- LIPME, INRAE, CNRS, Université de Toulouse, Castanet-Tolosan, France
In a local environment, plant networks include interactions among individuals of different species and among genotypes of the same species. While interspecific interactions are recognized as main drivers of plant community patterns, intraspecific interactions have recently gained attention in explaining plant community dynamics. However, an overview of intraspecific genotype-by-genotype interaction patterns within wild plant species is still missing. From the literature, we identified 91 experiments that were mainly designed to investigate the presence of positive interactions based on two contrasting hypotheses. Kin selection theory predicts partisan help given to a genealogical relative. The rationale behind this hypothesis relies on kin/non-kin recognition, with the positive outcome of kin cooperation substantiating it. On the other hand, the elbow-room hypothesis supports intraspecific niche partitioning leading to positive outcome when genetically distant genotypes interact. Positive diversity-productivity relationship rationalizes this hypothesis, notably with the outcome of overyielding. We found that both these hypotheses have been highly supported in experimental studies despite their opposite predictions between the extent of genetic relatedness among neighbors and the level of positive interactions. Interestingly, we identified a highly significant effect of breeding system, with a high proportion of selfing species associated with the presence of kin cooperation. Nonetheless, we identified several shortcomings regardless of the species considered, such as the lack of a reliable estimate of genetic relatedness among genotypes and ecological characterization of the natural habitats from which genotypes were collected, thereby impeding the identification of selective drivers of positive interactions. We therefore propose a framework combining evolutionary ecology and genomics to establish the eco-genomic landscape of positive GxG interactions in wild plant species.
Introduction
During the course of its life cycle, a plant can interact directly or indirectly – consecutively and/or concurrently – with multiple neighboring plants. Plant social networks include interactions among individuals of different species (i.e., interspecific interactions) and among genotypes of the same species (i.e., intraspecific interactions) in a local environment. Plant–plant interactions play an important role in regulating the diversity and structure of plant communities and ultimately ecosystem functioning, through their effects on resource availability and habitat structure (Brooker, 2006; Martorell and Freckleton, 2014). Studying the mechanisms underlying plant–plant interactions is therefore essential to understand the dynamics of plant communities, which may in turn help to predict the resilience of plant species to anthropogenic-related global changes (Subrahmaniam et al., 2018). For instance, ongoing climate warming has resulted in modifications of plant assemblages due to increase of plant biomass, reduced diversity (Baldwin et al., 2014) and shifts in the distribution areas of plant species (Gilman et al., 2010; Singer et al., 2013).
Plant–plant interactions can be divided into four main categories depending on the net benefit and cost associated with the interaction (Subrahmaniam et al., 2018). Firstly, competitive interactions (–/–) are those that come with a significant cost for both partners (benefit < 0, cost > 0 for both partners). Competition is characterized by reciprocal negative effects on plant growth or fitness caused by the presence of neighbors (Keddy, 2015). Since all plants share few basic requirements, limitations of resources such as the availability of nutrients, water or light could drive competition among plants (Turkington and Harper, 1979; Chaney and Baucom, 2014). Second, asymmetric interactions (–/+) yield benefit to one of the partners, at the cost of the other interactor (benefit < 0 and cost > 0 for the helper; benefit > 0 and cost < 0 for the receiver). Parasitic plants are the prime example of this kind of behavior. In addition, plants releasing allelochemicals, which negatively influence the physiology of their neighbors, can be grouped under this category. Third, commensal interactions (0/+) are those that are beneficial for one of the partners, but there is no cost associated with providing such aid (benefit = 0 and cost = 0 for helper; benefit > 0 and cost = 0 for the receiver of the help). Many examples of such interactions exist at the interspecific level, like nurse plant effects in deserts or climbing plants that use the stems of other plants to avoid shade (Padilla and Pugnaire, 2006; Gianoli, 2015). Lastly, individuals can also reciprocally benefit (+/+) from being associated with a partner (such that the net benefit > 0 for each plant partner). One of the first examples demonstrating this mutually beneficial relationship came from a leguminous shrub, Retama sphaerocarpa and its understory species Marrubium vulgare (Pugnaire et al., 1996b). Increasing resource availability leading to facilitative benefits of being underneath the shrub, was reciprocated by improved water relationships due to the understory species in this study.
Estimating the relative importance of these broad categories in explaining patterns of plant communities is still under debate and mainly focused on interactions at the interspecific level. Interspecific competitive interactions have been traditionally recognized as the major factors driving the structure (Goldberg and Barton, 1992), diversity (Chesson, 2000) and dynamics of plant communities (Tilman, 1985). However, the role of positive interactions among species (including both commensal interactions and reciprocal helping interactions) in regulating the composition of communities, has recently gained attention (Bertness and Callaway, 1994; Callaway, 1995; Brooker and Callaghan, 1998; Dormann and Brooker, 2002; Bruno et al., 2003; Kotowska et al., 2010; Wendling et al., 2017). In particular, positive interactions among species have been put forward to explain overyielding, a phenomenon which corresponds to the increase in productivity of species when grown in a mixture, as opposed to monoculture (Harper, 1977; Vandermeer, 1981; Loreau, 2004; Schmid et al., 2008). Also being increasingly recognized is the notion that studying Genotype-by-Genotype (GxG) interactions (also referred as intergenotypic interactions) at the intraspecific level might be a prerequisite for understanding eco-evolutionary patterns of plant communities (Hughes et al., 2008). Indeed, a huge number of genotypes of varying levels of genetic relatedness can co-exist within a local population, even in the case of highly selfing species. For instance, a recent study on Arabidopsis thaliana revealed that the genetic diversity observed within a local population can represent almost one-sixth of the genetic diversity at the worldwide scale (Frachon et al., 2017). Consequently, the patterns of interactions between different genotypes within one population are bound to vary as well.
Several meta-analyses have been carried out to understand patterns of GxG interactions at the interspecific level in herbaceous wild plant species (Maestre et al., 2005) as well as in trees (Piotto, 2008; Zhang et al., 2012). However, to our knowledge, an overview of GxG interaction patterns at the intraspecific level within wild plant species is still missing from literature. We therefore aimed to make a synthesis on such interactions. By surveying the literature, we quickly observed that most studies have been dedicated to testing the presence of positive interactions within different plant species. Moreover, studies on these positive GxG interactions have been based upon testing two main non-exclusive hypotheses with opposite relationships between the extent of genetic relatedness among neighbors and the level of positive interactions (File et al., 2012): kin/non-kin recognition or elbow-room. Rooted in evolutionary biology concepts, the first hypothesis is based on the kin selection theory advocating that individuals increase their inclusive fitness by modifying their behavior to help a relative, thereby leading to kin cooperation (Hamilton, 1964). On the other hand, rooted in ecological concepts, the elbow-room hypothesis assumes that intraspecific resource partitioning occurs and increases as the genetic distance between neighbors increases (Argyres and Schmitt, 1992). The resulting expected positive relationship between genotypic diversity and productivity corresponds to overyielding at the intraspecific level.
After highlighting the nature of the data of 78 identified articles and identifying the main caveats before extracting trends from these data, we tested whether kin cooperation and overyielding observed within our sample dataset was dependent upon specific features of a species (e.g., breeding system, mode of seed dispersal, life cycle etc.). Finally, we introduce several avenues that deserve to be explored to obtain a thorough picture of GxG interaction patterns within wild species. We particularly stress the need to integrate genomics and evolutionary ecology to fully understand the complexity of intraspecific positive genetic interactions in wild plant populations.
Literature Survey and Hypotheses Tested
For this review, we only focused on studies looking at intraspecific interactions within wild populations of herbaceous species. We made this choice because the number of generations of wild herbaceous species is clearly lesser than the one of trees. Therefore, fitness proxies can be better estimated during their life cycle. Several keywords were used to gather these studies, i.e., ‘GxG interactions,’ ‘intraspecific interactions,’ ‘intraspecific variation,’ ‘intra/inter-population variation,’ and ‘group selection.’ The websites inspected included Google Scholar, Web of Science, Sci-hub, as well as the web application Researcher1. Although we tried to do a comprehensive analysis to include a maximum number of studies reporting intraspecific GxG interactions, the list is certainly not exhaustive, and some studies may have been overlooked. We gathered a list of 78 articles published in the last 36 years and including 91 experiments (Supplementary Table 1). Interestingly, we observed a sharp increase in cumulative number of experimental papers published over the years, notably in the last 15 years (Figure 1A). This illustrates the rising interest in examining intraspecific GxG interactions within wild plant species.
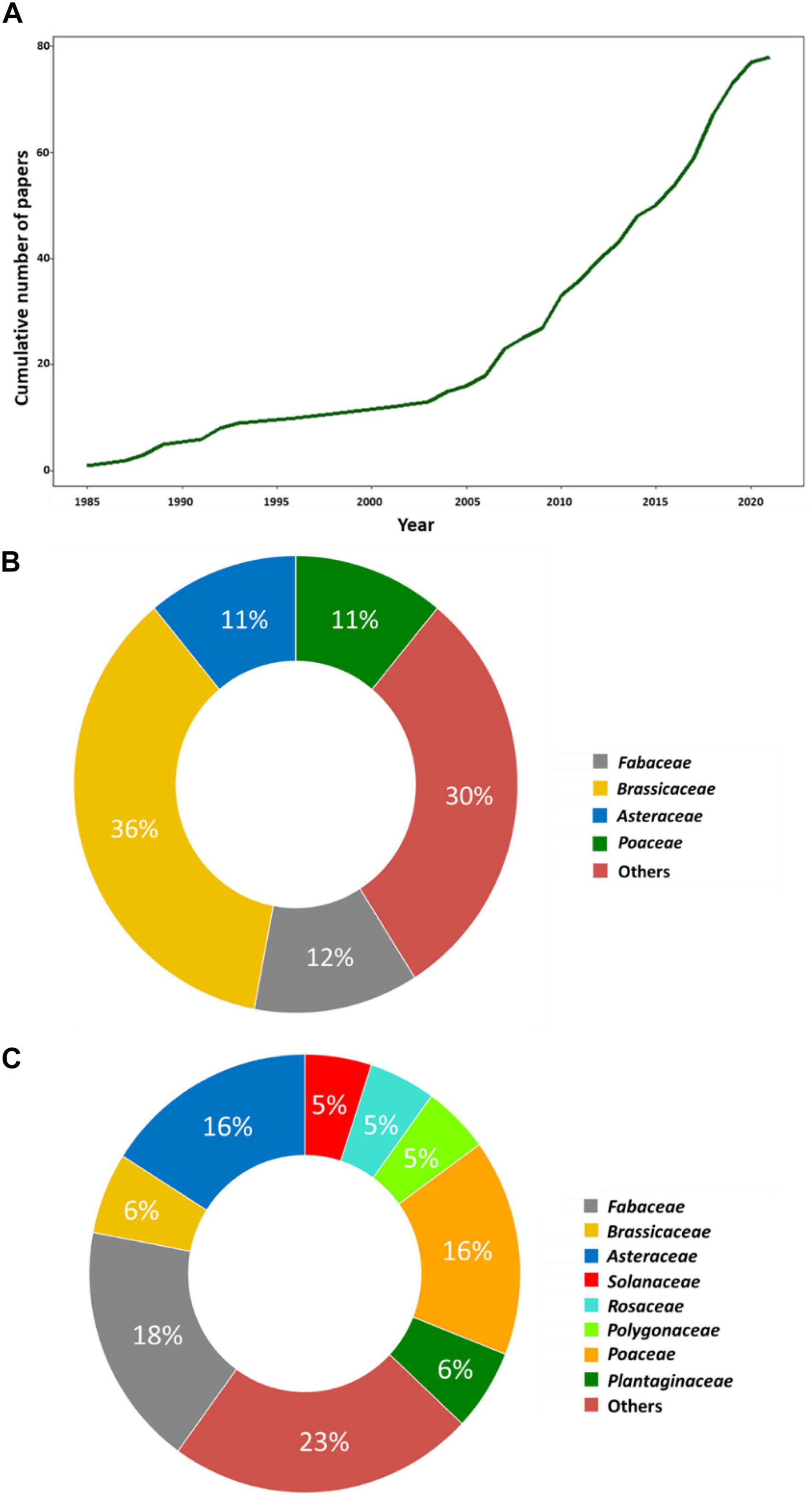
Figure 1. Surveying GxG interactions within herbaceous wild species. (A) Cumulative number of papers on GxG interactions in wild plant species from 1985 to 2021. (B) Doughnut plots describing the distribution of the major botanical families used in the study of intraspecific GxG interactions. (C) Doughnut plots describing the distribution of the major botanical families used in the study of intraspecific GxG interactions upon removal of Arabidopsis thaliana.
The list includes 47 species belonging to 19 botanical families (Figure 1B). The most commonly studied botanical families are the Brassicaceae (36%), Fabaceae (12%), Asteraceae (11%), and Poaceae (11%). However, there is a significant bias in the family Brassicaceae toward A. thaliana as it constitutes about ∼88% of the studies from this family. Upon removing this species, the relative proportion of botanical families studied is more consistent with the number of studied species within each family (Figure 1C and Supplementary Table 1).
The reported experiments can be categorized upon testing either the ‘kin/non-kin recognition’ hypothesis (∼58% of the experiments) where the differential response of a genotype was tested in the presence/absence of a relative genotype (kin) vs. a stranger genotype in pairwise experiments, or the elbow-room (or ‘genotypic diversity-productivity relationship’) hypothesis (31% of the experiments) where fitness proxies were compared between monocultures using multiple kin individuals (compound intra-genotypic interactions) and mixtures of different genotypes (compound inter-genotypic interactions) (Figure 2A and Supplementary Table 1).
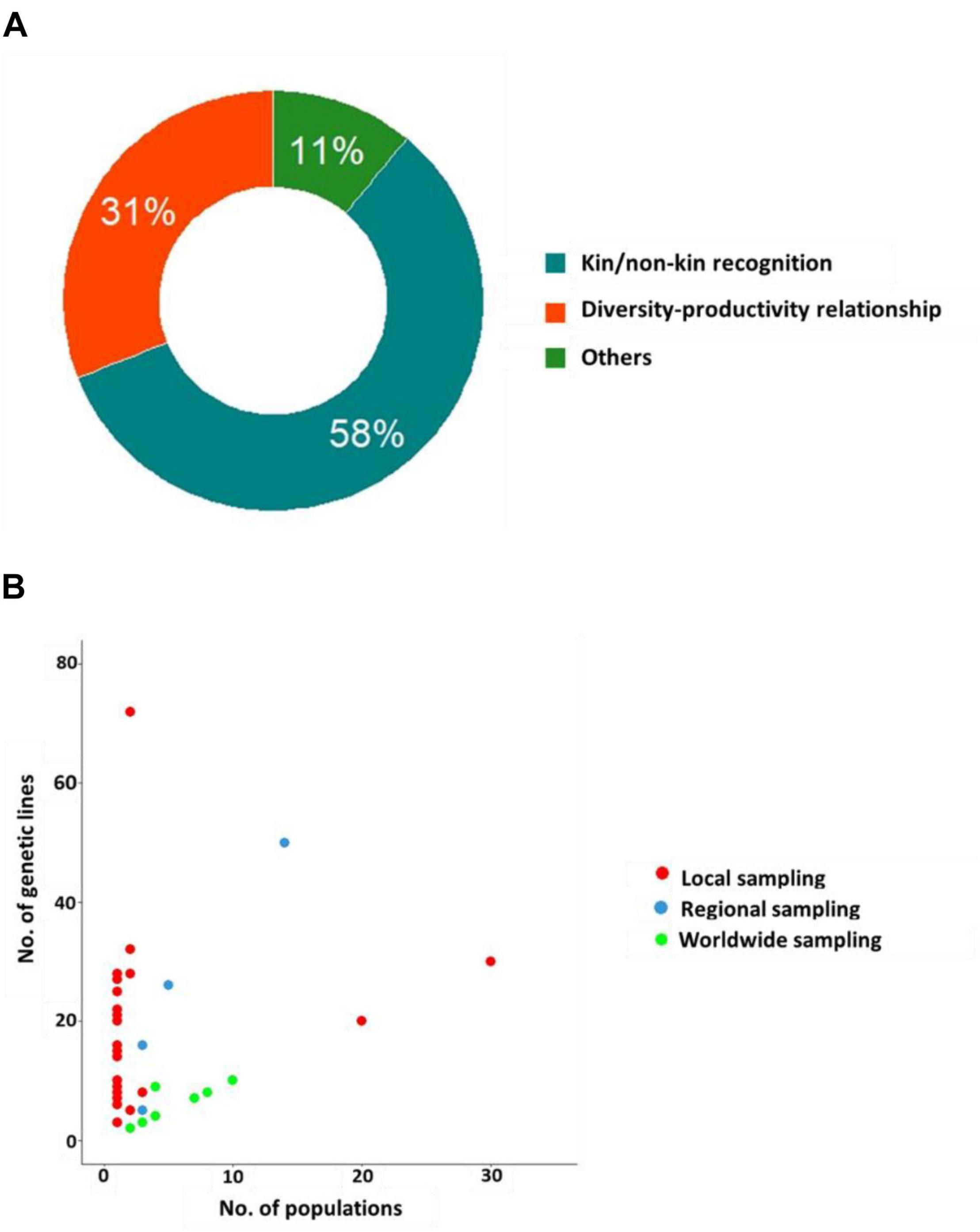
Figure 2. Classification of experimental studies on GxG interactions. (A) Doughnut plot describing the distribution of the papers according to the rationale of studying GxG interactions. (B) Relationship between the number of genetic lines and the number of populations according to three geographic scales. For better illustration, the two outliers with 60 genotypes each from a different population, are not incorporated in the figure (Fitzpatrick et al., 2019; Turner et al., 2020).
The remaining experiments (∼11%) were grouped under the category ‘Others’. These experiments aimed at (i) characterizing the genetic architecture underlying GxG interactions (Botto and Coluccio, 2007; Mutic and Wolf, 2007), (ii) studying the effects of GxG interactions on intra-individual traits such as genome size variation (Šmarda et al., 2010) and transcriptomic profiles (Bowsher et al., 2017), (iii) studying extended phenotypes such as root exudate profiles (Badri et al., 2012) and soil microbial communities (Burghardt et al., 2019; Fitzpatrick et al., 2019), (iv) testing the effect of adding a neighbor plant on genotype-by-environment interactions (i.e., GxExG instead of GxGxE) (Cahill et al., 2010), (v) examining local adaptation of genotypes (Linhart, 1988; Espeland and Rice, 2007), and (vi) investigating individual vs. group selection in wild plant populations (Goodnight, 1985; Donohue, 2003). For the latter, we need to stress that the existence of group selection is still controversial (Nowak, 2006; Nowak et al., 2010; Rousset and Lion, 2011; Kramer and Meunier, 2016) and it will not be addressed in this review.
Highlights of the Nature of the Data and Identification of Caveats
Life-History Traits
The list of 47 species is divided roughly equally between annuals (∼46%) and perennials (∼54%) (Supplementary Table 1) and is dominated by selfing species that make up about 44% of the dataset. The remaining species represent species with mixed breeding systems (∼27%), outcrossing species (∼19%) and clonal species (∼10%). Allochory (assisted seed dispersal) is predominant in this list as about ∼60% of the listed species exhibited this mode of seed dispersal, while only ∼40% of listed species have an autochorous (self) mode of seed dispersal (Supplementary Table 1).
Nature and Extent of Genetic Variation
Around 86% of the experiments listed were based on genotypes collected in natural populations (Supplementary Table 1). With the main goal of dissecting the underlying genetic and molecular mechanisms of GxG interactions, the remaining experiments were based on (i) experimental populations such as F2 populations, families of Recombinant Inbred Lines (RILs) or Introgression lines (IL, ∼9%) (Goodnight, 1985; Griffing, 1989; Botto and Coluccio, 2007; Mutic and Wolf, 2007; Willis et al., 2010; Latzel et al., 2013; Wuest and Niklaus, 2018; Hauri et al., 2021), and (ii) mutant lines (∼5%) (Cahill et al., 2005; Crepy and Casal, 2015; Wagg et al., 2015; Zhang and Tielbörger, 2019; Supplementary Table 1). Unsurprisingly, given the great amount of genetic resources publicly available, all these experiments concern A. thaliana, with the exception of Trifolium pratense (Wagg et al., 2015) and Solanum lycopersicum (Hauri et al., 2021).
For experiments based on genotypes collected in natural populations, we observed a clear tradeoff between the number of genotypes used and the number of populations sampled (Figure 2B). The number of natural genetic lines used to evaluate GxG interactions was highly variable among experiments, ranging from 2 to 72 (mean ∼12) (Figure 2B). On average, these lines have been collected from about four populations (min = 1, max = 60) (Figure 2B). In wild plant populations, intraspecific neighbors share common space over generations, and this increases their probability for repeated interactions. Consequently, positive interactions are more likely to evolve between members of a single population rather than between members of different populations (Nowak, 2006). Accordingly, most experiments that do not employ A. thaliana are based on natural genetic lines from a single population (∼75%) or sampled at a regional scale (between 2 and 14 populations, ∼11%). On the other hand, an opposite trend is observed across the 29 experiments performed on A. thaliana. Almost 90% of the experiments on this species utilized genotypes coming from worldwide collections. The main hypothesis to explain this bias in using worldwide genotypes in A. thaliana might be related to its predominantly selfing breeding system, which initially suggested that most populations were monomorphic (Platt et al., 2010). Therefore, the large public collections of genotypes that are available for A. thaliana mostly correspond to one representative genotype per population. However, progressively, a substantial number of studies challenged this view by revealing extensive genetic diversity within populations (Le Corre, 2005; Jorgensen and Emerson, 2008; Bomblies et al., 2010; Platt et al., 2010; Kronholm et al., 2012; Brachi et al., 2013; Roux and Bergelson, 2016; Frachon et al., 2017, 2018, 2019; Fulgione et al., 2018), thereby giving an opportunity of studying more relevant GxG interactions in A. thaliana at the local scale.
Based on all the experiments listed in this survey, we nonetheless identified a major shortcoming of the plant material used to study GxG interactions, regardless of the identity of the species considered. As previously mentioned, testing both the kin/non-kin recognition and elbow-room hypotheses requires an estimation of the degree of genetic relatedness among interacting genotypes. Kin selection theory predicts partisan help given to close relatives. By contrast, according to the elbow-room hypothesis, genetically close relatives will compete for the same resources and increasing genetic distance between genotypes can translate into increasing niche partitioning. Testing for these contrasting predictions would require integrating information about the extent of genetic relatedness among interacting genotypes. However, this crucial information has been poorly considered in these experiments (but see Crutsinger et al., 2006, 2008), which may question the realism of having tested interactions between different genotypes in some studies, in particular when genotypes were originating from a single population (Supplementary Table 1).
Relevant Growth Conditions?
Performing experiments in controlled and field conditions are complementary (Bergelson and Roux, 2010; Brachi et al., 2010). Experiments conducted under controlled conditions drastically reduce environmental noise, thereby allowing the establishment of a direct link between phenotypic observations and genotype performance under a given set of stable environmental conditions. On the other hand, in the field, plants are exposed to a greater but more ecologically realistic range of abiotic and biotic fluctuations than typically encountered in controlled conditions. Nonetheless, encompassing all these environmental fluctuations requires the field experiments to be repeated over several years.
In our survey, ∼82% of the experiments were conducted under controlled conditions (Supplementary Table 1). Out of these, ∼55 and ∼20% of the experiments were conducted in greenhouse conditions and growth chambers (including root chambers and growth tunnels), respectively. The remaining experiments (∼7%) were performed under in vitro conditions. On the other hand, few experiments (∼14%) had been conducted under field conditions, fewer in the native habitats (only two reported experiments, Supplementary Table 1). Finally, four experiments (∼4%) were conducted in both greenhouse and field conditions (Espeland and Rice, 2007; Andersson, 2014; Ehlers et al., 2016). The type of growth conditions used to study intraspecific GxG interactions is therefore strongly biased in favor of controlled conditions, notably when compared to other types of biotic interactions. For instance, in a recent review on Genome-Wide Association studies (GWAS) performed on plant – pathogen interactions, 60% of the studies were conducted in controlled conditions (greenhouse/growth chambers) and 40% under field conditions (Bartoli and Roux, 2017).
Of particular note, around 68% of the experiments in our survey tested the effect of a particular environmental factor on GxG interactions, either in controlled or field conditions (Supplementary Table 1). Around 55% of these experiments tested the effect of abiotic factors, including light quality, nutrient status, CO2 concentration and drought. On the other hand, ∼50% of these experiments tested the effect of biotic factors, which mainly encompassed density and soil conditioning by one or a mixture of genotypes, in particular to investigate the diversity-productivity relationship (Bukowski and Petermann, 2014; Semchenko et al., 2017; Bukowski et al., 2018). Unfortunately, the treatments applied may not have been ecologically relevant. Indeed, in our survey, only ∼19% of the experiments loosely described the ecology of the populations used in the experiments (Supplementary Table 1). At most, only a rough description of habitats from which the genotypes were collected, was given. Since interspecific positive plant–plant interactions have been postulated to evolve in natural settings including a certain level of abiotic and/or biotic stress (i.e., ‘stress gradient hypothesis’; Bertness and Callaway, 1994; Brooker and Callaghan, 1998; Bruno et al., 2003), this is of particular importance to finely describe the ecology of native habitats from which genotypes are collected. This may in turn allow testing the stress gradient hypothesis at the intraspecific level.
A Large Diversity of Phenotyped Traits
All the traits measured across the 91 experiments are listed in Supplementary Table 2. To assess GxG interactions, an average of 3.5 traits per study have been directly measured on plants (min = 1, max = 10). We divided the list of traits into four broad categories, each relating to a distinct eco-function of the plant (i.e., root related traits, shoot related traits, life history traits and seed production related traits). While ∼34% of the experiments scored life history related traits (e.g., germination and flowering timing), ∼53 and ∼76% of the experiments measured root (e.g., root length and biomass) and shoot (e.g., plant height and dry biomass) related traits, respectively. Seed production related traits (e.g., number of fruits and number of seeds per fruit) were measured in ∼40% of the experiments. Interestingly, most experiments focused on collecting phenotypic information using either two (∼46%) or three (∼20%) categories. About 40% of the experiments looked at both root and shoot related phenotypes, while ∼29% of experiments focused on both shoot and life history related traits. Only four experiments (∼4%) focused on all four categories (Willson et al., 1987; Linhart, 1988; Masclaux et al., 2010).
Another interesting category of traits corresponds to extended phenotypes, which contains traits that were not directly phenotyped on the interacting individuals but rather looked at alteration of the immediate environment in GxG interactions. In our survey, ∼14% of studies examined such extended phenotypes, which mainly focused on either studying soil microbial communities (File et al., 2012; Wagg et al., 2015; Abdala-Roberts et al., 2017; Burghardt et al., 2019; Tomiolo et al., 2020), tritrophic interactions (Crutsinger et al., 2006, 2008; Kotowska et al., 2010; Karban et al., 2013; Abdala-Roberts et al., 2017; Hauri et al., 2021) or biochemical properties of plant secreted compounds (Badri et al., 2012; Kalske et al., 2019).
When considering experiments testing the kin/non-kin recognition hypothesis, root related traits were phenotyped significantly more often than in experiments testing the diversity-productivity relationship (contingency chi-squared test, χ2 = 154.59, df = 3, P = 2.68 E-33, Figure 3). A potential explanation for this stems from the extensive literature on kin recognition studies, where many studies have demonstrated kin recognition by roots and subsequent nepotistic response across the plant kingdom (reviewed in Supplementary Tables 1, 2). On the other hand, experiments testing the diversity-productivity relationship examined more shoot and seed production related traits than experiments pertaining to kin/non-kin recognition (Figure 3). A potential explanation for this observation could be linked to the difficulties associated with measuring individual specific root response in experiments testing for the diversity-productivity relationship. Often, to measure specific root traits, the total root biomass of several genotypes present in a pot has been considered as a dependent measure, which obviously impedes the estimation of the relative contribution of each individual genotype in the pot. Therefore, aboveground traits could be an easier measure for quantifying productivity.
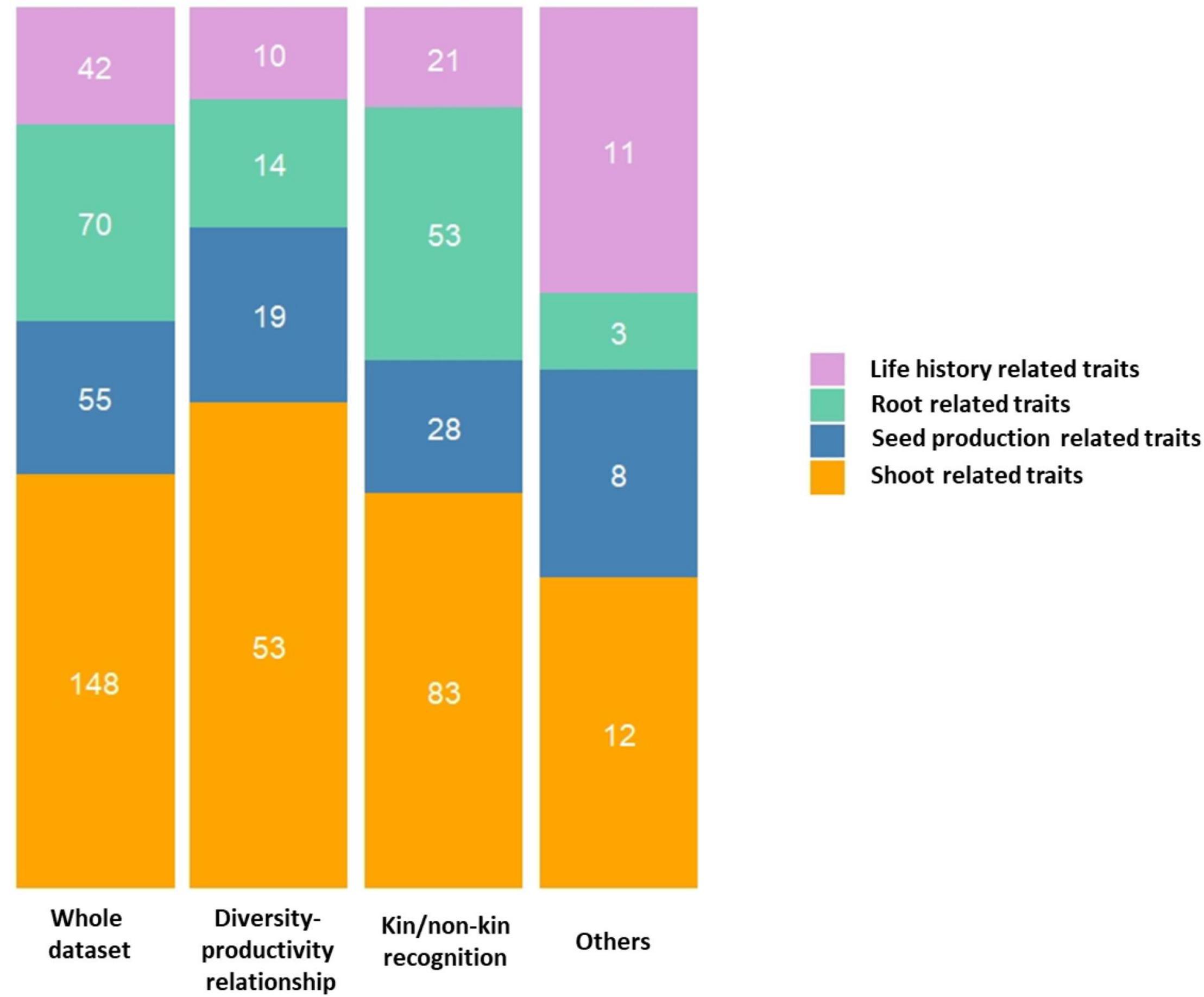
Figure 3. Stacked barplots illustrating the distribution of studies based on trait categories according to different rationales. The number of traits studied within each category are indicated within each stack.
Plant neighbor interactions are often dynamic in nature, and can switch from competitive to positive interactions depending on (i) the environmental stress perceived (Bertness and Callaway, 1994), (ii) life stages of interacting species (Walker and Vitousek, 1991; Kellman and Kading, 1992; Chapin et al., 1994; Pugnaire et al., 1996a), or (iii) the physiological capacity of the interacting species (e.g., facilitation by improving plant water uptake under canopy shade or soil oxygenation; Callaway and King, 1996; Holmgren et al., 1997). In all the experiments listed in our survey, this potential switch of interactions at the intraspecific level has been overlooked. Testing for such complex dynamic GxG interactions would require phenotypic measurements of various plant compartments at multiple time points.
Direct vs. Indirect GxG Interactions
We further categorized the experiments depending on whether the interactions tested were direct or indirect. We classified the interactions to be indirect when the experiment did not include placing at least two genotypes in a pot together or to allow any direct contact between them. In our survey, such indirect interactions have been addressed in ∼10% of the 91 experiments (Supplementary Table 1). Indirect interactions between genotypes may occur by two main processes. Firstly, soil conditioning can mediate indirect interactions between genotypes. There is mounting evidence suggesting the role of root exudates in identity recognition of neighbors. In our survey, two studies confirmed kin recognition by showing alteration of root response upon external root exudate application in indirect GxG interactions (Biedrzycki et al., 2010; Semchenko et al., 2014). Furthermore, the composition of root exudates, which is genotype specific, may define the assembly of distinct soil microbial communities in the rhizosphere. Consequently, different genotypes in one population can be associated with different microbiota. Therefore, similar to soil microbiota mediated plant soil feedback (PSF) that has been abundantly documented at interspecific level and recognized in driving ecosystem processes (Klironomos, 2002; Kardol et al., 2006, 2007; Reinhart and Callaway, 2006; Petermann et al., 2008), soil microbiota may have a central role in mediating GxG interactions in natural plant communities. In our survey, four experiments reported indirect interactions for which the effect of soil microbiota conditioning by one genotype is tested on the growth response of another genotype (Bukowski and Petermann, 2014; Wagg et al., 2015; Bukowski et al., 2018; Burghardt et al., 2019). One of these studies suggested a role in maintaining genotypes coexistence (Bukowski et al., 2018), which is in line with recent theoretical demonstration of the putative role of microbes in altruism between organisms (Lewin-Epstein et al., 2017). Yet, the genuine role of PSF in shaping evolution of plant populations is largely unexplored (Wagg et al., 2015), and investigation of the role of microbiota on GxG interactions represents a future challenge.
Secondly, indirect interactions between genotypes can also be mediated by volatile organic compounds (VOCs) produced by plants both aboveground and underground. VOCs are known to act in neighbor detection (Ninkovic et al., 2019) and to affect intraspecific competition in crops (Ninkovic, 2003) and trees (Ormeño et al., 2007). In our survey, only three experiments focused on interactions through aerial VOCs (Karban et al., 2013 in Artemisia tridentata; Kalske et al., 2019 in Solidago altissima; and Shimola and Bidart, 2019 in A. thaliana). The cocktail of VOCs has been found to be highly variable among individuals and has been proposed to help distinguishing kin from stranger genotypes (Karban et al., 2014). Still, there is a caveat in understanding the exact role of VOCs in indirect GxG interactions in wild species, and how they may affect intraspecific communication between genotypes under natural conditions.
Both Kin Cooperation and Overyielding Are Supported by Experiments
We divided the major conclusions from the 81 experiments underlying the two major rationales (kin/non-kin recognition vs. diversity-productivity relationship) into (i) kin cooperation (KC) for studies demonstrating positive interactions between kin genotypes, (ii) overyielding (OY) for studies where the mixture of genotypes was confirmed to be more productive than monocultures, and (iii) neutral for studies where no significant differences were observed between the treatments specified in the experiments. Among this set of 81 experiments, only ∼17% found no significant differences between treatments, while ∼50% reported KC and ∼33% studies reported OY (Supplementary Table 1). The relative proportion of KC, OY and neutral interactions in the survey can be matched to the two main rationales tested in the experiments. Experiments testing for kin/non-kin recognition confirmed more KC, while experiments aiming at testing diversity-productivity relationships, corroborated more cases of OY (contingency chi-squared test, χ2 = 31.68, df = 2, P = 1.32 E-07; Figure 4). However, we need to stress that the number of studies reporting no differences could be more than what is listed in this survey, owing to the high tendency of not publishing non-significant results.
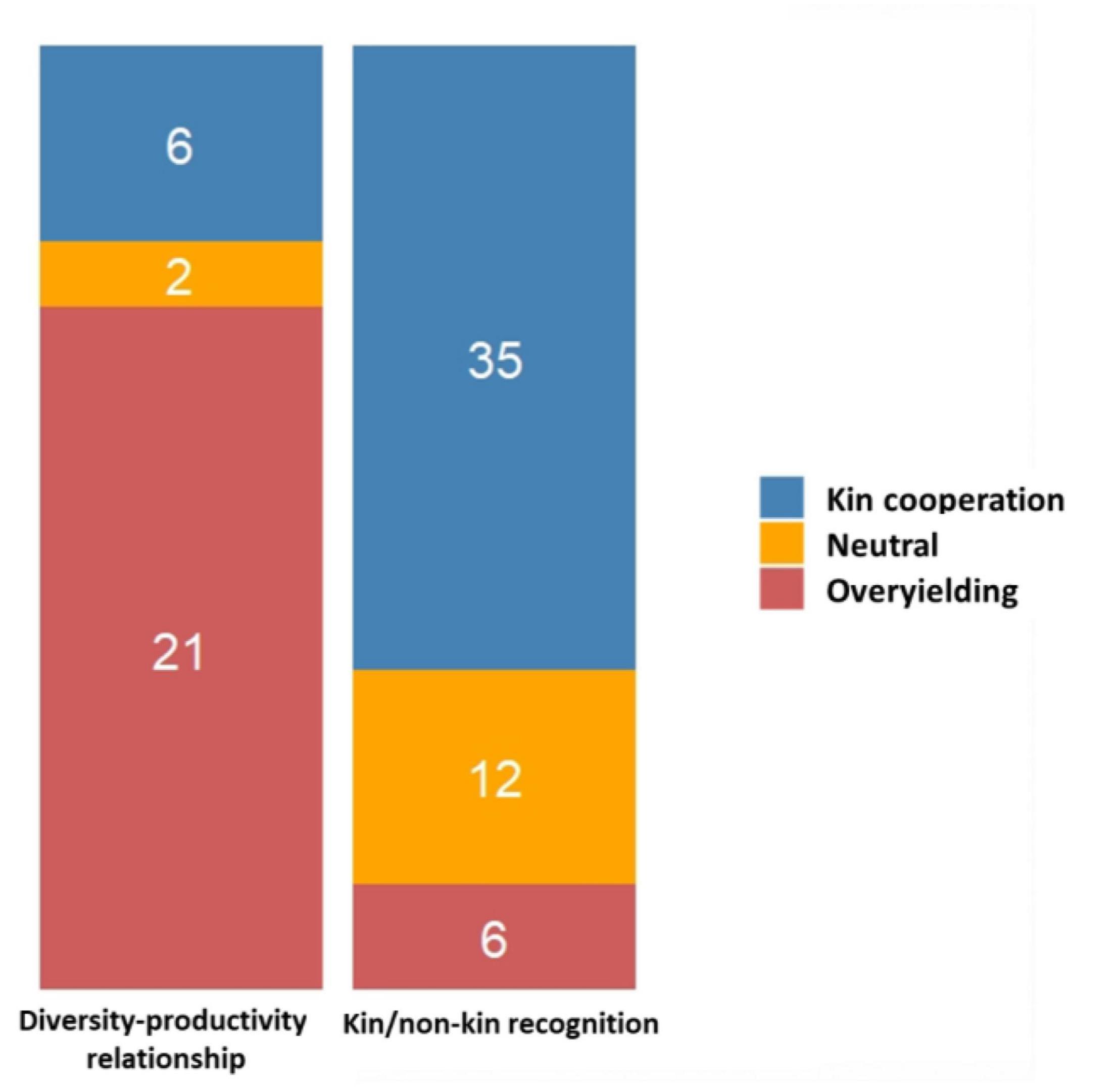
Figure 4. Stacked barplots illustrating the relative proportion of kin cooperation (KC), overyielding (OY) and neutral interactions between the ‘diversity-productivity relationship’ and ‘kin/non-kin recognition’ rationales. The number of studies supporting each type of interaction are indicated within each stack.
Species having low dispersal rate have been theoretically proposed to allow for evolution of kin selection mechanisms as interacting individuals are likely to be relatives (Hamilton, 1964). Moreover, species having strong spatial population structure have been poised to have positive frequency dependent interactions among genetically related individuals (Ehlers and Bilde, 2019). However, in our survey, when considering all the experiments, the relative proportion of KC, OY or neutral interactions was not found dependent on the mode of seed dispersal (autochory vs. allochory, contingency chi-squared test, χ2 = 2.03, df = 2, P = 0.362; Supplementary Figure 1) or on the geographical scale of sampling (one population vs. multiple populations, contingency chi-squared test, χ2 = 3.34, df = 2, P = 0.188; Supplementary Figure 2). Removing A. thaliana from the list yielded similar results for these proportions (autochory vs. allochory, contingency chi-squared test, χ2 = 0.67, df = 2, P = 0.714; one population vs. multiple populations, contingency chi-squared test, χ2 = 1.08, df = 2, P = 0.582; Supplementary Figures 1, 2).
In species with a high selfing rate, the presence of neighbors of high genetic relatedness in the vicinity of seed-dispersing mother plants is expected in natural populations, which may allow for the prevalence of kin selection mechanisms for increasing one’s inclusive fitness (Dudley et al., 2013; Ehlers and Bilde, 2019). The relative proportions between the three types of interactions (KC, OY or neutral) were significantly dependent on the breeding system of the plant species studied (selfing and clonal vs. outcrossing and mixed, χ2 = 22.23, P = 1.48 E-05). KC was detected in 56.9 and 31.8% of the experiments conducted on species with a selfing/clonal and outcrossing/mixed breeding system, respectively (Figure 5). This effect remained significant even upon the removal of A. thaliana from the list (χ2 = 15.04, P = 5.39 E-04).
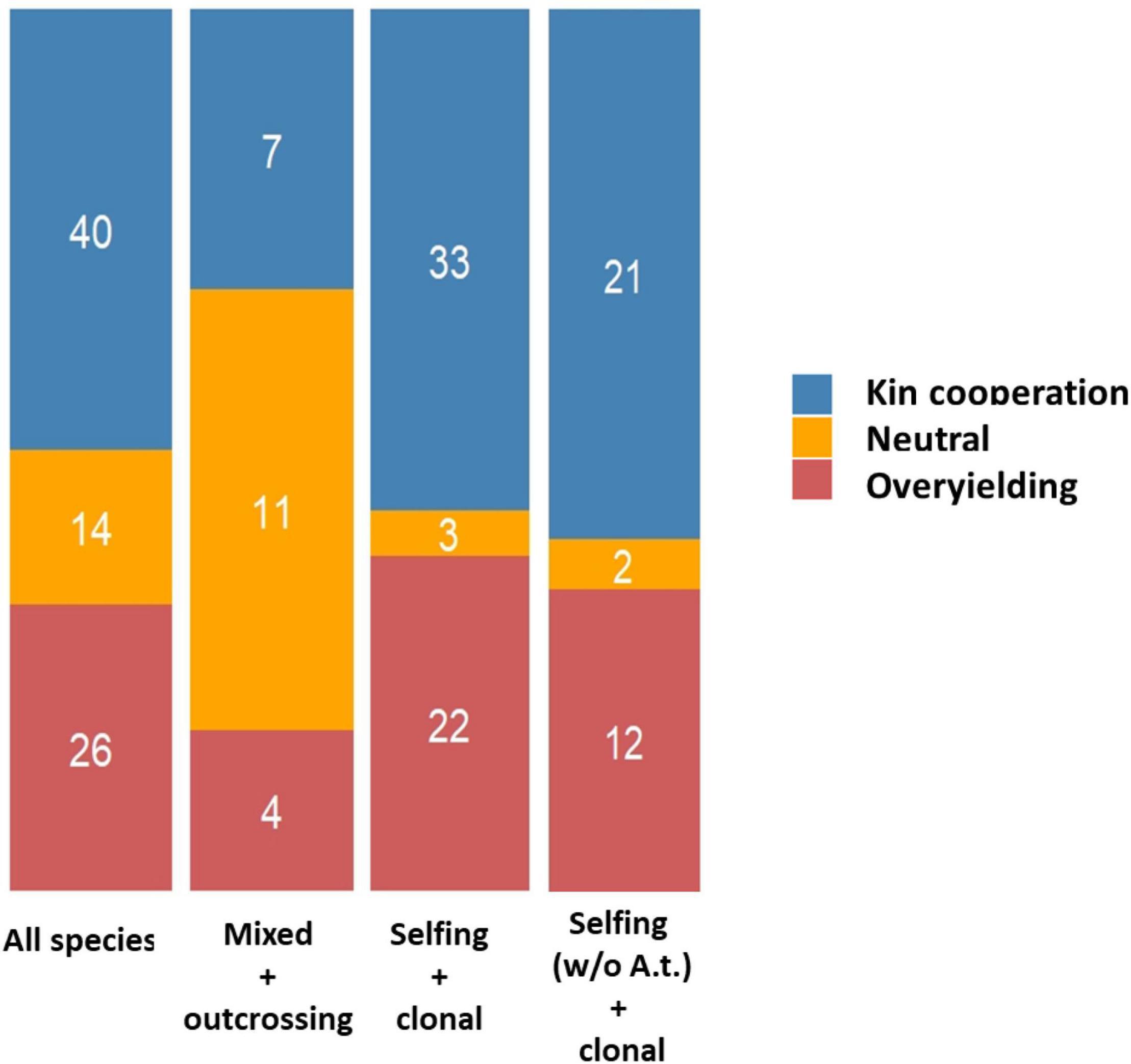
Figure 5. Stacked barplots illustrating the different outcomes of GxG according to the breeding system of species studied. MIXED + OUTCROSSING: species with an outcrossing breeding system or with a mixed breeding system. SELFING + CLONAL: species with a selfing breeding system or a vegetative reproduction. SELFING W/O A.T. + CLONAL: species with a selfing breeding system (without considering Arabidopsis thaliana) or a vegetative reproduction.
Species having an annual life cycle are known to be commonly composed of genetically related individuals, especially in cases of limited seed dispersal and self- fertilization (Cheplick and Kane, 2004). Therefore, kin selection mechanisms may prevail for annual plants. In our survey, the relative proportions between the three types of interactions (KC, OY or neutral) were found to be dependent on the annual vs. perennial life history strategy of the plant species considered (χ2 = 7.36, P = 0.025), with KC detected in 59.7 and 36.4% of the experiments conducted on annual and perennial species, respectively (Figure 6). This difference between these two life history strategies was however not significant upon removal of A. thaliana from the list of species considered (χ2 = 4.80, P = 0.091; Figure 6).
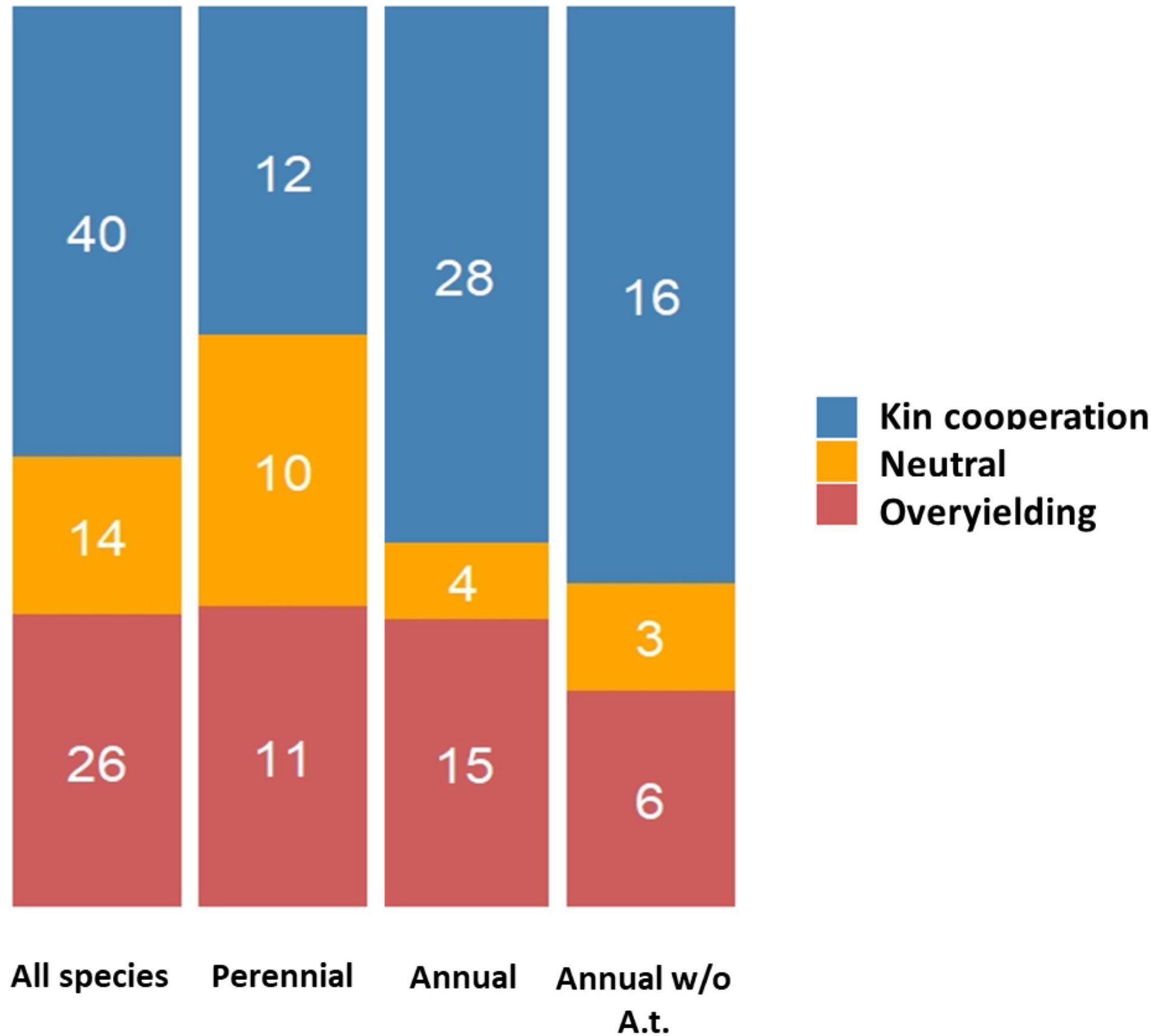
Figure 6. Stacked barplots illustrating the different outcomes of GxG interactions according to the annual vs. perennial strategy of species studied (with and without including Arabidopsis thaliana). The number of studies supporting each type of interaction are indicated within each stack.
At the interspecific level, increase in magnitude of positive interactions has been linked to varying levels of abiotic (Maestre et al., 2009) and biotic stresses (Smit et al., 2009) in various studies. In our survey, we identified two studies reporting a shift from KC to OY occurring between the same genotypes in response to a particular environmental factor. Firstly, in A. thaliana, genotypes performed better with kin under ambient CO2 concentrations, while genotypic mixtures were favored under elevated CO2 concentrations (Andalo et al., 2001). Secondly, in Phalaris arundinacea, genotypes performed better with kin in disturbed plots (with all native vegetation removed), while genotypic mixtures were confirmed to perform better in undisturbed plots (Collins et al., 2018). Both experiments provide evidence for the existence of similar dynamics of intraspecific positive interactions among different species in varying environments.
Future Avenues
Despite the demonstration of positive interactions in 36 species (Supplementary Table 1), the underlying adaptive genetic architecture is still an open question. In our survey, this challenge was addressed in only one study, with the identification of a single major QTL underlying kin cooperation in A. thaliana (Wuest and Niklaus, 2018). While informative, the low number of RILs used (N = 37) precluded a proper characterization of the genetic architecture (Keurentjes et al., 2007). In addition, remaining questions include (i) testing whether polymorphic genes involved in positive interactions have been shaped by natural selection and (ii) identifying the ecological factors driving adaptive KC or OY (Subrahmaniam et al., 2018; Frachon et al., 2019). Here, we therefore introduce a framework in order to establish a genomic map of local adaptive positive plant–plant interactions (Figure 7).
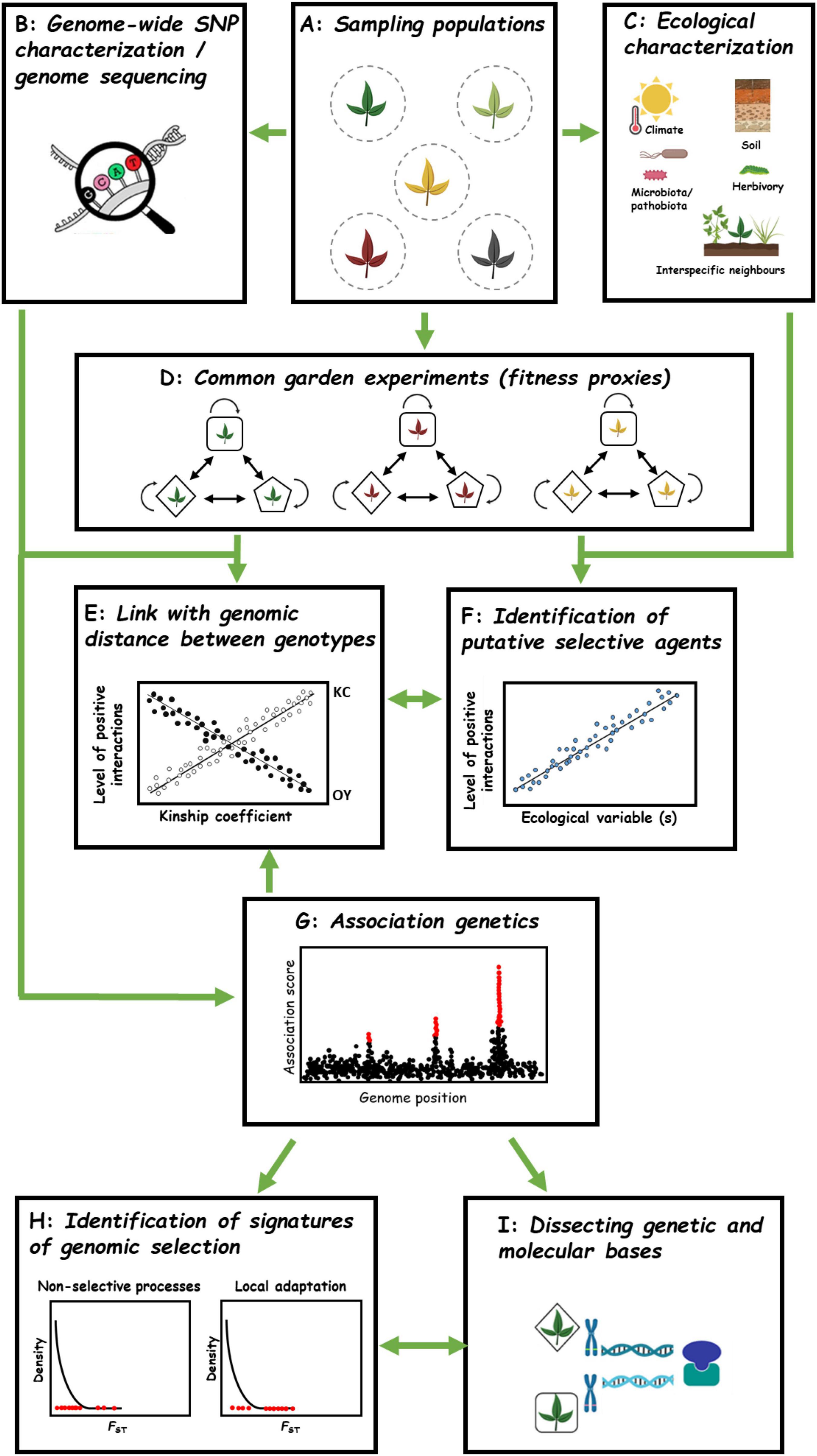
Figure 7. Future avenues for studying GxG interactions among wild plant populations. (A) Collecting multiple genotypes from several natural populations. (B) Genome sequencing/Genome-wide SNP characterization of all genotypes coming from different populations. (C) Characterizing abiotic and biotic ecological factors. (D) Conducting common garden experiments to dissect fitness outcomes in pairwise interactions among genotypes from one population (intragenotype and intra-population intergenotype combinations). Different genotypes are represented by different shapes. (E) Linking genomic distance with levels of positive interactions exhibited in pairwise GxG interactions. (F) Identifying putative ecological driver(s) associated with among-population variation of positive interactions. (G) Genome Wide Association mapping to identify genomic regions underlying positive interactions. (H) Enrichment analysis in signatures of selection, e.g., spatial genetic differentiation estimated by FST. (I) Cloning and functional characterization of genes underlying QTLs associated with positive interactions.
The first step would be collecting genetic material from multiple local populations (Figure 7A). Then, the genomic characterization of the plant material could be conducted either by genome-wide SNP characterization or whole genome sequencing methodologies. This will not only allow estimating the genetic relatedness among genotypes within and between local populations but will also be essential to run GWA mapping (see below, Figure 7B). Simultaneously, an extensive in situ characterization of both abiotic (e.g., climate, soil physico-chemical properties) and biotic (e.g., plant communities, herbivore communities, as well as microbiota including commensal, pathogenic and symbiotic microbes) factors should be conducted for each population (Figure 7C). While abiotic stress characterization is facilitated by publicly available databases for climate and dedicated platforms for soil physico-chemical properties, characterizing biotic factors can still be strenuous and time consuming. However, with the ever-decreasing cost of next-generation sequencing (NGS) technologies, describing the taxonomic and functional networks (microbes, plants, insects…) of hundreds of wild plant populations is not out of reach (Bartoli et al., 2018; Frachon et al., 2019). While collecting plant material and accumulating genomic, phenotypic and ecological data for tens to hundreds of populations represents a monumental task, this has already been achieved for a few wild plants including A. thaliana (Fournier-Level et al., 2011; Hancock et al., 2011; Brachi et al., 2013; Bartoli et al., 2018; Frachon et al., 2018, 2019) and sunflower (Todesco et al., 2020).
In order to estimate the natural variation of GxG interaction patterns among populations, the next step would be to conduct common garden experiments by estimating differences at the intra-population level between intra-genotypic interactions (monoculture) and inter-genotype pairwise interactions (Figure 7D). Although the best strategy would be to perform experiments in the native habitats from which the populations have been collected, this approach may not be manageable for a larger number of populations.
Kin recognition and elbow-room hypotheses make opposite predictions about the relationship between genetic distance and the extent of positive interactions between two genotypes (Figure 7E). Having genome sequence information of genotypes, linking level of positive interactions to the genomic distance of interacting genotypes could help validating either hypothesis for explaining positive interactions between populations (Figure 7E). Additionally, identification of ecological factors associated with positive interactions between populations could be of interest in understanding how positive interactions (KC or OY) may evolve under different environmental selection pressures in natural populations (Figure 7F) and designing experiments to test the relevance of the stress gradient hypothesis at the intraspecific level.
As a next step, a GWA mapping approach could be adopted to fine map genomic regions associated with natural variation in the level of positive interactions among populations (Figure 7G). GWA mapping has previously proven to be a powerful approach to identify QTLs associated with plant–plant interactions at the intraspecific and interspecific levels in many crop species (Subrahmaniam et al., 2018) as well as in A. thaliana (Baron et al., 2015; Frachon et al., 2017, 2019). The detected QTLs can in turn help to reliably discriminate between the three main hypotheses about the genetic architecture underlying positive interactions within species, that is (i) kin selection wherein a large chunk of the genome is shared among cooperating partners (polygenic architecture; Hamilton, 1964), (ii) Green beard effect with cooperative genotypes sharing the same allele at one or few QTLs identified (Dawkins, 1976), and (iii) complementarity where cooperative genotypes have different alleles at single or few QTLs identified (D’Souza et al., 2018).
Such QTLs can then allow identifying signatures of selection at the genomic level (Figure 7H). For instance, the type and strength of selection can be addressed by testing for any overlaps and enrichments between QTLs underlying positive interactions and genomic regions showing signatures of long-term (e.g., hard sweeps) or short-term selection (e.g., local adaptation with FST) (Fournier-Level et al., 2011; Hancock et al., 2011; Horton et al., 2012; Brachi et al., 2015; Frachon et al., 2017; Figure 7H).
After confirming the adaptive status of the genetic basis associated with natural variation of positive interactions, one could take the final steps toward understanding the genetic and molecular mechanisms (Figure 7I). For this, QTL cloning can be achieved by characterization of mutants (T-DNA, EMS…) or generation of transgenic lines with gain or loss of functions of the gene(s) of interest.
Altogether, combining ecology and evolutionary biology, along with quantitative genetics, genomics and molecular biology represents an unprecedented opportunity to establish the genetic and molecular picture of positive GxG interactions in wild plant species in their natural ecological landscape.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by a Ph.D. fellowship from the LABEX TULIP to HJS (ANR-10-LABX-41).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.683373/full#supplementary-material
Supplementary Table 1 | Survey of GxG interaction studies in herbaceous wild plant species. The first six columns (columns A–F) indicate the serial number, the list of different plant species studied, their botanical family, main breeding system, life cycle and natural seed dispersal mode. Next six columns (columns G–L) describe the genetic material used including the main type of genetic variation studied (here experimental variation indicates use of F2 population, Recombinant Inbred Lines or Introgression Lines, while artificial variation includes use of mutant lines), number of genetic lines sampled (here NA indicates not available), number of populations sampled (here NA indicates not available and NR indicates not relevant), the scale of sampling (here WW indicates worldwide sampling), whether genetic differences between the lines were tested or assumed, and the habitats from which these lines were sampled from. Following 15 columns (columns M–AA) are divided to describe the experimental design of the studies including the main rationale, whether the GxG interactions were tested directly or indirectly, the experimental design (here solo refers to treatments where genotypes were grown alone, intra refers to ‘intragenotype’ treatment where two individuals of the same genotype were placed together and inter refers to ‘intergenotype’ treatment where two different genotypes were placed together; in the ‘inter’ column, ‘x’ in red indicates a mixed polyculture treatment) and conditions under which the experiments were performed (here GH indicates greenhouse and GC indicates growth chamber). Additionally, details about the specific treatments applied and the traits measured are listed. Next three columns (columns AB–AD) detail the main ecological questions addressed in each study, major findings with respect to intraspecific plant–plant interactions and the main type of positive interaction observed in each experimental study. Here KC indicates kin cooperation, OY indicates overyielding, coop indicates studies where positive interactions were identified. However, due to lack of genetic information, these latter studies could not be categorized in either KC or OY. The last three columns (columns AE–AG) list the reference name, year of publication, and the journal of publication (Tonsor, 1989; Mahall and Callaway, 1991; Burt and Bell, 1992; Thomas and Bazzaz, 1993; Mahall and Callaway, 1996; Ronsheim and Anderson, 2001; Rautiainen et al., 2004; Falik et al., 2006; Dudley and File, 2007; Semchenko et al., 2007; Lankinen, 2008; Murphy and Dudley, 2009; Aguilera et al., 2011; Bhatt et al., 2011; Biernaskie, 2011; Markham and Halwas, 2011; Drummond and Vellend, 2012; Milla et al., 2012; Creissen et al., 2013; Tomimatsu et al., 2014; Yang et al., 2015; Corliss and Sultan, 2016; Huber et al., 2016; Palmer et al., 2016; Abd El-Gawad et al., 2017; Jie et al., 2017; Li and Xu, 2018; Torices et al., 2018; Baker et al., 2019; Smith et al., 2019; Tomiolo et al., 2019; Zhang et al., 2019; Goddard et al., 2020; Yamawo and Mukai, 2020).
Supplementary Table 2 | List of different traits measured in GxG interaction studies listed in the literature survey. The serial number and list of species studied in the 91 experiments are listed in the first two columns (columns A and B). The next four columns (columns C–F) describe the measured trait categories that have been divided into root, shoot, life history and seed production related traits. Following this, the columns (columns G–J) summarize the total number of traits studied in each trait category within the experiments. The next two columns detail the extended phenotypes, if recorded within the experiment (columns K and L). Following this, the total number of traits recorded within the experiment are listed (column M). Lastly, the two columns (columns N and O) list the reference name, and the year of publication.
Footnotes
References
Abd El-Gawad, A. M., Zotti, M., Sarker, T. C., Mazzoleni, S., and Bonanomi, G. (2017). Does a plant detect its neighbor if it is kin or stranger? Evidence from a common garden experiment. Commun. Ecol. 18, 305–310. doi: 10.1556/168.2017.18.3.9
Abdala-Roberts, L., Pratt, R., Pratt, J. D., and Mooney, K. A. (2017). Traits underlying community consequences of plant intra-specific diversity. PLoS One 12:e0183493. doi: 10.1371/journal.pone.0183493
Aguilera, A. G., Colón-Carmona, A., Kesseli, R., and Dukes, J. S. (2011). No accession-specific effect of rhizosphere soil communities on the growth and competition of Arabidopsis thaliana accessions. PLoS One 6:e0027585. doi: 10.1371/journal.pone.0027585
Andalo, C., Goldringer, I., and Godelle, B. (2001). Inter- and intragenotypic competition under elevated carbon dioxide in Arabidopsis thaliana. Ecology 82, 157–164. doi: 10.2307/2680093
Andersson, S. (2014). Indirect genetic effects from competition in the clonal herb Sedum album (Crassulaceae). PLoS One 9:e0106104. doi: 10.1371/journal.pone.0106104
Argyres, A. Z., and Schmitt, J. (1992). Neighbor relatedness and competitive performance in Impatiens capensis (Balsaminaceae): a test of the resource partitioning hypothesis. Am. J. Bot. 79, 181–185. doi: 10.2307/2445106
Badri, D. V., de-la-Peña, C., Lei, Z., Manter, D. K., Chaparro, J. M., Guimarães, R. L., et al. (2012). Root secreted metabolites and proteins are involved in the early events of plant-plant recognition prior to competition. PLoS One 7:e0046640. doi: 10.1371/journal.pone.0046640
Baker, B. H., Sultan, S. E., Lopez-Ichikawa, M., and Waterman, R. (2019). Transgenerational effects of parental light environment on progeny competitive performance and lifetime fitness. Philos. Trans. R. Soc. B Biol. Sci. 374:20180182. doi: 10.1098/rstb.2018.0182
Baldwin, A. H., Jensen, K., and Schönfeldt, M. (2014). Warming increases plant biomass and reduces diversity across continents, latitudes, and species migration scenarios in experimental wetland communities. Glob. Change Biol. 20, 835–850. doi: 10.1111/gcb.12378
Baron, E., Richirt, J., Villoutreix, R., Amsellem, L., and Roux, F. (2015). The genetics of intra- and interspecific competitive response and effect in a local population of an annual plant species. Funct. Ecol. 29, 1361–1370. doi: 10.1111/1365-2435.12436
Bartoli, C., Frachon, L., Barret, M., Rigal, M., Huard-Chauveau, C., Mayjonade, B., et al. (2018). In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 12, 2024–2038. doi: 10.1038/s41396-018-0152-7
Bartoli, C., and Roux, F. (2017). Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front. Plant Sci. 8:763. doi: 10.3389/fpls.2017.00763
Bergelson, J., and Roux, F. (2010). Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat. Rev. Genet. 11, 867–879. doi: 10.1038/nrg2896
Bertness, M. D., and Callaway, R. (1994). Positive interactions in communities. Trends Ecol. Evol. 9, 187–191. doi: 10.1016/0169-5347(94)90087-6
Bhatt, M. V., Khandelwal, A., and Dudley, S. A. (2011). Kin recognition, not competitive interactions, predicts root allocation in young Cakile edentula seedling pairs. New Phytol. 189, 1135–1142. doi: 10.1111/j.1469-8137.2010.03548.x
Biedrzycki, M. L., Jilany, T. A., Dudley, S. A., and Bais, H. P. (2010). Root exudates mediate kin recognition in plants. Commun. Integr. Biol. 3, 28–35. doi: 10.4161/cib.3.1.10118
Biernaskie, J. M. (2011). Evidence for competition and cooperation among climbing plants. Proc. R. Soc. B Biol. Sci. 278, 1989–1996. doi: 10.1098/rspb.2010.1771
Bomblies, K., Yant, L., Laitinen, R. A., Kim, S. T., Hollister, J. D., Warthmann, N., et al. (2010). Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet. 6:e1000890. doi: 10.1371/journal.pgen.1000890
Botto, J. F., and Coluccio, M. P. (2007). Seasonal and plant-density dependency for quantitative trait loci affecting flowering time in multiple populations of Arabidopsis thaliana. Plant Cell Environ. 30, 1465–1479. doi: 10.1111/j.1365-3040.2007.01722.x
Bowsher, A. W., Shetty, P., Anacker, B. L., Siefert, A., Strauss, S. Y., and Friesen, M. L. (2017). Transcriptomic responses to conspecific and congeneric competition in co-occurring Trifolium. J. Ecol. 105, 602–615. doi: 10.1111/1365-2745.12761
Brachi, B., Faure, N., Horton, M., Flahauw, E., Vazquez, A., Nordborg, Bergelson, J., et al. (2010). Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6:e1000940. doi: 10.1371/journal.pgen.1000940
Brachi, B., Meyer, C. G., Villoutreix, R., Platt, A., Morton, T. C., Roux, F., et al. (2015). Co-selected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc. Natl. Acade. Sci. U.S.A. 112, 4032–4037. doi: 10.1073/pnas.1421416112
Brachi, B., Villoutreix, R., Faure, N., Hautekèete, N., Piquot, Y., Pauwels, M., et al. (2013). Investigation of the geographical scale of adaptive phenological variation and its underlying genetics in Arabidopsis thaliana. Mol. Ecol. 22, 4222–4240. doi: 10.1111/mec.12396
Brooker, R. W. (2006). Plant – plant interactions and environmental change. New Phytol. 171, 271–284. doi: 10.1111/j.1469-8137.2006.01752.x
Brooker, R. W., and Callaghan, T. V. (1998). The balance between positive and negative plant interactions and its relationship to environmental gradients: a model. Oikos 81, 196–207.
Bruno, J. F., Stachowicz, J. J., and Bertness, M. D. (2003). Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 18, 119–125. doi: 10.1016/S0169-5347(02)00045-9
Bukowski, A. R., and Petermann, J. S. (2014). Intraspecific plant-soil feedback and intraspecific overyielding in Arabidopsis thaliana. Ecol. Evolut. 4, 2533–2545. doi: 10.1002/ece3.1077
Bukowski, A. R., Schittko, C., and Petermann, J. S. (2018). The strength of negative plant–soil feedback increases from the intraspecific to the interspecific and the functional group level. Ecol. Evol. 8, 2280–2289. doi: 10.1002/ece3.3755
Burghardt, L. T., Epstein, B., and Tiffin, P. (2019). Legacy of prior host and soil selection on rhizobial fitness in planta. Evolution 73, 2013–2023. doi: 10.1111/evo.13807
Burt, A., and Bell, G. (1992). Tests of sib diversification theories of outcrossing in Impatiens capensis: effects of inbreeding and neighbour relatedness on production and infestation. J. Evol. Biol. 5, 575–588. doi: 10.1046/j.1420-9101.1992.5040575.x
Cahill, J. F., Kembel, S. W., and Gustafson, D. J. (2005). Differential genetic influences on competitive effect and response in Arabidopsis thaliana. J. Ecol. 93, 958–967. doi: 10.1111/j.1365-2745.2005.01013.x
Cahill, J. F., McNickle, G. G., Haag, J. J., Lamb, E. G., Nyanumba, S. M., and Clair, C. C. S. (2010). Plants integrate information about nutrients and neighbors. Science 328:1657. doi: 10.1126/science.1189736
Callaway, R. M., and King, L. (1996). Oxygenation of the soil rhizosphere by Typha latifolia and its facilitative effects on other species. Ecology 77, 1189–1195.
Chaney, L., and Baucom, R. S. (2014). The costs and benefits of tolerance to competition in Ipomoea purpurea, the common morning glory. Evolution 68, 1698–1709. doi: 10.1111/evo.12383
Chapin, E. S. III, Walker, L. R., Fastie, C. L., and Sharman, L. C. (1994). Mechanisms of primary succession following deglaciation at Glacier Bay. Alaska. Ecol. Monogr 64, 149–175.
Cheplick, G. P., and Kane, K. H. (2004). Genetic relatedness and competition in Triplasis purpurea (Poaceae): resource partitioning or kin selection? Int. J. Plant Sci. 165, 623–630.
Chesson, P. (2000). Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. doi: 10.1146/annurev.ecolsys.31.1.343
Collins, A. R., Beckage, B., and Molofsky, J. (2018). Small-scale genotypic richness stabilizes plot biomass and increases phenotypic variance in the invasive grass Phalaris arundinacea. J. Plant Ecol. 11, 47–55. doi: 10.1093/jpe/rtx056
Corliss, C. T., and Sultan, S. E. (2016). Evolutionary potential for increased invasiveness: high-performance Polygonum cespitosum genotypes are competitively superior in full sun. Am. J. Bot. 103, 348–354. doi: 10.3732/ajb.1500306
Creissen, H. E., Jorgensen, T. H., and Brown, J. K. M. (2013). Stabilization of yield in plant genotype mixtures through compensation rather than complementation. Ann. Bot. 112, 1439–1447. doi: 10.1093/aob/mct209
Crepy, M. A., and Casal, J. J. (2015). Photoreceptor-mediated kin recognition in plants. New Phytol. 205, 329–338. doi: 10.1111/nph.13040
Crutsinger, G. M., Collins, M. D., Fordyce, J. A., Gompert, Z., Nice, C. C., and Sanders, N. J. (2006). Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 313, 966–968.
Crutsinger, G. M., Souza, L., and Sanders, N. J. (2008). Intraspecific diversity and dominant genotypes resist plant invasions. Ecol. Lett. 11, 16–23. doi: 10.1111/j.1461-0248.2007.01118.x
Donohue, K. (2003). The influence of neighbor relatedness on multilevel selection in the Great Lakes sea rocket. Am. Natural. 162, 77–92. doi: 10.1086/375299
Dormann, C. F., and Brooker, R. W. (2002). Facilitation and competition in the high Arctic: the importance of the experimental approach. Acta Oecol. 23, 297–301. doi: 10.1016/S1146-609X(02)01158-X
Drummond, E. B. M., and Vellend, M. (2012). Genotypic diversity effects on the performance of Taraxacum officinale populations increase with time and environmental favorability. PLoS One 7:e0030314. doi: 10.1371/journal.pone.0030314
D’Souza, G., Shitut, S., Preussger, D., Yousif, G., Waschina, S., and Kost, C. (2018). Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488. doi: 10.1039/c8np00009c
Dudley, S. A., and File, A. L. (2007). Kin recognition in an annual plant. Biol. Lett. 3, 435–438. doi: 10.1098/rsbl.2007.0232
Dudley, S. A., Murphy, G. P., and File, A. L. (2013). Kin recognition and competition in plants. Funct. Ecol. 27, 898–906. doi: 10.1111/1365-2435.12121
Ehlers, B. K., and Bilde, T. (2019). Inclusive fitness, asymmetric competition and kin selection in plants. Oikos 128, 765–774. doi: 10.1111/oik.06390
Ehlers, B. K., David, P., Damgaard, C. F., and Lenormand, T. (2016). Competitor relatedness, indirect soil effects and plant coexistence. J. Ecol. 104, 1126–1135. doi: 10.1111/1365-2745.12568
Espeland, E. K., and Rice, K. J. (2007). Facilitation across stress gradients: the importance of local adaptation. Ecology 88, 2404–2409. doi: 10.1890/06-1217.1
Falik, O., de Kroon, H., and Novoplansky, A. (2006). Physiologically-mediated self/nonself root discrimination in Trifolium repens has mixed effects on plant performance. Plant Signal. Behav. 1, 116–121. doi: 10.4161/psb.1.3.2639
File, A. L., Klironomos, J., Maherali, H., and Dudley, S. A. (2012). Plant kin recognition enhances abundance of symbiotic microbial partner. PLoS One 7:e0045648. doi: 10.1371/journal.pone.0045648
Fitzpatrick, C. R., Mustafa, Z., and Viliunas, J. (2019). Soil microbes alter plant fitness under competition and drought. J. Evol. Biol. 32, 438–450. doi: 10.1111/jeb.13426
Fournier-Level, A., Korte, A., Cooper, M. D., Nordborg, M., Schmitt, J., and Wilczek, A. M. (2011). A map of local adaptation in Arabidopsis thaliana. Science 334, 86–89. doi: 10.1126/science.1209271
Frachon, L., Bartoli, C., Carrère, S., Bouchez, O., Chaubet, A., Gautier, M., et al. (2018). A genomic map of climate adaptation in Arabidopsis thaliana at a micro-geographic scale. Front. Plant Sci. 9:967. doi: 10.3389/fpls.2018.00967
Frachon, L., Libourel, C., Villoutreix, R., Carrère, S., Glorieux, C., Huard-Chauveau, C., et al. (2017). Intermediate degrees of synergistic pleiotropy drive adaptive evolution in ecological time. Nat. Ecol. Evol. 1, 1551–1561. doi: 10.1038/s41559-017-0297-1
Frachon, L., Mayjonade, B., Bartoli, C., Hautekèete, N. C., and Roux, F. (2019). Adaptation to plant communities across the genome of Arabidopsis thaliana. Mol. Biol. Evol. 36, 1442–1456. doi: 10.1093/molbev/msz078
Fulgione, A., Koornneef, M., Roux, F., Hermisson, J., and Hancock, A. M. (2018). Madeiran Arabidopsis thaliana reveals ancient long-range colonization and clarifies demography in Eurasia. Mol. Biol. Evol. 35, 564–574. doi: 10.1093/molbev/msx300
Gianoli, E. (2015). The behavioural ecology of climbing plants. AoB PLANTS 7, 1–11. doi: 10.1093/aobpla/plv013
Gilman, S. E., Urban, M. C., Tewksbury, J., Gilchrist, G. W., and Holt, R. D. (2010). A framework for community interactions under climate change. Trends Ecol. Evol. 25, 325–331. doi: 10.1016/j.tree.2010.03.002
Goddard, E. L., Varga, S., John, E. A., and Soulsbury, C. D. (2020). Evidence for root kin recognition in the clonal plant species Glechoma hederacea. Front. Ecol. Evol. 8:578141. doi: 10.3389/fevo.2020.578141
Goldberg, D. E., and Barton, A. M. (1992). Patterns and consequences of interspecific competition in natural communities - a review of field experiments with plants. Am. Nat. 139, 771–801. doi: 10.1086/285357
Goodnight, C. J. (1985). The influence of environmental variation on group and individual selection in a cress. Evolution 39, 545–558. doi: 10.2307/2408652
Hamilton, W. D. (1964). The genetical evolution of social behavior. I. J. Theor. Bio. 7, 1–16. doi: 10.1016/0022-5193(64)90038-4
Hancock, A. M., Brachi, B., Faure, N., Horton, M. W., Jarymowycz, L. B., Sperone, F. G., et al. (2011). Adaptation to climate across the Arabidopsis thaliana genome. Science 334, 83–86. doi: 10.1126/science.1209244
Hauri, K. C., Glassmire, A. E., and Wetzel, W. C. (2021). Chemical diversity rather than cultivar diversity predicts natural enemy control of herbivore pests. Ecol. Appl. 31:e2289. doi: 10.1002/eap.2289
Holmgren, M., Scheffer, M., and Huston, M. A. (1997). The interplay of facilitation and competition in plant communities. Ecology 78, 1966–1975.
Horton, M. W., Hancock, A. M., Huang, Y. S., Toomajian, C., Atwell, S., Auton, A., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44, 212–216. doi: 10.1038/ng.1042.Genome-wide
Huber, H., During, H. J., de Bruin, F. B., Vermeuien, P. J., and Anten, N. P. R. (2016). Genotypic and phenotypic diversity does not affect productivity and drought response in competitive stands of Trifolium repens. Front. Plant Sci. 7:364. doi: 10.3389/fpls.2016.00364
Hughes, A. R., Inouye, B. D., Johnson, M. T. J., Underwood, N., and Vellend, M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. doi: 10.1111/j.1461-0248.2008.01179.x
Jie, L., Xingliang, X., and Tao, L. (2017). Kin interactions of Arabidopsis based on the integrated performance of plants. J. Resour. Ecol. 8, 185–190. doi: 10.5814/j.issn.1674-764x.2017.02.009
Jorgensen, T. H., and Emerson, B. C. (2008). Functional variation in a disease resistance gene in populations of Arabidopsis thaliana. Mol. Ecol. 17, 4912–4923. doi: 10.1111/j.1365-294X.2008.03960.x
Kalske, A., Shiojiri, K., Uesugi, A., Sakata, Y., Morrell, K., and Kessler, A. (2019). Insect herbivory selects for volatile-mediated plant-plant communication. Curr. Biol. 29, 3128.e3–3133.e3. doi: 10.1016/j.cub.2019.08.011
Karban, R., Shiojiri, K., Ishizaki, S., Wetzel, W. C., and Evans, R. Y. (2013). Kin recognition affects plant communication and defence. Proc. R. Soc. B Biol. Sci. 280, 1–5. doi: 10.1098/rspb.2012.3062
Karban, R., Wetzel, W. C., Shiojiri, K., Ishizaki, S., Ramirez, S. R., and Blande, J. D. (2014). Deciphering the language of plant communication: volatile chemotypes of sagebrush. J. Physiol. 204, 380–385. doi: 10.1111/nph.12887
Kardol, P., Bezemer, T. M., and Van der Putten, W. H. (2006). Temporal variation in plant–soil feedbacks controls succession. Ecol. Lett. 9, 1080–1088.
Kardol, P., Cornips, N. J., Van Kempen, M. M. L., Bakx-Schotman, J. M. T., and Van der Putten, W. H. (2007). Microbe-mediated plant–soil feedbacks cause historical contingency effects in plant community assembly. Ecol. Monogr. 77, 147–162.
Kellman, M., and Kading, M. (1992). Facilitation of tree seed-ling establishment in a sand dune succession. J. Veg. Sci. 3, 679–688.
Keurentjes, J. J. B., Bentsink, L., Alonso-Blanco, C., Hanhart, C. J., de Vries, H. B., Effgen, S., et al. (2007). Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175, 891–905. doi: 10.1534/genetics.106.066423
Klironomos, J. N. (2002). Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70.
Kotowska, A. M., Cahill, J. F., and Keddie, B. A. (2010). Plant genetic diversity yields increased plant productivity and herbivore performance. J. Ecol. 98, 237–245. doi: 10.1111/j.1365-2745.2009.01606.x
Kramer, J., and Meunier, J. (2016). Kin and multilevel selection in social evolution: a never-ending controversy? F1000Res. 5:F1000FacultyRev-776.
Kronholm, I., Picó, F. X., Alonso-Blanco, C., Goudet, J., and de Meaux, J. (2012). Genetic basis of adaptation in Arabidopsis thaliana: local adaptation at the seed dormancy QTL DOG1. Evolution 66, 2287–2302. doi: 10.1111/j.1558-5646.2012.01590.x
Lankinen, Å. (2008). Root competition influences pollen competitive ability in Viola tricolor: effects of presence of a competitor beyond resource availability? J. Ecol. 96, 756–765. doi: 10.1111/j.1365-2745.2008.01387.x
Latzel, V., Allan, E., Bortolini Silveira, A., Colot, V., Fischer, M., and Bossdorf, O. (2013). Epigenetic diversity increases the productivity and stability of plant populations. Nat. Commun. 4, 1–7. doi: 10.1038/ncomms3875
Le Corre, V. (2005). Variation at two flowering time genes within and among populations of Arabidopsis thaliana: comparison with markers and traits. Mol. Ecol. 14, 4181–4192. doi: 10.1111/j.1365-294X.2005.02722.x
Lewin-Epstein, O., Aharonov, R., and Hadany, L. (2017). Microbes can help explain the evolution of host altruism. Nat. Commun. 8, 1–7. doi: 10.1038/ncomms14040
Li, J., and Xu, X. (2018). Relatives growing together: pair density and kinship. Plant Ecol. 219, 159–168. doi: 10.1007/s11258-017-0785-z
Linhart, Y. B. (1988). Intrapopulation differentiation in annual plants. III. The contrasting effects of intra- and interspecific competition. Evolution 42, 1047–1064. doi: 10.2307/2408920
Maestre, F. T., Callaway, R. M., Valladares, F., and Lortie, C. J. (2009). Refining the stress-gradient hypothesis for competition and facilitation in plant communities. J. Ecol. 97, 199–205. doi: 10.1111/j.1365-2745.2008.01476.x
Maestre, F. T., Valladares, F., and Reynolds, J. F. (2005). Is the change of plant-plant interactions with abiotic stress predictable? A meta-analysis of field results in arid environments. J. Ecol. 93, 748–757. doi: 10.1111/j.1365-2745.2005.01017.x
Mahall, B. E., and Callaway, R. M. (1991). Root communication among desert shrubs. Proc. Natl. Acad. Sci. U.S.A. 88, 874–876. doi: 10.1073/pnas.88.3.874
Mahall, B. E., and Callaway, R. M. (1996). Effects of regional origin and genotype on intraspecific root communication in the desert shrub Ambrosia dumosa (Asteraceae). Am. J. Bot. 83, 93–98. doi: 10.2307/2445959
Markham, J., and Halwas, S. (2011). Effect of neighbour presence and soil volume on the growth of Andropogon gerardii Vitman. Plant Ecol. Divers. 4, 265–268. doi: 10.1080/17550874.2011.618515
Martorell, C., and Freckleton, R. P. (2014). Testing the roles of competition, facilitation and stochasticity on community structure in a species-rich assemblage. J. Ecol. 102, 74–85. doi: 10.1111/1365-2745.12173
Masclaux, F., Hammond, R. L., Meunier, J., Gouhier-Darimont, C., Keller, L., and Reymond, P. (2010). Competitive ability not kinship affects growth of Arabidopsis thaliana accessions. New Phytol. 185, 322–331. doi: 10.1111/j.1469-8137.2009.03057.x
Milla, R., Del Burgo, A. V., Escudero, A., and Iriondo, J. M. (2012). Kinship rivalry does not trigger specific allocation strategies in Lupinus angustifolius. Ann. Bot. 110, 165–175. doi: 10.1093/aob/mcs093
Murphy, G. P., and Dudley, S. A. (2009). Kin recognition: competition and cooperation in Impatiens (Balsaminaceae). Am. J. Bot. 96, 1990–1996. doi: 10.3732/ajb.0900006
Mutic, J. J., and Wolf, J. B. (2007). Indirect genetic effects from ecological interactions in Arabidopsis thaliana. Mol. Ecol. 16, 2371–2381. doi: 10.1111/j.1365-294X.2007.03259.x
Ninkovic, V. (2003). Volatile communication between barley plants affects biomass allocation. J. Exp. Bot. 54, 1931–1939. doi: 10.1093/jxb/erg192
Ninkovic, V., Rensing, M., Dahlin, I., and Markovic, D. (2019). Who is my neighbor? Volatile cues in plant interactions. Plant Signal. Behav. 14:1634993. doi: 10.1080/15592324.2019.1634993
Nowak, M. A. (2006). Five rules for the evolution of cooperation. Science 314, 1560–1563. doi: 10.1126/science.1133755.Five
Nowak, M. A., Tarnita, C. E., and Wilson, E. O. (2010). The evolution of eusociality. Nature 466, 1057–1062. doi: 10.1038/nature09205
Ormeño, E., Bousquet-Mélou, A., Mévy, J. P., Greff, S., Robles, C., Bonin, G., et al. (2007). Effect of intraspecific competition and substrate type on terpene emissions from some Mediterranean plant species. J. Chem. Ecol. 33, 277–286. doi: 10.1007/s10886-006-9219-4
Padilla, F. M., and Pugnaire, F. I. (2006). The role of nurse plants in restoration. Front. Ecol. Environ. 4:196–202. doi: 10.1890/1540-92952006004[0196:TRONPI]2.0.CO;2
Palmer, A. G., Ali, M., Yang, S., Parchami, N., Bento, T., Mazzella, A., et al. (2016). Kin recognition is a nutrient-dependent inducible phenomenon. Plant Signal. Behav. 11:e1224045. doi: 10.1080/15592324.2016.1224045
Petermann, J. S., Fergus, A. J. F., Turnbull, L. A., and Schmid, B. (2008). Janzen Connell effects are widespread and strong enough to maintain diversity. Ecology 89, 2399–2406. doi: 10.1890/07-2056.1
Piotto, D. (2008). A meta-analysis comparing tree growth in monocultures and mixed plantations. For. Ecol. Manag. 255, 781–786. doi: 10.1016/j.foreco.2007.09.065
Platt, A., Horton, M., Huang, Y. S., Li, Y., Anastasio, A. E., Mulyati, N. W., et al. (2010). The scale of population structure in Arabidopsis thaliana. PLoS Genet. 6:e1000843. doi: 10.1371/journal.pgen.1000843
Pugnaire, F. I., Haase, P., Puigdefabregas, J., Cueto, M., Clark, S. C., and Incoll, L. D. (1996a). Facilitation and succession under the canopy of a leguminous shrub in a semiarid environment in southeast Spain. Oikos 76, 455–464.
Pugnaire, F. I., Haase, P., and Puigdefabregas, J. (1996b). Facilitation between higher plant species in a semiarid environment. Ecology 77, 1420– 1426.
Rautiainen, P., Koivula, K., and Hyvärinen, M. (2004). The effect of within-genet and between-genet competition on sexual reproduction and vegetative spread in Potentilla anserina ssp. egedii. J. Ecol. 92, 505–511. doi: 10.1111/j.0022-0477.2004.00878.x
Reinhart, K. O., and Callaway, R. M. (2006). Soil biota and invasive plants. New Phytol. 170, 445–457. doi: 10.1111/j.1469-8137.2006.01715.x
Ronsheim, M. L., and Anderson, S. E. (2001). Population-level specificity in the plant-mycorrhizae association alters intraspecific interactions among neighboring plants. Oecologia 128, 77–84. doi: 10.1007/s004420000625
Rousset, F., and Lion, S. (2011). Much ado about nothing: nowak et al.’s charge against inclusive fitness theory. J. Evol. Biol. 24, 1386–1392.
Roux, F., and Bergelson, J. (2016). The genetics underlying natural variation in the biotic interactions of Arabidopsis thaliana: the challenges of linking evolutionary genetics and community ecology. Curr. Top. Dev. Biol. 119, 111–156. doi: 10.1016/bs.ctdb.2016.03.001
Schmid, B., Hector, A., Saha, P., and Loreau, M. (2008). Biodiversity effects and transgressive overyielding. J. Plant Ecol. 1, 95–102. doi: 10.1093/jpe/rtn011
Semchenko, M., John, E. A., and Hutchings, M. J. (2007). Effects of physical connection and genetic identity of neighbouring ramets on root-placement patterns in two clonal species. New Phytol. 176, 644–654. doi: 10.1111/j.1469-8137.2007.02211.x
Semchenko, M., Saar, S., and Lepik, A. (2014). Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytol. 204, 631–637. doi: 10.1111/nph.12930
Semchenko, M., Saar, S., and Lepik, A. (2017). Intraspecific genetic diversity modulates plant-soil feedback and nutrient cycling. New Phytol. 216, 90–98. doi: 10.1111/nph.14653
Shimola, J., and Bidart, M. G. (2019). Herbivory and plant genotype influence fitness-related responses of Arabidopsis thaliana to indirect plant-plant interactions. Am. J. Plant Sci. 10, 1287–1299. doi: 10.4236/ajps.2019.108093
Singer, A., Travis, J. M. J., and Johst, K. (2013). Interspecific interactions affect species and community responses to climate shifts. Oikos 122, 358–366. doi: 10.1111/j.1600-0706.2012.20465.x
Šmarda, P., Horová, L., Bureš, P., Hralová, I., and Marková, M. (2010). Stabilizing selection on genome size in a population of Festuca pallens under conditions of intensive intraspecific competition. New Phytol. 187, 1195–1204. doi: 10.1111/j.1469-8137.2010.03335.x
Smit, C., Rietkerk, M., and Wassen, M. J. (2009). Inclusion of biotic stress (consumer pressure) alters predictions from the stress gradient hypothesis. J. Ecol. 97, 1215–1219. doi: 10.1111/j.1365-2745.2009.01555.x
Smith, A. L., Atwater, D. Z., and Callaway, R. M. (2019). Early sibling conflict may ultimately benefit the family. Am. Nat. 194, 482–487. doi: 10.1086/704773
Subrahmaniam, H. J., Libourel, C., Journet, E. P., Morel, J. B., Muños, S., Niebel, A., et al. (2018). The genetics underlying natural variation of plant–plant interactions, a beloved but forgotten member of the family of biotic interactions. Plant J. 93, 747–770. doi: 10.1111/tpj.13799
Thomas, S. C., and Bazzaz, F. A. (1993). The genetic component in plant size hierarchies: norms of reaction to density in a Polygonum species. Ecol. Monogr. 63, 231–249.
Todesco, M., Owens, G. L., Bercovich, N., Légaré, J. S., Soudi, S., Burge, D. O., et al. (2020). Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584, 602–607. doi: 10.1038/s41586-020-2467-6
Tomimatsu, H., Nakano, K., Yamamoto, N., and Suyama, Y. (2014). Effects of genotypic diversity of Phragmites australis on primary productivity and water quality in an experimental wetland. Oecologia 175, 163–172. doi: 10.1007/s00442-014-2896-8
Tomiolo, S., Damgaard, C., Zhang, S., Kelly, S., Tao, K., Ronfort, J., et al. (2020). Identity and provenance of neighbors, genotype-specific traits and abiotic stress affect intraspecific interactions in the annual legume Medicago truncatula. bioRxiv [Preprint]. doi: 10.1101/2020.10.08.330944
Tomiolo, S., Thomas, C., Jespersen, M. K., Damgaard, C. F., and Ehlers, B. K. (2019). Intraspecific interactions in the annual legume Medicago minima are shaped by both genetic variation for competitive ability and reduced competition among kin. bioRxiv [Preprint]. doi: 10.1101/695908
Tonsor, S. J. (1989). Relatedness and intraspecific competition in Plantago lanceolata. Am. Nat. 134, 897–906. doi: 10.1086/285020
Torices, R., Gómez, J. M., and Pannell, J. R. (2018). Kin discrimination allows plants to modify investment towards pollinator attraction. Nat. Commun. 9, 1–6. doi: 10.1038/s41467-018-04378-3
Turkington, R., and Harper, J. L. (1979). The growth, distribution and neighbour relationships of Trifolium repens in a permanent pasture. IV. Fine-scale biotic differentiation. J. Ecol. 67, 245–254.
Turner, K. G., Lorts, C. M., Haile, A. T., and Lasky, J. R. (2020). Effects of genomic and functional diversity on stand-level productivity and performance of non-native Arabidopsis. Proc. R. Soc. B Biol. Sci. 287:20202041. doi: 10.1098/rspb.2020.2041
Vandermeer, J. (1981). The interference production principle: ecological theory for agriculture. BioScience 31, 361–364.
Wagg, C., Boller, B., Schneider, S., Widmer, F., and van der Heijden, M. G. A. (2015). Intraspecific and intergenerational differences in plant-soil feedbacks. Oikos 124, 994–1004. doi: 10.1111/oik.01743
Walker, L. R., and Vitousek, P. M. (1991). An invader alters germination and growth of a native dominant tree in Hawaï. Ecology 72, 1449–1455.
Wendling, M., Büchi, L., Amossé, C., Jeangros, B., Walter, A., and Charles, R. (2017). Specific interactions leading to transgressive overyielding in cover crop mixtures. Agric Ecosyst. Environ. 241, 88–99. doi: 10.1016/j.agee.2017.03.003
Willis, C. G., Brock, M. T., and Weinig, C. (2010). Genetic variation in tolerance of competition and neighbour suppression in Arabidopsis thaliana. J. Evol. Biol. 23, 1412–1424. doi: 10.1111/j.1420-9101.2010.02003.x
Willson, M. F., Hoppes, W. G., Goldman, D. A., Thomas, P. A., and Bothwell, J. L. (1987). Sibling competition in plants: an experimental study. Am. Nat. 129, 304–311.
Wuest, S. E., and Niklaus, P. A. (2018). A plant biodiversity effect resolved to a single chromosomal region. Nat. Ecol. Evol. 2, 1933–1939. doi: 10.1038/s41559-018-0708-y
Yamawo, A., and Mukai, H. (2020). Outcome of interspecific competition depends on genotype of conspecific neighbours. Oecologia 193, 415–423. doi: 10.1007/s00442-020-04694-w
Yang, L., Callaway, R. M., and Atwater, D. Z. (2015). Root contact responses and the positive relationship between intraspecific diversity and ecosystem productivity. AoB Plants 7, 1–8. doi: 10.1093/aobpla/plv053
Zhang, R., and Tielbörger, K. (2019). Facilitation from an intraspecific perspective – stress tolerance determines facilitative effect and response in plants. New Phytol. 221, 2203–2212. doi: 10.1111/nph.15528
Zhang, Y., Chen, H. Y. H., and Reich, P. B. (2012). Forest productivity increases with evenness, species richness and trait variation: a global meta-analysis. J. Ecol. 100, 742–749. doi: 10.1111/j.1365-2745.2011.01944.x
Keywords: intraspecific variation, genotype-by-genotype interactions, kin cooperation, diversity-productivity relationship, overyielding
Citation: Subrahmaniam HJ, Roby D and Roux F (2021) Toward Unifying Evolutionary Ecology and Genomics to Understand Positive Plant–Plant Interactions Within Wild Species. Front. Plant Sci. 12:683373. doi: 10.3389/fpls.2021.683373
Received: 20 March 2021; Accepted: 10 June 2021;
Published: 09 July 2021.
Edited by:
Pierre-Emmanuel Courty, INRA Centre Dijon Bourgogne Franche-Comté, FranceReviewed by:
Leopoldo Javier Iannone, CONICET Institute for Biomedical Research (BIOMED), ArgentinaTom Bennett, University of Cambridge, United Kingdom
Copyright © 2021 Subrahmaniam, Roby and Roux. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabrice Roux, ZmFicmljZS5yb3V4QGlucmFlLmZy
 Harihar Jaishree Subrahmaniam
Harihar Jaishree Subrahmaniam Dominique Roby
Dominique Roby Fabrice Roux
Fabrice Roux