Abstract
Accurate estimation of mangrove biomass is significant for ensuring the mangrove ecosystem’s productivity and global carbon cycling. Although well-known deep neural networks (DNNs) have been successfully applied in mangrove biomass estimation using remote sensing data, the key problem of data scarcity is not addressed very well for existing methods. Thus, a novel DNN called self-supervised disturbing feature reconstruction network (SSDFRN) is constructed in this article for mangrove biomass estimation with limited data. First, a disturbing feature reconstruction-based self-supervised learning (DFRSSL) method based on random feature shuffle and disturbing feature reconstruction is proposed for solving the data scarcity problem. In addition, a multi-view convolutional neural network (MVCNN) is constructed by stacking several multi-view cascaded convolution modules (MVCCMs), which effectively enhances feature learning performance and improves mangrove biomass estimation accuracy. The mangrove biomass dataset obtained from Ximen Island (28° 21′ N, 121° 10′ E) is used in this study to verify the outperformance of SSDFRN. The experimental results illustrate that SSDFRN is effective in deep feature learning and mangrove biomass estimation with limited data.
1 Introduction
Mangrove plays a significant role in maintaining biodiversity, carbon sequestration, and carbon storage (Tran et al., 2022). Accurate estimation of aboveground biomass (AGB) is an important part of the mangrove ecosystem carbon cycle, which is conducive for assessing the carbon sink potential of the mangrove ecosystem (Sliva et al., 2024). The traditional mangrove survey method is destructive, costly, and inefficient, which greatly restricts the efficiency of estimating and monitoring mangrove biomass distribution (Morais et al., 2021; Zhang et al., 2022). Due to the interference of various external environmental factors (e.g., growing environment, climatic factors, and geographical position), it is difficult to promptly and accurately estimate mangrove biomass using the traditional survey method.
Remote sensing technology has the advantages of a large spatial scale, strong timeliness, and high efficiency (Zhao et al., 2023; Hazmy et al., 2024), which greatly saves the manpower and material resources required by traditional investigation (Gao et al., 2022) (Muhd-Ekhzarizal et al., 2018). employed simple and multilinear regression methods for the estimation of AGB in the entire study area using remote sensing images (Pandey et al., 2019). utilized logarithmic and polynomial (second degree) models for mangrove biomass estimation, and the study shows that normalized difference vegetation index (NDVI) and enhanced vegetation index (EVI) derived from satellite images are effective indexes for biomass estimation. However, traditional remote sensing-based linear regression and non-linear regression models have poor performance in estimating mangrove biomass and are not suitable for practical application.
Machine learning (ML) [e.g., support vector machine (SVM) (Li et al., 2023), random forest (RF) (Xiao et al., 2024), and support vector regression (SVR) (Rahimikhoob et al., 2023)] learns the relationship between input and output by fitting a flexible model (Teshome et al., 2023), which has been widely used in mangrove biomass estimation (Tian et al., 2021; Hao et al., 2024) (Selvaraj and Perez, 2023). developed an RF-based spatial estimation approach to assess mangrove AGB using the Google Earth Engine (GEE) platform (Bui et al., 2024). proposed an ML-based (i.e., LightGBM and XGBoost) AGB estimation method, and the hyperparameters were tuned by Bayesian-based optimizers and a novel Tasmanian Devil optimization algorithm (Tian et al., 2022). analyzed the quantitative relationship between invasive mangrove biomass and hydrological units using different ML algorithms (Do et al., 2022). proposed a principal component analysis-based ML technique for estimating the mangrove AGB (Rijal et al., 2023). developed a novel ML-based mangrove aboveground carbon (AGC) estimation technique based on extreme gradient boosting and genetic algorithm analyses (Hu et al., 2024). proposed an SVM-based AGB estimation method using remote sensing data (Luo et al., 2024). developed a novel AGB estimation method using SVR, and the parameters of SVR were optimized by the global best particle swarm algorithm. However, the feature learning ability of traditional ML methods is limited due to their shallow network structures. In addition, RF-based methods typically cannot make accurate estimations when the training samples are limited, and the performance of SVM and SVR models depends heavily on the choice of kernel function (He et al., 2024). Thus, effective feature learning techniques are needed for mangrove biomass estimation based on remote sensing data.
Recently, deep learning (DL) techniques have been widely used in various domains due to their outstanding feature learning capacity (Li et al., 2024; Miao and Yu, 2024). Typical deep neural networks (DNNs) [e.g., deep belief network (DBN), convolutional neural network (CNN), and long short-term memory (LSTM) network] have been successfully applied in mangrove biomass estimation (Akkem et al., 2023) (Chen et al., 2022). proposed a generative adversarial network for data augmentation using Sentinel-2 images and a DBN for deep feature learning and obtaining the salt marsh distribution (Nakajima et al., 2023). proved that the CNN-based estimation technique is outstanding and can be applied for monitoring crop AGB in diverse cultivars (Tian et al., 2024). proposed a novel AGB estimation method based on CNN and LSTM using different remote sensing image data (Talebiesfandarani and Shamsoddini, 2022). developed a novel global-scale biomass estimation method by learning features using CNN, and RF and SVR are used for feature selection (Song and Wang, 2023). proposed a recurrent neural network (RNN)-based method for forest energy estimation (Zhang et al., 2024). proposed a novel framework for AGB estimation using Sentinel-1 synthetic aperture radar (SAR) and Sentinel-2 optical data, where the bidirectional long short-term memory (BiLSTM) neural network is implemented for deep feature learning (Liu et al., 2024). proposed a residual neural network (ResNet)-based model to extract phenological information from wheat and implemented the AGB estimation. Nevertheless, these methods always assume that training samples are sufficient and rely heavily on the quantity and quality of data. When the data are limited, these models are prone to overfitting. In the actual scenario, the data scarcity problem is inescapable due to the difficulty of obtaining high-quality mangrove biomass data, which greatly limits the application of these methods.
In order to address the above problems, a novel DNN, called self-supervised disturbing feature reconstruction network (SSDFRN), is proposed for mangrove biomass estimation with limited data in this study. The main contributions of this study are summarized as follows: 1) a self-supervised disturbing feature reconstruction network is proposed for deep feature learning, 2) a disturbing feature reconstruction-based self-supervised learning (DFRSSL) method based on random feature shuffle and disturbing feature reconstruction is developed for solving the data scarcity problem, and 3) a multi-view convolutional neural network (MVCNN) is constructed by stacking several multi-view cascaded convolution modules (MVCCMs), which effectively enhances the feature learning performance and improves the mangrove biomass estimation accuracy. The experimental results on the mangrove biomass dataset obtained from Ximen Island (28° 21′ N, 121° 10′ E) demonstrate the outperformance of SSDFRN for mangrove biomass estimation with limited data.
The remainder of this article is organized as follows. The details about SSDFRN are given in Section 2. The experimental analysis of SSDFRN-based mangrove biomass estimation is implemented in Section 3. Finally, the conclusions are given in Section 4.
2 Self-supervised disturbing feature reconstruction network
In this study, SSDFRN is proposed for mangrove biomass estimation with limited data. In particular, first, Landsat 8 remote sensing data and Digital Elevation Model (DEM) data are used for extracting 22 features (i.e., band information, vegetation indexes, texture features, and elevation features). Then, the shuffle window is masked on partial input features randomly for generating auxiliary data and residual data. MVCNN is constructed for deep feature learning from residual data, and learned features are combined with auxiliary features for disturbing feature reconstruction. In the process of DFRSSL, the feature representation ability of the deep network will be greatly enhanced with limited data.
2.1 Network structure
The SSDFRN-based mangrove biomass estimation method is shown in Figure 1, which includes two stages: DFRSSL and fine-tuning. In Figure 1, L denotes the total number of input features, and W is the length of the shuffle window. In the stage of DFRSSL, first, limited samples are used to generate plenty of auxiliary data and residual data. Then, MVCNN is implemented for deep feature learning from sufficient residual data. Finally, disturbing feature reconstruction is implemented based on deep features and auxiliary data for solving the problem of feature learning with limited data. In the stage of fine-tuning, learned representations are fed into the biomass estimator for mangrove biomass estimation.
Figure 1
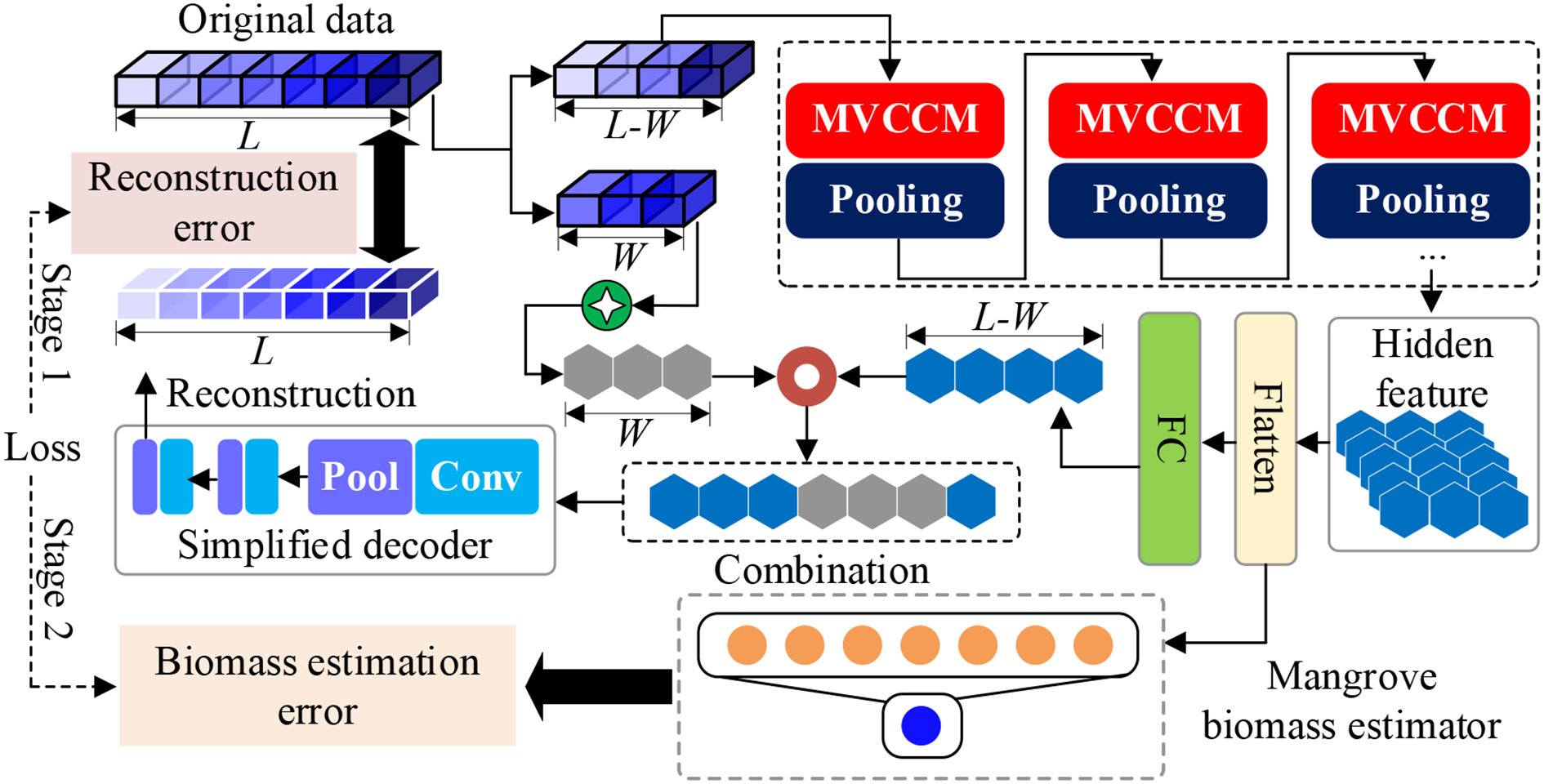
Network structure of SSDFRN. SSDFRN, self-supervised disturbing feature reconstruction network.
2.2 Disturbing feature reconstruction-based self-supervised learning
In the actual scenario of mangrove biomass estimation, the problem of data scarcity is inevitable due to the difficulty of data collection. Traditional mangrove biomass estimation methods highly depend on data quantity and quality, which could limit their applications in a real environment. Thus, DFRSSL is developed in this study for solving the problem of feature learning and mangrove biomass estimation with limited data.
2.2.1 Generation of auxiliary data and residual data
In the stage of DFRSSL, first, Landsat 8 remote sensing image and DEM elevation data are used for extracting 22 features (i.e., band information, vegetation indexes, texture features, and elevation features). Then, the shuffle window is masked on partial input features randomly for generating auxiliary data and residual data. This operation has two main functions: 1) the masking method can generate data pairs that are not limited by the scarcity of original data so as to solve the problem of limited data availability. 2) This method largely reduces redundancy and creates a challenging SSL task that requires a holistic understanding of the relationship between all input features (i.e., band information, vegetation indexes, texture features, and elevation features). The generation process of auxiliary data and residual data is shown in Figure 2. In particular, a shuffle window with length W is used to randomly select W features for feature disturbance. In the meantime, Gaussian noise is used to mask the W features after the disturbance to simulate the interference of the external environment.
Figure 2
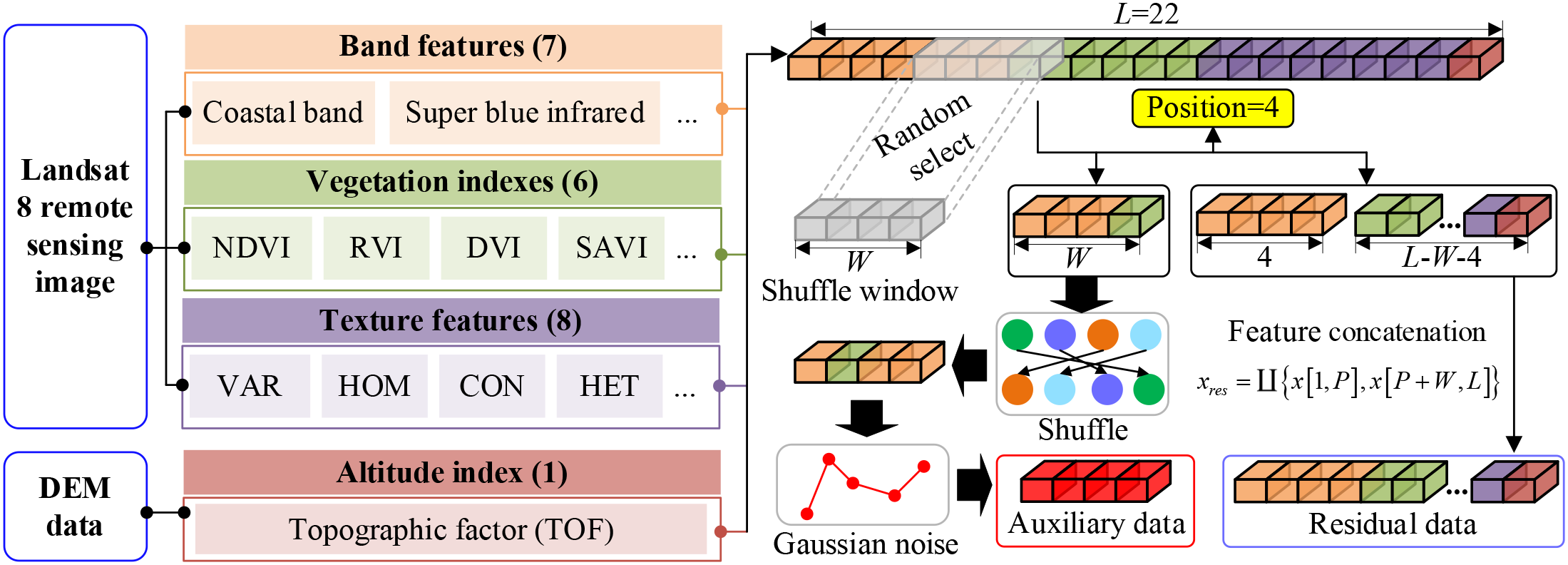
Generation process of auxiliary data and residual data.
The generation process of auxiliary data is represented as follows:
where P is the randomly selected feature position, indicates that an integer is randomly selected from a to b, L is the total number of input features (L = 22 in this study), W is the length of shuffle window, denotes the generated auxiliary data, represents the Gaussian noise, x[a, b] represents features from segments a to b of the original data, and is the feature disturbance operation. For the unselected feature segments, the residual data are obtained by data concatenation, which is expressed as follows:
where xresindicates the residual data, and is the data concatenation operation. It should be noted that the number of auxiliary data and that of residual data are not limited by the size of the original samples, and the original limited data will be greatly enhanced by generating multiple auxiliary and residual data.
2.2.2 Disturbing feature reconstruction-based self-supervised learning
In this study, MVCNN is constructed for deep feature learning from residual data. The structure of MVCNN is shown in Figure 3, which is composed of multiple MVCCMs. In Figure 3, the numbers of circles in MVCCM signify different sliding strides.
Figure 3
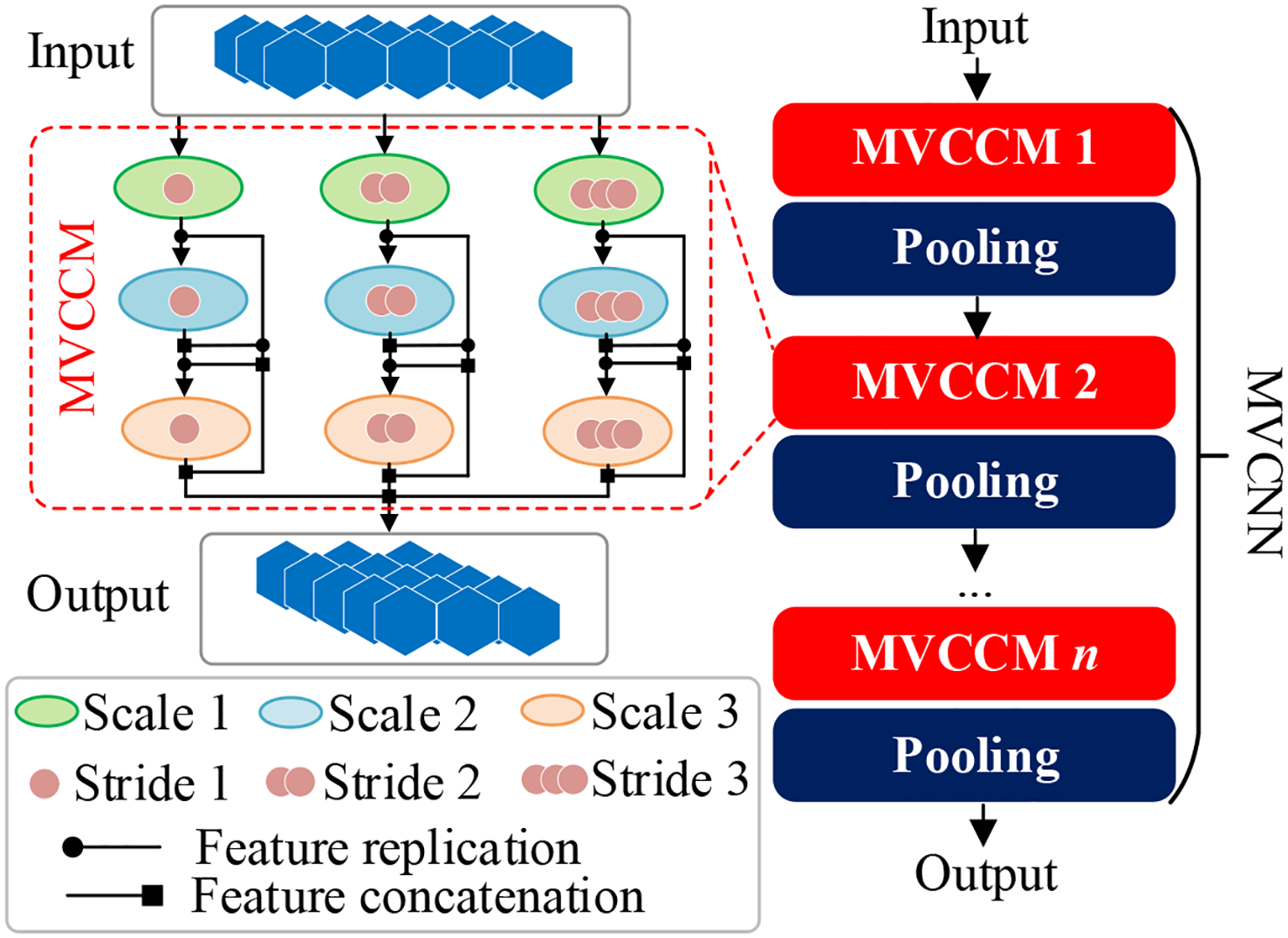
Structure of MVCCM and MVCNN. MVCCM, multi-view cascaded convolution module; MVCNN, multi-view convolutional neural network.
Taking the first MVCCM as an example, the small-scale convolution is first used to obtain small-field features from residual data xres using different convolution steps, which are calculated as follows:
where denotes the ith small-view feature and is the small-scale convolution with step size i. After obtaining the small-view features, medium-scale convolution is used to obtain medium-view features with different convolution steps, as follows:
where denotes the ith medium-view feature, and represents the medium-scale convolution with step size i. After obtaining the small-view features and medium-view features, large-scale convolution is used to obtain the big-view features based on the cascading convolution, which is calculated as follows:
where is the ith big-view feature, denotes the large-scale convolution with step size i, and represents the feature concatenation operation. Finally, the output feature of the first MVCCM is obtained as follows:
where OMVCCM represents the output feature of the first MVCCM. MVCNN is constructed by cascading multiple MVCCMs and pooling layers, and the output feature of MVCNN is represented as follows:
where OMVCNN denotes the learned hidden representation, indicates the cascading operation of n MVCCMs, and Pool and MVCCM represent the pooling layer and MVCCM, respectively.
After obtaining hidden representations OMVCNN, the simplified decoder is implemented for disturbing feature reconstruction (He et al., 2022) based on auxiliary data and corresponding feature position P. First, the hidden representation OMVCNN learned by MVCNN is flattened and then spliced with auxiliary data as the input of simplified decoder. It should be noted that the feature disturbing position is considered in the splicing process. The input of the simplified decoder is obtained as follows:
where xde denotes the input feature of the simplified decoder and Flat () indicates the feature flatten operation. In this study, multiple one-dimensional convolution layers and pooling layers are cascaded to construct the simplified decoder, which is represented as follows:
where Ode represents output feature of the simplified decoder. Finally, the reconstructed feature is obtained as follows:
where is the output reconstructed feature and FC indicates the fully connected layer.
2.3 Mangrove biomass estimation
In the stage of fine-tuning, the biomass estimator is constructed based on two fully connected layers for mangrove biomass estimation, as follows:
where denotes the estimated mangrove biomass and FCi indicates the ith fully connection layer.
2.4 Loss function of SSDFRN
In this study, the training process of SSDFRN consists of two stages: DFRSSL and fine-tuning. In the stage of DFRSSL, the loss of SSDFRN is calculated as follows:
where Ls1 is the loss of SSDFRN in the DFRSSL stage; and denote the reconstructed value and actual value of the ith feature for the jth mangrove sample, respectively; and M is the total number of samples.
In the stage of fine-tuning, the loss of SSDFRN is calculated as follows:
where Ls2 is the loss of SSDFRN in the fine-tuning stage, and and represent the estimated mangrove biomass and the actual mangrove biomass of the jth sample, respectively. The training process of SSDFRN is shown in Table 1.
Table 1
| Training of SSDFRN |
|---|
| Input: x: 22 features extracted from Landsat 8 remote sensing image and DEM elevation data; |
| y: actual mangrove biomass value. |
| Init parameters of SSDFRN. |
| Stage 1: DFRSSL |
| For each training epoch: |
| Randomly select W features from x and record the position P by Equation 1; |
| Generate auxiliary data by Equation 2; |
| Combine remaining features as the residual data xres by Equation 3; |
| Obtain the hidden representation OMVCNN from xres by Equations 4–8; |
| Combine auxiliary data and hidden representation OMVCNN by Equation 9; |
| Obtain reconstructed feature by Equations 10, 11. |
| Compute loss of SSDFRN in the stage of DFRSSL by Equation 13; |
| Update parameters of SSDFRN. |
| End |
| Stage 2: Fine-tuning |
| For each training epoch: |
| Obtain deep features by MVCNN optimized in stage 1; |
| Obtain estimated mangrove biomass by Equation 12; |
| Compute mangrove biomass estimation error by Equation 14; |
| Update parameters of mangrove biomass estimator. |
| End |
Training process of SSDFRN.
SSDFRN, self-supervised disturbing feature reconstruction network; DFRSSL, disturbing feature reconstruction-based self-supervised learning; MVCNN, multi-view convolutional neural network.
3 Experimental analysis
In this section, the mangrove biomass dataset obtained from Ximen Island (28° 21′ N, 121° 10′ E) is used for verifying the effectiveness of SSDFRN in mangrove biomass estimation. The research area is located north of Wenzhou City, which experiences a subtropical oceanic monsoon climate. The study area is mild and humid throughout the year, with abundant rainfall. The experiment hardware environment is as follows: CPU, Intel® i7-10875H; GPU, RTX2060 6G. The software environment is as follows: programming language, Python3.7; compiler, Pycharm2022.3.2; framework, Tensorflow-gpu2.6.0+Cuda10.0.
3.1 Experimental description
3.1.1 Data description
Ximen Island covers a land area of 6.98 km2 and a mudflat area of 15.11 km2. The average annual temperature is approximately 18.3°C, with an annual precipitation of 1,595.7 mm and an average annual sunshine duration of 1,714.6 hours. The mangrove wetland in this region serves as the ecological restoration project area for the coastal mangrove wetlands of Leqing City, encompassing an area of approximately 28.96 hectares for mangrove planting and 4.57 hectares for introduced mangrove maintenance (Hao et al., 2024). The plant species in this area is the Kandelia obovata. The location of the study area and sample distribution are shown in Figure 4. Landsat 8 remote sensing image data (with a spatial resolution of 30 m) from August 2022 are used in this study to map the mangrove biomass. The 21 features, including band information, vegetation indexes, texture features, and elevation features, are extracted from Landsat 8 remote sensing images. In addition, DEM data are derived from Aster GDEM with a resolution of 30 m, which is used to extract the altitude index (i.e., topographic factor). All 22 input features are presented in Table 2.
Figure 4
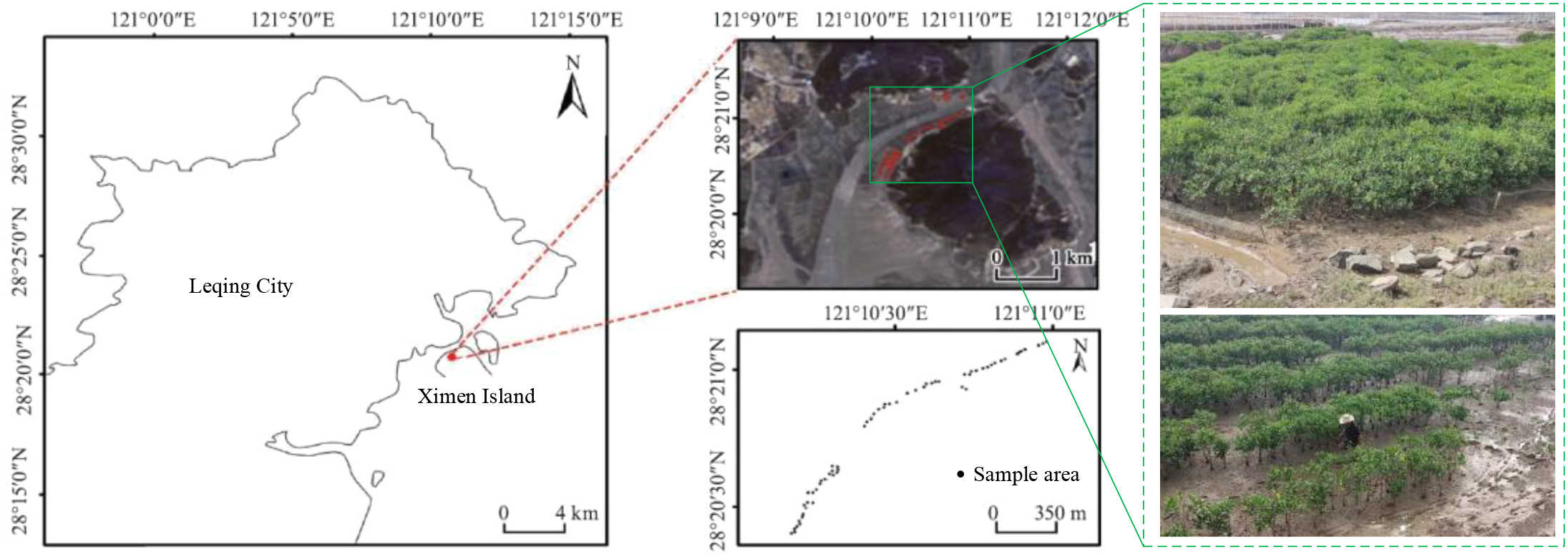
The specific location and distribution of the study area (Hao et al., 2024).
Table 2
| Data type | Feature description | Detailed features |
|---|---|---|
| Landsat 8 remote sensing data | Band | Coastal band, super blue infrared, sum green index, red band, near-infrared wave, short-wave infrared 1, short-wave infrared 2 |
| Vegetation indexes | Normalized difference vegetation index (NDVI), ratio vegetation index (RVI), difference vegetation index (DVI), soil-adjusted vegetation index (SAVI), enhanced vegetation index (EVI), green normalized difference vegetation index (GNDVI) | |
| Texture features | Variance (VAR), homogeneity (HOM), contrast (CON), heterogeneity (HET), entropy (ENT), angular second moment (ASM), correlation (COR), mean (MEA) | |
| Digital elevation model data | Altitude index | Topographic factor (TOF) |
Details about all 22 input features.
The biomass equation (Equation 15) based on the stem diameter of the near-ground branches at the base of Kandelia candel is used to calculate the biomass:
where y is the total biomass of kandelia samples and D is the branch trunk diameter near the ground of kandelia.
3.1.2 Parameter setting
The network structure and parameters of SSDFRN are listed in Table 3, where W is the length of the shuffle window; V1, V2, and V3 are convolution kernel size of small-, medium-, and large-view branches, respectively; S1, S2, and S3 represent different sliding strides; K refers to the number of convolution kernels; PW and PS are the pooling window and the pooling stride, respectively; N is the number of neurons; and Vde and Sde are decoder convolution kernel size and convolution stride, respectively.
Table 3
| Module | Structure | Parameters |
|---|---|---|
| Input | – | – |
| Feature disturbing | – | W = 4 |
| MVCNN | MVCCM 1 | V 1 = 2, V2 = 3, V3 = 4, S1 = 1, S2 = 2, S3 = 3, K = 16 |
| Pooling 1 | PW = 2, PS = 2 | |
| MVCCM 2 | V 1 = 2, V2 = 3, V3 = 4, S1 = 1, S2 = 2, S3 = 3, K = 32 | |
| Pooling 2 | PW = 2, PS = 2 | |
| Flatten | Fully connected layer | N = 928 − 18 |
| Simplified decoder | Convolution 1 | K = 16, Vde = 3, Sde = 1 |
| Pooling 1 | PW = 2, PS = 2 | |
| Convolution 2 | K = 32, Vde = 3, Sde = 1 | |
| Pooling 2 | PW = 2, PS = 2 | |
| Biomass estimator | Fully connected layer | N = 200 − 1 |
| Batch size = 20, learning rate = 0.0001 | ||
Detailed structure and parameters of SSDFRN.
SSDFRN, self-supervised disturbing feature reconstruction network; MVCCM, multi-view cascaded convolution module.
3.2 Experimental result
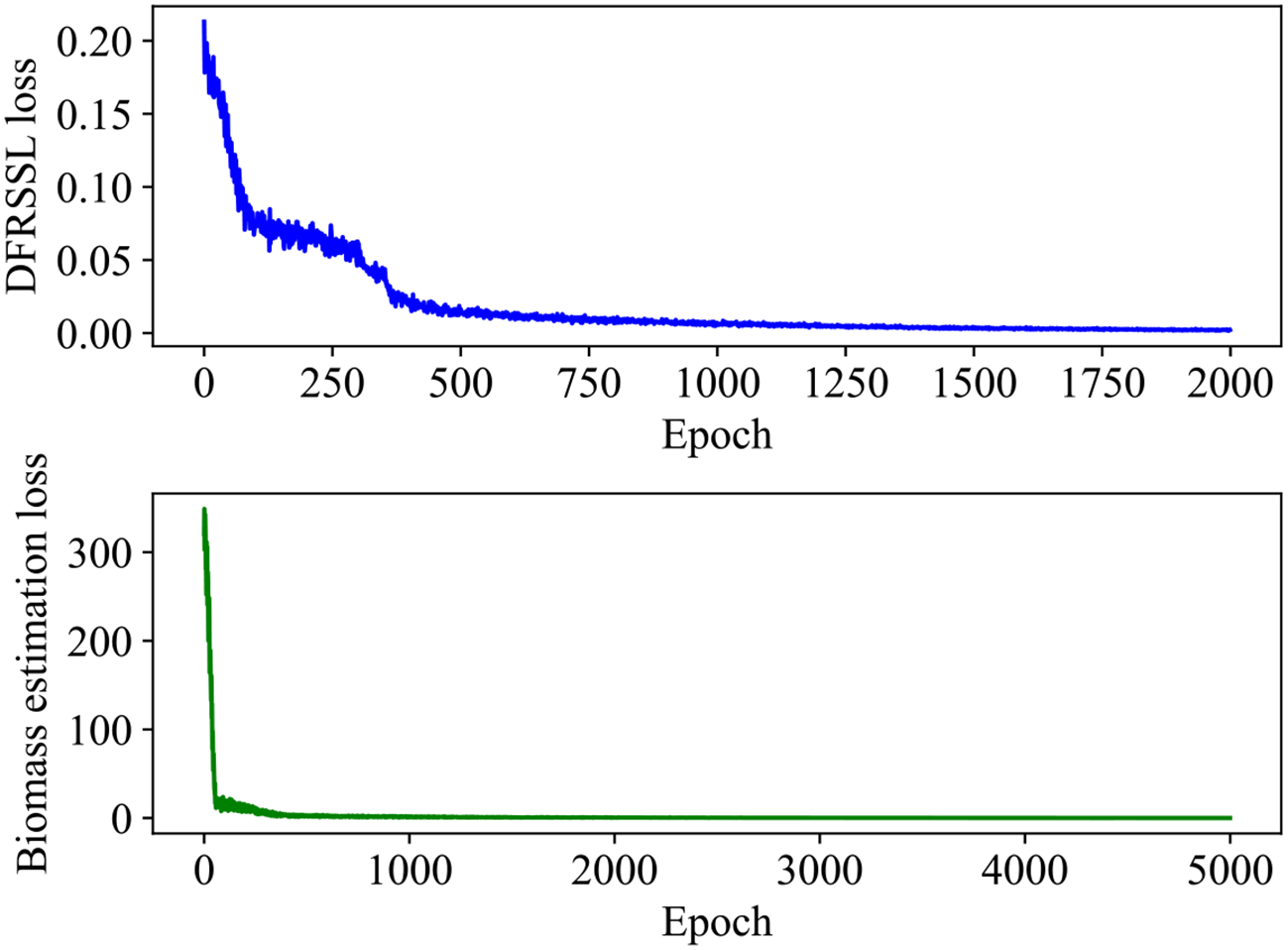
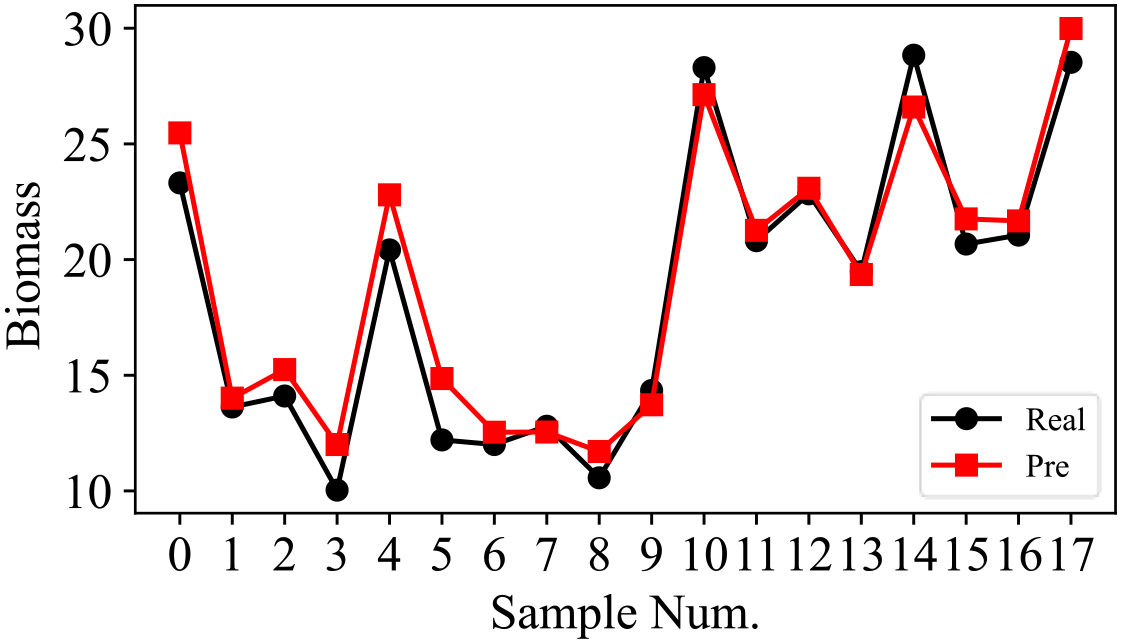
In this study, a total of 58 samples are considered in the experiment, where 70% of the samples are used for training and the remaining 30% of the samples are used for testing. Details about the dataset are presented in Table 4. The training process of SSDFRN is shown in Figure 5. It is clear that with the increase in training epochs, the DFRSSL loss and mangrove biomass estimation loss are decreasing. When the training epoch reaches 1,000, all losses are nearly zero, which means that SSDFRN exhibits excellent biomass estimation performance on the training set. The mangrove biomass estimation results on the testing set are shown in Figure 6. The detailed estimation results are presented in Table 5, where AE, MAE, and RMSE denote the absolute error, mean absolute error, and root mean square error, respectively. It is obvious that the estimated mangrove biomass values are close to the actual mangrove biomass values, which indicates the outperformance of SSDFRN on deep feature learning and mangrove biomass estimation with limited samples.
Table 4
| Samples | Sample number | Geographical position | Data shape |
|---|---|---|---|
| Total samples | 58 | (28° 21′ N, 121° 10′ E) | [1×22] |
| Training | 40 | ||
| Testing | 18 |
Detailed information about the mangrove biomass dataset.
Figure 5
The training process of SSDFRN. SSDFRN, self-supervised disturbing feature reconstruction network.
Figure 6
The SSDFRN-based mangrove biomass estimation results of testing samples. SSDFRN, self-supervised disturbing feature reconstruction network.
Table 5
| Biomass | Sample Num. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| Actual | 23.314 | 13.628 | 14.103 | 10.036 | 20.418 | 12.205 | 12.006 | 12.796 | 10.568 | 14.34 | 28.298 | 20.841 | 22.841 | 19.479 | 28.834 | 20.671 | 21.057 | 28.526 |
| Estimation | 25.475 | 14.01 | 15.244 | 12.005 | 22.799 | 14.859 | 12.537 | 12.546 | 11.705 | 13.724 | 27.128 | 21.265 | 23.081 | 19.353 | 26.593 | 21.758 | 21.673 | 29.987 |
| AE | 2.161 | 0.382 | 1.141 | 1.969 | 2.381 | 2.654 | 0.531 | 0.25 | 1.137 | 0.616 | 1.17 | 0.424 | 0.24 | 0.126 | 2.241 | 1.087 | 0.616 | 1.461 |
| MAE | 1.145 | |||||||||||||||||
| RMSE | 1.396 | |||||||||||||||||
Detailed testing results.
AE, absolute error; MAE, mean absolute error; RMSE, root mean square error.
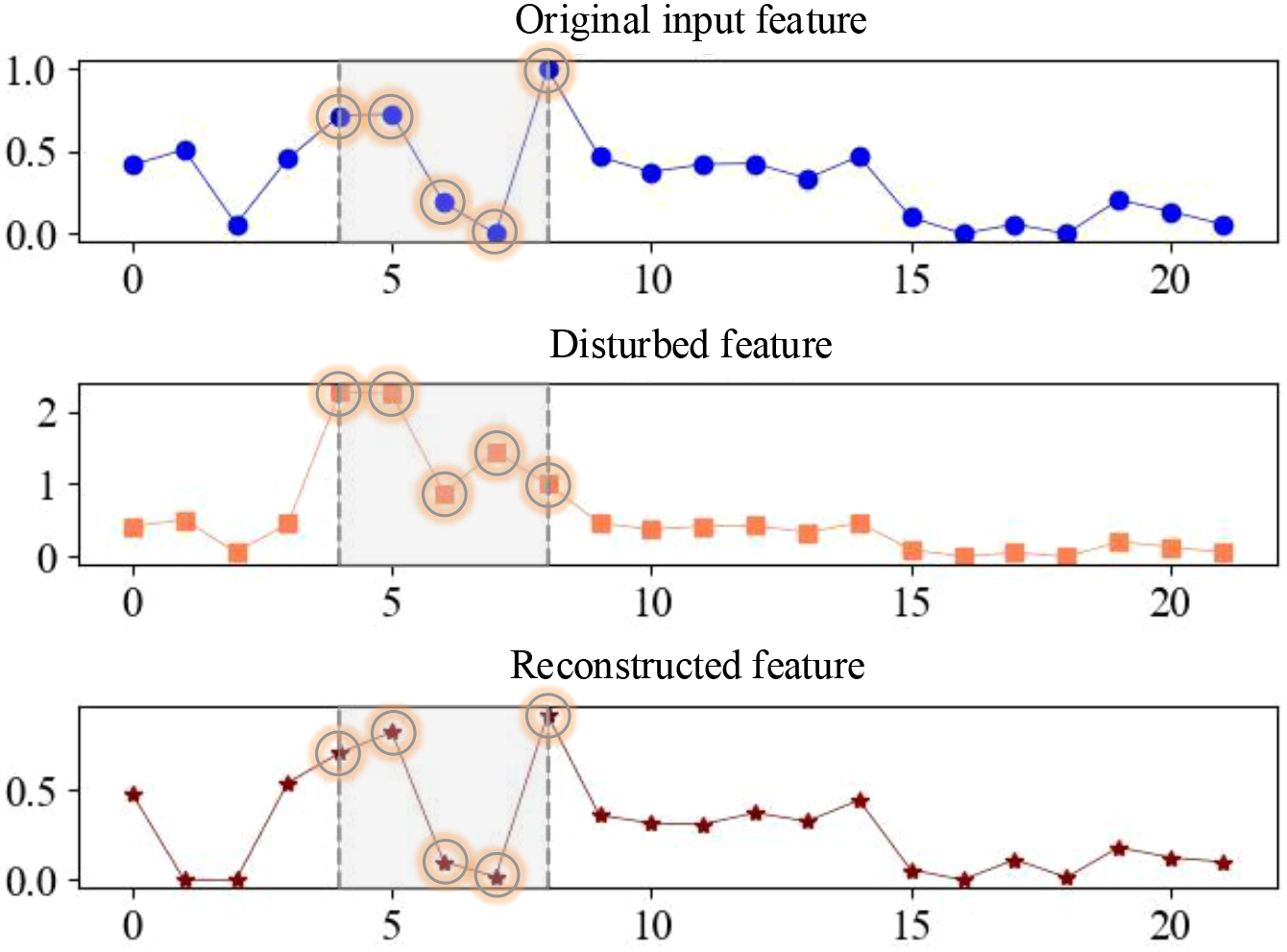
In order to verify the effectiveness of DFRSSL, the original input features, disturbed features, and the corresponding reconstructed features obtained by DFRSSL are visualized in this study, as shown in Figure 7. It can be found that parts with original input features are shuffled and drowned by strong noise (gray parts) after the feature disturbance. After DFRSSL, noise features are greatly reconstructed by SSDFRN, which demonstrates the generalizability of SSDFRN under external interference. It indicates that SSDFRN is good at deep feature learning from limited samples based on DFRSSL.
Figure 7
The visualization of input features, disturbed features, and reconstructed features.
3.3 Ablation study
In this section, the ablation study is implemented to verify the effectiveness of DFRSSL and MVCNN. The description of different tasks is given in Table 6, where “√” and “X” denote that the corresponding module is included and excluded in the task, respectively. The testing results are shown in the last two columns of Table 6. It is clear that the mangrove biomass estimation errors (i.e., MAE and RMSE) increase significantly when DFRSSL or MVCNN is removed from SSDFRN. It indicates that DFRSSL and MVCNN greatly enhance the feature learning and mangrove biomass estimation performance of SSDFRN.
Table 6
| Task no. | Modules | Results | ||
|---|---|---|---|---|
| DFRSSL | MVCCM | MAE | RMSE | |
| T1 | ✓ | ✓ | 1.145 | 1.396 |
| T2 | ✓ | X | 1.367 | 1.660 |
| T3 | X | ✓ | 1.250 | 1.626 |
Different tasks in ablation study.
DFRSSL, disturbing feature reconstruction-based self-supervised learning; MVCCM, multi-view cascaded convolution module; MAE, mean absolute error; RMSE, root mean square error.
3.4 Result comparison and discussion
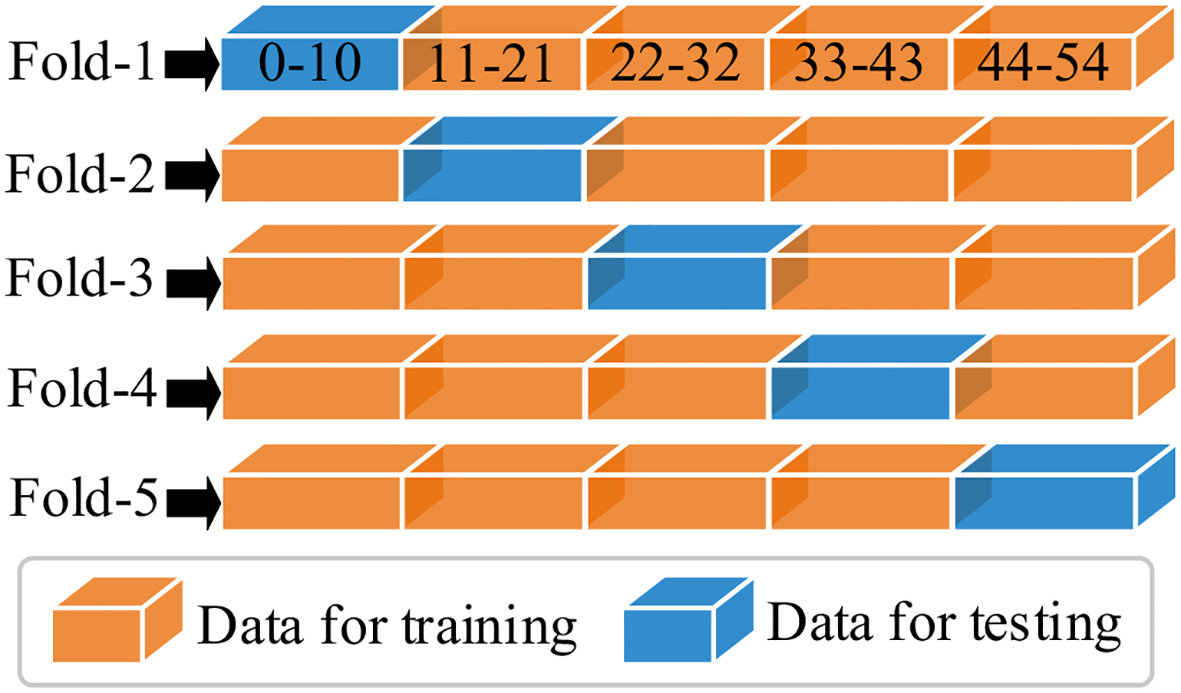
In this section, the feature learning and mangrove biomass estimation performance of SSDFRN are compared with that of state-of-the-art methods [i.e., SVR (Luo et al., 2024), CNN (Nakajima et al., 2023), RNN (Song and Wang, 2023), BiLSTM (Zhang et al., 2024), ResNet (Liu et al., 2024), multi-branch convolutional neural network (MBCNN) (Zhao et al., 2021), and densely connected convolutional network (DenseNet) (Huang et al., 2017)]. As presented in Figure 8, fivefold cross-validation is used for data separation, where 80% of the original samples are used for training and the remaining 20% of the samples are used for testing. The computational efficiency analysis is implemented for different DNNs, and the comparison results are listed in Table 7. It is evident that the running time of SSDFRN is a little higher than that of other methods, and the parameter complexity and memory occupation are comparable to those of most methods, which is satisfactory for practical applications. The comparison results based on the fivefold cross-validation method are shown in Table 8. Taking fold-5 as an example, the mangrove biomass estimation results are given in Figure 9, and the estimation errors are shown in Figure 10. It is obvious that the mangrove biomass values estimated by SSDFRN are closer to the actual values compared with those of other methods. In particular, the mangrove biomass estimation error of SSDFRN on samples 6 and 10 is significantly smaller than that of other methods, which demonstrates the outperformance of SSDFRN on deep feature learning and mangrove biomass estimation with limited samples.
Figure 8
Data separation for different folds.
Table 7
| Model | Runtime (s) | Parameter complexity (Mega Floating-Point Operations Per Second (MFLOPs)) | Memory footprint (MB) | GPU utilization (%) |
|---|---|---|---|---|
| SSDFRN | 8.71 | 10.54 | 2,242.58 | 31.55 |
| CNN | 6.89 | 6.72 | 2,203.80 | 10.50 |
| RNN | 6.62 | 4.09 | 1,403.93 | 14.36 |
| LSTM | 7.27 | 10.75 | 1,424.23 | 18.84 |
| MBCNN | 7.36 | 10.17 | 2,209.74 | 20.33 |
| ResNet | 7.33 | 7.17 | 2,211.09 | 21.87 |
| DenseNet | 7.35 | 10.97 | 2,204.02 | 17.00 |
Computational efficiency comparison of different DNNs.
DNNs, deep neural networks; SSDFRN, self-supervised disturbing feature reconstruction network; CNN, convolutional neural network; RNN, recurrent neural network; LSTM, long short-term memory; MBCNN, multi-branch convolutional neural network; ResNet, residual neural network; DenseNet, densely connected convolutional network.
Table 8
| Method | Fold-1 | Fold-2 | Fold-3 | Fold-4 | Fold-5 | Avg | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | MAE | RMSE | |
| SSDFRN | 1.236 | 1.535 | 1.231 | 1.632 | 1.007 | 1.201 | 1.281 | 1.487 | 1.123 | 1.467 | 1.176 | 1.464 |
| SVM | 2.146 | 2.323 | 2.83 | 3.675 | 2.233 | 2.86 | 2.32 | 2.905 | 2.574 | 3.571 | 2.421 | 3.067 |
| CNN | 1.352 | 1.776 | 1.599 | 1.878 | 1.6 | 1.889 | 1.667 | 1.997 | 1.921 | 2.391 | 1.628 | 1.986 |
| RNN | 1.753 | 1.863 | 2.204 | 2.832 | 1.352 | 1.518 | 1.877 | 2.368 | 2.281 | 2.997 | 1.893 | 2.316 |
| LSTM | 1.345 | 1.572 | 1.716 | 2.208 | 1.643 | 1.872 | 1.55 | 1.766 | 1.53 | 2 | 1.557 | 1.884 |
| MBCNN | 1.663 | 1.958 | 1.911 | 2.298 | 1.124 | 1.353 | 1.659 | 1.956 | 2.302 | 2.916 | 1.732 | 2.096 |
| ResNet | 1.459 | 1.677 | 1.745 | 2.185 | 1.092 | 1.330 | 1.497 | 1.637 | 1.666 | 2.117 | 1.492 | 1.789 |
| DenseNet | 1.321 | 1.605 | 1.706 | 2.037 | 1.468 | 1.782 | 1.528 | 1.738 | 1.430 | 1.766 | 1.491 | 1.786 |
Fivefold cross-validation-based comparison results.
MAE, mean absolute error; RMSE, root mean square error; SSDFRN, self-supervised disturbing feature reconstruction network; SVM, support vector machine; RNN, recurrent neural network; LSTM, long short-term memory; MBCNN, multi-branch convolutional neural network; ResNet, residual neural network; DenseNet, densely connected convolutional network.
Figure 9
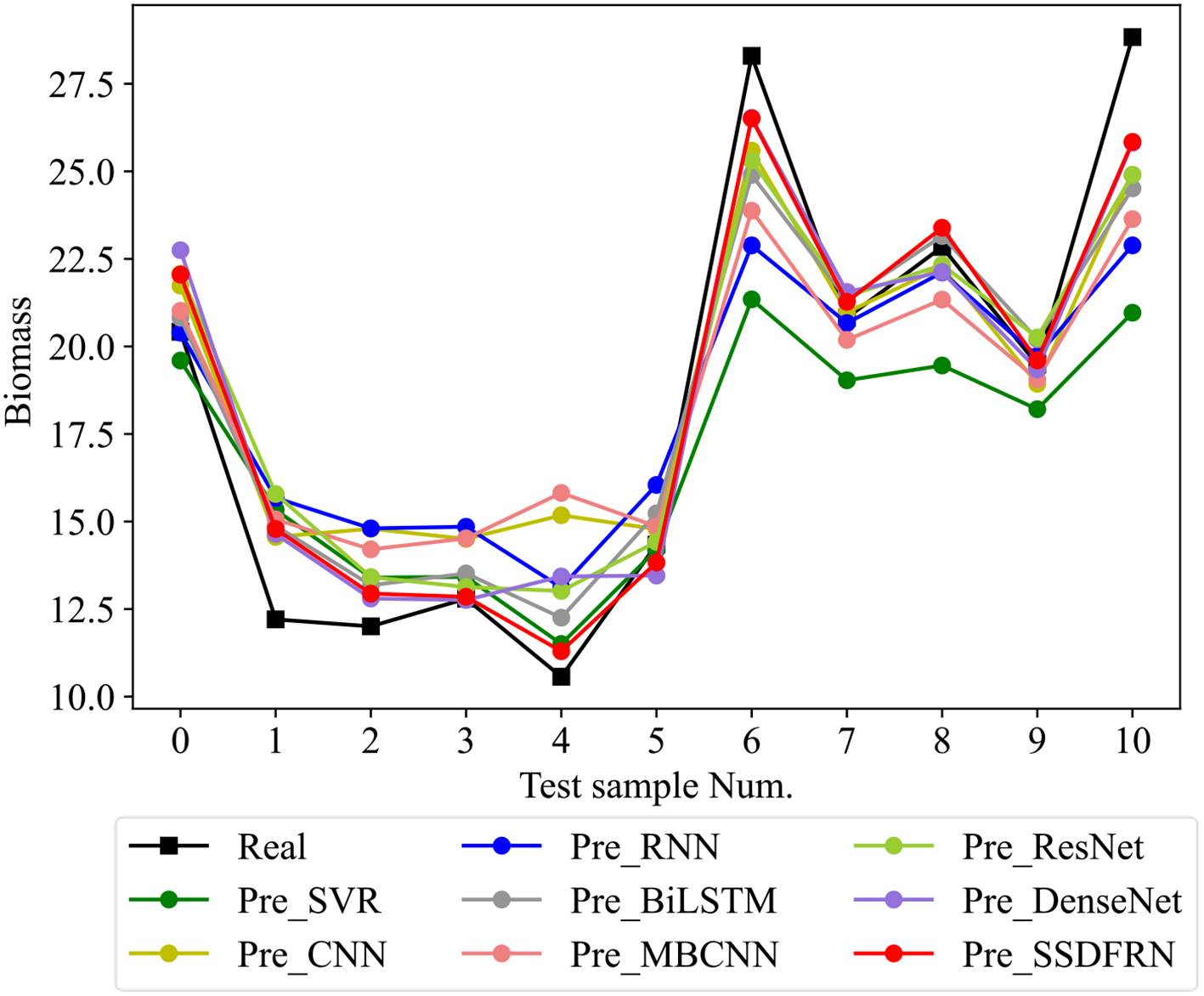
Mangrove biomass estimation results of different methods.
Figure 10
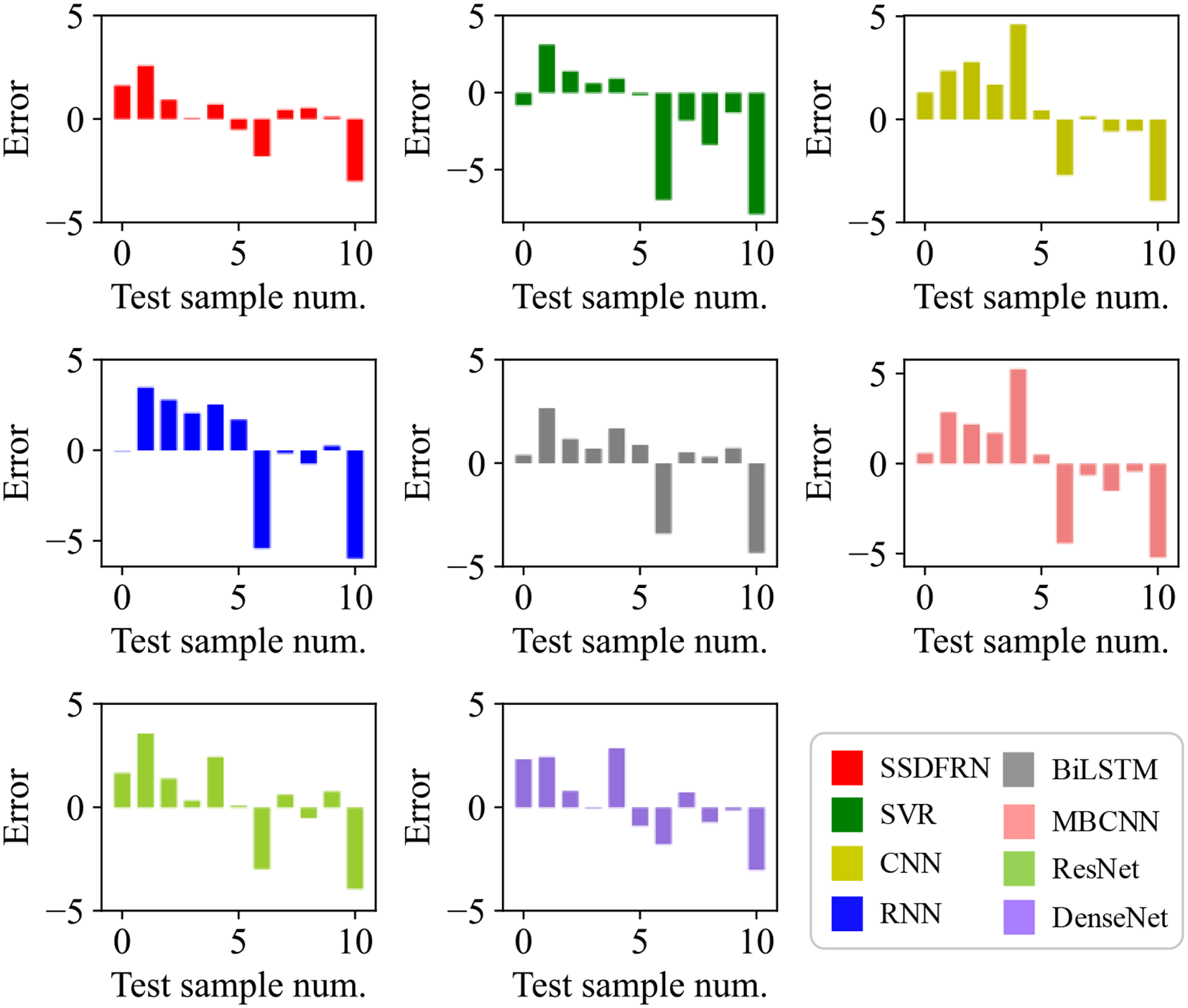
Mangrove biomass estimation errors.
4 Conclusions
In this study, a novel DNN, i.e., SSDFRN, is developed for mangrove biomass estimation with limited data. DFRSSL is implemented by random feature shuffle and disturbing feature reconstruction, which effectively solves the key problem of data scarcity. In particular, the shuffle window is masked on partial input features randomly for generating sufficient auxiliary data and residual data. The network is pre-trained by disturbing feature reconstruction for deep feature learning using sufficient auxiliary data and residual data. In addition, a novel feature extractor, i.e., MVCNN, is constructed by stacking several MVCCMs, which effectively enhances the feature learning performance and improves the mangrove biomass estimation accuracy. The outperformance of SSDFRN is verified on the mangrove biomass dataset obtained from Ximen Island (28° 21′ N, 121° 10′ E). The testing results illustrate that SSDFRN can effectively perform deep feature learning and mangrove biomass estimation with limited data. However, the hyperparameters of SSDFRN are determined manually in this study, which is inconvenient and needs to be improved in the future. Moreover, the length of the masking window in SSDFRN is fixed, which may result in the excessive destruction of key features and may increase the difficulty of feature learning. Concurrently, the contribution of non-key features to the masking operation is inefficient, which requires further study. The future work will strive to collect more data from different study areas to underpin subsequent research and focus on improving the generalization performance of SSDFRN for different mangrove species and geographical positions simultaneously.
Statements
Data availability statement
The datasets presented in this article are not readily available because they are confidential. Requests to access the datasets should be directed to the corresponding author.
Author contributions
JH: Writing – original draft. XX: Writing – original draft, Conceptualization, Formal analysis. HX: Writing – review & editing, Funding acquisition. GX: Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was supported by the Open Fund of Wenzhou Future City Research Institute (No. WL2023006), Wenzhou Science and Technology Bureau (No. S2023030), and Zhejiang Province Department of Education (No. Y202351948).
Acknowledgments
All researchers would like to express their gratitude to all the participants for taking their precious time to participate in this study. They also thank Mengqi Miao and Jianbo Yu for the idea discussions, verification, and data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Akkem Y. Biswas S. Varanasi A. (2023). Smart farming using artificial intelligence: A review. Eng. Appl. Artif. Intell.120, 105899. doi: 10.1016/j.engappai.2023.105899
2
Bui Q. Pham Q. Pham V. Tran V. Nguyen D. Nguyen Q. et al . (2024). Hybrid machine learning models for aboveground biomass estimations. Ecol. Inf.79, 102421. doi: 10.1016/j.ecoinf.2023.102421
3
Chen C. Ma Y. Ren G. Wang J. (2022). Aboveground biomass of salt-marsh vegetation in coastal wetlands: Sample expansion of in situ hyperspectral and Sentinel-2 data using a generative adversarial network. Remote Sens. Environ.270, 112885. doi: 10.1016/j.rse.2021.112885
4
Do A. Tran H. Ashley M. Nguyen A. (2022). Monitoring landscape fragmentation and aboveground biomass estimation in Can Gio Mangrove Biosphere Reserve over the past 20 years. Ecol. Inf.70, 101743. doi: 10.1016/j.ecoinf.2022.101743
5
Gao L. Chai G. Zhang X. (2022). Above-ground biomass estimation of plantation with different tree species using airborne LiDAR and hyperspectral data. Remote Sens.14, 2568. doi: 10.3390/rs14112568
6
Hao J. Lyu K. Hu T. Wang Y. Xu G. (2024). Remote sensing inversion of mangrove biomass based on machine learning. For. Grassland Resour. Res.01, 65–72. doi: 10.13466/j.cnki.lczyyj.2024.01.009
7
Hazmy A. Hawbani A. Wang X. Al-Dubai A. Ghannami A. Yahya A. et al . (2024). Potential of satellite-airborne sensing technologies for agriculture 4.0 and climate-resilient: A review. IEEE Sensors J.24, 4161–4180. doi: 10.1109/JSEN.2023.3343428
8
He K. Chen X. Xie S. Li Y. Dollar P. Girshick R. (2022). Masked autoencoders are scalable vision learners,” in 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR 2022). New Orleans, LA, 15979-15988. IEEE Computer Society. doi: 10.1109/CVPR52688.2022.01553
9
He Y. Yin H. Chen Y. Xiang R. Zhang Z. Chen H. (2024). Soil salinity estimation based on sentinel-1/2 texture features and machine learning. IEEE Sensors J.24, 15302–15310. doi: 10.1109/JSEN.2024.3377682
10
Hu H. Zhou H. Cao K. Lou W. Zhang G. Gu Q. et al . (2024). Biomass estimation of milk vetch using UAV hyperspectral imagery and machine learning. Remote Sens.16, 2183. doi: 10.3390/rs16122183
11
Huang G. Liu Z. Maaten L. Weinberger K. (2017). “Densely connected convolutional networks,” in 30th Proceedings of the IEEE conference on computer vision and pattern recognition, Honolulu, HI, IEEE Computer Society. 2261–2269. doi: 10.1109/CVPR.2017.243
12
Li Z. He Q. Li J. (2024). A survey of deep learning-driven architecture for predictive maintenance. Eng. Appl. Artif. Intell.133, 108285. doi: 10.1016/j.engappai.2024.108285
13
Li R. Sun C. Liu Y. Mei Y. Tan J. (2023). Prediction of the parallelism error and unbalance of aero-engine rotors based on intelligent algorithm. IEEE Trans. Instrumentation Measurement72, 1006510. doi: 10.1109/TIM.2023.3289542
14
Liu T. Yang T. Zhu S. Mou N. Zhang W. Wu W. et al . (2024). Estimation of wheat biomass based on phenological identification and spectral response. Comput. Electron. Agric.222, 109076. doi: 10.1016/j.compag.2024.109076
15
Luo H. Qin S. Li J. Lu C. Yue C. Ou G. (2024). High-density forest AGB estimation in tropical forest integrated with PolInSAR multidimensional features and optimized machine learning algorithms. Ecol. Indic.160, 111878. doi: 10.1016/j.ecolind.2024.111878
16
Miao M. Yu J. (2024). Deep feature interactive network for machinery fault diagnosis using multi-source heterogeneous data. Reliability Eng. System Saf.242, 109795. doi: 10.1016/j.ress.2023.109795
17
Morais T. Teixeira R. Figueiredo M. Domingos T. (2021). The use of machine learning methods to estimate aboveground biomass of grasslands: A review. Ecol. Indic.130, 108081. doi: 10.1016/j.ecolind.2021.108081
18
Muhd-Ekhzarizal M. E. Mohd-Hasmadi I. Hamdan O. Mohamad-Roslan M. K. Noor-Shaila S. (2018). Estimation of aboveground biomass in mangrove forests using vegetation indices from SPOT-5 image. J. Trop. For. Sci.30, 224–233. doi: 10.26525/jtfs2018.30.2.224233
19
Nakajima K. Tanaka Y. Katsura K. Yamaguchi T. Watanabe T. Shiraiwa T. (2023). Biomass estimation of World rice (Oryza sativa L.) core collection based on the convolutional neural network and digital images of canopy. Plant Production Sci.26, 187–196. doi: 10.1080/1343943X.2023.2210767
20
Pandey P. Anand A. Srivastava P. (2019). Spatial distribution of mangrove forest species and biomass assessment using field inventory and earth observation hyperspectral data. Biodiversity Conserv.28, 2143–2162. doi: 10.1007/s10531-019-01698-8
21
Rahimikhoob H. Delshad M. Habibi R. (2023). Leaf area estimation in lettuce: Comparison of artificial intelligence-based methods with image analysis technique. Measurement222, 113636. doi: 10.1016/j.measurement.2023.113636
22
Rijal S. Pham T. Noer’Aulia S. Putera M. Saintilan N. (2023). Mapping mangrove above-ground carbon using multi-source remote sensing data and machine learning approach in Loh Buaya, Komodo National Park, Indonesia. Forest14, 94. doi: 10.3390/f14010094
23
Selvaraj J. Perez B. (2023). Estimating mangrove aboveground biomass in the Colombian Pacific coast: A multisensor and machine learning approach. Heliyon9, e20745. doi: 10.1016/j.heliyon.2023.e20745
24
Sliva G. Neves J. Marcatti G. Soares C. Calegario N. Araujo Junior C. et al . (2024). Use of artificial neural networks with the physiological principles to predict growth model. Eng. Appl. Artif. Intell.136, 108914. doi: 10.1016/j.engappai.2024.108914
25
Song Y. Wang Y. (2023). A big-data-based recurrent neural network method for forest energy estimation. Sustain. Energy Technol. Assessments55, 102910. doi: 10.1016/j.seta.2022.102910
26
Talebiesfandarani S. Shamsoddini A. (2022). Global-scale biomass estimation based on machine learning and deep learning methods. Remote Sens. Applications-Society Environ.28, 100868. doi: 10.1016/j.rsase.2022.100868
27
Teshome F. Bayabil H. Hoogenboom G. Schaffer B. Singh A. Ampatzidis Y. (2023). Unmanned aerial vehicle (UAV) imaging and machine learning applications for plant phenotyping. Comput. Electron. Agric.212, 108064. doi: 10.1016/j.compag.2023.108064
28
Tian Y. Huang H. Zhou G. Zhang Q. Tao J. Zhang Y. et al . (2021). Aboveground mangrove biomass estimation in Beibu Gulf using machine learning and UAV remote sensing. Sci. Total Environ.781, 146816. doi: 10.1016/j.scitotenv.2021.146816
29
Tian X. Li J. Zhang F. Zhang H. Jiang M. (2024). Forest aboveground biomass estimation using multisource remote sensing data and deep learning algorithms: A case study over hangzhou area in China. Remote Sens.16, 1074. doi: 10.3390/rs16061074
30
Tian Y. Zhang Q. Huang H. Huang Y. Tao J. Zhou G. et al . (2022). Aboveground biomass of typical invasive mangroves and its distribution patterns using UAV-LiDAR data in a subtropical estuary: Maoling River estuary. Guangxi China Ecol. Indic.136, 108694. doi: 10.1016/j.ecolind.2022.108694
31
Tran T. Reef R. Zhu X. (2022). A review of spectral indices for mangrove remote sensing. Remote Sens.14, 4868. doi: 10.3390/rs14194868
32
Xiao Y. Jin M. Qi G. Shi W. Li K. Du X. (2024). Interpreting the influential factors in ship detention using a novel random forest algorithm considering dataset imbalance and uncertainty. Eng. Appl. Artif. Intell.133, 108369. doi: 10.1016/j.engappai.2024.108369
33
Zhang Y. Li M. Li G. Li J. Zheng L. Zhang M. et al . (2022). Multi-phenotypic parameters extraction and biomass estimation for lettuce based on point clouds. Measurement204, 112094. doi: 10.1016/j.measurement.2022.112094
34
Zhang J. Zhou C. Zhang G. Yang Z. Pang and Y. Luo Z. (2024). A novel framework for forest above-ground biomass inversion using multi-source remote sensing and deep learning. Forests15, 456. doi: 10.3390/f15030456
35
Zhao W. Kang Y. Chen H. Zhao Z. Zhao Z. Zhai Y. (2023). Adaptively attentional feature fusion oriented to multiscale object detection in remote sensing images. IEEE Trans. Instrumentation Measurement72, 5008111. doi: 10.1109/TIM.2023.3246536
36
Zhao H. Mao Z. Zhang J. Zhang X. Zhao N. Jiang Z. (2021). Multi-branch convolutional neural networks with integrated cross-entropy for fault diagnosis in diesel engines. Measurement Sci. Technol.32, 045103. doi: 10.1088/1361-6501/abcefb
Summary
Keywords
mangrove biomass estimation, self-supervised learning, disturbed feature reconstruction, multi-view convolution neural network, deep learning
Citation
Hao J, Xu X, Xu H and Xu G (2025) Self-supervised disturbing feature reconstruction network for mangrove biomass estimation with limited data. Front. Plant Sci. 16:1623458. doi: 10.3389/fpls.2025.1623458
Received
22 May 2025
Accepted
31 July 2025
Published
03 September 2025
Volume
16 - 2025
Edited by
Christian Joshua Sanders, Southern Cross University, Australia
Reviewed by
Heng Dong, Fuzhou Institute of Technology, China
Zhenhui Sun, Tianjin Chengjian University, China
Updates
Copyright
© 2025 Hao, Xu, Xu and Xu.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Xu, 20096342@zjcst.edu.cn
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.