- 1Department of Plant Science and Landscape Architecture, University of Maryland, College Park, MD, United States
- 2School of Plant Sciences and Food Security, Tel Aviv University, Tel Aviv, Israel
FK506-BINDING PROTEIN 42/TWISTED DWARF 1 (FKBP42/TWD1) directly regulates cellular trafficking and activation of multiple ATP-BINDING CASSETTE (ABC) transporters from the ABCB and ABCC subfamilies. abcb1 abcb19 double mutants exhibit remarkable phenotypic overlap with twd1 including severe dwarfism, stamen elongation defects, and compact circinate leaves; however, twd1 mutants exhibit greater loss of polar auxin transport and additional helical twisting of roots, inflorescences, and siliques. As abcc1 abcc2 mutants do not exhibit any visible phenotypes and TWD1 does not interact with PIN or AUX1/LAX auxin transporters, loss of function of other ABCB auxin transporters is hypothesized to underly the remaining morphological phenotypes. Here, gene expression, mutant analyses, pharmacological inhibitor studies, auxin transport assays, and direct auxin quantitations were used to determine the relative contributions of loss of other reported ABCB auxin transporters (4, 6, 11, 14, 20, and 21) to twd1 phenotypes. From these analyses, the additional reduction in plant height and the twisted inflorescence, root, and silique phenotypes observed in twd1 compared to abcb1 abcb19 result from loss of ABCB6 and ABCB20 function. Additionally, abcb6 abcb20 root twisting exhibited the same sensitivity to the auxin transport inhibitor 1-napthalthalamic acid as twd1 suggesting they are the primary contributors to these auxin-dependent organ twisting phenotypes. The lack of obvious phenotypes in higher order abcb4 and abcb21 mutants suggests that the functional loss of these transporters does not contribute to twd1 root or shoot twisting. Analyses of ABCB11 and ABCB14 function revealed capacity for auxin transport; however, their activities are readily outcompeted by other substrates, suggesting alternate functions in planta, consistent with a spectrum of relative substrate affinities among ABCB transporters. Overall, the results presented here suggest that the ABCB1/19 and ABCB6/20 pairs represent the primary long-distance ABCB auxin transporters in Arabidopsis and account for all reported twd1 morphological phenotypes. Other ABCB transporters appear to participate in highly localized auxin streams or mobilize alternate transport substrates.
Introduction
A unique group of FK506-BINDING PROTEINs (FKBPs) function in the folding of ATP-BINDING CASSETTE (ABC) transporter proteins (Geisler et al., 2003, 2004; Banasavadi-Siddegowda et al., 2011). These “long” FKBPs contain between one and three FK506-binding domains (FKBD), a tetratricopeptide repeat (TPR) domain, a calmodulin binding domain (CaM-BD), and an in-plane membrane anchor (IPM) and are found ubiquitously in eukaryotes (Geisler and Bailly, 2007). A prominent example is mammalian FKBP8/38 that functions in the post-translational folding of the ATP-gated chloride channel ABCC7/Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) on the endoplasmic reticulum (ER) surface to promote trafficking to the plasma membrane (PM; Banasavadi-Siddegowda et al., 2011; Hutt et al., 2012).
In Arabidopsis, FK506-BINDING PROTEIN 42 (FKBP42) functions in the processing of ATP-BINDING CASSETTE subfamily B (ABCB) and C (ABCC) transporters at the ER and is required for localization and subsequent functionality of these proteins on the plasma and vacuolar membranes, respectively (Geisler et al., 2003, 2004; Bouchard et al., 2006; Bailly et al., 2008; Wu et al., 2010; Wang et al., 2013). Loss of FKBP42 underlies the pronounced phenotypes of the Arabidopsis twisted dwarf 1 (twd1) mutant including dwarfism, stamen defects and reduced fertility, and compact circinate leaves, as well as helical twisting of roots, inflorescences, and siliques (Pérez-Pérez et al., 2002; Geisler et al., 2003). In roots, twd1 helical cell file twisting is non-handed, occurring in both left- and right-handed orientations (Weizbauer et al., 2011; Wang et al., 2013). In Medicago trunculata, loss of the TWD1 ortholog SSP1 also results in defective auxin-dependent seedpod spine formation (Zhao et al., 2021).
Initial molecular interaction studies using yeast two-hybrid identified ABCB1/19 and ABCC1/2 as strong TWD1 interactors (Geisler et al., 2003, 2004). Subsequent studies using fluorescent protein fusions and bioluminescence resonance energy transfer (BRET) validated these interactions for ABCB1/19 and identified ABCB4 as an additional TWD1 interactor (Wu et al., 2010; Kubeš et al., 2012; Wang et al., 2013; Hao et al., 2020). These reports showed that TWD1 functions at the ER and is required for trafficking and activity of the most abundant ABCB isoforms (1, 4, and 19) at the PM. TWD1 interactions with the C-termini of ABCB and ABCC protein subfamily members are mechanistically distinct, as ABCBs associate with the FKBP domain of TWD1 while ABCCs interact with a distinct tetratricopeptide repeat domain (Geisler et al., 2003, 2004). Additional direct and genetic associations of TWD1 with other proteins including PINOID (Henrichs et al., 2012), ACTIN7 (Zhu et al., 2016), and BRI1 (Chaiwanon et al., 2016; Zhao et al., 2016) have also been identified.
Several twd1 phenotypes are also observed in mutants lacking functional ABCB auxin transporters. abcb1 abcb19 double mutants (hereafter abcb1/19) also exhibit severe dwarf stature, circinate leaves, and reduced auxin-dependent stamen elongation and dehiscence defects associated with reduced seed set (Noh et al., 2001; Blakeslee et al., 2007; Titapiwatanakun et al., 2009; Cecchetti et al., 2015). However, helical twisting of roots, stems, and siliques in twd1 are not observed in abcb1/19. Despite the strong interaction of TWD1 with ABCC1 and ABCC2, abcc1 abcc2 double mutants do not show any obvious morphological defects (Geisler et al., 2004; Song et al., 2010). Therefore, a compelling rationale for the phenotypic differences between twd1 and abcb1/19 is the activation of other ABCB auxin transporters by TWD1 (Geisler et al., 2017). This is corroborated by the additional ~10–15% loss in rootward auxin transport in twd1 compared to abcb1/19 (Geisler et al., 2003). However, abcb mutational analyses are complicated by multiple gene duplication events observed in the plant ABCB gene family (Verrier et al., 2008; Knöller et al., 2010; Kang et al., 2011; Banasiak and Jasiński, 2021). As a result, any mutational analysis of ABCB transporters must consider overlapping function and sometimes compensatory expression of highly similar gene products, most notably Arabidopsis ABCB1/19, 4/21, 6/20, 13/14, and 11/12 (Noh et al., 2001; Geisler et al., 2003; Lin and Wang, 2005; Kaneda et al., 2011; Kamimoto et al., 2012; Zhang et al., 2018; Jenness et al., 2019). High degrees of sequence similarity and linkage disequilibria resulting from gene duplication with ABCB gene clusters further complicate mutational and induced loss of function analyses. Further, light regimes appear to regulate TWD1 activation of ABCBs (Christie et al., 2011).
TWD1 is ubiquitous, but abundance is relatively low, with typical transcript levels <10% of ABCB1 and ABCB19 combined (Geisler et al., 2003; Klepikova et al., 2016). The observed stoichiometry is consistent with proposed and experimentally demonstrated activities of TWD1 (Geisler et al., 2016), the limited increases in auxin transport activity observed with TWD1 and ABCB combined overexpression (Bouchard et al., 2006), and evidence that only overexpression of TWD1 without its hydrophobic membrane association domain leads to increased auxin transport and enhanced growth due to increased ABCB activity (Bailly et al., 2013). Cumulatively, these reports also suggest that any loss of additional ABCB transporter function associated with twd1 phenotypes occurs in discrete tissues where the specific isoforms are more abundant.
Plant ABCB transporters appear to be selective for aromatic and aliphatic organic acids, with enhanced specificity evident when ABCBs, PINs, and TWD1 co-occur (Geisler et al., 2005; Bouchard et al., 2006; Titapiwatanakun et al., 2009; Yang and Murphy, 2009; Bailly et al., 2012, 2013; Deslauriers and Spalding, 2021). Recently, a C-terminal proline residue (P1008 in ABCB1) present in a subgroup of ABCB transporters that have been associated with auxin transport (ABCB1, 19, 6, 20, 4, 21, 15–18, 22) was identified (Hao et al., 2020). The authors imply that this residue is an identifier for ABCBs that may be activated by the peptidyl-prolyl cis-trans isomerase (PPIase) activity of TWD1 and consequent specificity for auxin efflux. However, the importance of this proline residue is equivocal since attempts to demonstrate TWD1 PPIase activity have been unsuccessful (Geisler et al., 2003), and the conserved proline residue was not essential for ABCB1-TWD1 interaction (Hao et al., 2020).
Systematic biochemical characterization of TWD1 interactions with the full range of 21 Arabidopsis ABCB transporters has been limited by intrinsic qualities of the ABCB proteins. Some ABCB proteins have proven to be non-functional or highly mis-localized when expressed in commonly used mammalian cell systems, tobacco BY-2 cells, or Saccharomyces cerevisiae (Noh et al., 2001; Geisler et al., 2005; Kubeš et al., 2012). As a result, yeast two-hybrid assays can only be performed with soluble protein fragments (Geisler et al., 2003). Other plant and animal cell systems previously used to study the auxin transport activity of some ABCBs (Petrášek et al., 2006; Blakeslee et al., 2007) have proven to be impractical for systematic protein interaction studies. A BRET-based system has been used to study TWD1 interactions with some ABCBs, but has not been useful for evaluation of Arabidopsis ABCB19 (Bailly et al., 2008; Wang et al., 2013; Hao et al., 2020). Partially functional ABCB-fluorescent protein fusions have been used to demonstrate cellular trafficking defects of ABCB1, 4, and 19 (Wu et al., 2010; Wang et al., 2013; Yang et al., 2013). However, application of this methodology to low abundance ABCB members has limited utility, as overexpression invariably results in distributions in the endomembrane compartments observed when trafficking of ABCB19 is perturbed (Yang et al., 2013).
A similar limitation has been observed in comparative proteomic studies. Arabidopsis ABCBs are not amenable to many standard protein biochemistry techniques (Noh et al., 2001; Terasaka et al., 2005; Blakeslee et al., 2007; Titapiwatanakun et al., 2009; Yang and Murphy, 2009), and only abundant isoforms are reported in proteomic identifications of PM fractions or interaction studies if present at all (Alexandersson et al., 2004; Borner et al., 2005; Titapiwatanakun et al., 2009; Henrichs et al., 2012; Demir et al., 2013; Zhu et al., 2016). As a result, differential distributions of less abundant ABCBs between twd1 and Col-0 PMs are not convincing. However, affinity chromatography using the ABCB/TWD1 binding auxin efflux inhibitor 1-naphthylphthalamic acid (NPA) identified ABCB1, 4, and 19 as well as at least two other unresolved ABCBs when analyzed by mass spectroscopy (Bernasconi et al., 1996; Butler et al., 1998; Murphy et al., 2002).
Here we engaged in a phenotypic analysis of known and putative ABCB auxin transporters under uniform conditions and a single ecotypic background to ascertain that all the major phenotypes associated with twd1 are observed with combined loss of four ABCB transporters. Analysis of ABCB transporter expression identified ABCB1/19, and ABCB6/20 as the primary candidates underlying twd1 phenotypes in aerial tissues and ABCB1/19, 4/21, 6/20, and 11 in the root. Mutant analysis showed that combined loss of ABCB1/19, and ABCB6/20 function is sufficient to confer all the major phenotypes associated with twd1. Further investigation reveals ABCB11 and ABCB14 can transport auxin but likely have alternate substrates in planta.
Materials and Methods
Plant Material and Growth Conditions
Mutants used in this study were in the Col-0 background and previously described: abcb1-100 (Lin and Wang, 2005), abcb19-101 (Lin and Wang, 2005), abcb1-100 abcb19-101 (Jenness et al., 2019), abcb4-1 (Terasaka et al., 2005), abcb21-2 (Jenness et al., 2019), abcb4-1 abcb21-2 (Jenness et al., 2019), amiRNA1334 (Zhang et al., 2018), abcb6-1 (Zhang et al., 2018), abcb20-1 (Zhang et al., 2018), abcb20-2 (Zhang et al., 2018), abcb6-1 abcb20-2 (Zhang et al., 2018), twd1-3 (Geisler et al., 2003), abcb11-1 (SALK_057628; this study), abcb11-2 (SALK_037942; this study), abcb14-1 (Kaneda et al., 2011), pin1-7 (Furutani et al., 2004). All triple mutants were generated by crossing the alleles designated in the text and figure legends into abcb1-100 abcb19-101. Specific alleles in each figure and table are listed in Supplementary Table 3. For seedling growth, surface-sterilized seeds were plated on ¼ MS medium containing 1 g L−1 MES, 0.5% sucrose, and 0.8% agar (pH 5.6). Seeds were stratified 2 days in the dark at 4°C, then grown vertically under continuous 100 μmol m−2 s−1 light at 22°C for the times indicated. For mature plants, seeds were sown on soil and stratified 2–4 days in the dark at 4°C. Plants were grown in growth chambers with fluorescent light supplemented with incandescent bulbs set to 100 μmol m−2 s−1, 16 h photoperiod, and 21°C for the times specified.
Seedling and Mature Plant Imaging and Measurements
Images of mature plants were taken with a Cannon Stylus 1010 digital camera set on a tripod for stability. Close-up leaf, inflorescence, and silique images were collected on a Zeiss Stemi-2000 stereo microscope (Carl Zeiss, Germany) using Lumenera Infinity2 software (Lumenera Corp.). Seedling and root cell file images were collected on a Leica DMI6000B using LAS imaging software (Leica Co., Germany) with the exception of amiRNA1334 and abcb6-1 abcb20-1 which were collected on a Zeiss LSM 780 using a 10× objective. Root length measurements were made from high-resolution scanned images. All seedling measurements were made using ImageJ (Schneider et al., 2012). Measurements for mature aerial tissues were done manually. Brightness and contrast were adjusted using Adobe Photoshop to enhance visualization of the cell profiles equally within each image set as noted in the respective figure legends. All assays were conducted at least three times with similar results.
Quantitative Real-Time PCR
Total RNA was extracted using ZR Plant RNA Mini Prep kit (Zymo Research) followed by treatment with DNaseI (New England Biolabs). Total RNA (1.5 μg) was used for first-strand synthesis using SuperScript III reverse transcriptase (Thermo Fisher Scientific). Real-time PCR was performed on a CFX Connect (Bio-Rad Laboratories) using EvaGreen qPCR master mix (Biotium). Primers used are listed in Supplementary Table 1. Transcript levels normalized against PP2A (AT1G69960) or ACT2 (AT3G18780) produced similar results.
Auxin Quantitations
Auxin quantitations were conducted as previously described (Novák et al., 2012; Jenness et al., 2019). Ions and mass transitions are found in Supplementary Table 2.
Auxin Transport Assays
Inflorescence transport assays were conducted as described in Kaneda et al. (2011) with some modifications. Briefly, ~15 cm inflorescences were excised at the base and the uppermost floral cluster with supporting stalk and peduncles was removed. All remaining floral organs and peduncles were removed, and the stalks were cut into 2 cm segments. The apical 2 cm and 4–6 cm segments were placed inverted (or upright) in 1:1 ratio of cold IAA: 3H-IAA (500 nM each; 22 Ci mmol−1, American Radiolabeled Chemicals) in 5 mM MES buffer with 1% sucrose (pH 5.5) for 1 h at room temperature in the dark, then washed and incubated in blank buffer for an additional 12 h. The distal 2 mm were then collected and measured for radioactivity by liquid scintillation counting. For seedling assays, 6% agarose beads (Colloidal Science Solutions, AMB-0601-0010) soaked in a 1:1 ratio of cold IAA: 3H-IAA (50 nM each; 24 Ci mmol−1, American Radiolabeled Chemicals) were placed over the shoot apex/cotyledonary node or the root–shoot transition zone (RSTZ) of 5.5 days seedlings transferred to a discontinuous filter paper system (Peer and Murphy, 2007). For hypocotyl transport, after incubation 2 mm segments were excised from the RSTZ (10 segments were pooled per replicate). For root transport, 3 mm segments starting from the root tip were collected and pooled separately (10 segments were pooled per replicate). Root tip transport assays were conducted as in Kubeš et al. (2012). Segments were incubated 24 h in 5 ml scintillation cocktail and the amount of radioactivity measured by liquid scintillation counting.
Histochemical Staining
The 1.650 kb ABCB11 promoter fragment upstream of the start codon was amplified by PCR then cloned into pENTR/D-TOPO (Thermo Fisher Scientific; primers specified in Supplementary Table 1). The fragment was then transferred into the Gateway compatible vector pGWB3 (Nakagawa et al., 2007) upstream of the β-glucuronidase (GUS) coding sequence by LR reaction (Thermo Fisher Scientific). Constructs were transformed into Col-0 via floral dip (Clough and Bent, 1998). For GUS staining, tissues were incubated in 90% acetone at 4°C for 20 min, then immersed in staining solution (50 mM sodium phosphate buffer, pH 7.0, 0.1% triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 1 mM X-gluc) and incubated in the dark at 37°C for 5 h, unless otherwise specified. Stained samples were incubated with 70% ethanol at 4°C overnight before imaging. proABCB11:GUS inflorescence stems were GUS stained prior to hand sectioning. Cantils were hand sectioned from inflorescence stems and stained using Toluidine Blue O (TBO) prior to imaging a Zeiss Stemi-2000 stereo microscope (Carl Zeiss, Germany) using Lumenera Infinity2 software (Lumenera Corp.).
Transport Assays in Schizosaccharomyces pombe
Yeast assays were conducted using 40 nM 3H-IAA (20 Ci mmol−1, American Radiolabeled Chemicals) or 50 nM 3H-benzoic acid (20 Ci mmol−1, American Radiolabeled Chemicals) as previously described (Yang and Murphy, 2009). Assays conducted with 3H-benzoic acid alone or 1:1 IAA:benzoic acid (40 nM each) were conducted over 8 min. The ABCB11 expression construct was created by amplifying ABCB11 using primers containing NcoI and XmaI restriction sites (Supplementary Table 1). The digested ABCB11 product was cloned into the corresponding NcoI and XmaI sites of the pREP41 yeast expression vector. Expression vectors were transformed into S. pombe by electroporation.
Protein Sequence Analysis
Protein sequence relationships were inferred by using the Maximum Likelihood method and JTT matrix-based model (Jones et al., 1992). Initial protein alignments were conducted using MUSCLE (Edgar, 2004) and analyses were conducted in MEGA11 (Tamura et al., 2021). The tree with the highest log likelihood (−32,080.72) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 21 amino acid sequences. There were a total of 1,447 positions in the final dataset. Structure-based sequence conservation was determined from the MUSCLE sequence alignment and the AlphaFold2 ABCB19 prediction (Jumper et al., 2021; Varadi et al., 2022) using the ConSurf server (Ashkenazy et al., 2016). Colors indicating conserved and flexible residues were applied to the ABCB19 structure by running the ConSurf script in Pymol.
Statistics
All statistical analyses were performed using JMP PRO 14 (SAS Institute Inc.).
Results
Identification of Candidate ABCB Genes
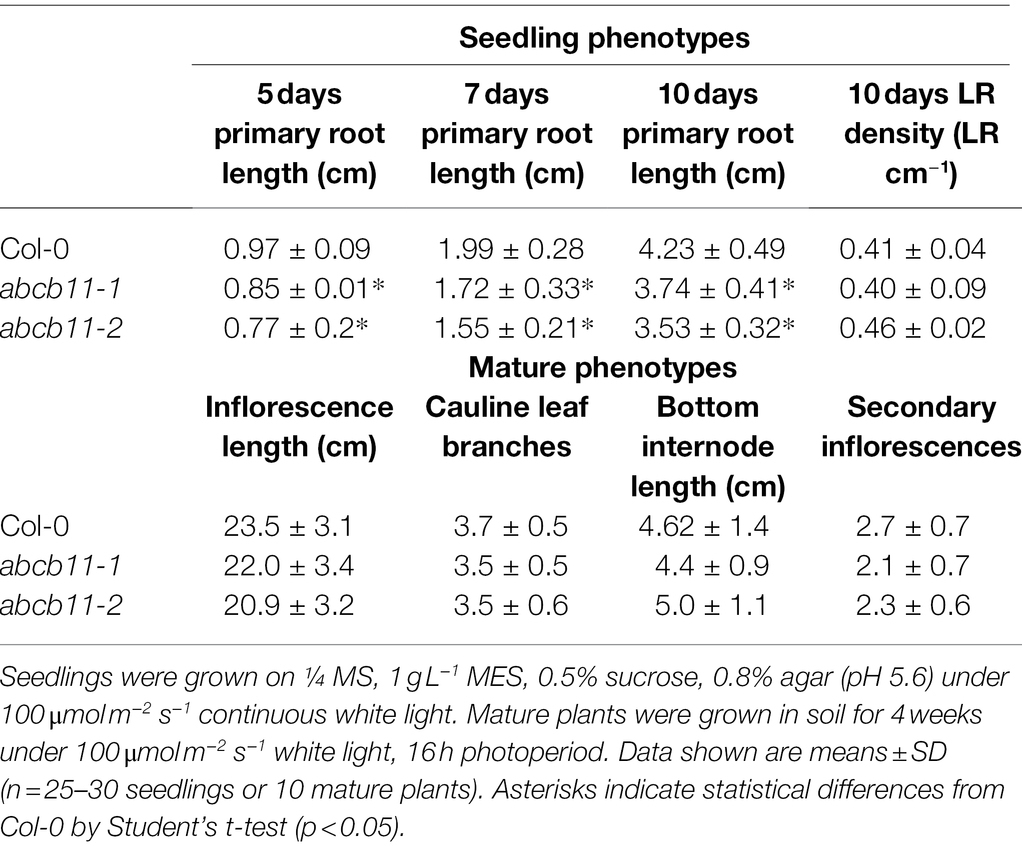
The demonstrated dependence of ABCB1 and 19 on TWD1 for proper trafficking and functionality at the PM is apparent in the significant phenotypic overlap between twd1 and abcb1/19 double mutants including severe dwarf inflorescence phenotypes and compact and curled rosette and cauline leaves (Figures 1A,B,D; Noh et al., 2001; Geisler et al., 2003; Pérez-Pérez et al., 2004; Blakeslee et al., 2007). twd1 mutants, however, display additional helical twisting of inflorescence stems and siliques that is not observed in abcb1/19 double mutants (Figures 1A,B,D; Geisler et al., 2003; Pérez-Pérez et al., 2004). twd1 mutants also show helical super-twisting roots (Figure 1C; Geisler et al., 2003). This phenotype has been reported both as present and absent in abcb1/19 mutants which may reflect differences in ecotypic background, mutant alleles, and growth conditions (Geisler et al., 2003; Bouchard et al., 2006; Bailly et al., 2008; Wu et al., 2010; Wang et al., 2013). In the abcb1/19 double mutant used in this study (abcb1-100 abcb19-101) roots exhibit increased waving and turning but not super-twisting cell files (Figure 1C). Additionally, reproducible twd1-like cell file twisting was not observed in abcb1/19 under varying media or sucrose concentrations, light fluencies, or photoperiod (Supplementary Figures 1A–C).
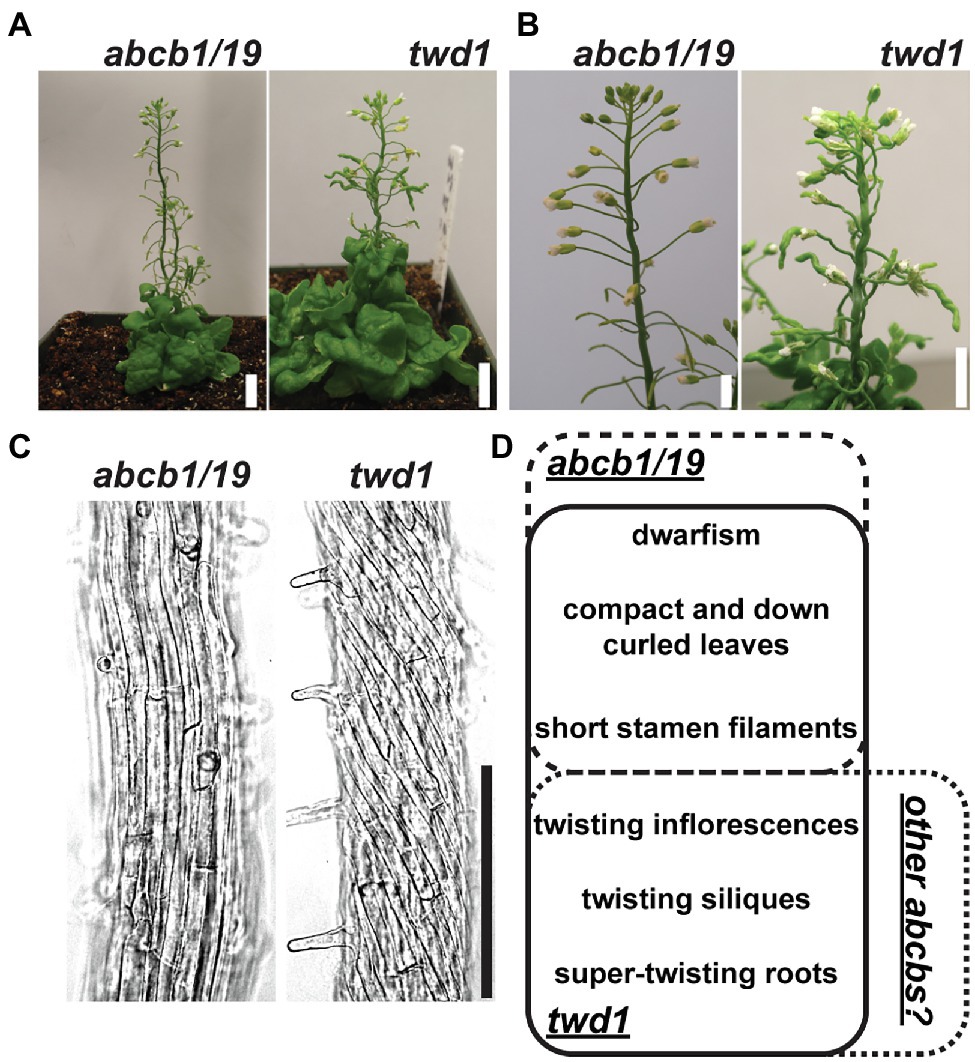
Figure 1. twd1 exhibits additional root, shoot, and silique twisting not observed in abcb1/19. (A) Comparison of mature plant phenotypes of abcb1/19 and twd1. (B) Close-up of twisting inflorescence stems and siliques observed in twd1 but not in abcb1/19. (C) Bright field images abcb1/19 and twd1 roots. Images were converted to black and white, and brightness and contrast were adjusted to show cell files. (D) Venn diagram of the major phenotypes of abcb1/19 and twd1, and twd1 phenotypes that may be accounted for in other abcb mutants. Scale bars: (A) 1 cm; (B) 500 μm; (C) 200 μm.
The Arabidopsis genome encodes 21 full-length ABCB isoforms (and the non-transcribed pseudogene ABCB8) that form highly conserved paralogous pairs and sets from gene duplication events (Supplementary Figures 2A–C; Verrier et al., 2008). The auxin transporters ABCB1, 4, and 19 have the highest levels of expression in auxin conducting tissues (Supplementary Figure 3; Klepikova et al., 2016) and have high affinity for TWD1 (Geisler et al., 2003; Wu et al., 2010; Bailly et al., 2013; Wang et al., 2013; Hao et al., 2020). Additionally, previous studies have identified auxin transport contributions from ABCB6 and 20 in shoots, and ABCB21 in roots (Kamimoto et al., 2012; Zhang et al., 2018; Jenness et al., 2019). Although ABCB2 expression is relatively high, it does not interact with TWD1 (Geisler et al., 2003) and does not transport auxin in yeast (Yang and Murphy, 2009). ABCB11 was previously suggested to contribute to rootward auxin transport in inflorescence stems (Kaneda et al., 2011). However, ABCB11 is expressed primarily in roots (Supplementary Figure 3) where contribution to auxin transport and twd1-like root phenotypes seems more likely. ABCB15-18, and 22 were recently categorized as Auxin Transporting ABCBs (ATAs) based on sequence analysis (Hao et al., 2020). However, based on the moderate expression levels of TWD1 and stoichiometric requirement for TWD1 activation of more highly expressed ABCBs, the extent to which this cluster of ABCBs requires TWD1 activation will require further experimental evidence. Taken together and considering functional redundancy, these results suggest that the ABCB4/21, 6/20, and 11/12 paralogs are the best candidates to account for the remaining twd1 twisting phenotypes not observed in abcb1/19.
abcb4 and abcb21 Are Not Primary Contributors to the Most Obvious twd1 Phenotypes
Due to high levels of ABCB4 expression in the root (Supplementary Figure 3; Klepikova et al., 2016) and interaction with TWD1 (Wu et al., 2010; Hao et al., 2020), it was hypothesized that loss of ABCB4 in addition to ABCB1 and ABCB19 could account for the twd1 root super-twisting phenotype. abcb4 mutants share partially overlapping root phenotypes with twd1 including defects in root hair length and orientation and conditional effects on primary root length (Santelia et al., 2005; Terasaka et al., 2005; Cho et al., 2007; Kubeš et al., 2012). Higher order mutants of ABCB4 and its paralog ABCB21 were therefore examined for contribution to twd1 phenotypes. ABCB4 and ABCB21 do not have compensatory expression in their reciprocal mutant backgrounds, their expression domains do not substantially overlap (Supplementary Figure 3; Santelia et al., 2005; Terasaka et al., 2005; Kamimoto et al., 2012; Kubeš et al., 2012; Klepikova et al., 2016), and no twisting phenotypes were reported in abcb4/21 aerial tissues (Jenness et al., 2019). Consistent with this, abcb4/21 did not exhibit any of the highly reproducible twd1 primary root growth or twisting phenotypes under any growth conditions (Figures 2A,B). It was then hypothesized that additional loss of ABCB4 or ABCB21 in addition to abcb1/19 could recapitulate the twd1 root phenotypes. No twd1-like root growth or twisting was observed in abcb1/4/19 or abcb1/19/21 roots (Figures 2C,D). abcb1/19/21 mutants were previously reported to exhibit slight alterations in rosette leaf morphology and epidermal cell size compared to abcb1/19 (Jenness et al., 2019). However, abcb1/4/19 and abcb1/19/21 triple mutant rosettes and inflorescences were visibly indistinguishable from abcb1/19 and no twd1-like twisting phenotypes were observed (Supplementary Figures 4A,B). Together, these results suggested that ABCB4 and ABCB21 are not primary contributors to twd1 phenotypes and that mis-regulation of other ABCB auxin transporters likely account for the additional root and shoot twisting phenotypes.
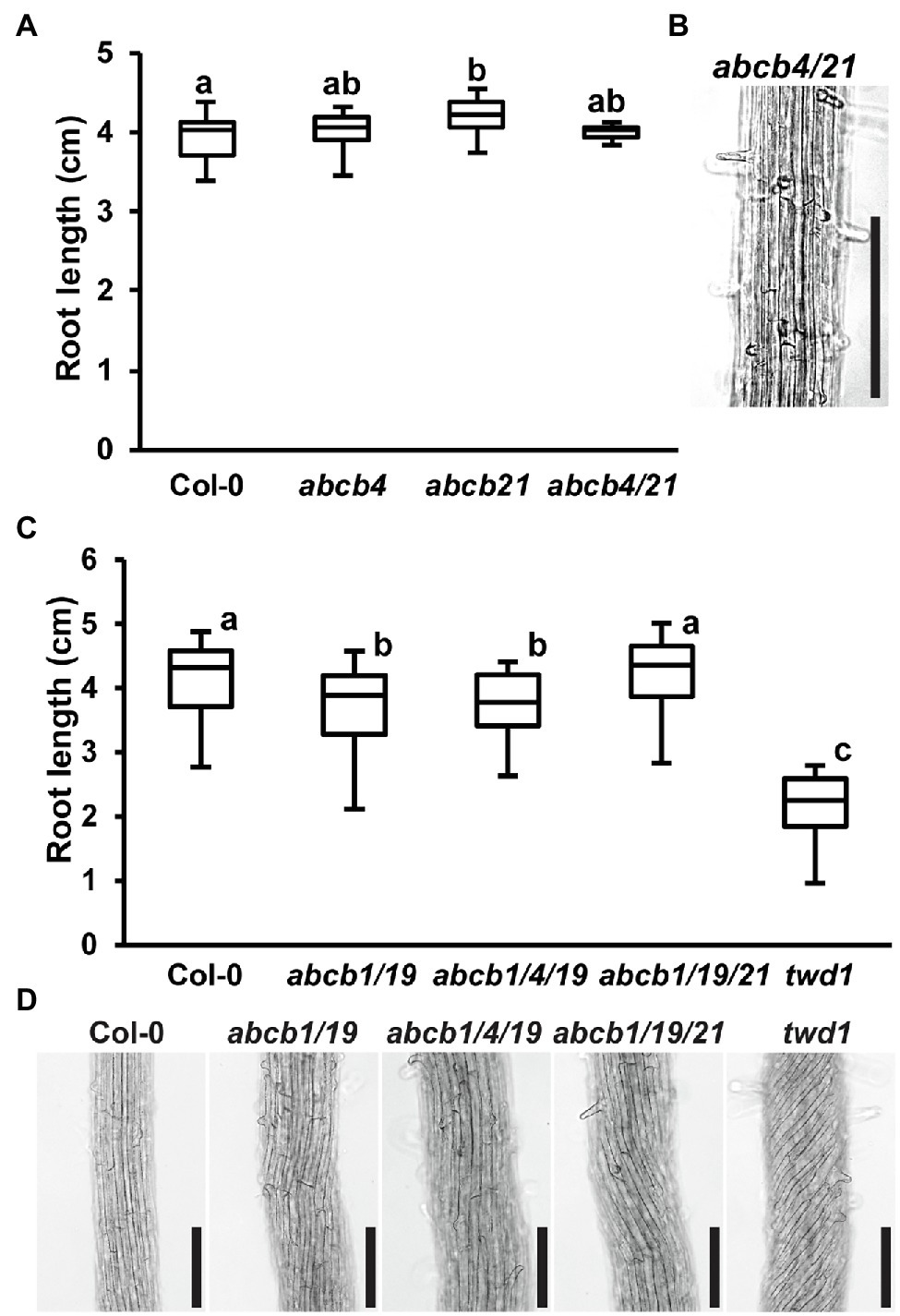
Figure 2. Loss of abcb4 and abcb21 does not result in twd1-like root phenotypes. (A) Root lengths of 7-day light-grown Col-0, abcb4, abcb21, and abcb4/21 seedlings. (B) Root cell files of 7-day light-grown abcb4/21 seedlings. (C) Root lengths of 7-day light-grown Col-0, abcb1/19, abcb1/4/19, abcb1/19/21, and twd1 seedlings. (D) Root cell files of seedlings in (C). Images in (B,C) were converted to black and white, and brightness and contrast were adjusted to show cell files. Lowercase letters indicate statistical difference by pairwise Student’s t-test (p < 0.05; n = 3 replicates of 10–13). Scale bars: 200 μm.
Loss of ABCB6 and ABCB20 Accounts for twd1-Like Rosette and Inflorescence Phenotypes
ABCB6 and ABCB20 are the most highly expressed ABCB auxin transporters in aerial tissues after ABCB1 and 19 (Supplementary Figure 3; Klepikova et al., 2016). Single and double mutants of these transporters were therefore analyzed for defects in leaf, stem, and silique morphology. At the pre-flowering stage, both abcb1/19 and twd1 exhibited compact rosettes with severe downward leaf curling (Figures 3A,B; Noh et al., 2001; Geisler et al., 2003; Pérez-Pérez et al., 2004; Blakeslee et al., 2007). These phenotypes can be attributed primarily to loss of ABCB19 function as it is the only single abcb mutant to show substantial defects in leaf morphology reported to date (Figure 3A; Supplementary Figure 5; Noh et al., 2001; Zhao et al., 2013; Jenness et al., 2020). Single abcb1, abcb6, and abcb20 mutants did not exhibit morphological leaf defects (Figure 3A; Supplementary Figure 5) in part due to compensatory expression and redundant function between paralogous transporters as previously described (Zhang et al., 2018; Jenness et al., 2019). Double abcb6/20 rosettes were compact, tended to laterally twist and skew, and had increased leaf down curling and wrinkling which resembled abcb19 (Figure 3B; Supplementary Figure 5). These results point to the loss of ABCB1 and ABCB19 as the primary contributors to twd1 leaf phenotypes with additional contributions from ABCB6 and ABCB20.

Figure 3. Aerial tissue phenotypes observed in twd1 can be attributed to loss of ABCB1, 6, 19, and 20 function. (A) Single abcb mutant rosettes 21 days after germination. (B) twd1 and double abcb mutant rosettes 21 days after germination. (C) Mature single and double abcb mutants 35 days after germination. (D) Twisting of inflorescence stems in abcb6/20 double mutants. (E,F) Close-ups images of abcb6/20 showing (E) twisted cell files and (F) twisted siliques. (G) Occasional stem–pedicel separation defects observed in abcb6/20 mutants. (H) Cantil spur formation in abcb1/19 grown under 50 μmol m−2 s−1, 16-h photoperiod. (I) Toluidine blue O staining of hand sectioned abcb1/19 cantil spur from (H). Scale bars: (A) 1 cm; (B,I) 0.5 cm; (C) 5 cm; (D,H) 1 cm; (E–G) 1 mm.
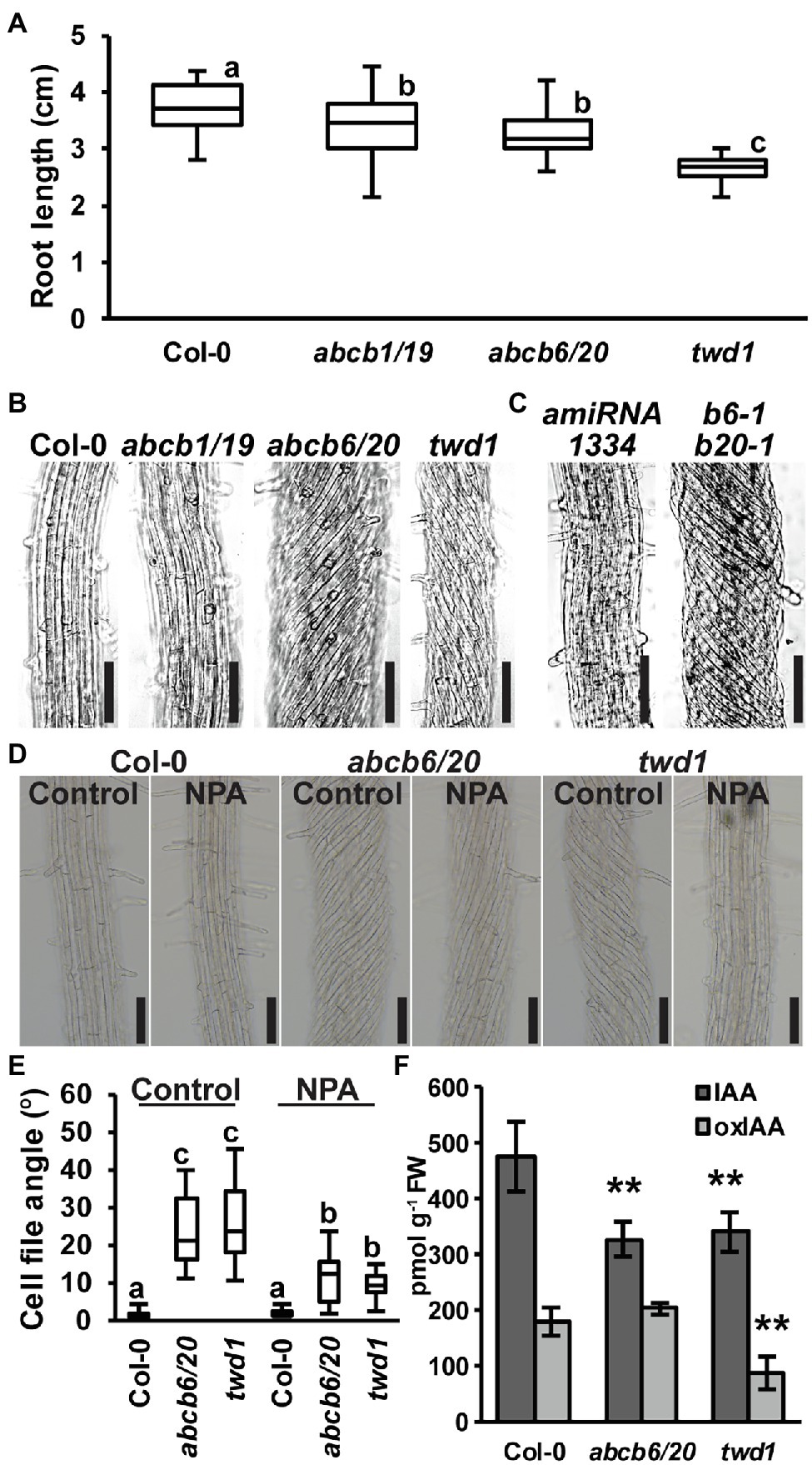
Figure 4. Root phenotypes observed in twd1 can be attributed to loss of ABCB1/19 and 6/20 function. (A) 7-day primary root lengths (n = 3 replicates of 10–13). (B,C) Bright field images of the distal elongation and maturation zone in 7-day seedling roots. Images in (B,C) were converted to black and white, and brightness and contrast were adjusted equally within each set to show cell files. (D) Treatment with 1 μM NPA suppressed root twisting in abcb6/20 and twd1. Brightness and contrast were adjusted equally to show cell files. (E) Quantification of cell file angles in (D). Data shown are means ± SE (n = 3 replicates of 10–11). (F) Quantification of IAA and oxIAA in 5.5-day light-grown roots. Data shown are means ± SD (n = 4–5 independent pools of 25 mg tissue). Lowercase letters indicate statistical difference by all pairwise Student’s t-test (p < 0.05). Asterisks indicate statistical differences by Student’s t-test (**p < 0.01). Scale bars: 100 μm.
After flowering, abcb1/19 and twd1 developed short inflorescence stems (Figure 1A; Noh et al., 2001; Geisler et al., 2003; Pérez-Pérez et al., 2004; Blakeslee et al., 2007). Consistent with previous reports, abcb1 and abcb19 single mutants exhibited slightly decreased inflorescence lengths, which were exaggerated in abcb1/19 (Figure 3C; Table 1; Noh et al., 2001; Blakeslee et al., 2007). While abcb6 and abcb20 inflorescences were not different and slightly longer than Col-0, respectively, abcb6/20 double mutants were severely dwarfed (Figure 3C; Table 1). Single and double abcb1 and abcb19 mutants had reduced cauline leaf branches (Table 1). Examination of internodes in abcb1/19 and abcb6/20 revealed that both double mutants had drastically reduced internode lengths (Table 1). Additionally, abcb6/20 mutants had fewer secondary inflorescences (Table 1). Comparison of twd1 inflorescence phenotypes was not possible due to the severe delay in bolting.
abcb1/19 exhibited increased stem and silique waving and skewing but twd1-like helical twisting of these organs was not observed (Figures 1A,B). Instead, these twisting phenotypes could be attributed to loss of ABCB6 and ABCB20 function (Figures 3D–F). While not observed in the single mutants, abcb6/20 exhibited twisted inflorescence stems (Figures 3D,E), twisted siliques (Figure 3F), and lateral organ separation defects (Figure 3G) which resemble the fused branches and pedicels observed in abcb19 (Zhao et al., 2013). Long-day-grown abcb6/20 and abcb1/19 mutants also developed expanded pedicel–stem junctions that resembled the recently described cantil cuffs observed in Col-0 under short-day conditions (Figure 3E; Supplementary Figures 6A–C; Gookin and Assmann, 2021). These phenotypes were exaggerated in abcb1/19 under low light (Figure 3H) forming cantil spurs that were accompanied by ectopic vasculature formation (Figure 3I).
Overall, the leaf and inflorescence phenotypes associated with loss of ABCB6 and 20 are consistent with those previously reported (Zhang et al., 2018). Together these results point to the loss of ABCB6 and 20 as the primary contributors to the aerial organ twisting observed in twd1, and the combinatorial effects of loss of ABCB1, 19, 6, and 20 function in the severe compact leaf and dwarf inflorescence phenotypes. It was hypothesized that an abcb1/19/6/20 quadruple mutant would completely recapitulate the twd1 phenotype but the isolation of abcb1/19/6/20, abcb1/19/6, and abcb1/6 mutants was not successful by crossing due to the close proximity of ABCB1 and ABCB6 on chromosome 2. Analysis of abcb1/19/20 did not reveal any visible differences in rosettes or inflorescence from abcb1/19 (Supplementary Figures 7A,B).
Loss of ABCB6 and ABCB20 Accounts for twd1-Like Helical Root Twisting
Previous analyses found that abcb6/20 mutants exhibit minor root elongation defects, but root twisting phenotypes were not reported (Zhang et al., 2018). Consistent with these results, measurement of abcb6/20 double mutants revealed slight reductions in primary root elongation similar to abcb1/19 but not as severe as twd1 (Figure 4A). However, examination of cell files in the distal elongation and maturation zones also revealed extreme twisting of the epidermis in abcb6/20 that resembled twd1 that was not present in abcb1/19 (Figure 4B). To further validate the root twisting phenotype, the previously characterized amiRNA1334 ABCB6/20 knock-down and abcb6-1 abcb20-1 double mutant lines were also examined (Zhang et al., 2018). While amiRNA1334 showed increased cell file irregularity which was consistent with reduced ABCB6 and ABCB20 expression, the abcb6-1 abcb20-1 double mutant displayed the same non-handed cell file twisting as abcb6/20 (Figure 4C). Further loss of abcb20 in addition to abcb1/19 (abcb1/19/20) enhanced root growth compared to abcb1/19 (Supplementary Figures 7C,D) but no cell file twisting was observed. These results demonstrate that cell file twisting only occurs with loss of both abcb6 and abcb20, which is consistent with functional redundancy between these paralogs as described (Zhang et al., 2018). Previous reports have shown that exogenous treatment with NPA inhibits cellular auxin efflux in part by disrupting TWD1 interactions with ABCB auxin transporters (Bailly et al., 2008; Christie et al., 2011) and NPA can suppress cell file twisting in twd1 roots (Wu et al., 2010; Wang et al., 2013). Treatment with 1 μM NPA suppressed cell file twisting in abcb6/20 to angles that were not significantly different from NPA-treated twd1 (Figures 4D,E). Some abcb6/20 roots showed some residual twisting that may reflect minor contributions from ABCB1/19. Quantification of free IAA levels in roots revealed both abcb6/20 and twd1 have significantly reduced auxin levels (Figure 4F). Additionally, twd1 also contained reduced levels of the auxin oxidation catabolite oxindole-3-acetic acid (oxIAA; Figure 4F) suggesting additional auxin homeostasis mechanisms are differentially activated in twd1 and abcb6/20. These results suggest the combined loss of ABCB1, 19, 6, and 20 function contributes to the root elongation defects observed in twd1, and that abcb6 and 20 are the primary contributors to the NPA-sensitive root cell file twisting phenotype.
ABCB11 Does Not Contribute to Long-Distance Auxin Transport Streams
Although combined loss of abcb1/19 and abcb6/20 account for all the major twd1 phenotypes, ABCB11 has also been implicated in auxin transport in inflorescence stems (Kaneda et al., 2011) and is the next highest ABCB transporter expressed in the root after ABCB19, 4, and 1 (Supplementary Figure 3). Tissue-specific expression was confirmed by histochemical analysis of a 2.104 kb ABCB11 promoter fragment fused to β-Glucuronidase (GUS) in Col-0. Strong GUS staining was observed in the root tip and moderate expression observed in the mature root outside of the vasculature (Figures 5A,B). No staining was detected in the hypocotyl, shoot apex, cotyledons, rosette leaves, or apical regions of the inflorescence stem including mature flowers (Figures 5C–F). Occasional staining was observed in young, unopened flowers (Figure 5F). Previously, strong ABCB11 expression associated with the vasculature was reported in the upper 3 cm of the inflorescence stem (Kaneda et al., 2011). However, no observable GUS staining could be detected in this region with the ABCB11 promoter construct used in this study (Figure 5G). The results presented here are more consistent with the expression patterns found in RNA-seq and microarray databases (Zimmermann, 2004; Klepikova et al., 2016).

Figure 5. ABCB11 is primarily expressed in the root and root tip. (A) Strong proABCB11:GUS expression is observed in the root tip. Roots were stained for 1 h. (B) Moderate proABCB11:GUS expression is observed in the epidermis, cortex, and endodermis of the mature root. (C–G) proABCB11:GUS expression is not observed in the (C) hypocotyl, shoot apex, or petioles, (D) cotyledons, (E) rosette leaves, (F) apical regions of the inflorescence stem or mature flowers, or (G) upper 3–4 cm of the inflorescence stem. Occasional staining could be observed in young unopened flowers (E, inset). Stems in (G) were GUS stained prior to hand sectioning. Scale bars: (A,F,G) 500 μm, (B,F) 1 mm, (C,E) 2 mm, (D) 100 μm.
Previous reports suggested that ABCB11 contributes to rootward auxin transport in inflorescence stems (Kaneda et al., 2011). However, according to the Arabidopsis Biological Resource Center (ABRC) the T-DNA insertion line used in those studies (SALK_094249), understandably misreported as an insertion in ABCB11, instead contains an insertion in the adjacent, paralogous, and highly similar gene ABCB12. Therefore, two new alleles of abcb11 (SALK_057628 and SALK_037942), designated abcb11-1 and abcb11-2, were isolated and confirmed to have transcript levels below the limit of detection (Supplementary Figures 8A,B). Additionally, no change in ABCB12 expression was observed in abcb11-1 suggesting ABCB12 does not compensate for loss of ABCB11 function (Supplementary Figure 8C). Based on the lack of compensation and extremely low ABCB12 expression in all tissues (Supplementary Figure 3), ABCB12 was not examined any further.
Phenotypic analysis of abcb11-1 and abcb11-2 mutants revealed slight defects in primary root elongation, but no other phenotypic differences from Col-0 were observed under stable conditions (Figures 6A,B; Table 2). However, abcb11-1 seedlings grown on vertical plates did exhibit increased incidence of root waving and skewing in response to mechanical perturbation or directional light. Root waving and skewing phenotypes are primarily regulated by auxin transport in the root tip (Okada and Shimura, 1990; Mullen et al., 1998; Hu et al., 2021) where ABCB11 is primarily expressed and can be quantified in seedlings grown on vertical media set to varying inclination angles (Okada and Shimura, 1990; Oliva and Dunand, 2007). To quantify the contribution of ABCB11 to root waving seedlings were grown on media set at varying inclination angles. When plates were set to 15 degrees from vertical, abcb11-1 showed increased root waving compared to Col-0, but no significant difference in primary root length (Supplementary Figures 9A–C).
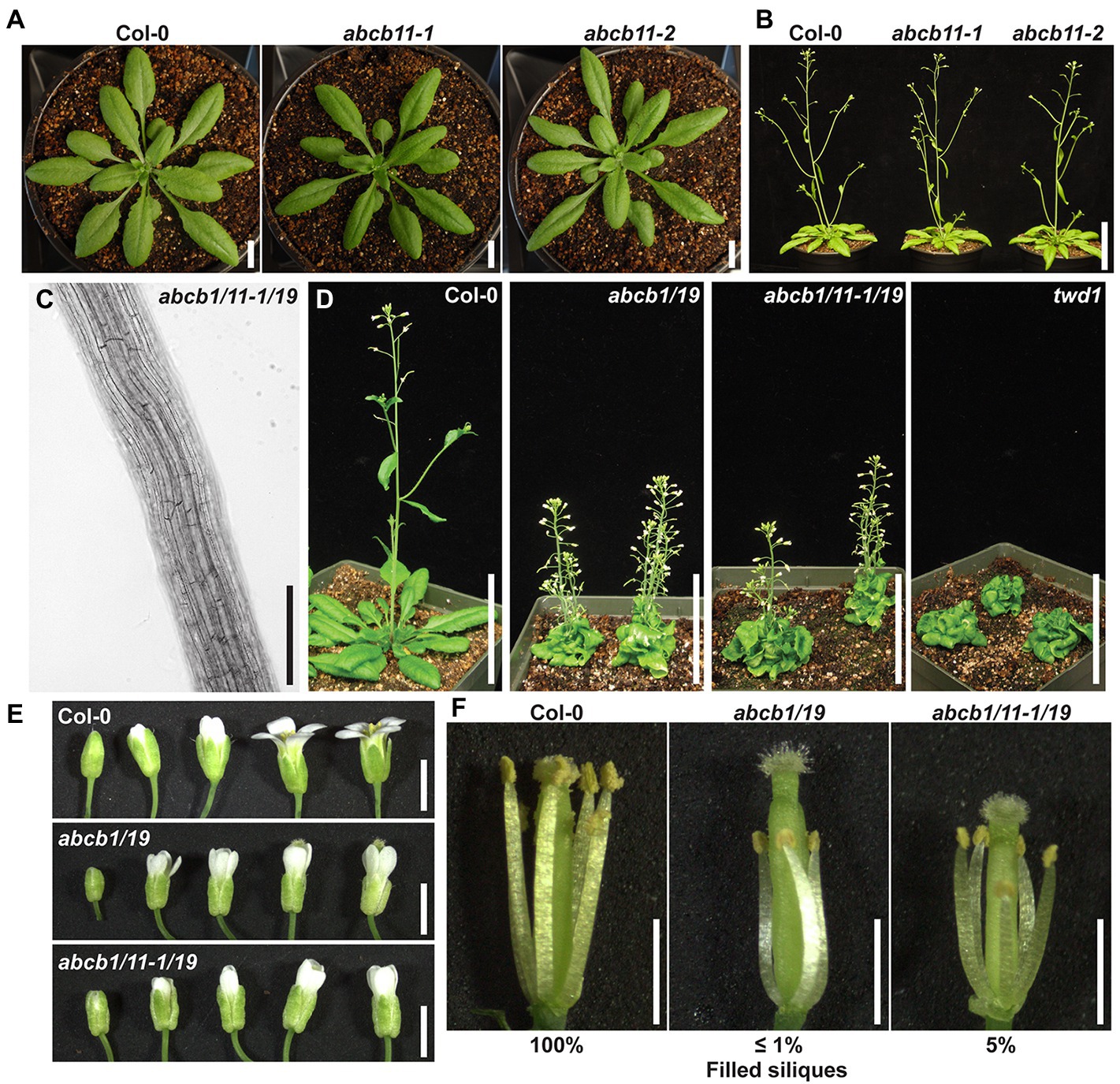
Figure 6. ABCB11 contributes to auxin phenotypes supplementary to ABCB1/19. (A) Col-0 and abcb11 rosettes at time of bolting. (B) Col-0 and abcb11 mature plants. (C) Root cell files of 7-day light-grown abcb1/11-1/19 seedlings. Images were converted to black and white, and brightness and contrast were adjusted to show cell files. (D) 35-day rosettes and inflorescences of Col-0, abcb1/19, abcb1/11-1/19, and twd1. (E) Flower series of Col-0, abcb1/19, and abcb1/11-1/19. (F) Col-0, abcb1/19, and abcb1/11-1/19 flowers with petals removed. Scale bars: (A) 1 cm, (B,C) 5 cm, (D) 200 μm, (E,F) 1 mm.
Further analysis showed abcb1/11/19 triple mutants, generated using abcb11-1, phenotypically resembled abcb1/19 and did not exhibit any root, inflorescence, or silique twisting (Figures 6C,D). In aerial tissues, a slight increase in silique filling from self-pollination was observed in abcb1/11/19 compared to abcb1/19. Decreased auxin transport in abcb1/19 anthers results in short anther filament length and reduced self-pollination (Figures 6E,F; Noh et al., 2001; Cecchetti et al., 2008; Titapiwatanakun et al., 2009). In addition to the short anther phenotype associated with abcb1/19, abcb1/11/19 also exhibited shorter pistils that allowed for a small amount of self-pollination (Figures 6E,F). This resulted in ~5% filled siliques by the time flowering was complete compared to 100% in Col-0 and ≤1% in abcb1/19. This suggests ABCB11 regulates auxin transport required for pistil elongation.
To see if abcb11-1 exhibited defects in rootward auxin transport, inflorescence transport assays were conducted using abcb11-1 according to Kaneda et al. (2011) with some modifications as defined in the Materials and Methods section. Col-0, abcb1, abcb14, abcb19, and pin1 were included as controls. In the apical 2 cm segments of abcb19 and pin1 auxin transport was reduced by ~52% and ~28%, respectively (Figure 7A). In the 4–6 cm segments, transport was reduced in abcb19 and pin1 by ~15% and ~32% (Figure 7A). A slight reduction in transport was observed in abcb14 lower segments (Figure 7A), which is consistent with previous reports (Kaneda et al., 2011). abcb11-1 mutants did not have any differences in auxin transport with either stem segment. No differences were observed when stems were placed upright in the 3H-IAA solution (Figure 7B). These results are consistent with the lack of ABCB11 expression in inflorescence stems (Figure 5G; Supplementary Figure 3; Zimmermann, 2004; Klepikova et al., 2016) and suggest ABCB11 does not participate in auxin transport in these tissues. This function can instead be attributed to ABCB12 as previously described (Kaneda et al., 2011).
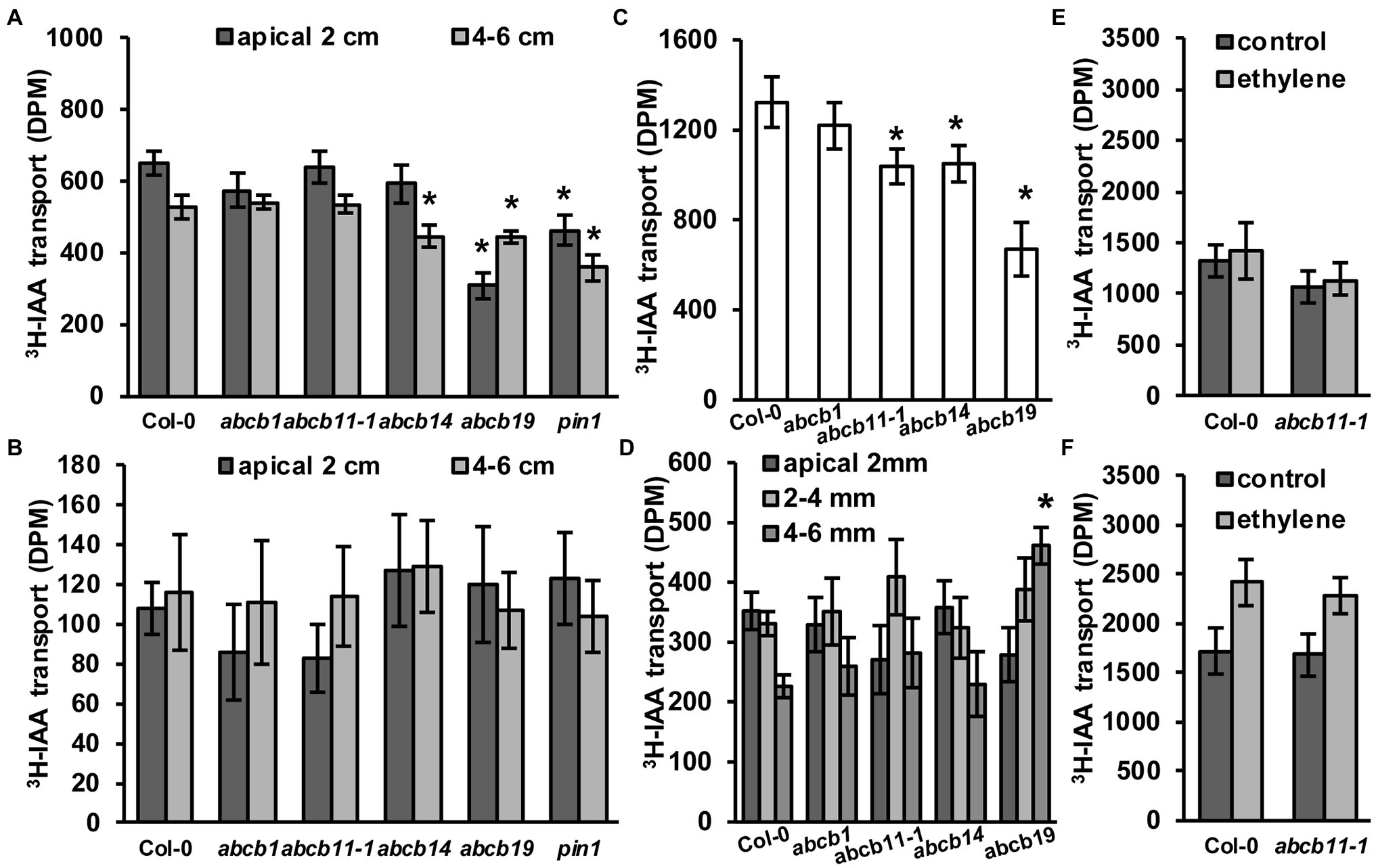
Figure 7. abcb11-1 mutants exhibit minor defects in root auxin transport. (A,B) 3H-IAA transport in (A) upright (rootward/basipetal) and (B) inverted Col-0, abcb1, abcb11, abcb14, abcb19, and pin1 inflorescence stems segments. Assays were conducted as in Kaneda et al. (2011) with modifications as noted in the Materials and Methods section. Data shown are means ± SD (n = 3 pools independent of five segments). (C,D) 3H-IAA transport in Col-0, abcb11, abcb14, and abcb19 seedlings from the shoot apex to (C) the root–shoot transition zone (RSTZ) or (D) the root tip. Data shown are means ± SD (n = 3 independent pools of 10). (E,F) Shootward auxin transport from the root tip with 3H-IAA placed on the (E) columella or (F) quiescent center. Shootward assays were conducted as in Kubeš et al. (2012). Data shown are means ± SD (n = 3 independent pools of 10). Asterisks indicate statistical difference from Col-0 by Student’s t-test (*p < 0.05).
Rootward auxin transport in abcb11-1 seedlings was also analyzed (Figure 7C). The greatest reduction (~43%) was observed in abcb19, while transport in abcb1 and abcb11-1 was reduced ~16% and ~17%, respectively. Auxin transport from the shoot apex to the root–shoot transition zone was significantly reduced in abcb11, abcb14, and abcb19 (Figure 7C). Since ABCB14 was initially identified as a malate/citrate transporter (Lee et al., 2008), auxin transport was also assessed when malate was co-applied to the seedling shoot apex. Competition with a 5:1 molar ratio of malate:3H-IAA reduced auxin transport ~24% in Col-0 (Supplementary Figure 10). Together these results suggested competitive inhibition of other auxin transport components in these respective mutants contribute to rootward auxin flux. Conversely, abcb14 exhibited close to zero reduction. This suggests that competition of 3H-IAA with malate reduces ABCB14 mediated auxin transport. Transport assays with radiolabeled malate were not feasible due to rapid metabolism of applied malate.
As ABCB11 is moderately expressed in the seedling root (Figures 5A,B; Supplementary Figure 3) and slight defects are observed in abcb11-1 and abcb11-2 root growth (Table 2), auxin transport from the RSTZ to the root apex was assayed. Transport in abcb1 and abcb19 was assayed as a positive control, and in abcb14, which is not expressed in the root, as a negative control. Only abcb19 exhibited a reduction in auxin transport to the root apex, but abcb11-1 exhibited a slightly increased accumulation at 2–4 mm from the apex (Figure 7D). These alterations in hypocotyl and root transport in abcb11-1 mutants were reminiscent of the reduced transport in abcb4 hypocotyls due to the reduced auxin sink in the root (Terasaka et al., 2005). This suggested ABCB11 may have some limited function in auxin transport at the root tip where it is primarily expressed (Figure 5A; Supplementary Figure 3). However, shootward transport of auxin from the columella or cells surrounding the quiescent center was not different from Col-0 (Figures 7E,F). Shootward auxin transport from these sites in abcb11-1 in the presence of ethylene was also found to be non-different from Col-0, similar to what was observed in abcb4 (Kubeš et al., 2012).
Since no clear role for ABCB11-mediated auxin transport was found in planta it was hypothesized that ABCB11 may transport alternate substrates. To test this, ABCB11 was analyzed for auxin transport activity when expressed in S. pombe. Cells expressing ABCB11 accumulated ~18% less 3H-IAA after 4 min and ~27% less after 6 min compared to empty vector controls (Figure 8A). However, expression of ABCB11 also reduced 3H-benzoic acid accumulation (Figure 8B). Competition of 3H-IAA with benzoic acid (1:1 molar ratio) reduced 3H-IAA transport to control levels in cells expressing ABCB11 but not ABCB19 (Figure 8C). This is consistent with previous reports showing benzoic acid is not a substrate for ABCB19 (Yang and Murphy, 2009). These results, along with the lack of auxin related phenotypes and lack of consistent auxin transport defects in planta, suggest that ABCB11 has the capacity to transport auxin but likely transports alternate substrates in vivo.
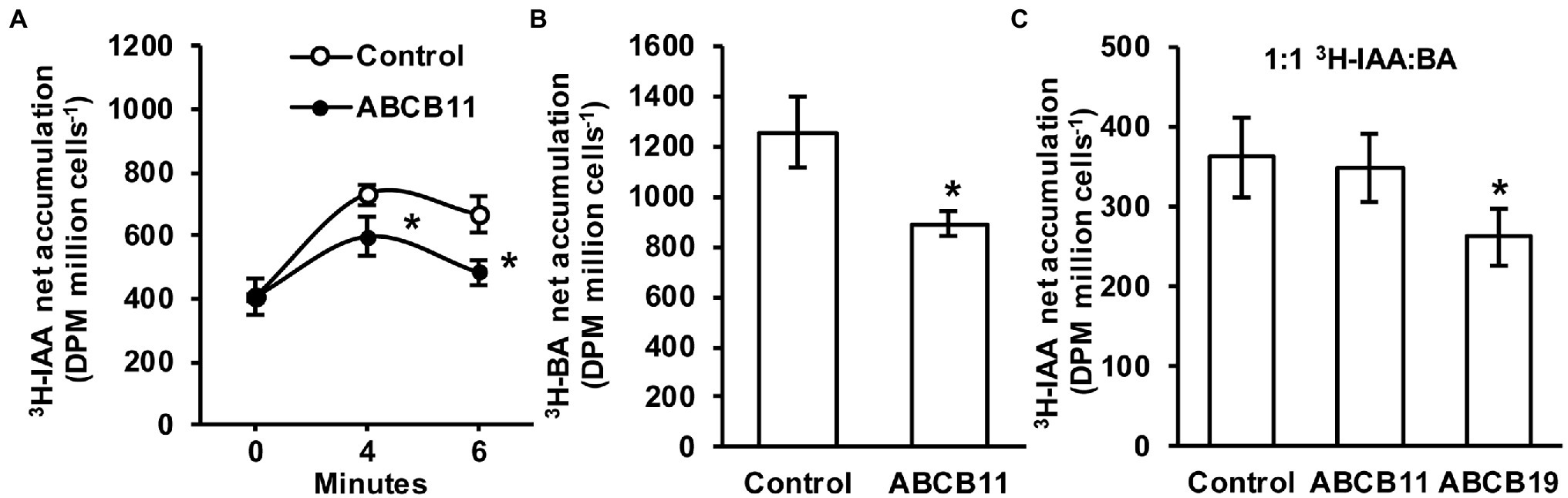
Figure 8. Transport activity of ABCB11 expressed in Schizosaccharomyces pombe. (A) 3H-IAA accumulation in S. pombe expressing ABCB11 or empty vector control plasmid. Data shown are means ± SD (n = 4 independent transformations). (B) 3H-benzoic acid (BA) accumulation in S. pombe expressing ABCB11 or empty vector control plasmid. Data shown are means ± SD (n = 4 independent transformations). (C) Competition of 3H-IAA with cold benzoic acid (BA; 1:1 molar ratio) in S. pombe expressing ABCB11, ABCB19, or empty vector control plasmid. Data shown are means ± SD (n = 4 independent transformations). Asterisks indicate statistical difference by Student’s t-test (*p < 0.05).
Discussion
The twisted dwarf 1 (twd1) and double abcb1/19 mutants exhibit significant overlap in morphology including reduced primary root elongation, severely compact and curled leaves, and dwarfed inflorescence height due to reduced long-distance auxin transport (Geisler et al., 2017). However, twd1 also exhibits twisting roots, inflorescence stems, and siliques that are not observed in abcb1/19. The results presented here suggest that all major twd1 phenotypes are accounted for with the loss of ABCB1, 6, 19, and 20 function and that loss of other well-documented TWD1 interactions with other cellular components provide minor contributions to morphological phenotypes.
Several reports have consistently found that abcb19 and abcb1/19 mutants exhibit ≥50% reductions in polar auxin transport that result in decreased root length, decreased inflorescence height, short anther filaments, and altered leaf morphology (Noh et al., 2001; Geisler et al., 2003, 2005; Blakeslee et al., 2007; Wu et al., 2007; Christie et al., 2011; Sukumar et al., 2013; Zhao et al., 2013; Bennett et al., 2016; Jenness et al., 2019, 2020). The lack of phenotypes associated with abcb1 single mutants is attributed to compensatory expression of ABCB19 (Blakeslee et al., 2007; Jenness et al., 2019). Although organ separation defects have been described in an abcb19-5 allele (Zhao et al., 2013) that are not observed in other abcb19 null mutants, inflorescence and silique twisting has not been reported in abcb1, abcb19, or abcb1/19.
The results presented here suggest the combined loss of ABCB1/19 and ABCB6/20 contribute to the severe dwarf and twisting rosette and inflorescence phenotypes associated with twd1. Rosettes of abcb1/19 were almost indistinguishable from twd1 suggesting ABCB1 and ABCB19 have a primary function in rosette leaf development. abcb6/20 rosettes were also compact, although not to the same extent as abcb1/19 or twd1. While abcb1/19 leaves were more downward curled, abcb6/20 tended to curl and skew laterally. Similar trends were observed in inflorescence stems, where loss of ABCB1/19 and ABCB6/20 was associated with decreased inflorescence height, but helical stem, and silique twisting was only observed in abcb6/20.
In the root, helical twisting has been reported as present (Geisler et al., 2003; Bouchard et al., 2006; Bailly et al., 2008) and missing (Wu et al., 2010) in abcb1/19. These differences have been attributed to differences in alleles and ecotypic backgrounds (Wang et al., 2013). The abcb1/19 double mutants used in this study displayed reduced root elongation with increased root waving and irregular growth, but epidermal cell file twisting was never observed. Instead, abcb6/20 mutants displayed helical non-handed cell file twisting. Treatment with NPA suppressed twisting to a similar extent in abcb6/20 and twd1, and was consistent with previous reports with twd1 (Wu et al., 2010; Wang et al., 2013). Although a subset of ABCBs and TWD1 bind NPA directly (Murphy et al., 2002; Bailly et al., 2008; Kim et al., 2010), NPA rescues twisting in the absence of TWD1 and ABCB6/20 suggesting the rescue is independent of TWD1 and ACTIN-dependent ABCB trafficking. Although the exact mechanism is not known it is likely that ABCB6/20 or TWD1 loss of function leads to helical twisting via ectopic auxin accumulations, differential cell expansion, and geometric constraints as has been described for other twisting mutants (Weizbauer et al., 2011). This twisted growth likely also involves PIN upregulation and PIN-directed transport as overexpression of PINs leads to hypocotyl twisting that can be rescued by NPA treatment (Mravec et al., 2008).
Attempts were made to generate an abcb1/19/6/20 quadruple mutant as it would be expected to fully resemble twd1. However, no combinations of abcb1/6 were isolated presumably due to infrequent recombination between ABCB1 and ABCB6, which are separated by only 983,313 nucleotides on chromosome 2. Analysis of abcb1/19/20 mutants did not reveal any additional phenotypes to abcb1/19 double mutants, which is consistent with the functional redundancy of ABCB6 and 20 reported here and previously (Zhang et al., 2018). Generation of the abcb1/19/6/20 quadruple mutant will likely require use of a CRISPR-CasX approach of extraordinary finesse, as attempts to generate such mutants so far have inactivated a larger number of ABCB genes.
Phenotypic characterizations provide little insight into ABCB11 function. ABCB11 expression is primarily limited to the root, however, and no major differences in auxin transport in the root were observed in abcb11 mutants. In flowers, combined loss of ABCB1, 11, and 19 contributed to reduce pistil elongation, suggesting ABCB11 may contribute to auxin transport supplementary to ABCB1 and 19 in this tissue. ABCB11 is also expressed in anthers (Klepikova et al., 2016), however, loss of ABCB11 did not enhance the filament elongation defects associated with abcb1/19, again pointing to a supplementary role in auxin transport. These results support a role for TWD1 in regulating auxin-dependent floral organ elongation (Pérez-Pérez et al., 2004) via ABCB1, 11, and 19. Additional roles for TWD1 in regulating BRI1 and brassinosteroid-dependent pollen development are likely (Vogler et al., 2014; Zhao et al., 2016). When expressed in S. pombe, ABCB11 does increase auxin export, but this activity was competitively eliminated by benzoic acid with relative ease. This suggests ABCB11 may function in transport of additional amphipathic or aromatic organic acid substrates in planta. Less specific auxin transport activity is observed with the malate/citrate transporter ABCB14 (Lee et al., 2008), which appears to mediate transport of exogenously applied auxin (Kaneda et al., 2011), but exhibits preferential transport of simple organic acids (Supplementary Figure 10; Lee et al., 2008; Yang and Murphy, 2009). Since endogenous malate/citrate levels are several fold higher than auxin, ABCB14 contribution to auxin transport is expected to be minimal.
The exact mechanism by which such a small amount of TWD1 maintains the trafficking and activity of this relatively abundant group of ABCB transporters is still unclear, but could involve its role in mediating actin bundling (Zhu et al., 2016). Emerging methods including single molecule interaction visualizations and new tools to detect ABC transporter function in real time may be able to provide further insight in the future.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
MJ and AM planned and designed the research and wrote the manuscript with input from all authors. MJ, RT, GB, WT, YZ, CP, and AM performed the experiments. MJ, RT, WT, and AM analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Department of Energy, Basic Energy Sciences, grant no. DE-FG02-06ER15804 to AM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Markus Geisler, Wendy Peer, Burkhard Schulz, Haibing Yang, and Joshua Blakeslee for ongoing discussions of TWD1-ABCB interactions. Jesica Reemmer is thanked for generating preliminary analyses of ABCB11 function. Eilon Shani is also thanked for ABCB6/20 discussions and help to validate mutant phenotypes.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.840260/full#supplementary-material
References
Alexandersson, E., Saalbach, G., Larsson, C., and Kjellbom, P. (2004). Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 45, 1543–1556. doi: 10.1093/pcp/pch209
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. doi: 10.1093/nar/gkw408
Bailly, A., Sovero, V., Vincenzetti, V., Santelia, D., Bartnik, D., Koenig, B. W., et al. (2008). Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 283, 21817–21826. doi: 10.1074/jbc.M709655200
Bailly, A., Wang, B., Zwiewka, M., Pollmann, S., Schenck, D., Lüthen, H., et al. (2013). Expression of TWISTED DWARF1 lacking its in-plane membrane anchor leads to increased cell elongation and hypermorphic growth. Plant J. 77, 108–118. doi: 10.1111/tpj.12369
Bailly, A., Yang, H., Martinoia, E., Geisler, M., and Murphy, A. S. (2012). Plant lessons: exploring ABCB functionality through structural modeling. Front. Plant Sci. 2:108. doi: 10.3389/fpls.2011.00108
Banasavadi-Siddegowda, Y. K., Mai, J., Fan, Y., Bhattacharya, S., Giovannucci, D. R., Sanchez, E. R., et al. (2011). FKBP38 peptidylprolyl isomerase promotes the folding of cystic fibrosis transmembrane conductance regulator in the endoplasmic reticulum. J. Biol. Chem. 286, 43071–43080. doi: 10.1074/jbc.M111.269993
Banasiak, J., and Jasiński, M. (2021). ATP-binding cassette transporters in nonmodel plants. New Phytol. 233, 1597–1612. doi: 10.1111/nph.17779
Bennett, T., Hines, G., van Rongen, M., Waldie, T., Sawchuk, M. G., Scarpella, E., et al. (2016). Connective auxin transport in the shoot facilitates communication between shoot apices. PLoS Biol. 14:e1002446. doi: 10.1371/journal.pbio.1002446
Bernasconi, P., Patel, B. C., Reagan, J. D., and Subramanian, M. V. (1996). The N-1-naphthylphthalamic acid-binding protein is an integral membrane protein. Plant Physiol. 111, 427–432. doi: 10.1104/pp.111.2.427
Blakeslee, J. J., Bandyopadhyay, A., Ok, R. L., Mravec, J., Titapiwatanakun, B., Sauer, M., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19, 131–147. doi: 10.1105/tpc.106.040782
Borner, G. H. H., Sherrier, D. J., Weimar, T., Michaelson, L. V., Hawkins, N. D., MacAskill, A., et al. (2005). Analysis of detergent-resistant membranes in arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116. doi: 10.1104/pp.104.053041
Bouchard, R., Bailly, A., Blakeslee, J. J., Oehring, S. C., Vincenzetti, V., Lee, O. R., et al. (2006). Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 281, 30603–30612. doi: 10.1074/jbc.M604604200
Butler, J. H., Hu, S., Brady, S. R., Dixon, M. W., and Muday, G. K. (1998). In vitro and in vivo evidence for actin association of the naphthylphthalamic acid-binding protein from zucchini hypocotyls. Plant J. 13, 291–301. doi: 10.1046/j.1365-313X.1998.00017.x
Cecchetti, V., Altamura, M. M., Falasca, G., Costantino, P., and Cardarelli, M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20, 1760–1774. doi: 10.1105/tpc.107.057570
Cecchetti, V., Brunetti, P., Napoli, N., Fattorini, L., Altamura, M. M., Costantino, P., et al. (2015). ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. J. Integr. Plant Biol. 57, 1089–1098. doi: 10.1111/jipb.12332
Chaiwanon, J., Garcia, V. J., Cartwright, H., Sun, Y., and Wang, Z. Y. (2016). Immunophilin-like FKBP42/TWISTED DWARF1 interacts with the receptor kinase BRI1 to regulate brassinosteroid signaling in Arabidopsis. Mol. Plant 9, 593–600. doi: 10.1016/j.molp.2016.01.008
Cho, M., Sang, H. L., and Cho, H. T. (2007). P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19, 3930–3943. doi: 10.1105/tpc.107.054288
Christie, J. M., Yang, H., Richter, G. L., Sullivan, S., Thomson, C. E., Lin, J., et al. (2011). Phot1 inhibition of ABCB19 primes lateral Auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 9:e1001076. doi: 10.1371/journal.pbio.1001076
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Demir, F., Horntrich, C., Blachutzik, J. O., Scherzer, S., Reinders, Y., Kierszniowska, S., et al. (2013). Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. U. S. A. 110, 8296–8301. doi: 10.1073/pnas.1211667110
Deslauriers, S. D., and Spalding, E. P. (2021). Electrophysiological study of Arabidopsis ABCB4 and PIN2 auxin transporters: evidence of auxin activation and interaction enhancing auxin selectivity. Plant Direct 5:e361. doi: 10.1002/pld3.361
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030. doi: 10.1242/dev.01388
Geisler, M., Aryal, B., di Donato, M., and Hao, P. (2017). A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 58, 1601–1604. doi: 10.1093/pcp/pcx104
Geisler, M., and Bailly, A. (2007). Tête-à-tête: the function of FKBPs in plant development. Trends Plant Sci. 12, 465–473. doi: 10.1016/j.tplants.2007.08.015
Geisler, M., Bailly, A., and Ivanchenko, M. (2016). Master and servant: regulation of auxin transporters by FKBPs and cyclophilins. Plant Sci. 245, 1–10. doi: 10.1016/j.plantsci.2015.12.004
Geisler, M., Blakeslee, J. J., Bouchard, R., Lee, O. R., Vincenzetti, V., Bandyopadhyay, A., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44, 179–194. doi: 10.1111/j.1365-313X.2005.02519.x
Geisler, M., Girin, M., Brandt, S., Vincenzetti, V., Plaza, S., Paris, N., et al. (2004). Arabidopsis immunophilin-like TWD1 functionally interacts with vacuolar ABC transporters. Mol. Biol. Cell 15, 3393–3405. doi: 10.1091/mbc.e03-11-0831
Geisler, M., Kolukisaoglu, H. U., Bouchard, R., Billion, K., Berger, J., Saal, B., et al. (2003). TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14, 4238–4249. doi: 10.1091/mbc.e02-10-0698
Gookin, T. E., and Assmann, S. M. (2021). Cantil: A previously unreported organ in wild-type Arabidopsis regulated by FT, ERECTA and heterotrimeric G proteins. Development 148:dev195545. doi: 10.1242/dev.195545
Hao, P., Xia, J., Liu, J., di Donato, M., Pakula, K., Bailly, A., et al. (2020). Auxin-transporting ABC transporters are defined by a conserved D/E-P motif regulated by a prolylisomerase. J. Biol. Chem. 295, 13094–13106. doi: 10.1074/jbc.RA120.014104
Henrichs, S., Wang, B., Fukao, Y., Zhu, J., Charrier, L., Bailly, A., et al. (2012). Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J. 31, 2965–2980. doi: 10.1038/emboj.2012.120
Hu, Y., Omary, M., Hu, Y., Doron, O., Hoermayer, L., Chen, Q., et al. (2021). Cell kinetics of auxin transport and activity in Arabidopsis root growth and skewing. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-021-21802-3
Hutt, D. M., Roth, D. M., Chalfant, M. A., Youker, R. T., Matteson, J., Brodsky, J. L., et al. (2012). FK506 binding protein 8 peptidylprolyl isomerase activity manages a late stage of cystic fibrosis transmembrane conductance regulator (CFTR) folding and stability. J. Biol. Chem. 287, 21914–21925. doi: 10.1074/jbc.M112.339788
Jenness, M. K., Carraro, N., Pritchard, C. A., and Murphy, A. S. (2019). The arabidopsis ATP-BINDING CASSETTE transporter ABCB21 regulates auxin levels in cotyledons, the root pericycle, and leaves. Front. Plant Sci. 10:806. doi: 10.3389/fpls.2019.00806
Jenness, M. K., Tayengwa, R., and Murphy, A. S. (2020). An ATP-binding cassette transporter, ABCB19, regulates leaf position and morphology during phototropin1-mediated blue light responses. Plant Physiol. 184, 1601–1612. doi: 10.1104/pp.20.00223
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kamimoto, Y., Terasaka, K., Hamamoto, M., Takanashi, K., Fukuda, S., Shitan, N., et al. (2012). Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol. 53, 2090–2100. doi: 10.1093/pcp/pcs149
Kaneda, M., Schuetz, M., Lin, B. S. P., Chanis, C., Hamberger, B., Western, T. L., et al. (2011). ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 62, 2063–2077. doi: 10.1093/jxb/erq416
Kang, J., Park, J., Choi, H., Burla, B., Kretzschmar, T., Lee, Y., et al. (2011). Plant ABC transporters. Am. Soc. Plant Biol. 9:e0153. doi: 10.1199/tab.0153
Kim, J.-Y., Henrichs, S., Bailly, A., Vincenzetti, V., Sovero, V., Mancuso, S., et al. (2010). Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. J. Biol. Chem. 285, 23309–23317. doi: 10.1074/jbc.M110.105981
Klepikova, A. V., Kasianov, A. S., Gerasimov, E. S., Logacheva, M. D., and Penin, A. A. (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070. doi: 10.1111/tpj.13312
Knöller, A. S., Blakeslee, J. J., Richards, E. L., Peer, W. A., and Murphy, A. S. (2010). Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J. Exp. Bot. 61, 3689–3696. doi: 10.1093/jxb/erq180
Kubeš, M., Yang, H., Richter, G. L., Cheng, Y., Młodzińska, E., Wang, X., et al. (2012). The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 69, 640–654. doi: 10.1111/j.1365-313X.2011.04818.x
Lee, M., Choi, Y., Burla, B., Kim, Y.-Y., Jeon, B., Maeshima, M., et al. (2008). The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 10, 1217–1223. doi: 10.1038/ncb1782
Lin, R., and Wang, H. (2005). Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 138, 949–964. doi: 10.1104/pp.105.061572
Mravec, J., Kubes, M., Bielach, A., Gaykova, V., Petrasek, J., Skupa, P., et al. (2008). Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135, 3345–3354. doi: 10.1242/dev.021071
Mullen, J. L., Turk, E., Johnson, K., Wolverton, C., Ishikawa, H., Simmons, C., et al. (1998). Root-growth behavior of the Arabidopsis mutant rgr: roles of gravitropism and circumnutation in the waving/coiling phenomenon. Plant Physiol. 118, 1139–1145. doi: 10.1104/pp.118.4.1139
Murphy, A. S., Hoogner, K. R., Peer, W. A., and Taiz, L. (2002). Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 128, 935–950. doi: 10.1104/pp.010519
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Noh, B., Murphy, A. S., and Spalding, E. P. (2001). Multidrug resistance-like genes of arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13, 2441–2454. doi: 10.1105/tpc.010350
Novák, O., Hényková, E., Sairanen, I., Kowalczyk, M., Pospíšil, T., and Ljung, K. (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72, 523–536. doi: 10.1111/j.1365-313X.2012.05085.x
Okada, K., and Shimura, Y. (1990). Reversible root tip rotation in arabidopsis seedlings induced by obstacle-touching stimulus. Science 250, 274–276. doi: 10.1126/science.250.4978.274
Oliva, M., and Dunand, C. (2007). Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytol. 176, 37–43. doi: 10.1111/j.1469-8137.2007.02184.x
Peer, W. A., and Murphy, A. S. (2007). Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12, 556–563. doi: 10.1016/j.tplants.2007.10.003
Pérez-Pérez, J. M., Ponce, M. R., and Micol, J. L. (2002). The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242, 161–173. doi: 10.1006/dbio.2001.0543
Pérez-Pérez, J. M., Ponce, M. R., and Micol, J. L. (2004). The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol. 134, 101–117. doi: 10.1104/pp.103.032524
Petrášek, J., Mravec, J., Bouchard, R., Blakeslee, J. J., Abas, M., Seifertová, D., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918. doi: 10.1126/science.1123542
Santelia, D., Vincenzetti, V., Azzarello, E., Bovet, L., Fukao, Y., Düchtig, P., et al. (2005). MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 579, 5399–5406. doi: 10.1016/j.febslet.2005.08.061
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Song, W. Y., Park, J., Mendoza-Cózatl, D. G., Suter-Grotemeyer, M., Shima, D., Hörtensteiner, S., et al. (2010). Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. U. S. A. 107, 21187–21192. doi: 10.1073/pnas.1013964107
Sukumar, P., Maloney, G. S., and Muday, G. K. (2013). Localized induction of the ATP-binding cassette B19 Auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 162, 1392–1405. doi: 10.1104/pp.113.217174
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Terasaka, K., Blakeslee, J. J., Titapiwatanakun, B., Peer, W. A., Bandyopadhyay, A., Makam, S. N., et al. (2005). PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17, 2922–2939. doi: 10.1105/tpc.105.035816
Titapiwatanakun, B., Blakeslee, J. J., Bandyopadhyay, A., Yang, H., Mravec, J., Sauer, M., et al. (2009). ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 57, 27–44. doi: 10.1111/j.1365-313X.2008.03668.x
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Verrier, P. J., Bird, D., Burla, B., Dassa, E., Forestier, C., Geisler, M., et al. (2008). Plant ABC proteins--a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. doi: 10.1016/j.tplants.2008.02.001
Vogler, F., Schmalzl, C., Englhart, M., Bircheneder, M., and Sprunck, S. (2014). Brassinosteroids promote Arabidopsis pollen germination and growth. Plant Reprod. 27, 153–167. doi: 10.1007/s00497-014-0247-x
Wang, B., Bailly, A., Zwiewka, M., Henrichs, S., Azzarello, E., Mancuso, S., et al. (2013). Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell 25, 202–214. doi: 10.1105/tpc.112.105999
Weizbauer, R., Peters, W. S., and Schulz, B. (2011). Geometric constraints and the anatomical interpretation of twisted plant organ phenotypes. Front. Plant Sci. 2:62. doi: 10.3389/fpls.2011.00062
Wu, G., Lewis, D. R., and Spalding, E. P. (2007). Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19, 1826–1837. doi: 10.1105/tpc.106.048777
Wu, G., Otegui, M. S., and Spalding, E. P. (2010). The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell 22, 3295–3304. doi: 10.1105/tpc.110.078360
Yang, H., and Murphy, A. S. (2009). Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 59, 179–191. doi: 10.1111/j.1365-313X.2009.03856.x
Yang, H., Richter, G. L., Wang, X., Młodzińska, E., Carraro, N., Ma, G., et al. (2013). Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J. 74, 37–47. doi: 10.1111/tpj.12103
Zhang, Y., Nasser, V., Pisanty, O., Omary, M., Wulff, N., di Donato, M., et al. (2018). A transportome-scale amiRNA-based screen identifies redundant roles of Arabidopsis ABCB6 and ABCB20 in auxin transport. Nat. Commun. 9:4204. doi: 10.1038/s41467-018-06410-y
Zhao, Z., Chai, M., Sun, L., Cong, L., Jiang, Q., Zhang, Z., et al. (2021). Identification of a gene responsible for seedpod spine formation and other phenotypic alterations using whole-genome sequencing analysis in Medicago truncatula. J. Exp. Bot. doi: 10.1093/jxb/erab359
Zhao, H., Liu, L., Mo, H., Qian, L., Cao, Y., Cui, S., et al. (2013). The ATP-binding cassette transporter ABCB19 regulates postembryonic organ separation in Arabidopsis. PLoS One 8:e60809. doi: 10.1371/journal.pone.0060809
Zhao, B., Lv, M., Feng, Z., Campbell, T., Liscum, E., and Li, J. (2016). TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol. Plant 9, 582–592. doi: 10.1016/j.molp.2016.01.007
Zhu, J., Bailly, A., Zwiewka, M., Sovero, V., di Donato, M., Ge, P., et al. (2016). TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 28, 930–948. doi: 10.1105/tpc.15.00726
Keywords: ABCB transporter, auxin, organ twisting, FKBP42/TWD1, Arabidopsis thaliana
Citation: Jenness MK, Tayengwa R, Bate GA, Tapken W, Zhang Y, Pang C and Murphy AS (2022) Loss of Multiple ABCB Auxin Transporters Recapitulates the Major twisted dwarf 1 Phenotypes in Arabidopsis thaliana. Front. Plant Sci. 13:840260. doi: 10.3389/fpls.2022.840260
Edited by:
Junpei Takano, Osaka Prefecture University, JapanReviewed by:
Takashi Hashimoto, Nara Institute of Science and Technology (NAIST), JapanSatoshi Naramoto, Hokkaido University, Japan
Copyright © 2022 Jenness, Tayengwa, Bate, Tapken, Zhang, Pang and Murphy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angus S. Murphy, YXNtdXJwaHlAdW1kLmVkdQ==
 Mark K. Jenness
Mark K. Jenness Reuben Tayengwa
Reuben Tayengwa Gabrielle A. Bate
Gabrielle A. Bate Wiebke Tapken1
Wiebke Tapken1 Yuqin Zhang
Yuqin Zhang Changxu Pang
Changxu Pang Angus S. Murphy
Angus S. Murphy