- 1Faculty of Agriculture, Forestry and Food Engineering, Yibin University, Yibin, Sichuan, China
- 2Crop Research Institute, Shandong Academy of Agricultural Sciences, Jinan, Shandong, China
- 3State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, Sichuan, China
- 4Key Laboratory of Aquatic Genomics, Ministry of Agriculture and Rural Affairs, Beijing Key Laboratory of Fishery Biotechnology, Chinese Academy of Fishery Sciences, Beijing, China
Actin-depolymerizing factors (ADFs) play crucial roles in cytoskeletal dynamics and stress adaptation in plants. In this study, we identified nine ADF genes (TbADF1 to TbADF9) in Triticum monococcum L. subsp. aegilopoides. Chromosomal distribution analysis revealed that these genes are unevenly distributed across five chromosomes, with evidence of tandem duplication events. Phylogenetic analysis clustered the TbADFs into four subfamilies, indicating evolutionary conservation among wheat relatives. Gene structure and motif analyses confirmed the presence of a conserved ADF domain. Additionally, promoter region analysis revealed a variety of cis-regulatory elements associated with hormone signaling and stress responses. Predictions of binding pockets and protein–protein interaction networks indicated potential functional sites and interactions with cytoskeletal regulators. Codon usage bias analysis showed a preference for GC-rich codons, which may enhance translation efficiency under stress. Codon usage bias analysis indicated GC-rich optimization, potentially enhancing translation efficiency under stress. Promoter methylation levels ranged from 0.0907 to 0.3053, suggesting that epigenetic regulation may contribute to the control of gene expression. Transcriptomic profiling across six tissues and under cold stress conditions (4°C for 24 hours) revealed both tissue-specific expression patterns and differential cold responses. Notably, TbADF1, TbADF4, TbADF6, and TbADF7 were upregulated, with TbADF6 exhibiting the strongest induction, as its TPM value increased from 29.07 to 300.01. Furthermore, co-expression and gene ontology enrichment analyses of the upregulated genes identified key biological pathways involved in membrane integrity, phosphorylation, ribosome maturation, and lipid signaling. These findings highlight the central role of TbADF6 in cold adaptation.
Introduction
Triticum monococcum L. subsp. aegilopoides is recognized as one of the earliest domesticated cereal crops, with its cultivation dating back approximately 10,000 years to the Fertile Crescent (Ahmed et al., 2023). As an ancient wheat species, it exhibits remarkable resilience compared to modern common wheat, particularly in its tolerance to abiotic stresses such as drought, low temperatures, and nutrient-deficient soil (Vanková et al., 2014). This resilience is largely attributed to its rich genetic diversity, making wild einkorn wheat a valuable resource for introgressing stress-tolerance traits into modern wheat cultivars (Liu et al., 2015; Tounsi et al., 2016; Zhang et al., 2021a). Although its cultivation has substantially declined in modern agriculture, wild einkorn wheat remains a critical component of global wheat germplasm collections. It provides essential genetic material for breeding programs aimed at improving yield stability and environmental adaptability, particularly in the face of ongoing climate change (Mondal et al., 2016).
Actin-depolymerizing factor (ADF) is a highly conserved, small actin-binding protein ubiquitously present in eukaryotic organisms. Initially identified in chicken brain tissue (Bamburg et al., 1980), ADF has since been found to be widely distributed across plant species (Inada, 2017). ADF proteins are central to regulating actin cytoskeleton dynamics by binding to actin monomers (G-actin) and filaments (F-actin), thereby promoting severing, depolymerization, and bundling (Bernstein and Bamburg, 2010; Du et al., 2016). These activities are essential for maintaining cellular structure and support diverse physiological processes, including cell polarity, proliferation, division, intracellular trafficking, signal transduction, and pathogen recognition (Drøbak et al., 2004). Furthermore, ADFs play important roles in plant responses to abiotic stresses such as cold and drought, underscoring their significance in enhancing plant adaptability and resilience under adverse environmental conditions.
Low-temperature stress is a major constraint on crop productivity, as it inhibits plant growth, delays development, and, in severe cases, causes tissue damage or regression, ultimately leading to yield losses (Song et al., 2017; Hassan et al., 2021). Increasing evidence indicates that ADF genes play a critical role in mediating plant responses to cold stress through dynamic regulation of the cytoskeleton. In maize, for instance, 15 ADF genes exhibit tissue-specific and stimulus-responsive expression patterns, suggesting functional diversification (Yang et al., 2024). In A. thaliana, knockout mutants lacking AtADF5 show significantly reduced survival under low-temperature conditions, highlighting the essential role of ADFs in cold tolerance (Zhang et al., 2021b). Similar patterns have been observed in wheat, where cold-tolerant cultivars display specific upregulation of ADF genes in response to low temperatures, whereas cold-sensitive varieties show minimal expression changes (Ouellet et al., 2001). A genome-wide analysis of wheat identified 25 ADF genes, seven of which are significantly regulated by cold stress. Notably, heterologous expression of the wheat gene TaADF16 in Arabidopsis enhanced cold tolerance, suggesting its potential utility in transgenic approaches to improve stress resilience (Xu et al., 2021).
Although modern wheat breeding has substantially increased yield and grain quality, it has also led to a significant reduction in the genetic diversity of cultivated varieties, often compromising stress-resistance alleles. To address this limitation, growing attention has been directed toward wheat wild relatives and landraces, such as wild einkorn wheat, which serve as reservoirs of untapped genetic variation (Riar et al., 2021). In this study, we utilized the reference genome of wild einkorn wheat to identify nine ADF genes. These genes were systematically characterized with respect to their gene structures, conserved motifs, and expression patterns across different tissues and under cold stress. Our findings not only enhance the understanding of ADF gene family evolution and function in early domesticated wheat but also offer valuable genomic resources for breeding programs targeting improved cold tolerance. Ultimately, these insights contribute to the broader goal of promoting sustainable wheat production under increasingly variable environmental conditions.
Materials and methods
Plant materials
Triticum monococcum L. subsp. aegilopoides, accession G52, an ancient and resilient crop with considerable potential for improving stress tolerance in modern wheat, was employed to investigate the role of ADF genes in response to cold stress. Seeds were kindly provided by Professor Lianquan Zhang (Sichuan Agricultural University). To break dormancy, seeds were stratified at 4°C for 24 hours, followed by germination at room temperature for 7 days. Subsequently, ten uniform seedlings were subjected to cold treatment at 4°C for 24 hours, while another set of ten seedlings was maintained under control conditions. Leaf tissues from both treated and control groups were collected, with three biological replicates per group. Samples were immediately flash-frozen in liquid nitrogen and stored at −80°C for subsequent analyses.
Identification of ADF genes in Triticum monococcum L. subsp. aegilopoides
The reference genome of Triticum monococcum L. subsp. aegilopoides TA299 was obtained from the DRYAD repository (https://doi.org/10.5061/dryad.v41ns1rxj). The ADF domain (Pfam accession: PF00241) was retrieved from the Pfam database (http://pfam.xfam.org/). A Hidden Markov Model (HMM) search was performed using HMMER v3.0 (Mistry et al., 2013) with a stringent E-value threshold of ≤ 1e−10 to identify candidate ADF protein sequences. The presence of the ADF domain in these sequences was further validated using the NCBI Conserved Domain Database (CDD; https://www.ncbi.nlm.nih.gov/cdd/), SMART (http://smart.embl-heidelberg.de/), and Pfam. Physicochemical properties of the identified TbADFs, including molecular weight, isoelectric point, and instability index, were calculated using the ExPASy ProtParam tool (https://web.expasy.org/protparam/). Subcellular localization of TbADFs were predicted using WoLF PSORT (https://wolfpsort.hgc.jp/) and Cell-PLoc 2.0(http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/).
Chromosomal localization, phylogenetic analysis, and collinearity analysis of the TbADFs
Positional information for ADF genes was extracted from the GFF3 annotation file of the Triticum monococcum L. subsp. aegilopoides reference genome. Chromosomal locations of the identified ADF genes were visualized using TBtools v2.097 (Chen et al., 2023). A phylogenetic tree was constructed based on full-length ADF protein sequences from eight plant species—Arabidopsis thaliana, Aegilops tauschii, Oryza sativa, Zea mays, Triticum urartu, Triticum turgidum, Triticum aestivum, and Triticum monococcum L. subsp. aegilopoides—using the Maximum Likelihood (ML) method in IQ-TREE v2.0 (Nguyen et al., 2015). The best-fit amino acid substitution model was determined automatically using ModelFinder within IQ-TREE, and tree robustness was assessed with 1,000 bootstrap replicates. To investigate evolutionary relationships and genomic conservation, collinearity analysis between Triticum monococcum L. subsp. aegilopoides and three related wheat species (T. urartu, T. turgidum, and T. aestivum) was performed using MCScanX (Wang et al., 2012) with default parameters.
Analysis of conserved motifs and cis-acting elements in the TbADFs
Gene structure information for the ADF genes was extracted from the GFF3 annotation file of the Triticum monococcum L. subsp. aegilopoides reference genome. Conserved motifs within the TbADFs protein sequences were identified using the MEME Suite (http://meme-suite.org/meme), with the maximum number of motifs set to 10 and other parameters kept at default settings. To investigate potential regulatory mechanisms, the 2,000 bp upstream regions from the transcription start sites of the TbADFs were extracted as putative promoter sequences. These promoter regions were analyzed for cis-acting regulatory elements using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Protein 3D structure modeling and binding pocket prediction
The tertiary structure of the TbADFs were predicted using AlphaFold2. The predicted 3D structure was subsequently visualized and analyzed using PyMOL v2.5.5 (DeLano, 2002), facilitating detailed examination of the protein’s spatial conformation. To further investigate the surface topology and potential functional sites, the CASTp 3.0 web server (http://sts.bioe.uic.edu/castp/index.html?2pk9) (Tian et al., 2018) was utilized to identify and characterize surface pockets and cavities at multiple scales, offering insights into possible ligand-binding regions and structural features relevant to TbADFs.
Codon usage bias was assessed using CodonW (version 1.4.2; http://codonw.sourceforge.net/), with several key indices calculated to characterize codon usage patterns. These included the effective number of codons (ENC), codon adaptation index (CAI), relative synonymous codon usage (RSCU), overall genomic GC content, GC content at the third codon position (GC3s), and nucleotide frequencies at the third synonymous codon position (T3s, C3s, A3s, G3s). To further investigate the factors shaping codon usage bias, Parity Rule 2 (PR2) analysis was performed to explore the relative contributions of mutational pressure and natural selection. Additionally, ENC-GC3s plots were generated to evaluate the extent of codon bias across genes. All visualizations, including the ENC and PR2 plots, were created using the Matplotlib library (Barrett et al., 2005) to ensure clear graphical representation of the results.
Protein interaction network prediction
The TbADFs amino acid sequences from Triticum monococcum L. subsp. aegilopoides were submitted to the STRING database (http://www.string-db.org/) (Szklarczyk et al., 2021) to construct a protein-protein interaction (PPI) network, with A. thaliana selected as the reference organism due to its well-characterized protein interaction data. The analysis was performed using default parameters, including a medium confidence score threshold (0.6) to predict functional associations based on evidence such as co-expression, experimental data, and conserved co-occurrence. The resulting PPI network was visualized to identify potential interacting partners of TbADFs and to infer their functional roles in cellular processes.
DNA methylation frequency calculation
To evaluate DNA methylation levels in TbADFs, we quantified methylation frequencies in both promoter and gene body regions using publicly available whole-genome bisulfite sequencing data for Triticum monococcum L. subsp. aegilopoides accession TA299, obtained from the Dryad Digital Repository (https://datadryad.org/dataset/doi:10.5061/dryad.v41ns1rxj). Promoter regions were defined as the 2 kilobases upstream of the transcription start site (TSS), while gene body regions included the entire transcribed sequence excluding the promoter.
For each TbADFs and genomic region, methylation frequency was calculated as the ratio of methylated CpG sites to the total number of CpG sites (methylated + unmethylated), using the following formula:
Methylation calls were extracted from the downloaded data files, and region-specific methylation frequencies were computed using custom Python scripts. These scripts parsed CpG methylation count data and calculated the average methylation level across all CpG sites within each defined region for each gene.
Tissue-specific and cold stress-induced expression analysis of TbADFs
Transcriptome data for Triticum monococcum L. subsp. aegilopoides from various tissues and cold stress treatments were retrieved from the NCBI Sequence Read Archive (SRA) under accession numbers PRJEB61155 and PRJEB21284. Adapter sequences and low-quality reads were removed using Trimmomatic (v0.39) (Bolger et al., 2014). The resulting high-quality reads were pseudo-aligned to the reference genome and quantified using Kallisto (v0.46.2) (Bray et al., 2016) to estimate gene expression levels. A heatmap was generated using TBtools (v2.097) (Chen et al., 2023) to visualize expression patterns. To explore gene co-expression relationships, Pearson correlation analysis was performed between four upregulated TbADFs and all other expressed genes. Raw p-values from these pairwise correlation tests were adjusted for multiple comparisons using the Benjamini-Hochberg procedure to control the false discovery rate (FDR) at 5%. Genes with a correlation coefficient greater than 0.9 and a p-value less than 0.001 were first identified as candidate highly co-expressed, these were subsequently subjected to Benjamini-Hochberg correction, and pairs with an FDR-adjusted p-value less than 0.05 were considered highly co-expressed. Co-expression networks and conserved motif distributions were visualized using Cytoscape (v3.10.2) (Shannon et al., 2003). Gene Ontology (GO) enrichment analysis was conducted to infer the potential biological functions of the co-expressed genes, with GO annotations obtained from the KOBAS database (http://bioinfo.org/kobas) (Xie et al., 2011). The enrichment results were visualized using appropriate R packages.
Real-time RT-PCR
Total RNA was extracted from Triticum monococcum L. subsp. aegilopoides using the RNAprep Pure Plant Kit (Tiangen, China), following the manufacturer’s instructions. First-strand cDNA was synthesized from 1 µg of total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). Gene-specific primers for the TbADF gene family (Supplementary Table S1) were designed using Primer 5 software. Quantitative real-time PCR (qRT-PCR) was conducted on a Veriti 96-Well Thermal Cycler (Applied Biosystems, USA), with actin used as the internal reference gene for normalization. Each 10 µl reaction contained 5 µl of 2× TB Green Premix Ex Taq II (Tli RNaseH Plus; Takara, Japan), 0.4 µl of each primer (10 µM), 1 µl of cDNA, and 3.2 µl of sterile double-distilled water. The amplification protocol consisted of an initial denaturation at 95°C for 3 min, followed by 39 cycles of 95°C for 10 s and 58°C for 30 s. A melt curve analysis was performed to verify amplification specificity, using a ramp from 65°C to 95°C in 0.5°C increments after denaturation at 95°C for 5 s. Relative expression levels of TbADFs were calculated using the 2−ΔΔCt method.
Subcellular localization
Clone the CDS sequence of TbADF6 from Triticum monococcum L. subsp. aegilopoides cDNA. Digest the pCAMBIA2300-GFP vector with BamHI, and insert the amplified product into the linearized pCAMBIA2300-GFP vector using the pEASY®-Basic Seamless Cloning and Assembly Kit. Verify the recombinant construct by DNA sequencing. Transform the construct into Agrobacterium. Resuspend and mix the Agrobacterium cultures, then infiltrate them into tobacco leaves. After 36 hours of incubation, observe the fluorescence using a confocal laser scanning microscope.
Results
Identification of ADFs in Triticum monococcum L. subsp. aegilopoides
To identify ADF genes in Triticum monococcum L. subsp. aegilopoides, BLASTP searches were performed using known ADF protein sequences from A. thaliana, O. sativa, and Zea mays as queries against the Triticum monococcum L. subsp. aegilopoides. Redundant sequences were filtered using HMM profiling, resulting in the identification of nine unique ADF proteins, designated TbADF1 through TbADF9 (Supplementary Table S2). These proteins ranged in length from 138 to 270 amino acids, with molecular weights between 16.08 and 30.19 kDa and isoelectric points (pI) from 4.81 to 9.44. Six of the proteins were acidic (pI< 7). The instability index ranged from 33.17 to 56.08, indicating that six proteins may be unstable (instability index > 40). The aliphatic index varied from 58.17 to 80.52, and all nine proteins had negative GRAVY (grand average of hydropathicity) scores, suggesting a hydrophilic nature. Subcellular localization predictions indicated that all TbADFs are cytoplasmic.
Chromosomal distribution and evolutionary analysis of the TbADFs
Chromosomal mapping revealed that the nine TbADFs are unevenly distributed across five chromosomes. Specifically, chromosomes 2A, 4A, and 6A each contain one gene, while chromosome 1A has two genes, and chromosome 5A carries four (Figure 1A). Notably, TbADF6, TbADF7, and TbADF8 are clustered in adjacent genomic regions, suggesting possible tandem duplication events or localized selective pressures. To assess the evolutionary relationships among TbADFs, a phylogenetic tree was constructed using 97 ADF protein sequences from eight plant species. These ADFs clustered into four subfamilies (subgroups A–D), each containing 13 to 46 members. The TbADFs are represented in all four subgroups: one gene in subgroup A, two genes each in subgroups B and C, and four genes in subgroup D (Figure 1B).
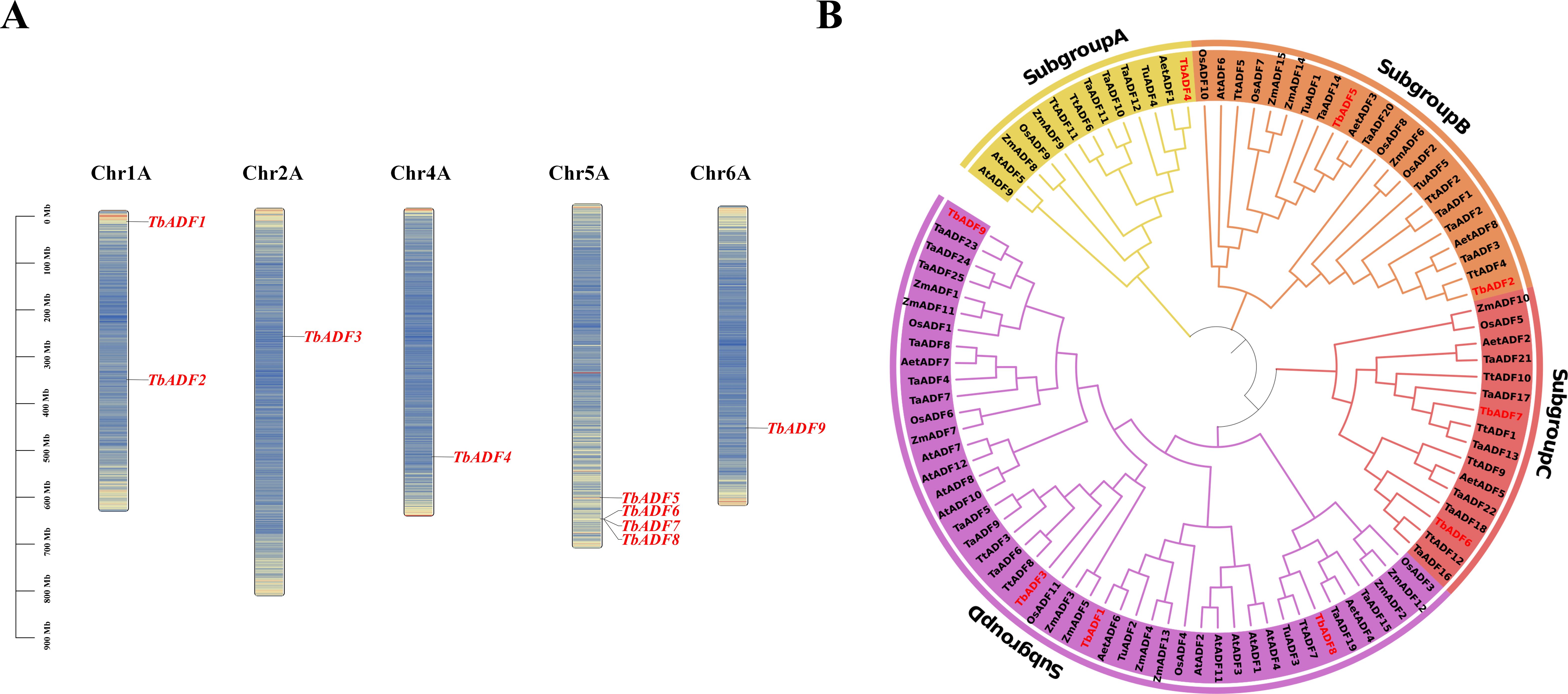
Figure 1. Localization and phylogenetic analysis of ADF genes. (A) Distribution of ADF genes across chromosomes, with positions indicated by colored bars. (B) Phylogenetic tree of ADF family proteins from multiple species.
To further explore ADF gene conservation, synteny analysis was performed between Triticum monococcum L. subsp. aegilopoides and three wheat relatives of varying ploidy levels: diploid Triticum urartu, tetraploid Triticum turgidum, and hexaploid Triticum aestivum. This analysis identified 3, 12, and 16 orthologous gene pairs, respectively (Figures 2A–C), suggesting that ADF genes have been conserved through wheat evolution and polyploidization, likely under purifying selection.
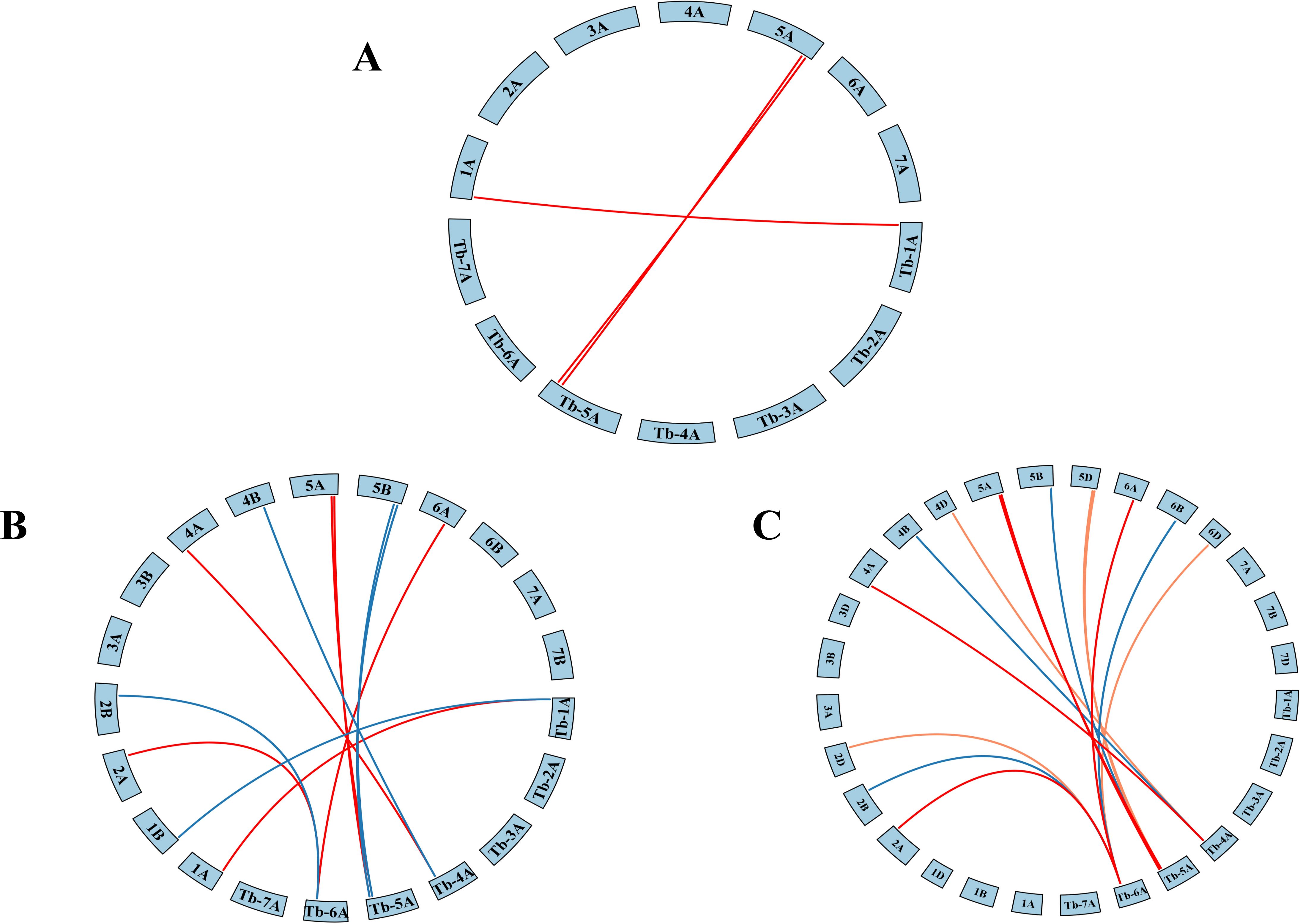
Figure 2. Collinearity analysis of Triticum monococcum L. subsp. aegilopoides with different ploidy levels of wheat: (A) Triticum monococcum L. subsp. aegilopoides-Triticum urartu, (B) Triticum monococcum L. subsp. aegilopoides-Triticum turgidum, (C) Triticum monococcum L. subsp. aegilopoides -Triticum aestivum.
Analysis of conserved motifs and cis-acting regulatory elements of TbADFs
Gene structure analysis revealed a relatively conserved exon-intron architecture among the nine TbADFs. Three genes contained two exons, five had three exons, and TbADF7 exhibited the most complex structure with five exons (Figure 3A). This conserved structural architecture suggests evolutionary stability within the ADF gene family. Motif analysis using the MEME Suite identified four conserved motifs shared by all TbADFs, predominantly located within the canonical ADF domain (Figure 3A). Multiple sequence alignment and tertiary structure modeling further confirmed the conserved positioning of the ADF domain across all family members (Figure 3B). Structural predictions generated by AlphaFold2 and visualized with PyMOL reinforced this conserved spatial configuration, highlighting the structural integrity of the ADF domain among TbADFs (Figure 3C).
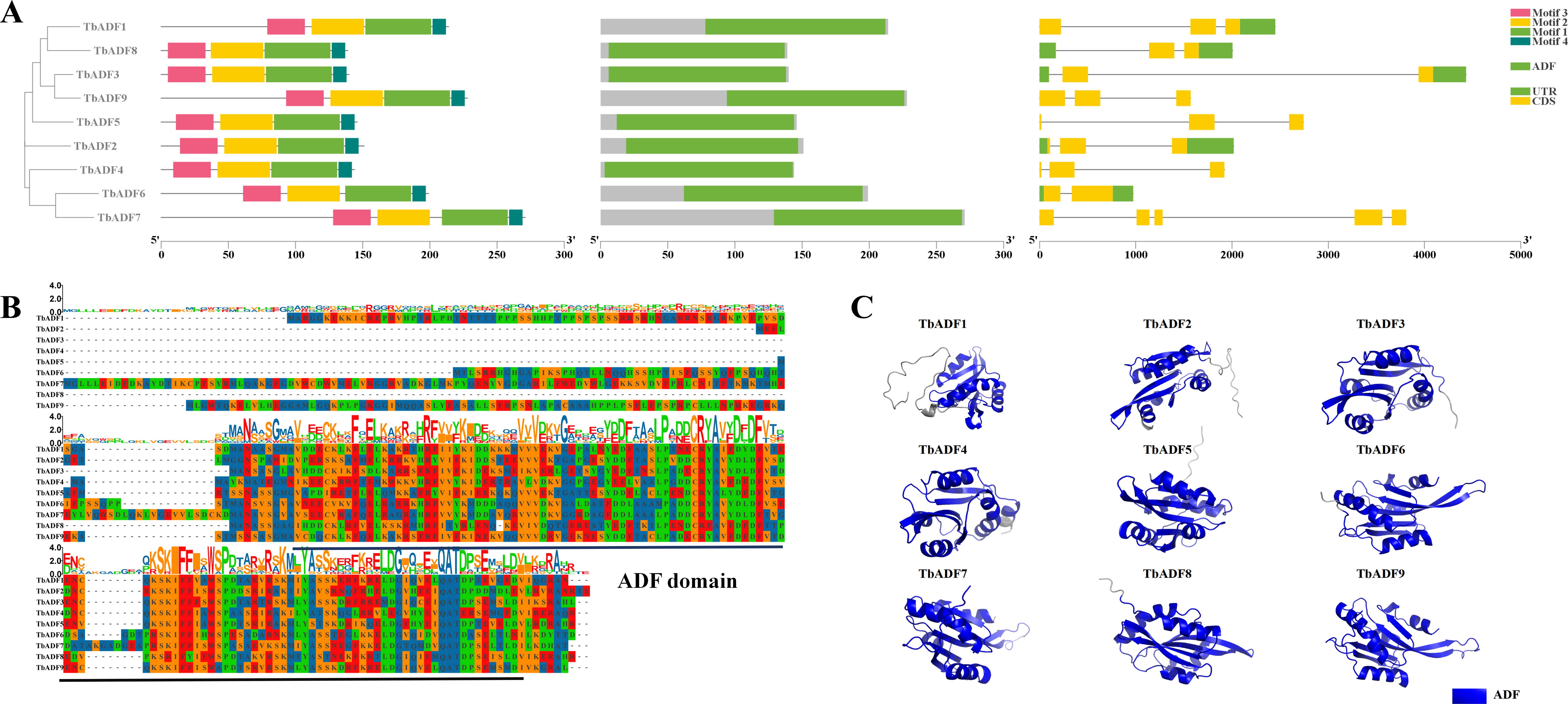
Figure 3. Conservation of ADF gene protein motifs, domains, and gene structure in Triticum monococcum L. subsp. aegilopoides (A); Protein sequence alignment (B); Protein tertiary structure (C).
To explore regulatory potential, the 2,000 bp upstream promoter regions of TbADFs were analyzed using PlantCARE. Numerous cis-acting regulatory elements were detected, including those associated with growth, hormone signaling, and stress response (Figure 4; Supplementary Table S3). Notable elements included hormone-responsive motifs such as the abscisic acid response element (ABRE), jasmonic acid-responsive motifs (CGTCA-motif and TGACG-motif), auxin-responsive TGA-element, and gibberellin-responsive P-box. Additionally, stress-associated elements such as the low-temperature response element (LTR), drought-responsive DRE, and hypoxia-responsive motifs (GC-motif, ARE) were also identified. Collectively, these findings suggest that TbADFs are not only structurally conserved but are also likely involved in complex regulatory networks mediating hormonal and environmental stress responses. This highlights their potential roles in enhancing wheat adaptability to adverse environmental conditions.
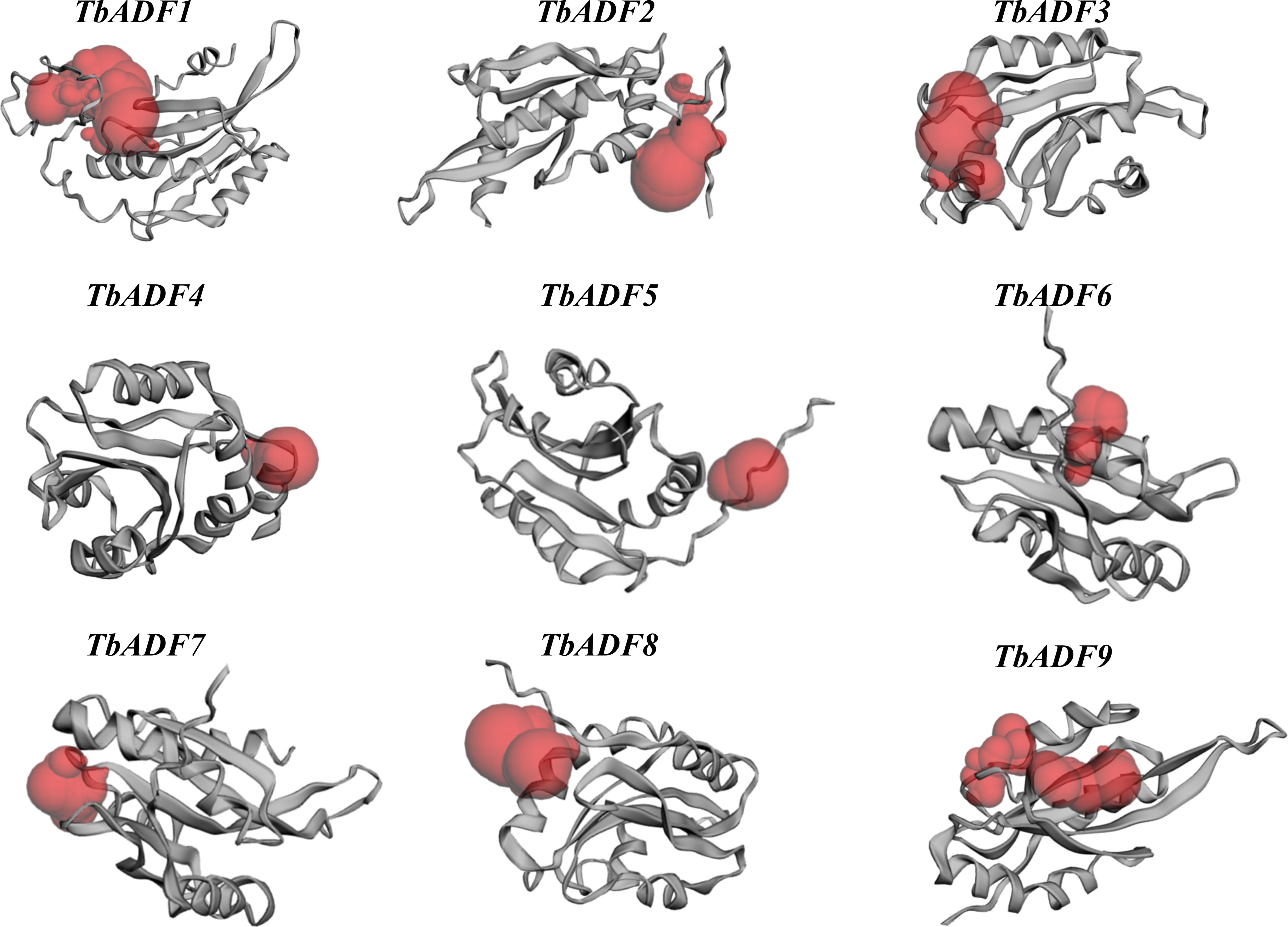
Prediction of catalytic active sites and binding pockets in TbADFs
The detection of protein binding pockets is critical for understanding structural diversity and interaction specificity, particularly in predicting functional sites such as ligand-binding regions. To explore functional diversity among TbADFs, potential catalytic and ligand-binding sites were predicted using CASTp 3.0. Red-highlighted regions indicated the predicted binding pockets (Figure 5), and their surface areas and volumes were calculated (Supplementary Table S4). These binding sites are surrounded by key amino acid residues such as tyrosine (Tyr), phenylalanine (Phe), valine (Val), asparagine (Asn), lysine (Lys), alanine (Ala), aspartic acid (Asp), glutamic acid (Glu), isoleucine (Ile), and leucine (Leu), which are likely involved in mediating protein–ligand interactions and catalytic activity. The conservation of these residues supports their functional importance and potential roles in the stress-responsive behavior of ADFs.
Prediction of protein-protein interactions in the TbADFs
To elucidate the protein interaction network of the ADF gene family in Triticum monococcum L. subsp. aegilopoides, nine TbADF protein sequences were analyzed using the STRING database with A. thaliana as the reference organism. (Figure 6A). These interacting proteins are implicated in critical cellular processes, including cytoskeletal dynamics, cell elongation, shape modulation, division, and migration, which collectively influence transpiration regulation, pathogen defense responses, and overall plant growth and development. The high connectivity of TbADFs with these partners, supported by a confidence score threshold of 0.7, suggests a conserved functional module that may underpin stress adaptation and developmental plasticity in wild einkorn wheat. This interaction network provides a foundation for future functional studies.
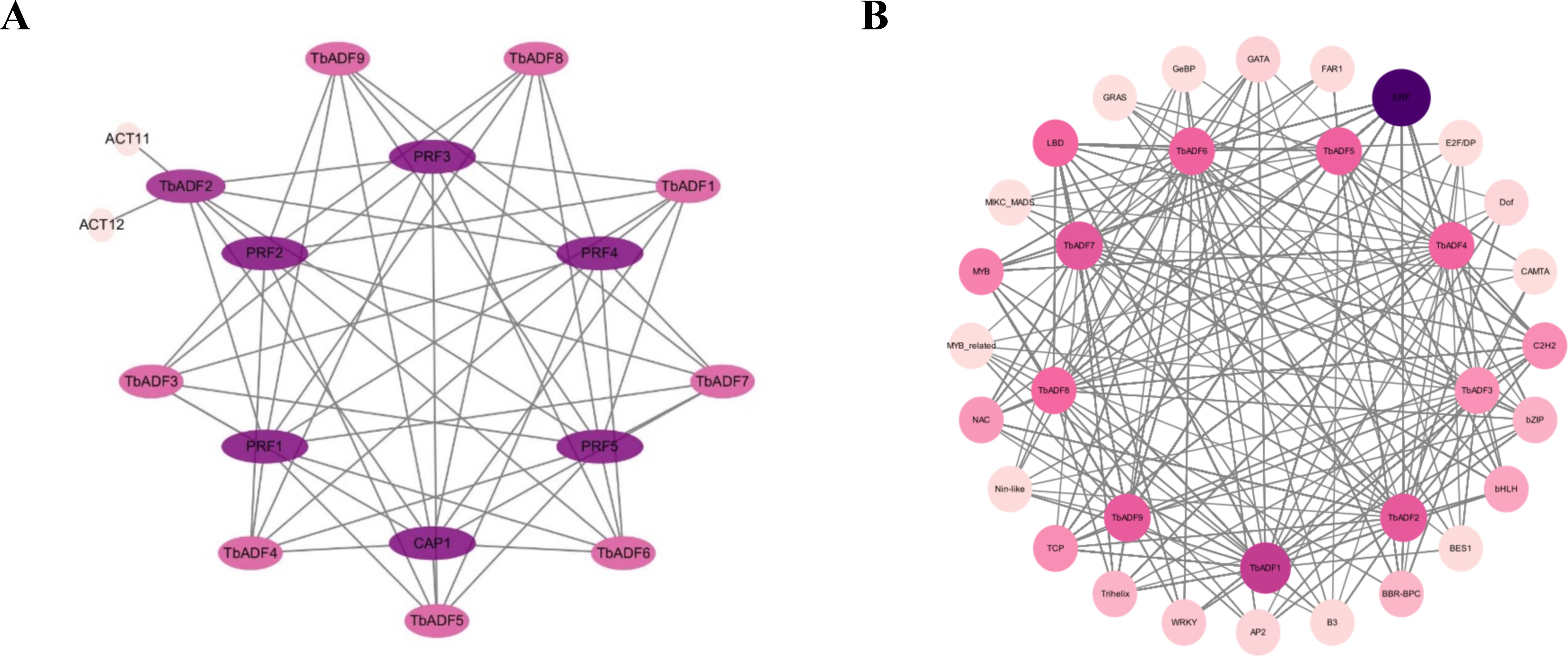
Figure 6. Predicted protein-protein interactions and transcription factor binding networks of TbADFs in Triticum monococcum L. subsp. aegilopoides. (A) Protein-protein interaction network of TbADFs with interacting partners. (B) Transcription factor binding network highlighting interactions between TbADFs and various transcription factor families.
To further explore transcriptional regulation, promoter regions of the TbADFs genes were scanned for TF binding motifs using the FIMO (Figure 6B). This analysis revealed a complex regulatory network characterized by diverse TF interactions. Notably, members of the ERF family exhibited the highest binding site frequency, with a total of 2516 sites identified across multiple TbADFs, particularly in TbADF1 and TbADF7. This suggests that ERF transcription factors may play a dominant role in modulating ADF gene expression, likely in response to abiotic stress signals, given the GC-rich nature of their target motifs. In addition, several other TF families, including LBD, MYB, TCP, and C2H2, also contribute to the regulatory architecture. Although these families are less abundant than ERFs, their presence suggests auxiliary roles in fine-tuning ADF gene expression under specific developmental or stress-related conditions.
Codon usage bias analysis of TbADFs
The codon usage bias of the ADF gene family in Triticum monococcum L. subsp. aegilopoides was analyzed to explore the evolutionary pressures shaping gene sequence composition. The ENC plot (Figure 7A) showed that ADF genes exhibit ENC values ranging from 39.7 to 52.3. Most values fall below the expected standard curve under neutral evolution, indicating a moderate codon usage bias predominantly influenced by GC content at the GC3s, which ranged from 0.644 to 0.829 (Supplementary Table S5).
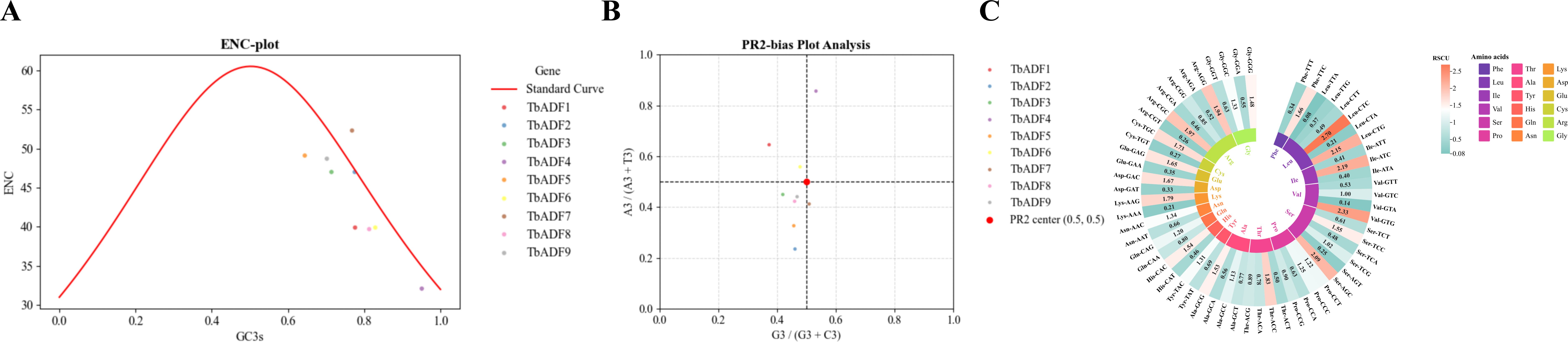
Figure 7. Codon usage bias analysis of TbADFs in Triticum monococcum L. subsp. aegilopoides. (A) ENC-plot showing the ENC versus GC3s, with the standard curve indicating expected ENC under neutral evolution; data points represent TbADF1 to TbADF9, highlighting moderate codon usage bias. (B) PR2-bias plot illustrating the relationship between A3/(A3+T3) and G3/(G3+C3) for each TbADFs, with the central point (0.5, 0.5) indicating balanced nucleotide usage; data points cluster near this center, suggesting combined effects of mutation and selection. (C) RSCU heatmap displaying RSCU values for TbADFs, with color gradients indicating codon preferences (red for high, blue for low); amino acids are labeled with their corresponding codons, revealing a preference for G/C-ending codons.
The PR2-bias plot (Figure 7B) demonstrated an approximately balanced distribution between A3 and T3, and between G3 and C3, with most data points clustering near the central point (0.5, 0.5). This pattern suggests that both mutational pressure and natural selection contribute to shaping codon usage in TbADFs.
A heatmap of RSCU values (Figure 7C) further highlighted a preference for G/C-ending codons across the ADF family, consistent with the GC-rich genomic context of T. monococcum subsp. aegilopoides. Notably, codons such as GGC (Glycine) and UGC (Cysteine) were overrepresented, implying potential optimization for translation efficiency. This bias may be linked to tRNA abundance or a selective advantage for efficient protein synthesis, particularly under stress conditions, aligning with the known role of ADF proteins in cytoskeletal dynamics and stress response.
Analysis of DNA methylation levels in TbADFs
This analysis was based on high-quality, genome-wide methylation data for Triticum monococcum L. subsp. aegilopoides accession TA299, which we obtained from Ahmed et al. (2023). The final dataset comprised 221,159,329 CpG sites with a total sequencing depth of 19,524,541,735 and an average coverage of 88.28× per site. Using this high-coverage dataset, we calculated average methylation frequencies for all CpG sites within each defined gene region. The analysis of CpG methylation frequencies in the promoter and gene body regions of nine ADF genes (TbADF1 to TbADF9) revealed significant variation in methylation levels (Supplementary Table S6), with promoter methylation ranging from 0.0907 (TbADF2) to 0.3053 (TbADF9) and gene body methylation from 0.0797 (TbADF2) to 0.2740 (TbADF1). The mean promoter methylation (0.1923) was slightly higher than the mean gene body methylation (0.1823), suggesting a potential regulatory role of promoter methylation in gene expression. Notably, TbADF9 exhibited the highest methylation levels in both regions (0.3053 and 0.2644, respectively), which may indicate a repressive epigenetic state, while TbADF2 displayed the lowest levels (0.0907 and 0.0797).
Gene expression analysis of TbADFs in different tissues
Transcriptomic analysis across six tissues of Triticum monococcum L. subsp. aegilopoides revealed distinct tissue-specific expression patterns among the nine TbADFs (Figure 8A). TbADF2 and TbADF7 exhibited predominant expression in the glumes, whereas TbADF3 and TbADF9 were highly expressed in the grains, with low expression in other tissues. The remaining genes showed broader expression profiles: TbADF1 was mainly expressed in both flag leaves and grains; TbADF4 displayed elevated expression in flag leaves, glumes, and grains; and TbADF5 and TbADF8 were primarily expressed in flag leaves, glumes, and roots. These expression patterns indicate that TbADFs are functionally diversified, contributing to tissue-specific roles during the growth and development of wild einkorn wheat.
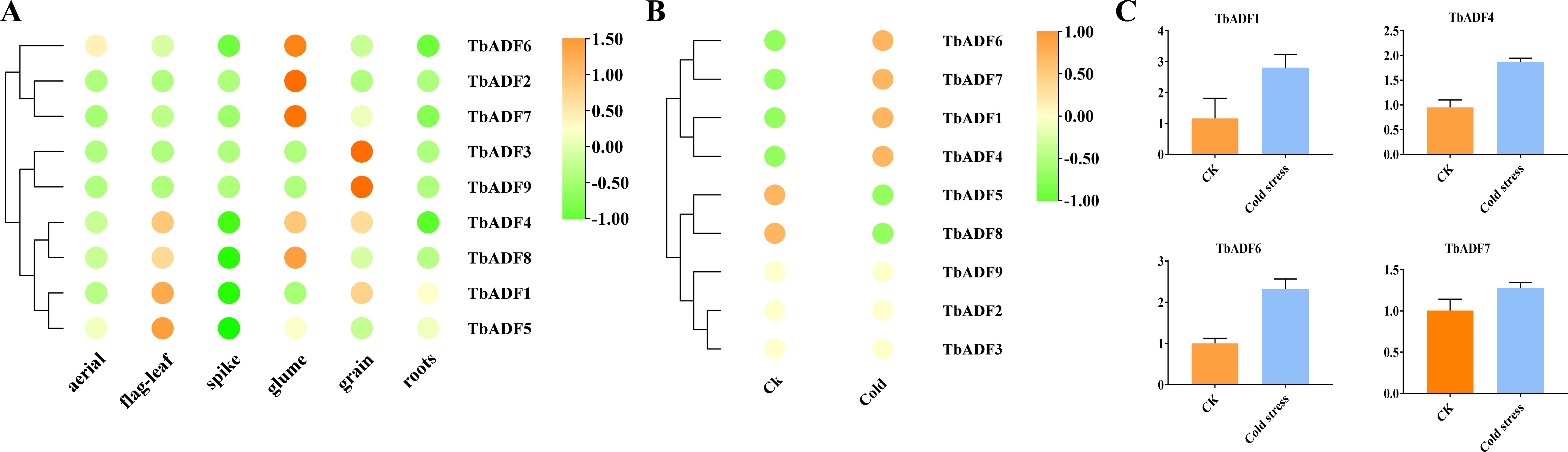
Figure 8. Expression profiles and cold stress response of TbADF genes in Triticum monococcum L. subsp. aegilopoides. (A) Expression profiles of TbADF genes across six tissues: aerial, flag leaf, spike, glume, grain, and roots. (B) Expression changes of TbADF genes under cold stress treatment based on transcriptome data. (C) qPCR validation of four selected TbADF genes under cold stress conditions.
Transcriptomic response of TbADFs to cold stress
Transcriptomic profiling following a 24-hour cold treatment at 4°C revealed distinct differential expression patterns among the TbADFs. Notably, TbADF2, TbADF3, and TbADF9 exhibited no detectable expression under either control or cold-stress conditions, suggesting their non-involvement in the immediate cold response (Figure 8B). In contrast, the remaining six ADF genes were cold-responsive: TbADF5 and TbADF8 showed downregulation, whereas TbADF1, TbADF4, TbADF6, and TbADF7 were significantly upregulated. Among them, TbADF6 displayed the most dramatic induction, with TPM increasing from 29.07 to 300.01, highlighting its potential central role in cold stress adaptation. To validate these transcriptomic findings, expression analysis of TbADF1, TbADF4, TbADF6, and TbADF7 was conducted, confirming consistent upregulation of all four genes under cold stress conditions (Figure 8C). This concordance between transcriptome data and expression validation strongly supports their involvement in the cold response of wild einkorn wheat.
To further dissect the molecular mechanisms underlying this response, we focused on the four upregulated genes, TbADF1, TbADF4, TbADF6, and TbADF7. Pearson correlation analysis (r > 0.9, p< 0.001) identified strongly co-expressed genes under cold stress: 103 for TbADF1, 1,269 for TbADF4, 1,706 for TbADF6, and 1,657 for TbADF7 (Figure 9A). To elucidate the biological relevance of these co-expression networks, we performed GO enrichment analysis (Figure 9B).
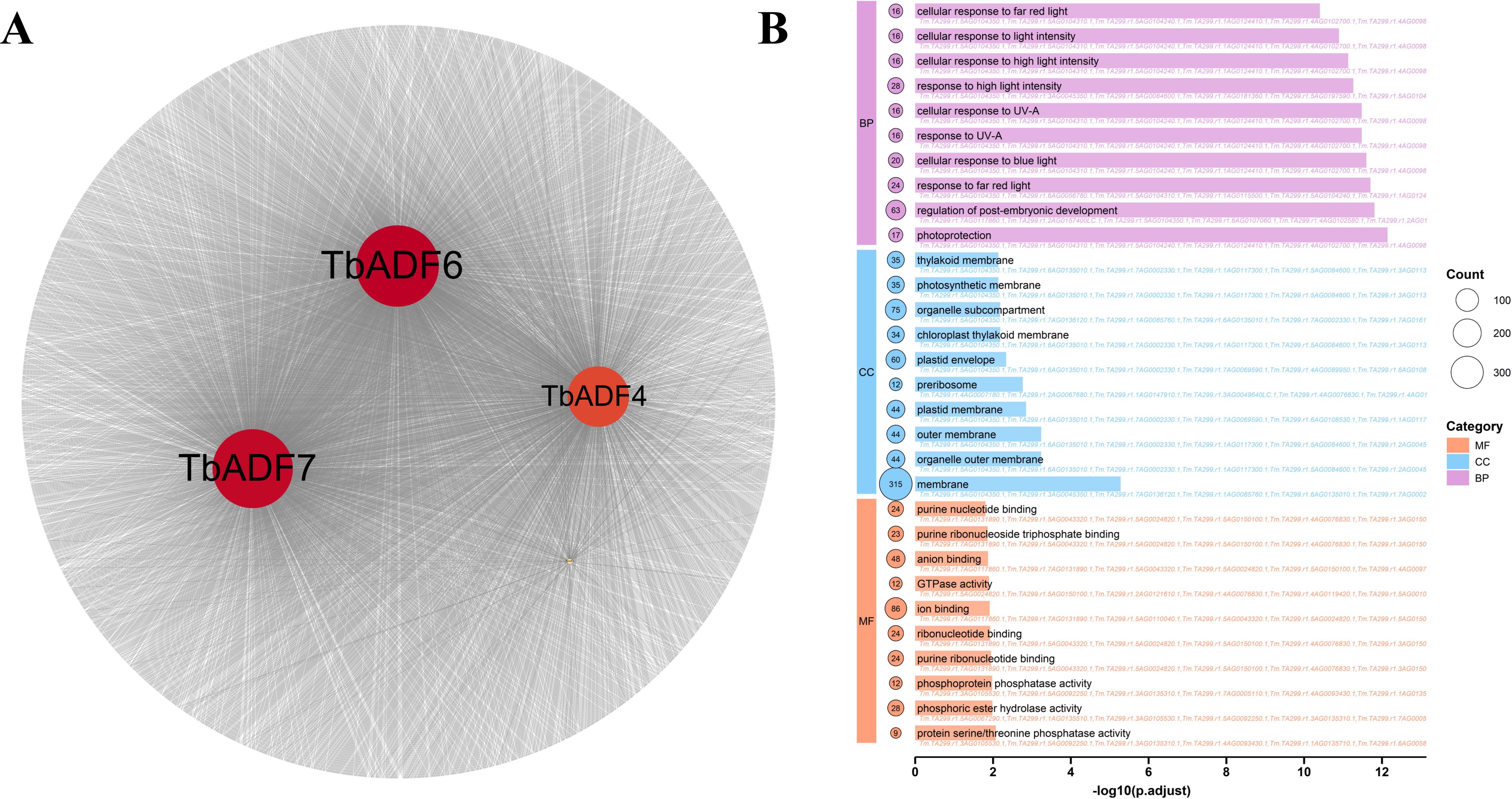
Figure 9. Co-expression network and GO enrichment analysis of genes highly correlated with four ADF genes under cold stress in Triticum monococcum L. subsp. aegilopoides. (A) Co-expression network illustrating interactions of genes highly correlated with TbADF1, TbADF4, TbADF6, and TbADF7 (highlighted in red), with node size and connectivity reflecting the strength of associations. (B) Bar plot of GO enrichment analysis showing significantly enriched biological processes (BP), cellular components (CC), and molecular functions (MF) among co-expressed genes, with -log10(p-adjust) values indicating statistical significance and color coding by category.
In the MF category, terms such as purine ribonucleoside triphosphate binding (GO:0035639) and phosphoprotein phosphatase activity (GO:0004721) were significantly enriched, implicating roles in energy metabolism and phosphorylation-dependent signal transduction during cold stress. In the CC category, enrichments for membrane (GO:0016020) and organelle outer membrane (GO:0031968) suggest localization of co-expressed proteins to structural membrane sites, possibly mediating signal transmission and maintaining cellular integrity under stress. BP terms such as maturation of LSU-rRNA (GO:0000470) and intracellular chemical homeostasis (GO:0055082) point to enhanced ribosome biogenesis and metabolic homeostasis, key mechanisms for sustaining protein synthesis and stability during stress. Furthermore, enrichments in response to fatty acid (GO:0070542) and cellular response to endogenous stimulus (GO:0071495) underscore the involvement of lipid signaling and hormone-regulated pathways, both critical for cold adaptation through modulation of membrane fluidity and internal signaling cascades. Collectively, these findings highlight the intricate regulatory and functional roles of cold-inducible ADF genes, particularly TbADF6, in orchestrating a coordinated molecular response to low-temperature stress in wild einkorn wheat.
Subcellular localization of TbADF6 protein
To investigate the subcellular localization of TbADF6, a GFP-tagged fusion construct was generated and transiently expressed in tobacco leaves via Agrobacterium-mediated transformation. Confocal laser scanning microscopy showed that the GFP-ADF fusion protein was mainly localized in the nucleus and cytoplasm (Figure 10), which is consistent with the subcellular localization prediction (Supplementary Table S2). These results suggest that TbADF6 protein may function in both compartments.
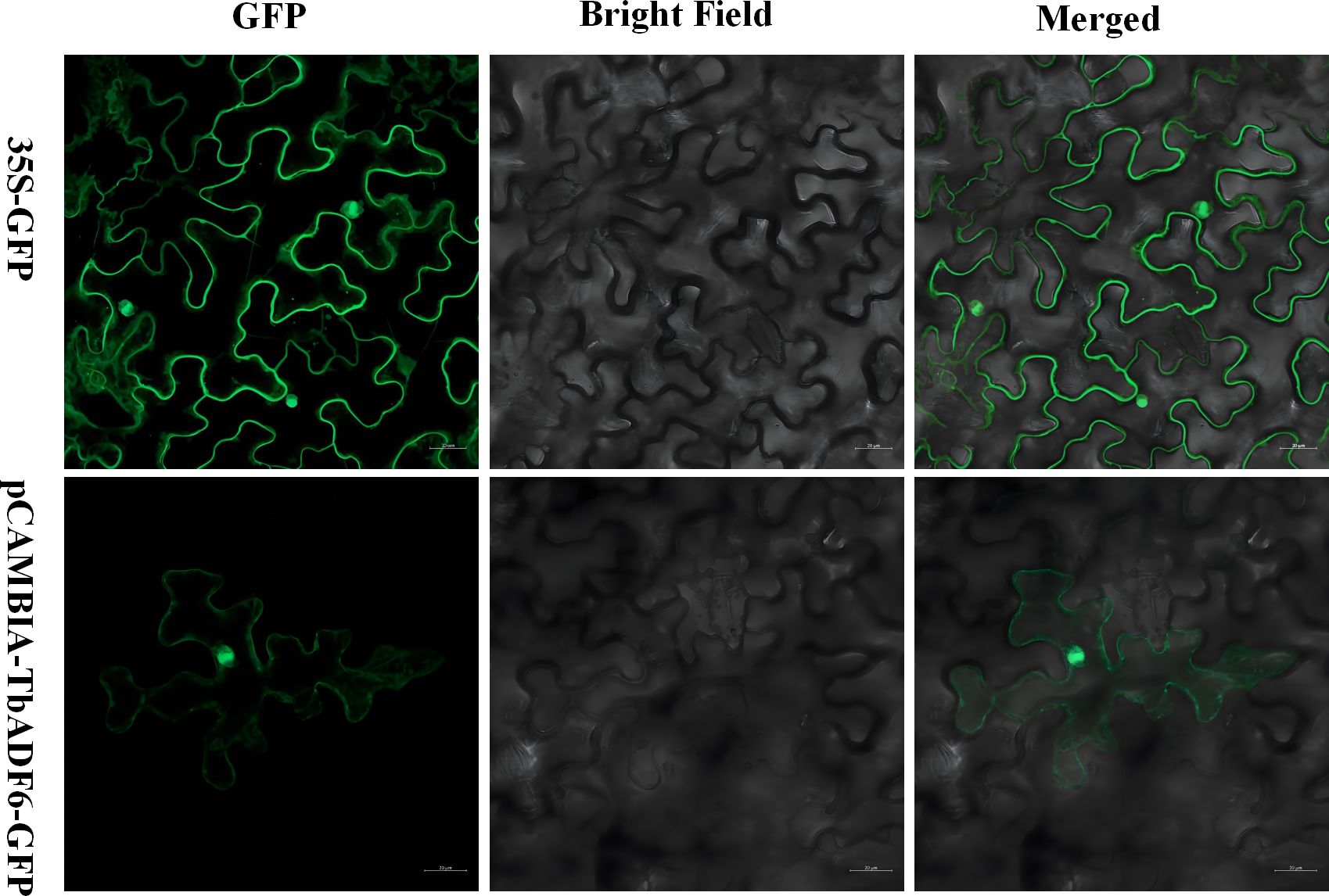
Figure 10. Subcellular localization analysis of TbADF6 in Nicotiana tabacum leaves; 35S-GFP is the negative control. Scale bar: 20 µm.
Discussion
ADFs are key regulators of cytoskeletal dynamics in plants, influencing essential cellular processes such as morphogenesis, intracellular trafficking, and responses to abiotic stresses. Increasing evidence highlights their critical role in enhancing plant resilience under cold stress by modulating actin filament organization and signaling pathways (Staiger, 2000; Staiger and Blanchoin, 2006). In this study, we present a comprehensive characterization of the ADF gene family in Triticum monococcum L. subsp. aegilopoides, a wild diploid wheat species valued for its inherent stress tolerance. Through an integrative approach combining genomic, structural, and transcriptomic analyses, we identified nine TbADFs and explored their potential involvement in cold stress adaptation. Our findings provide new insights into the evolutionary conservation and functional diversification of TbADFs and lay the groundwork for future studies on cytoskeletal regulation under stress conditions.
The discovery of nine TbADFs in T. monococcum subsp. aegilopoides is consistent with the ADF gene family sizes reported in other monocots, such as rice and maize, which harbor 11 and 15 ADF genes, respectively (Yang et al., 2024). Phylogenetic analysis classified these TbADFs into four subgroups, a pattern conserved across plant species, suggesting an ancient diversification of ADF functions prior to monocot-dicot divergence (Ruzicka et al., 2007). The chromosomal distribution, with a notable cluster of TbADF6, TbADF7, and TbADF8 on chromosome 5Ab, hints at tandem duplication events, a common mechanism for gene family expansion and functional specialization in plants (Cannon et al., 2004). Such duplication events can provide raw genetic material for evolutionary innovation, allowing duplicated genes to diverge in expression patterns or molecular function (Mondal et al., 2016). The physical proximity of TbADF6, TbADF7, and TbADF8 suggests they arose from local duplication, and preliminary expression data indicate that these genes exhibit differential tissue-specific expression, supporting the hypothesis of functional divergence. This likely reflects subfunctionalization or neofunctionalization processes that enable more refined regulation of actin dynamics in response to developmental or environmental cues (Panchy et al., 2016). Collinearity analysis with T. urartu, T. turgidum, and T. aestivum further revealed a high degree of conservation across wheat species, despite differing ploidy levels. This preservation through polyploidization events underscores the indispensable role of ADFs in cytoskeletal regulation and stress responses, likely maintained by purifying selection (Adams and Wendel, 2005).
Structural analyses revealed that the TbADFs possess conserved exon–intron structures and motif organizations, particularly within the actin-binding ADF domain, which is central to actin filament severing and depolymerization (Lappalainen et al., 1998). 3D structure prediction and binding site analysis via AlphaFold2 and CASTp identified conserved residues, such as tyrosine and lysine, within potential actin-interaction pockets, mirroring functional features described in AtADFs (Dong et al., 2001). Additionally, codon usage analysis revealed a strong preference for G/C-ending codons, which may enhance translation efficiency under stress conditions, an adaptive feature commonly observed in stress-responsive genes (Sharp and Li, 1987).
Transcriptomic analysis across six tissues of Triticum monococcum L. subsp. aegilopoides revealed distinct tissue-specific expression patterns among the nine TbADFs, underscoring their functional diversification in wild einkorn wheat. These genes encode proteins critical for regulating actin cytoskeleton dynamics, which are essential for key cellular processes such as division, expansion, and responses to abiotic stress. Notably, TbADF2 and TbADF7 exhibiting high expression in glumes, suggesting specialized roles in cytoskeletal remodeling during the development of these protective structures, which are vital for safeguarding reproductive tissues under fluctuating environmental conditions (Whitehead and Crawford, 2005). TbADF3 and TbADF9 were more abundant in grains, likely by facilitating actin-mediated nutrient transport and cell wall biosynthesis (Hussey et al., 2006). TbADF1 was primarily expressed in flag leaves and grains, implying a role in coordinating cytoskeletal dynamics to support resource allocation during grain maturation (Hussey et al., 2002). TbADF4 displayed elevated expression in flag leaves, glumes, and grains, suggesting a versatile function in metabolically active tissues. TbADF5 and TbADF8 were predominantly expressed in flag leaves, glumes, and roots, likely contributing to root growth and nutrient transport (Jiang et al., 1997). Under cold stress conditions, TbADF1, TbADF4, TbADF6, and TbADF7 were significantly upregulated, with TbADF6 showing the most prominent induction. Given the known function of ADFs in actin remodeling, these genes may help stabilize cytoskeletal structure, support vesicle trafficking, or preserve membrane organization during cold-induced cytoplasmic changes (Örvar et al., 2000).
Co-expression analysis of cold-induced TbADFs revealed networks enriched in genes associated with energy metabolism, signal transduction, and membrane integrity. Enriched GO terms such as purine ribonucleoside triphosphate binding and membrane organization underscore the cytoskeleton’s central role in energy-dependent stress signaling. PPI predictions using STRING uncovered potential interactions with profilins (PRFs) and cyclase-associated proteins (CAP1), key components of actin turnover and cytoskeletal reorganization (Pollard and Cooper, 2009). These interactions highlight the functional integration of TbADFs within broader stress adaptation networks.
The transcriptional response of the TbADFs to cold stress indicates that specific members are tightly regulated in response to low-temperature signals, suggesting functional divergence within this family. This variability aligns with previous findings in wheat, where ADFs exhibit differential expression under abiotic stresses (Xu et al., 2021). The complete transcriptional silence of TbADF2, TbADF3, and TbADF9 under both control and cold-treated conditions may reflect either spatial or developmental specificity, pseudogenization, or functional redundancy that renders them unresponsive to acute cold stimuli. Such silencing could be indicative of evolutionary adaptations, as observed in other plant species where non-responsive gene family members serve niche roles (Moore and Purugganan, 2005). In contrast, the upregulation of TbADF1, TbADF4, TbADF6, and TbADF7 underscores their potential involvement in early cold stress signaling or adaptation processes. Among these, TbADF6 exhibited the most striking induction, suggesting it may act as a primary cold-responsive regulator within the ADF family in wild einkorn wheat.
Our integrative analysis suggests that multiple TbADFs collaboratively contribute to Triticum monococcum L. subsp. aegilopoides’s cold stress adaptation through both shared and distinct regulatory functions. GO enrichment of highly co-expressed partners for TbADF1, TbADF4, TbADF6 and TbADF7 indicates their involvement in diverse biological processes including ribonucleoprotein complex biogenesis, hormone metabolic pathways, photosynthetic membrane organization, and intercellular signaling. For example, TbADF1 shows strong associations with nucleolar and ribonucleoprotein complex functions, suggesting roles in fundamental growth regulation, while TbADF4 and TbADF7 are enriched in pathways related to phosphatase activity, RNA helicase activity, and photoprotection. TbADF6 is embedded in a dense regulatory network, sharing expression patterns with over 1,700 genes, implying its participation in broader stress-responsive pathways. This network density aligns with findings in rice, where hub genes under cold stress coordinate extensive gene interactions (Byun et al., 2021). GO enrichment of these co-expressed genes unveiled biological processes related to membrane structure, phosphorylation, ribosome maturation, and intracellular homeostasis—core components of cellular resilience to cold stress (Thomashow, 1999).
Similarly, recent studies in wheat identified 25 ADF genes, with TaADF16 strongly upregulated under cold acclimation and freezing. Overexpression of TaADF16 in Arabidopsis improved freezing tolerance by reducing ion leakage and increasing survival through better ROS scavenging and membrane protection. It also activated cold-responsive genes like CBF1, COR15A, and RD22, indicating involvement in the conserved ICE-CBF-COR pathway. Other TaADFs, such as TaADF13, TaADF17, and TaADF22, also respond to cold stress, suggesting a coordinated ADF gene network in cold adaptation (Xu et al., 2021). Notably, TbADF6 is located on chromosome 5Ab in Triticum monococcum L. subsp. aegilopoides and shows conserved synteny with TaADF16 on 5A, TaADF18 on 5B, and TaADF22 on 5D in hexaploid wheat. This collinearity indicates that TbADF6 is the orthologous counterpart of these cold-responsive TaADFs, suggesting functional conservation and evolutionary retention of this locus across wheat genomes. The presence of homologous genes on chromosomes 5B and 5D reflects the two rounds of polyploidization events in wheat evolution (Ramírez-González et al., 2018), which generated additional homeologous copies of TbADF6 in the B and D subgenomes. The syntenic relationship underscores the importance of TbADF6 as a likely key regulator in cold adaptation, maintained across multiple wheat subgenomes through evolutionary history. Additionally, terms related to fatty acid response and endogenous stimulus perception suggest that lipid-mediated signaling and hormonal regulation may work in concert with TbADFs-mediated actin remodeling to maintain membrane integrity and initiate adaptive signaling under cold conditions. These processes are known to be critical in plants for maintaining cellular homeostasis under low temperatures. Together, these findings indicate that cold-inducible TbADFs, particularly TbADF6, play central roles in orchestrating cytoskeletal and signaling reprogramming during early cold response. This highlights ADF genes as potential targets for genetic manipulation aimed at improving cold tolerance in wheat and other cereals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XL: Writing – original draft, Data curation, Conceptualization, Visualization, Validation, Formal analysis. MZ: Visualization, Formal analysis, Investigation, Writing – review & editing. JS: Writing – review & editing, Conceptualization, Funding acquisition. LW: Investigation, Writing – review & editing. MS: Investigation, Writing – review & editing. QW: Writing – original draft, Investigation, Software, Visualization. YZ: Conceptualization, Writing – review & editing, Funding acquisition. LZ: Writing – review & editing, Supervision, Conceptualization. HL: Conceptualization, Writing – review & editing, Supervision, Funding acquisition. GC: Writing – review & editing, Software, Conceptualization, Validation, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was financially supported by the Open Foundation of The National Key Laboratory of Wheat Improvement (KFKT202504 to XL); Yibin University Research Projects (2025XJQH16 to XL); Open Foundation of Key Laboratory of Wuliangye-flavor Liquor Solid-state Fermentation, China National Light Industry (2023JJ008 to MZ); Sichuan Science and Technology Program (2025ZNSFSC1010; MZGC20240023 to MZ); the Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2025C01 to GC); Younth Taishan Scholar Project of Shandong Province (tsqn202408302 to YZ); The Shandong Postdoctoral Science Foundation (SDCX-ZG-202400117 to CG).
Acknowledgments
We thank Jingye Fu from Sichuan Agricultural University for editing the manuscript and providing valuable suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1649202/full#supplementary-material
References
Adams, K. L. and Wendel, J. F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141. doi: 10.1016/j.pbi.2005.01.001
Ahmed, H. I., Heuberger, M., Schoen, A., Koo, D.-H., Quiroz-Chavez, J., Adhikari, L., et al. (2023). Einkorn genomics sheds light on history of the oldest domesticated wheat. Nature 620, 830–838. doi: 10.1038/s41586-023-06389-7
Bamburg, J., Harris, H., and Weeds, A. (1980). Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 121, 178–182. doi: 10.1016/0014-5793(80)81292-0
Barrett, P., Hunter, J., Miller, J. T., Hsu, J.-C., and Greenfield, P. (2005). Matplotlib–A portable python plotting package. In Astronomical data analysis software and systems XIV. 347, 91.
Bernstein, B. W. and Bamburg, J. R. (2010). ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20, 187–195. doi: 10.1016/j.tcb.2010.01.001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Byun, M. Y., Cui, L. H., Lee, A., Oh, H. G., Yoo, Y.-H., Lee, J., et al. (2021). Abiotic stress-induced actin-depolymerizing factor 3 from deschampsia Antarctica enhanced cold tolerance when constitutively expressed in rice. Front. Physiol. 12, 734500. doi: 10.3389/fpls.2021.734500
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 1–21. doi: 10.1186/1471-2229-4-10
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
DeLano, W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92.
Dong, C.-H., Xia, G.-X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N.-H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. doi: 10.1105/TPC.010051
Drøbak, B., Franklin-Tong, V., and Staiger, C. (2004). The role of the actin cytoskeleton in plant cell signaling. New Phytol. 163, 13–30. doi: 10.1111/j.1469-8137.2004.01076.x
Du, J., Wang, X., Dong, C.-H., Yang, J. M., and Yao, X. J. (2016). Computational study of the binding mechanism of actin-depolymerizing factor 1 with actin in Arabidopsis thaliana. PloS One 11, e0159053. doi: 10.1371/journal.pone.0159053
Hassan, M. A., Xiang, C., Farooq, M., Muhammad, N., Yan, Z., Hui, X., et al. (2021). Cold stress in wheat: plant acclimation responses and management strategies. Front. Plant Sci. 12, 676884. doi: 10.3389/fpls.2021.676884
Hussey, P. J., Allwood, E. G., and Smertenko, A. P. (2002). Actin–binding proteins in the Arabidopsis genome database: properties of functionally distinct plant actin–depolymerizing factors/cofilins. Philos. Trans. R. Soc. London Ser. B: Biol. Sci. 357, 791–798. doi: 10.1098/rstb.2002.1086
Hussey, P. J., Ketelaar, T., and Deeks, M. J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57, 109–125. doi: 10.1146/annurev.arplant.57.032905.105206
Inada, N. (2017). Plant actin depolymerizing factor: actin microfilament disassembly and more. J. Plant Res. 130, 227–238. doi: 10.1007/s10265-016-0899-8
Jiang, C. J., Weeds, A. G., and Hussey, P. J. (1997). The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. 12, 1035–1043. doi: 10.1046/j.1365-313X.1997.12051035.x
Lappalainen, P., Kessels, M. M., Cope, M. J. T., and Drubin, D. G. (1998). The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol. Biol. Cell 9, 1951–1959. doi: 10.1091/mbc.9.8.1951
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, H., Sultan, M. A. R. F., Liu, X., Zhang, J., Yu, F., and Zhao, H. X. (2015). Physiological and comparative proteomic analysis reveals different drought responses in roots and leaves of drought-tolerant wild wheat (Triticum boeoticum). PloS One 10, e0121852. doi: 10.1371/journal.pone.0121852
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121–e121. doi: 10.1093/nar/gkt263
Mondal, S., Rutkoski, J. E., Velu, G., Singh, P. K., Crespo-Herrera, L. A., Guzman, C., et al. (2016). Harnessing diversity in wheat to enhance grain yield, climate resilience, disease and insect pest resistance and nutrition through conventional and modern breeding approaches. Front. Plant Sci. 7, 991. doi: 10.3389/fpls.2016.00991
Moore, R. C. and Purugganan, M. D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. doi: 10.1016/j.pbi.2004.12.001
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Örvar, B. L., Sangwan, V., Omann, F., and Dhindsa, R. S. (2000). Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J. 23, 785–794. doi: 10.1046/j.1365-313x.2000.00845.x
Ouellet, F., Carpentier, E., Cope, M. J. T., Monroy, A. F., and Sarhan, F. (2001). Regulation of a wheat actin-depolymerizing factor during cold acclimation. Plant Physiol. 125, 360–368. doi: 10.1104/pp.125.1.360
Panchy, N., Lehti-Shiu, M., and Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Pollard, T. D. and Cooper, J. A. (2009). Actin, a central player in cell shape and movement. Science 326, 1208–1212. doi: 10.1126/science.1175862
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, 6403. doi: 10.1126/science.aar6089
Riar, A. K., Chhuneja, P., Keller, B., and Singh, K. (2021). Mechanism of leaf rust resistance in wheat wild relatives, Triticum monococcum L. and T. boeoticum L. Plant Genet. Resour. 19, 320–327. doi: 10.1017/S147926212100037X
Ruzicka, D. R., Kandasamy, M. K., McKinney, E. C., Burgos-Rivera, B., and Meagher, R. B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52, 460–472. doi: 10.1111/j.1365-313X.2007.03257.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharp, P. M. and Li, W.-H. (1987). The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295. doi: 10.1093/nar/15.3.1281
Song, G., Zhang, R., Zhang, S., Li, Y., Gao, J., Han, X., et al. (2017). Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genomics 18, 1–15. doi: 10.1186/s12864-017-3556-2
Staiger, C. J. (2000). Signaling to the actin cytoskeleton in plants. Annu. Rev. Plant Biol. 51, 257–288. doi: 10.1146/annurev.arplant.51.1.257
Staiger, C. J. and Blanchoin, L. (2006). Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 9, 554–562. doi: 10.1016/j.pbi.2006.09.013
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612. doi: 10.1093/nar/gkab835
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Tian, W., Chen, C., Lei, X., Zhao, J., and Liang, J. (2018). CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367. doi: 10.1093/nar/gky473
Tounsi, S., Ben Amar, S., Masmoudi, K., Sentenac, H., Brini, F., and Véry, A.-A. (2016). Characterization of two HKT1;4 transporters from Triticum monococcum to elucidate the determinants of the wheat salt tolerance Nax1 QTL. Plant Cell Physiol. 57, 2047–2057. doi: 10.1093/pcp/pcw123
Vanková, R., Kosová, K., Dobrev, P., Vítámvás, P., Trávníčková, A., Cvikrová, M., et al. (2014). Dynamics of cold acclimation and complex phytohormone responses in Triticum monococcum lines G3116 and DV92 differing in vernalization and frost tolerance level. Environ. Exp. Bot. 101, 12–25. doi: 10.1016/j.envexpbot.2014.01.002
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Whitehead, A. and Crawford, D. L. (2005). Variation in tissue-specific gene expression among natural populations. Genome Biol. 6, 1–14. doi: 10.1186/gb-2005-6-2-r13
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, K., Zhao, Y., Zhao, S., Liu, H., Wang, W., Zhang, S., et al. (2021). Genome-wide identification and low temperature responsive pattern of actin depolymerizing factor (ADF) gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 12, 618984. doi: 10.3389/fpls.2021.618984
Yang, R., Wang, F., Luo, P., Xu, Z., Wang, H., Zhang, R., et al. (2024). The role of the adf gene family in maize response to abiotic stresses. Agronomy 14, 717. doi: 10.3390/agronomy14040717
Zhang, M., Liu, X., Peng, T., Wang, D., Liang, D., Li, H., et al. (2021a). Identification of a recessive gene YrZ15–1370 conferring adult plant resistance to stripe rust in wheat-Triticum boeoticum introgression line. Theor. Appl. Genet. 134, 2891–2900. doi: 10.1007/s00122-021-03866-3
Keywords: Triticum monococcum L. subsp. aegilopoides, ADF, evolution, expression profiles, cold stress
Citation: Liu X, Zhang M, Su J, Wu L, Shen M, Wang Q, Zhuang Y, Zhang L, Li H and Chen G (2025) Genome-wide identification and comprehensive characterization of the ADF gene family in Triticum monococcum L. subsp. aegilopoides with insights into structure, evolution and cold stress response. Front. Plant Sci. 16:1649202. doi: 10.3389/fpls.2025.1649202
Received: 18 June 2025; Accepted: 18 July 2025;
Published: 04 August 2025.
Edited by:
Yogeshwar Vikram Dhar, Ruhr University Bochum, GermanyReviewed by:
Fang Liu, Anhui Academy of Agricultural Sciences, ChinaRuisi Yang, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2025 Liu, Zhang, Su, Wu, Shen, Wang, Zhuang, Zhang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Chen, Y2cxOTkxMDkwNUBnbWFpbC5jb20=
 Xin Liu
Xin Liu Minghu Zhang1
Minghu Zhang1 Lianquan Zhang
Lianquan Zhang Haosheng Li
Haosheng Li Gang Chen
Gang Chen