- 1State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Research Center for Perennial Rice Engineering and Technology of Yunnan, School of Agriculture, Yunnan University, Kunming, China
- 2State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China
- 3Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 4College of Agriculture and Biology Science, Dali University, Dali, China
- 5BGI-Baoshan, Baoshan, China
- 6Agricultural Biotechnology Directorate of the Ethiopian Biotechnology Institute, Addis Ababa, Ethiopia
The genetic control of plant architecture in crops is critical for agriculture and understanding morphological evolution. This study showed that an open reading frame (ORF) of the rice domestication gene PROG1 appeared 3.4–3.9 million years ago (Mya). Subsequently, it acquired a novel protein-coding gene function in the genome of O. rufipogon (~0.3–0.4 Mya). This extremely young gene and its paralogous C2H2 genes located nearby define the prostrate architecture of O. rufipogon and, thus, are of adaptive significance for wild rice in swamp and water areas. However, selection for dense planting and high yield during rice domestication silenced the PROG1 gene and caused the loss of the RPAD locus containing functional C2H2 paralogs; hence, domesticated lines exhibit an erect plant architecture. Analysis of the stepwise origination process of PROG1 and its evolutionary genetics revealed that this zinc-finger coding gene may have rapidly evolved under positive selection and promoted the transition from non- or semi-prostrate growth to prostrate growth. A transgenic assay showed that PROG1 from O. rufipogon exerts a stronger function compared with PROG1 sequences from other Oryza species. However, the analysis of the expression levels of PROG1 in different Oryza species suggests that the transcriptional regulation of PROG1 has played an important role in its evolution. This study provides the first strong case showing how a fundamental morphological trait evolved in Oryza species driven by a gene locus.
Introduction
Unlike ancient genes, which often perform critical functions in species, newly evolved genes have been considered to be dispensable or to have minor biological functions (Miklos and Rubin, 1996; Zhang et al., 1999; Krylov et al., 2003). Previously, de novo origin of a protein-coding gene from non-coding sequences was even generally considered impossible (Jacob, 1977). Although recent works have reported that the existence of physiologically essential de novo genes and novel genes from gene duplication, to date, there are no reports of such genes controlling fundamental morphological traits (Chen et al., 2010; Li C.Y. et al., 2010; Li D. et al., 2010).
With the increasing amounts of genome data for Oryza species being reported, there species have become good model species for plant comparative genomics and phenotype studies, and the relationships between genotype and phenotype can be studied systematically in these taxa. Although a recent study identified several de novo genes based on expression at the RNA or protein level in Oryza sativa (Zhang L. et al., 2019), domestication genes that have been fixed in cultivated rice via a loss of function and their evolution progress have not been detected.
Asian cultivated rice (O. sativa) was domesticated ∼8,000–10,000 years ago (Sharma et al., 2000; Vaughan et al., 2008; Fuller et al., 2010). In the course of domestication, some traits, such as shattering (Konishi et al., 2006; Li et al., 2006), panicle architecture (Ishii et al., 2013; Zhu et al., 2013) and pericarp and hull colors (Sweeney et al., 2006; Zhu et al., 2011), were changed. In particular, plant architecture underwent extensive changes associated with efficient agricultural use, including the change from prostrate growth in the cultivated rice progenitor to an erect structure in both Asian and African cultivars. In previous studies, the monogenetic domestication gene PROG1 in O. sativa and its paralog in Oryza rufipogon were cloned and identified as transcription factors based on their ∼90 bp C2H2-type zinc-finger motifs (Jin et al., 2008; Tan et al., 2008). These paralogs were found to have undergone strong artificial selection during the history of rice domestication (Jin et al., 2008; Tan et al., 2008; Wu et al., 2018). Although other genes controlling tiller angle and branching that play important roles in rice architecture, such as Tiller Angle Controlling (TAC1), LA1 (LAZY1), IDEAL PLANT ARCHITECTURE1 (IPA1), and OsTb2, have been cloned in O. sativa, these genes have undergone selection only via artificial selection for high-density planting during domestication (Li et al., 2007; Yoshihara and Iino, 2007; Yu et al., 2007; Jiang et al., 2012; Lu et al., 2013; Lyu et al., 2020), with no evidence of a history of both natural and artificial selection.
In this study, the domestication gene PROG1 was analyzed and identified as a young gene in Oryza that has driven the evolution of plant architecture. The open reading frame (ORF) of PROG1 arose in O. punctata and evolved via natural selection into a prostrate-growth gene in O. rufipogon. More interestingly, PROG1 was then functionally lost in O. sativa through artificial selection, which accompanied locus deletions (RICE PLANT ARCHITECTURE DOMESTICATION, RPAD) linked to the PROG1 gene during artificial selection on architecture in the domestication of cultivated rice (Wu et al., 2018). Therefore, we hypothesize that the successive gain and loss of function of PROG1 locus under natural and artificial selection, respectively, could result in variation of plant architecture during Oryza evolution.
Materials and Methods
PROG1 Locus Sequence Alignment and Origin Analysis
Ten released genomes of Oryza species [O. sativa (Sasaki and International Rice Genome Sequencing Project, 2005), O. glaberrima (Wang et al., 2014), O. longistaminata (Zhang et al., 2015), O. meridionalis (Zhang et al., 2014), O. glumaepatula (Zhang et al., 2014)], O. brachyantha (Chen et al., 2013), O. rufipogon (Stein et al., 2018), O. nivara (Stein et al., 2018), O. barthii (Stein et al., 2018), O. punctata (Stein et al., 2018), and the B. distachyon genome (Vogel et al., 2010) were used in this study. The PROG1 locus and its neighboring genes (two upstream and two downstream) were extracted from the O. sativa genome, and BLAST (Altschul et al., 1997) was used to obtain the genome sequences of remaining species, which were then annotated and aligned by using MEGA6 (Tamura et al., 2013).
Phylogenetic Tree Construction and Divergence Time Estimation for 10 Oryza Species
Blastall (v2.2.21) (Altschul et al., 1997) with a threshold of “-e 1e-5” was used to align peptide sequences from the 10 Oryza species, and gene families were clustered by OrthoMCL (v1.4) (Li et al., 2003). From the identified single-copy gene families, 4-fold degenerated (4D) sites in the coding sequences of the genes were extracted and concatenated. Multiple sequence alignments were performed by MUSCLE (v3.7) (Edgar, 2004), and a phylogenetic tree with settings nst = 6, rates = invgamma and ngen = 1,000,000 was reconstructed using MrBayes (v3.1.2) (Ronquist and Huelsenbeck, 2003). To estimate divergence times among the 10 species, the program MCMCTree in PAML9 (v4.4) (Yang, 1997) with the parameters “clock = 3 and RootAge ≤ 0.1” was used. The divergence times were constrained by the fossil calibration times from TimeTree (0.4 Mya between O. sativa and O. rufipogon, 0.6–2.0 Mya between O. punctata and O. meridionalis and 9–15 Mya between O. punctata and O. brachyantha) (Hedges et al., 2006).
Vector Construction and Rice Transformation
The PROG1 promoter (1.5 kb) from O. rufipogon and coding sequences (CDSs) from O. sativa, O. rufipogon (Yuanjiang), O. nivara, O. longistaminata, O. meridionalis, O. glumaepatula, and O. punctata were amplified and inserted into the expression cassette pCAMBIA1300, and the ProPROG1:PROG1-NOS vectors were constructed. The recombinant plasmids were transferred into calli of the japonica rice cultivar Zhonghua11 (ZH11) by an Agrobacterium tumefaciens-mediated transformation method. The forward primer for the PROG1 promoter was 5′-AATCAGCTCGAGCTAGGTCTTTG-3′, and the reverse primer for the PROG1 promoter was 5′-GAAAGGAAAATGGGACAAGCTAT-3′. The forward primer for the PROG1 CDS was 5′-ATGGATCCCTCATCGGCTTC-3′, and the reverse primer for the PROG1 CDS was 5′-CTAGAGGCCGAGCTCGAGGA-3′.
Prog1 Locus Expression Analysis
Eight transcriptomes of O. sativa (Zhang et al., 2010), two transcriptomes of O. nivara and O. barthii (Wang et al., 2014), two transcriptomes of O. punctata (SRR1171006 and SRR1171007 in NCBI), two transcriptomes of O. brachyantha (Chen et al., 2013) and 11 transcriptomes of B. distachyon (Davidson et al., 2012) were downloaded from NCBI. Eight transcriptomes of O. longistaminata were obtained previously work (Zhang et al., 2015). RNA-seq reads from each sample were mapped to the corresponding reference genome with TopHat 2.0.3 with default parameters, and Cufflinks was then used to evaluate the FPKM values (Trapnell et al., 2012) of the PROG1 locus and the internal control gene Actin1. To investigate the expression of the PROG1 locus at the tiller base in O. rufipogon, O. nivara, O. barthii, O. longistaminata and O. glumaepatula, total RNA was extracted using TRIzol reagent (Invitrogen, United States) and reverse transcribed using the Revert Aid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. qRT-PCR of PROG1 was performed following the manufacturer’s instructions, and the Tubulin gene was used as the internal control. The forward primer for PROG1 was 5′-GATCCCTCATCGGCTTCTT-3′, and the reverse primer for PROG1 was 5′-GGAACAGCCTCACTTGCTTG-3′. The forward primer for Tubulin was 5′-GCTCCGTGGCGGTATCAT-3′, and the reverse primer for Tubulin was 5′-CGGCAGTTGACAGCCCTAG -3′.
Field Experiment and Plant Architecture Survey
To investigate the plant architectures of Oryza species, 10 species (O. sativa, O. rufipogon, O. nivara, O. glaberrima, O. barthii, O. longistaminata, O. meridionalis, O. glumaepatula, O. punctata, and O. brachyantha) were grown in Xishuangbanna, Southwest China. The plant architectures were surveyed after 3 months.
Tests of Selection on PROG1 in O. rufipogon Populations
PROG1 population data (Tan et al., 2008) for O. rufipogon were downloaded from NCBI and aligned using MEGA6 (Tamura et al., 2013). Tajima’s D test and Fu and Li’s test were conducted using DnaSP 5 (Librado and Rozas, 2009).
Monte Carlo Simulations
Monte Carlo simulations were performed to determine the possibility that a random sequence could produce a C2H2 gene. Random sequences of 90 bp length (seed sequences) were generated accordance with the A, T, G, and C frequencies of the rice genome. The total number of seed sequences per simulation was a quotient of genome size and seed sequence length (T). A set of 100,000 simulations run with T seed sequences in each simulation was used. The number of C2H2 motifs for each seed sequence was counted by searching the motif pattern “Ø-X-C-X2,4,5-C-X3-Ø-X5-Ø -X2-H-X3,4-H” in all six reading frames (Klug and Schwabe, 1995). The distribution of the observed number of C2H2 genes was illustrated using kernel density estimation as implemented in R. The p-value (observed ≥ expected) was calculated by counting the frequencies of observed C2H2 genes that were equal to or larger than the expected C2H2 genes.
Clustering of C2H2 Transcription Factor Genes in the Genome of O. sativa
The amino acid sequences of 189 C2H2 transcription factor genes (Agarwal et al., 2007) were divided into three types: full-length sequences, sequences with only the C2H2 motif, and sequences without the C2H2 motif. Multiple sequence alignments were performed using MUSCLE (v3.7) (Edgar, 2004). A phylogenetic tree was constructed with FastTree (2.1.10) (Price et al., 2009) and viewed with ETE (Huerta-Cepas et al., 2016).
Results
The ORF of PROG1 Appeared in O. punctata and Has Experienced Different Evolutionary Fates in Different Oryza Species
The PROG1 locus was analyzed by using the available genome sequences of eight AA genome Oryza species, i.e., O. sativa (Sasaki and International Rice Genome Sequencing Project, 2005), O. glaberrima (Wang et al., 2014), O. longistaminata (Zhang et al., 2015), O. glumaepatula (Zhang et al., 2014), O. meridionalis (Zhang et al., 2014), O. rufipogon (Stein et al., 2018), O. nivara (Stein et al., 2018), and O. barthii (Stein et al., 2018); one BB genome from O. punctata (Stein et al., 2018); and one FF genome from O. brachyantha (Chen et al., 2013). One non-Oryza Gramineae species, Brachypodium distachyon (Vogel et al., 2010), was used as an outgroup. Based on genomic alignment of the syntenic PROG1 sequence, O. rufipogon shares homologous syntenic regions with the other species assessed in this study (Supplementary Table S1). Highly similar flanking genes, such as Os07g0153300 and Os07g0153400 in the 5′ upstream region of PROG1 and Os07g0154100 and Os07g0154300 downstream were found in the eight AA genome Oryza species (Figure 1A). Although annotated genes in the neighboring loci were found, sequences homologous to PROG1 but without traces of the short C2H2-type zinc-finger motif sequence (∼90 bp) were identified in the two distant species O. brachyantha and B. distachyon. The absence of the C2H2 motif in non-Oryza-species results in the lack of any homology to the peptide sequence. In the BB genome of O. punctata, the homologous OPUNC07G03350.1 coding sequence, which was similar to that of O. rufipogon in length, showed four in-frame deletions, two in-frame insertions and more than 44 non-synonymous mutations (Supplementary Table S2). There was also a frameshift deletion in O. barthii or a frameshift insertion in both O. nivara and O. meridionalis (Figure 1B and Supplementary Figures S1, S2). Although the remaining three AA genomes (O. sativa, O. longistaminata, and O. glumaepatula) showed several mutations, they appeared to contain intact ORF sequences homologous to the O. rufipogon PROG1 gene including a number of non-synonymous substitutions, two deletions and two insertions in O. longistaminata and one deletion and two insertions in O. glumaepatula (Supplementary Table S2 and Supplementary Figures S1, S2). Considering the fully prostrate architecture of O. rufipogon and the fact that the O. rufipogon PROG1 gene is different from those in other Oryza species based on the sequence alignment, PROG1 of O. rufipogon may have a highly effective function in controlling the prostrate phenotype. Therefore, we speculate that PROG1 is a new gene, the ORF of which appeared in O. punctata, and may have undergone functionalization only in O. rufipogon.
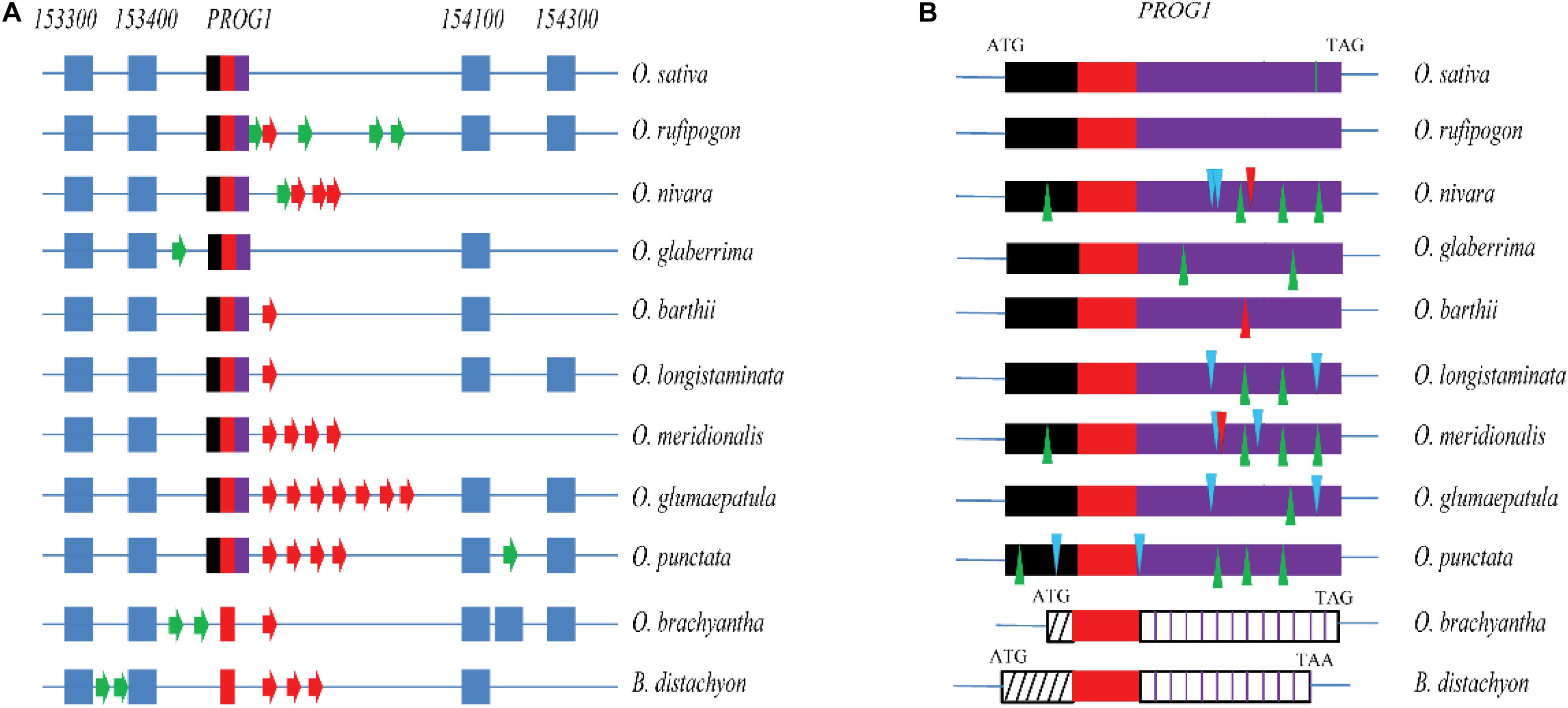
Figure 1. Alignment of the PROG1 locus in different Oryza species and the outgroup B. distachyon. (A) Collinearity of the PROG1 region in different species. 153300, 153400, 154100, and 154300 represent the flanking genes Os07g0153300, Os07g0153400, Os07g0154100, and Os07g0154300, respectively. Blue blocks and tricolor (black, red, and purple) blocks represent neighboring genes and PROG1 (the red blocks represent C2H2-type zinc-finger sequences). Arrows indicate other annotated genes in different genomes, green arrows indicate ORFs without homology with PROG1, and red arrows represent ORFs with C2H2 type zinc-finger sequences. (B) Alignment of the sequence of the PROG1 locus in different species. Black, red, and purple blocks represent alignable 5′-termini, C2H2 type zinc-finger and 3′-terminal sequences, respectively; black and purple striped blocks represent unalignable 5′-termini and 3′-terminal sequences, respectively; green and blue triangles represent in-frame deletions and insertions, respectively. The red triangles (O. barthii and O. meridionalis) represent frameshift indels (red triangles above the lines represent frameshift insertions, and red triangles represent frameshift deletions), and the green vertical line represents the M6 site in O. sativa.
PROG1 May Have Arose via Gene Duplication
Considering the sequence mutation data, PROG1 appears to have arisen via gene duplication in O. rufipogon. To test this hypothesis, the possibility that C2H2 genes could originate in the rice genome by random chance was analyzed. In this analysis, Monte Carlo simulations of C2H2 motifs with repeated sampling for 100,000 iterations were conducted, and the expected number of C2H2 motifs was compared with the observed number (Figure 2A). Previous studies have identified 189 C2H2 transcription factors in the rice genome (Agarwal et al., 2007). Before clustering these genes, the conserved C2H2 motifs were deleted. Based on the computed distance matrices, the 189 genes were clustered into five groups (Figure 2B). This observation showed that these 189 C2H2 transcription factor genes were likely duplicated from five ancestral genes, or “starter gene.” We then performed Monte Carlo simulations with 100,000 iterations, using the number of starter genes as the expected number and the number of simulated C2H2 motifs as the observed number. We found that the chance of C2H2 motifs originating in the rice genome by random chance was 99.055%, suggesting that ab initio origination of a C2H2 gene is possible in the rice genome. These observations indicated that a C2H2 gene can be easily generated by random mutations in the rice genome. Moreover, the assessment of large gene families in the rice genome, such as PROG1 (LOC_Os07g05900), showed an incidental gene duplication stemming from of a proto-PROG1 gene (Figure 2B). The RPAD locus, which also participates in domesticated plant architecture in both Asian and African cultivated rice, harbors a tandem repeat of zinc-finger genes (including the PROG1 gene) controlling plant architecture in wild rice (Wu et al., 2018). To determine whether the tandem repeats of zinc-finger genes originated from the same proto-PROG1 gene, further phylogenetic tree analysis was performed, which clustered eight C2H2 genes, including PROG1, into one group (Supplementary Figure S3). This finding implies that the tandem repeats of the zinc-finger genes were produced by gene duplication. These results indicate that PROG1 may have been produced from other C2H2-containing paralogous genes by gene duplication.
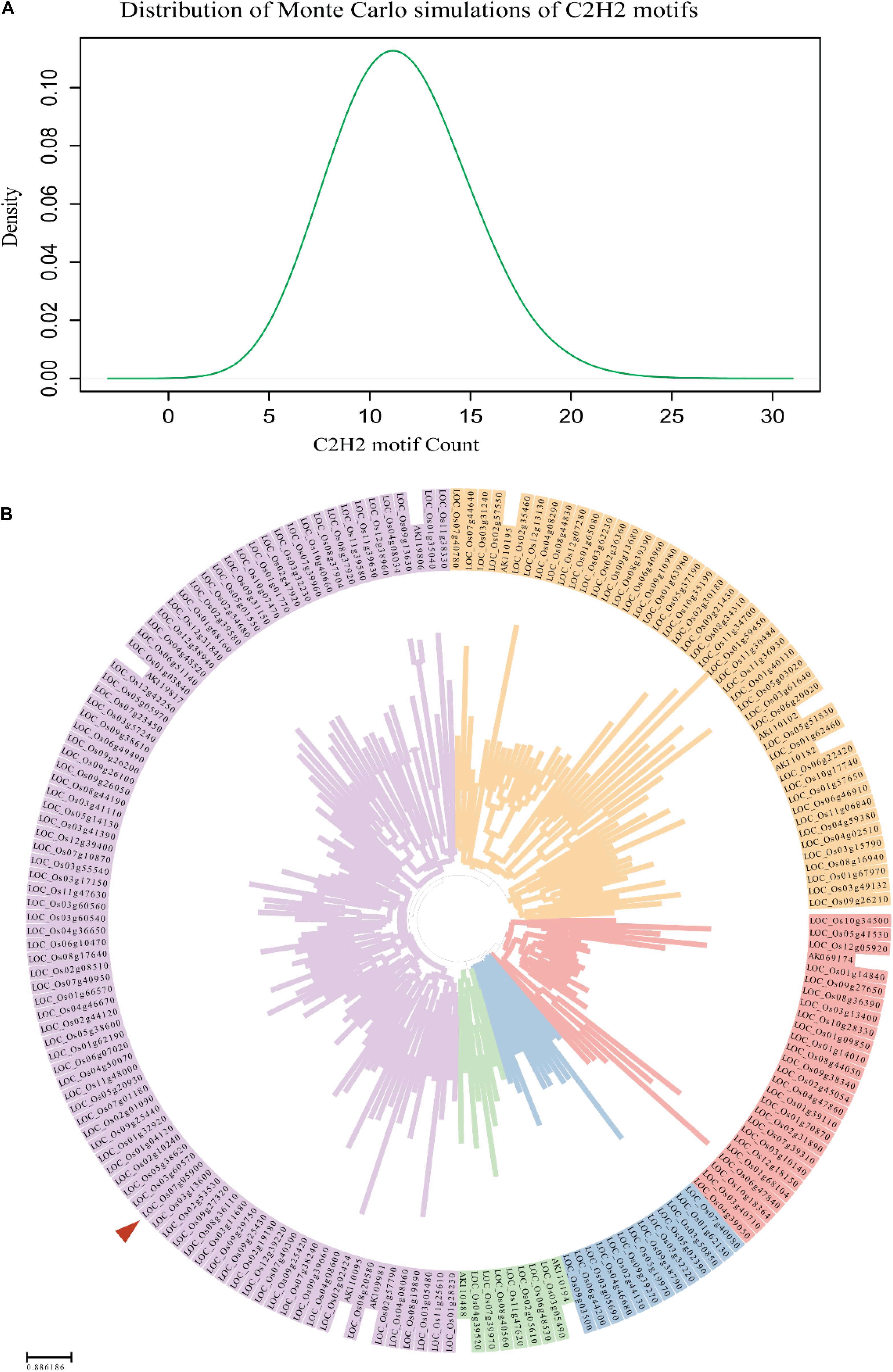
Figure 2. Monte Carlo simulation results for the random de novo origination of a C2H2 motif. (A) Distribution of Monte Carlo simulations of C2H2 motifs. (B) Clustering relationships of the 189 C2H2 transcription factor genes (Agarwal et al., 2007) in the genome of O. sativa. A red triangle indicates the PROG1 (LOC_Os07g05900) of O. sativa.
PROG1 of O. rufipogon Has a Strong Function in the Prostrate Plant Architecture
Interestingly, PROG1 of O. sativa was found to be a pseudogene selected by a strong artificial selection (Jin et al., 2008; Tan et al., 2008; Xu et al., 2011; Huang et al., 2012). The transgenic experiments demonstrated that although PROG1 in O. sativa has lost its function, it is actively involved in the prostrate phenotype of O. rufipogon (Figure 3); this finding is consistent with previous studies (Jin et al., 2008; Tan et al., 2008). All PROG1-homologous coding sequences from other Oryza species (except for that of O. barthii) driven by PROG1 promoter of O. rufipogon were transformed into the Zhonghua 11 variety of O. sativa to verify their functions. The results suggested that O. rufipogon PROG1 clearly has a function in producing a prostrate phenotype (Figure 3). The PROG1 homologs of other Oryza species have no function or only weakly affect plant architecture. Interestingly, some transgenic lines expressing O. longistaminata PROG1 showed divergent architecture (Figure 3).
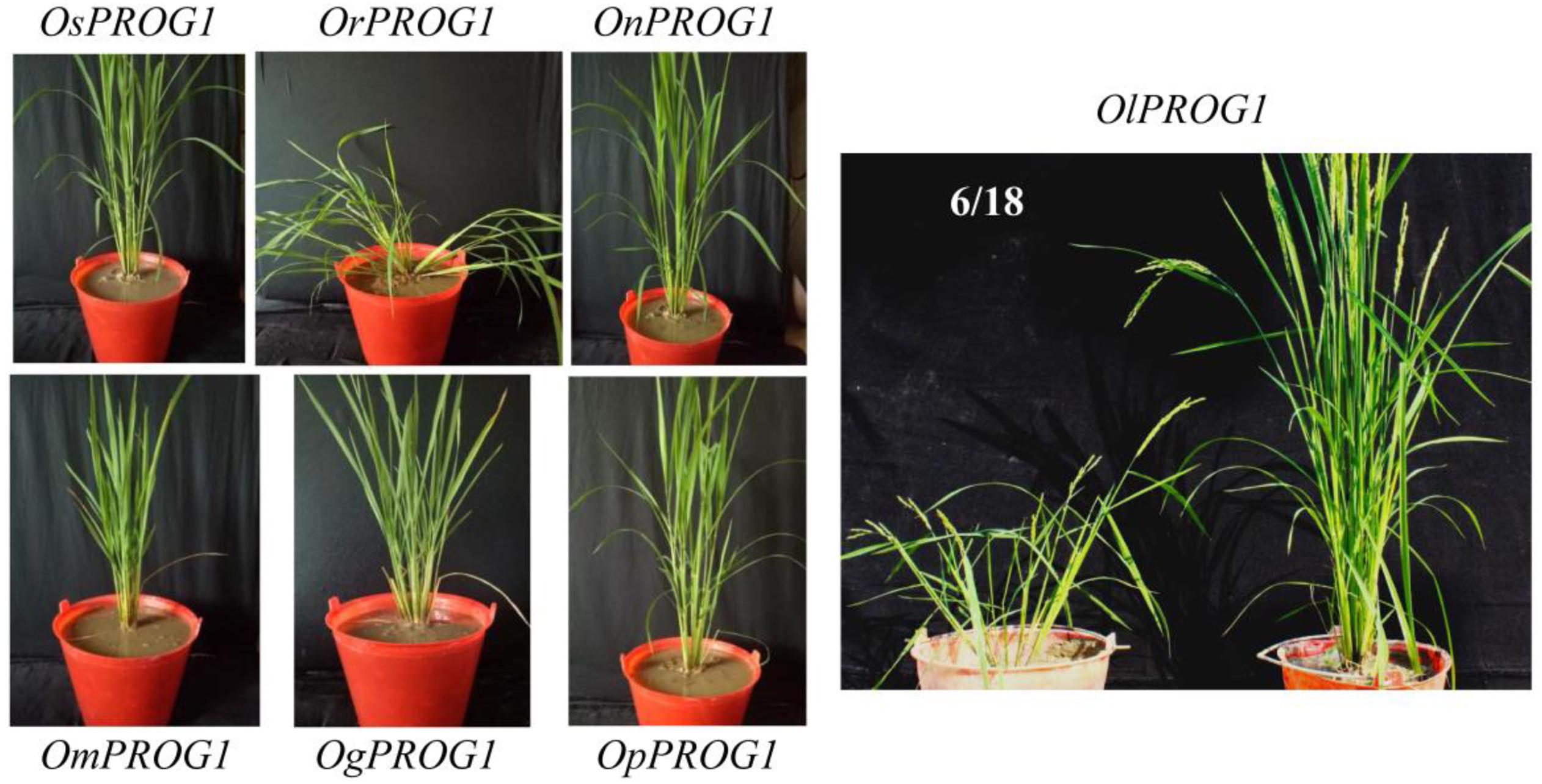
Figure 3. Role of Oryza PROG1s in controlling prostrate plant architecture driven by the O. rufipogon PROG1 promoter. Transgenic verification of various PROG1 alleles in O. sativa (Zhonghua 11). OsPROG1, PROG1 from O. sativa. OrPROG1, PROG1 from O. rufipogon. OnPROG1, PROG1 from O. nivara. OmPROG1, PROG1 from O. meridionalis. OgPROG1, PROG1 from O. glumaepatula. OpPROG1, PROG1 from O. punctata. OlPROG1, PROG1 from O. longistaminata. Transgenic lines for each gene, n >15. The 6/18 ratio indicates that 6 lines exhibit a prostrate phenotype among 18 transgenic lines.
Because PROG1 functions in the tiller base of O. rufipogon and determines plant architecture, real-time PCR (qRT-PCR) was conducted to determine the expression of the PROG1 homologs of O. rufipogon, O. nivara, O. barthii, O. longistaminata, and O. glumaepatula. This expression analysis showed that the PROG1 gene was expressed in the unelongated basal tiller internodes of O. rufipogon, O. longistaminata, and O. glumaepatula (Figure 4). Previous studies also reported high expression of PROG1 in the unelongated basal tiller internodes of O. rufipogon (Jin et al., 2008). However, an extremely low level of expression in the unelongated basal internodes was detected in O. sativa (Jin et al., 2008), O. nivara, and O. barthii (Figure 4). In two transcriptomes (leaf and panicle) of O. nivara and O. barthii (Wang et al., 2014), only an extremely low level of expression of a pseudo-PROG1 gene was detected in the panicle transcriptome of O. barthii, and no expression was detected in O. nivara. In eight transcriptomes from the rhizome, stem, rhizome tips, stem tips, stamens, pistils, hybrid line stamens, and hybrid line pistils of O. longistaminata (Zhang et al., 2015), low levels of expression were detected in only the rhizome tips and stamens (Supplementary Table S3). The transgenic test in this study revealed PROG1 as an expressible pseudogene in O. glumaepatula that possibly plays a role in regulating rhizome transverse elongation in O. longistaminata. In the NCBI database of the BB genome of O. punctata, no expression of OPUNC07G03350.1 was detected in the two transcriptomes (root and panicle) (Supplementary Table S3). In the two transcriptomes of O. brachyantha (Chen et al., 2013) and 11 transcriptomes of B. distachyon (two transcriptomes from 20-day leaves, two transcriptomes from embryos 25 days after pollination, endosperm 25 days after pollination, early inflorescence, emerging inflorescences, pistils, seeds 5 days after pollination, anthers, and seeds 10 days after pollination) (Davidson et al., 2012), no expression of annotated non-homologous genes at the PROG1 locus was detected (Supplementary Table S4). Except in O. rufipogon, the collective expression results of the Oryza species did not indicate the PROG1 locus as a unique gene related to tiller development. The very low expression of the PROG1 locus in the various tissues of different Oryza species suggests the possibility that the PROG1 gene evolved as a functional gene until the appearance of the most recent O. rufipogon ancestor.
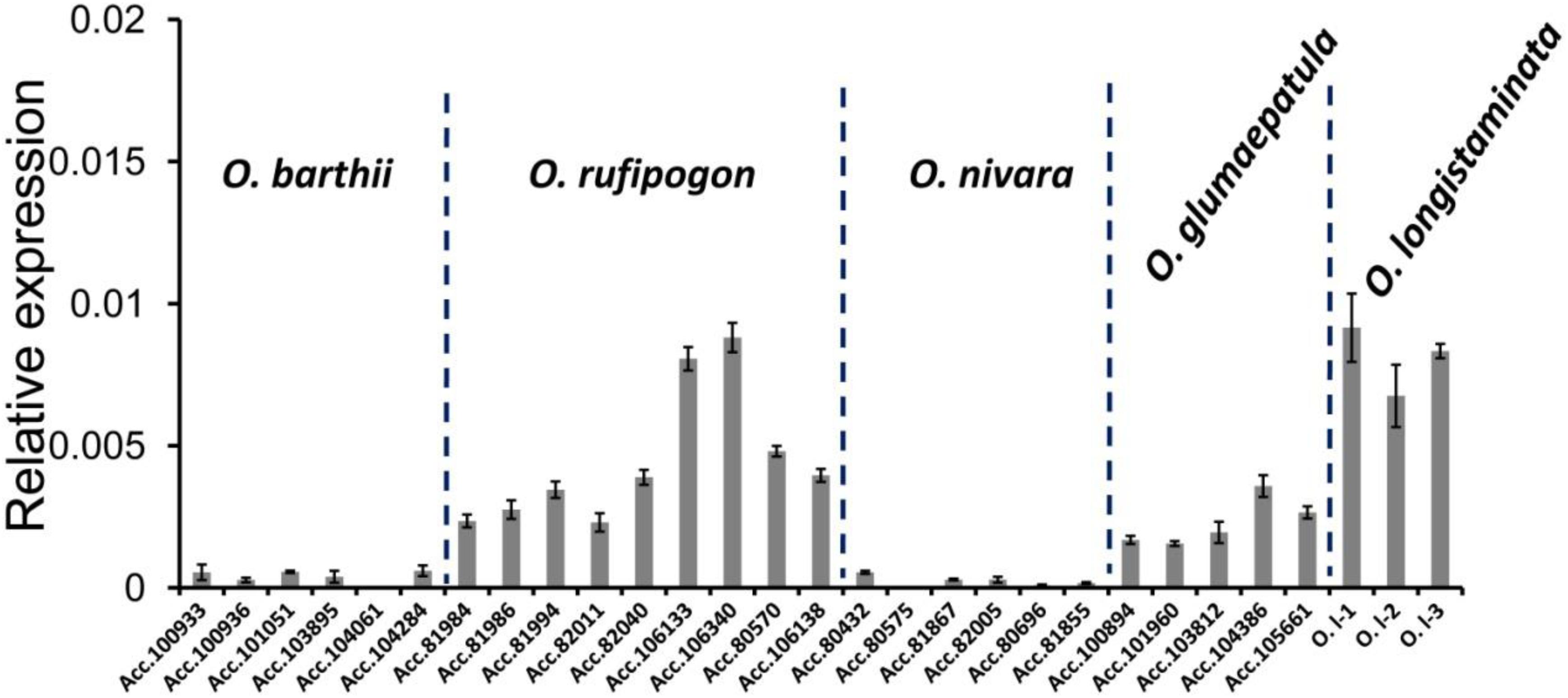
Figure 4. The different expression patterns of PROG1 in different Oryza species. qRT-PCR analysis of PROG1 in unelongated basal tiller internodes of various accessions (Supplementary Table S5) of Oryza species. Error bars indicate the standard deviation (SD) of three biological repeats.
The Young PROG1 Gene Underwent Strong Natural Selection in O. rufipogon
Using the whole genome, a phylogenetic tree of 10 Oryza species was constructed, and their divergence times were estimated using all the single-copy genes in their genomes. The phylogenetic results of this study are similar to those shown in TimeTree (Zhu and Ge, 2005; Hedges et al., 2006; Figure 5). For instance, 11.4 Mya in TimeTree and 12.1 Mya in our results were the suggested divergence times of O. brachyantha. Likewise, O. rufipogon diverged from other species at 0.4 Mya in TimeTree and 0.3 Mya in this study. The first homologous ORF of PROG1 appeared in the BB genome of O. punctata at ~3.4–3.9 Mya. These results suggest that the PROG1 gene encoding a plant architecture regulatory protein in O. rufipogon was notably young (0.3–0.4 Mya).
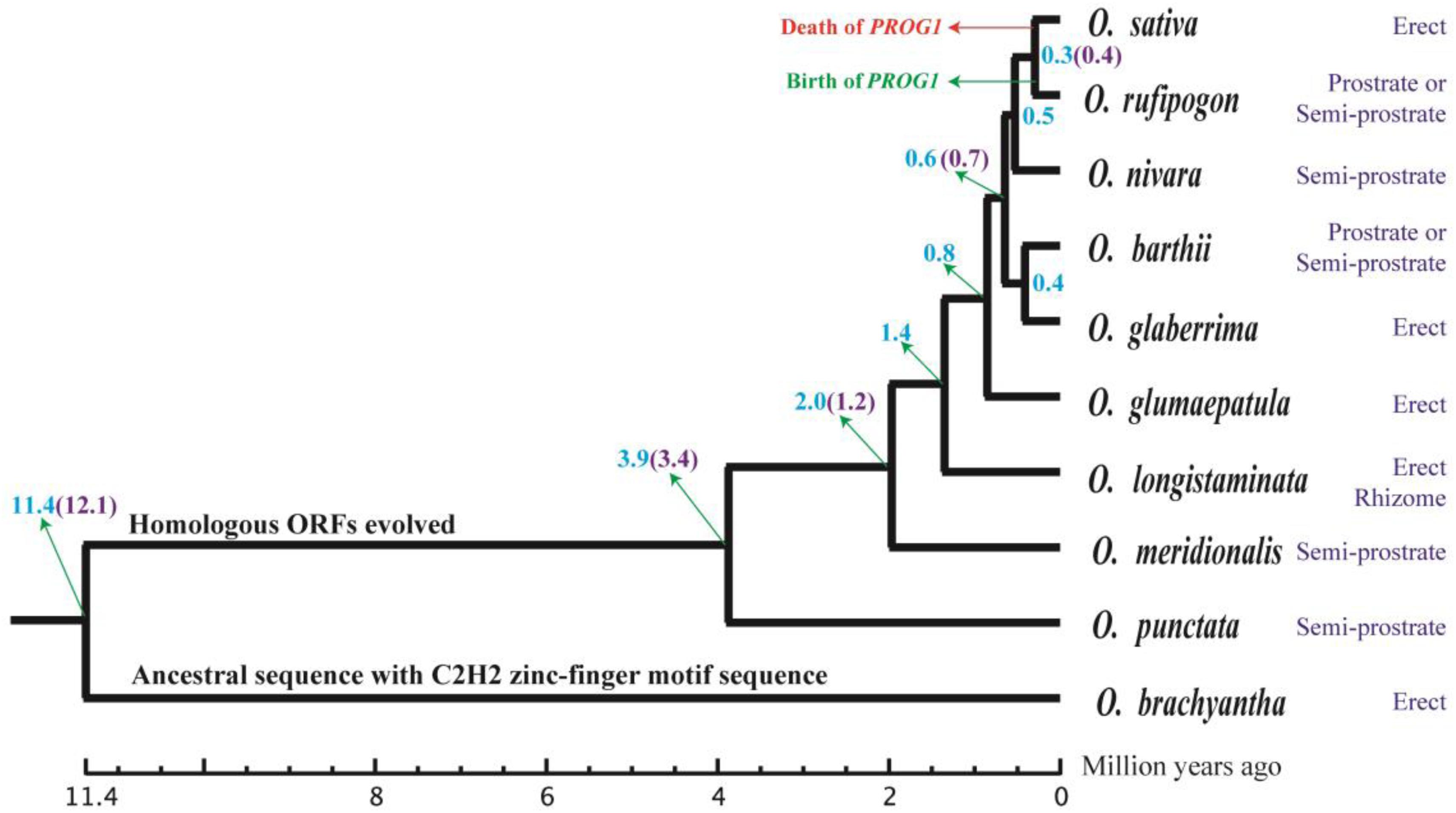
Figure 5. Evolution of plant architecture in Oryza species. The evolutionary process of the young orphan gene PROG1 was labeled on the corresponding branches, and the plant architecture phenotypes of 10 Oryza species are shown to the right of the phylogenetic tree. The estimated divergence time is labeled at each node. The numbers marked with blue were estimated with whole-genome data, and those marked with purple are from TimeTree.
To examine PROG1 as a gene controlling the prostrate plant architecture in O. rufipogon, we grew O. rufipogon, O. nivara, O. glaberrima, O. barthii, O. longistaminata, O. glumaepatula, and O. punctata to check their architectures. During this architecture assessment, a fully prostrate plant architecture was observed only in O. rufipogon. The other Oryza species showed non-prostrate or semi-prostrate architectures that were either strictly erect or semi-erect with some angled tillers (Supplementary Table S5 and Figure 5). The published population data of PROG1 in O. rufipogon (Tan et al., 2008) were analyzed to assess the hypothesis of a selective sweep of PROG1 in O. rufipogon. Tajima’s D test detected a significant departure from neutrality in this gene (3.03762, P < 0.001) and suggested strong natural selection on PROG1 in O. rufipogon. Furthermore, Fu and Li’s test was conducted using the outgroup sequences of PROG1 loci from four Oryza species (O. punctata, O. glumaepatula, O. longistaminata, and O. sativa). A significant signal of positive selection was detected in O. rufipogon in relation to either of the outgroup sequences, with a p < 0.05 for both the D and F statistics of Fu and Li’s test (Supplementary Table S6). These results provide evidence that the prostrate architecture is a derived trait that has undergone strong natural selection in O. rufipogon and that the trait is conferred or enhanced by the PROG1 gene.
Discussion
Plant architecture plays important roles in plant survival and adaptation to diverse conditions. It has been reported that O. rufipogon inhabits swamps with moderately deep water (Grillo et al., 2009), and thus, the prostrate plant architecture that evolved in O. rufipogon likely allowed it to spread across the water to achieve greater access to light and chemical nutrients and confers structural tenacity on variable surfaces. Our study demonstrated that the critical PROG1 gene locus, which regulates plant architecture in O. rufipogon, emerged through a process of de novo origination from 3.4 to 3.9 Mya in O. punctata and evolved into a functional gene strongly affecting phenotype in O. rufipogon between 0.3 and 0.4 Mya. Because PROG1 evolved recently, the adaptive evolution and selection of PROG1 in O. rufipogon populations may still be ongoing. Published population data for PROG1 in O. rufipogon (Tan et al., 2008) were analyzed to examine the hypothesis of a selective sweep of PROG1 in O. rufipogon. Tajima’s D test revealed a significant departure from neutrality for this gene (3.03762, P < 0.001) and suggested the existence of strong natural selection on PROG1 in O. rufipogon. The sequence evolution in the PROG1 gene, together with the derived prostrate plant architecture of O. rufipogon, suggests strong adaptive evolution that started with the fixation of this young gene in ancestral O. rufipogon populations and continues in an extant population.
Causative mutations including SNPs (Konishi et al., 2006; Li et al., 2006; Lin et al., 2007), structural variations such as small indels (Sweeney et al., 2006; Hua et al., 2015; Bessho-Uehara et al., 2016; Jin et al., 2016), large structural variations (Wu et al., 2018), and mobile DNA elements (Studer et al., 2011) play important roles during crop domestication and usually result in dysfunctions and/or alterations of the expression patterns of domestication-related genes. New genes can be produced in multiple ways, including gene duplication and de novo origination from previously non-coding sequences (Ding et al., 2012). The PROG1 gene may have originated in the BB genome of O. punctata as a C2H2 gene with an unknown function and been neo-functionalized by gaining a function related to prostrate architecture in O. rufipogon. Similar tandem repeats of zinc-finger protein-coding genes have been found in the collinear chromosomal region of the RPAD locus and might be recognized as an ancient zinc-finger gene cluster with a conserved functional role in the regulation of plant growth habits. Because several C2H2 genes, including PROG1, ZnF5, ZnF7, and ZnF8, function in the control of the prostrate-growth trait (Wu et al., 2018), causative mutations cannot be found in PROG1. A second hypothesis is that this gene was not a functional gene before the evolution of O. rufipogon, which is supported by evidence from transgenic experiments, gene expression, phenotyping and assessments of repeated ORF disruptions and remarkable sequence variability in different species.
Interestingly, the PROG1 expression level in O. longistaminata is as high as that in O. rufipogon. Considering that some transgenic lines expressing O. longistaminata PROG1 exhibit a semi-prostrate phenotype, whether O. longistaminata PROG1 functions in the lateral elongation of rhizomes needs further investigation. The deletion site at the RPAD locus was also a target of artificial selection during domestication in both Asian and African rice (Wu et al., 2018). Although it remains unclear whether the causative mutations within the PROG1 promoter and coding sequence are associated with prostrate function, variations in protein sequence and expression were selected by rice breeding (Jin et al., 2008; Tan et al., 2008).
Domestication-related genes such as fw2.2, fascinated (fas), teosinte glume architecture (tga1), teosinte branched1 (tb1), IPA1, and OsTb2, are responsible for agricultural advances and morphological improvements during rice, tomato and maize domestication (Doebley et al., 1997; Wang et al., 1999, 2005; Frary et al., 2000; Cong et al., 2008; Lu et al., 2013; Lyu et al., 2020). Mutations in genes such as tb1, tin1, IPA1, and OsTb2 brought about the change from wild Mexican grass (teosinte) and O. rufipogon to the cultivate type architecture mainly for branch numbers in maize and rice domestication (Doebley et al., 1997; Wang et al., 1999; Lu et al., 2013; Zhang X. et al., 2019; Lyu et al., 2020). Tiller angle controlling genes TAC1 and LA1 (LAZY1) and PROG1 also play important roles in rice architecture for high-density planting during rice domestication (Li et al., 2007; Yoshihara and Iino, 2007; Yu et al., 2007; Jin et al., 2008; Tan et al., 2008; Jiang et al., 2012; Lyu et al., 2020). These observations lead us to speculate that these genes contributed to survival and adaptation in the wild ancestor species. A similar transition from prostrate to erect growth occurred during the domestication of wheat (Waisel, 1987). This finding provides another piece of evidence that a gene influencing plant architecture can be adaptive in the ancestor species, whereas its mutation can improve plant architecture and yield in domesticated crops (Doebley et al., 1997; Clark et al., 2006; Wang et al., 2018).
Conclusion
PROG1 is a new functional gene that was likely generated through gene duplication, and its predicted young age could be a result of a loss of sequence identity due to a high level of substitution in an ancient gene. Natural selection in a swamp habitat led to PROG1 functionalization to produce fully prostrate plant architecture, and artificial domestication aimed at maximizing yield via the high-density planting of rice with an erect plant architecture led to the pseudogenization of this gene and deletion of the RPAD locus.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: SRR1171006 and SRR1171007.
Author Contributions
LH and YZ designed and performed the experiments, designed the experiments, and wrote the manuscript. HL and JW analyzed the sequence data. RZ and YL generated the transgenic materials. YB and MN provided assistance in vector construction. SZ, GH, BC, QH, YG, and JZ provided assistance in the phenotyping experiment. GM revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation for Young Scientists of China (31701094 and 31601274) and grants from the Yunnan Provincial Science and Technology Department (2018IC096, 2019ZG013, and 2019HC028).
Conflict of Interest
YL and YZ were employed by BGI-Baoshan.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Manyuan Long of the University of Chicago, Prof. Fengyi Hu of the Yunnan University and Profs. Wen Wang, Song Ge, and Mingsheng Chen of the Chinese Academy of Sciences for providing suggestions for this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00876/full#supplementary-material
References
Agarwal, P., Arora, R., Ray, S., Singh, A. K., Singh, V. P., Takatsuji, H., et al. (2007). Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2020 65, 467–485. doi: 10.1007/s11103-007-9199-y
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J. H., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Bessho-Uehara, K., Wang, D. R., Furuta, T., Minami, A., Nagai, K., Gamuyao, R., et al. (2016). Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. U.S.A. 113, 8969–8974. doi: 10.1073/pnas.1604849113
Chen, J. F., Huang, Q. F., Gao, D. Y., Wang, J. Y., Lang, Y. S., Liu, T. Y., et al. (2013). Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat. Commun. 4:1595. doi: 10.1038/ncomms2596
Chen, S. D., Zhang, Y. E., and Long, M. Y. (2010). New genes in Drosophila quickly become essential. Science 330, 1682–1685. doi: 10.1126/science.1196380
Clark, R., Wagler, T., Quijada, P., and Doebley, J. (2006). A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38, 594–597. doi: 10.1038/ng1784
Cong, B., Barrero, L. S., and Tanksley, S. D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40, 800–804. doi: 10.1038/ng.144
Davidson, R. M., Gowda, M., Moghe, G., Lin, H. N., Vaillancourt, B., Shiu, S. H., et al. (2012). Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 71, 492–502. doi: 10.1111/j.1365-313X.2012.05005.x
Ding, Y., Zhou, Q., and Wang, W. (2012). Origins of new genes and evolution of their novel functions. Annu. Rev. Ecol. Evol. Syst. 43, 345–363. doi: 10.1146/annurev-ecolsys-110411-160513
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E. V. D., Cong, B., Liu, J. P., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. doi: 10.1126/science.289.5476.85
Fuller, D. Q., Sato, Y. I., Castillo, C., Qin, L., Weisskopf, A. R., Kingwell-Banham, E. J., et al. (2010). Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthrop. Sci. 2, 115–131. doi: 10.1007/s12520-010-0035-y
Grillo, M. A., Li, C. B., Fowlkes, A. M., Briggeman, T. M., Zhou, A. L., Schemske, D. W., et al. (2009). Genetic architecture for the adaptive origin of annual wild rice, Oryza nivara. Evolution 63, 870–883. doi: 10.1111/j.1558-5646.2008.00602.x
Hedges, S. B., Dudley, J., and Kumar, S. (2006). TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972. doi: 10.1093/bioinformatics/btl505
Hua, L., Wang, D. R., Tan, L. B., Fu, Y. C., Liu, F. X., Xiao, L. T., et al. (2015). LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27, 1875–1888. doi: 10.1105/tpc.15.00260
Huang, X. H., Kurata, N., Wei, X. H., Wang, Z. X., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Ishii, T., Numaguchi, K., Miura, K., Yoshida, K., Thanh, P. T., Htun, T. M., et al. (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45, 462–465. doi: 10.1038/ng.2567
Jiang, J. H., Tan, L. B., Zhu, Z. F., Fu, Y. C., Liu, F. X., Cai, H. W., et al. (2012). Molecular evolution of the TAC1 gene from rice (Oryza sativa L.). J. Genet. Genomics 39, 551–560. doi: 10.1016/j.jgg.2012.07.011
Jin, J., Hua, L., Zhu, Z. F., Tan, L. B., Zhao, X. H., Zhang, W. F., et al. (2016). GAD1 encodes a secreted peptide that regulates grain number, grain length, and Awn development in rice domestication. Plant Cell 28, 2453–2463. doi: 10.1105/tpc.16.00379
Jin, J., Huang, W., Gao, J. P., Yang, J., Shi, M., Zhu, M. Z., et al. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369. doi: 10.1038/ng.247
Klug, A., and Schwabe, J. W. (1995). Protein motifs 5. Zinc fingers. FASEB J. 9, 597–604. doi: 10.1096/fasebj.9.8.7768350
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Krylov, D. M., Wolf, Y. I., Rogozin, I. B., and Koonin, E. V. (2003). Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13, 2229–2235. doi: 10.1101/gr.1589103
Li, C. B., Zhou, A. L., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi: 10.1126/science.1123604
Li, C. Y., Zhang, Y., Wang, Z. B., Zhang, Y., Cao, C. M., Zhang, P. W., et al. (2010). A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput. Biol. 6:e1000734. doi: 10.1371/journal.pcbi.1000734
Li, D., Dong, Y., Jiang, Y., Jiang, H. F., Cai, J., and Wang, W. (2010). A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 20, 408–420. doi: 10.1038/cr.2010.31
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Li, P. J., Wang, Y. H., Qian, Q., Fu, Z. M., Wang, M., Zeng, D. L., et al. (2007). LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. doi: 10.1038/cr.2007.38
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, Z. W., Griffith, M. E., Li, X. R., Zhu, Z. F., Tan, L. B., Fu, Y. C., et al. (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226, 11–20. doi: 10.1007/s00425-006-0460-4
Lu, Z., Yu, H., Xiong, G., Wang, J., Jiao, Y., Liu, G., et al. (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759. doi: 10.1105/tpc.113.113639
Lyu, J., Huang, L. Y., Zhang, S. L., Zhang, Y. S., He, W. M., Zeng, P., et al. (2020). Neo-functionalization of a Teosinte branched 1 homologue mediates adaptations of upland rice. Nat. Commun. 11:725. doi: 10.1038/s41467-019-14264-1
Miklos, G. L., and Rubin, G. M. (1996). The role of the genome project in determining gene function: insights from model organisms. Cell 86, 521–529. doi: 10.1016/S0092-8674(00)80126-9
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sasaki, T., and International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Sharma, S., Tripathy, S., and Biswal, J. (2000). “Origin of O. sativa and its ecotypes,” in Rice Breeding and Genetics: Research Priorities and Challenges, ed. J. S. Nanda (Enfield, NH: Science Publications), 349–369.
Stein, J. C., Yu, Y., Copetti, D., Zwickl, D. J., Zhang, L., Zhang, C. J., et al. (2018). Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296. doi: 10.1038/s41588-018-0261-2
Studer, A., Zhao, Q., Ross-Ibarra, J., and Doebley, J. (2011). Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163. doi: 10.1038/ng.942
Sweeney, M. T., Thomson, M. J., Pfeil, B. E., and McCouch, S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294. doi: 10.1105/tpc.105.038430
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, L. B., Li, X. R., Liu, F. X., Sun, X. Y., Li, C. G., Zhu, Z. F., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi: 10.1038/ng.197
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Vaughan, D. A., Lu, B. R., and Tomooka, N. (2008). The evolving story of rice evolution. Plant Sci. 174, 394–408. doi: 10.1016/j.plantsci.2008.01.016
Vogel, J. P., Garvin, D. F., Mockler, T. C., Schmutz, J., Rokhsar, D., Bevan, M. W., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Waisel, Y. (1987). Evolution of erect growth forms in domesticated wheats: possible effects of grazing. Oecologia 73, 630–632. doi: 10.1007/BF00379428
Wang, B., Smith, S. M., and Li, J. Y. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Wang, H., Nussbaum-Wagler, T., Li, B. L., Zhao, Q., Vigouroux, Y., Faller, M., et al. (2005). The origin of the naked grains of maize. Nature 436, 714–719. doi: 10.1038/nature03863
Wang, M. H., Yu, Y., Haberer, G., Marri, P. R., Fan, C., Goicoechea, J. L., et al. (2014). The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46, 982–988. doi: 10.1038/ng.3044
Wang, R. L., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. doi: 10.1038/18435
Wu, Y. Z., Zhao, S. S., Li, X. R., Zhang, B., Jiang, L. Y., Tang, Y. Y., et al. (2018). Deletions linked to PROG1 gene participate in plant architecture domestication in Asian and African rice. Nat. Commun. 9:4157. doi: 10.1038/s41467-018-06509-2
Xu, X., Liu, X., Ge, S., Jensen, J. D., Hu, F. Y., Li, X., et al. (2011). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30, 105–111. doi: 10.1038/nbt.2050
Yang, Z. H. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556. doi: 10.1093/bioinformatics/13.5.555
Yoshihara, T., and Iino, M. (2007). Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 48, 678–688. doi: 10.1093/pcp/pcm042
Yu, B. S., Lin, Z. W., Li, H. X., Li, X. J., Li, J. Y., Wang, Y. H., et al. (2007). TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. doi: 10.1111/j.1365-313X.2007.03284.x
Zhang, G. J., Guo, G. W., Hu, X. D., Zhang, Y., Li, Q. Y., Li, R. Q., et al. (2010). Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 20, 646–654. doi: 10.1101/gr.100677.109
Zhang, L., Ren, Y., Yang, T., Li, G. W., Chen, J. H., Gschwend, A. R., et al. (2019). Rapid evolution of protein diversity by de novo origination in Oryza. Nat. Ecol. Evol. 3, 679–690. doi: 10.1038/s41559-019-0822-5
Zhang, Q. J., Zhu, T., Xia, E. H., Shi, C., Liu, Y. L., Zhang, Y., et al. (2014). Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. U.S.A. 111, E4954–E4962. doi: 10.1073/pnas.1418307111
Zhang, X., Lin, Z. L., Wang, J., Liu, H. Q., Zhou, L. N., Zhong, S. Y., et al. (2019). The tin1 gene retains the function of promoting tillering in maize. Nat. Commun. 10:5608. doi: 10.1038/s41467-019-13425-6
Zhang, Y. Q., Roote, J., Brogna, S., Davis, A. W., Barbash, D. A., Nash, D., et al. (1999). stress sensitive B encodes an adenine nucleotide translocase in Drosophila melanogaster. Genetics 153, 891–903.
Zhang, Y. S., Zhang, S. L., Liu, H., Fu, B. Y., Li, L. J., Xie, M., et al. (2015). Genome and comparative transcriptomics of african wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility. Mol. Plant 8, 1683–1686. doi: 10.1016/j.molp.2015.08.006
Zhu, B. F., Si, L. Z., Wang, Z. X., Zhou, Y., Zhu, J. J., Shangguan, Y. Y., et al. (2011). Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 155, 1301–1311. doi: 10.1104/pp.110.168500
Zhu, Q. H., and Ge, S. (2005). Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 167, 249–265. doi: 10.1111/j.1469-8137.2005.01406.x
Keywords: rice, Oryza species, PROG1, plant architecture, evolution, domestication
Citation: Huang L, Liu H, Wu J, Zhao R, Li Y, Melaku G, Zhang S, Huang G, Bao Y, Ning M, Chen B, Gong Y, Hu Q, Zhang J and Zhang Y (2020) Evolution of Plant Architecture in Oryza Driven by the PROG1 Locus. Front. Plant Sci. 11:876. doi: 10.3389/fpls.2020.00876
Received: 05 March 2020; Accepted: 28 May 2020;
Published: 23 June 2020.
Edited by:
Angelica Cibrian-Jaramillo, Centro de Investigación y Estudios Avanzados, Instituto Politécnico Nacional de México (CINVESTAV), MexicoReviewed by:
Mingsheng Chen, Institute of Genetics and Developmental Biology (CAS), ChinaEnrique Maguilla, Universidad de Sevilla, Spain
Copyright © 2020 Huang, Liu, Wu, Zhao, Li, Melaku, Zhang, Huang, Bao, Ning, Chen, Gong, Hu, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liyu Huang, bHlodWFuZ0B5bnUuZWR1LmNu; Yesheng Zhang, emhhbmd5ZXNoZW5nMTk4N0AxNjMuY29t
†These authors have contributed equally to this work
 Liyu Huang
Liyu Huang Hui Liu
Hui Liu Junjie Wu4†
Junjie Wu4†