- Biotechnology Regulatory Services, Animal and Plant Health Inspection Service, United States Department of Agriculture, Riverdale, MD, United States
Biotechnology can provide a valuable tool to meet UN Sustainable Development Goals and U.S. initiatives to find climate solutions and improve agricultural sustainability. The literature contains hundreds of examples of crops that may serve this purpose, yet most remain un-launched due to high regulatory barriers. Recently the USDA revised its biotechnology regulations to make them more risk-proportionate, science-based, and streamlined. Here, we review some of the promising leads that may enable agriculture to contribute to UN sustainability goals. We further describe and discuss how the revised biotechnology regulation would hypothetically apply to these cases.
Introduction
The UN Sustainable Development Goals (SDG) are critically important for humanity and the planet (United Nations, 2015). Agricultural innovation can positively contribute to many of these goals such as ending hunger, promoting sustainable agriculture or clean energy, to name a few (Secretary-General, 2019). Through Executive Order #14008 (Executive Office of the President, 2021), the Biden Administration prioritized building a modern sustainable infrastructure and an equitable clean energy future demonstrating a commitment to many of these same goals.
Biotechnology has contributed to agricultural sustainability through traits that reduce over the top insecticide application. (Brookes and Barfoot, 2017). Regulatory barriers have limited both diverse trait development and developers who use biotechnology (Bradford et al., 2005; Hoffman, 2021). In May 2020, the U.S. Department of Agriculture (USDA) issued revised biotechnology regulations that offer a more risk-proportionate approach and are expected to spur innovation while ensuring products are safe for agriculture and the environment. Below, we review key changes in the revised regulations, illustrate their application using examples of promising leads in the literature, and show how they are likely to spur trait development that hold promise for improving agricultural sustainability and addressing some climate challenges.
Changes in USDA’s revised biotech regulation
Under the revised regulations, and using Plant Protection Act authority, USDA considers whether an organism developed using genetic engineering poses an increased plant pest risk relative to a suitable comparator (USDA-APHIS, 2020). Several key changes contribute to more risk-proportionate regulation for plants created using genetic engineering. First, the revised regulations establish three exemptions for certain modifications a plant developed using genetic engineering may contain.1
These exemptions are based on types of modifications that commonly occur during conventional breeding (USDA-APHIS, 2020; Hoffman, 2021). Basing the exemptions on specifically described modifications, rather than a risk assessment, allows developers to objectively assess whether their plants meet the criteria exemption.
Second, the revised regulations establish an exemption for a plant-trait-mechanism of action (MOA) combination that USDA previously reviewed and determined not to pose a plant pest risk. This provision eliminates the burden of unnecessary re-reviews of plants whose risks were already considered.2
Third, the revised regulations establish a risk-based approach called regulatory status review (RSR) to determine whether a non-exempt plant is regulated. A key difference between RSR and the previous petition process is that RSR uses problem formulation and risk assessment to evaluate the characteristics and risk of the plant prior to a formal determination to continue to exercise oversight (Hoffman, 2021). Under the revised regulation, a developer can elect to undergo the RSR process prior to field testing. USDA expects that the RSR process will be an effective means to winnow the potentially riskier plants developed using genetic engineering from the less risky ones without imposing substantial regulatory burdens on the less risky ones (Hoffman, 2021). Among other things, USDA considers whether a genetic modification to a plant will increase the likelihood of harm to non-target species beneficial to agriculture or will increase the distribution or exacerbate the impact of plant pests that may be associated with that plant. USDA will undertake an initial review to efficiently distinguish plants developed using genetic engineering that do not pose plausible pathways to increased plant pest risk from those that do and, thus, require further evaluation. This initial review, which is based on a description of the plant, the trait, and the MOA, does not initially require field data. When USDA finds a plant does not pose plausible pathways to increased plant pest risk during the initial review phase, developers can attain regulatory certainty early in product development, which helps developers raise venture capital to see a product through to launch.3
Biotechnology crop development under the legacy regulation
In 2008-2012, the mean cost for discovery, development, and authorization of a new crop created using genetic engineering was $136 M, where regulatory requirement costs averaged $35.1M, and the mean duration to bring a crop to market was 13.1 years (McDougall, 2011). The high-cost developments and long duration reduce return on investment. Consequently, the technology has principally been used on large acreage crops (corn, soybean, cotton, alfalfa, sugarbeet, potato, and canola) engineered with relatively few traits. Of the 136 petitions granted non-regulated status by the USDA, 109 were from those 7 major crops, and 80 had no traits other than either herbicide or insect resistance4. Under the legacy regulations, not all crops created with genetic engineering fell under the regulations. In 2010, USDA instituted a process known as “Am I Regulated” (AIR process), which provided a voluntary mechanism for developers to obtain USDA’s opinion about whether a plant was subject to regulation5. During the last 10 years (2011-2020), the number of organizations using the AIR process increased nearly 4 fold relative to the petition process, while the number of different crops considered increased greater than 5 fold (Hoffman, 2021), suggesting the legacy regulation created a significant barrier to market development of any crop that fell under the regulations.
We expect that the new more risk-appropriate regulation will lead to the commercialization of additional crops and traits. Current literature shows hundreds of proofs of concept for traits with potential to meet SDGs. The examples we provide: exclude plants currently subject to regulation as most have associated Confidential Business Information claims; only include crop plants tested either in the field or greenhouse; and include one representative example when the same plant-trait-MOA was discussed in more than one paper. For each example, we considered whether the plant would likely qualify for an exemption (Table 1) or likely be evaluated under the RSR process (Table 2) based on the information in the paper (Supplementary Table 1 provides additional information on the MOA). Importantly, these considerations are meant to give an approximation of traits relevant to sustainability goals that are ripe for development; they are not and should not be construed as regulatory decisions since we may be missing key details.

Table 1 Crops that may contribute to sustainable agriculture that could potentially qualify for USDA regulatory exemption.
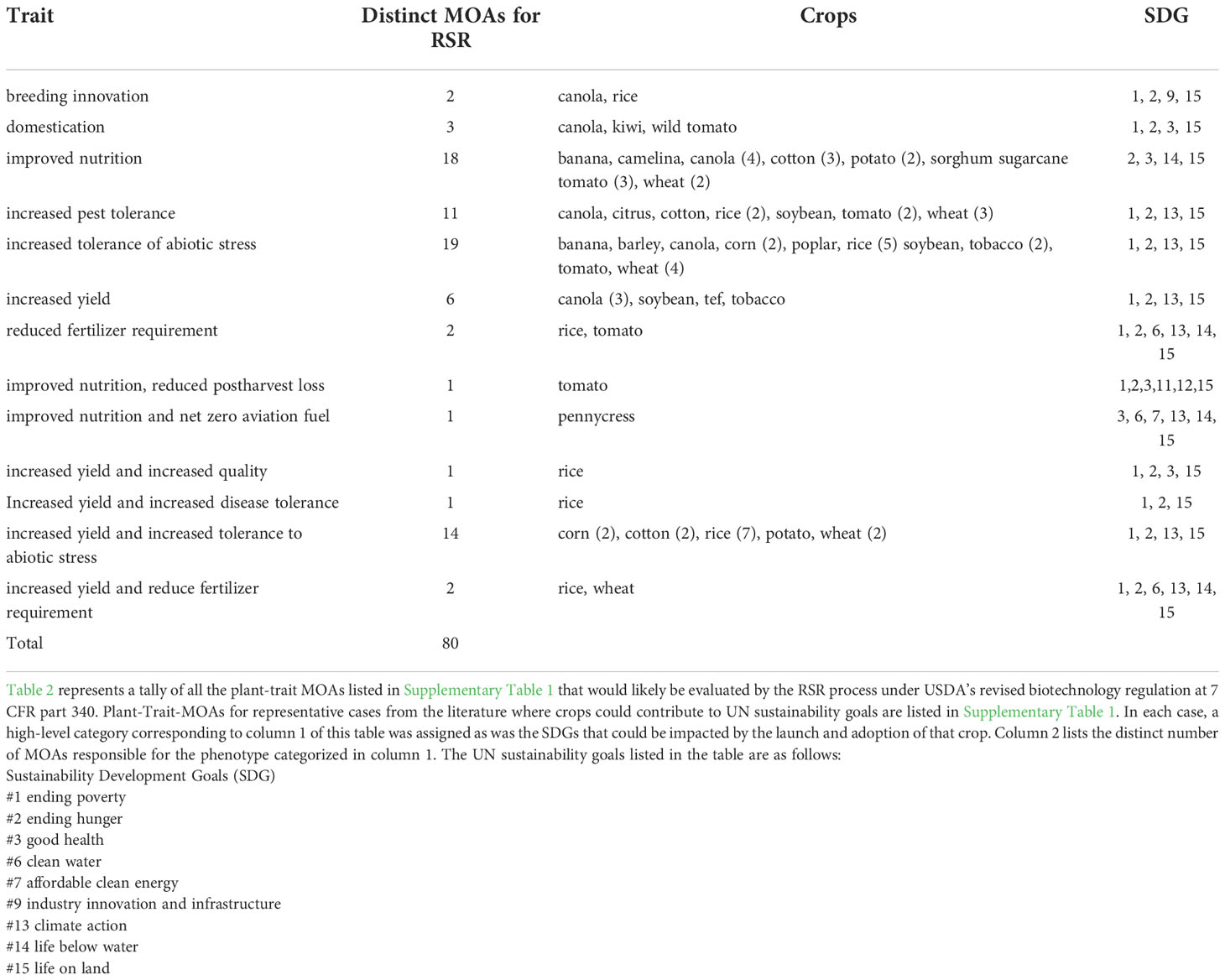
Table 2 Crops that could contribute to sustainable agriculture that would undergo regulatory status review.
Traits that may promote sustainable agriculture and/or mitigate adverse impacts of climate change
Producing more with fewer resources
Producing more food with fewer resources directly supports SDGs associated with reducing poverty, ending hunger, climate action, and conservation (SDGs #1, #2, #6, #13, and #15).
Knockout (KO) strategies in corn, rice, soybean, tomato, and wheat have been used to delete quantitative trait loci (QTL) negatively associated with yield (Zhou et al., 2019), a subfamily of ABA receptors (Miao et al., 2018), or genes that change plant architecture to allow denser planting (Tian et al., 2019), changes in spike inflorescence architecture (Wang et al., 2022), simultaneous increases in panicle number and tiller number (Song et al., 2022; Huang et al., 2018), timing of flowering (Cai et al., 2020), or result in more fruit and less shoot per plant (Rodríguez-Leal et al., 2017). Such plants are likely exempt from regulation and have been shown to increase yield in initial studies without increasing inputs.
USDA would evaluate crops developed with transgenic modifications under the RSR process, as in the case of tobacco engineered for increased production through reduced photorespiration (Cavanagh et al., 2022) or acceleration of the relaxation of non-photochemical quenching during sun-shade transitions (De Souza et al., 2022). Likewise, KOs of all homoeologous alleles in polyploids that increase production would be evaluated under the RSR process, as in the case of canola (Yang et al., 2018; Karunarathna et al., 2020; Zheng et al., 2020) and tef (Beyene et al., 2022). Diploid rice with a KO of 3 different cytochrome P-450 genes to increase production (Usman et al., 2020b) may qualify for exemption depending on whether the individually exempt traits are stacked by breeding (which could be exempt) or molecularly (which would require RSR). Overexpression of a transcription factor in rice was shown to increase both yield and resistance to blight by Xanthomonas (Liu et al., 2019).
Reduced postharvest losses
The Food and Agriculture Organization (FAO) estimates that 14% of the world’s food is lost from production before reaching the retail level (FAO, 2019). Reducing postharvest losses could increase food availability and could free land for conservation (SDGs #12, #2, #15). Developers have used genome editing to increase a tomato’s shelf life by either KO of pectate lyase (Uluisik et al., 2016), polygalacturonse (Nie et al., 2022) or by recreating the allele of a known delayed fruit deterioration mutation (Yu et al., 2017). Each of these tomato varieties would likely qualify for exemption.
Increased disease tolerance
FAO estimates that annually 20-40 percent of global crop production is lost to pests (FAO, 2019). Increasing disease tolerance could result in increased food and could reduce the volume of crop protection chemicals applied for disease control supporting SDGs related to protecting human health, the land, and water related ecosystems (SDGs #2, #3, #15, #6).
Pathogens often exploit susceptible plant genes to facilitate their infection (Van Schie and Takken, 2014). KOs have been used on a variety of susceptible genes to increase disease tolerance in apple (Pompili et al., 2020), barley (Hoffie et al., 2022), canola with the susceptibility gene only in the A genome (Pröbsting et al., 2020), cassava (Gomez et al., 2019), corn (Liu et al., 2022), cucumber (Chandrasekaran et al., 2016), rice (Zhou et al., 2018), tomato (Nekrasov et al., 2017), and watermelon (Zhang et al., 2020a) (Table 1). Increased resistance to wheat stem rust Ug99 was conferred into wheat (T. aestivum) by introducing resistance genes from either einkorn wheat (T. monococcum) (Chen et al., 2018) or durham wheat T. turgidum (Zhang et al., 2017b), both of which are in the wheat gene pool. These examples and others in banana, rice and tomato (Tripathi et al. 2019; Macovei et al., 2018; Ortigosa et al., 2019; Zhou et al 2022b) would likely be exempt
In other instances, developers have used strategies that involve multiple modifications or the use of transgenes to increase disease resistance, which USDA would likely evaluate under the RSR process. KOs of susceptibility genes in canola (Sun et al., 2018), citrus (Peng et al., 2017), cotton (Zhang et al., 2018b) and wheat (Zhang et al., 2017c; Koller et al., 2019; Li et al., 2022b) conferred resistance to various diseases. Multiple promoter deletions in several sugar transport genes conferred broad spectrum resistance in rice to several races of bacterial blight (Oliva et al., 2019). With respect to transgenes, broad resistance to three rice diseases was accomplished by transcriptional and translational controlled expression of the Arabidopsis NPR1 gene (Xu et al., 2017). Expression of transgenes to confer host induced gene silencing has shown promise in controlling fungi, nematodes (Kong et al., 2022), sap sucking and chewing insects, and viruses (Koch and Wassenegger, 2021) and Crispr-Cas lines targeting virus coat proteins have conferred resistance to viruses (Tashkandi et al. 2018).
Increased tolerance of abiotic stress
Climate change is expected to have a net negative impact on agricultural productivity (Raza et al., 2019). Traits that could help crops adapt to climate change include tolerance to heat, drought, and salinity.
Table 1 lists 4 cases for increased abiotic tolerance, all in rice, that would likely qualify for an exemption based on partial deletions (Liu et al., 2020a; Santosh Kumar et al., 2020) or KO (Zhang et al., 2019) of endogenous transcription factors or addition of an allele of Sub1, a transcription factor within the rice gene pool that confers tolerance to flooding (Xu et al., 2006). KO of an ABA receptor increased both yield and tolerance to drought (Usman et al., 2020a) and a triple KO of a putative auxin transport protein, a QTL negatively associated with yield, and a MYB30 transcription factor increased both yield and cold tolerance (Zeng et al., 2019).
Tolerance to environmental stress has been conferred by overexpression of transcription factors that turn on genes in the stress response (Casaretto et al., 2016). Other strategies with successful proof of concept include reducing stomatal opening (Głowacka et al., 2018), increasing specific protein turnover (Alfatih et al., 2020), overexpressing glutaredoxins (Sprague et al., 2022), and modulating stress related signaling (Zang et al., 2018). In all these cases, crops yield better than the comparator under abiotic stress. In some cases, the modified plants yield better than controls even in the absence of stress (Beznec et al., 2021; Esmaeili et al., 2021; Yu et al., 2021; Sprague et al., 2022). Examples likely requiring an RSR are included in Table 2 representing 27 cases from banana (Sreedharan et al., 2013), barley (Hughes et al., 2017), canola (Wu et al., 2020), corn (Shi et al., 2017; Nuccio et al., 2015), cotton (Mishra et al. 2017), poplar (Li et al., 2018b), potato (Yu et al., 2021), rice (Shim et al., 2018; El-Esawi and Alayafi 2019; Liu et al. 2020b; Caine et al., 2019; Zhang et al., 2018a; Jiang et al., 2019; Park et al., 2020; Joshi et al., 2019; Selvaraj et al. 2017; Selvaraj et al., 2020, Usman et al., 2020a), soybean (Ribichich et al., 2020), tobacco (Sharma et al., 2019), tomato (Yin et al. 2018), and wheat (El-Esawi et al., 2019; Zhang et al., 2017a; Mega et al., 2019; Zhou et al. 2022a; González et al., 2019).
Reduced fertilizer requirement
Fertilizer costs are among the most expensive inputs for a farmer (Langemeier et al., 2019) and they represent one of the largest energy expenditures for agriculture (Amenumey and Capel, 2014). Agriculture nutrient runoff is a significant contributor to impairment in assessed rivers and streams (US-EPA, 2018), and the single largest source of nutrient pollution to the Gulf of Mexico’s “dead zone” (Ribaudo et al., 2011). Unabsorbed nitrogen by crops leads to increased production of nitrous oxide, a greenhouse gas which is 300 times more potent than carbon dioxide (Sisharmini et al., 2019). Crops that require less fertilizer input could contribute to clean water and streams, climate action, life below water, life on land, and to ending poverty by reducing fertilizer costs (SDGs #6, #13, #15, #1). Expression of rice GR4 transcription factor from its own promoter results in increases in nitrogen use efficiency (NUE) and yield (Li et al., 2018c). Rice with C-terminal indels in the gene lonely guy (OsLOG5), which catalyzes the formation of active cytokinin from inactive forms, led to increased yields under well-watered, drought, normal nitrogen and low nitrogen (Wang et al., 2020). Both may qualify for exemption.
Examples likely to require RSR evaluation include increased NUE in rice by expression of a cucumber alanine aminotransferase under the control of a rice root specific promoter (Sisharmini et al., 2019), increased NUE in rice through co-overexpression of the rice nitrate transporter (OsNRT2.3a) and its partner protein (NAR2.1a) under the control of the CaMV35S promoter (Chen et al., 2020), increased tolerance to potassium deficiency in rice by overexpression of a rice peroxiredoxin gene (Mao et al., 2018), and increased tolerance to phosphate deficiency in tomato by expression of choline oxidase from the bacteria, Arthrobacter globiformis (Li et al., 2019).
Improved nutrition
Increasing the nutritional value of crops is consistent with good health and well-being (SDG #3) and has been accomplished through KO strategies likely qualifying for exemption in diploids and RSR in polyploids. Healthier fatty acids have been made in oil crops (eg. (Haun et al., 2014; Jarvis et.al, 2021; Okuzaki et al. 2018; Jiang et al., 2017). Other improvements in nutrition include low reducing sugars (potato) (Clasen et al., 2016), high amylose (rice) (Sun et al., 2017), high gamma aminobutyric acid (tomato) (Nonaka et al., 2017), increased vitamin D3 (tomato) (Li et al., 2022a), increased ascorbic acid (tomato) (Do et al., 2022; Zheng et al., 2022), high isoflavone (soybean) (Zhang et al., 2020b), high carotenoid (banana, rice, and tomato) (Paul et al., 2017; Li et al., 2018d), increased protein (canola) (Xie et al., 2020; Zhai et al., 2020), high anthocyanin (tomato) (Butelli et al., 2008), and high iron (Wheat) (Connorton et al., 2017).
In some cases, the crop was made healthier by reducing an anti-nutrient. KO of key transporter proteins resulted in plants that had low uptake of cadmium (Tang et al., 2017) or cesium (Nieves-Cordones et al., 2017). Cotton seed was engineered to be gossypol free thereby creating a new food source by making the meal and oil suitable for human and animal consumption (Sunilkumar et al., 2006; Janga et al., 2019; Li et al., 2021). Other toxic substances eliminated or reduced from crops include steroidal glycosides in potato (Nakayasu et al., 2018), erucic acid in pennycress (Mcginn et al., 2019), lignin in sugarcane (Kannan et al., 2018), kafirin (a difficult protein to digest) in sorghum (Li et al., 2018a), reduced phytic acid in canola (which increases the bioavailability of phosphate in feed) (Sashidhar et al., 2020), reduced cyanide in cassava (Juma et al., 2022), and reduced amylose (rice) and reduced gluten wheat which is of benefit to some on restricted diets (Sánchez-León et al., 2018; Yunyan et al., 2019).
Domestication
Domestication of wild crops through centuries of breeding and selection has inadvertently reduced genetic diversity (Smýkal et al., 2018), limiting traits beneficial for sustainable agriculture. With an understanding of domestication traits, genome editing can rapidly improve agronomic performance of wild relatives by reuniting lost but desirable traits, such as stress tolerance, with agronomically valuable characteristics (Zsögön et al., 2018). For example, in a wild relative of tomato, an initial attempt has been made to make a new tomato variety by introducing six domestication traits that resulted in improvements in fruit number, size, shape, nutrient content and plant architecture (Zsögön et al., 2018). Similar examples are seen in alternative crops such as pennycress (Mcginn et al., 2019), ground cherry (Lemmon et al., 2018) and kiwi (Varkonyi-Gasic et al., 2019), and in standard crops such as canola and tomato (Braatz et al., 2017; Zhai et al., 2019; Kwon et al., 2020). The KO strategies used in (Lemmon et al., 2018; Zhai et al., 2019; Kwon et al., 2020) might qualify for exemption while the RSR process is more likely for the others named above.
Breeding innovations
Hybrid corn seed is almost exclusively grown in the U.S. because of its dramatic increases in yield and vigor (Nielsen, 2020). Widespread use of hybrid seed technology occurred first in corn because the crop’s separate male and female flowers simplifies the hybridization procedure. In crops where hybrid seed is not economically viable because flowers have both male and female organs (perfect flowers), yield gains have typically languished relative to hybrid seed crops (Perez-Prat and Van Lookeren Campagne, 2002). In crops with perfect flowers, KO strategies have been used to introduce male sterility and/or eliminate self-incompatibility (Li et al., 2016; Li et al., 2017; Ye et al., 2018; Ma et al., 2019; Okada et al., 2019; Dai et al., 2022; Ye et al., 2022; Chen et al., 2019; Cigan et al., 2017) to enable efficient hybrid seed production. All female plants were produced in cucumber by KO of a gene required for carpel development (Hu et al., 2017) all of these would likely qualify for exemption. Potato, normally a tetraploid, is being reinvented into a diploid inbred line-based crop that will help achieve yield and vigor gains seen in other hybrid crops (Jansky et al., 2016; Hosaka and Sanetomo, 2020). Recently, apomixis was engineered in rice (Khanday et al., 2019). This trait will enable hybrid seeds to be propagated clonally thereby dramatically reducing the cost of hybrid seed. It involves the KO of three genes and the expression of a normally pollen-specific gene in the egg cell so it would be evaluated under the RSR process. These outcomes contribute to industry innovations (SDG #9) and are expected to facilitate the development of new crops with increased yields and tolerance to abiotic and biotic stresses consistent with numerous other SDGs.
Urban agriculture
Urban vertical farming has been touted as a means to increase agricultural sustainability through demonstrated increases in agricultural productivity, food safety, biosecurity and reduced inputs (water, fertilizer, and pesticides), land use, and transportation costs (SDG #11) relative to outdoor agriculture (Benke and Tomkins, 2017) (SDGS #2, #3, #6, #11, #13-15). Using controlled environments with recycling of nutrients and water, renewable energy, and automation, vertical farming may also offer solutions to climate change and labor shortages that have plagued outdoor agriculture.
Genome editing has been used to create tomato varieties more suitable to controlled environment production (Klap et al., 2017; Soyk et al., 2017; Ueta et al., 2017; Tomlinson et al., 2019; Kwon et al., 2020). The tomato varieties described in the urban agriculture section would likely be eligible for exemption provided the traits were combined by breeding.
Discussion
In this paper, we provide over 140 examples of crops that were created with biotechnology that could contribute to UN sustainability goals. This survey represents just a fraction of the traits being developed to improve sustainability and/or other purposes. Furthermore, traits that show promise in one species frequently prove to be valuable in improving closely related species. Considering that there are hundreds of crops related to those in Tables 1 and 2 (Khoshbakht and Hammer, 2008), it implies that tens of thousands of new crop varieties can be created based on the examples listed in Tables 1 and 2 alone. We estimate that over 60 of the described crop varieties would likely qualify for exemption from USDA oversight. For the remaining crops that would likely be evaluated through the RSR process, the regulatory pathway will be more risk-proportionate, science-based, product-based and streamlined compared to the former petition process (Hoffman, 2021). We already see academics interested in developing products that could successfully navigate the revised regulations. For example, although the Martin lab published the successful creation of a high anthocyanin tomato in 2008 (Butelli et al., 2008), they did not seek regulatory approval in the United States until April 2021 when the RSR process became first available for tomatoes and they received regulatory clearance September 2022.
Based on the large number of promising crop-trait-MOA combinations that have been discovered and the more streamlined, risk-proportionate, and science-based oversight in USDA’s revised regulation, we fully expect to see diverse developers advance more traits that may help promote sustainability. Whether they are ultimately commercialized may depend on overcoming other obstacles including requirements from other regulatory authorities, social understanding and acceptance, and on their economic viability. Both United States Food and Drug Administration (FDA) and Environmental Protection Agency (EPA) continue to play a role in the approval of biotechnology crops in the United States and are actively considering how to streamline their regulations regarding genome editing (OSTP, 2017). EPA has proposed an exemption for certain plant incorporated protectants created through biotechnology that could have otherwise been created through conventional breeding (US-EPA, 2012), while FDA has produced a plant and animal biotechnology innovation action plan and intends to update existing procedures for voluntary premarket consultations (US-Food and Drug Administration, 2018). Several countries (Brazil, Columbia, Argentina, Chile, Israel, Australia, and Japan) do not regulate some genome edited crops lacking foreign DNA as Genetically Modified Organisms (GMOs) and several other countries are considering adopting a similar approach (Schmidt et al., 2020). As the number of like-minded countries grow, regulatory obstacles are expected to diminish.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by the U.S. Department of Agriculture, Animal and Plant Health Inspection Service.
Acknowledgments
The author gratefully acknowledges the critical review of the manuscript and valuable feedback by Bernadette Juarez, Alan Pearson, and Subray Hegde.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1055529/full#supplementary-material
Footnotes
- ^ The SECURE rule (15) exempts plants containing a single modification where1) “the genetic modification is a change resulting from cellular repair of a targeted DNA break in the absence of an externally provided repair template; or2) the genetic modification is a targeted single base pair substitution; or3) the genetic modification introduces a gene known to occur in the plant’s gene pool or makes changes in a targeted sequence to correspond to a known allele of such a gene or to a known structural variation present in the gene pool.
- ^ Developers can request a confirmation from APHIS that a modified plant qualifies for an exemption and is not subject to the regulations in 7 CFR part 340. USDA APHIS | Confirmation Letters accessed 09.27.22
- ^ USDA APHIS | Plant-Trait-Mechanism of Action (MOA) combinations that have been determined by APHIS not to require regulation under 7 CFR part 340.accessed 09.27.22. USDA APHIS | Confirmation Letters accessed 09.27.22
- ^ USDA APHIS | Petitions for Determination of Nonregulated Status accessed 09.27.22
- ^ USDA APHIS | Regulated Article Letters of Inquiry accessed 09.27.22
- ^ q-a-confirmation-process.pdf (usda.gov) p.3-4.
- ^ CFR part 340.1(b)(4)
References
Alfatih, A., Wu, J., Jan, S. U., Zhang, Z. S., Xia, J. Q., Xiang, C. B. (2020). Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 43, 2743–2754. doi: 10.1111/pce.13856
Amenumey, S. E., Capel, P. D. (2014). Fertilizer consumption and energy input for 16 crops in the united states. Natural Resour. Res. 23, 299–309. doi: 10.1007/s11053-013-9226-4
Benke, K., Tomkins, B. (2017). Future food-production systems: vertical farming and controlled-environment agriculture. Sustainability: Science Pract. Policy 13, 13–26. doi: 10.1080/15487733.2017.1394054
Beyene, G., Chauhan, R. D., Villmer, J., Husic, N., Wang, N., Gebre, E., et al. (2022). CRISPR/Cas9-mediated tetra-allelic mutation of the ‘Green revolution’ SEMIDWARF-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef). Plant Biotechnol. J. 20, 1716–1729. doi: 10.1111/pbi.13842
Beznec, A., Faccio, P., Miralles, D. J., Abeledo, L. G., Oneto, C. D., Garibotto, M. B., et al. (2021). Stress-induced expression of IPT gene in transgenic wheat reduces grain yield penalty under drought. J. Genet. Eng. Biotechnol. 19, 67. doi: 10.1186/s43141-021-00171-w
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. doi: 10.1104/pp.17.00426
Bradford, K. J., Van Deynze, A., Gutterson, N., Parrott, W., Strauss, S. H. (2005). Regulating transgenic crops sensibly: lessons from plant breeding, biotechnology and genomics. Nat. Biotechnol. 23, 439–444. doi: 10.1038/nbt1084
Brookes, G., Barfoot, P. (2017). Environmental impacts of genetically modified (GM) crop use 1996-2015: Impacts on pesticide use and carbon emissions. GM Crops Food 8, 117–147. doi: 10.1080/21645698.2017.1309490
Butelli, E., Titta, L., Giorgio, M., Mock, H.-P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi: 10.1038/nbt.1506
Caine, R. S., Yin, X., Sloan, J., Harrison, E. L., Mohammed, U., Fulton, T., et al. (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 221, 371–384. doi: 10.1111/nph.15344
Cai, Y., Wang, L., Chen, L., Wu, T., Liu, L., Sun, S., et al. (2020). Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18, 298–309. doi: 10.1111/pbi.13199
Casaretto, J. A., El-Kereamy, A., Zeng, B., Stiegelmeyer, S. M., Chen, X., Bi, Y. M., et al. (2016). Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17, 312. doi: 10.1186/s12864-016-2659-5
Cavanagh, A. P., South, P. F., Bernacchi, C. J., Ort, D. R. (2022). Alternative pathway to photorespiration protects growth and productivity at elevated temperatures in a model crop. Plant Biotechnol. J. 20, 711–721. doi: 10.1111/pbi.13750
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chen, J., Liu, X., Liu, S., Fan, X., Zhao, L., Song, M., et al. (2020). Co-Overexpression of OsNAR2.1 and OsNRT2.3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11, 1245. doi: 10.3389/fpls.2020.01245
Chen, F., Yang, Y., Li, B., Liu, Z., Khan, F., Zhang, T., et al. (2019). Functional analysis of m-locus protein kinase revealed a novel regulatory mechanism of self-incompatibility in brassica napus l. Int. J. Mol. Sci. 20, 3303. doi: 10.3390/ijms20133303
Chen, S., Zhang, W., Bolus, S., Rouse, M. N., Dubcovsky, J. (2018). Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PloS Genet. 14, e1007287. doi: 10.1371/journal.pgen.1007287
Cigan, A. M., Singh, M., Benn, G., Feigenbutz, L., Kumar, M., Cho, M. J., et al. (2017). Targeted mutagenesis of a conserved anther-expressed P450 gene confers male sterility in monocots. Plant Biotechnol. J. 15, 379–389. doi: 10.1111/pbi.12633
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. doi: 10.1111/pbi.12370
Connorton, J. M., Jones, E. R., Rodríguez-Ramiro, I., Fairweather-Tait, S., Uauy, C., Balk, J. (2017). Wheat vacuolar iron transporter TaVIT2 transports fe and Mn and is effective for biofortification. Plant Physiol. 174, 2434–2444. doi: 10.1104/pp.17.00672
Dai, X., Han, H., Huang, W., Zhao, L., Song, M., Cao, X., et al. (2022). Generating novel Male sterile tomatoes by editing respiratory burst oxidase homolog genes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.817101
De Souza, A. P., Burgess, S. J., Doran, L., Hansen, J., Manukyan, L., Maryn, N., et al. (2022). Soybean photosynthesis and crop yield are improved by accelerating recovery from photoprotection. Science 377, 851–854. doi: 10.1126/science.adc9831
Do, J. H., Park, S. Y., Park, S. H., Kim, H. M., Ma, S. H., Mai, T. D., et al. (2022). Development of a genome-edited tomato with high ascorbate content during later stage of fruit ripening through mutation of SlAPX4. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.836916
El-Esawi, M. A., Alayafi, A. A. (2019). Overexpression of rice Rab7 gene improves drought and heat tolerance and increases grain yield in rice (Oryza sativa l.). Genes 10, 56. doi: 10.3390/genes10010056
El-Esawi, M. A., Al-Ghamdi, A. A., Ali, H. M., Ahmad, M. (2019). Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum l.). Genes 10, 163. doi: 10.3390/genes10020163
Esmaeili, N., Cai, Y., Tang, F., Zhu, X., Smith, J., Mishra, N., et al. (2021). Towards doubling fibre yield for cotton in the semiarid agricultural area by increasing tolerance to drought, heat and salinity simultaneously. Plant Biotechnol. J. 19, 462–476. doi: 10.1111/pbi.13476
Executive Office of the President (2021). Tackling the climate crisis at home and abroad (Washington DC: Federal Register).
FAO (2019). The state of food and agriculture. moving forward on food loss and food reduction (Rome: FAO).
Głowacka, K., Kromdijk, J., Kucera, K., Xie, J., Cavanagh, A. P., Leonelli, L., et al. (2018). Photosystem II subunit s overexpression increases the efficiency of water use in a field-grown crop. Nat. Commun. 9, 868. doi: 10.1038/s41467-018-03231-x
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
González, F. G., Capella, M., Ribichich, K. F., Curín, F., Giacomelli, J. I., Ayala, F., et al. (2019). Field-grown transgenic wheat expressing the sunflower gene HaHB4 significantly outyields the wild type. J. Exp. Bot. 70, 1669–1681. doi: 10.1093/jxb/erz037
Haun, W., Coffman, A., Clasen, B. M., Demorest, Z. L., Lowy, A., Ray, E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. doi: 10.1111/pbi.12201
Hoffie, R. E., Perovic, D., Habekuß, A., Ordon, F., Kumlehn, J. (2022). Novel resistance to the bymovirus BaMMV established by targeted mutagenesis of the PDIL5-1 susceptibility gene in barley. Plant Biotechnol. J. doi: 10.1111/pbi.13948
Hoffman, N. E. (2021). Revisions to USDA biotechnology regulations: The SECURE rule. Proc. Natl. Acad. Sci. 118, e2004841118. doi: 10.1073/pnas.2004841118
Hosaka, K., Sanetomo, R. (2020). Creation of a highly homozygous diploid potato using the s locus inhibitor ( sli ) gene. Euphytica 216, 1–16. doi: 10.1007/s10681-020-02699-3
Huang, L., Zhang, R., Huang, G., Li, Y., Melaku, G., Zhang, S., et al. (2018). Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system. Crop J. 6, 475–481. doi: 10.1016/j.cj.2018.05.005
Hughes, J., Hepworth, C., Dutton, C., Dunn, J. A., Hunt, L., Stephens, J., et al. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776–787. doi: 10.1104/pp.16.01844
Hu, B., Li, D., Liu, X., Qi, J., Gao, D., Zhao, S., et al. (2017). Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 10, 1575–1578. doi: 10.1016/j.molp.2017.09.005
Janga, M. R., Pandeya, D., Campbell, L. M., Konganti, K., Villafuerte, S. T., Puckhaber, L., et al. (2019). Genes regulating gland development in the cotton plant. Plant Biotechnol. J. 17, 1142–1153. doi: 10.1111/pbi.13044
Jansky, S. H., Charkowski, A. O., Douches, D. S., Gusmini, G., Richael, C., Bethke, P. C., et al. (2016). Reinventing potato as a diploid inbred line–based crop. Crop Sci. 56, 1412–1422. doi: 10.2135/cropsci2015.12.0740
Jarvis, B. A., Romsdahl, T. B., Mcginn, M. G., Nazarenus, T. J., Cahoon, E. B., Chapman, K. D., et al. (2021). CRISPR/Cas9-induced fad2 and rod1 mutations stacked with fae1 confer high oleic acid seed oil in pennycress (Thlaspi arvense l.). Front. Plant Sci. 12, 652319. doi: 10.3389/fpls.2021.652319
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., Weeks, D. P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi: 10.1111/pbi.12663
Jiang, D., Zhou, L., Chen, W., Ye, N., Xia, J., Zhuang, C. (2019). Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in rice via ABA-mediated pathways. Rice (N Y) 12, 76. doi: 10.1186/s12284-019-0334-6
Joshi, R., Sahoo, K. K., Singh, A. K., Anwar, K., Pundir, P., Gautam, R. K., et al. (2019). Enhancing trehalose biosynthesis improves yield potential in marker-free transgenic rice under drought, saline, and sodic conditions. J. Exp. Bot. 71, 653–668. doi: 10.1093/jxb/erz462
Juma, B. S., Mukami, A., Mweu, C., Ngugi, M. P., Mbinda, W. M. (2022). Targeted mutagenesis of the CYP79D1 gene via Crispr/Cas9 mediated genome editing results in lower leels of cyanide in cassava. Front. Plant Science. 13, 1009860. doi: 10.3389/fpls.2022.1009860
Kannan, B., Jung, J. H., Moxley, G. W., Lee, S.-M., Altpeter, F. (2018). TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 16, 856–866. doi: 10.1111/pbi.12833
Karunarathna, N. L., Wang, H., Harloff, H.-J., Jiang, L., Jung, C. (2020). Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 18, 2251–2266. doi: 10.1111/pbi.13381
Khanday, I., Skinner, D., Yang, B., Mercier, R., Sundaresan, V. (2019). A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95. doi: 10.1038/s41586-018-0785-8
Khoshbakht, K., Hammer, K. (2008). How many plant species are cultivated? Genet. Resour. Crop Evol. 55, 925–928. doi: 10.1007/s10722-008-9368-0
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2017). Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi: 10.1111/pbi.12662
Koch, A., Wassenegger, M. (2021). Host-induced gene silencing - mechanisms and applications. New Phytol. 231, 54–59. doi: 10.1111/nph.17364
Koller, T., Brunner, S., Herren, G., Sanchez-Martin, J., Hurni, S., Keller, B. (2019). Field grown transgenic Pm3e wheat lines show powdery mildew resistance and no fitness costs associated with high transgene expression. Transgenic Res. 28, 9–20. doi: 10.1007/s11248-018-0099-5
Kong, L., Shi, X., Chen, D., Yang, N., Yin, C., Yang, J., et al. (2022). Host-induced silencing of a nematode chitin synthase gene enhances resistance of soybeans to both pathogenic heterodera glycines and fusarium oxysporum. Plant Biotechnol. J. 20, 809–811. doi: 10.1111/pbi.13808
Kwon, C. T., Heo, J., Lemmon, Z. H., Capua, Y., Hutton, S. F., Van Eck, J., et al. (2020). Rapid customization of solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 38, 182–188. doi: 10.1038/s41587-019-0361-2
Langemeier, M. R., Dobbins, C. L., Nielsen, B., Vyn, T. J., Casteel, S., Johnson, B. (2019) 2020 purdue crop cost and return guide. Available at: https://ag.purdue.edu/commercialag/Documents/Resources/Mangagement-Strategy/Crop-Economics/id166_2020%20-%20september%202019%20projections.pdf (Accessed 08.11.2021).
Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., et al. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018a). Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 177, 1425–1438. doi: 10.1104/pp.18.00200
Li, B., Liang, S., Alariqi, M., Wang, F., Wang, G., Wang, Q., et al. (2021). The application of temperature sensitivity CRISPR/LbCpf1 (LbCas12a) mediated genome editing in allotetraploid cotton (G. hirsutum) and creation of nontransgenic, gossypol-free cotton. Plant Biotechnol. J. 19, 221–223. doi: 10.1111/pbi.13470
Li, S., Lin, Y.-C. J., Wang, P., Zhang, B., Li, M., Chen, S., et al. (2018b). The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in populus trichocarpa. Plant Cell 31, 663–686.
Li, S., Lin, D., Zhang, Y., Deng, M., Chen, Y., Lv, B., et al. (2022b). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460. doi: 10.1038/s41586-022-04395-9
Li, J., Scarano, A., Gonzalez, N. M., D’orso, F., Yue, Y., Nemeth, K., et al. (2022a). Biofortified tomatoes provide a new route to vitamin d sufficiency. Nat. Plants 8, 611–616. doi: 10.1038/s41477-022-01154-6
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018c). Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Liu, C., Kong, M., Yang, F., Zhu, J., Qi, X., Weng, J., et al. (2022). Targeted generation of null mutants in ZmGDIα confers resistance against maize rough dwarf disease without agronomic penalty. Plant Biotechnol. J. 20, 803–805. doi: 10.1111/pbi.13793
Liu, X.-H., Lyu, Y.-S., Yang, W., Yang, Z.-T., Lu, S.-J., Liu, J.-X. (2020a). A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 18, 1317–1329. doi: 10.1111/pbi.13297
Liu, M., Shi, Z., Zhang, X., Wang, M., Zhang, L., Zheng, K., et al. (2019). Inducible overexpression of ideal plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants 5, 389–400. doi: 10.1038/s41477-019-0383-2
Liu, X., Wu, D., Shan, T., Xu, S., Qin, R., Li, H., et al. (2020b). The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 103, 545–560. doi: 10.1007/s11103-020-01010-1
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018d). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00559
Li, Q., Zhang, D., Chen, M., Liang, W., Wei, J., Qi, Y., et al. (2016). Development of japonica photo-sensitive genic Male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genomics 43, 415–419. doi: 10.1016/j.jgg.2016.04.011
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., Liu, J., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genomics 44, 465–468. doi: 10.1016/j.jgg.2017.02.002
Li, D., Zhang, T., Wang, M., Liu, Y., Brestic, M., Chen, T. H. H., et al. (2019). Genetic engineering of the biosynthesis of glycine betaine modulates phosphate homeostasis by regulating phosphate acquisition in tomato. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01995
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Mao, X., Zheng, Y., Xiao, K., Wei, Y., Zhu, Y., Cai, Q., et al. (2018). OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem. Biophys. Res. Commun. 495, 461–467. doi: 10.1016/j.bbrc.2017.11.045
Ma, C., Zhu, C., Zheng, M., Liu, M., Zhang, D., Liu, B., et al. (2019). CRISPR/Cas9-mediated multiple gene editing in brassica oleracea var. capitata using the endogenous tRNA-processing system. Horticulture Res. 6, 20. doi: 10.1038/s41438-018-0107-1
McDougall, P. (2011). The cost and time involved in the discovery, developmentand authorization of a new plant biotechnology derived trait: A consultancy study for CropLife international (Midlothian, UK: Crop Life America).
Mcginn, M., Phippen, W. B., Chopra, R., Bansal, S., Jarvis, B. A., Phippen, M. E., et al. (2019). Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnol. J. 17, 776–788. doi: 10.1111/pbi.13014
Mega, R., Abe, F., Kim, J. S., Tsuboi, Y., Tanaka, K., Kobayashi, H., et al. (2019). Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 5, 153–159. doi: 10.1038/s41477-019-0361-8
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2018). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. U.S.A. 115, 6058–6063. doi: 10.1073/pnas.1804774115
Mishra, N., Sun, L., Zhu, X., Smith, J., Prakash, A., Srivastava, Yang, X., et al. (2017). Overexpression of the rice SUMO E3 ligase gene OsSIZ1 in cotton enhances drought and heat tolerance, and substantially improves fiber yields in the field under reduced irrigation and rainfed conditions. Plant Cell Physiol. 58, 735–746. doi: 10.1093/pcp/pcx032
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 131, 70–77. doi: 10.1016/j.plaphy.2018.04.026
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. doi: 10.1038/s41598-017-00578-x
Nielsen, R. L. (2020) Historical corn grain yields in the US. Available at: https://www.agry.purdue.edu/ext/corn/news/timeless/YieldTrends.html#:~:text=Historical%20Corn%20Grain%20Yields%20in%20the%20U.S.%201,influenced%20primarily%20by%20year-to-year%20variability%20in%20growing%20conditions (Accessed 08.12.21).
Nie, H., Shi, Y., Geng, X., Xing, G. (2022). CRISRP/Cas9-mediated targeted mutagenesis of tomato polygalacturonase gene (SlPG) delays fruit softening. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.729128
Nieves-Cordones, M., Mohamed, S., Tanoi, K., Kobayashi, N. I., Takagi, K., Vernet, A., et al. (2017). Production of low-cs+ rice plants by inactivation of the k+ transporter OsHAK1 with the CRISPR-cas system. Plant J. 92, 43–56. doi: 10.1111/tpj.13632
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., Ezura, H. (2017). Efficient increase of -aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7, 7057. doi: 10.1038/s41598-017-06400-y
Nuccio, M. L., Wu, J., Mowers, R., Zhou, H. P., Meghji, M., Primavesi, L. F., et al. (2015). Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 33, 862–869. doi: 10.1038/nbt.3277
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17, 1905–1913. doi: 10.1111/pbi.13106
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
OSTP (2017) Modernizing the regulatory system for biotechnology products: Final version of the 2017 update to the coordinated framework for the regulation of biotechnology. Available at: https://usbiotechnologyregulation.mrp.usda.gov/biotechnologygov/home/modernizing/modernizing_biotechnology_framework.
Park, S.-I., Kim, J.-J., Shin, S.-Y., Kim, Y.-S., Yoon, H.-S. (2020). ASR enhances environmental stress tolerance and improves grain yield by modulating stomatal closure in rice. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01752
Paul, J.-Y., Khanna, H., Kleidon, J., Hoang, P., Geijskes, J., Daniells, J., et al. (2017). Golden bananas in the field: elevated fruit pro-vitamin a from the expression of a single banana transgene. Plant Biotechnol. J. 15, 520–532. doi: 10.1111/pbi.12650
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Perez-Prat, E., Van Lookeren Campagne, M. M. (2002). Hybrid seed production and the challenge of propagating male-sterile plants. Trends Plant Sci. 7, 199–203. doi: 10.1016/S1360-1385(02)02252-5
Pompili, V., Dalla Costa, L., Piazza, S., Pindo, M., Malnoy, M. (2020). Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 18, 845–858. doi: 10.1111/pbi.13253
Pröbsting, M., Schenke, D., Hossain, R., Häder, C., Thurau, T., Wighardt, L., et al. (2020). Loss of function of CRT1a (calreticulin) reduces plant susceptibility to verticillium longisporum in both arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2328–2344. doi: 10.1111/pbi.13394
Raza, A., Razzaq, A., Mehmood, S. S., Zou, X., Zhang, X., Lv, Y., et al. (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants (Basel) 8, 34–63. doi: 10.3390/plants8020034
Ribaudo, M., Delgado, J., Hansen, L., Livingston, M. (2011). Nitrogen in agricultural systems: Implications for conservation policy. (U.S: Department of Agriculture) ERS.
Ribichich, K. F., Chiozza, M., Ávalos-Britez, S., Cabello, J. V., Arce, A. L., Watson, G., et al. (2020). Successful field performance in warm and dry environments of soybean expressing the sunflower transcription factor HB4. J. Exp. Bot. 71, 3142–3156. doi: 10.1093/jxb/eraa064
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e478. doi: 10.1016/j.cell.2017.08.030
Sánchez-León, S., Gil-Humanes, J., Ozuna, C. V., Giménez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi: 10.1111/pbi.12837
Santosh Kumar, V. V., Verma, R. K., Yadav, S. K., Yadav, P., Watts, A., Rao, M. V., et al. (2020). CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 26, 1099–1110. doi: 10.1007/s12298-020-00819-w
Sashidhar, N., Harloff, H. J., Potgieter, L., Jung, C. (2020). Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 18, 2241–2250. doi: 10.1111/pbi.13380
Schmidt, S. M., Belisle, M., Frommer, W. B. (2020). The evolving landscape around genome editing in agriculture: Many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO Rep. 21, e50680. doi: 10.15252/embr.202050680
Secretary-General (2019). Global sustainable development report 2019: The future is now – science for Achieving sustainable development (New York:: United Nations).
Selvaraj, M. G., Ishizaki, T., Valencia, M., Ogawa, S., Dedicova, B., Ogata, T., et al. (2017). Overexpression of an arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 15, 1465–1477. doi: 10.1111/pbi.12731
Selvaraj, M. G., Jan, A., Ishizaki, T., Valencia, M., Dedicova, B., Maruyama, K., et al. (2020). Expression of the CCCH-tandem zinc finger protein gene OsTZF5 under a stress-inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotechnol. J. 18, 1711–1721. doi: 10.1111/pbi.13334
Sharma, V., Goel, P., Kumar, S., Singh, A. K. (2019). An apple transcription factor, MdDREB76, confers salt and drought tolerance in transgenic tobacco by activating the expression of stress-responsive genes. Plant Cell Rep. 38, 221–241. doi: 10.1007/s00299-018-2364-8
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shim, J. S., Oh, N., Chung, P. J., Kim, Y. S., Choi, Y. D., Kim, J.-K. (2018). Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00310
Sisharmini, A., Apriana, A., Khumaida, N., Trijatmiko, K. R., Purwoko, B. S. (2019). Expression of a cucumber alanine aminotransferase2 gene improves nitrogen use efficiency in transgenic rice. J. Genet. Eng. Biotechnol. 17, 9. doi: 10.1186/s43141-019-0010-7
Smýkal, P., Nelson, M. N., Berger, J. D., Von Wettberg, E. J. B. (2018). The impact of genetic changes during crop domestication. Agronomy 8, 119. doi: 10.3390/agronomy8070119
Song, X., Meng, X., Guo, H., Cheng, Q., Jing, Y., Chen, M., et al. (2022). Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 40, 1403–1411. doi: 10.1038/s41587-022-01281-7
Soyk, S., Lemmon, Z. H., Oved, M., Fisher, J., Liberatore, K. L., Park, S. J., et al. (2017). Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 169, 1142–1155.e1112. doi: 10.1016/j.cell.2017.04.032
Sprague, S. A., Tamang, T. M., Steiner, T., Wu, Q., Hu, Y., Kakeshpour, T., et al. (2022). Redox-engineering enhances maize thermotolerance and grain yield in the field. Plant Biotechnol. J. 20, 1819–1832. doi: 10.1111/pbi.13866
Sreedharan, S., Shekhawat, U. K. S., Ganapathi, T. R. (2013). Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1;2 display high tolerance levels to different abiotic stresses. Plant Biotechnol. J. 11, 942–952. doi: 10.1111/pbi.12086
Sunilkumar, G., Campbell, L. M., Puckhaber, L., Stipanovic, R. D., Rathore, K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. U.S.A. 103, 18054–18059. doi: 10.1073/pnas.0605389103
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00298
Sun, Q., Lin, L., Liu, D., Wu, D., Fang, Y., Wu, J., et al. (2018). CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in brassica napus l. Int. J. Mol. Sci. 19, 2716–2735. doi: 10.3390/ijms19092716
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low cd-accumulating indica rice without compromising yield. Sci. Rep. 7, 14438. doi: 10.1038/s41598-017-14832-9
Tashkandi, M., Ali, Z., Aljedaani, F., Shami, A., Mahfouz, M. M. (2018). Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signaling & Behavior 13, e1525996.
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365, 658–664. doi: 10.1126/science.aax5482
Tomlinson, L., Yang, Y., Emenecker, R., Smoker, M., Taylor, J., Perkins, S., et al. (2019). Using CRISPR/Cas9 genome editing in tomato to create a gibberellin-responsive dominant dwarf DELLA allele. Plant Biotechnol. J. 17, 132–140. doi: 10.1111/pbi.12952
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the b genome of musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 46. doi: 10.1038/s42003-019-0288-7
Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., et al. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 7, 507. doi: 10.1038/s41598-017-00501-4
Uluisik, S., Chapman, N. H., Smith, R., Poole, M., Adams, G., Gillis, R. B., et al. (2016). Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 34, 950–952. doi: 10.1038/nbt.3602
USDA-Aphis (2020). Movement of certain genetically engineered organisms. Federal Register 85, 29790–29838.
US-EPA. (2012). Summaries of Water Pollution Reporting Categories. (Washington DC: US-EPA). Available at: adapted from doc no. EPA841-R-12-104. https://www.epa.gov/system/files/documents/2022-04/34parentattainsdescriptions.pdf Washington DC: US-EPA.
US-EPA (2020). Pesticides; exemptions of certain plant-incorporated protectants (PIPs) derived from newer technologies (Federal Register).
US Food and Drug Administration (2018). Plant and animal biotechnology innovation action plan. (Maryland: Silver Spring)
Usman, B., Nawaz, G., Zhao, N., Liao, S., Liu, Y., Li, R. (2020a). Precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa l.) by regulating circadian rhythm and abiotic stress responsive proteins. Int. J. Mol. Sci. 21, 7854–7883. doi: 10.3390/ijms21217854
Usman, B., Nawaz, G., Zhao, N., Liu, Y., Li, R. (2020b). Generation of high yielding and fragrant rice (Oryza sativa l.) lines by CRISPR/Cas9 targeted mutagenesis of three homoeologs of cytochrome P450 gene family and OsBADH2 and transcriptome and proteome profiling of revealed changes triggered by mutations. Plants 9, 788–815. doi: 10.3390/plants9060788
Usman, B., Zhao, N., Nawaz, G., Qin, B., Liu, F., Liu, Y., et al. (2021). CRISPR/Cas9 guided mutagenesis of grain size 3 confers increased rice (Oryza sativa l.) grain length by regulating cysteine proteinase inhibitor and ubiquitin-related proteins. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22063225
Van Schie, C. C. N., Takken, F. L. W. (2014). Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Varkonyi-Gasic, E., Wang, T., Voogd, C., Jeon, S., Drummond, R. S. M., Gleave, A. P., et al. (2019). Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 17, 869–880. doi: 10.1111/pbi.13021
Wang, Y., Du, F., Wang, J., Wang, K., Tian, C., Qi, X., et al. (2022). Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nat. Plants 8, 930–939. doi: 10.1038/s41477-022-01197-9
Wang, C., Wang, G., Gao, Y., Lu, G., Habben, J. E., Mao, G., et al. (2020). A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol. Biol. 102, 373–388. doi: 10.1007/s11103-019-00952-5
Wu, J., Yan, G., Duan, Z., Wang, Z., Kang, C., Guo, L., et al. (2020). Roles of the brassica napus DELLA protein BnaA6.RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10.ABF2. Front. Plant Sci. 11, 577. doi: 10.3389/fpls.2020.00577
Xie, T., Chen, X., Guo, T., Rong, H., Chen, Z., Sun, Q., et al. (2020). Targeted knockout of BnTT2 homologues for yellow-seeded brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68, 5676–5690. doi: 10.1021/acs.jafc.0c01126
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. doi: 10.1038/nature04920
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., et al. (2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. doi: 10.1038/nature22372
Yang, Y., Zhu, K., Li, H., Han, S., Meng, Q., Khan, S. U., et al. (2018). Precise editing of CLAVATA genes in brassica napus l. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. doi: 10.1111/pbi.12872
Ye, Q., Meng, X., Chen, H., Wu, J., Zheng, L., Shen, C., et al. (2022). Construction of genic male sterility system by CRISPR/Cas9 editing from model legume to alfalfa. Plant Biotechnol. J. 20, 613–615. doi: 10.1111/pbi.13770
Ye, M., Peng, Z., Tang, D., Yang, Z., Li, D., Xu, Y., et al. (2018). Generation of self-compatible diploid potato by knockout of s-RNase. Nat. Plants 4, 651–654. doi: 10.1038/s41477-018-0218-6
Yin, Y., Qin, K., Song, X., Zhang, Q., Zhou, Y., Xia, X., et al. (2018). BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 59, 2239–2254. doi: 10.1093/pcp/pcy146
Yin, W., Xiao, Y., Niu, M., Meng, W., Li, L., Zhang, X., et al. (2020). ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 32, 2292–2306. doi: 10.1105/tpc.19.00542
Yu, Q., Liu, S., Yu, L., Xiao, Y., Zhang, S., Wang, X., et al. (2021). RNA Demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 39, 1581–1588. doi: 10.1038/s41587-021-00982-9
Yunyan, F., Jie, Y., Fangquan, W., Fangjun, F., Wenqi, L., Jun, W., et al. (2019). Production of two elite glutinous rice varieties by editing wx gene. Rice Sci. 26, 118–124. doi: 10.1016/j.rsci.2018.04.007
Yu, Q.-H., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7, 11874. doi: 10.1038/s41598-017-12262-1
Zang, X., Geng, X., He, K., Wang, F., Tian, X., Xin, M., et al. (2018). Overexpression of the wheat (Triticum aestivum l.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and arabidopsis. Front. Plant Sci. 9, 1710. doi: 10.3389/fpls.2018.01710
Zeng, Y., Wen, J., Zhao, W., Wang, Q., Huang, W. (2019). Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR-Cas9 system. Front. Plant Sci. 10, 1663. doi: 10.3389/fpls.2019.01663
Zhai, Y., Cai, S., Hu, L., Yang, Y., Amoo, O., Fan, C., et al. (2019). CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in brassica napus l. Theor. Appl. Genet. 132, 2111–2123. doi: 10.1007/s00122-019-03341-0
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in brassica napus l. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017c). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, W., Chen, S., Abate, Z., Nirmala, J., Rouse, M. N., Dubcovsky, J. (2017b). Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl. Acad. Sci. U.S.A. 114, E9483–e9492. doi: 10.1073/pnas.1706277114
Zhang, P., Du, H., Wang, J., Pu, Y., Yang, C., Yan, R., et al. (2020b). Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 18, 1384–1395. doi: 10.1111/pbi.13302
Zhang, Z., Ge, X., Luo, X., Wang, P., Fan, Q., Hu, G., et al. (2018b). Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00842
Zhang, A., Liu, Y., Wang, F., Li, T., Chen, Z., Kong, D., et al. (2019). Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 39, 47. doi: 10.1007/s11032-019-0954-y
Zhang, M., Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., et al. (2020a). CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to fusarium oxysporum f.sp. niveum. Plant Cell Rep. 39, 589–595. doi: 10.1007/s00299-020-02516-0
Zhang, N., Yin, Y., Liu, X., Tong, S., Xing, J., Zhang, Y., et al. (2017a). The E3 ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant Physiol. 175, 1878–1892. doi: 10.1104/pp.17.01319
Zhang, J., Zhang, H., Srivastava, A. K., Pan, Y., Bai, J., Fang, J., et al. (2018a). Knockdown of rice MicroRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 176, 2082–2094. doi: 10.1104/pp.17.01432
Zheng, X., Yuan, Y., Huang, B., Hu, X., Tang, Y., Xu, X., et al. (2022). Control of fruit softening and ascorbic acid accumulation by manipulation of SlIMP3 in tomato. Plant Biotechnol. J. 20, 1213–1225. doi: 10.1111/pbi.13804
Zheng, M., Zhang, L., Tang, M., Liu, J., Liu, H., Yang, H., et al. (2020). Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus l.). Plant Biotechnol. J. 18, 644–654. doi: 10.1111/pbi.13228
Zhou, X., Liao, H., Chern, M., Yin, J., Chen, Y., Wang, J., et al. (2018). Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. 115, 3174–3179. doi: 10.1073/pnas.1705927115
Zhou, Y., Liu, J., Guo, J., Wang, Y., Ji, H., Chu, X., et al. (2022a). GmTDN1 improves wheat yields by inducing dual tolerance to both drought and low-n stress. Plant Biotechnol. J. 20, 1606–1621. doi: 10.1111/pbi.13836
Zhou, J., Xin, X., He, Y., Chen, H., Li, Q., Tang, X., et al. (2019). Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 38, 1–11. doi: 10.1007/s00299-018-2340-3
Zhou, Y., Xu, S., Jiang, N., Zhao, X., Bai, Z., Liu, J., et al. (2022b). Engineering of rice varieties with enhanced resistances to both blast and bacterial blight diseases via CRISPR/Cas9. Plant Biotechnol. J. 20, 876–885. doi: 10.1111/pbi.13766
Keywords: genome editing, regulatory policy, genetic engineering, plant biotechnology, environmental protection
Citation: Hoffman NE (2022) USDA’s revised biotechnology regulation’s contribution to increasing agricultural sustainability and responding to climate change. Front. Plant Sci. 13:1055529. doi: 10.3389/fpls.2022.1055529
Received: 27 September 2022; Accepted: 28 October 2022;
Published: 23 November 2022.
Edited by:
Karthikeyan Adhimoolam, Jeju National University, South KoreaReviewed by:
Pankaj Kumar Bhowmik, National Research Council Canada (NRC), CanadaJianxin Shi, Shanghai Jiao Tong University, China
Copyright © 2022 Hoffman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neil E. Hoffman, bmVpbC5lLmhvZmZtYW5AdXNkYS5nb3Y=
 Neil E. Hoffman
Neil E. Hoffman