- 1Laboratory of Plant Molecular Genetics & Crop Gene Editing, School of Life Sciences, Linyi University, Linyi, China
- 2Key Laboratory of Molecular Epigenetics of MOE, Northeast Normal University, Changchun, China
Auxin is one of the traditional plant hormones, whereas peptide hormones are peptides with hormone activities. Both auxin and plant peptide hormones regulate multiple aspects of plant growth and development, and there are cross-talks between auxin and plant peptide hormones. PAMP-INDUCED SECRETED PEPTIDES (PIPs) and PIP-LIKEs (PIPLs) are a new family of plant peptide hormone, and PIPL3/TARGET OF LBD SIXTEEN 2 (TOLS2) has been shown to regulate lateral root formation in Arabidopsis. We report here the identification of PIP2 as an auxin response gene, and we found it plays a role in regulating root and hypocotyl development in Arabidopsis. By using quantitative RT-PCR, we found that the expression of PIP2 but not PIP1 and PIP3 was induced by auxin, and auxin induced expression of PIP2 was reduced in nph4-1 and arf19-4, the lost-of-function mutants of Auxin Response Factor 7 (ARF7) and ARF19, respectively. By generating and characterizing overexpressing transgenic lines and gene edited mutants for PIP2, we found that root length in the PIP2 overexpression plant seedlings was slightly shorter when compared with that in the Col wild type plants, but root length of the pip2 mutant seedlings remained largely unchanged. For comparison, we also generated overexpressing transgenic lines and gene edited mutants for PIP3, as well as pip2 pip3 double mutants. Surprisingly, we found that root length in the PIP3 overexpression plant seedlings is shorter than that of the PIP2 overexpression plant seedlings, and the pip3 mutant seedlings also produced short roots. However, root length in the pip2 pip3 double mutant seedlings is largely similar to that in the pip3 single mutant seedlings. On the other hand, hypocotyl elongation assays indicate that only the 35S:PIP2 transgenic plant seedlings produced longer hypocotyls when compared with the Col wild type seedlings. Further analysis indicates that PIP2 promotes cell division as well as cell elongation in hypocotyls. Taken together, our results suggest that PIP2 is an auxin response gene, and PIP2 plays a role in regulating root and hypocotyl elongation in Arabidopsis likely via regulating cell division and cell elongation.
Introduction
The plant hormone auxin regulates multiple aspects of plant growth and development largely by activating the expression of auxin response genes (Davies, 1995; Chapman and Estelle, 2009). The activation of auxin response genes is mainly regulated by the TRANSPORT INHIBITOR RESPONSE 1 (TIR1) auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2005), and two different families of transcription factors, i.e., the AUXIN RESPONSE FACTOR (ARF) family and the AUXIN (Aux)/INDOLE-3-ACETIC ACID (IAA) protein family (Guilfoyle et al., 1998; Reed, 2001; Guilfoyle and Hagen, 2007). Five of the ARFs, including ARF5, ARF6, ARF7, ARF8, and ARF19 function as transcription activators and are able to activate the expression of auxin response genes (Tiwari et al., 2003; Wang et al., 2005). However, when the level of cellular auxin is low, Aux/IAA proteins, the transcription repressors in auxin signaling (Tiwari et al., 2004), can form dimmers with ARF activators and inhibit their activities (Tiwari et al., 2003). When the level of cellular auxin is elevated, auxin are able to bind and activate the TIR1 auxin receptor, leading to degradation of Aux/IAA proteins via 26S proteasome, therefore release the inhibition of Aux/IAA proteins on ARF activators, resulting in activation of auxin response genes (Guilfoyle and Hagen, 2007; Tan et al., 2007; Hayashi, 2012).
So far several different gene families such as Aux/IAAs, GRETCHEN HAGENs (GH3s), and SMALL AUXIN-UP RNAs (SAURs; Hagen and Guilfoyle, 2002), and some other genes such as ASYMMETRIC LEAVES2-LIKE/LATERAL ORGAN BOUNDARIES DOMAIN (ASL/LBD), PACLOBUTRAZOL RESISTANCE 6 (PRE6) and LATERAL ROOT PRIMORDIUM1 (LRP1; Lee et al., 2009; Coudert et al., 2013; Zheng et al., 2017; Singh et al., 2020), have been identified as auxin response genes. However, considering that auxin is involved in the regulation of almost all the aspects of plant growth and development, large numbers of auxin response genes should still remain unidentified (Kieffer et al., 2010). On the other hand, exploration of the functions of the auxin response genes is still on going, as an example, the SAURs were identified as an auxin response gene family about 25 years ago (Gil et al., 1994), yet it is only in recent years that SAURs have been identified to regulate several different aspects of plant growth and development, such as cell expansion (Spartz et al., 2012; Kong et al., 2013; Qiu, et al., 2020), pollen tube growth (He et al., 2018), apical hook development (Kathare et al., 2018), hypocotyl and stamen filament elongation (Chae et al., 2012), and leaf senescence (Hou et al., 2013; Wen et al., 2020).
Peptide hormones are peptides with hormone activities in animal, bacteria and yeast (Edlund and Jessell, 1999). The first plant peptide hormone, systemin, was identified about 30 years ago (Pearce et al., 1991), and more than 20 different types of plant peptide hormones have been identified since then (Hirakawa et al., 2017; Hirakawa and Sawa, 2019). Plant peptide hormones are also involved in the regulation of different aspects of plant growth and development. As examples, CLAVATA3/ENDOSPERM SURROUNDING REGIONs (CLEs) regulate the maintains of shoot and root apical meristem (Kinoshita et al., 2007; Jun et al., 2010; Katsir et al., 2011; Guo et al., 2015), POLARIS (PLS), AUXIN-RESPONSICE ENDOFENOUS POLYPEPTIDE 1(AREP1) and GROWTH FACTOR/CLE LIKE/GOLVEN (RGF/CLEL/GLV) regulate root growth (Casson et al., 2002; Matsuzaki et al., 2010; Meng et al., 2012a; Fernandez et al., 2013; Yang et al., 2014), RGF/CLEL/GLV regulates lateral root formation (Matsuzaki et al., 2010; Meng et al., 2012a; Fernandez et al., 2013), PLS regulates vascular development (Casson et al., 2002), EPIDERMAL PATTERNING FACTORs (EPFs) regulate stomata development (Hara et al., 2007; Hunt and Gray, 2009; Sugano et al., 2010), DEVIL (DVL1) and ROTUNDIFOLIA4 (ROT4) regulate leaf and fruit development (Narita et al., 2004; Wen et al., 2004), and INFLORESCENCE DEFICIENT IN ABSCISSION LIKEs (IDLs) regulate floral organ abscission (Butenko et al., 2003; Cho et al., 2008; Stenvik et al., 2008).
At least some of the aspects of plant growth and development are regulate by both auxin response genes and plant peptide hormones. For example, both the plant peptide hormones PLS, AREP1 and RGF/CLEL/GLV and some Aux/IAA proteins such as IAA9 are able to regulate root growth (Casson et al., 2002; Matsuzaki et al., 2010; Meng et al., 2012a; Fernandez et al., 2013; Yang et al., 2014), and auxin is involved in CLE regulated vascular proliferation (Whitford et al., 2008). Some other experiments have also indicated that there are cross-talk between auxin and some of plant peptide hormones. For example, the expression of PLS, AREP1 and RGF/CLEL/GLV genes are induced by auxin, whereas PLS and RGF/CLEL/GLV peptides are able to regulate auxin transport (Casson et al., 2002; Chilley et al., 2006; Meng et al., 2012b; Whitford et al., 2012; Yang et al., 2014).
PAMP-INDUCED SECRETED PEPTIDES (PIPs) and PIP-LIKEs (PIPLs) are a new family of plant peptide hormone identified in Arabidopsis in recent years (Hou et al., 2014; Vie et al., 2015). Both PIP and PIPL propeptides have an N-terminal signal peptide and a C-terminal SGPS motif, which is part of the biologically active peptides, with an exception of PIP2 and PIP3 prepropeptides, which have two SGPS motifs (Hou et al., 2014; Vie et al., 2015). The PIP peptides including PIP1, PIP2 and PIP3 have been shown to modulate immunity (Hou et al., 2014; Najafi et al., 2020), and the expression of several PIPs and PIPLs family genes is induced by biotic and/or abiotic stresses (Hou et al., 2014; Vie et al., 2015). On the other hand, it has been reported that the PIPL3/TARGET OF LBD SIXTEEN 2(TOLS2) is able to regulate lateral root formation (Toyokura, et al., 2019). Here, we report the identification of PIP2 as an auxin response gene, and we found that PIP2 is involved in the regulation of root and hypocotyl development in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
The Columbia-0 (Col) ecotype Arabidopsis (Arabidopsis thaliana) was used as wild type for plant transformation and auxin response analysis of the PIP genes, and as a control for root length, hypocotyl length, cell number and cell length analysis. The nph4-1 and arf19-4 mutants are in the Col wild type background (Harper et al., 2000; Wang et al., 2005). The 35S:PIP2 and 35S:PIP3 overexpress plants were generated by transforming Col wild type plants, and the pip2 and pip3 single and the pip2 pip3 double mutants were obtained by editing PIP2 and PIP3 genes in the Col wild type plants via CRISPR/Cas9 gene editing techniques.
For plant transformation, the Col wild type seeds were sown directly into the soil pots and grown in a growth chamber. To obtain seedlings for auxin treatment and phenotypic analysis, seeds of the Col wild type, the nph4-1, arf19-4, pip2, pip3, and pip2 pip3 mutants, and the 35S:PIP2 and 35S:PIP3 overexpress plants were surface sterilized with 25% (v/v) bleach for 10 min, washed with sterile deionized water for four times, and then sown on 1/2 Murashige and Skoog (MS) petri plates, containing vitamins (Plant Media), 1% (w/v) sucrose, pH 5.8, and solidified with 0.6% (w/v) phytoagar (Plant Media). The plates were then kept in 4°C for 2 days, and then moved to a growth chamber.
The growth condition in the growth chamber was set as 23°C temperature, 60% relative humidity conditions, and photon density set at ~120 μmol m−2 s −1 under a 16 h light/8 h dark photoperiod unless indicated otherwise.
Auxin Treatment, RNA Isolation, and Quantitative RT-PCR
To examine the expression of PIP2 and PIP3 in response to auxin, 10-day-old Col wild type seedlings were transferred to petri plates containing 10 μM IAA and shaked on a shaker in dark for 4 h. To examine auxin regulated epression of PIP2 and IAA19 in nph4-1 and arf19-4 mutants, 10-day-old Col wild type, and nph4-1 and arf19-4 mutant seedlings were treated with 10 μM IAA for 4 h. Seedlings were collected, total RNA was isolated, cDNA was synthesized as described previously (Wang et al., 2015a), and used to detect the expression of PIP2, PIP3 and IAA19 with a process described previously (Wang et al., 2015b), and the expression of ACTIN2 (ACT2) gene was used as an internal control. The primers used for quantitative RT-PCR (qRT-PCR) analysis of IAA19 and ACT2 have been described previously (Liu et al., 2015; Wang et al., 2015a,b), and analyzed by using delta delta method (∆∆Ct). The primers used for qRT-PCR analysis of PIP2 and PIP3 were 5'-GGAGAAGTTCGTGGCTAGTTTAT-3' and 5'-CTTCCTGTCCACGACCTTATG-3', 5'-AGAGAACCTCGTGGCTAAGT-3' and 5'-GGGACCTGAATGCTTACCATATT-3' respectively.
Constructs
To generate pPZP-35S:PIP2 and pPZP-35S:PIP3 constructs for plant transformation, the full length open-reading frame (ORF) sequences of PIP2 and PIP3 were amplified and inserted, respectively into the pUC19 vector with an N-terminal HA tag using NdeI and SacI restriction sites (Tiwari et al., 2004; Tian et al., 2015). The 35S:PIP2 and 35S:PIP3 fragments in the pUC19-35S:PIP2 and pUC19-35S:PIP3 constructs were then digested with Pst1 and Sac1 enzymes and sub-cloned into the binary vector pPZP211 (Hajdukiewicz et al., 1994). The primers used to amplify PIP2 were 5'-CAACATATGATGATGAACAAAAACGTTCTG-3' and 5'-CAAGAGCTCTTAGTGGCCCGGTCCG-3', to amplify PIP3 were, 5'-CAACATATGATGATGAACAAAGTTGTTTTGG-3', and, 5'-CAAGAGCTCTTAGTGACCGGGTCCACTC-3'.
To generate CRISPR/Cas9 constructs for gene editing of PIP2 and PIP3, exon sequences of PIP2 and PIP3 were evaluated on CRISPRscan1 for potential target sequences. Target specificity was then assessed on Cas-OFFinder.2 The cas9 targeted sequences selected for PIP2 were 5'-GTTCTTCATGTTGATTGGTT(CGG)-3' and 5'-GCTTGGTCTAACAAAGACCG(AGG)-3', for PIP3 were 5'-GTGGTGGAGGCTCGTCCTTT(GGG)-3' and 5'-GAAGGCTGAAGAGAACCTCG(TGG)-3'. The target sequences were inserted into the pHEE-FT vector (Cheng et al., 2019). The primer used to generate CRISPR/Cas9 constructs for editing PIP2 were DT1-BsF (PIP2), 5'-ATATATGGTCTCGATTGTTCTTCATGTTGATTGGTTGTT-3', DT1-F0 (PIP2), 5'- TGTTCTTCATGTTGATTGGTTGTTTTAGAGCTAGAAATAGC-3', DT2-R0 (PIP2), 5'-AACCGGTCTTTGTTAGACCAAGCAATCTCTTAGTCGACTCTAC-3, DT2-BsR (PIP2), 5'- ATTATTGGTCTCGAAACCGGTCTTTGTTAGACCAAGCAA-3'; for editing PIP3 were DT1-BsF (PIP3), 5'-ATATATGGTCTCGATTGTGGTGGAGGCTCGTCCTTTGTT-3',
DT1-F0 (PIP3), 5'-TGTGGTGGAGGCTCGTCCTTTGTTTTAGAGCTAGAAATAGC-3',
DT2-R0 (PIP3), 5'-AACCGAGGTTCTCTTCAGCCTTCAATCTCTTAGTCGACTCTAC-3', DT2-BsR (PIP3), 5'-ATTATTGGTCTCGAAACCGAGGTTCTCTTCAGCCTTCAA-3'; for editing both PIP2 and PIP3 were DT1-BsF (PIP2&PIP3), 5'-ATATATGGTCTCGATTGTGGTGGAGGCTCGTCCTTTGTT-3',
DT1-F0 (PIP2&PIP3), 5'-TGTGGTGGAGGCTCGTCCTTTGTTTTAGAGCTAGAAATAGC-3', DT2-R0 (PIP2&PIP3), 5' AACAACCAATCAACATGAAGAACAATCTCTTAGTCGACTCTAC-3',
DT2-BsR (PIP2&PIP3), 5'-ATTATTGGTCTCGAAACAACCAATCAACATGAAGAACAA -3'. U6-26-IDF and U6-29-IDR primers used for colony PCR and sequencing of the CRISPR/Cas9 constructs have been described previously (Chen et al., 2019a).
Plants Transformation, Transgenic Plants Selection, and Cas9-Free Mutant Isolation
To generate overexpress plants and Cas9 free mutants, about 1-month-old Col wild type plants with several mature flowers were transformed with pPZP211-35S:PIP2, pPZP211-35S:PIP3, and the CRISPR/Cas9 constructs respectively, via Agrobacterium tumefaciens (GV3101) mediated floral dip method (Clough and Bent, 1998).
The 35S:PIP2 and 35S:PIP3 overexpression plants were selected as described previously (Wang et al., 2020). Multiple homozygous lines were obtained and two lines with high expression levels of PIP2 and PIP3, respectively were used for the experiments.
Gene edited mutants were selected by germinating the T1 seeds on 1/2 MS plates containing 50 μg/ml Kanamycin and 100 μg/ml Carbenicillin, examining gene editing status in the early flowering plants by amplifying and sequencing the genomic sequence PIP2 and PIP3, respectively, and then selecting homozygous mutants from normal flowering T2 plants. The absent of T-DNA insertion in the homozygous mutants were confirmed by PCR amplification of Cas9 gene fragment as described previously (Cheng et al., 2019).
DNA Isolation and PCR
To check the editing status of PIP2 and PIP3, DNA was isolated from the leaves of T1 or T2 transgenic plants. The extracted DNA was used as a template for PCR amplification using genomic primers specific to PIP2 and PIP3, respectively. To obtain Cas9 free mutant plants, DNA was isolated from the leaves of T2 progeny of the edited T1 plants, and used as template for PCR amplification using Cas9 specific primer. The primers used for PCR amplification of Cas9 gene have been described previously (Chen et al., 2019a).
Primary Root Length Assays
Primary root length of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plant seedlings, and the pip2, pip3, pip2 pip3 mutant seedlings were assayed as described previously (Wang et al., 2019). For each line, 21–25 seedlings were used for the experiments.
Hypocotyl Length Assays
Seeds of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants were sterilized and sown on 1/2 MS plates, kept at 4°C in the dark for 2 days, and then moved to a growth room with dim light (~60 μmol m−2 s −1). Four-day-old seedlings were used for hypocotyl length assays as reported previously (Wang et al., 2007; Gao et al., 2008). Pictures were taken by using a Nikon digital camera, and the hypocotyl length was calculated by using Image J software. For each line, 29–42 seedlings were used for the experiments.
Hypocotyl Cell Number and Cell Length Assays
Hypocotyl cell number and cell length were measured as described previously with some modifications (Scheres et al., 1994; Wang et al., 2007; Gao et al., 2008; Qu et al., 2017). In brief, 4-day-old dim light-grown seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants were mounted in a film of water on a glass slide and covered with a cover slip to prevent dehydration. Cell number was counted under an OLYMPUS BX53 microscope, at the distance between the top of the root hairs around the collet, and the base of the “V” made by the petioles of the cotyledon (Scheres et al., 1994). The second row cells from the top to the base of the hypocotyls epidermis in longitudinal direction were used for cell length measurement (Qu et al., 2017). Pictures were taken under an OLYMPUS BX53 microscope, and cell length was measured by using Image J. For each line, 22–29 seedlings were used for the experiments.
Results
PIP2 Is an Auxin Response Genes
It has been previously reported that the expression of some plant peptide hormone genes including PLS and RGF/CLEL/CLV was regulated by auxin (Casson et al., 2002; Chilley et al., 2006; Meng et al., 2012b; Whitford et al., 2012; Guo et al., 2015). The PIPLs peptide hormone gene PIPL3 has recently been shown to regulate lateral root initiation in Arabidopsis, a process controlled by auxin (Toyokura et al., 2019), inducing a cross talk between PIP peptide hormone and auxin.
To examine if PIP peptide hormones may be involved in the regulation of auxin regulated plant growth and development. We first examined the expression of PIP genes including PIP1, PIP2, and PIP3 in response to auxin. Seedlings of the Col wild type Arabidopsis were treated with IAA for 4 h and qRT-PCR was used to examine the expression of the PIP genes. As shown in Figure 1A, the expression level of PIP2 increased about 10 folds in response to auxin treatment, whereas the expression level of PIP1 and PIP3 remained largely unchanged, suggest that PIP2 is an auxin response gene, but PIP1 and PIP3 are not.
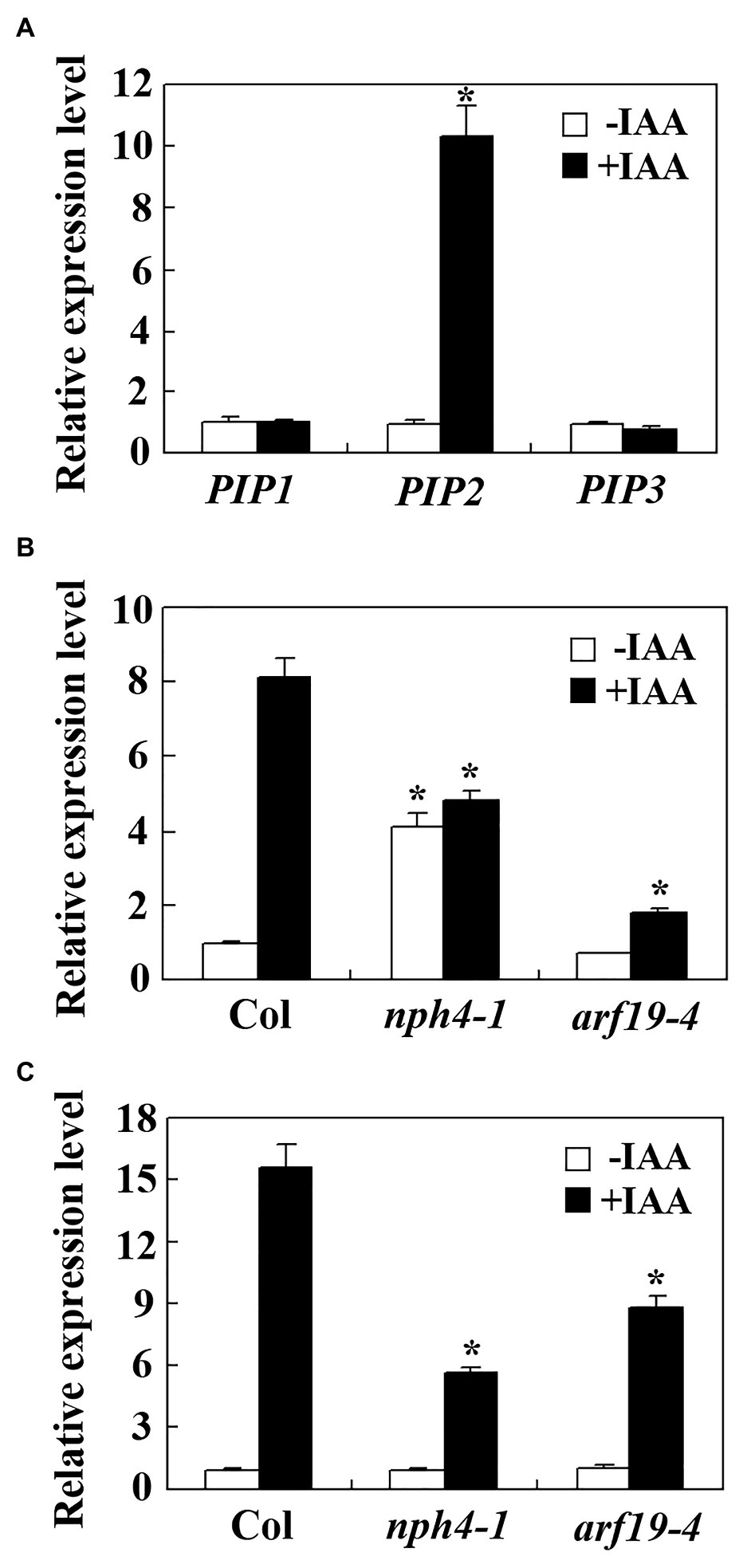
Figure 1. PIP2 is an auxin response gene. (A) Expression of PAMP-INDUCED SECRETED PEPTIDES (PIPs) in response to auxin treatment. Ten-days-old Col wild type seedlings were treated with 10 μM INDOLE-3-ACETIC ACID (IAA) for 4 h, total RNA was isolated and quantitative RT-PCR (qRT-PCR) was used to examine the expression of PIPs. Expression of ACTIN2 (ACT2) was used as an inner control, and the expression level of corresponding PIP genes in the control seedlings was set as 1. Data represent mean ± SD of three repeats. *significantly different from absent of IAA (student’s t test, p < 0.001). Expression of PIP2 (B) and IAA19 (C) in the nph4-1 and arf19-4 mutants in response to auxin treatment. Ten-day-old Col wild type, nph4-1 and arf19-4 mutant seedlings were treated with 10 μM IAA for 4 h. Total RNA was isolated and qRT-PCR was used to examine the expression of PIP2 or IAA19. Expression of ACT2 was used as an inner control, and the expression level of PIP2 or IAA19 in control seedlings of the Col wild type was set as 1. Data represent mean ± SD of three repeats. *significantly different from the corresponding expression level in the Col wild type seedlings (student’s t-test, p < 0.001). The experiments were repeated three times with similar results.
It has been shown that five of the ARFs, including ARF5, ARF6, ARF7, ARF8, and ARF19 are activators that positively regulating the expression of some auxin response genes (Tiwari et al., 2003; Wang et al., 2005), to examine if they may involve in the regulation of PIP2, we examine auxin response of PIP2 in nph4-1/arf7 and arf19-4, two ARF activator gene mutants in hand by using qRT-PCR. We found that the auxin response of PIP2 was decreased in both nph4-1 and arf19-4 mutants (Figure 1B), suggest that ARF7 and ARF19 may regulate the expression of PIP2. To our surprise, we found that the basal expression level of PIP2, i.e., in the absence of auxin was increased about 4-fold in the nph4-1 mutant (Figure 1B). As a control, auxin response of IAA19 was reduced in the nhp4-1 and arf19 mutants, but their basal expression levels remained largely unchanged in both mutants (Figure 1C), a result similar as reported previously (Wang et al., 2005).
Generation of PIP2 Gene Mutants by CRISPR-Cas9 Gene Editing
To examine the functions of PIP2, we generated plants overexpressing PIP2, and gene edited mutants of PIP2 gene via CRISPR/Cas9 mediated gene editing. Overexpression plants were generated by transforming Col wild type Arabidopsis with pPZP211-35S:PIP2 construct, selecting homozygous plants in T3 generation, and examining the expression level of PIP2 in the homozygous transgenic plants (Figure 2A). We also generated PIP3 overexpression plants (Figure 2B), in order to compare the functions of auxin responsive and non-responsive PIP genes. Two independent lines with similar expression levels of PIP genes were selected for further experiments.
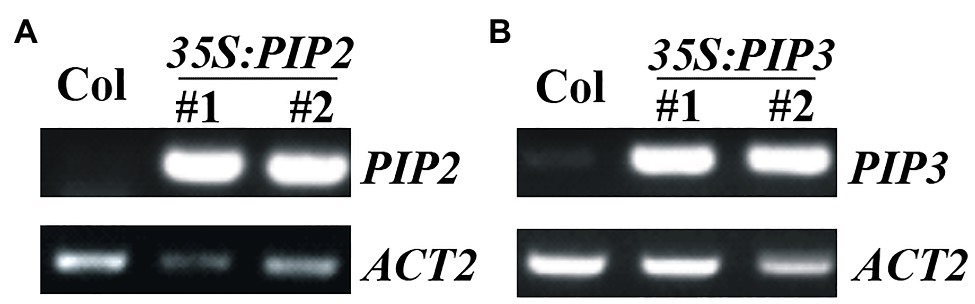
Figure 2. Expression of PIP2 and PIP3 in the 35S:PIP2 and 35S:PIP3 transgenic plants. (A) Expression of PIP2 in the 35S:PIP2 transgenic plants. (B) Expression of PIP3 in the 35S:PIP3 transgenic plants. Total RNA was isolated from 10-day-old homozygous transgenic plants and RT-PCR was used to examine the expression of PIP2 or PIP3. Expression of ACT2 was used as a control.
Gene edited mutants of PIP2 gene was generated by transforming Col wild type Arabidopsis with PIP2 targeting CRISPR/Cas9 construct generated by using a pHEE-FT vector (Cheng et al., 2019), checking gene editing status in early flowering T1 plants, selecting Cas9-free homozygous mutants in normal flowering T2 generations. For comparison, we generated gene edited mutant for PIP3 gene by transforming Col wild type Arabidopsis with PIP3 targeting CRISPR/Cas9 construct, as well as mutants with both PIP2 and PIP3 genes were edited by transforming Col wild type Arabidopsis with CRISPR/Cas9 construct targeting both PIP2 and PIP3.
Two independent single mutants for PIP2 and PIP3 genes respectively, i.e., pip2-c1, pip2-c2, pip3-c1, and pip3-c2, and two independent double mutants, i.e., pip2 pip3-c1, pip2 pip3-c2 were obtained and used for the experiments. In the pip2 mutants, either a single nucleotide insertion or a small fragment deletion was occurred (Figure 3A). For both the pip3 mutants, a small fragment deletion was occurred (Figure 3B). Whereas in the pip2 pip3 double mutants, a single nucleotide insertion was occurred for PIP2 (Figure 3A), and either a single nucleotide insertion or a small fragment deletion was occurred for PIP3 (Figure 3B). All the nucleotide insertion or small fragment deletion led to amino substitution and premature stop of the ORF, as a result, the predicated amino acid sequences for PIP2 and PIP3 genes in the single and double mutants leak the amino acids of the mature PIP2 and PIP3 peptides (Figure 4).
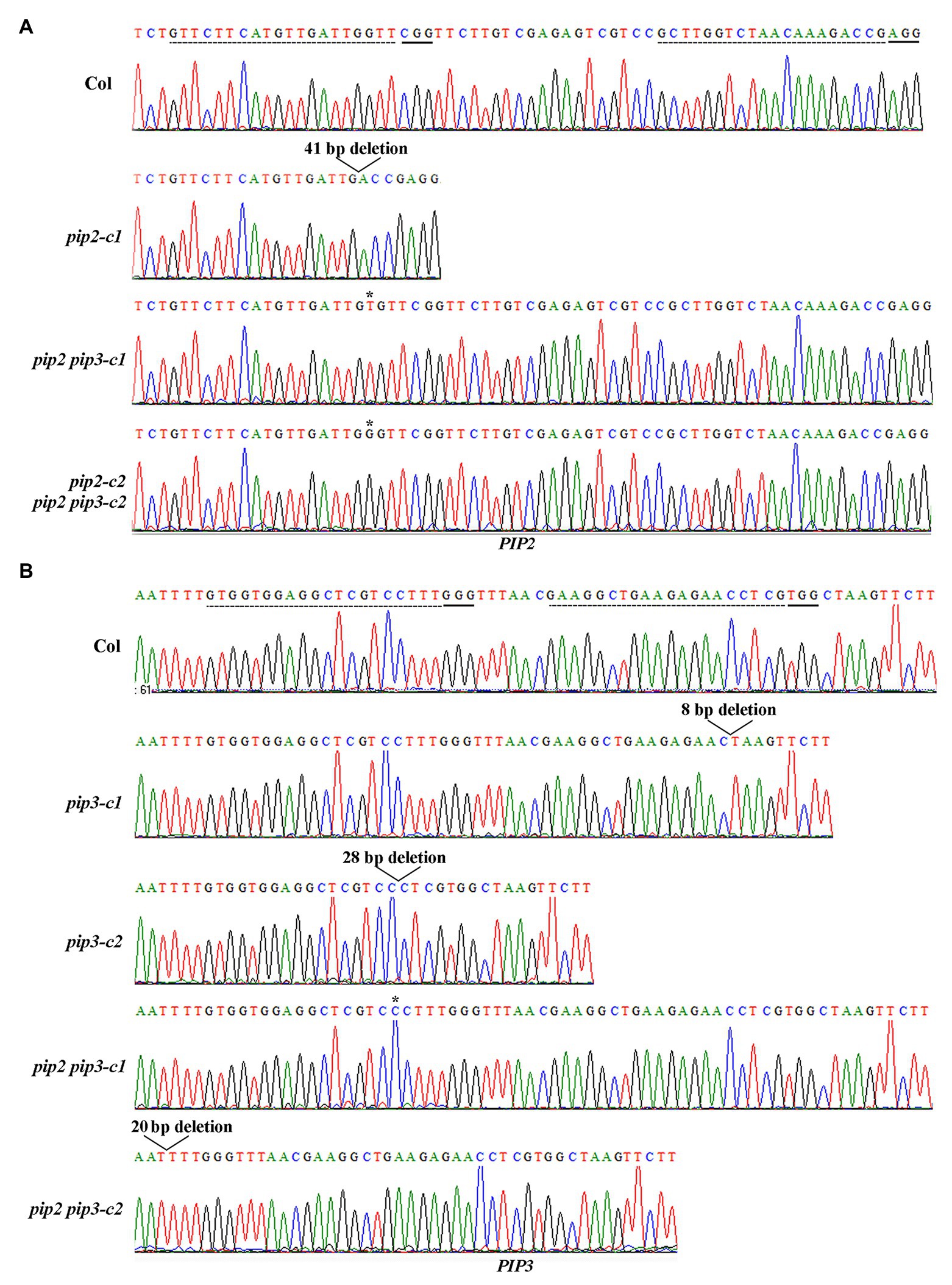
Figure 3. Generation of the pip2, pip3, and pip2 pip3 mutants by CRISPR/Cas9 gene editing. (A) Editing status of PIP2 in the pip2 and pip2 pip3 mutants. (B) Editing status of PIP3 in the pip3 and pip2 pip3 mutants. DNA was isolated from leaves collected from early bolting T1 plants or normal bolting T2 plants, and PCR was used to amplify the coding sequence of PIP2 and/or PIP3. The PCR products were recovered and sequenced, and sequencing results were compared with genome sequence of PIP2 or PIP3 to check the editing status. Dash lines indicate the target sequences, and solid lines indicate the PAM sites.

Figure 4. Amino acid alignment of PIP2 and PIP3 in the Col wild type and the pip2, pip3 and pip2 pip3 mutants. The open-reading frame (ORF) of PIP2 and PIP3 sequences in the pip2 and pip2 pip3 mutants were identified by using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/), and corresponding amino acid sequences were used for alignment with the amino acid sequences of PIP2 and PIP3, respectively. Under lines indicate the mature PIP2 and PIP3 peptides.
PIP2 and PIP3 Affect Root Elongation in Arabidopsis Seedlings
As regulating root elongation is one of the characterized functions of auxin (Rehman et al., 2007), we examine the possible roles of PIP2 in root elongation by using the overexpression plants and gene edited mutants generated. Sterilized seeds of the Col wild type, the 35S:PIP2 transgenic plants and the pip2 mutants were plated on 1/2 MS plates, and grown vertically for root elongation observation. As shown in Figure 5A, the 35S:PIP2 transgenic plant seedlings produced short roots when compared with the Col wild type seedlings, whereas that in the pip2 mutant seedlings remained largely unchanged. Quantitative analysis showed that the root length of the 35S:PIP2 transgenic plant seedlings were about 90% of the Col wild type (Figure 5B). On the other hand, the transgenic plant seedlings expressing PIP3, the non-auxin responsive PIP gene, produced much shorter roots when compared with that in the Col wild type seedlings, and the root length in the pip3 mutant seedlings was also reduced (Figure 5A). The root length in both the 35S:PIP3 transgenic plant seedlings and the pip3 mutant seedlings was about 60% of the Col wild type seedlings (Figure 5B). We also found that root length in the pip2 pip3 double mutant seedlings is largely similar to that in the pip3 single mutant seedlings (Figure 5).
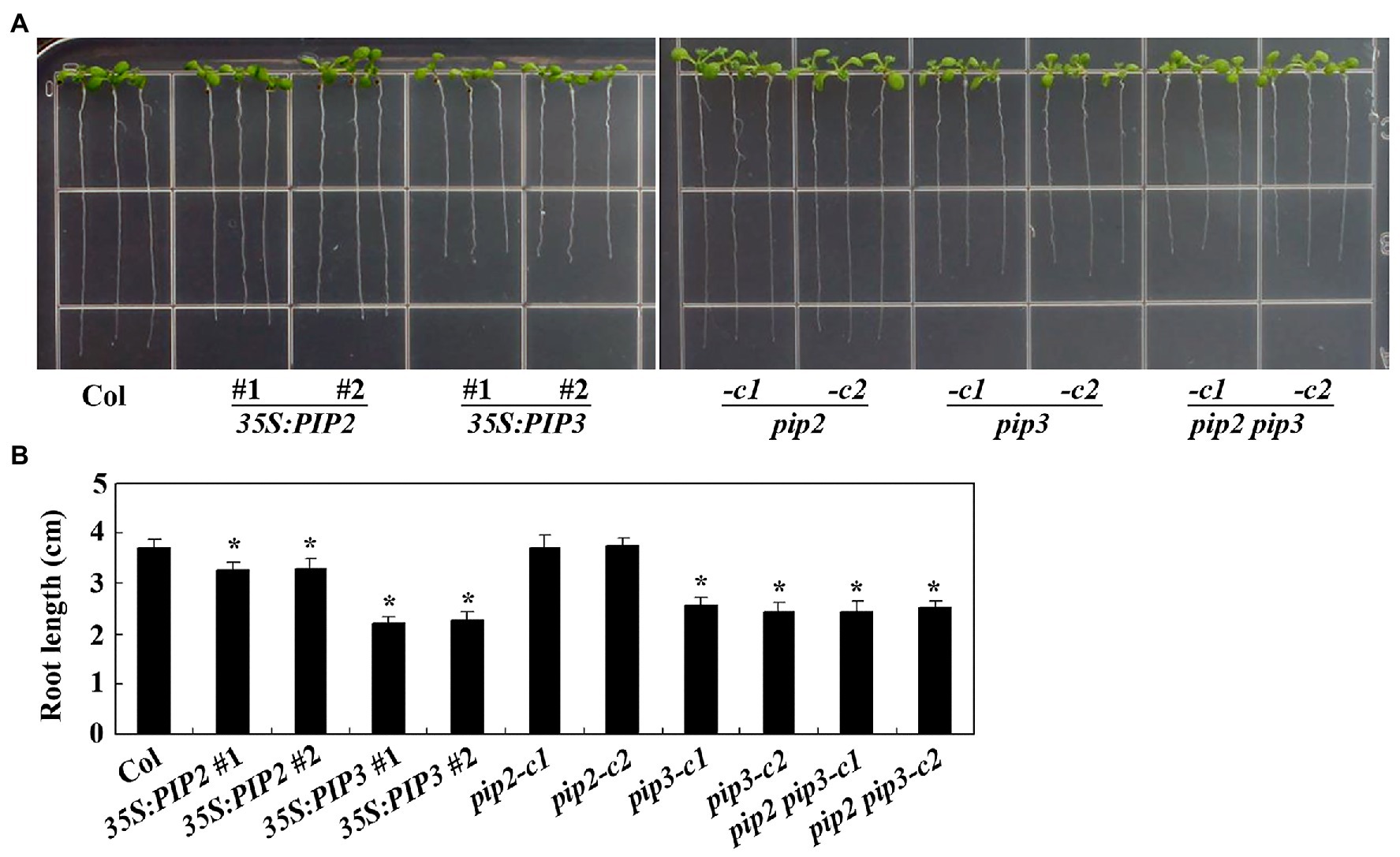
Figure 5. PIP2 and PIP3 affect root elongation. (A) Root elongation in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3 and pip2 pip3 mutants. Seeds were sterilized and plated on 1/2 Murashige and Skoog (MS) plates, kept at 4°C and in darkness for 2 days before transferred to a growth room and grown vertically for 8 days. Pictures were taken by using a digital camera. (B) Root length in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants. Root length of 8-day-old seedlings was measured. Data represent the mean ± SD of 21–25 seedlings. *significantly different from that of the Col wild type seedlings (student’s t-test, p < 0.001). The experiments were repeated three times with similar results.
PIP2 Affects Hypocotyl Elongation
Having shown that PIP2 is involved in the regulation of root elongation, we want further examine the cellular basis of PIP2 in regulating root elongation, i.e., if PIP2 may affects cell division and cell elongation. Considering that cell division and cell elongation in root may vary at different development stages, we sought to examine cell division and cell elongation of epidermis cells in hypocotyls, where the number of epidermis cells is pre-determined during embryogenesis (Gendreau et al., 1997), and has been shown to be a reliable and robust system for simultaneously detect defects in cell division and cell elongation (Ullah et al., 2001, 2003; Gao et al., 2008).
To examine the effects of PIP2 in cell division and cell elongation, we first examined hypocotyl elongation in the Col wild type, the 35S:PIP2 transgenic plant and the pip2 mutant seedlings. Sterilized seeds the Col wild type, the 35S:PIP2 transgenic plants and the pip2 mutants were plated on 1/2 MS plates grown vertically under dim light for hypocotyl length assays. We found that, unlike that observed in root elongation, seedlings of the 35S:PIP2 transgenic plant produced longer hypocotyls (Figure 6A), i.e., an ~15% longer compared with the Col wild type seedlings (Figure 6B), whereas that in the pip2 mutant seedlings remained similar to the Col wild type (Figure 6). On the other hand, although root length was affected in both the 35S:PIP3 transgenic plant and the pip3 mutant seedlings (Figure 6), the hypocotyl length in the seedlings of these plants is largely unaffected, and the hypocotyl length in the pip2 pip3 double mutants is also indistinguishable from the Col wild type seedlings (Figure 6).
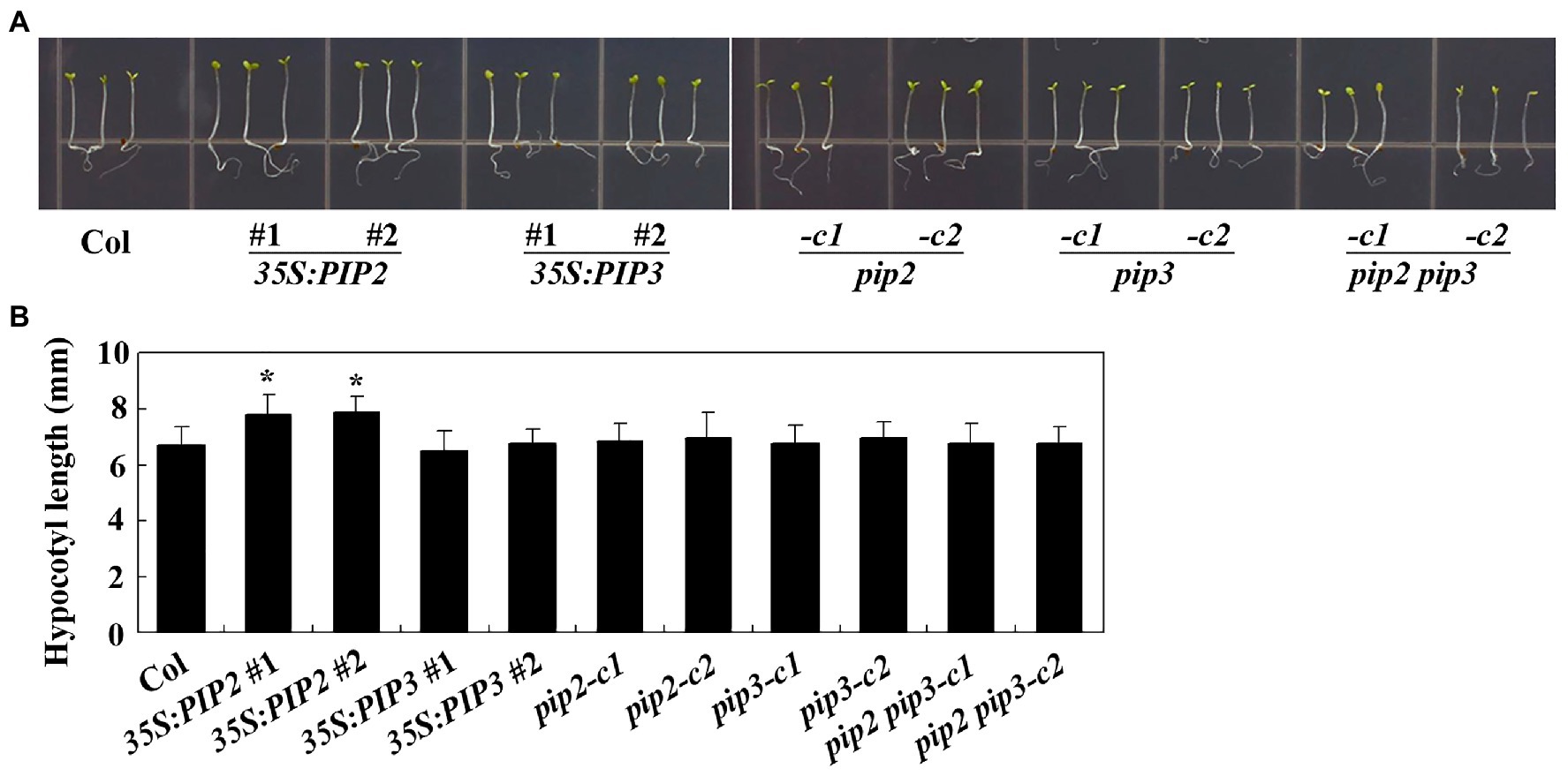
Figure 6. PIP2 affects hypocotyl elongation. (A) Hypocotyl elongation in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants. Seeds were sterilized and plated on 1/2 MS plates, kept at 4°C and in darkness for 2 days before transferred to a growth room and grown under dim light. Pictures were taken 4 days after the transfer by using a digital camera. (B) Hypocotyl length in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants. Hypocotyl length of 4-day-old seedlings grown under dim light were measured. Data represent the mean ± SD of 29–42 seedlings. *significantly different from that of the Col wild type seedlings (student’s t-test, p < 0.001). The experiments were repeated three times with similar results.
PIP2 Affects Cell Division and Elongation in Hypocotyls
We then examined cell division and cell elongation of epidermis cells in hypocotyls of dim light grown seedlings of the Col wild type, the 35S:PIP2 transgenic plants and the pip2 mutants. As shown in Figure 7A, the overall morphology of the epidermis cells in the 35S:PIP2 transgenic plants and the pip2 mutant seedlings are largely indistinguishable from that in the Col wild type plants. However, quantitative analysis shows that the hypocotyls of the 35S:PIP2 transgenic plants produced more epidermis cells, i.e., ~36 cells in a single cell file in the 35S:PIP2 transgenic plant seedlings compared to ~30 cells in the Col wild type seedlings (Figure 7B). In addition, epidermis cell length in the hypocotyls of the 35S:PIP2 transgenic plant seedlings was also increased, i.e., ~150 μM in the 35S:PIP2 transgenic plant seedlings compared to ~120 in the Col wild type seedlings. Consistent with hypocotyl length, no changes in epidermis cell number and cell length were observed in hypocotyls of the 35S:PIP3 transgenic plant seedlings, the pip2 and the pip2 single and the pip2 pip3 double mutant seedlings (Figure 7).
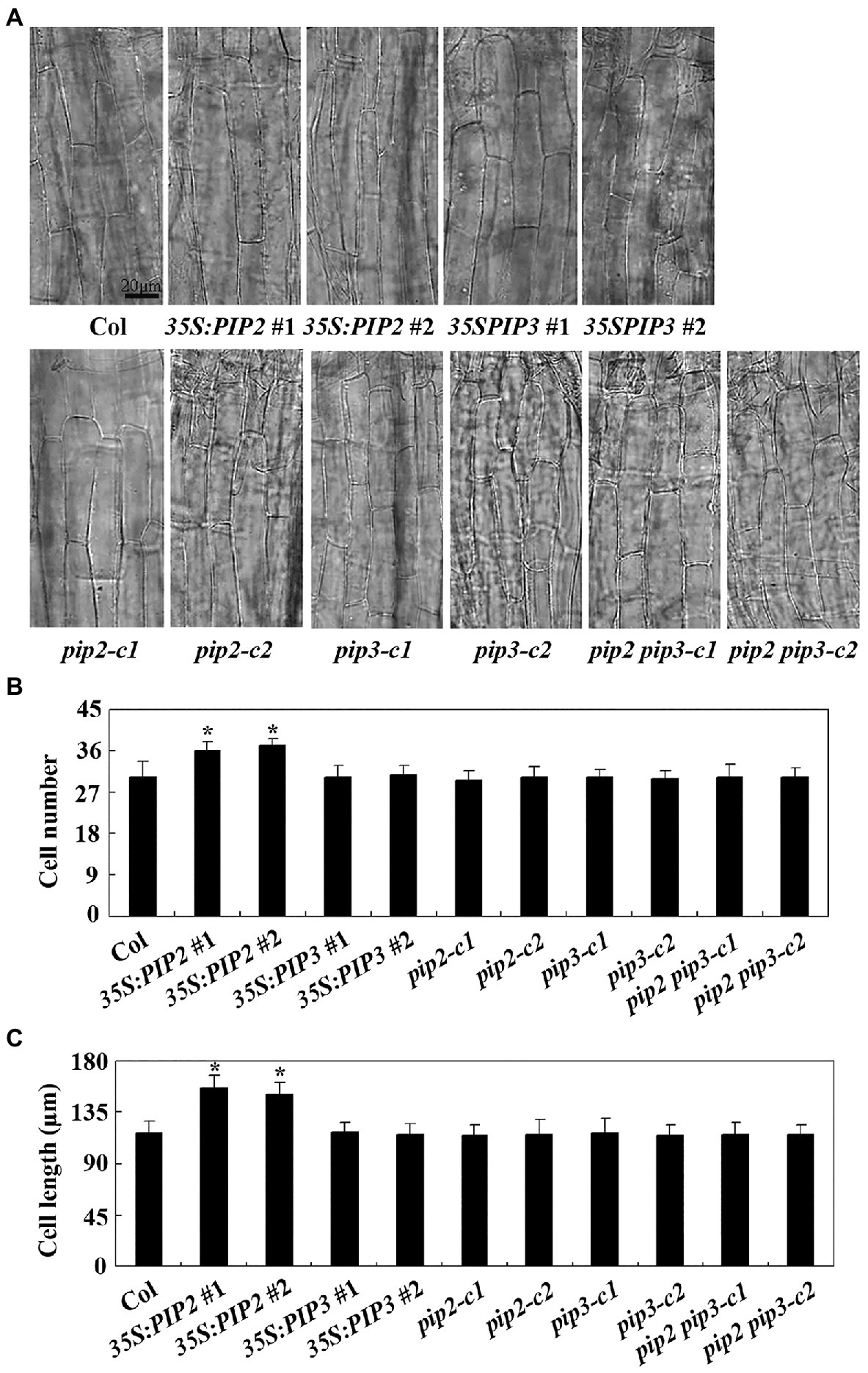
Figure 7. PIP2 affects cell prolification and cell elongation in hypocotyls. (A) Hypocotyl cells in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants. Four-day-old dim light-grown seedlings were fixed and pictures were taken under an OLYMPUS BX53 microscope. (B) Number of hypocotyl cell in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3 and pip2 pip3 mutants. Cell number of a single cell line of the 4-day-old seedlings grown under dim light was counted under an OLYMPUS BX53 microscope. Data represent the mean ± SD of 10–12 seedlings. *significantly different from that of the Col wild type seedlings (student’s t-test, p < 0.001). The experiments were repeated three times with similar results. (C) Hypocotyl cell length in seedlings of the Col wild type, the 35S:PIP2 and 35S:PIP3 transgenic plants, and the pip2, pip3, and pip2 pip3 mutants. Length of the second row cells from the top to the base of the hypocotyls epidermis in longitudinal direction were was measured by using Image J. Data represent the mean ± SD of 22–29 seedlings. *significantly different from that of the Col wild type seedlings (student’s t-test, p < 0.001). The experiments were repeated three times with similar results.
Discussion
Accumulated experiment evidence suggest that there are cross talks between the plant hormone auxin and the plant peptide hormones. It has been shown that auxin is able to regulated the expression of some plant peptide hormone genes (Casson et al., 2002; Chilley et al., 2006; Meng et al., 2012b; Whitford et al., 2012; Yang et al., 2014), and some plant peptide hormones are able to regulate auxin transport (Casson et al., 2002; Chilley et al., 2006; Meng et al., 2012b; Whitford et al., 2012; Yang et al., 2014). Consistent with the presence of cross talks between plant hormones and plant peptide hormones, some plant peptide hormones and auxin response genes have been shown to be able to regulate the same specific aspects of plant growth and development (Casson et al., 2002; Matsuzaki et al., 2010; Meng et al., 2012a; Fernandez et al., 2013; Yang et al., 2014).
PIPL3, a member of the PIPs and PIPLs, a plant peptide hormone family identified in recent years (Hou et al., 2014; Vie et al., 2015), has recently shown to regulate lateral root formation (Toyokura, et al., 2019). In the gLBD16-SRDX transgenic plants, the expression of the TOLS2pro:GUS reporter is induced by auxin (Toyokura, et al., 2019), indicating a cross talk between PIPL3 and auxin. At least two pieces of evidence suggest that there is also cross talk between PIP2 and auxin. One is that the expression of PIP2 was induced by auxin, and auxin induced expression of PIP2 was reduced in ARF activator gene mutants nph4-1 and arf19-4 (Figure 1). Another is that both root elongation and hypocotyl elongation, two of many aspects of plant growth and development regulated by auxin (Chapman and Estelle, 2009), are affected in the PIP2 overexpression plant seedlings (Figures 5, 6). Yet it is possible that the PIPs and/or PIPLs whose expression is not regulated by auxin may also have cross talks with auxin, as root elongation was affected in the PIP3 overexpression plant and pip3 mutant seedlings (Figure 5). Generation of overexpressing plants and/or gene edited mutants for PIP2 and PIP3 in auxin signaling mutants may able to examine directly if there is cross talk between PIPs/PIPLs and auxin in regulating root and hypocotyl elongation.
Different from that of PIP2, the expression levels of PIP1 and PIP3 remind largely unchanged in response to auxin treatment (Figure 1), suggest that other signaling pathways may also regulate the expression of PIPs. As a matter of fact, previously reports showed that the expression of several genes of the PIPs and PIPLs family is induced by biotic and/or abiotic stress (Hou et al., 2014; Vie et al., 2015), suggest that other plant hormones such salicylic acid and abscisc acid may regulate the expression of PIPs and/or PIPLs. Available evidence suggest that PIP1 and PIP2 play an important role in regulating plant response to biotic stresses (Hou et al., 2014; Vie et al., 2015), eventhough the expression of both PIP2 and PIP3 was not affected by ABA treatment (Vie et al., 2015), considering that the expression of PIP2 and PIP3 was affected by salt and cold (Vie et al., 2015), it is very likely that PIP2 and PIP3 may also involve in the regulation of plant response to abiotic stresses.
To our surprise, we found that root length was reduced in both PIP3 overexpression plant and pip3 mutant seedlings (Figure 5), indicating that right amount of PIP3 peptides may be critical for proper root elongation. We also found that both root length and hypocotyl length in the pip3 single and the pip2 pip3 double mutants are indistinguishable (Figures 5, 6), suggest that they may not have redundant functions in regulating root and hypocotyl elongation. However, considering that there are three PIP and eight PIPL genes in Arabidopsis (Hou et al., 2014; Vie et al., 2015; Toyokura, et al., 2019), we could not rule out the possibility that PIP and/or PIPL peptide hormones may function redundantly to regulate plant growth and development. It is possible that PIP2 and PIP3 may have different functions and/or functional mechanisms in regulating plant growth and development. As a matter of fact, it has been shown that PIP2 and PIP3 regulate plant biotic response in different ways, PIP2 regulates antipathogen activity by regulating the expression of some PTI-related genes, WRKY genes, flg22-induced receptor-like kinase 1 (FRK1) and the SAR marker gene PR-1 (Chen et al., 2019b), whereas PIP3 regulates immunity by regulating the biosynthesis and signaling of SA and JA in Arabidopsis (Najafi et al., 2020). On the other hand, both PIP2 and PIP3 contain two conserved SGPS motifs (Vie et al., 2015), and may able to produce two mature peptides, which may have different functions. As an example, CLAVATA3/ESR-RELATED 18 (CLE18) produces two peptides, one functions as an inhibitor of tracheal element differentiation and root growth (Ito et al., 2006), whereas the other promotes root growth (Meng et al., 2012a).
By examining cell numbers and cell length in hypocotyls, we found that PIP2 may regulate cell division as well as cell elongation (Figure 7), therefore to regulate root and hypocotyl elongation. However, further efforts are required to explore the functional mechanism of PIP2 in regulating root and hypocotyl elongation. First, it will be of interest to identify the receptors of PIP2. Both PIP1 and PIP2 regulate plant immunity, and RECEPTOR-LIKE KINASE 7 (RLK7) has been identified as a receptor of PIP1 (Hou et al., 2014). Interestingly, RLK7 is also a receptor of TOLS2/PIPL3, (Toyokura, et al., 2019), therefore it is worthwhile to examine if RLK7 may serve as a receptor of PIP2. Second, it will be of interest to examine how the expression of PIP2 is regulated. The expression level of PIP2 was increased in response to auxin treatment, and auxin induced expression of PIP2 was reduced in the nph4-1 and arf19-4 mutants (Figure 1). Considering that ARF activators are responsible for the activation of auxin response genes (Tiwari et al., 2003; Wang et al., 2005), these results suggest that ARF activators may regulate the expression of PIP2, yet more experiments are required to examine if ARF activators may directly regulate the expression of PIP2. Considering that TOLS2/PIPL3 is a direct target of LBD16, and TOLS2/PIPL3 functions through an auxin-SLR/IAA14-ARF7/19-LBD16-TOLS2/PIPL3-RLK7-PUCHI pathway to regulate lateral root founder cell formation (Toyokura, et al., 2019), it is also possible that PIP2 is directly regulated by LBD16 or some other regulator downstream of ARF7/ARF19. Third, identification of PIP2 regulated genes may also help reveal the functional mechanisms of PIP2. It has been reported that among the four auxin-induced LR-related reporter genes (De Rybel et al., 2010), only two were induced by TOLS2/PIPL3, suggest that TOLS2/PIPL3 may induce gene expression in a pathway independent of auxin signaling (Toyokura, et al., 2019), but this cannot rule out the possibility that TOLS2/PIPL3 may play a feedback regulating role in auxin signaling. This may be also the case of PIP2. Last but not least, our genetic evidence suggest that PIP2 play a different role in regulating root and hypocotyl elongation, i.e., inhibit root elongation (Figure 5), but promote hypocotyl elongation (Figure 6), which may cause by different sensitivities of different parts of the plant in response to the peptides produced by overexpressing PIP2. Considering that PIPs/PIPLs are peptide hormone, and similar to overexpress PIP1 and PIP2 genes in plants, exogenous application of synthetic PIP1 and PIP2 peptides are able to enhanced immune responses in Arabidopsis (Hou et al., 2014), it is still worthwhile to example if exogenous application synthetic PIP2 and PIP3 peptides may able to, and how to regulate plant growth and development.
Nevertheless, our results show that PIP2 is an auxin response gene, and that PIP2 may regulate root and hypocotyl elongation via regulating cell division and cell elongation.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
SW conceived the study. SH, WW, and SW designed the experiments and drafted the manuscript. SH, WW, SA, XW, Adnan, YC, CW, YW, NZ, HT, and SC did the experiments. XH, WW, TW, and SW analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (32071938), the National Key R&D Program of China (2016YFD0101900) and a startup funding from Linyi University (LYDX2019BS039). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Butenko, M. A., Patterson, S. E., Grini, P. E., Stenvik, G. E., Amundsen, S. S., Mandal, A., et al. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15, 2296–2307. doi: 10.1105/tpc.014365
Casson, S. A., Chilley, P. M., Topping, J. F., Evans, I. M., Souter, M. A., and Lindsey, K. (2002). The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14, 1705–1721. doi: 10.1105/tpc.002618
Chae, K., Isaacs, C. G., Reeves, P. H., Maloney, G. S., Muday, G. K., Nagpal, P., et al. (2012). Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 71, 684–697. doi: 10.1111/j.1365-313X.2012.05024.x
Chapman, E. J., and Estelle, M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rew. Genet. 43, 265–285. doi: 10.1146/annurev-genet-102108-134148
Chen, Y. L., Fan, K. T., Hung, S. C., and Chen, Y. R. (2019b). The role of peptides cleaved from protein precursors in eliciting plant stress reactions. New Phytol. 225, 2267–2282. doi: 10.1111/nph.16241
Chen, S., Zhang, N., Zhang, Q., Zhou, G., Tian, H., Hussain, S., et al. (2019a). Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 20:2695. doi: 10.3390/ijms20112695
Cheng, Y., Zhang, N., Hussain, S., Ahmed, S., Yang, W., and Wang, S. (2019). Integration of a FT expression cassette into CRISPR/Cas9 construct enables fast generation and easy identification of transgene-free mutants in Arabidopsis. PLoS One 14:e0218583. doi: 10.1371/journal.pone.0218583
Chilley, P. M., Casson, S. A., Tarkowski, P., Hawkins, N., Wang, K. L., Hussey, P. J., et al. (2006). The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 18, 3058–3072. doi: 10.1105/tpc.106.040790
Cho, S. K., Larue, C. T., Chevalier, D., Wang, H., Jinn, T. L., Zhang, S., et al. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 105, 15629–15634. doi: 10.1073/pnas.0805539105
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coudert, Y., Dievart, A., Droc, G., and Gantet, P. (2013). ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol. Biol. Evol. 30, 569–572. doi: 10.1093/molbev/mss250
Davies, P. J. (1995). Plant Hormones: Physiology, Biochemistry and Molecular Biology. Dordrecht: Kluwer.
De Rybel, B., Vassileva, V., Parizot, B., Demeulenaere, M., Grunewald, W., Audenaert, D., et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706. doi: 10.1016/j.cub.2010.09.007
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Edlund, T., and Jessell, T. M. (1999). Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell 96, 211–224. doi: 10.1016/S0092-8674(00)80561-9
Fernandez, A., Drozdzecki, A., Hoogewijs, K., Nguyen, A., Beeckman, T., Madder, A., et al. (2013). Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol. 161, 954–970. doi: 10.1104/pp.112.206029
Gao, Y., Wang, S., Asami, T., and Chen, J.-G. (2008). Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein alpha subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant Cell Physiol. 49, 1013–1024. doi: 10.1093/pcp/pcn078
Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Höfte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. doi: 10.1104/pp.114.1.295
Gil, P., Liu, Y., Orbović, V., Verkamp, E., Poff, K. L., and Green, P. J. (1994). Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 104, 777–784. doi: 10.1104/pp.104.2.777
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Guilfoyle, T. J., Ulmasov, T., and Hagen, G. (1998). The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 54, 619–627. doi: 10.1007/s000180050190
Guo, H., Zhang, W., Tian, H., Zheng, K., Dai, X., Liu, S., et al. (2015). An auxin responsive CLE gene regulates shoot apical meristem development in Arabidopsis. Front. Plant Sci. 6:295. doi: 10.3389/fpls.2015.00295
Hagen, G., and Guilfoyle, T. J. (2002). Auxin-responsive gene expression: genes, promoters, and regulatory factors. Plant Mol. Biol. 49, 373–485. doi: 10.1023/A:1015207114117
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/BF00014672
Hara, K., Kajita, R., Torii, K. U., Bergmann, D. C., and Kakimoto, T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725. doi: 10.1101/gad.1550707
Harper, R. M., Stowe-Evans, E. L., Luesse, D. R., Muto, H., Tatematsu, K., Watahiki, M. K., et al. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. doi: 10.1105/tpc.12.5.757
Hayashi, K.-I. (2012). The interaction and integration of auxin signaling components. Plant Cell Physiol. 53, 965–975. doi: 10.1093/pcp/pcs035
He, S. L., Hsieh, H. L., and anf Jauh, G. Y. (2018). SMALL AUXIN UP RNA62/75 are required for the translation of transcripts essential for pollen tube growth. Plant Physiol. 178, 626–640. doi: 10.1104/pp.18.00257
Hirakawa, Y., and Sawa, S. (2019). Diverse function of plant peptide hormones in local signaling and development. Curr. Opin. Plant Biol. 51, 81–87. doi: 10.1016/j.pbi.2019.04.005
Hirakawa, Y., Torii, K. U., and Uchida, N. (2017). Mechanisms and strategies shaping plant peptide hormones. Plant Cell Physiol. 58, 1313–1318. doi: 10.1093/pcp/pcx069
Hou, S., Wang, X., Chen, D., Yang, X., Wang, M., Turrà, D., et al. (2014). The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 10:e1004331. doi: 10.1371/journal.ppat.1004331
Hou, K., Wu, W., and Gan, S. S. (2013). SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 161, 1002–1009. doi: 10.1104/pp.112.212787
Hunt, L., and Gray, J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864–869. doi: 10.1016/j.cub.2009.03.069
Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., Dohmae, N., et al. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. doi: 10.1126/science.1128436
Jun, J., Fiume, E., Roeder, A. H., Meng, L., Sharma, V. K., Osmont, K. S., et al. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154, 1721–1736. doi: 10.1104/pp.110.163683
Kathare, P. K., Dharmasiri, S., and Dharmasiri, N. (2018). SAUR53 regulates organ elongation and apical hook development in Arabidopsis. Plant Signal. Behav. 13:e1514896. doi: 10.1080/15592324.2018.1514896
Katsir, L., Davies, K. A., Bergmann, D. C., and Laux, T. (2011). Peptide signaling in plant development. Curr. Biol. 21, 356–364. doi: 10.1016/j.cub.2011.03.012
Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi: 10.1038/nature03542
Kieffer, M., Neve, J., and Kepinski, S. (2010). Defining auxin response contexts in plant development. Curr. Opin. Plant Biol. 13, 12–20. doi: 10.1016/j.pbi.2009.10.006
Kinoshita, A., Nakamura, Y., Sasaki, E., Kyozuka, J., Fukuda, H., and Sawa, S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821–1825. doi: 10.1093/pcp/pcm154
Kong, Y., Zhu, Y., Gao, C., She, W., Lin, W., Chen, Y., et al. (2013). Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 54, 609–621. doi: 10.1093/pcp/pct028
Lee, H. W., Kim, N. Y., Lee, D. J., and Kim, J. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151, 1377–1389. doi: 10.1104/pp.109.143685
Liu, S., Hu, Q., Luo, S., Li, Q., Yang, X., Wang, X., et al. (2015). Expression of wild-type PtrIAA14.1, a poplar aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 6:388. doi: 10.3389/fpls.2015.00388
Matsuzaki, Y., Ogawa-Ohnishi, M., Mori, A., and Matsubayashi, Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067. doi: 10.1126/science.1191132
Meng, L., Buchanan, B. B., Feldman, L. J., and Luan, S. (2012a). CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 109, 1760–1765. doi: 10.1073/pnas.1119864109
Meng, L., Buchanan, B. B., Feldman, L. J., and Luan, S. (2012b). A putative nuclear CLE-like (CLEL) peptide precursor regulates root growth in Arabidopsis. Mol. Plant 5, 955–957. doi: 10.1093/mp/sss060
Najafi, J., Brembu, T., Vie, A. K., Viste, R., Winge, P., Somssich, I. E., et al. (2020). PAMP-INDUCED SECRETED PEPTIDE 3 modulates immunity in Arabidopsis. J. Exp. Bot. 71, 850–864. doi: 10.1093/jxb/erz482
Narita, N. N., Moore, S., Horiguchi, G., Kubo, M., Demura, T., Fukuda, H., et al. (2004). Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 38, 699–713. doi: 10.1111/j.1365-313X.2004.02078.x
Pearce, G., Strydom, D., Johnson, S., and Ryan, C. A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–898. doi: 10.1126/science.253.5022.895
Qiu, T., Qi, M., Ding, X., Zheng, Y., Zhou, T., Chen, Y., et al. (2020). The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 125, 805–819. doi: 10.1093/aob/mcz160
Qu, X., Zhao, Z., and Tian, Z. (2017). ERECTA regulates cell elongation by activating auxin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 8:1688. doi: 10.3389/fpls.2017.01688
Reed, J. W. (2001). Roles and activities of aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. doi: 10.1016/S1360-1385(01)02042-8
Rehman, A., Bannigan, A., Sulaman, W., Pechter, P., Blancaflor, E. B., and Baskin, T. I. (2007). Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J. 50, 514–528. doi: 10.1111/j.1365-313X.2007.03068.x
Scheres, B., Wolkenfelt, H., Willemsen, V., Terlouw, M., Lawson, E., Dean, C., et al. (1994). Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120, 2475–2487.
Singh, S., Yadav, S., Singh, A., Mahima, M., Singh, A., Gautam, V., et al. (2020). Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. 101, 87–100. doi: 10.1111/tpj.14520
Spartz, A. K., Lee, S. H., Wenger, J. P., Gonzalez, N., Itoh, H., Inzé, D., et al. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70, 978–990. doi: 10.1111/j.1365-313X.2012.04946.x
Stenvik, G. E., Tandstad, N. M., Guo, Y., Shi, C. L., Kristiansen, W., Holmgren, A., et al. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817. doi: 10.1105/tpc.108.059139
Sugano, S. S., Shimada, T., Imai, Y., Okawa, K., Tamai, A., Mori, M., et al. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241–244. doi: 10.1038/nature08682
Tan, X., Calderon-Villalobos, L. I. A., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitun ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Tian, H., Guo, H., Dai, X., Cheng, Y., Zheng, K., Wang, X., et al. (2015). An ABA down-regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis. Sci. Rep. 5:17587. doi: 10.1038/srep17587
Tiwari, S. B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. doi: 10.1105/tpc.008417
Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. doi: 10.1105/tpc.017384
Toyokura, K., Goh, T., Shinohara, H., Shinoda, A., Kondo, Y., Okamoto, Y., et al. (2019). Lateral inhibition by a peptide hormone-receptor cascade during Arabidopsis lateral root founder cell formation. Dev. Cell 48, 64–75. doi: 10.1016/j.devcel.2018.11.031
Ullah, H., Chen, J. G., Temple, B., Boyes, D. C., Alonso, J. M., Davis, K. R., et al. (2003). The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15, 393–409. doi: 10.1105/tpc.006148
Ullah, H., Chen, J. G., Young, J. C., Im, K. H., Sussman, M. R., and Jones, A. M. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science 292, 2066–2069. doi: 10.1126/science.1059040
Vie, A. K., Najafi, J., Liu, B., Winge, P., Butenko, M. A., Hornslien, K. S., et al. (2015). The IDA/IDA-LIKE and PIP/PIP-LIKE gene families in Arabidopsis: phylogenetic relationship, expression patterns, and transcriptional effect of the PIPL3 peptide. J. Exp. Bot. 66, 5351–5365. doi: 10.1093/jxb/erv285
Wang, S., Chang, Y., Guo, J., and Chen, J.-G. (2007). Arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872. doi: 10.1111/j.1365-313X.2007.03096.x
Wang, X., Liu, S., Tian, H., Wang, S., and Chen, J.-G. (2015b). The small ethylene response factor ERF96 is involved in the regulation of the abscisic acid response in Arabidopsis. Front. Plant Sci. 6:1064. doi: 10.3389/fpls.2015.01064
Wang, S., Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2005). AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17, 1979–1993. doi: 10.1105/tpc.105.031096
Wang, X., Wang, X., Hu, Q., Dai, X., Tian, H., Zheng, K., et al. (2015a). Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 83, 300–311. doi: 10.1111/tpj.12887
Wang, W., Wang, X., Wang, X., Ahmed, S., Hussain, S., Zhang, N., et al. (2019). Integration of RACK1 and ethylene signaling regulates plant growth and development in Arabidopsis. Plant Sci. 280, 31–40. doi: 10.1016/j.plantsci.2018.11.009
Wang, X., Wang, W., Wang, Y., Zhou, G., Liu, S., Li, D., et al. (2020). AIW1 and AIW2, two ABA-induced WD40 repeat-containing transcription repressors function redundantly to regulate ABA and salt responses in Arabidopsis. J. Plant Interact. 15, 196–206. doi: 10.1080/17429145.2020.1778110
Wen, J., Lease, K. A., and Walker, J. C. (2004). DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J. 37, 668–677. doi: 10.1111/j.1365-313X.2003.01994.x
Wen, Z., Mei, Y., Zhou, J., Cui, Y., Wang, D., and Wang, N. N. (2020). SAUR49 can positively regulate leaf senescence by suppressing SSPP in Arabidopsis. Plant Cell Physiol. 61, 644–658. doi: 10.1093/pcp/pcz231
Whitford, R., Fernandez, A., De Groodt, R., Orteqa, E., and Hilson, P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. U.S.A. 105, 18625–19630. doi: 10.1073/pnas.0809395105
Whitford, R., Fernandez, A., Tejos, R., Pérez, A. C., Kleine-Vehn, J., Vanneste, S., et al. (2012). GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev. Cell 22, 678–685. doi: 10.1016/j.devcel.2012.02.002
Yang, F., Song, Y., Yang, H., Liu, Z., Zhu, G., and Yang, Y. (2014). An auxin-responsive endogenous peptide regulates root development in Arabidopsis. J. Integr. Plant Biol. 56, 635–647. doi: 10.1111/jipb.12178
Keywords: auxin, peptide hormone, PIP2, PIP3, root elongation, Arabidopsis
Citation: Hussain S, Wang W, Ahmed S, Wang X,, Cheng Y, Wang C, Wang Y, Zhang N, Tian H, Chen S, Hu X, Wang T and Wang S (2021) PIP2, An Auxin Induced Plant Peptide Hormone Regulates Root and Hypocotyl Elongation in Arabidopsis . Front. Plant Sci. 12:646736. doi: 10.3389/fpls.2021.646736
Edited by:
Ling Huang, Salk Institute for Biological Studies, United StatesReviewed by:
Dior Rose Kelley, Iowa State University, United StatesWei Zhang, Shandong University, China
Copyright © 2021 Hussain, Wang, Ahmed, Wang, Adnan, Cheng, Wang, Wang, Zhang, Tian, Chen, Hu, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shucai Wang, d2FuZ3NodWNhaUB5YWhvby5jb20=
†These authors have contributed equally to this work
 Saddam Hussain
Saddam Hussain Wei Wang
Wei Wang Sajjad Ahmed
Sajjad Ahmed Xutong Wang
Xutong Wang Adnan2
Adnan2 Hainan Tian
Hainan Tian Tianya Wang
Tianya Wang Shucai Wang
Shucai Wang