- 1Quantitative Biosciences & Engineering, Colorado School of Mines, Golden, CO, United States
- 2Chemical & Biological Engineering, Colorado School of Mines, Golden, CO, United States
Genome-scale metabolic models (GEMs) provide a systems-level framework for understanding and engineering microalgal metabolism. This review explores the evolution of GEMs in microalgae, highlighting advances in light modeling, automation, and multi-omics integration. Special emphasis is placed on Chlamydomonas reinhardtii as a model species. Limitations of current models, particularly for microalgae, are discussed, alongside promising developments in dynamic modeling and machine learning. Together, these innovations chart a path toward more predictive, adaptable GEMs that can accelerate biotechnological applications of microalgae in sustainable production systems.
Introduction
Microalgae have demonstrated significant potential for the sustainable production of biofuels and other valuable products. As cell factories, microalgae can be optimized for biofuel production (Makareviciene and Sendzikiene, 2022), wastewater processing (Ahmed et al., 2022), and the creation of a wide variety of high-value bioproducts such as nutraceuticals and pharmaceuticals (Abu-Ghosh et al., 2021; Khanra et al., 2022) (see Table 1). Microalgae have also been shown to have a solar conversion efficiency of 4.4% (Huntley and Redalje, 2007), considerably higher than the solar conversion efficiency of terrestrial plants which is typically between 1-2% (Vasudevan and Briggs, 2008). The advantage in solar conversion efficiency for microalgae then translates to higher growth rates and annual yields compared to terrestrial plants (Chung et al., 2011). Since many bioproducts produced by microalgae are intracellular, their yields are closely tied to biomass accumulation, meaning that higher growth rates generally result in greater overall production of desired bioproducts.
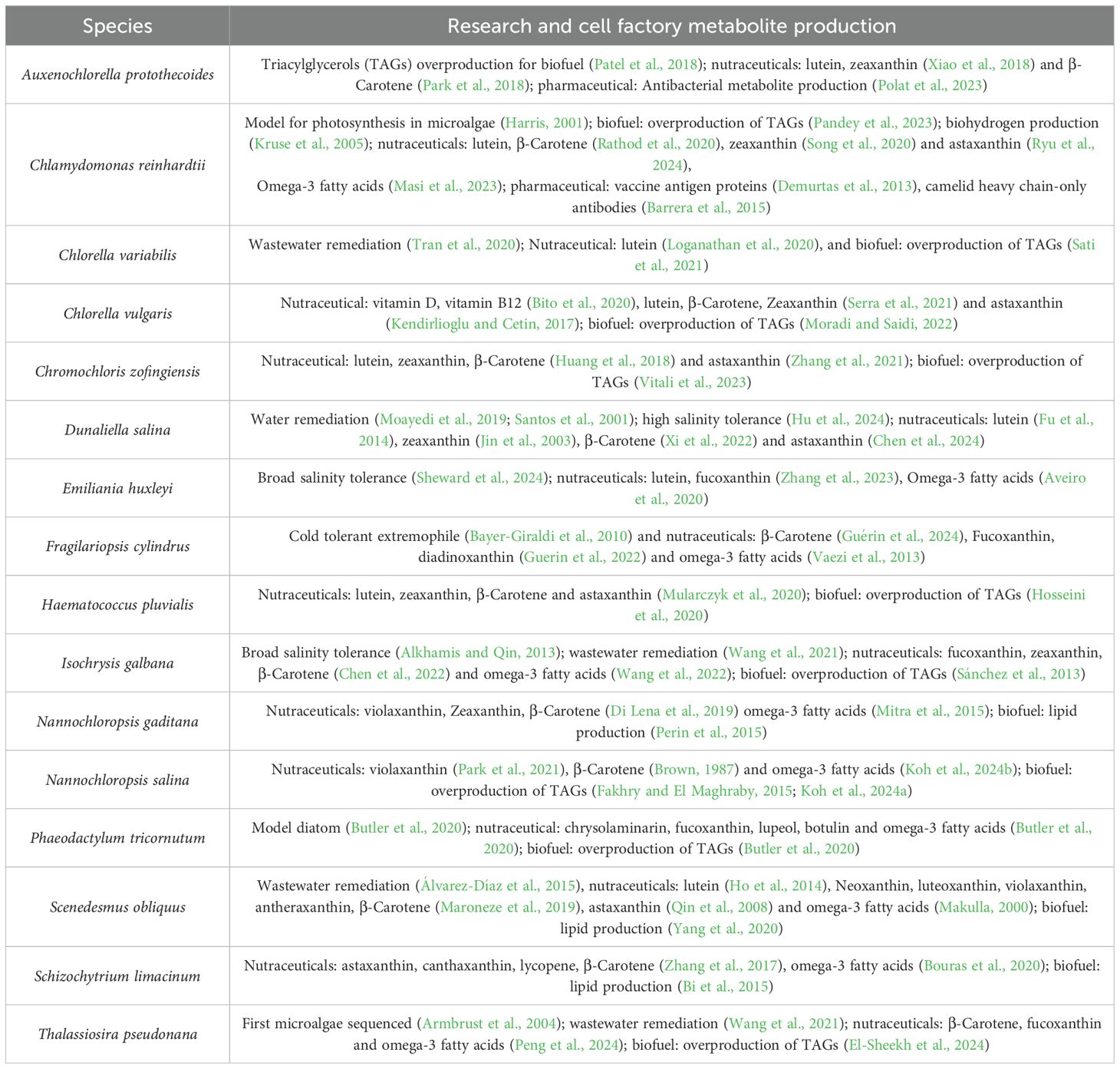
Table 1. Algae species that currently have GEMs reconstructed for them as well as research and cell factory applications of each species.
Unfortunately, algae have not fully realized their potential as cellular factories due to a number of challenges associated with economical production at large scale (Bošnjaković and Sinaga, 2020). A main driver of the overall cost of production is the productivity of the algae (growth rate x production rate) which influences the choice of photobioreactors, separation and labor costs (Acién et al., 2012; Awasthi et al., 2020; Stichnothe et al., 2016). Maximizing productivity can lead to lower downstream costs, and one tool that has been proven to be successful in rerouting carbon in metabolism is metabolic engineering, specifically the use of metabolic models, to predict and implement genetic changes that can improve overall productivity (Hu et al., 2023) and product specific productivity (Yan et al., 2019; Song et al., 2020). For example, metabolic models have been used to guide the overexpression of acetyl-CoA carboxylase to increase lipid accumulation for biodiesel production (Yan et al., 2019) and redirect carbon flux toward carotenoid biosynthesis by optimizing the isoprenoid pathway (Song et al., 2020). These efforts demonstrate how metabolic models can enable precise identification of limiting steps in target pathways and support design strategies to improve yields of economically valuable compounds.
Computational tools provide powerful means to investigate the complexities of metabolism. Among the computational methods employed in metabolic engineering, genome-scale metabolic models (GEMs) stand out due to their relative ease of implementation and comprehensive, systems level approach. GEMs are in silico representations of an organism’s metabolic capacity based on the organism’s sequenced genome, enumerating all reactions and metabolites encoded within. Experimental data, such as carbon uptake and excretion, biomass composition and growth rate can be used to constrain the model (Bernstein et al., 2021). Dramatically decreasing costs for high quality genome sequencing has led to increased sequence data for GEM reconstruction (Pareek et al., 2011), and advances in genome annotation have enabled more complete simulations of metabolic processes. GEMs can be used to identify gene knockouts that lead to increased yield or productivity. They can also be used to predict changes in yield due to the incorporation of heterologous metabolic pathways, narrowing the potential mutants to be screened in the lab and drastically decreasing research and development investment (Mekanik et al., 2023). By representing the entire metabolic capacity of an organism, GEMs have also been used to identify genetic targets that are not easy to predict a priori as having an impact on the productivity of a specific product (Levering et al., 2016; Yang et al., 2018). The utilization of GEMs is not limited to screening genetic changes, GEMs can additionally be applied to understand how an organism will respond to environmental changes. These applications include media optimization and predictions on the most crucial nutrients for growth (Van Tol and Armbrust, 2021). GEMs also can be utilized to rapidly provide predictions on the changes that varying growth conditions will have phenotypically (Zuniga et al., 2016). In silico studies provide an effective method to aid in target selection for traditional experiments, enabling researchers to investigate the impact of thousands of genetic or environmental changes in a fraction of the time it takes to create and characterize in the lab (Ofaim et al., 2021; Nocon et al., 2014).
GEMs have been extensively employed to study metabolism across a wide range of organisms, with the majority of existing literature and models focused on heterotrophic systems such as bacteria and yeast (Orth et al., 2011; Monk et al., 2017; Lu et al., 2019). This emphasis is reflected in the greater availability of heterotrophic GEMs on public repositories such as BioModels (Malik-Sheriff et al., 2020) and BiGG Models (Norsigian et al., 2020). Although some algal GEMs are hosted on these platforms, the listings are not comprehensive and often require manual literature searches to identify additional models. Nonetheless, GEMs in both heterotrophic and autotrophic organisms have proven highly effective for simulating metabolic fluxes, identifying genetic engineering targets and optimizing growth conditions.
Applying GEMs to photoautotrophic organisms, particularly eukaryotic microalgae, presents a distinct set of challenges. These include the need to simulate light-dependent metabolism, diel cycling, and shifting cellular objectives across changing environmental conditions, all within a framework that traditionally assumes steady-state behavior. In this review, we examine the specific difficulties encountered when constructing and utilizing GEMs for photoautotrophic microalgae, as well as the current limitations that hinder their broader adoption and predictive accuracy. A dedicated section explores the role of Chlamydomonas reinhardtii, which has emerged as a cornerstone species in algal systems biology and a model for developing and refining GEMs in microalgae. Finally, we highlight future directions in GEM research, including the integration of dynamic modeling, multi-omics data, and machine learning techniques, all of which are poised to significantly advance the utility of GEMs in both fundamental research and applied biotechnology.
Chlamydomonas reinhardtii: a keystone species for microalga GEM reconstruction
Chlamydomonas reinhardtii has received extensive attention in scientific research (Harris, 2001), emerging as a pivotal organism for studying microalgae and aquatic photosynthetic systems (Calatrava et al., 2023). As a model green microalga, C. reinhardtii has served as the foundation for GEMs in algal species. The first GEM for C. reinhardtii was developed by Boyle and Morgan in 2009 (Boyle and Morgan, 2009), marking the first GEM constructed for any algal species (see Figure 1). Another noteworthy GEM is iCre1355 (Imam et al., 2015), which has served as a foundational platform for subsequent models. Derived like many of the currently available GEMs from the earlier iRC1080 (Chang et al., 2011), iCre1355 (Imam et al., 2015) incorporates updates based on improvements made to the annotation of the genome, rectifying inaccuracies in gene-protein reaction associations. This improved model has been utilized to predict growth under varying light conditions (Shene et al., 2018). iCre1355 (Imam et al., 2015) was also utilized in the development of the first diurnal metabolic model in microalgae developed by Metcalf and Boyle (Metcalf Alex and Boyle Nanette, 2022). This diurnal model is a type of transient metabolic model (TMM). TMMs are computational models that capture dynamic changes in metabolism under varying environmental conditions. The Metcalf and Boyle TMM incorporated quantitative, time dependent transcriptomic data to constrain the availability of the associated gene products and metabolic reactions and more accurately predict growth in diurnal conditions.

Figure 1. Historical perspective on the generation of algal GEMs, organized by species and year of publication.
The GEM developed by Yao et al. (2023), merged the iCre1355 and iGR774 models, replacing the chloroplast reactions in iCre1355 (Imam et al., 2015) with the more detailed iGR774 (Bjerkelund Rokke et al., 2020) chloroplast specific model. This integration allowed for a more biologically accurate depiction of chloroplast metabolism, improving compartmental resolution, gene-reaction mapping, and the model’s ability to simulate light-driven and plastid-localized processes. Yao et al. additionally utilized protein constrained flux balance analysis (PC-FBA), an extension of traditional FBA that integrates enzyme capacity and proteome allocation to better reflect cellular limitations. This approach allows for context-specific flux predictions informed by transcriptomic data and represents the first implementation of a protein-constrained model (PC-Model) for a microalgal GEM.
More recently, Arend et al (Arend et al., 2023). continued this advancement by directly integrating quantitative proteomic data to constrain enzyme usage, offering a more accurate representation of in vivo metabolic states. This proteomics-driven approach narrows the solution space of the model, leading to improved predictions of enzyme allocation and flux distributions. With these advancements, C. reinhardtii’s GEMs continue to be at the forefront of advancing algal biotechnology, significantly contributing to the understanding of microalgal metabolism and algal GEM reconstructions.
Challenges and limitations of algal genome-scale metabolic models
GEMs are a powerful and rapidly advancing tool for understanding cellular metabolism, however, like any complex modeling approach, there are challenges that researchers continue to address to unlock their full potential. One challenge across all organisms but particularly in non-model species is inaccurate or incomplete genome annotations, which leads to gaps that need to be manually filled. This issue is particularly pronounced in photoautotrophic organisms such as microalge, as fewer well-annotated reference genomes are available for comparison. C. reinhardtii largely represents an exception as its genome has undergone extensive sequencing and curation (Craig et al., 2023), as well as support by databases such as Phytozome (Goodstein et al., 2012), ChlamyCyc (May et al., 2009) and AlgePath (Zheng et al., 2014). Not all organisms have this extensive research with many inaccuracies arising from the need for homology-based annotations, which while faster than manual curation, can assign functions without biochemical validation. Based on how automated annotation algorithms work, poor annotations can be carried through to new organisms. An additional challenge with annotation is that many metabolic pathways and reactions, particularly in non-model organisms, are still being discovered or refined, which can create gaps in the models that require extensive manual curation or assumptions to fill (Karp et al., 2018). Beyond annotation issues, GEMs also face limitations due to their reliance on stoichiometric reactions rather than reaction kinetics. By ignoring reaction kinetics, the entire metabolic network can be modeled; but it comes at a cost because the level of detail is greatly reduced. Kinetic models have been developed for well-studied organisms such as Escherichia coli (Khodayari et al., 2014), but they include far fewer reactions than GEMs due to the requirement for detailed kinetic data. For microalgae, such data is especially scarce, making GEMs the most practical framework for modeling their metabolism. To enhance their accuracy, GEMs can integrate omics data such as transcriptomics and proteomics. This data provides crucial insights into cellular states and responses. However, aligning diverse omics datasets with GEMs is another challenge, requiring sophisticated computational techniques. Fortunately, advancements in data integration and computational methods are allowing GEMs to incorporate omics data more effectively and enhance their predictive power (Sen and Orešič, 2023). However, even with these advancements in annotation and omics integration, GEMs still face limitations due to key assumptions most notably the reliance on steady state conditions that pose unique challenges in photosynthetic organisms.
Adopting a steady-state assumption poses significant challenges for GEMs in photosynthetic microalgae, where complex diel fluctuations and regulatory mechanisms make strict steady-state models less representative of metabolic dynamics. While this assumption is important mathematically, converting a set of ordinary differential equations to a set of linear equations, it limits the application of GEMs to steady growth conditions. This is particularly pronounced in photosynthetic organisms due to typical growth in diel light conditions which results in substantial fluctuations in metabolism (Fisher et al., 2023) due to the shift from day to night and vice versa. Photosynthesis also involves numerous regulatory mechanisms, such as photoprotection (Goss and Jakob, 2010), photosynthetic quenching (Schubert et al., 2006), and variations in photon flux (Schnurr et al., 2016), all of which are difficult to represent with static models. To account for regulatory elements such as enzyme capacity constraints and gene expression control, the integration of proteomic and transcriptomic data into GEMs is essential. Transcriptomics can be used to infer active pathways by adjusting reaction constraints based on gene expression levels, while proteomics enables more accurate estimation of enzyme abundances and capacities to constrain the solution space of the model. However, such genome-wide data sets remain scarce for most microalgae due to limited experimental and financial investment. Chlamydomonas reinhardtii stands out in this regard, as it benefits from available transcriptomic and proteomic data. An extension of this problem is the use of a single objective function (most often to maximize biomass). While this objective function matches the cellular objective for heterotrophic bacteria quite well (Orth et al., 2011), this objective is especially problematic in photosynthetic organisms due to the decoupling of carbon and energy inputs and the time-dependent nature of cellular division in diel light. Additionally, the biomass function for algae is more complex and dynamic than those seen in heterotrophic organisms, as many can grow in autotrophic, mixotrophic, and heterotrophic states. Each of these trophic states requires a distinct biomass formulation to reflect the underlying physiological differences (Matos et al., 2017). Moreover, algal cells must continuously optimize their metabolism in response to environmental conditions. These can vary such as minimizing energy usage when light is not present or the formation of storage products in preparation for environmental changes. Additionally autotrophic and mixotrophic growth results in biomass composition are more dependent on the environment, changing with light intensity throughout the day under diel conditions (Jallet et al., 2016).
These challenges have motivated the development of more sophisticated GEMs that better capture the complexity of photosynthetic microalgae. Recent models have begun to incorporate multiple objective functions, simulate compartmentalized metabolism and account for trophic flexibility. Other innovations address environmental responsiveness, such as stress adaptation and diel regulation. The following sections highlight these advancements through examples of automated reconstruction tools, light modeling, omics integration, and dynamic modeling (see Figure 2).
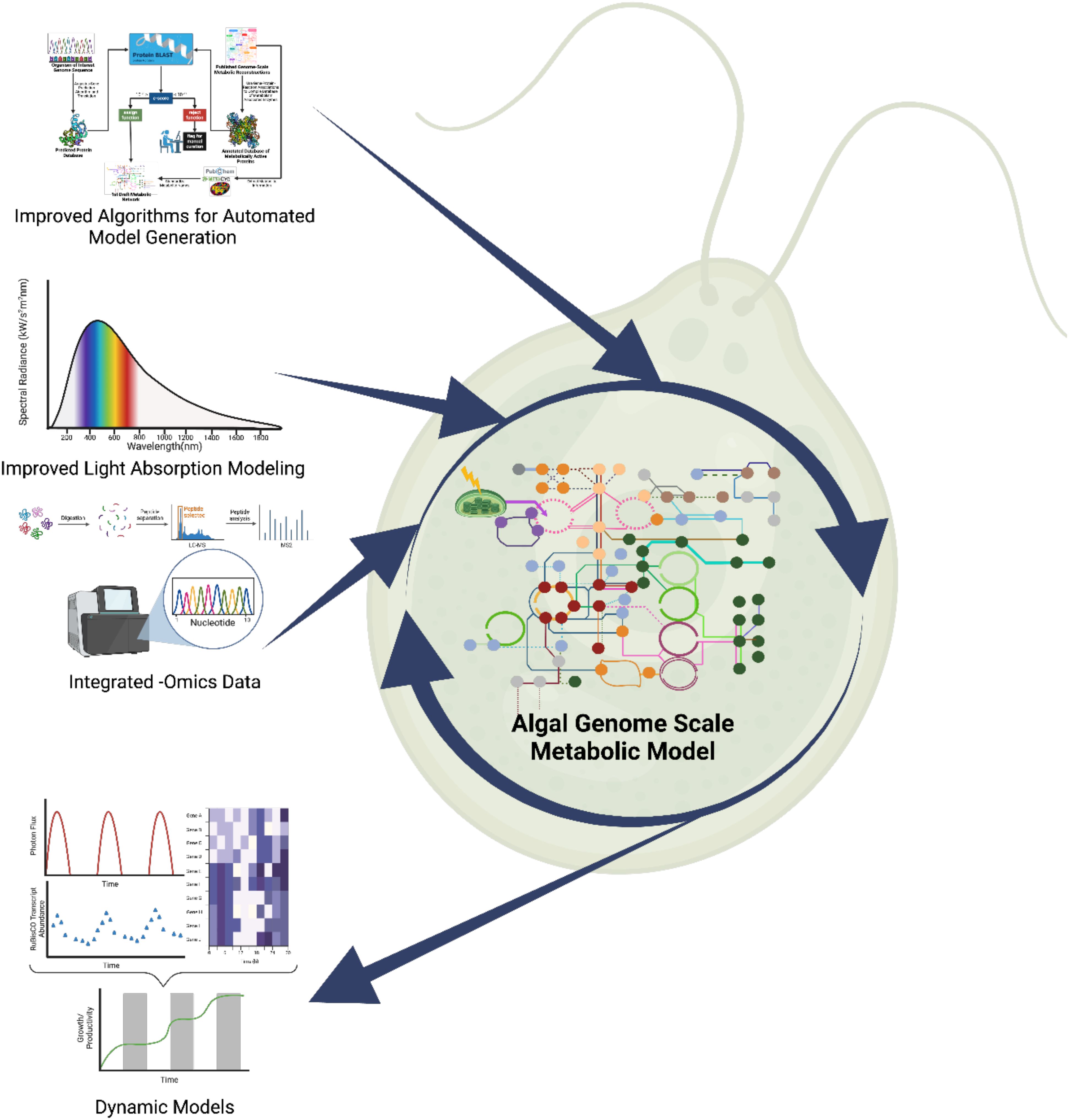
Figure 2. Challenges for algal GEM reconstruction include the need for automated algorithms for model reconstruction specifically for algae, improved light modeling and integrating -omics data to improve predictability. Addressing these will enable the design of dynamic models that can better predict growth in dynamic conditions, such as day/night cycles.
Automation of model reconstruction
Automated reconstruction of GEMs helps address the time-intensive nature of model development by streamlining the reconstruction process, making it feasible to generate high-quality models for a wider range of organisms. The GEM iChr1915 (Meagher et al., 2024) for Chromochloris zofingiensis represent significant advancement in the automatic curation of photosynthetic metabolic networks. iChr1915 (Meagher et al., 2024) utilized an algorithm called Rapid Annotation of Photosynthetic Systems (RAPS) (Metcalf et al., 2020) to automate much of the process. Other GEM automation tools exist such as model SEED (Devoid et al., 2013) and CarveMe (MaChado et al., 2018), however these automation tools are not tailored for use on algae. The model SEED (Devoid et al., 2013) framework plantSEED (Seaver et al., 2014) is, as its name would suggest, better suited for the reconstruction of plant GEMs as it carries over many highly conserved reactions in plants to avoid issues with gap filling. Including these conserved reactions in algal GEM reconstructions doesn’t properly represent the diversity of microalgal metabolism (Catalanotti et al., 2013) and variation from plant metabolism (Tamoi and Shigeoka, 2015). CarveMe (MaChado et al., 2018) additionally is primarily for the reconstruction of prokaryotes and bacterial communities with reactions pulled from the BiGG database (Schellenberger et al., 2010) excluding reactions unique to eukaryotic organisms. The use of RAPS (Metcalf et al., 2020) enabled the development of a high quality first draft network in only 20 minutes; the resulting model only required minimal manual curation. RAPS (Metcalf et al., 2020) facilitates the automated curation of GEMs for photosynthetic algae by leveraging manual curation efforts already invested in published models and using these to generate new models.
Another automation tool that has been utilized in GEM reconstruction is RAVEN toolbox (Agren et al., 2013), which was utilized in the reconstruction of iLB1027_lipid and iLB1025 (Levering et al., 2016). The original RAVEN toolbox provided a MATLAB-based framework to facilitate semi-automated draft reconstruction of metabolic networks through homology-based mapping from annotated genomes to template models. In this case, RAVEN was used to generate an initial draft network by identifying homologous genes based on previously published models from photosynthetic organisms. This draft network served as the foundation, which was further refined using updated genome annotations, subcellular localization predictions, and biochemical validation. Although significant manual effort was required to correct compartmentalization, balance reactions, and incorporate complex eukaryotic features, the automated steps provided by RAVEN accelerated the initial reconstruction process and ensured alignment with known gene–reaction relationships. A newer version of the RAVEN toolbox, RAVEN 2.0 (Wang et al., 2018), has since been developed with expanded capabilities, including integration of MetaCyc-based reconstruction (Caspi et al., 2020) and improved model interoperability.
These method addresses a key challenge in GEM development: the time-consuming nature of manual curation and annotation gaps. By using RAPS and RAVEN, researchers can streamline the initial stages of model development, allowing them to focus on gap-filling and other manual curation efforts that will lead to a high-quality network. This hybrid approach reduces the time-intensive nature of fully manual curation by automating the initial draft creation and filling metabolic gaps while still incorporating the precision of expert intervention where needed.
Modeling light harvesting
Because microalgae are photosynthetic organisms incorporating light dynamics such as wavelength, intensity and spectral composition into GEMs is crucial for accurately capturing their metabolism and improving model predictions. The first GEM for microalgae to account for different wavelengths of photons in its metabolic network was iRC1080 (Chang et al., 2011), a model for C. reinhardtii allowing for variations in light conditions to influence the model. iRC1080 (Chang et al., 2011) achieved this by defining spectral ranges associated with all the photon-utilizing reactions in the network connecting and allowing for 11 distinct light sources such as solar light as well as halogen and LED lights to be modeled. The metabolic network was also verified with over 90% of transcripts predicted by iRC1080 (Chang et al., 2011) being found in experimental transcriptomic data. Additionally, iRC1080 (Chang et al., 2011) accurately predicted solar conversion efficiency to be 2%, matching experimental results. The coupling of light wavelengths with reactions marked a substantial improvement on previous models and allows for the optimization of light sources as well as elucidating the phenotypic results of varying light conditions. Similarly, the Chlorella variabilis model iAJ526 (Juneja et al., 2016) accounts for varying light conditions by simulating the effects of twelve different light sources on growth rate and uptake rates. These light sources were like those modeled in iRC1080 (Chang et al., 2011) representing light sources that have been utilized in algal growth, but had a greater focus on modeling different combinations of LED light and didn’t include sunlight. Three of these light conditions were experimentally validated, confirming predictions made by the model that white light would provide the best growth followed by red/blue light then red light. iAJ526 (Juneja et al., 2016) predicts higher growth rates than those observed experimentally under all light conditions with the authors attributing the differences to issues with the model’s lack of growth kinetics and photoinhibition. These models advance GEM reconstruction in algae and other photosynthetic organisms by offering a more robust representation of the effects light intensity and composition have on metabolism.
Innovations in GEMs for microalgae have also addressed other limitations traditionally seen in GEMs, particularly those affecting photosynthetic organisms. For instance, the Thalassiosira pseudonana model iTps1432 (Van Tol and Armbrust, 2021) incorporates the application of photon loss reactions to simulate photosynthetic quenching. By including these reactions, iTps1432 (van Tol and Armbrust, 2021) offers valuable insight into photon loss reactions with particular interest coming from predictions around cyclic electron flow at low light intensities. At these lower light intensities, the model predicts that a significant portion of total electron flow is made up of cyclic electron flow supporting other findings highlighted in the paper that cyclic electron flow is important for ATP generation at low light (Bailleul et al., 2015). Cyclic electron flow is not only important for ATP generation and modeling light dynamics but has also been demonstrated to be important in lipid biosynthesis pathways in algae (Chen et al., 2015). This highlights the potential improvements adding light dynamics reaction within GEMs can provide. With this added insight these models can be better applied to determine targets for improving metabolic engineering outcomes under autotrophic conditions.
Models with a focus on reactions outside of carbon metabolism
GEMs can be applied to explore algal production of value-added compounds beyond traditional targets like biomass and hydrocarbons. One such emerging application is the modeling of green hydrogen production, which has gained significant interest in recent years (Borges et al., 2024). Models such as iMM627 (Mekanik et al., 2019) for Auxenochlorella protothecoides and iRJ1321 (Shah et al., 2017) for Nannochloropsis gaditana incorporate predictions of hydrogen production. The iMM627 (Mekanik et al., 2019) model integrates two objective functions maximizing both biomass and hydrogen production. By incorporating multiple objectives, the model can more completely utilize the GEMs metabolic network and better represent reactions outside of central carbon metabolism. Additionally, although not originally designed for hydrogen production, the AlgaGEM model (Gomes De Oliveira Dal’molin et al., 2011) for Chlamydomonas reinhardtii was used to maximize hydrogen synthesis through modification of its objective function, demonstrating that any genome-scale metabolic model can, in principle, be adapted to study hydrogen production or any other product based on the set objective function.
Beyond expanding product scope, recent GEMs have also improved pathway resolution for key metabolic processes such as nitrogen metabolism. The Nannochloropsis salina model iNS934 (Loira et al., 2017) provides a more detailed representation of nitrogen metabolism, capturing the intricate balance between carbon fixation and nitrogen assimilation, while also incorporating a variety of nitrogen sources. This allows iNS934 (Loira et al., 2017) to integrate essential reactions not directly tied to carbon metabolic pathways, addressing gaps present in earlier models and offering more flexibility when optimizing media recipes. Such refinements enhance the model’s utility for strain engineering under nutrient-limited conditions and support the development of cost-effective cultivation strategies.
Model robustness
Enhancing the robustness of GEMs improves their ability to simulate organismal responses to environmental stress and genetic perturbations, making them more reliable tools for predictive modeling and metabolic engineering. The Lavoie et al. Fragilariopsis cylindrus model (Lavoie et al., 2020) focuses on reaction robustness to help analyze how metabolic networks maintain stability under stress or environmental shifts. This robustness analysis, combines flux balance analysis (FBA) with minimization of metabolic adjustment (MOMA) (Segrè et al., 2002) allows for better prediction of how networks respond to perturbations made by knock outs. In contrast to flux variability analysis (FVA) (Gudmundsson and Thiele, 2010), which assesses the flexibility of individual reactions by calculating the range of fluxes consistent with optimal growth, MOMA evaluates robustness based on the assumption that, following a perturbation, the network minimizes its deviation from the wild-type flux distribution without immediately reoptimizing for a new objective. This makes MOMA particularly useful for modeling short-term or acute responses, when the organism has not yet had time to adapt through regulation or evolution. This improves the GEM’s ability to simulate stress responses, addressing a significant aspect of how F. cylindrus survives well in its very dynamic environment (Yoshida et al., 2020).
The Recht et al. model (Recht et al., 2014) for Haematococcus pluvialis further incorporates variability flux sampling (VFS), an additional step on the commonly used FVA. VFS enables more accurate flux predictions and a deeper analysis of metabolic pathways as it not only predicts the range of possible fluxes, as is done in FVA, but also includes determinations about the probabilities of various fluxes. Incorporating VFS allows for better understanding of pathways that are activated as a stress response as demonstrated in the models focus on exploring the shift toward fatty acid synthesis under nitrogen starvation. Variability Flux Sampling (VFS) enhances interpretation of flux flexibility by generating probability distributions of feasible flux values through random sampling and constrained optimization, rather than assuming a single optimal flux solution. This allows models to reflect the range and likelihood of alternative flux states under given physiological constraints. While not inherently dynamic, the application of VFS across time-resolved datasets such as in H. pluvialis under nitrogen deprivation captures experimentally observed shifts in metabolism, including the transition from carbohydrate accumulation to fatty acid biosynthesis (Recht et al., 2012), underscoring the need for models that can represent metabolic plasticity under stress. Although demonstrated here in the context of specific GEMs, VFS and MOMA are generalizable approach that can be applied to any GEM to enhance the characterization of condition-dependent metabolic states.
Integration of additional omics data and dynamic modeling
Integrating omics data into GEMs enhances their predictive power by capturing regulatory and physiological constraints that are not represented by purely stoichiometrically models. The Yao et al. model (Yao et al., 2023) for C. reinhardtii does this by incorporating RNA sequencing data to assume the proteome of the organism as well as enzyme data to create a protein-constrained metabolic model (PC-model). This allows for the model to better represent the dynamics that are lost in the conventional approach of representing metabolism only stoichiometrically. However, while transcriptomics provides useful insights into gene expression, it does not fully reflect metabolic activity due to regulatory layers such as translation, protein turnover, and post-translational modifications. The model by Arend et al (Arend et al., 2023). published shortly after the Yao et al. model advances this framework by directly incorporating quantitative proteomic measurements. The data collected was used to calculate in vivo apparent turnover numbers (kapp) for 568 reactions, providing a more accurate basis for constraining enzyme usage within the model. Of the 1460 enzymes, 936 (64%) were quantified in at least one experimental condition, representing the most extensive proteome coverage achieved for C. reinhardtii to date. This allowed the model to more accurately constrain enzyme usage by grounding flux predictions in measured protein abundances, thereby significantly reducing the solution space and increasing the physiological relevance of the predicted flux distributions. By aligning enzyme usage with what is actually present in the cell, the model more faithfully captures metabolic capabilities. Models that incorporate omics data have great potential in better representing the complex regulatory mechanisms present around metabolism (Carthew, 2021) as well as applications under varying growth conditions (Gim et al., 2016).
Another model, iEH410 (Knies et al., 2015) for Emiliania huxleyi, introduces diurnal FBA (diuFBA), significantly improving the simulation of internal regulation of metabolic reactions by moving beyond static flux distributions and better reflecting real-time cellular responses. diuFBA simulates the organism’s metabolism under alternating light and dark conditions. This approach partitions a 24-hour diurnal cycle into discrete light and dark phases, assuming quasi-steady-state conditions within each phase. Another important feature of this model is that it allows for dynamic optimization of storage metabolites, such as mannitol and lipids, rather than relying on fixed concentrations set by the biomass function, as is standard. To achieve this dynamic optimization, diuFBA extends the stoichiometric matrix to include duplicated networks for the light and dark periods, which are connected through reversible transfer reactions for storage metabolites. The model integrates fluxes over each phase duration using explicit Euler integration, enabling the calculation of net concentration changes across the full cycle. This formulation preserves the structure of classical FBA, allowing for efficient convex optimization while capturing the temporal redistribution of metabolic resources that occurs in response to circadian environmental changes. By solving for metabolite accumulation across light and dark periods within a single optimization problem, diuFBA offers a more biologically relevant representation of photosynthetic metabolism without the computational complexity of fully dynamic simulations. However, while this approach captures resource allocation across day-night transitions, it still assumes steady-state behavior within each phase and cannot represent short-term metabolic fluctuations. This limitation has motivated the development of transient metabolic models (TMMs), which aim to simulate cellular metabolism at finer temporal resolution under continuously changing environmental conditions.
TMMs offers a promising avenue for future research utilizing the value of GEMs while offering a dynamic model. While dynamic models have been developed for heterotrophic organisms such as E. coli (Yang et al., 2019) and photosynthetic species like Synechocystis sp (Rügen et al., 2015), similar models have been largely absent in microalgae. The first TMM for microalgae was developed by Metcalf and Boyle (Metcalf Alex and Boyle Nanette, 2022) in C. reinhardtii to model growth in diel light. The model was based on experimental transcriptomics data based on growth in 12:12 hour day:night cycles as this data was used to constrain the availability of the associated enzymatic reactions based on gene expression data. Additionally, the TMM also decoupled the biomass objective functions from the standard static biomass equation allowing it to better simulate the cells adapting to the changing environmental conditions over a day. This is a substantial improvement on GEMs, addressing one of their key challenges: that they are generally static stoichiometric representations of metabolism. The dynamics of the TMM also allow for better targeting for metabolic engineering as these models better represents the fluctuations in metabolism over the course of a day rather than at a single point in the day. Despite these advantages, the implementation of TMMs depends on high-resolution, time-series transcriptomic data, the generation and integration of which are both labor-intensive and expensive. However, the ability to simulate time-resolved shifts in gene expression and metabolism makes this investment particularly valuable, especially for photosynthetic organisms where diel dynamics are fundamental to metabolic function.
Prospects for future microalgal genome-scale metabolic models
Future advances in GEM formulation will enable more sophisticated models that will be better suited to predicting the dynamic and complex metabolism of microalgae. While GEMs have traditionally relied on steady-state assumptions using FBA, incorporating regulatory constraints has successfully been demonstrated in the Yao et al. model and iEH410. While both these GEMs incorporated transcriptomics data, there are further advancements that can be made to the reconstruction of future GEMs incorporating multi-omics data. Tools such as GECKO 2.0 (Domenzain et al., 2022) allow for pipelines for the implementation of enzyme kinetic parameters and proteomic data into GEMs which has already been utilized in multiple species of yeast, E. coli and Homo sapiens. By adding additional layers of omics data GEMs can address limitations that are presented in many of the currently available static stoichiometric models.
Another emerging direction for algal GEMs is the application of microbial community models (MCMs), which have garnered considerable interest in recent years (Tarzi et al., 2024). MCMs capture the complex inter-specific interactions that microalgae experience in both natural and engineered environments. Rather than existing in isolation, algae typically coexist with diverse microbial partners that influence their metabolism through nutrient exchange, competition, and metabolic cross-feeding. To simulate these interactions, community-scale modeling tools such as SteadyCom (Chan et al., 2017), MICOM (Diener et al., 2020), and the Microbiome Modeling Toolbox (Heinken and Thiele, 2022) enable constraint-based simulations that consider the growth and resource allocation strategies of multiple interacting species. Building on this, dynamic models including dOptCom (Zomorrodi et al., 2014) and COMETS (Harcombe et al., 2014) incorporate spatial and temporal variation, making them especially suited for studying nutrient shifts and microbial succession. Incorporating algal GEMs into these MCM frameworks could improve predictive accuracy under realistic conditions, uncover emergent properties such as division of labor and metabolite sharing, and support the design of more productive algal–bacterial consortia for biotechnology applications. While data availability remains a constraint for many microalgal species, machine learning offers exciting opportunities. In particular, deep learning, which uses neural networks to perform multi-level predictions (Lecun et al., 2015), has already improved genome annotations in bacterial metagenomes (Boer et al., 2024). Applying similar approaches to microalgae could enable the reconstruction of GEMs for the vast number of algal species that remain unculturable (Sharma and Rai, 2010). This could not only improve our understanding of these organisms but also help design more effective cultivation strategies.
Another promising avenue is the integration of GEMs with Transient Metabolic Models (TMMs), which simulate metabolic changes over time and under varying environmental conditions. While a TMM has been developed for Chlamydomonas reinhardtii (Metcalf Alex and Boyle Nanette, 2022), other microalgae including those with GEMs (see Table 2) currently lack such dynamic models. Expanding TMMs to include additional species and conditions such as UV radiation, temperature fluctuations, and nutrient availability could dramatically enhance the applicability of GEMs in modeling real-world scenarios (El-Sheekh et al., 2021; Al Jabri et al., 2021; Ikaran et al., 2015).
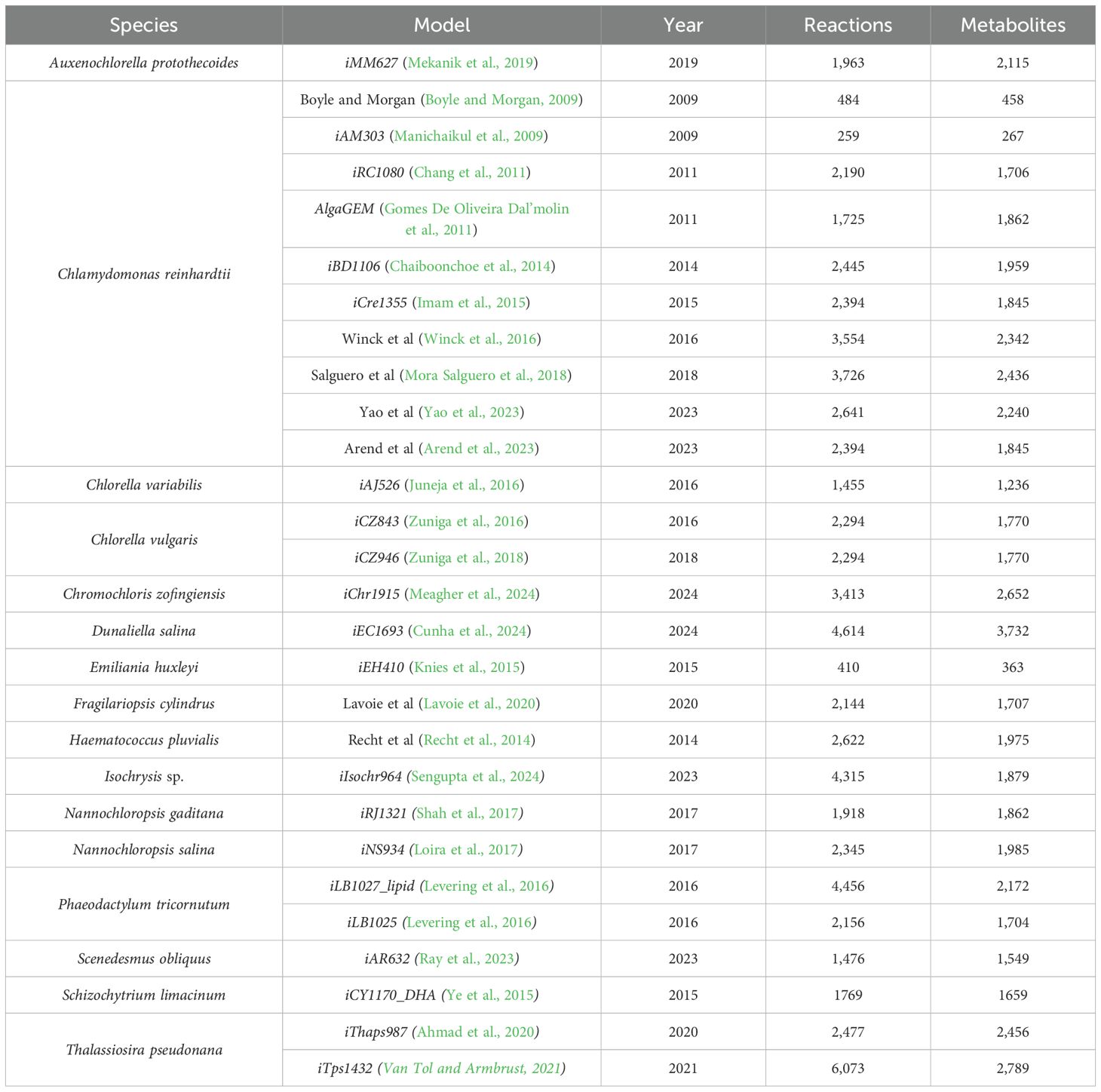
Table 2. The table displays the GEMs currently published, organized by species and year of publication from oldest to newest.
Altogether, these innovations including multi-omics integration, machine learning, and dynamic modeling represent the future of microalgal GEMs. They offer a more comprehensive understanding of algal metabolism, particularly under diel cycles and photosynthetic fluctuations, moving the field closer to realizing the full potential of microalgae in biotechnology and sustainability applications.
Conclusion
Microalgae hold immense potential for contributing to a sustainable future through their applications in biofuels, bioremediation, and the production of high-value products. The development of GEMs has emerged as a powerful tool in understanding the complex metabolic networks of these organisms, enabling researchers to optimize their metabolic pathways effectively. However, while GEMs have made significant strides, they are not without limitations. Issues related to incomplete genome annotations, static assumptions, and the integration of multi-omics data continue to pose challenges for GEMs to more accurately simulate metabolism. To address these limitations and fully harness the capabilities of microalgae, there is a pressing need for the creation of more GEMs across a diverse array of algal species. Expanding the repertoire of GEMs will enhance our understanding of algal metabolism and facilitate the development of tailored strategies for metabolic engineering. By addressing the existing challenges and improving GEM methodologies, we can pave the way for a more environmentally friendly future, ultimately contributing to a more sustainable and productive bioproduct landscape.
Author contributions
JT: Writing – review & editing, Writing – original draft, Conceptualization, Visualization, Formal analysis. NB: Visualization, Conceptualization, Resources, Writing – review & editing, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. JT and NB were both supported by DOE Office of Science, Office of Biological and Environmental Research (BER), grant no. DE-SC0023027.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu-Ghosh, S., Dubinsky, Z., Verdelho, V., and Iluz, D. (2021). Unconventional high-value products from microalgae: A review. Bioresour Technol. 329, 124895. doi: 10.1016/j.biortech.2021.124895
Acién, F. G., Fernández, J. M., Magán, J. J., and Molina, E. (2012). Production cost of a real microalgae production plant and strategies to reduce it. Biotechnol. Adv. 30, 1344–1353. doi: 10.1016/j.biotechadv.2012.02.005
Agren, R., Liu, L., Shoaie, S., Vongsangnak, W., Nookaew, I., and Nielsen, J. (2013). The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PloS Comput. Biol. 9, e1002980. doi: 10.1371/journal.pcbi.1002980
Ahmad, A., Tiwari, A., and Srivastava, S. (2020). A genome-scale metabolic model of thalassiosira pseudonana CCMP 1335 for a systems-level understanding of its metabolism and biotechnological potential. Microorganisms 8, 1396. doi: 10.3390/microorganisms8091396
Ahmed, S. F., Mofijur, M., Parisa, T. A., Islam, N., Kusumo, F., Inayat, A., et al. (2022). Progress and challenges of contaminate removal from wastewater using microalgae biomass. Chemosphere 286, 131656. doi: 10.1016/j.chemosphere.2021.131656
Al Jabri, H., Taleb, A., Touchard, R., Saadaoui, I., Goetz, V., and Pruvost, J. (2021). Cultivating microalgae in desert conditions: evaluation of the effect of light-temperature summer conditions on the growth and metabolism of nannochloropsis QU130. Appl. Sci. 11, 3799. doi: 10.3390/app11093799
Alkhamis, Y. and Qin, J. G. (2013). Cultivation of isochrysis galbana in phototrophic, heterotrophic, and mixotrophic conditions. BioMed. Res. Int. 2013, 983465. doi: 10.1155/2013/983465
Álvarez-Díaz, P. D., Ruiz, J., Arbib, Z., Barragán, J., Garrido-Pérez, M., and Perales, J. (2015). Wastewater treatment and biodiesel production by Scenedesmus obliquus in a two-stage cultivation process. Bioresource Technol. 181, 90–96. doi: 10.1016/j.biortech.2015.01.018
Arend, M., Zimmer, D., Xu, R., Sommer, F., Mühlhaus, T., and Nikoloski, Z. (2023). Proteomics and constraint-based modelling reveal enzyme kinetic properties of Chlamydomonas reinhardtii on a genome scale. Nat. Commun. 14, 4781. doi: 10.1038/s41467-023-40498-1
Armbrust, E. V., Berges, J. A., Bowler, C., Green, B. R., Martinez, D., Putnam, N. H., et al. (2004). The genome of the diatom thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. doi: 10.1126/science.1101156
Aveiro, S. S., Melo, T., Figueiredo, A., Domingues, P., Pereira, H., Maia, I. B., et al. (2020). The polar lipidome of cultured emiliania huxleyi: A source of bioactive lipids with relevance for biotechnological applications. Biomolecules 10, 1434. doi: 10.3390/biom10101434
Awasthi, M. K., Sarsaiya, S., Patel, A., Juneja, A., Singh, R. P., Yan, B., et al. (2020). Refining biomass residues for sustainable energy and bio-products: An assessment of technology, its importance, and strategic applications in circular bio-economy. Renewable Sustain. Energy Rev. 127, 109876. doi: 10.1016/j.rser.2020.109876
Bailleul, B., Berne, N., Murik, O., Petroutsos, D., Prihoda, J., Tanaka, A., et al. (2015). Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. doi: 10.1038/nature14599
Barrera, D. J., Rosenberg, J. N., Chiu, J. G., Chang, Y.-N., Debatis, M., Ngoi, S.-M., et al. (2015). Algal chloroplast produced camelid VHH antitoxins are capable of neutralizing botulinum neurotoxin. Plant Biotechnol. J. 13, 117–124. doi: 10.1111/pbi.2014.13.issue-1
Bayer-Giraldi, M., Uhlig, C., John, U., Mock, T., and Valentin, K. (2010). Antifreeze proteins in polar sea ice diatoms: diversity and gene expression in the genus Fragilariopsis. Environ. Microbiol. 12, 1041–1052. doi: 10.1111/j.1462-2920.2009.02149.x
Bernstein, D. B., Sulheim, S., Almaas, E., and Segrè, D. (2021). Addressing uncertainty in genome-scale metabolic model reconstruction and analysis. Genome Biol. 22, 64. doi: 10.1186/s13059-021-02289-z
Bi, Z., He, B. B., and Mcdonald, A. G. (2015). Biodiesel Production from Green Microalgae Schizochytrium limacinum via in Situ Transesterification. Energy Fuels 29, 5018–5027. doi: 10.1021/acs.energyfuels.5b00559
Bito, T., Okumura, E., Fujishima, M., and Watanabe, F. (2020). Potential of chlorella as a dietary supplement to promote human health. Nutrients 12, 2524. doi: 10.3390/nu12092524
Bjerkelund Rokke, G., Hohmann-Marriott, M. F., and Almaas, E. (2020). An adjustable algal chloroplast plug-and-play model for genome-scale metabolic models. PloS One 15, e0229408. doi: 10.1371/journal.pone.0229408
Boer, M. D., Melkonian, C., Zafeiropoulos, H., Haas, A. F., Garza, D. R., and Dutilh, B. E. (2024). Improving genome-scale metabolic models of incomplete genomes with deep learning. iScience 27, 111349. doi: 10.1016/j.isci.2024.111349
Borges, P. T., Sales, M. B., César Guimarães, C. E., De França Serpa, J., De Lima, R. K. C., Sanders Lopes, A. A., et al. (2024). Photosynthetic green hydrogen: Advances, challenges, opportunities, and prospects. Int. J. Hydrogen Energy 49, 433–458. doi: 10.1016/j.ijhydene.2023.09.075
Bošnjaković, M. and Sinaga, N. (2020). The perspective of large-scale production of algae biodiesel. Appl. Sci. 10, 8181. doi: 10.3390/app10228181
Bouras, S., Katsoulas, N., Antoniadis, D., and Karapanagiotidis, I. T. (2020). Use of biofuel industry wastes as alternative nutrient sources for DHA-yielding schizochytrium limacinum production. Appl. Sci. 10, 4398. doi: 10.3390/app10124398
Boyle, N. R. and Morgan, J. A. (2009). Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst. Biol. 3, 4. doi: 10.1186/1752-0509-3-4
Brown, J. S. (1987). Functional organization of chlorophyll a and carotenoids in the alga, Nannochloropsis salina. Plant Physiol. 83, 434–437. doi: 10.1104/pp.83.2.434
Butler, T., Kapoore, R. V., and Vaidyanathan, S. (2020). Phaeodactylum tricornutum: A diatom cell factory. Trends Biotechnol. 38, 606–622. doi: 10.1016/j.tibtech.2019.12.023
Calatrava, V., Tejada-Jimenez, M., Sanz-Luque, E., Fernandez, E., Galvan, A., and Llamas, A. (2023). Chlamydomonas reinhardtii, a reference organism to study algal-microbial interactions: why can’t they be friends? Plants (Basel) 12, 788. doi: 10.20944/preprints202301.0223.v1
Carthew, R. W. (2021). Gene regulation and cellular metabolism: an essential partnership. Trends Genet. 37, 389–400. doi: 10.1016/j.tig.2020.09.018
Caspi, R., Billington, R., Keseler, I. M., Kothari, A., Krummenacker, M., Midford, P. E., et al. (2020). The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 48, D445–D453. doi: 10.1093/nar/gkz862
Catalanotti, C., Yang, W., Posewitz, M. C., and Grossman, A. R. (2013). Fermentation metabolism and its evolution in algae. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00150
Chaiboonchoe, A., Dohai, B. S., Cai, H., Nelson, D. R., Jijakli, K., and Salehi-Ashtiani, K. (2014). Microalgal metabolic network model refinement through high-throughput functional metabolic profiling. Front. Bioengineering Biotechnol. 2. doi: 10.3389/fbioe.2014.00068
Chan, S. H. J., Simons, M. N., and Maranas, C. D. (2017). SteadyCom: Predicting microbial abundances while ensuring community stability. PloS Comput. Biol. 13, e1005539. doi: 10.1371/journal.pcbi.1005539
Chang, R. L., Ghamsari, L., Manichaikul, A., Hom, E. F., Balaji, S., Fu, W., et al. (2011). Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 7, 518. doi: 10.1038/msb.2011.52
Chen, H., Hu, J., Qiao, Y., Chen, W., Rong, J., Zhang, Y., et al. (2015). Ca2+-regulated cyclic electron flow supplies ATP for nitrogen starvation-induced lipid biosynthesis in green alga. Sci. Rep. 5, 15117. doi: 10.1038/srep15117
Chen, H.-H., Wu, J.-X., Huang, R., Dai, J.-L., Liang, M.-H., and Jiang, J.-G. (2024). Enhancing astaxanthin accumulation through the expression of the plant-derived astaxanthin biosynthetic pathway in Dunaliella salina. Plant Physiol. Biochem. 211, 108697. doi: 10.1016/j.plaphy.2024.108697
Chen, D., Yuan, X., Zheng, X., Fang, J., Lin, G., Li, R., et al. (2022). Multi-omics analyses provide insight into the biosynthesis pathways of fucoxanthin in isochrysis galbana. Genomics Proteomics Bioinf. 20, 1138–1153. doi: 10.1016/j.gpb.2022.05.010
Chung, I. K., Beardall, J., Mehta, S., Sahoo, D., and Stojkovic, S. (2011). Using marine macroalgae for carbon sequestration: a critical appraisal. J. Appl. Phycology 23, 877–886. doi: 10.1007/s10811-010-9604-9
Craig, R. J., Gallaher, S. D., Shu, S., Salomé, P. A., Jenkins, J. W., Blaby-Haas, C. E., et al. (2023). The Chlamydomonas Genome Project, version 6: Reference assemblies for mating-type plus and minus strains reveal extensive structural mutation in the laboratory. Plant Cell 35, 644–672. doi: 10.1093/plcell/koac347
Cunha, E., Sousa, V., Vicente, A., Geada, P., and Dias, O. (2024). Towards a genome-scale metabolic model of Dunaliella salina. IFAC-PapersOnLine 58, 37–42. doi: 10.1016/j.ifacol.2024.10.007
Demurtas, O. C., Massa, S., Ferrante, P., Venuti, A., Franconi, R., and Giuliano, G. (2013). A chlamydomonas-derived human papillomavirus 16 E7 vaccine induces specific tumor protection. PloS One 8, e61473. doi: 10.1371/journal.pone.0061473
Devoid, S., Overbeek, R., Dejongh, M., Vonstein, V., Best, A. A., and Henry, C. (2013). “Automated genome annotation and metabolic model reconstruction in the SEED and model SEED,” in Systems metabolic engineering: methods and protocols. Ed. Alper, H. S. (Humana Press, Totowa, NJ).
Diener, C., Gibbons Sean, M., and Resendis-Antonio, O. (2020). MICOM: metagenome-scale modeling to infer metabolic interactions in the gut microbiota. mSystems 5, 10.1128/msystems.00606–19. doi: 10.1128/msystems.00606-19
Di Lena, G., Casini, I., Lucarini, M., and Lombardi-Boccia, G. (2019). Carotenoid profiling of five microalgae species from large-scale production. Food Res. Int. 120, 810–818. doi: 10.1016/j.foodres.2018.11.043
Domenzain, I., Sánchez, B., Anton, M., Kerkhoven, E. J., Millán-Oropeza, A., Henry, C., et al. (2022). Reconstruction of a catalogue of genome-scale metabolic models with enzymatic constraints using GECKO 2.0. Nat. Commun. 13, 3766. doi: 10.1038/s41467-022-31421-1
El-Sheekh, M. M., Alwaleed, E. A., Ibrahim, A., and Saber, H. (2021). Detrimental effect of UV-B radiation on growth, photosynthetic pigments, metabolites and ultrastructure of some cyanobacteria and freshwater chlorophyta. Int. J. Radiat. Biol. 97, 265–275. doi: 10.1080/09553002.2021.1851060
El-Sheekh, M. M., Galal, H. R., Mousa, A. S. H. H., and Farghl, A. A. M. (2024). Improving the biodiesel production in the marine diatom Thalassiosira pseudonana cultivated in nutrient deficiency and sewage water. Environ. Sci. pollut. Res. 31, 63764–63776. doi: 10.1007/s11356-024-35409-w
Fakhry, E. M. and El Maghraby, D. M. (2015). Lipid accumulation in response to nitrogen limitation and variation of temperature in Nannochloropsis salina. Bot. Stud. 56, 6. doi: 10.1186/s40529-015-0085-7
Fisher, N. L., Halsey, K. H., Suggett, D. J., Pombrol, M., Ralph, P. J., Lutz, A., et al. (2023). Light-dependent metabolic shifts in the model diatom Thalassiosira pseudonana. Algal Res. 74, 103172. doi: 10.1016/j.algal.2023.103172
Fu, W., Paglia, G., Magnúsdóttir, M., Steinarsdóttir, E. A., Gudmundsson, S., Palsson, B.Ø., et al. (2014). Effects of abiotic stressors on lutein production in the green microalga Dunaliella salina. Microbial Cell factories 13, 1–9. doi: 10.1186/1475-2859-13-3
Gim, G. H., Ryu, J., Kim, M. J., Kim, P. I., and Kim, S. W. (2016). Effects of carbon source and light intensity on the growth and total lipid production of three microalgae under different culture conditions. J. Ind. Microbiol. Biotechnol. 43, 605–616. doi: 10.1007/s10295-016-1741-y
Gomes De Oliveira Dal’molin, C., Quek, L.-E., Palfreyman, R. W., and Nielsen, L. K. (2011). AlgaGEM – a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome. BMC Genomics 12, S5. doi: 10.1186/1471-2164-12-S4-S5
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Goss, R. and Jakob, T. (2010). Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynthesis Res. 106, 103–122. doi: 10.1007/s11120-010-9536-x
Gudmundsson, S. and Thiele, I. (2010). Computationally efficient flux variability analysis. BMC Bioinf. 11, 489. doi: 10.1186/1471-2105-11-489
Guérin, S., Bruyant, F., Gosselin, M., Babin, M., and Lavaud, J. (2024). Photoperiodic dependent regulation of photosynthesis in the polar diatom Fragilariopsis cylindrus. Front. Photobiol. 2, 1387119. doi: 10.3389/fphbi.2024.1387119
Guerin, S., Raguenes, L., Croteau, D., Babin, M., and Lavaud, J. (2022). Potential for the production of carotenoids of interest in the polar diatom fragilariopsis cylindrus. Mar. Drugs 20, 491. doi: 10.3390/md20080491
Harcombe, W. R., Riehl William, J., Dukovski, I., Granger Brian, R., Betts, A., Lang Alex, H., et al. (2014). Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 7, 1104–1115. doi: 10.1016/j.celrep.2014.03.070
Harris, E. H. (2001). Chlamydomonasas A model organism. Annu. Rev. Plant Biol. 52, 363–406. doi: 10.1146/annurev.arplant.52.1.363
Heinken, A. and Thiele, I. (2022). Microbiome Modelling Toolbox 2.0: efficient, tractable modelling of microbiome communities. Bioinformatics 38, 2367–2368. doi: 10.1093/bioinformatics/btac082
Ho, S.-H., Chan, M.-C., Liu, C.-C., Chen, C.-Y., Lee, W.-L., Lee, D.-J., et al. (2014). Enhancing lutein productivity of an indigenous microalga Scenedesmus obliquus FSP-3 using light-related strategies. Bioresource Technol. 152, 275–282. doi: 10.1016/j.biortech.2013.11.031
Hosseini, A., Jazini, M., Mahdieh, M., and Karimi, K. (2020). Efficient superantioxidant and biofuel production from microalga Haematococcus pluvialis via a biorefinery approach. Bioresource Technol. 306, 123100. doi: 10.1016/j.biortech.2020.123100
Hu, J., Wang, D., Chen, H., and Wang, Q. (2023). Advances in genetic engineering in improving photosynthesis and microalgal productivity. Int. J. Mol. Sci. 24, 1898. doi: 10.3390/ijms24031898
Hu, H., Wu, B.-L., Wei, D., Yu, L., Li, W.-H., and Zhu, S.-G. (2024). Salinity controlling enhanced high-salinity pickle wastewater treatment coupling with high-value fatty acid production by Dunaliella salina. J. Cleaner Production 448, 141732. doi: 10.1016/j.jclepro.2024.141732
Huang, W., Lin, Y., He, M., Gong, Y., and Huang, J. (2018). Induced high-yield production of zeaxanthin, lutein, and β-carotene by a mutant of chlorella zofingiensis. J. Agric. Food Chem. 66, 891–897. doi: 10.1021/acs.jafc.7b05400
Huntley, M. E. and Redalje, D. G. (2007). CO2 mitigation and renewable oil from photosynthetic microbes: A new appraisal. Mitigation Adaptation Strategies Global Change 12, 573–608. doi: 10.1007/s11027-006-7304-1
Ikaran, Z., Suárez-Alvarez, S., Urreta, I., and Castañón, S. (2015). The effect of nitrogen limitation on the physiology and metabolism of chlorella vulgaris var L3. Algal Res. 10, 134–144. doi: 10.1016/j.algal.2015.04.023
Imam, S., Schauble, S., Valenzuela, J., Lopez Garcia De Lomana, A., Carter, W., Price, N. D., et al. (2015). A refined genome-scale reconstruction of Chlamydomonas metabolism provides a platform for systems-level analyses. Plant J. 84, 1239–1256. doi: 10.1111/tpj.2015.84.issue-6
Jallet, D., Caballero, M. A., Gallina, A. A., Youngblood, M., and Peers, G. (2016). Photosynthetic physiology and biomass partitioning in the model diatom Phaeodactylum tricornutum grown in a sinusoidal light regime. Algal Res. 18, 51–60. doi: 10.1016/j.algal.2016.05.014
Jin, E., Feth, B., and Melis, A. (2003). A mutant of the green alga Dunaliella salina constitutively accumulates zeaxanthin under all growth conditions. Biotechnol. bioengineering 81, 115–124. doi: 10.1002/bit.10459
Juneja, A., Chaplen, F. W. R., and Murthy, G. S. (2016). Genome scale metabolic reconstruction of Chlorella variabilis for exploring its metabolic potential for biofuels. Bioresour Technol. 213, 103–110. doi: 10.1016/j.biortech.2016.02.118
Karp, P. D., Weaver, D., and Latendresse, M. (2018). How accurate is automated gap filling of metabolic models? BMC Syst. Biol. 12, 73. doi: 10.1186/s12918-018-0593-7
Kendirlioglu, G. and Cetin, A. K. (2017). Effect of different wavelengths of light on growth, pigment content and protein amount of Chlorella vulgaris. Fresenius Environ. Bull. 26, 7974–7980.
Khanra, A., Vasistha, S., Rai, M. P., Cheah, W. Y., Khoo, K. S., Chew, K. W., et al. (2022). Green bioprocessing and applications of microalgae-derived biopolymers as a renewable feedstock: Circular bioeconomy approach. Environ. Technol. Innovation 28, 102872. doi: 10.1016/j.eti.2022.102872
Khodayari, A., Zomorrodi, A. R., Liao, J. C., and Maranas, C. D. (2014). A kinetic model of Escherichia coli core metabolism satisfying multiple sets of mutant flux data. Metab. Eng. 25, 50–62. doi: 10.1016/j.ymben.2014.05.014
Knies, D., Wittmuss, P., Appel, S., Sawodny, O., Ederer, M., and Feuer, R. (2015). Modeling and Simulation of Optimal Resource Management during the Diurnal Cycle in Emiliania huxleyi by Genome-Scale Reconstruction and an Extended Flux Balance Analysis Approach. Metabolites 5, 659–676. doi: 10.3390/metabo5040659
Koh, H. G., Chang, Y. K., and Kang, N. K. (2024a). Enhancing lipid productivity in Nannochloropsis salina by overexpression of endogenous glycerol-3-phosphate dehydrogenase. J. Appl. Phycology 36, 73–85. doi: 10.1007/s10811-023-03141-6
Koh, H. G., Jeon, S., Kim, M., Chang, Y. K., Park, K., Park, S. H., et al. (2024b). Optimization and mechanism analysis of photosynthetic EPA production in Nannochloropsis salina: Evaluating the effect of temperature and nitrogen concentrations. Plant Physiol. Biochem. 211, 108729. doi: 10.1016/j.plaphy.2024.108729
Kruse, O., Rupprecht, J., Bader, K.-P., Thomas-Hall, S., Schenk, P. M., Finazzi, G., et al. (2005). Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 280, 34170–34177. doi: 10.1074/jbc.M503840200
Lavoie, M., Saint-Beat, B., Strauss, J., Guerin, S., Allard, A., Falciatore, A., et al. (2020). Genome-scale metabolic reconstruction and in silico perturbation analysis of the polar diatom fragilariopsis cylindrus predicts high metabolic robustness. Biol. (Basel) 9, 30. doi: 10.3390/biology9020030
Lecun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Levering, J., Broddrick, J., Dupont, C. L., Peers, G., Beeri, K., Mayers, J., et al. (2016). Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PloS One 11, e0155038. doi: 10.1371/journal.pone.0155038
Loganathan, B. G., Orsat, V., and Lefsrud, M. (2020). Evaluation and interpretation of growth, biomass productivity and lutein content of Chlorella variabilis on various media. J. Environ. Chem. Eng. 8, 103750. doi: 10.1016/j.jece.2020.103750
Loira, N., Mendoza, S., Paz Cortes, M., Rojas, N., Travisany, D., Genova, A. D., et al. (2017). Reconstruction of the microalga Nannochloropsis salina genome-scale metabolic model with applications to lipid production. BMC Syst. Biol. 11, 66. doi: 10.1186/s12918-017-0441-1
Lu, H., Li, F., Sánchez, B. J., Zhu, Z., Li, G., Domenzain, I., et al. (2019). A consensus S. cerevisiae metabolic model Yeast8 and its ecosystem for comprehensively probing cellular metabolism. Nat. Commun. 10, 3586. doi: 10.1038/s41467-019-11581-3
MaChado, D., Andrejev, S., Tramontano, M., and Patil, K. R. (2018). Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 46, 7542–7553. doi: 10.1093/nar/gky537
Makareviciene, V. and Sendzikiene, E. (2022). Application of microalgae biomass for biodiesel fuel production. Energies 15, 4178. doi: 10.3390/en15114178
Makulla, A. (2000). Fatty acid composition of Scenedesmus obliquus: Correlation to dilution rates. Limnologica 30, 162–168. doi: 10.1016/S0075-9511(00)80011-0
Malik-Sheriff, R. S., Glont, M., Nguyen, T. V. N., Tiwari, K., Roberts, M. G., Xavier, A., et al. (2020). BioModels—15 years of sharing computational models in life science. Nucleic Acids Res. 48, D407–D415. doi: 10.1093/nar/gkz1055
Manichaikul, A., Ghamsari, L., Hom, E. F. Y., Lin, C., Murray, R. R., Chang, R. L., et al. (2009). Metabolic network analysis integrated with transcript verification for sequenced genomes. Nat. Methods 6, 589–592. doi: 10.1038/nmeth.1348
Maroneze, M. M., Zepka, L. Q., Lopes, E. J., Pérez-Gálvez, A., and Roca, M. (2019). Chlorophyll oxidative metabolism during the phototrophic and heterotrophic growth of scenedesmus obliquus. Antioxidants 8, 600. doi: 10.3390/antiox8120600
Masi, A., Leonelli, F., Scognamiglio, V., Gasperuzzo, G., Antonacci, A., and Terzidis, M. A. (2023). Chlamydomonas reinhardtii: A factory of nutraceutical and food supplements for human health. Molecules 28, 1185. doi: 10.3390/molecules28031185
Matos, Â.P., Cavanholi, M. G., Moecke, E. H. S., and Sant’anna, E. S. (2017). Effects of different photoperiod and trophic conditions on biomass, protein and lipid production by the marine alga Nannochloropsis gaditana at optimal concentration of desalination concentrate. Bioresource Technol. 224, 490–497. doi: 10.1016/j.biortech.2016.11.004
May, P., Christian, J.-O., Kempa, S., and Walther, D. (2009). ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics 10, 209. doi: 10.1186/1471-2164-10-209
Meagher, M., Metcalf, A., Vigliotti, M., Ramsey, S. A., Prentice, W., Cohen, L., et al. (2024). Genome-scale metabolic model accurately predicts fermentation of glucose by Chromochloris zofingiensis. Algal Res. 84, 103805. doi: 10.1016/j.algal.2024.103805
Mekanik, M., Fotovat, R., Motamedian, E., and Jafarian, V. (2023). Improvement of lutein production in auxenochlorella protothecoides using its genome-scale metabolic model and a system-oriented approach. Appl. Biochem. Biotechnol. 195, 889–904. doi: 10.1007/s12010-022-04186-y
Mekanik, M., Motamedian, E., Fotovat, R., and Jafarian, V. (2019). Reconstruction of a genome-scale metabolic model for Auxenochlorella protothecoides to study hydrogen production under anaerobiosis using multiple optimal solutions. Int. J. Hydrogen Energy 44, 2580–2591. doi: 10.1016/j.ijhydene.2018.12.049
Metcalf, A. J., Nagygyor, A., and Boyle, N. R. (2020). Rapid Annotation of Photosynthetic Systems (RAPS): automated algorithm to generate genome-scale metabolic networks from algal genomes. Algal Res. 50, 101967. doi: 10.1016/j.algal.2020.101967
Metcalf Alex, J. and Boyle Nanette, R. (2022). Rhythm of the night (and day): predictive metabolic modeling of diurnal growth in chlamydomonas. mSystems 7, e00176–e00122. doi: 10.1128/msystems.00176-22
Mitra, M., Patidar, S. K., George, B., Shah, F., and Mishra, S. (2015). A euryhaline Nannochloropsis gaditana with potential for nutraceutical (EPA) and biodiesel production. Algal Res. 8, 161–167. doi: 10.1016/j.algal.2015.02.006
Moayedi, A., Yargholi, B., Pazira, E., and Babazadeh, H. (2019). Investigated of desalination of saline waters by using dunaliella salina algae and its effect on water ions. Civil Eng. J. 5, 2450–2460. doi: 10.28991/cej-2019-03091423
Monk, J. M., Lloyd, C. J., Brunk, E., Mih, N., Sastry, A., King, Z., et al. (2017). iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 35, 904–908. doi: 10.1038/nbt.3956
Moradi, P. and Saidi, M. (2022). Biodiesel production from Chlorella Vulgaris microalgal-derived oil via electrochemical and thermal processes. Fuel Process. Technol. 228, 107158. doi: 10.1016/j.fuproc.2021.107158
Mora Salguero, D. A., Fernández-Niño, M., Serrano-Bermúdez, L. M., Páez Melo, D. O., Winck, F. V., Caldana, C., et al. (2018). Development of a Chlamydomonas reinhardtii metabolic network dynamic model to describe distinct phenotypes occurring at different CO2 levels. PeerJ 6, e5528. doi: 10.7717/peerj.5528
Mularczyk, M., Michalak, I., and Marycz, K. (2020). Astaxanthin and other Nutrients from Haematococcus pluvialis-Multifunctional Applications. Mar. Drugs 18, 459. doi: 10.3390/md18090459
Nocon, J., Steiger, M. G., Pfeffer, M., Sohn, S. B., Kim, T. Y., Maurer, M., et al. (2014). Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab. Eng. 24, 129–138. doi: 10.1016/j.ymben.2014.05.011
Norsigian, C. J., Pusarla, N., Mcconn, J. L., Yurkovich, J. T., Dräger, A., Palsson, B. O., et al. (2020). BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 48, D402–D406. doi: 10.1093/nar/gkz1054
Ofaim, S., Sulheim, S., Almaas, E., Sher, D., and Segre, D. (2021). Dynamic allocation of carbon storage and nutrient-dependent exudation in a revised genome-scale model of prochlorococcus. Front. Genet. 12, 586293. doi: 10.3389/fgene.2021.586293
Orth, J. D., Conrad, T. M., Na, J., Lerman, J. A., Nam, H., Feist, A. M., et al. (2011). A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 7, 535. doi: 10.1038/msb.2011.65
Pandey, S., Kumar, P., Dasgupta, S., Archana, G., and Bagchi, D. (2023). Gradient strategy for mixotrophic cultivation of chlamydomonas reinhardtii: small steps, a large impact on biofuel potential and lipid droplet morphology. Bioenergy Res. 16, 163–176. doi: 10.1007/s12155-022-10454-w
Pareek, C. S., Smoczynski, R., and Tretyn, A. (2011). Sequencing technologies and genome sequencing. J. Appl. Genet. 52, 413–435. doi: 10.1007/s13353-011-0057-x
Park, S.-H., Kyndt, J., Chougule, K., Park, J.-J., and Brown, J. K. (2018). Low-phosphate-selected Auxenochlorella protothecoides redirects phosphate to essential pathways while producing more biomass. PloS One 13, e0198953. doi: 10.1371/journal.pone.0198953
Park, S.-B., Yun, J.-H., Ryu, A. J., Yun, J., Kim, J. W., Lee, S., et al. (2021). Development of a novel nannochloropsis strain with enhanced violaxanthin yield for large-scale production. Microbial Cell Factories 20, 43. doi: 10.1186/s12934-021-01535-0
Patel, A., Matsakas, L., Rova, U., and Christakopoulos, P. (2018). Heterotrophic cultivation of Auxenochlorella protothecoides using forest biomass as a feedstock for sustainable biodiesel production. Biotechnol. Biofuels 11, 169. doi: 10.1186/s13068-018-1173-1
Peng, M., Lin, S., Shen, Y., Peng, R., Li, S., Jiang, X., et al. (2024). Effects of light quality on the growth, productivity, fucoxanthin accumulation, and fatty acid composition of Thalassiosira pseudonana. J. Appl. Phycology 36, 1667–1678. doi: 10.1007/s10811-024-03245-7
Perin, G., Bellan, A., Segalla, A., Meneghesso, A., Alboresi, A., and Morosinotto, T. (2015). Generation of random mutants to improve light-use efficiency of Nannochloropsis gaditana cultures for biofuel production. Biotechnol. Biofuels 8, 161. doi: 10.1186/s13068-015-0337-5
Polat, E., Yavuztürk-Gül, B., Ünver, H., and Altinbaş, M. (2023). Biotechnological product potential of Auxenochlorella protothecoides including biologically active compounds (BACs) under nitrogen stress conditions. World J. Microbiol. Biotechnol. 39, 198. doi: 10.1007/s11274-023-03642-z
Qin, S., Liu, G.-X., and Hu, Z.-Y. (2008). The accumulation and metabolism of astaxanthin in Scenedesmus obliquus (Chlorophyceae). Process Biochem. 43, 795–802. doi: 10.1016/j.procbio.2008.03.010
Rathod, J. P., Vira, C., Lali, A. M., and Prakash, G. (2020). Metabolic engineering of chlamydomonas reinhardtii for enhanced β-carotene and lutein production. Appl. Biochem. Biotechnol. 190, 1457–1469. doi: 10.1007/s12010-019-03194-9
Ray, A., Kundu, P., and Ghosh, A. (2023). Reconstruction of a Genome-Scale Metabolic Model of Scenedesmus obliquus and Its Application for Lipid Production under Three Trophic Modes. ACS Synth Biol. 12, 3463–3481. doi: 10.1021/acssynbio.3c00516
Recht, L., Topfer, N., Batushansky, A., Sikron, N., Gibon, Y., Fait, A., et al. (2014). Metabolite profiling and integrative modeling reveal metabolic constraints for carbon partitioning under nitrogen starvation in the green algae Haematococcus pluvialis. J. Biol. Chem. 289, 30387–30403. doi: 10.1074/jbc.M114.555144
Recht, L., Zarka, A., and Boussiba, S. (2012). Patterns of carbohydrate and fatty acid changes under nitrogen starvation in the microalgae Haematococcus pluvialis and Nannochloropsis sp. Appl. Microbiol. Biotechnol. 94, 1495–1503. doi: 10.1007/s00253-012-3940-4
Rügen, M., Bockmayr, A., and Steuer, R. (2015). Elucidating temporal resource allocation and diurnal dynamics in phototrophic metabolism using conditional FBA. Sci. Rep. 5, 15247. doi: 10.1038/srep15247
Ryu, Y.-K., Lee, W.-K., Park, G.-H., Kim, T., Lee, Y., Koh, E.-J., et al. (2024). Preliminary assessment of astaxanthin production in a new Chlamydomonas strain. Algal Res. 82, 103629. doi: 10.1016/j.algal.2024.103629
Sánchez, Á., Maceiras, R., Cancela, Á., and Pérez, A. (2013). Culture aspects of Isochrysis galbana for biodiesel production. Appl. Energy 101, 192–197. doi: 10.1016/j.apenergy.2012.03.027
Santos, C. A., Vieira, A. M., Fernandes, H. L., Empis, J. A., and Novais, J. M. (2001). Optimisation of the biological treatment of hypersaline wastewater from Dunaliella salina carotenogenesis. J. Chem. Technol. Biotechnology: Int. Res. Process Environ. Clean Technol. 76, 1147–1153. doi: 10.1002/jctb.497
Sati, H., Chokshi, K., Soundarya, R., Ghosh, A., and Mishra, S. (2021). Seaweed-based biostimulant improves photosynthesis and effectively enhances growth and biofuel potential of a green microalga Chlorella variabilis. Aquaculture Int. 29, 963–975. doi: 10.1007/s10499-021-00667-9
Schellenberger, J., Park, J. O., Conrad, T. M., and Palsson, B.Ø. (2010). BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinf. 11, 213. doi: 10.1186/1471-2105-11-213
Schnurr, P. J., Espie, G. S., and Allen, G. D. (2016). The effect of photon flux density on algal biofilm growth and internal fatty acid concentrations. Algal Res. 16, 349–356. doi: 10.1016/j.algal.2016.04.001
Schubert, H., Andersson, M., and Snoeijs, P. (2006). Relationship between photosynthesis and non-photochemical quenching of chlorophyll fluorescence in two red algae with different carotenoid compositions. Mar. Biol. 149, 1003–1013. doi: 10.1007/s00227-006-0265-9
Seaver, S. M. D., Gerdes, S., Frelin, O., Lerma-Ortiz, C., Bradbury, L. M. T., Zallot, R., et al. (2014). High-throughput comparison, functional annotation, and metabolic modeling of plant genomes using the PlantSEED resource. Proc. Natl. Acad. Sci. 111, 9645–9650. doi: 10.1073/pnas.1401329111
Segrè, D., Vitkup, D., and Church, G. M. (2002). Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. 99, 15112–15117. doi: 10.1073/pnas.232349399
Sen, P. and Orešič, M. (2023). Integrating omics data in genome-scale metabolic modeling: A methodological perspective for precision medicine. Metabolites 13, 855. doi: 10.3390/metabo13070855
Sengupta, A., Gupta, T., Chakraborty, A., Kulshrestha, S., Redhu, R., Bhattacharjya, R., et al. (2024). A novel draft genome-scale reconstruction model of isochrysis sp: exploring metabolic pathways for sustainable aquaculture innovations. Microbiol. Biotechnol. Lett. 52, 141–151. doi: 10.48022/mbl.2309.09011
Serra, A. T., Silva, S. D., Pleno De Gouveia, L., Alexandre, A. M. R. C., Pereira, C. V., Pereira, A. B., et al. (2021). A single dose of marine chlorella vulgaris increases plasma concentrations of lutein, β-carotene and zeaxanthin in healthy male volunteers. Antioxidants 10, 1164. doi: 10.3390/antiox10081164
Shah, A. R., Ahmad, A., Srivastava, S., and Jaffar Ali, B. M. (2017). Reconstruction and analysis of a genome-scale metabolic model of Nannochloropsis gaditana. Algal Res. 26, 354–364. doi: 10.1016/j.algal.2017.08.014
Sharma, N. K. and Rai, A. K. (2010). Biodiversity and biogeography of microalgae: progress and pitfalls. Environ. Rev. 19, 1–15. doi: 10.1139/a10-020
Shene, C., Asenjo, J. A., and Chisti, Y. (2018). Metabolic modelling and simulation of the light and dark metabolism of Chlamydomonas reinhardtii. Plant J. 96, 1076–1088. doi: 10.1111/tpj.2018.96.issue-5
Sheward, R. M., Gebühr, C., Bollmann, J., and Herrle, J. O. (2024). Short-term response of Emiliania huxleyi growth and morphology to abrupt salinity stress. Biogeosciences 21, 3121–3141. doi: 10.5194/bg-21-3121-2024
Song, I., Kim, J., Baek, K., Choi, Y., Shin, B., and Jin, E. (2020). The generation of metabolic changes for the production of high-purity zeaxanthin mediated by CRISPR-Cas9 in Chlamydomonas reinhardtii. Microbial Cell Factories 19, 220. doi: 10.1186/s12934-020-01480-4
Stichnothe, H., Storz, H., Meier, D., De Bari, I., and Thomas, S. (2016). “Chapter 2 - development of second-generation biorefineries,” in Developing the global bioeconomy. Eds. Lamers, P., Searcy, E., Hess, J. R., and & Stichnothe, H. (Cambridge, Massachusetts, USA: Academic Press).
Tamoi, M. and Shigeoka, S. (2015). Diversity of regulatory mechanisms of photosynthetic carbon metabolism in plants and algae. Bioscience Biotechnology Biochem. 79, 870–876. doi: 10.1080/09168451.2015.1020754
Tarzi, C., Zampieri, G., Sullivan, N., and Angione, C. (2024). Emerging methods for genome-scale metabolic modeling of microbial communities. Trends Endocrinol. Metab. 35, 533–548. doi: 10.1016/j.tem.2024.02.018
Tran, D. T., Van Do, T. C., Nguyen, Q. T., and Le, T. G. (2020). Simultaneous removal of pollutants and high value biomaterials production by Chlorella variabilis TH03 from domestic wastewater. Clean Technol. Environ. Policy 23, 3–17. doi: 10.1007/s10098-020-01810-5
Vaezi, R., Napier, J. A., and Sayanova, O. (2013). Identification and functional characterization of genes encoding omega-3 polyunsaturated fatty acid biosynthetic activities from unicellular microalgae. Mar. Drugs 11, 5116–5129. doi: 10.3390/md11125116
Van Tol, H. M. and Armbrust, E. V. (2021). Genome-scale metabolic model of the diatom Thalassiosira pseudonana highlights the importance of nitrogen and sulfur metabolism in redox balance. PloS One 16, e0241960. doi: 10.1371/journal.pone.0241960
Vasudevan, P. T. and Briggs, M. (2008). Biodiesel production—current state of the art and challenges. J. Ind. Microbiol. Biotechnol. 35, 421–421. doi: 10.1007/s10295-008-0312-2
Vitali, L., Lolli, V., Sansone, F., Kumar, A., Concas, A., and Lutzu, G. A. (2023). Lipid content and fatty acid methyl ester profile by Chromochloris zofingiensis under chemical and metabolic stress. Biomass Conversion Biorefinery. 15, 4941–4954. doi: 10.1007/s13399-023-04153-5
Wang, H., Marcišauskas, S., Sánchez, B. J., Domenzain, I., Hermansson, D., Agren, R., et al. (2018). RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PloS Comput. Biol. 14, e1006541. doi: 10.1371/journal.pcbi.1006541
Wang, H., Qi, M., Bo, Y., Zhou, C., Yan, X., Wang, G., et al. (2021). Treatment of fishery wastewater by co-culture of Thalassiosira pseudonana with Isochrysis galbana and evaluation of their active components. Algal Res. 60, 102498. doi: 10.1016/j.algal.2021.102498
Wang, Y.-Y., Xu, S.-M., Cao, J.-Y., Wu, M.-N., Lin, J.-H., Zhou, C.-X., et al. (2022). Co-cultivation of Isochrysis galbana and Marinobacter sp. can enhance algal growth and docosahexaenoic acid production. Aquaculture 556, 738248. doi: 10.1016/j.aquaculture.2022.738248
Winck, F. V., Melo, D. O., Riano-Pachon, D. M., Martins, M. C., Caldana, C., and Barrios, A. F. (2016). Analysis of Sensitive CO2 Pathways and Genes Related to Carbon Uptake and Accumulation in Chlamydomonas reinhardtii through Genomic Scale Modeling and Experimental Validation. Front. Plant Sci. 7, 43. doi: 10.3389/fpls.2016.00043
Xi, Y., Bian, J., Luo, G., Kong, F., and Chi, Z. (2022). Enhanced β-carotene production in Dunaliella salina under relative high flashing light. Algal Res. 67, 102857. doi: 10.1016/j.algal.2022.102857
Xiao, Y., He, X., Ma, Q., Lu, Y., Bai, F., Dai, J., et al. (2018). Photosynthetic Accumulation of Lutein in Auxenochlorella protothecoides after Heterotrophic Growth. Mar. Drugs 16, 283. doi: 10.3390/md16080283
Yan, J., Kuang, Y., Gui, X., Han, X., and Yan, Y. (2019). Engineering a Malic enzyme to enhance lipid accumulation in Chlorella protothecoides and direct production of biodiesel from the microalgal biomass. Biomass Bioenergy 122, 298–304. doi: 10.1016/j.biombioe.2019.01.046
Yang, L., Ebrahim, A., Lloyd, C. J., Saunders, M. A., and Palsson, B. O. (2019). DynamicME: dynamic simulation and refinement of integrated models of metabolism and protein expression. BMC Syst. Biol. 13, 2. doi: 10.1186/s12918-018-0675-6
Yang, J. E., Park, S. J., Kim, W. J., Kim, H. J., Kim, B. J., Lee, H., et al. (2018). One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat. Commun. 9, 79. doi: 10.1038/s41467-017-02498-w
Yang, L., Ren, L., Tan, X., Chu, H., Chen, J., Zhang, Y., et al. (2020). Removal of ofloxacin with biofuel production by oleaginous microalgae Scenedesmus obliquus. Bioresour Technol. 315, 123738. doi: 10.1016/j.biortech.2020.123738
Yao, H., Dahal, S., and Yang, L. (2023). Novel context-specific genome-scale modelling explores the potential of triacylglycerol production by Chlamydomonas reinhardtii. Microb. Cell Fact 22, 13. doi: 10.1186/s12934-022-02004-y
Ye, C., Qiao, W., Yu, X., Ji, X., Huang, H., Collier, J. L., et al. (2015). Reconstruction and analysis of the genome-scale metabolic model of schizochytrium limacinum SR21 for docosahexaenoic acid production. BMC Genomics 16, 799. doi: 10.1186/s12864-015-2042-y
Yoshida, K., Seger, A., Kennedy, F., Mcminn, A., and Suzuki, K. (2020). Freezing, Melting, and Light Stress on the Photophysiology of Ice Algae: Ex Situ Incubation of the Ice Algal diatom Fragilariopsis cylindrus (Bacillariophyceae) Using an Ice Tank. J. Phycology 56, 1323–1338. doi: 10.1111/jpy.13036
Zhang, K., Chen, L., Liu, J., Gao, F., He, R., Chen, W., et al. (2017). Effects of butanol on high value product production in Schizochytrium limacinum B4D1. Enzyme Microbial Technol. 102, 9–15. doi: 10.1016/j.enzmictec.2017.03.007
Zhang, J., Liu, F., Wang, Q., Gong, Q., and Gao, X. (2023). Effect of light wavelength on biomass, growth, photosynthesis and pigment content of emiliania huxleyi (Isochrysidales, cocco-lithophyceae). J. Mar. Sci. Eng. 11, 456. doi: 10.3390/jmse11020456
Zhang, Y., Ye, Y., Bai, F., and Liu, J. (2021). The oleaginous astaxanthin-producing alga Chromochloris zofingiensis: potential from production to an emerging model for studying lipid metabolism and carotenogenesis. Biotechnol. Biofuels 14, 119. doi: 10.1186/s13068-021-01969-z
Zheng, H.-Q., Chiang-Hsieh, Y.-F., Chien, C.-H., Hsu, B.-K. J., Liu, T.-L., Chen, C.-N. N., et al. (2014). AlgaePath: comprehensive analysis of metabolic pathways using transcript abundance data from next-generation sequencing in green algae. BMC Genomics 15, 196. doi: 10.1186/1471-2164-15-196
Zomorrodi, A. R., Islam, M. M., and Maranas, C. D. (2014). d-optCom: dynamic multi-level and multi-objective metabolic modeling of microbial communities. ACS Synthetic Biol. 3, 247–257. doi: 10.1021/sb4001307
Zuniga, C., Levering, J., Antoniewicz, M. R., Guarnieri, M. T., Betenbaugh, M. J., and Zengler, K. (2018). Predicting dynamic metabolic demands in the photosynthetic eukaryote chlorella vulgaris. Plant Physiol. 176, 450–462. doi: 10.1104/pp.17.00605
Zuniga, C., Li, C. T., Huelsman, T., Levering, J., Zielinski, D. C., Mcconnell, B. O., et al. (2016). Genome-Scale Metabolic Model for the Green Alga Chlorella vulgaris UTEX 395 Accurately Predicts Phenotypes under Autotrophic, Heterotrophic, and Mixotrophic Growth Conditions. Plant Physiol. 172, 589–602. doi: 10.1104/pp.16.00593
Keywords: metabolic flux, algae, photosynthesis, modeling, metabolism
Citation: Tamburro J and Boyle NR (2025) Charting the state of GEMs in microalgae: progress, challenges, and innovations. Front. Plant Sci. 16:1614397. doi: 10.3389/fpls.2025.1614397
Received: 18 April 2025; Accepted: 26 May 2025;
Published: 13 June 2025.
Edited by:
Jianping Yu, National Renewable Energy Laboratory (DOE), United StatesReviewed by:
Yantao Li, University of Maryland, College Park, United StatesAnna Santin, University of Padua, Italy
Anika Küken, University of Potsdam, Germany
Copyright © 2025 Tamburro and Boyle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nanette R. Boyle, bmJveWxlQG1pbmVzLmVkdQ==
 Jacob Tamburro
Jacob Tamburro Nanette R. Boyle
Nanette R. Boyle