- 1Research Base, Zhengzhou University, State Key Laboratory of Cotton Biology, Institute of Cotton Research of Chinese Academy of Agricultural Sciences, Anyang, China
- 2State Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Tai'an, China
The CLAVATA3/endosperm surrounding region-related (CLE) small peptides are a group of C-terminally encoded and post-translationally modified signal molecules involved in regulating the growth and development of various plants. However, the function and evolution of these peptides have so far remained elusive in cotton. In this study, 55, 56, and 86 CLE genes were identified in the Gossypium raimondii, Gossypium arboreum, and Gossypium hirsutum genomes, respectively, and all members were divided into seven groups. These groups were distinctly different in their protein characteristics, gene structures, conserved motifs, and multiple sequence alignment. Whole genome or segmental duplications played a significant role in the expansion of the CLE family in cotton, and experienced purifying selection during the long evolutionary process in cotton. Cis-acting regulatory elements and transcript profiling revealed that the CLE genes of cotton exist in different tissues, developmental stages, and respond to abiotic stresses. Protein properties, structure prediction, protein interaction network prediction of GhCLE2, GhCLE33.2, and GhCLE28.1 peptides were, respectively, analyzed. In addition, the overexpression of GhCLE2, GhCLE33.2, or GhCLE28.1 in Arabidopsis, respectively, resulted in a distinctive shrub-like dwarf plant, slightly purple leaves, large rosettes with large malformed leaves, and lack of reproductive growth. This study provides important insights into the evolution of cotton CLEs and delineates the functional conservatism and divergence of CLE genes in the growth and development of cotton.
Introduction
CLAVATA3/embryo surrounding region (CLE) peptides are one of the largest groups of post-translationally modified and short-secreted signaling peptides. They are post-translationally cleaved and then modified from their corresponding pre-propeptides to produce a ligand containing 12–13 amino acids. These peptides function by binding to a corresponding taxon of receptors and are primarily involved in the growth and development of plant meristematic tissues, including shoot apical meristem (SAM) (Fletcher et al., 1999), root apical meristem (RAM) (Stahl et al., 2009), vasculature (Etchells et al., 2015), and legume nodule meristem (Okamoto et al., 2009; Mortier et al., 2010; Lim et al., 2011), as well as respond to environmental stimuli (Wang et al., 2015). The CLE gene family was named following the first discovery of AtCLAVATA3 (CLV3) in Arabidopsis thaliana and ESR in Zea mays and it is considered to be structurally conserved group, CLV3 and ESR are similar in structure and function but unrelated. The protein sequences possess a small, conserved region of 14 amino acids (Opsahl-Ferstad et al., 1997; Cock and McCormick, 2001) and are expressed specifically in the embryo surrounding region (ESR) of the endosperm in Zea (Opsahl-Ferstad et al., 1997). Different ESR members exhibit varying levels of expression in the same region of the maize endosperm (Bonello et al., 2000).
Most plant CLE protein sequences are characterized by several common structural motifs, consisting of a signal motif of 45–90 nucleotides in length located at the N-terminus, a central changeable region ranging from 120 to 240 nucleotides (40–90 amino acids), and a highly conserved functional domain near the C-terminus (a conserved sequence of 14 amino acids: KRXVPXGPNPLHNR) (Cock and McCormick, 2001; Hastwell et al., 2015b, 2017a). Variable regions in CLE protein are essential to their function, and can be used to further clarify the differences in the family members of various species and subfamilies (Ni and Clark, 2006). In addition, some CLE genes encode an extra C-terminal domain (1–150 amino acids), not conserved among non-orthologous proteins, called a C-terminal extension. Members containing this type of domain have been found in some plant species, but little research has been done on their processing (Kinoshita et al., 2007; Oelkers et al., 2008; Strabala et al., 2014). In recent years, CLE-like peptides have gradually been found to share a similar structure but display unrelated activity in the functional domain (Meng et al., 2012). The tracheary element differentiation inhibitory factor (TDIF) is a CLE-like peptide, in which the functional domain consists of 12 amino acids containing two hydroxyproline residues (Hirakawa et al., 2010). The conserved functional domains of AtCLE41 and AtCLE44 are the same as that of TDIF. They interact with the TDIF receptor/phloem intercalated with xylem (LRR-RLK TDR/PXY) membrane protein kinase and are involved in promoting the proliferation of procambial cells and suppression of xylem differentiation (Hirakawa et al., 2008, 2010).
The biological functions of some CLE genes have been well-studied and characterized in model plants, such as Arabidopsis and rice. For example, CLV3 plays a critical role in SAM to regulate stem cell homeostasis (Fletcher et al., 1999). Overexpression of CLV3 inhibits organ initiation after the appearance of the first leaves, while CLV3 mutants display the opposite phenotype, including the build-up of stem cells in the center of shoot and floral meristems (Brand et al., 2000). In rice, floral organ number 4 (FON4), encodes a putative ortholog of AtCLV3, which is mainly expressed in a small group of cells at the apex of the SAMs. The FON4 mutants display abnormal enlargement of the embryonic and vegetative SAMs as well they increase in inflorescences and floral meristems (Chu et al., 2006). The ectopic expression of 18 different CLE genes (CLV3, CLE2, CLE3, CLE4, CLE5, CLE6, CLE7, CLE9, CLE10, CLE11, CLE13, CLE18, CLE19, CLE21, CLE25, CLE26, CLE42, and CLE44) in Arabidopsis resulted in similar levels of premature mortality or phenotypes with developmental timing delays (Strabala et al., 2006). Arabidopsis CLE genes that are involved in root development have also been fully described. For example, AtCLE40, encoding a peptide with close structural similarity to AtCLV3, plays an important role in the control of root tip movement and root length (Hobe et al., 2003). Furthermore, AtCLE1, 3, 4, and 7 are induced mainly in root pericycle cells under nitrogen deficiency, and overexpression of these genes suppresses the growth of lateral root primordia (Araya et al., 2014). AtCLE8 is expressed in seed embryos and endosperm and controls the expression of Wuschel-like homeobox 8 (WOX8), and together AtCLE8 and AtWOX8 form a signal transduction pathway involved in seed morphogenesis (Fiume and Fletcher, 2012). AtCLE41, AtCLE42, and AtCLE44 are known as tracheary element differentiation factors, and they control vascular meristematic tissue cell proliferation and differentiation by interacting with their receptor PXY/TDR (Hirakawa et al., 2008). However, many CLE gene family members have not yet been fully identified and their roles remained unclarified.
As a vital economic crop, cotton provides natural fibers to the world textile industry. Gossypium, the heterotetraploid, is the most widely planted cultivar that originated from a natural hybridization between the primitive G. arboreum (A-genome species) and native G. raimondii (D-genome species) (Hu et al., 2019). The biological mechanisms of CLE signal peptides in some simple plants have been well-studied. However, the evolution and functional differentiation of cotton CLE remain unknown. In this study, the members of the CLE family were identified genome-wide from three cotton genomes, and characterized for their sequence features, phylogenetic relationships, cis-elements, protein interaction, expression levels in tissue-specific, and in response to different abiotic stresses and functional verification. These results provide a treasurable resource to further dig out other functions and molecular mechanisms of the CLEs in cotton.
Materials and Methods
Sequence Search and Identification of Cotton CLE Genes
To identify the members of the CLE genes family in G. raimondii, G. arboreum, and G. hirsutum, protein sequences from A. rabidopsis and Populus trichocarpa in the published literature were used as query sequences (Han et al., 2016). They were downloaded from the Arabidopsis Information Resource (http://www.Arabidopsis.org) and Populus genome (http://www.phytozome.net/), respectively (Goodstein et al., 2012; Reiser et al., 2017). A BLAST (SequenceServer 2.0.) search was performed in the Cotton Gene database (http://www.cottongen.org) using BLASTP (Version 2.2) with e-value cutoff of 1e−10 (Yu et al., 2014; Li and Lu, 2019). Moreover, HMMER (version 2.41.1) model construction was also adopted for further retrieval (Potter et al., 2018). Candidate proteins without a conserved CLE motif (KRXVPXGPNPLHNR) were removed.
Analysis of Protein Features and Subcellular Localization Prediction
Using the website of CELLO v2.5 (http://cello.life.nctu.edu.tw/), the subcellular localization of the CLEs was predicted (Yu et al., 2004), and the molecular weight (Mw) and isoelectric point (pI) of the candidate members were calculated according to the ExPASy server (http://web.expasy.org/compute_pi/) (Bjellqvist et al., 1994; Wilkins et al., 1999). The CLE signal peptide cleavage sites were predicted using SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/) (Almagro Armenteros et al., 2019).
Multiple Sequence Alignment Analysis and Construction of Phylogenetic Trees
ClustalX 2.0 was used to perform multi-sequence alignment (Thompson et al., 1997). The full-length CLE sequences were aligned using ClustalW and then an unrooted phylogenetic tree was constructed using MEGA 7.0 with the neighbor-joining (NJ) method, which was further verified using the maximum likelihood (ML) method (Kumar et al., 2016). To support the presumed evolutionary relationships, the bootstrap method was used with 1,000 replicates. Weblogo3 (http://weblogo.threeplusone.com/) was used to predict the CLE functional domains and to characterize the protein features (Crooks et al., 2004).
Analysis of Chromosomal Locations, Gene Structures, and the Conservative Domain
The CDS and genomic sequences of the CLE genes in G. raimondii, G. arboreum, and G. hirsutum were searched in the Cotton Functional Genomics Database, using the known gene ID (https://cottonfgd.org/about/download.html) (Zhu et al., 2017). TBtools (https://github.com/CJ-Chen/TBtools) was employed locally to map the CLE genes on the chromosomes (Chen et al., 2020). The structure of the CLE genes was displayed using Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn), (Hu et al., 2015). The conserved sequence and number of conserved motifs were analyzed using the MEME Suite 5.3.3 (http://meme-suite.org/) (Bailey et al., 2009). The maximum number of conservative motifs was set to five, and then each subfamily was predicted one by one.
Analysis of Collinearity, Duplication, and Ka/Ks Values
Chromosomal location and gene duplication events were analyzed using the Advanced Circos of TBtools (https://github.com/CJ-Chen/TBtools). To exhibit segmentally duplicated pairs and orthologous pairs of CLE genes, the Multiple Synteny Plotter TBtools was used to draw collinearity maps (Chen et al., 2020). The Ka (non-synonymous substitution rate)/Ks (synonymous substitution rate) ratios of the CLE genes were calculated using KaKs_calculator 2.0 (Wang et al., 2010).
Analysis of cis-Acting Elements of Promoter Regions and Protein Interaction Analysis
The promoter sequences (2,000 bp upstream of the initiation codon “ATG”) of 86 gene members were filtered from the G. hirsutum genome (including GFF3 and FASTA sequence files) by TBtools. Putative cis-acting elements were identified using the online Plant CARE server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/search_CARE.html) and then visualized with TBtools (Lescot et al., 2002).
The secondary structure using PHYRE2 Protein Fold Recognition Server (http://www.sbg.bio.ic.ac.uk/phyre2/html/) (Kelley et al., 2015), hydrophobicity/hydrophilicity using ProtScale (https://web.expasy.org/protscale/) (Wilkins et al., 1999), signal peptide using the SignalP-5.0 Server (http://www.cbs.dtu.dk/services/SignalP/) (Almagro Armenteros et al., 2019), and transmembrane domain using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Chen et al., 2003) of GhCLE2, GhCLE33.2, and GhCLE28.1 peptides were predicted, respectively. Interaction network analysis was performed with STRING (Version 11.0) (https://string-db.org/cgi/input.pl) (Szklarczyk et al., 2019) on the foundation of the homologous proteins in Arabidopsis (Figure 1B1).
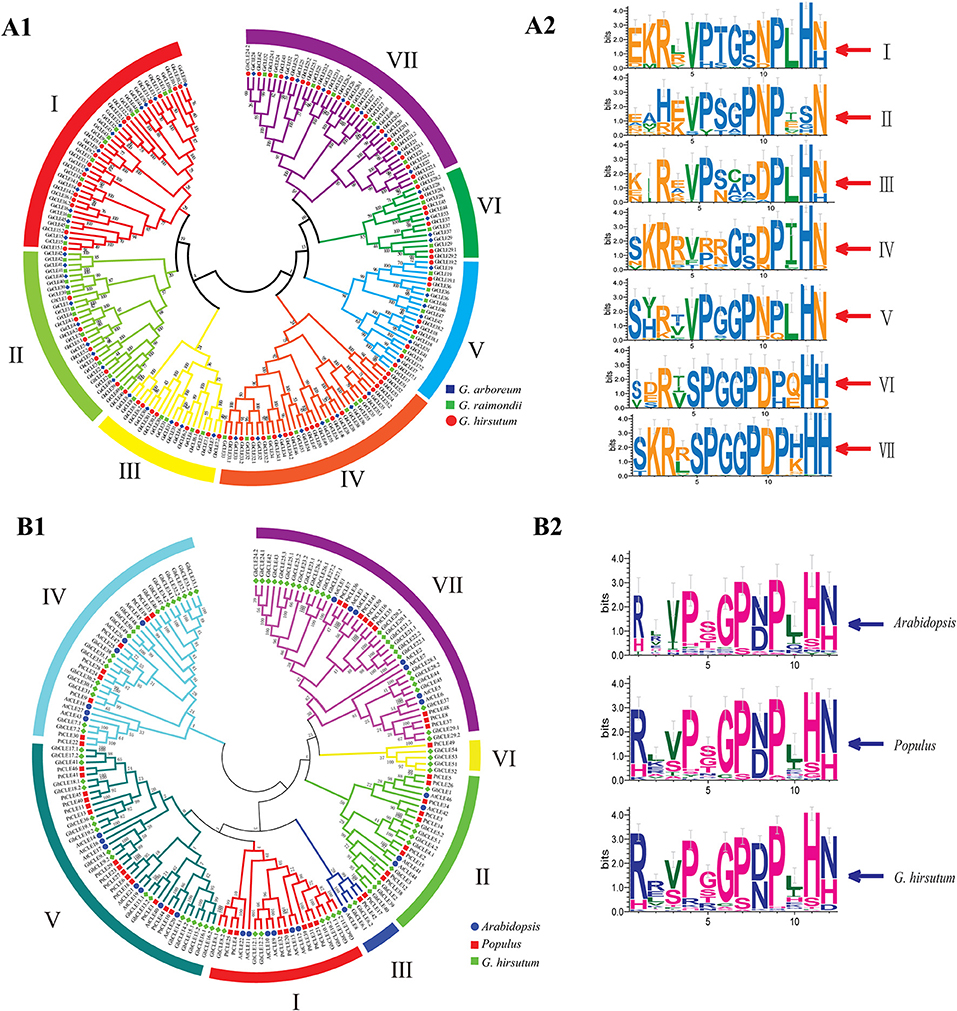
Figure 1. Phylogenetic trees were established using MEGA7 with the neighbor-joining (NJ) method. (A1) Phylogenetic tree of CLEs among G. raimondii, G. arboreum, and G. hirsutum. (A2) The weblogo represents CLE motifs (14 amino acids) of seven groups. (B1) Phylogenetic tree of CLEs among G. hirsutum, Arabidopsis, and Populus. (B2) The weblogo represents CLE motifs of Arabidopsis, poplar, and G.hirsutum (12 conserved amino acids).
Analysis of the Expression Patterns of Cotton CLE Genes
The RNA sequencing (RNA-seq) data of the CLE genes of G. hirsutum, in different tissues and under three abiotic stresses were downloaded from the NCBI Sequence Reading Archive (SRA: PRJNA248163). The expression pattern analyses were performed in roots, stems, leaves, petals, pistils, and stamens from intact mature flowers of G. hirsutum and different development stages of ovules. In addition, expression patterns were assessed under stresses including heat, drought, and salt treatments after 0, 1, 3, 6, and 12 h. The expression levels of the GhCLEs were calculated from fragments per kilobase million (FPKM) values and the expressed genes were identified using the default empirical abundance threshold of FPKM > 1 (Hart et al., 2013).
Plant Materials and Treatments
The cotton variety, TM-1 was provided by the Germplasm Bank of Cotton Research Institute, Chinese Academy of Agricultural Sciences. When the cotton seedlings had grown to three euphylla periods in seasonable growth conditions, the roots were irrigated with 20% polyethylene glycol 6000 (PEG6000), 150 mM NaCl solution, and ddH2O, respectively (ddH2O was used as a mock control). Seeding leaves were harvested after 0, 3, 6, 12, and 24 h under the drought and salt treatment, respectively. The samples were placed in liquid nitrogen immediately and stored at −80°C. Tissue-specific samples were taken from a field in Anyang, China. Roots, stems, leaves, petals, stamens, pistils, and ovules (0, 3, 5, 10, and 15 DPA) were also sampled and stored in the same way for RNA extraction. Three biological replicates of all the samples were used.
RNA Extraction and Quantitative Real-Time-PCR Analysis
Total RNA was extracted using an RNA prep Pure Plant Kit (Tiangen, Beijing, China). First-strand complementary DNA (cDNA) was obtained using a PrimeScript RT reagent kit (Perfect Real Time, Takara, Dalian, China). The cDNA samples were mixed with ddH2O at a ratio of 1:5 for quantitative real time PCR (qRT-PCR). SYBR Premix ExtaQTM II (TaKaRa, Japan) was used for qRT-PCR analysis on a 7,500 Fast Real-Time PCR system (Applied Biosystems, Inc., California, USA). The GhCLE expression levels in specific tissues under normal conditions and leaves under the different stress conditions were calculated using 2−ΔΔCT method. The cotton GhActin-7-like gene (Gene Bank ID: AY305733) acted as the endogenous control. All of the qRT-PCR primers were designed using Primer Premier 6.0 software, and are listed in Supplementary Table 9.
Vector Construction and Plant Transformation
The ORFs of GhCLE2, GhCLE33.2, and GhCLE28.1 were cloned under control of the 35S promoter in the plant expression vector pBI121. The constructed vectors were first transferred into Agrobacterium tumefaciens (strain GV3101) and later transferred into wild Arabidopsis (Col-0) using the floral-dip method by soaking the inflorescence for 1 min (Clough and Bent, 1998). The T1 seeds were screened in 1/2 MS culture medium containing 50 mg/L kanamycin. Homozygous T2 lines were obtained by self-pollination and T2 transgenic plants were used for phenotype analysis.
Results
Identification and Characterization Analysis of CLE Genes in Three Cotton Species
In total 55, 56, and 86 CLE genes were identified in G. ramondii, G. arboreum, and G. hirsutum genomes, respectively. Thirteen genes encoding proteins >15 kDa, and four encoding GhCLEs >20 kDa (GhCLE51/GhCLE52/GhCLE53/GhCLE54) were identified. There were 24% CLE proteins that do not have a signal peptide cleavage site, indicating that these may affect the formation of mature peptide. Most of the CLEs were localized outside the nucleus of which 65% CLEs gene members were located extracellularly (Supplementary Tables 1–3).
The CLE genes were unevenly distributed along the chromosomes. In G. hirsutum, 86 GhCLE members were distributed on all but four of the 26 chromosomes: A02, A13, D03, and D13 (Supplementary Figure 1A). In G. arboreum,56 GaCLE genes were localized on chromosomes, such as Ga01, Ga03, Ga05, Ga11 (six genes on each), Ga06, Ga10, Ga12 (three genes on each), Ga07 (four genes), Ga08 (nine genes), Ga09 (five genes), and Ga13 (two genes) (Supplementary Figure 1B). In G. raimondii, 55 GrCLE genes were distributed on all the 13 chromosomes with most residing on chromosome Gr04 (eight GrCLE genes). Gr12 contained only one member (Supplementary Figure 1C).
All the CLE genes in G. ramondii, G. arboreum, G. hirsutum, and poplar genomes were divided into seven groups (I–VII), except for Arabidopsis (Figure 1 and Supplementary Figure 2). The CLE sequence weblogos were drawn for each group to display the conserved sequences and some differences in the CLE domains among different groups or species were found (Figures 1A2,B2). For example, the last residue of Groups I–V mainly appeared as N (Asn), while the Groups VI and VII were presented as His (H) in cotton. The frequency of the RL (Arg and Leu) motif in Arabidopsis and poplar was higher than that of the RR motif, while the opposite was found in G. hirsutum (Figure 1B2).
All of the CLE protein sequences shared three characteristics: an N-terminal signal peptide, a conserved CLE motif containing 14 amino acids at or near the C-terminal, and a generally not conserved region between the signal peptide (Supplementary Figure 3). Apart from positions 12 and 13 in Group II and positions eight and nine in Group III, the CLE motifs (at amino acids 5–14) in each group were highly conserved in cotton, with 10 residues remaining almost unvaried. The CLE motifs of Group II lacked conservation of the His residue at position 13, which did not vary in the remaining groups. The Asn residues at position 14 in the Groups I–V were quite conserved, while in the Groups VI–VII, this position was taken by His (Figure 1A1,A2). The cotton CLE domain (12 residues) was highly conserved except 2, 3, 5, and 10 positions, which was similar to that seen in Arabidopsis and poplar (Figure 1B2).
Most of the CLE members did not contain introns, especially Groups V, VI, and VII and the remaining members contained two to three exons, except GaCLE38, which consisted of 11 exons and 10 introns. Five conserved domains were predicted in each monophyletic group. The motif map revealed that some genes may have lost or acquired conserved motifs during evolution. For example, GhCLE52 in Group IV may have lost motif 4 in comparison to its homologous gene, GhCLE51, while GhCLE31 in Group III may have acquired motif three compared to GrCLE31 and GaCLE31. GhCLE51–GhCLE54, encode CLE proteins carrying multiple identical motifs, containing 2, 5, 1, and 3 replicates of motifs 1 and 2, 2, 1, and 3 replicates of motif 5, respectively (Supplementary Figure 4).
Analysis of Collinearity, Duplication, and Ka/Ks Values of the CLEs Family
The collinearity of the CLE homologous genes in the three cotton genomes was analyzed. In total, 16, 21, and 46 pairs of CLEs intragenomic homologous genes were identified one by one in G. raimondii, G. arboreum, and G.hirsutum (Figure 2 and Supplementary Table 4). To understand the intergenomic collinearity, 73 and 38 collinear pairs of homologous genes were predicted between G. hirsutum and G. arboreum/G. raimondii, respectively. Interestingly, most of the GaCLE members corresponded to two GhCLE members, and one GrCLE member generally corresponds to only one GhCLE (Figure 3 and Supplementary Table 4). Furthermore, 39 pairs of genes derived from whole genome duplications or segmental duplications in G.hirsutum, and 40 or 47 pairs between G. hirsutum and G. arboreum/G.raimondii were identified. High-copy genes exist in most groups. The details of the type of duplication undergone by the collinear gene pairs are listed in Supplementary Tables 5–7.
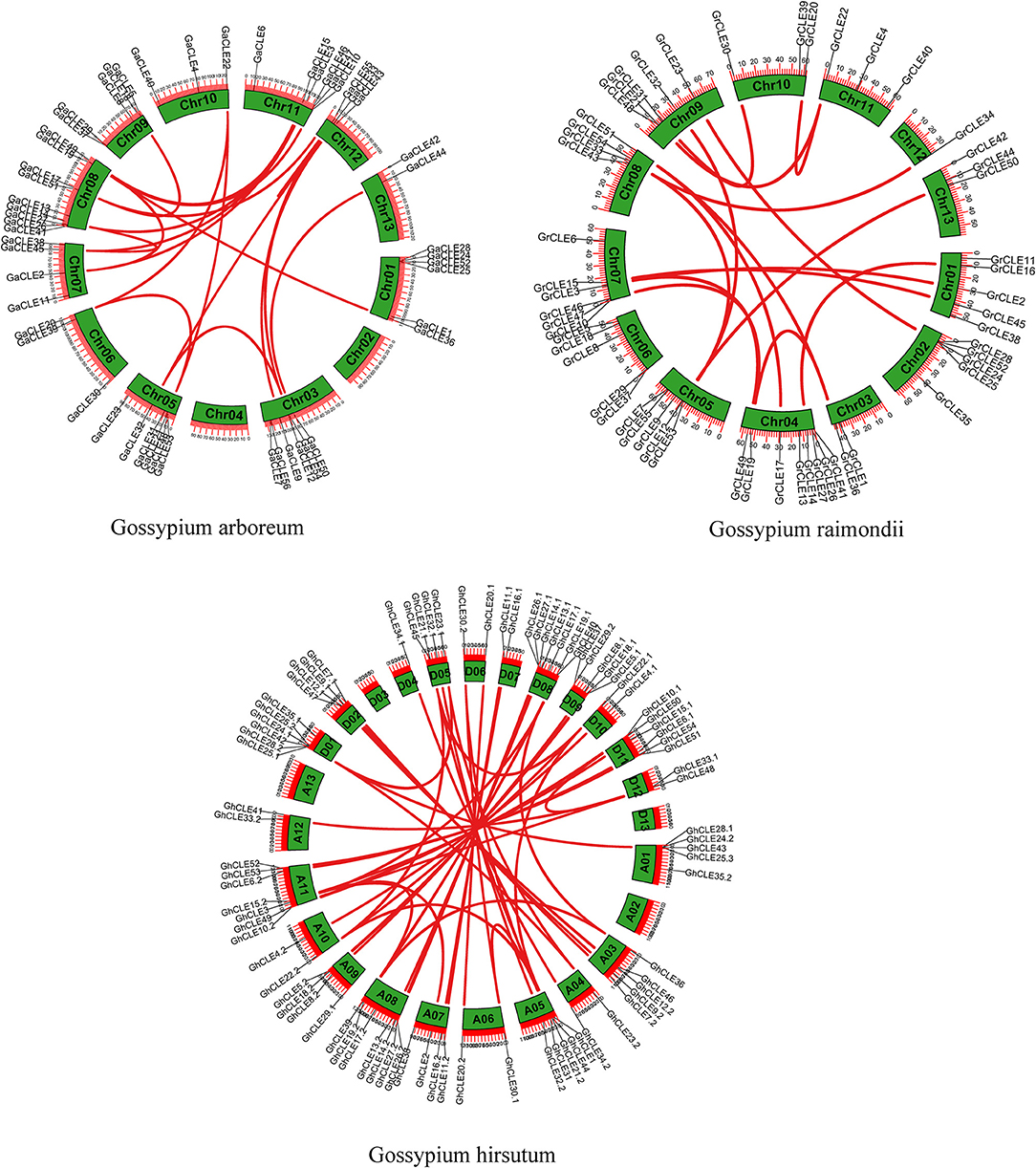
Figure 2. Distribution of CLE gene pairs on chromosomes in intra-genomics of G.arboreum, G. raimondii, and G. hirsutum. The green boxes represent chromosomes and the red lines represent CLE homologous pairs.
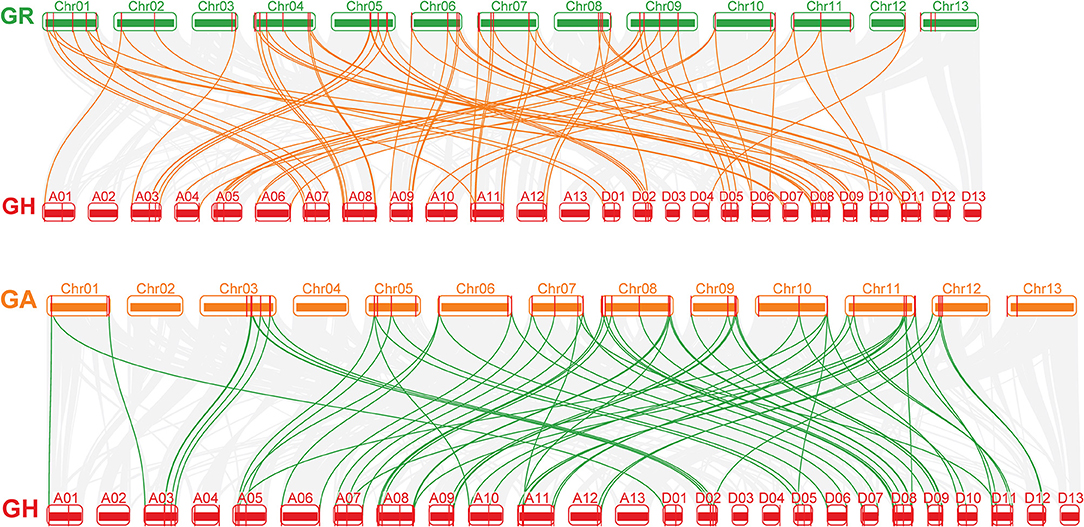
Figure 3. The collinear relationship between G. arboreum and G. hirsutum and between G. raimondii and G. hirsutum (from bottom to top). The gray lines: the collinearity of the whole genome among cotton. The yellow and green lines: the collinearity of CLE pairs in inter-genomics.
The selection pressure for CLE gene duplication was assessed using the Ka/Ks ratio (Supplementary Tables 5–7). Usually, a Ka/Ks value <1, indicates purifying selection, Ka/Ks = 1 indicates neutral evolution, and Ka/Ks > 1 indicates directional selection (Mayrose et al., 2007). Most of the Ka/Ks values were <1, indicating that the CLE gene family in cotton had undergone purifying selection during the long evolutionary process. Nevertheless, several gene pairs (GhCLE7.2/GhCLE7.1, GhCLE21.2/GhCLE21, GhCLE5.2/GaCLE5, GhCLE7.2/GaCLE7, GrCLE4/GhCLE4.1, and GrCLE18/GhCLE18.3) had Ka/Ks values >1, indicating that they may have been formed according to directional selection.
Analysis of cis-Acting Regulatory Elements of GhCLE Genes
To identify the potential function of cotton CLE genes, 32 types of cis-acting regulatory elements (CAREs) were obtained from GhCLE members, and grouped into four functional types as shown in Figure 4A. (i) Involved in stress responses, such as MYB binding sites (MBS) that result in drought-inducibility, anaerobic induction regulatory elements (ARE), low-temperature responsive elements (LTR), defense and stress responsive elements (TC-rich repeats), and anoxic specific inducibility (ARE) etc.; (ii) involved in hormonal regulation, including gibberellin-responsiveness (GA) elements (GARE-motif, TATC-box, P-box), abscisic acid (ABA) responsive elements (ABRE), auxin-responsive (IAA) elements (TGA, AuxRR-core), salicylic acid (SA) responsiveness (TCA), MeJA-responsive elements, and (CGTCA-motif, TCACG-motif); (iii)involved in plant development (CAAT-box, CAT-box, etc.); and (iv) involved in light responsive elements (GT1-motif, Box4, G-Box, etc.).
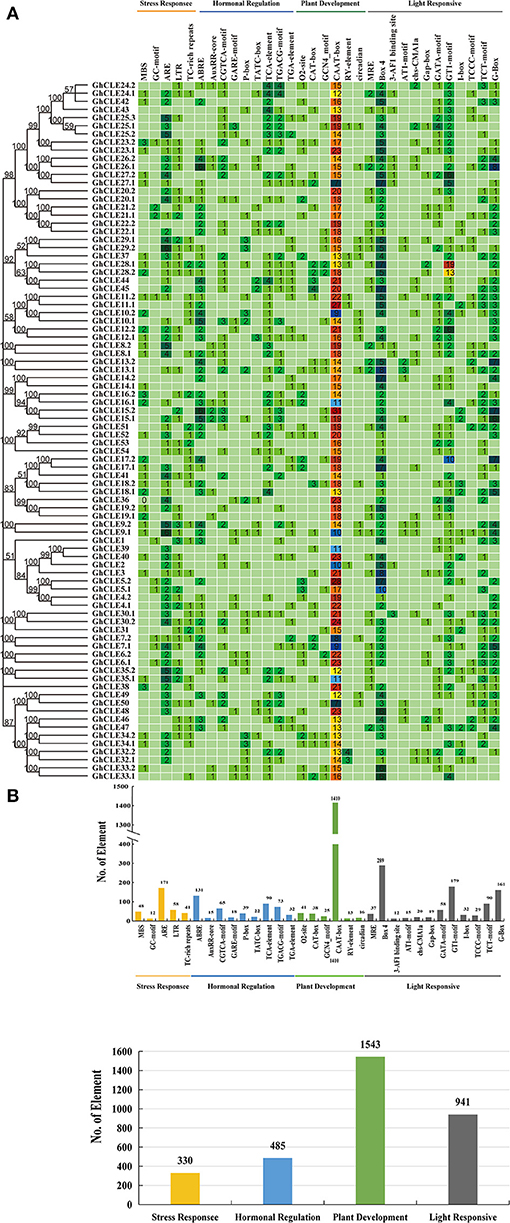
Figure 4. Analysis of Cis-acting element numbers in G. hirsutum CLE peptide signals. (A) The colors of the grid indicate the number of Cis-acting elements of the GhCLE genes. (B) The bar chart represents the sum of each type of Cis-acting elements.
Most CAREs were found to be involved in plant growth and development (all 1,543), especially the CAAT elements, accounting for 91% of them all. Almost all members contained 7 to 31 CAAT elements, except GhCLE1. The GCN4_motif is involved in endosperm expression, and the CAT-box element is related to meristem expression. Light responsiveness (941) and hormonal signaling (485) elements were the next to be identified. Nine and 19 types of hormonal signaling and light response- related CAREs were identified in GhCLE promoters, respectively. As for stress response elements (330), ARE was found to be the most abundant in the stress response group (accounting for 51%). More importantly, the CAREs, involved in drought-inducibility, only represented 14.5% of the total, while other stress-related cis-elements were identified and found to be involved in multiple abiotic stresses. More than one putative stress-related responsive element was identified in all GhCLE members (except GhCLE43, GhCLE22.2, GhCLE13.2, and GhCLE14.2) (Figure 4B).
In addition, highly similar types and quantities of CAREs among homologous genes were analyzed. Five pairs of ARE, involved in drought stress regulation (GhCLE9.1/GhCLE9.2, GhCLE35.1/GhCLE35.2, GhCLE8.1/GhCLE8.2, GhCLE29.1/GhCLE29.2, and GhCLE25.1/GhCLE25.2/GhCLE25.3) and GhCLE28.1 and GhCLE28.2 contained 19 and 13 GT1-motifs, involved in light responses, respectively, were identified (Figure 4).
GhCLE Expression Patterns Analysis in Specific Organs and Tissues
As important indicators of gene biological function, tissue- and organ-specific gene expression profiles were considered to be crucial. As shown in Figure 5A, GhCLE members were widely expressed in petals, stamens, pistils, and −3, 0, 3, 5, 10, and 20 days post-anthesis (DPA) ovules (reproductive organs) and in roots, stems, and leaves (vegetative organs). Many CLEs of Group III and IV (possess similar DPXHN motif in Figure 1A2) showed high expression levels in the reproductive tissues. For example, the homologous genes, GhCLE33.2, GhCLE33.1, and GhCLE47 showed significant levels of expression in the petals, stamens, and pistils. GhCLE6.1, GhCLE2, and GhCLE35.2 were highly expressed in all tested tissues. Most of the members of Groups V, VI, and VII (possess similar PGGP motif in Figure 1A2) were specifically expressed in root, especially GhCLE28.1, GhCLE28.2, GhCLE24.1, and GhCLE42, and GhCLE9.2 exhibited the highest expression level in 10 DPA ovules. Generally, the expression patterns of the GhCLE gene pairs showed high similarity. For example, GhCLE35.1 and GhCLE35.2 were significantly expressed in flower organs, leaves, and all stages of ovule development, while GhCLE28.1 and GhCLE28.2 were highly expressed in the roots. Nevertheless, the expression patterns of the two were not always the same between paralogous genes. For instance, GhCLE17.2 was predominantly expressed in roots, stems, and petals but GhCLE17.1 was not expressed at a low level in petals. GhCLE23.1 was highly expressed in roots and pistils but GhCLE23.2 was mainly expressed in roots and petals. GhCLE6.1 was highly expressed during ovule development, while GhCLE6.2 was not.
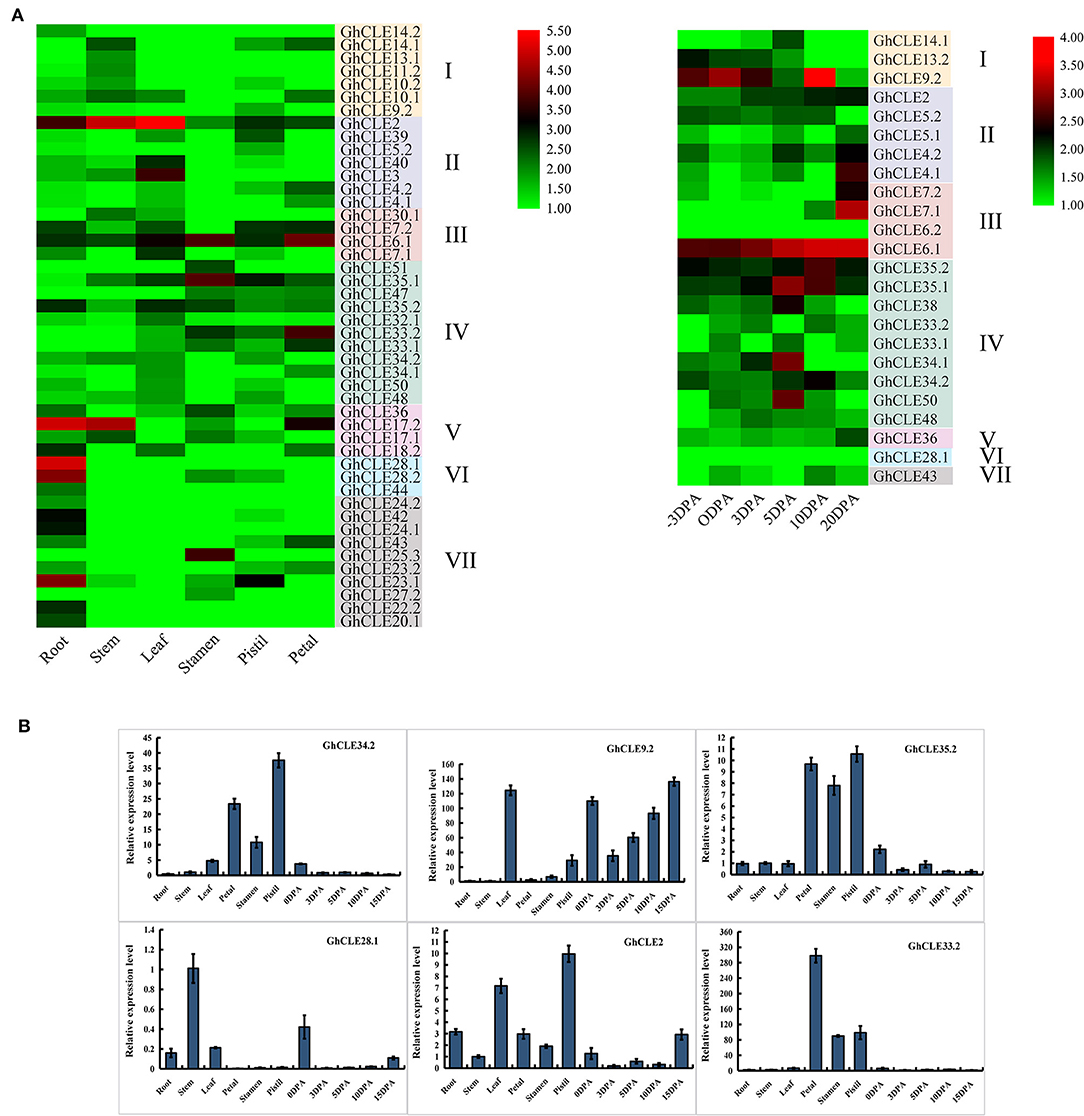
Figure 5. Expression patterns of GhCLEs in different tissues. (A) Tissue/organ-specific expression patterns of GhCLEs, including root, stem, leaf, petal, pistil, stamen, and ovule (−3, 0, 3, 5, 10, 20 DPA). Heat map was drawn according to calculation results of log2 of fragments per kilobase million (FPKM) RNA sequencing (RNA-seq) data. (B) Relative expression levels of six GhCLE genes in different tissues.
Based on the analysis of G. hirsutum expression profile, six genes (GhCLE34.2, GhCLE9.2, GhCLE35.2, GhCLE28.1, GhCLE2, and GhCLE33.2) were randomly selected for further RT-PCR verification. Four of six genes were highly expressed in the flower organs (petals, stamens, and pistils). GhCLE34.2, GhCLE35.2, and GhCLE33.2 of Group IV were significantly expressed in the flower organs, while GhCLE2 of Group II was mainly expressed in the pistil or leaves. GhCLE9.2 of Group I was distinctly expressed in leaves and ovules, while GhCLE28.1 of Group VI was prominently expressed in the stem (Figure 5B). The results of the qRT-PCR analyses were basically coincident with those obtained through RNA-seq (Figure 5).
GhCLE Expression Pattern Analysis in Various Abiotic Stresses
In order to understand the putative functions of GhCLE genes, a comprehensive analysis of the expression patterns was carried out under heat, salt, and drought treatments (Figure 6A). Forty-eight out of the 86 GhCLEs were barely expressed at all time points (these are not shown in the heat map). When the cotton plants were exposed to three stress treatments, more than half of the members were less or almost imperceptibly expressed, especially those in Groups IV, V, VI, and VII. GhCLE12.1 and GhCLE12.2 of Group I were significantly expressed for 1 h of drought treatment, and a relatively high expression was observed at 12 h under salt treatment. Among the Group III genes, GhCLE7.1 and GhCLE7.2 were notably expressed at the 3 h time point of drought stress and modestly expressed at other time points compared with the other members. GhCLE38 of Group IV was highly expressed at 1, 3, and 6 h of the salt and drought treatments, while GhCLE15.1 of Group I and GhCLE36 of Group V were only expressed at 1 h of heat treatment.
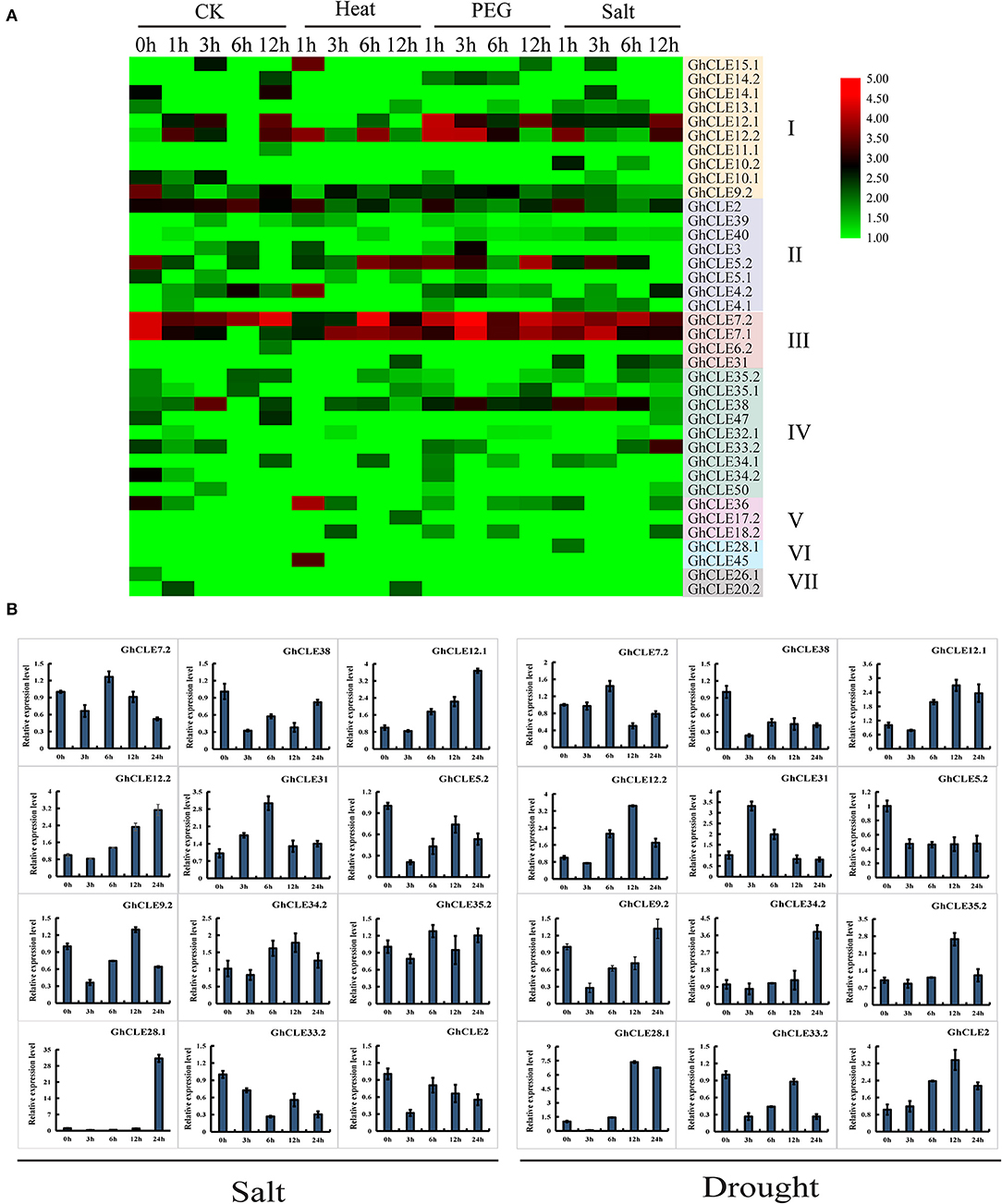
Figure 6. Expression patterns of GhCLEs under different stress. (A) Stress-induced expression patterns of GhCLEs under polyethylene glycol (PEG), and salt treatments for 1-, 3-, 6-,12-, and 24-h, respectively. Heat maps are drawn in the same way as in Figure 5. (B) Relative expression levels of 12 GhCLE genes under different stress.
According to the heatmap, the expression of 6 genes was increased at certain periods under the three stresses. This finding was further verified by qRT-PCR analysis with three technical repetitions. Three GhCLEs (GhCLE38, GhCLE5.2, and GhCLE33.2) were constantly downregulated under salt and drought treatment. The expression levels of GhCLE7.2, GhCLE31, and GhCLE35.2 were the highest at 6 h. Homologous genes, such as GhCLE12.1 and GhCLE12.2 showed continuous upward expression after salt stress and were predominantly expressed at 12 h after drought stress. GhCLE9.2 and GhCLE34.2 were significantly expressed at 12 h after salt treatment and 24 h after drought treatment, while GhCLE2 was continuously downregulated under salt treatment and showed the highest expression level at 12 h after drought stress. GhCLE35.2 and GhCLE28.1 were also highly expressed at 12 h after drought stress. Members of the same subfamily showed not only functional redundancy but also functional differentiation. For instance, GhCLE12.1, GhCLE12.2, and GhCLE9.2 in Group I were upregulated under salt and drought treatment. The expression levels of GhCLE34.2 and GhCLE35.2 in Group IV were upregulated under the two stresses, while GhCLE33.2 and GhCLE38 were downregulated (Figure 6B).
Analysis of Physical and Chemical Properties, Structure Prediction and Interaction Network of GhCLE Proteins
To understand the relationship between the structure and function of CLE proteins, GhCLE2, GhCLE33.2, and GhCLE28.1 peptides from different groups were analyzed. The three GhCLEs belonging to membrane proteins were found to form transmembrane region (close to N-terminal), extracellular region, and intracellular region (some proteins have one or more transmembrane regions) (Figure 7A). The number of hydrophilic amino acid residues was more than that for hydrophobic residues (a positive value represents hydrophobic residues and a negative value indicates hydrophilic residues) (Figure 7B). The signal peptide regions were analyzed at the N-terminals of three protein sequences, but GhCLE2 was less likely to have a signal peptide (42.21%) (Figure 7C). The secondary structures of the proteins were predicted as follows: theα-helix was dominant in the polypeptide chains of GhCLE28.1 and GhCLE33.2, followed by disordered structures and TM helices. The GhCLE2 protein contained 76% disordered structures, 49% α-helices, 16% TM helices, and 2% β-turn (Figure 7D).
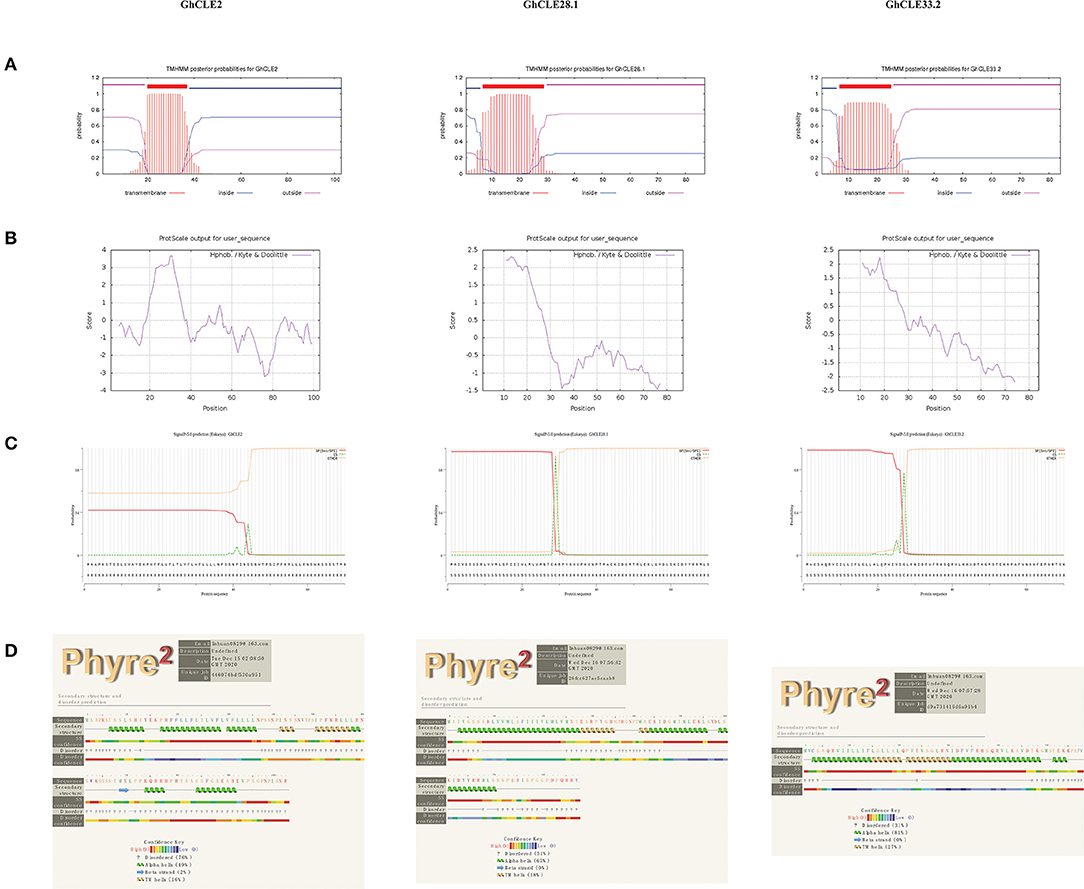
Figure 7. (A–D) Respectively represent transmembrane structure characteristics, hydrophobicity/hydrophilicity, signal peptides, and secondary structure of GhCLE2, GhCLE28.1, and GhCLE33.2.
Based on the findings in Arabidopsis, the interactions and regulatory pathways of homologous CLE peptides were predicted in cotton. There were three CLEs (GhCLE28.1, GhCLE2, and GhCLE33.2) in G. hirsutum corresponding to six genes in Arabidopsis. Ten high evident interactive proteins and co-expression proteins in Arabidopsis and other organisms were investigated in every query protein (Figure 8 and Supplementary Table 8). CLE5 and CLE6 were co-expressed with four genes (CLE2, CLE3, CLE4, and CLE7) within the family, respectively. They were closely related to GhCLE37, GhCLE28.1, and GhCLE28.2. While CLE41 (GhCLE2) co-expressed only with WOX4, CLE42 (GhCLE2) co-expressed with CLE43; even CLE44 (GhCLE2) and CLE45 (GhCLE33.2) had no co-expressed genes. Interestingly, CLE4 and CLE5 were co-expressed in Populus (Figure 8A). WOX4 and WOX14 played a role downstream of the PXY receptor kinase in the GhCLE2 (CLE41, CLE42, and CLE44) network (Figures 8C–E). Moreover, the interactive proteins involved in the GhCLE33.2 network included some receptor-like kinases (LRR-RLKs), such as BAM3, MAKR5, BRX, OPS, CLV3, CLE8, CLE22, CLE26, OPS, AT5G56040 (RGF1), and AT2G25790 (SKM1) (Figure 8F).
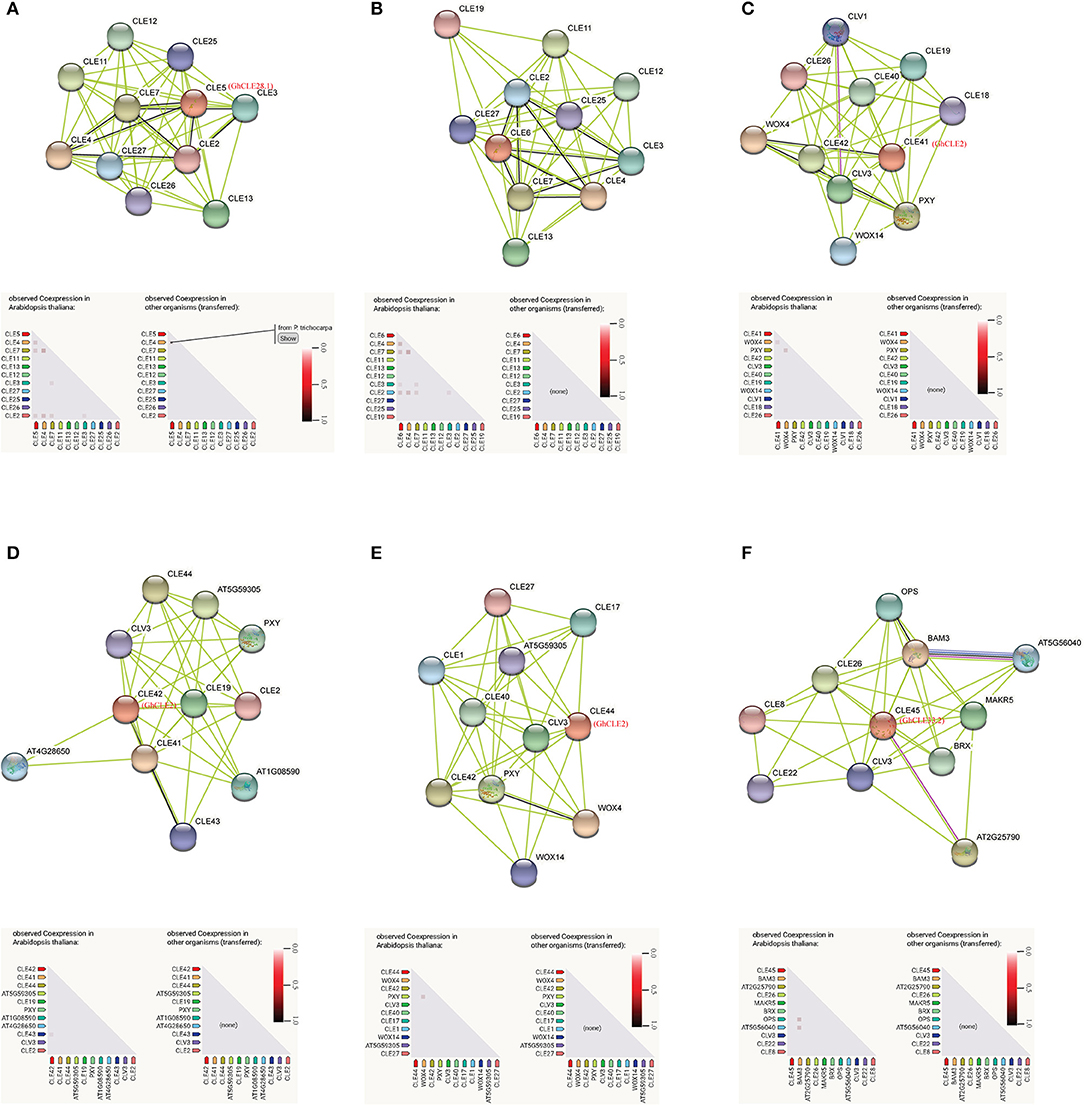
Figure 8. Prediction of interaction network between CLE signal peptide and other related proteins in G. hirsutum, with homologous protein in Arabidopsis as enquiry protein. Homologous proteins of Arabidopsis in G. hirsutum are shown in red. The black line represents the co-expressive relationship. The co-expression predicts functional association under the network diagram: in the triangle matrix, the intensity of the color indicates the level of confidence that two proteins are functionally associated. (A) CLE5; (B) CLE6; (C) CLE41; (D) CLE42; (E) CLE44; (F) CLE45.
Overexpression Analysis of GhCLE2, GhCLE33.2, and GhCLE28.1 in Arabidopsis
To clarify the biological functions of CLEs in cotton, three GhCLE genes were overexpressed in Arabidopsis. GhCLE2 was widely expressed in the most tested tissues, especially in leaves and stems; GhCLE33.2 was highly expressed in flower organs, notably petals; GhCLE28.1 was specifically expressed in roots. The successful transgenic lines were screened out in Figure 9. Compared with the wild-type (WT), the GhCLE2OE1 (L.1) and GhCLE2OE2 (L.2) lines grew into dense dwarf plants and produced a large number of small leaves and formed small rosettes, exhibiting a shrub-like phenotype. These lines also expressed short stems and small siliques (Figure 10A). Overexpression of the GhCLE33.2 gene in Arabidopsis was found to result in anthocyanin overproduction, especially in several old leaves and stems. The old leaves were light purple or even pink, while young leaves and stems were dark green or slightly purple. Moreover, line (L.3) showed mild developmental timing delays and formed small rosettes and dwarf phenotypes (Figure 10B). Unlike GhCLE2OE and GhCLE33.2OE, GhCLE28.1OE lines (L.4) displayed larger rosettes with leaves of misshapen character and abnormal enlargement, which also apparently result in short, flat-shaped stem with infertile flowers. In summary, the transgenic plants exhibited a significant decrease in plant height compared to the WT plants (Figure 10C). The above phenotypes of the overexpressed lines confirmed that the CLE family mainly regulates meristem and plays an important role in the growth and development of plants.
Figure 9. The transgenic lines were verified by Agarose gel electrophoresis. The red arrows point to successful transgenic lines.
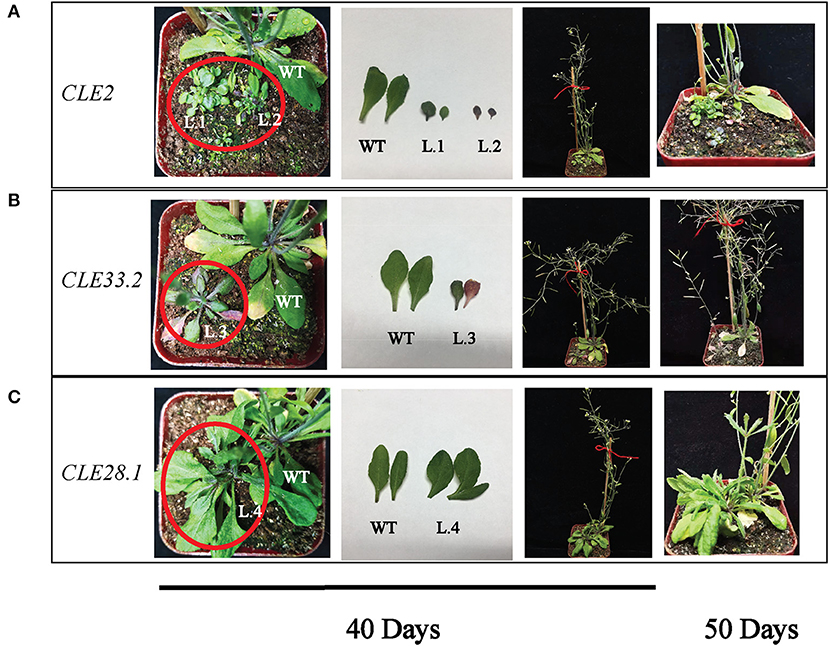
Figure 10. Phenotypes of GhCLE2 (A), GhCLE33.2 (B), and GhCLE28.1 (C) overexpressed in Arabidopsis. The pictures of the first three columns show the seedlings and corresponding leaves at 40 days after germination, and the last column shows seedlings at 50 days old.
Discussion
Over the past few years, CLE signal peptides have attracted attention from the scientific community. The biological function of peptides involved in plant growth, development, and responses to environmental stresses have become more evident, with a large number of peptide family members identified. Previous genome-wide studies of the CLE family have focused mainly on Arabidopsis, Oryza sativa, Glycine max, Zea, and Triticum aestivum (Opsahl-Ferstad et al., 1997; Suzaki et al., 2006; Oelkers et al., 2008; Hastwell et al., 2015a; Li et al., 2019). However, the completion of the cotton whole-genome sequencing project provides a chance to characterize the CLE gene family in this genus. Comprehensive identification of CLE peptides was performed in three cotton species. Eighty-six CLE members were identified in G. hirsutum, 56 in G. arboreum, and 55 in G. ramondii. Subsequently, a series of evolutionary analyses were performed, including chromosomal location, gene structure, identification of conservative domains, collinearity, protein properties and interaction, expression patterns, and functional verification.
Although the diploids, G. arboreum and G. ramondii are the ancestors of G. hirsutum, the distribution of CLE genes on their chromosomes are not greatly regular (Zhang et al., 2015). G. arboreum, the A-genome donor of G. hirsutum, has no CLE genes on chromosome Ga02 and Ga04. It is also found that there are no CLE genes on chromosome A13 of G. hirsutum while there are two genes on A04. In G. ramondii, the D-genome donor of G. hirsutum, all genes are irregularly distributed on all the 13 chromosomes, while there is no member on chromosomes, D03 and D13 of G. hirsutum. The number of CLE family genes in G. hirsutum theoretically should have been approximately twice that in diploid cotton, but it was found only to be 1.5-fold. Many of the CLE genes in G. hirsutum were considered double genes due to the synteny of the A and D subgenomes. Nevertheless, there were some exceptions that lacked one corresponding homologous gene. These counterparts may have been lost or became pseudogenes in their repeated regions during evolution process of G. hirsutum (Magadum et al., 2013; Iranzo et al., 2019). In addition, the number of cotton CLE genes was considerably more than that seen presently in grape (nine members) (Wang et al., 2019), Arabidopsis (at least 32) (Jun et al., 2010), common bean (44) (Hastwell et al., 2015a) and rice (47) (Suzaki et al., 2006; Kinoshita et al., 2007), but less than in hexaploid bread wheat (Tricticumaestivum L.) (104) (Li et al., 2019). However, the number of CLE genes (50 members) in Populus was similar to those in the A (G. arboreum) or D (G. ramondii) genomes (Liu et al., 2016). The quantity of CLE members in the allotetraploid species (G. hirsutum) was very close to that in soybean (84 CLEs) (Hastwell et al., 2015a). Apparently, if the genome size between species is significantly different, the number of CLEs is also different.
Segmental duplications appear most frequently in polyploid plants and reserve numerous duplicated chromosomal blocks within their genomes, such as diploid Arabidopsis (Cannon et al., 2004). In this study, 46 pairs of whole-genome or segmental duplications among 86 GhCLE genes, 73 pairs between G. hirsutum and G. arboreum, and 38 between G. hirsutum and G. raimondii were identified. Therefore, whole-genome or segmental duplications might have been one of the primary driving forces for CLE family expansion during the evolution of cotton, likely with SPL transcription factors (Cai et al., 2018). Group VII contained the most GhCLE members, including whole-genome or segmental duplications, thus introducing functional redundancy during the regulation of cotton development and responses to abiotic stress. However, the main expansion approach of the CLEs in wheat was tandem duplication (Li et al., 2019). Although wheat is of the same gene family, the way of amplification is probably different among species. These results may facilitate the characterization of new biological functions of new CLE genes (Cannon et al., 2004). Interestingly, previous studies found that the MW of all CLE family genes in Arabidopsis is <15 kDa, while 16 cotton CLE peptides are >15 kDa. In particular, four genes encode proteins >20 kDa (GhCLE51/GhCLE52/GhCLE53/GhCLE54 belonging to Group IV) in G. hirsutum (Cock and McCormick, 2001). These genes were speculated to be involved in C-terminal extension, as similar genes, such as AtCLV3, ZmESR3, PtCLE19, and OsCLE32, have been identified (Kinoshita et al., 2007; Oelkers et al., 2008; Wang and Fiers, 2010; Han et al., 2016). The proteins containing more than 200 amino acids along their length were located in the nucleus and had multiple CLE domains. GhCLE51/GhCLE54 and GhCLE52/GhCLE54 were two pairs of homologous genes located on the chromosomes, GhA11 and GhD11, respectively. These features indicated that they arose by segmental or whole-genome duplication and possess a quite close relationship. Therefore, their functions were thought to be redundant and different from other members but these are unknown.
Seven gene clusters were identified in cotton species. It was beneficial to elucidate similarities in the function of the latest CLE members and their related homologs and orthologs (Han et al., 2016; Hastwell et al., 2017b). Different numbers of gene clusters were found in different species in previous studies: three, four, five, and seven gene clusters were identified in grape, populus, wheat, and soybean, respectively (Hastwell et al., 2015a; Liu et al., 2016; Li et al., 2019; Wang et al., 2019). Residues of the CLE-conserved domain have undergone diverse changes during the evolution of species, resulting in different classifications and diverse functions. The CLE motif of Group I had the highest sequence similarity to the CLV3 motif (LRTVPSGPDPLHH), functioning in shoot and root meristems (Kondo et al., 2006). Group II showed the highest similarity to the TDIF motif (HEVPSGPNPISN), which acts in vasculature (Etchells et al., 2016). The CLE motif of the other groups was very diverse and showed significant differences to the motifs of the seven groups in soybean (Hastwell et al., 2015a). The biological functions of some cotton CLE genes that diversified during evolution are unknown. Therefore, whether the variation in residues of different groups contributed to functional differentiation needs to be further predicted and verified. Arabidopsis CLE peptides can be divided into two types, A-type and B-type, according to their biological functions. A-types affect cell differentiation in root and shoot apical meristem (AtCLE1, AtCLE4, AtCLE7, AtCLE19, AtCLE22, and AtCLE40) (Whitford et al., 2008; Yamaguchi et al., 2016), and B-types affect vascular development (AtCLE41-AtCLE44/TDIF) (Strabala et al., 2006; Oelkers et al., 2008; Whitford et al., 2008; Qiang et al., 2013). To determine the functional classification of cotton CLE peptides, the members of G. hirsutum and Populus were together grouped into seven gene clusters. In this study, nine GhCLE genes belonged to Group II which contained four AtCLEs (AtCLE41/AtCLE44/AtCLE42/AtCLE46) and nine PtCLEs. These CLE genes classified together with AtCLE41/TDIF, have the same CLE motif, especially GhCLE3, which suggests that GhCLE3 may affect vascular development (Whitford et al., 2008; Qiang et al., 2013).
An intriguing finding showed that some A-type CLE genes have distinctly specific expression patterns at certain development stages in the same tissues. CLE3, CLE5, CLE16, and CLE17 were differently expressed in the primary or lateral roots of not fully mature and fully differentiated plants (Jun et al., 2010). Certain members (such as GhCLE17.2, GhCLE28.1, GhCLE28.2, and GhCLE23.1) of Groups V and VII may have similar functions such as the above genes. Previous studies have indicated that the CLE5 and CLE6 peptides singly and together showed the rosette leaf shape phenotype. These two genes are also expressed at the base above the abscission zone of the flower organ (Jun et al., 2010; DiGennaro et al., 2018). GhCLE28.1 was found to be in the same subfamily as AtCLE5 and AtCLE6. Overexpression of GhCLE28.1 in Arabidopsis also shows enlarged rosettes and misshapen leaves, with a lack of apical meristem development. More importantly, the line did not form an inflorescence, so there was no progeny. The GhCLE28.1 was also mainly expressed in stems, 0DPA ovules, leaves, and roots. The GhCLE2 was widely expressed in most tested tissues, especially leaves, stems, and pistils. Overexpression of GhCLE2 in Arabidopsis mainly exhibited distinctive shrub-like dwarf plants lacking apical dominance, suggesting that the proliferation and differentiation of meristems may be inhibited, including root, stem, flower, and meristem, which were akin to the phenotypes of overexpressing AtCLE41, AtCLE42, and AtCLE44 (Strabala et al., 2006; Whitford et al., 2008). In this study, GhCLE2 not only changed the size and number of the leaves but also affected the accumulation of anthocyanin (L.2). Uniquely, GhCLE33.2OE showed that the overproduction of anthocyanin is significant; the rosettes were also small but bigger than those of the GhCLE2OE lines; bolting occurred later compared to the WT lines, and a handful of seeds were harvested. Similar to the effect of most CLE peptides, GhCLE33.2 also caused the entire line to be pygmean. Depuydt et al. (2013) showed that AtCLE45 and the specific receptor BAM3 affect proliferation and differentiation of protophloem, thereby influencing root meristem. The orthologous GhCLE33.2 and AtCLE45 have the same CLE motif (KRRVRRGSDPIHN) but the full-length protein sequence of CLE45 was longer than that of CLE33.2. However, GhCLE33.2 was hardly expressed in roots, suggesting that serious functional differentiation may have occurred in this member. To sum up, the overexpression of the three GhCLEs in this study showed the same or similar phenotypes to overexpression lines of certain CLEs in Arabidopsis or other species, such as AtCLE42, AtCLE44, and wheat TaCLE3 overexpression in Arabidopsis (Strabala et al., 2006; Li et al., 2019). To date, CLE peptides are proven to be involved in regulating stem cell homeostasis of the SAM and RAM, and they also regulate inflorescence development, lateral root growth, floral meristem formation, seed development and vascular formation (Clark et al., 1993; Strabala et al., 2006; Czyzewicz et al., 2013; Ingram and Gutierrez-Marcos, 2015). G. hirsutum serves as an ideal plant for different biological studies, such as genome evolution, polyploidization, and single-celled biological processes (Yin et al., 2021). Although some functions of GhCLE genes have been investigated, the more CLEs and their function need to be explored in the further.
The CLE peptide signaling pathway, which binds to phytohormone signaling pathways and perceives leucine-rich repeat receptor-like kinases (LRR-RLKs), is involved in the regulation of a variety of biological processes in response to environmental signals. As receptors, LRR-RLKs are perceived by CLE peptides, establishing the evolutionarily conserved CLE-RLK module, which can regulate plant development and responses to abiotic stresses by transmitting extracellular and intracellular signaling cascades (Betsuyaku et al., 2011; Murphy et al., 2012). This result was confirmed by the protein interaction analysis in this study. For instance, CLE (CLV1-like LRR-RLK) peptides play an eye-catching role in perceiving nitrate and controlling nodulation formation in legumes (Nishimura et al., 2002; Miyazawa et al., 2010; Miyawaki et al., 2013). Barely any meristem 3 (BAM3) acts as a receptor of CLE45 peptides to inhibit protophloem formation in Arabidopsis root meristems (Kang and Hardtke, 2016). In addition, AtCLE25 peptides convey water-deficiency signals via the vascular tissues of Arabidopsis and work with BAM receptors in leaves to influence abscisic acid synthesis and stomatal transpiration control (Takahashi et al., 2018). According to evolutionary analysis, the GhCLE38-BAM module is most likely to act in long-distance signal transmission as a conveyer during the dehydration response. Therefore, the evolutionary characteristic and biological functions of the specific receptors of CLE ligands have gradually been identified and studied, providing guidance for further research on the function of cotton CLE genes.
In summary, CLE members in G. raimondii, G. arboreum, and G. hirsutum were identified, analyzed using bioinformatics, and their expression patterns and functions verified. The present study may provide insight into their structures and enable further investigation into the functions of cotton CLEs according to previously known orthologs in model plants and published transcriptional evidence. In order to further understand how certain key peptides regulate plant development and responses to environmental stimuli, the functional domain and structural modifications of these mature peptides need to be determined. Since cotton is a heterotetraploid species with a large genome, there will have been a variety of changes during evolution. Even if there are many functionally redundant members in a gene family, some genes will still be functionally differentiated. Therefore, finding and mining these genes to improve plant or crop characteristics is a scientific mission.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HL, ZY, and WY conceived the research and designed the experiments. HL and WW conceived the structure of the manuscript and analyzed data and experiment results. HL wrote the initial manuscript. XC, XH, and SW participated in the experiments and modified the initial manuscript. ZS and YL participated in the second revision of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31201247), the Young Elite Scientists Sponsorship Program by CAST (2016QNRC001), and the National Key Research and Development Program of China (2016YFD0101400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to two reviewers for their constructive comments. We acknowledge Weidong Zhu (Agricultural Genomics Institute at Shenzhen, Shenzhen, China) for technical assistance. We also thank Li Yuan (Institute of Botany, the Chinese Academy of Sciences, Beijing, China) for the valuable comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.671626/full#supplementary-material
References
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Araya, T., Miyamoto, M., Wibowo, J., Suzuki, A., Kojima, S., Tsuchiya, Y. N., et al. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 2029–2034. doi: 10.1073/pnas.1319953111
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–208. doi: 10.1093/nar/gkp335
Betsuyaku, S., Sawa, S., and Yamada, M. (2011). The Function of the CLE peptides in plant development and plant-microbe interactions. Arabidopsis Book 9:e0149. doi: 10.1199/tab.0149
Bjellqvist, B., Basse, B., Olsen, E., and Celis, J. E. (1994). Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539. doi: 10.1002/elps.1150150171
Bonello, J. F., Opsahl-Ferstad, H. G., Perez, P., Dumas, C., and Rogowsky, P. M. (2000). Esr genes show different levels of expression in the same region of maize endosperm. Gene 246, 219–227. doi: 10.1016/s0378-1119(00)00088-3
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Cai, C., Guo, W., and Zhang, B. (2018). Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci. Rep. 8:762. doi: 10.1038/s41598-017-18673-4
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Yu, P., Luo, J., and Jiang, Y. (2003). Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 14, 859–865. doi: 10.1007/s00335-003-2296-6
Chu, H., Qian, Q., Liang, W., Yin, C., Tan, H., Yao, X., et al. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. doi: 10.1104/pp.106.086736
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cock, J. M., and McCormick, S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126, 939–942. doi: 10.1104/pp.126.3.939
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Czyzewicz, N., Yue, K., Beeckman, T., and De Smet, I. (2013). Message in a bottle: small signalling peptide outputs during growth and development. J. Exp. Bot. 64, 5281–5296. doi: 10.1093/jxb/ert283
Depuydt, S., Rodriguez-Villalon, A., Santuari, L., Wyser-Rmili, C., Ragni, L., and Hardtke, C. S. (2013). Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. U.S.A. 110, 7074–7079. doi: 10.1073/pnas.1222314110
DiGennaro, P., Grienenberger, E., Dao, T. Q., Jun, J. H., and Fletcher, J. C. (2018). Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct 2:e00103. doi: 10.1002/pld3.103
Etchells, J. P., Mishra, L. S., Kumar, M., Campbell, L., and Turner, S. R. (2015). Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055. doi: 10.1016/j.cub.2015.02.023
Etchells, J. P., Smit, M. E., Gaudinier, A., Williams, C. J., and Brady, S. M. (2016). A brief history of the TDIF-PXY signalling module: balancing meristem identity and differentiation during vascular development. New. Phytol. 209, 474–484. doi: 10.1111/nph.13642
Fiume, E., and Fletcher, J. C. (2012). Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24, 1000–1012. doi: 10.1105/tpc.111.094839
Fletcher, J. C., Brand, U., Running, M. P., Simon, R., and Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–1186. doi: 10.1093/nar/gkr944
Han, H., Zhang, G., Wu, M., and Wang, G. (2016). Identification and characterization of the Populus trichocarpa CLE family. BMC Genomics 17:174. doi: 10.1186/s12864-016-2504-x
Hart, T., Komori, H. K., LaMere, S., Podshivalova, K., and Salomon, D. R. (2013). Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics 14:778. doi: 10.1186/1471-2164-14-778
Hastwell, A. H., de Bang, T. C., Gresshoff, P. M., and Ferguson, B. J. (2017a). Author Correction: CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci. Rep. 7:15474. doi: 10.1038/s41598-017-14991-9
Hastwell, A. H., de Bang, T. C., Gresshoff, P. M., and Ferguson, B. J. (2017b). CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci. Rep. 7:9384. doi: 10.1038/s41598-017-09296-w
Hastwell, A. H., Gresshoff, P. M., and Ferguson, B. J. (2015a). Genome-wide annotation and characterization of CLAVATA/ESR (CLE) peptide hormones of soybean (Glycine max) and common bean (Phaseolus vulgaris), and their orthologues of Arabidopsis thaliana. J. Exp. Bot. 66, 5271–5287. doi: 10.1093/jxb/erv351
Hastwell, A. H., Gresshoff, P. M., and Ferguson, B. J. (2015b). The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Funct. Plant Biol. 42, 229–238. doi: 10.1071/fp14222
Hirakawa, Y., Kondo, Y., and Fukuda, H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629. doi: 10.1105/tpc.110.076083
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105, 15208–15213. doi: 10.1073/pnas.0808444105
Hobe, M., Müller, R., Grünewald, M., Brand, U., and Simon, R. (2003). Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 213, 371–381. doi: 10.1007/s00427-003-0329-5
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0:an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Ingram, G., and Gutierrez-Marcos, J. (2015). Peptide signalling during angiosperm seed development. J. Exp. Bot. 66, 5151–5159. doi: 10.1093/jxb/erv336
Iranzo, J., Wolf, Y. I., Koonin, E. V., and Sela, I. (2019). Gene gain and loss push prokaryotes beyond the homologous recombination barrier and accelerate genome sequence divergence. Nat. Commun. 10:5376. doi: 10.1038/s41467-019-13429-2
Jun, J., Fiume, E., Roeder, A. H., Meng, L., Sharma, V. K., Osmont, K. S., et al. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154, 1721–1736. doi: 10.1104/pp.110.163683
Kang, Y. H., and Hardtke, C. S. (2016). Arabidopsis MAKR5 is a positive effector of BAM3-dependent CLE45 signaling. EMBO Rep. 17, 1145–1154. doi: 10.15252/embr.201642450
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kinoshita, A., Nakamura, Y., Sasaki, E., Kyozuka, J., Fukuda, H., and Sawa, S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821–1825. doi: 10.1093/pcp/pcm154
Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., et al. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848. doi: 10.1126/science.1128439
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Y. C., and Lu, Y. C. (2019). BLASTP-ACC: parallel architecture and hardware accelerator design for BLAST-based protein sequence alignment. IEEE Trans. Biomed. Circuits Syst. 13, 1771–1782. doi: 10.1109/tbcas.2019.2943539
Li, Z., Liu, D., Xia, Y., Li, Z., Niu, N., Ma, S., et al. (2019). Identification and functional analysis of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) gene family in wheat. Int. J. Mol. Sci. 20:74319. doi: 10.3390/ijms20174319
Lim, C. W., Lee, Y. W., and Hwang, C. H. (2011). Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol. 52, 1613–1627. doi: 10.1093/pcp/pcr091
Liu, Z., Yang, N., Lv, Y., Pan, L., Lv, S., Han, H., et al. (2016). The CLE gene family in Populus trichocarpa. Plant Signal Behav. 11:e1191734. doi: 10.1080/15592324.2016.1191734
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., and Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet 92, 155–161. doi: 10.1007/s12041-013-0212-8
Mayrose, I., Doron-Faigenboim, A., Bacharach, E., and Pupko, T. (2007). Towards realistic codon models: among site variability and dependency of synonymous and non-synonymous rates. Bioinformatics 23, i319–327. doi: 10.1093/bioinformatics/btm176
Meng, L., Buchanan, B. B., Feldman, L. J., and Luan, S. (2012). A putative nuclear CLE-like (CLEL) peptide precursor regulates root growth in Arabidopsis. Mol. Plant 5, 955–957. doi: 10.1093/mp/sss060
Miyawaki, K., Tabata, R., and Sawa, S. (2013). Evolutionarily conserved CLE peptide signaling in plant development, symbiosis, and parasitism. Curr. Opin. Plant Biol. 16, 598–606. doi: 10.1016/j.pbi.2013.08.008
Miyazawa, H., Oka-Kira, E., Sato, N., Takahashi, H., Wu, G. J., Sato, S., et al. (2010). The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137, 4317–4325. doi: 10.1242/dev.058891
Mortier, V., Den Herder, G., Whitford, R., Van de Velde, W., Rombauts, S., D'Haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153, 222–237. doi: 10.1104/pp.110.153718
Murphy, E., Smith, S., and De Smet, I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24, 3198–3217. doi: 10.1105/tpc.112.099010
Ni, J., and Clark, S. E. (2006). Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140, 726–733. doi: 10.1104/pp.105.072678
Nishimura, R., Hayashi, M., Wu, G. J., Kouchi, H., Imaizumi-Anraku, H., Murakami, Y., et al. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429. doi: 10.1038/nature01231
Oelkers, K., Goffard, N., Weiller, G. F., Gresshoff, P. M., Mathesius, U., and Frickey, T. (2008). Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8:1. doi: 10.1186/1471-2229-8-1
Okamoto, S., Ohnishi, E., Sato, S., Takahashi, H., Nakazono, M., Tabata, S., et al. (2009). Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50, 67–77. doi: 10.1093/pcp/pcn194
Opsahl-Ferstad, H. G., Le Deunff, E., Dumas, C., and Rogowsky, P. M. (1997). ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J. 12, 235–246. doi: 10.1046/j.1365-313x.1997.12010235.x
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–w204. doi: 10.1093/nar/gky448
Qiang, Y., Wu, J., Han, H., and Wang, G. (2013). CLE peptides in vascular development. J. Integr. Plant Biol. 55, 389–394. doi: 10.1111/jipb.12044
Reiser, L., Subramaniam, S., Li, D., and Huala, E. (2017). Using the Arabidopsis Information Resource (TAIR) to Find Information About Arabidopsis Genes. Curr. Protoc. Bioinform. 60, 1.11.11–11.11.45. doi: 10.1002/cpbi.36
Stahl, Y., Wink, R. H., Ingram, G. C., and Simon, R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19, 909–914. doi: 10.1016/j.cub.2009.03.060
Strabala, T. J., O'Donnell, P. J., Smit, A. M., Ampomah-Dwamena, C., Martin, E. J., et al. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140, 1331–1344. doi: 10.1104/pp.105.075515
Strabala, T. J., Phillips, L., West, M., and Stanbra, L. (2014). Bioinformatic and phylogenetic analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE signal peptide genes in the Pinophyta. BMC Plant Biol. 14:47. doi: 10.1186/1471-2229-14-47
Suzaki, T., Toriba, T., Fujimoto, M., Tsutsumi, N., Kitano, H., and Hirano, H. Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47, 1591–1602. doi: 10.1093/pcp/pcl025
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–d613. doi: 10.1093/nar/gky1131
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238. doi: 10.1038/s41586-018-0009-2
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinform. 8, 77–80. doi: 10.1016/s1672-0229(10)60008-3
Wang, G., and Fiers, M. (2010). CLE peptide signaling during plant development. Protoplasma 240, 33–43. doi: 10.1007/s00709-009-0095-y
Wang, G., Zhang, G., and Wu, M. (2015). CLE peptide signaling and crosstalk with phytohormones and environmental stimuli. Front. Plant Sci. 6:1211. doi: 10.3389/fpls.2015.01211
Wang, P., Wang, Y., and Ren, F. (2019). Genome-wide identification of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) family in grape (Vitis vinifera L.). BMC Genomics 20:553. doi: 10.1186/s12864-019-5944-2
Whitford, R., Fernandez, A., De Groodt, R., Ortega, E., and Hilson, P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. U.S.A. 105, 18625–18630. doi: 10.1073/pnas.0809395105
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Yamaguchi, Y. L., Ishida, T., and Sawa, S. (2016). CLE peptides and their signaling pathways in plant development. J. Exp. Bot. 67, 4813–4826. doi: 10.1093/jxb/erw208
Yin, Z., Zhu, W., Zhang, X., Chen, X., Wang, W., Lin, H., et al. (2021). Molecular characterization, expression and interaction of MAPK, MAPKK and MAPKKK genes in upland cotton. Genomics 113(1 Pt 2), 1071–1086. doi: 10.1016/j.ygeno.2020.11.004
Yu, C. S., Lin, C. J., and Hwang, J. K. (2004). Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Yu, J., Jung, S., Cheng, C. H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–1236. doi: 10.1093/nar/gkt1064
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33, 531–537. doi: 10.1038/nbt.3207
Keywords: CLE peptide, molecular traits, functional analysis, Gossypium hirsutum, Gossypium raimondii, Gossypium arboreum
Citation: Lin H, Wang W, Chen X, Sun Z, Han X, Wang S, Li Y, Ye W and Yin Z (2021) Molecular Traits and Functional Analysis of the CLAVATA3/Endosperm Surrounding Region-Related Small Signaling Peptides in Three Species of Gossypium Genus. Front. Plant Sci. 12:671626. doi: 10.3389/fpls.2021.671626
Received: 24 February 2021; Accepted: 06 May 2021;
Published: 04 June 2021.
Edited by:
Guodong Wang, Shaanxi Normal University, ChinaReviewed by:
Haidong Yan, University of Georgia, United StatesDaniel Blaine Marchant, Stanford University, United States
Copyright © 2021 Lin, Wang, Chen, Sun, Han, Wang, Li, Ye and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zujun Yin, enVqdW55QDE2My5jb20=; Wuwei Ye, eWV3dXdlaUBjYWFzLmNu
†These authors have contributed equally to this work
 Huan Lin1†
Huan Lin1† Wei Wang
Wei Wang Zujun Yin
Zujun Yin