirunavukkarasu, N., Sharma, R., Singh, N., Shiriga, K., Mohan, S., Mittal, S., et al. (2017). Genome wide expression and functional interactions of genes under drought stress in maize. Int. J. Genomics 2017, .2568706. doi: 10.1155/2017/2568706
Van Aken, O. (2008). Methods and means for the production of plants with improved stress resistance.
Verslues, P. E., Lasky, J. R., Juenger, T. E., Liu, T. W., and Kumar, M. N. (2014). Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiology, 164(1), 144–159. doi: 10.1104/pp.113.224014
Waititu, J. K., Cai, Q., Sun, Y., Sun, Y., Li, C., Zhang, C., et al. (2021). Transcriptome profiling of maize (Zea mays L.) leaves reveals key cold-responsive genes, transcription factors, and metabolic pathways regulating cold stress tolerance at the seedling stage. Genes. 12, 1638. doi: 10.3390/genes12101638
Wang, Y., Guo, H., Wu, X., Wang, J., Li, H., and Zhang, R. (2022). Transcriptomic and physiological responses of contrasting maize genotypes to drought stress. Front. Plant Science. 13. doi: 10.3389/fpls.2022.928897
Wang, J., Xing, C., Wang, H., Zhang, H., Wei, W., Xu, J., et al. (2024). Identification of key modules and hub genes involved in regulating the feather follicle development of Wannan chickens using WGCNA. Poultry Science. 103, 103903. doi: 10.1016/j.psj.2024.103903
Wetterhorn, K. M. (2018). Enzymatic inactivation of Trichothecene mycotoxins associated with Fusarium head blight (Madison, WI: The University of Wisconsin-Madison).
Wold, S., Martens, H., and Wold, H. (1983). The multivariate calibration problem in chemistry solved by the PLS method. In A. Ruhe & B. Kågström (Eds.), Proceedings of the Conference on Matrix Pencils, 286–293. Springer-Verlag, Heidelberg. doi: 10.1007/978-3-642-61794-2_29
Xu, Y., Yu, Z. P., Zhang, D., Huang, J. G., Wu, C. G., Yang, G. D, et al. (2018). CYSTM, a novel non-secreted cysteine-rich peptide family, involved in environmental stresses in Arabidopsis thaliana. Plant Cell Physiology, 59(2), 423–438. doi: 10.1093/pcp/pcx204
Yang, M., Geng, M., Shen, P., Chen, X., Li, Y., and Wen, X. (2019). Effect of post-silking drought stress on the expression profiles of genes involved in carbon and nitrogen metabolism during leaf senescence in maize (Zea mays L.). Plant Physiology and Biochemistry, 135, 304–309. doi: 10.1016/j.plaphy.2018.12.025
Yao, Q. (2021). Crucial waterlogging-responsive genes and pathways revealed by comparative physiology and transcriptome in tropical and temperate maize (Zea mays L.) inbred lines. J. Plant Biol. 64, 313–325. doi: 10.1007/s12374-021-09298-2
Yennawar, N. H., Li, L.-C., Dudzinski, D. M., Tabuchi, A., and Cosgrove, D. J. (2006). Crystal structure and activities of EXPB1 (Zea m 1), a β-expansin and group-1 pollen allergen from maize. Proceedings of the National Academy of Sciences, 103 (40), 14664–14671. doi: 10.1073/pnas.0605979103
Zenda, T., Liu, S., Wang, X., Liu, G., Jin, H., Dong, A., et al. (2019). Key maize drought-responsive genes and pathways revealed by comparative transcriptome and physiological analyses of contrasting inbred lines. Int. J. Mol. Sci. 20, 1268. doi: 10.3390/ijms20061268
Zeng, R., Li, Z., Shi, Y., Fu, D., Yin, P., Cheng, J., et al. (2021). Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 12, 4713. doi: 10.1038/s41467-021-25001-y
Zhang, M., Kong, X., Xu, X., Li, C., Tian, H., and Ding, Z. (2015). Comparative transcriptome profiling of the maize primary, crown and seminal root in response to salinity stress. PloS One 10, e0121222. doi: 10.1371/journal.pone.0121222
Zhang, X., Liu, J., Huang, Y., Wu, H., Hu, X., Cheng, B., et al. (2022). Comparative transcriptomics reveal the molecular mechanism of the parental lines of maize hybrid An’nong876 in response to salt stress. Int. J. Mol. Sci. 23, 5231. doi: 10.3390/ijms23095231
Zhang, X., Liu, P., Qing, C., Yang, C., Shen, Y., and Ma, L. (2021). Comparative transcriptome analyses of maize seedling root responses to salt stress. PeerJ 9, e10765. doi: 10.7717/peerj.10765
Zhao, Y., Hu, F., Zhang, X., Wei, Q., Dong, J., Bo, C., et al. (2019). Comparative transcriptome analysis reveals important roles of nonadditive genes in maize hybrid An’nong 591 under heat stress. BMC Plant Biol. 19, 1–17. doi: 10.1186/s12870-019-1878-8
Keywords: a(biotic) stress, artificial intelligence, gene expression, maize, RNA-Seq
Citation: Pradhan AK, Gandham P, Rajasekaran K and Baisakh N (2025) Predictive prioritization of genes significantly associated with biotic and abiotic stresses in maize using machine learning algorithms. Front. Plant Sci. 16:1611784. doi: 10.3389/fpls.2025.1611784
Received: 14 April 2025; Accepted: 27 May 2025;
Published: 19 June 2025.
Edited by:
Gurjeet Singh, Texas A and M University, United StatesReviewed by:
Tanmaya Kumar Sahu, Indian Institute of Agricultural Biotechnology (ICAR), IndiaAli Babar, University of Florida, United States
Copyright © 2025 Pradhan, Gandham, Rajasekaran and Baisakh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niranjan Baisakh, bmJhaXNha2hAYWdjZW50ZXIubHN1LmVkdQ==
†ORCID: Niranjan Baisakh, orcid.org/0000-0001-8733-0923
 Anjan Kumar Pradhan
Anjan Kumar Pradhan Prasad Gandham
Prasad Gandham Kanniah Rajasekaran
Kanniah Rajasekaran Niranjan Baisakh
Niranjan Baisakh