- 1School of Molecular Sciences, The University of Western Australia, Perth, WA, Australia
- 2Australian Research Council (ARC) Centre of Excellence in Plant Energy Biology, The University of Western Australia, Perth, WA, Australia
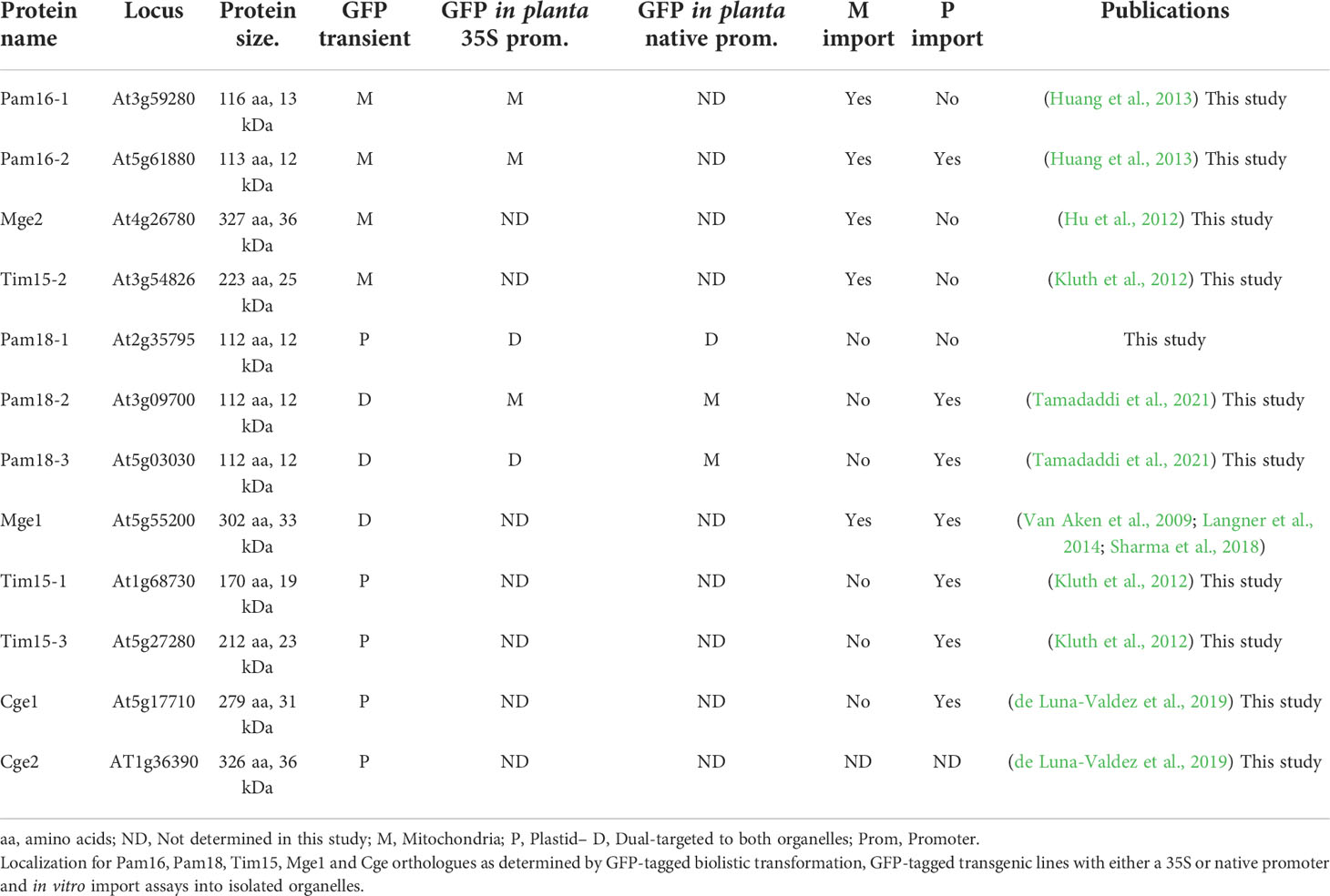
The import and assembly of most of the mitochondrial proteome is regulated by protein translocases located within the mitochondrial membranes. The Presequence Translocase-Associated Motor (PAM) complex powers the translocation of proteins across the inner membrane and consists of Hsp70, the J-domain containing co-chaperones, Pam16 and Pam18, and their associated proteins Tim15 and Mge1. In Arabidopsis, multiple orthologues of Pam16, Pam18, Tim15 and Mge1 have been identified and a mitochondrial localization has been confirmed for most. As the localization of Pam18-1 has yet to be determined and a plastid localization has been observed for homologues of Tim15 and Mge1, we carried out a comprehensive targeting analysis of all PAM complex orthologues using multiple in vitro and in vivo methods. We found that, Pam16 was exclusively targeted to the mitochondria, but Pam18 orthologues could be targeted to both the mitochondria and plastids, as observed for the PAM complex interacting partner proteins Tim15 and Mge1.
Introduction
Mitochondria and plastids evolved from two independent endosymbiotic events, approximately 2 and 1 billion years ago respectively (McFadden, 1999). Over the course of evolution, most of their genomes was lost, requiring organelle-specific protein import components and mechanisms to function. Protein import involves a multitude of chaperones, receptors, transporters and proteases that deliver nuclear encoded cytosolic proteins to the organelle, translocate them through membranes, process and sort the protein to its final intra-organellar destination (Ghifari et al., 2018). For most of the mitochondrial proteome, an N-terminal targeting signal delivers the precursor protein to the mitochondria, allowing it to translocate across the Translocase of the Outer Membrane (TOM) complex pore, further directing it to the TIM17:23 inner membrane complex. This complex consists of the inner membrane spanning channel proteins Tim17 and Tim23, and associated proteins such as Tim21 and Tim50 that stabilize the complex and the Presequence Assisted Motor (PAM) complex that powers the translocation of preproteins through the inner membrane channel (Neupert and Herrmann, 2007). In yeast, the PAM complex consists of mtHsp70, Tim44, Tim15, Pam18, Pam16, Pam17, and Mge1. MtHsp70 is the central motor that interacts with the precursor protein passing through TIM17:23 channel and powers translocation of proteins into the matrix by ATP hydrolysis requiring co-chaperones and nucleotide exchange factors (Kampinga and Craig, 2010). The co-chaperone Pam18, stimulates the ATPase activity of mtHsp70 (Truscott et al., 2003), whilst Pam16 tethers Pam18 to the PAM complex (Pais et al., 2011) and Pam17 stabilizes the complex (Sanjuán Szklarz et al., 2005; van der Laan et al., 2005). Mge1, catalyzes the exchange of ADP for ATP allowing release and reload of the substrate (Liu et al., 2003) (Figure 1). Tim44 acts as a scaffold protein to recruit mtHsp70 (De Los Rios et al., 2006; Schiller et al., 2008) whilst Tim15, (also named Zim17, ZR, HEP1) is thought to stabilize this complex (Sanjuán Szklarz et al., 2005; van der Laan et al., 2005).
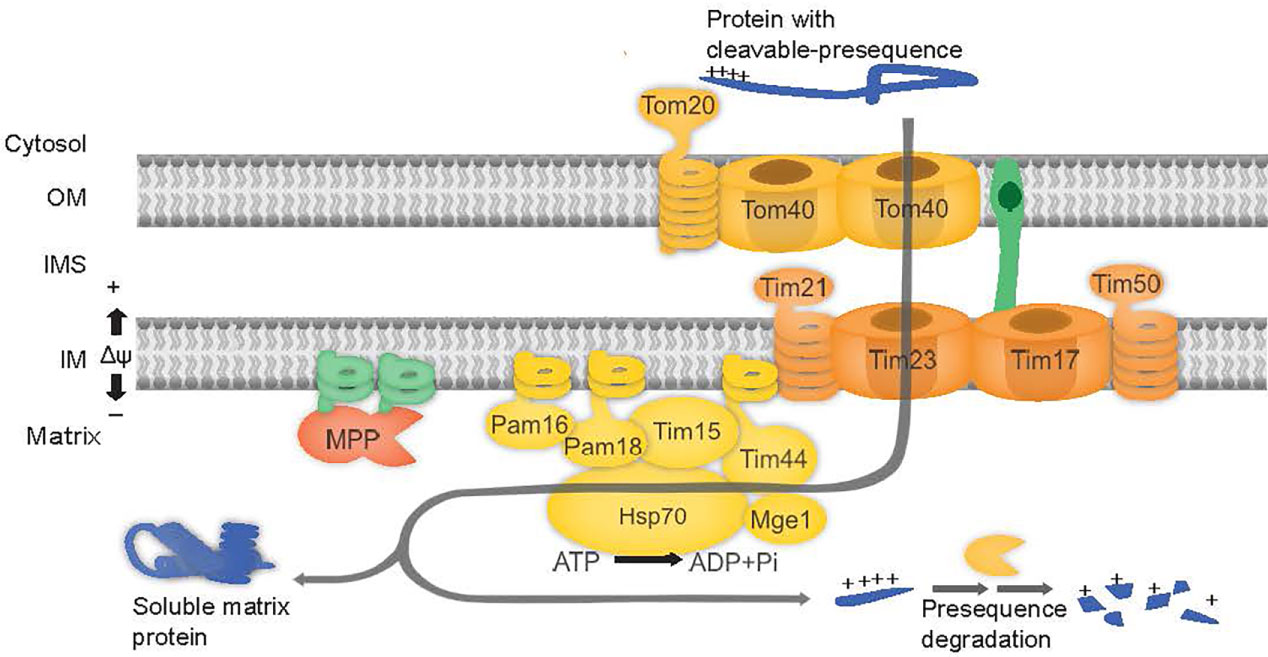
Figure 1 PAM complex subunits are involved in mitochondrial protein import. The preprotein binds to the outer membrane (OM) at the TOM complex and is translocated across Tom40. The preprotein is directed to the TIM17:23 complex, consisting of proteins Tim23, Tim17, Tim21, and Tim50. Release of proteins into matrix is powered by ATP hydrolysis via Hsp70 in the PAM complex consisting of proteins Tim44, Tim15, Pam16 and Pam18, Mge1 and Hsp70.
Pam18 and Pam16 belong to the DnaJ and DnaJ-like family of proteins respectively (Hennessy et al., 2005). J-domain proteins contain a canonical tripeptide HPD motif of Histidine (H), Proline (P), and Aspartate (D) residues essential for stimulating the ATPase activity of Hsp70 (Truscott et al., 2003; Hennessy et al., 2005). Pam16, lacking the HPD motif forms a heterodimer with Pam18 and in doing so, stimulates the activity of Pam18 (Liu et al., 2004). Both Pam16 and Pam18 areessential for yeast viability (Frazier et al., 2003; Truscott et al., 2003) and are anchored to the inner membrane via an N-terminal transmembrane domain (Mokranjac et al., 2006) (Figure 1).
The Arabidopsis thaliana (Arabidopsis) genome encodes for three putative homologues of Pam18 (Pam18-1, Pam18-2 and Pam18-3) and two of Pam16 (Pam16-1 and Pam16-2), whilst no Pam17 orthologue could be identified by sequence homology (Murcha et al., 2015). Pam16-1 was initially characterized by a forward genetic screen mutant, named muse5-1, which displays delayed growth and an enhanced stress response (Huang et al., 2013). A T-DNA insertion line for Pam16-2 was also characterized, which did not display any observable phenotype, and the double knock-out line muse5-1::pam16-2 was embryonically lethal suggesting functional overlap (Huang et al., 2013). Localization using biolistic transformation of GFP tagged Pam16-1 and Pam16-2 into onion epidermal cells identified both proteins to be targeted to mitochondria (Huang et al., 2013; Tamadaddi et al., 2021). GFP tagged constructs of Pam18-2 and Pam18-2 also identified both proteins to be targeted to the mitochondria in root cells of stable GFP fusion lines, whilst no GFP fluorescence targeting could be observed for Pam18-1 (Tamadaddi et al., 2021). Three genes encode for Tim15, Tim15-2 (also known as ZR3) shown to be localized to the mitochondria, is able to functionally complement a Tim15 yeast deletion strain (Kluth et al., 2012). Tim15-1/ZR1 and Tim15-3/ZR2 were shown to targeted to the chloroplasts (Kluth et al., 2012). In plants, two homologues exist for Mge1, Mge1 and Mge2 both shown to be mitochondrial proteins by GFP-tagged localization assays and proteomic analysis (Hu et al., 2012). Interestingly, Mge1 was shown to be dual-targeted to mitochondria and plastids in Arabidopsis cell suspensions and by protein uptake assays into isolated organelles (Van Aken et al., 2009; Langner et al., 2014). The undetermined localization of Pam18-1, the dual-targeting of Mge1 and the plastid localization of Tim15-1/3 proposes that PAM subunits may play a role in plastids. Here, we reexamined the targeting ability of all PAM complex homologues, Pam16-1, Pam16-2, Pam18-1, Pam18-2, Pam18-3, Tim15-1, Tim15-2, Tim15-3, Mge1 and Mge2 using GFP-tagged biolistic transformation stable transgenic GFP fusion lines and in vitro protein uptake assays into isolated organelles. We confirmed that Pam16-1 and Pam16-2 were exclusively targeted to the mitochondria, whilst Pam18-1, Pam18-2 and Pam18-3 could be dual-targeted to both the mitochondria and plastid, depending on the methodology used and tissue types investigated which suggests a possible role of the Pam18 co-chaperones in plastids.
Materials and methods
Bioinformatic analysis
The sequences for all Arabidopsis orthologues were obtained from TAIR (https://www.arabidopsis.org/). Homologues from Homo sapiens, Caenorhabditis elegans, Cyanobacteria, Oryza sativa, Zea mays, and Selaginella moellendorffii were obtained by BLAST in NCBI using standard parameters (https://www.ncbi.nlm.nih.gov/) (Supplementary Table 1). Protein sequences were aligned using ClustalW in MEGA7 (Kumar et al., 2016). Phylogenetic trees were constructed using the Maximum likelihood method based on the Jones-Taylor-Thornton (JTT) model and bootstrap consensus was inferred from 1000 replicates. Domains and motifs were analysed using InterPro (https://www.ebi.ac.uk/interpro/) and ScanProsite (https://prosite.expasy.org/scanprosite/) (Supplementary Table 1). Tertiary structures were predicted using Phyre2 (Kelley et al., 2015) and overlapped using Maestro (www.schrodinger.com).
Cloning and constructing of vectors
All clones were amplified from Arabidopsis cDNA with or without a stop codon using primers listed in Supplementary Table 2 and recombined into pDONR201 or pDONR207 using Gateway® Technology (Invitrogen). For GFP-tagged protein localization, cDNA clones were recombined into pDEST-cGFP (Carrie et al., 2007), for protein translation into pDEST14, for stable GFP fusion lines pGWB5 (Nakagawa et al., 2009). For radiolabelled protein translation, Pam16-1, Pam16-2, Pam18-1, Pam18-2, and Tim15-1 required additional methionines for visualization which were introduced to the N-terminus by PCR orusing the Agilent QuickChange II XL Site Directed Mutagenesis Kit with primers listed in Supplementary Table 2. Mge1 and Cge2 cDNA clones were both obtained from ABRC.
Plant material and growth conditions
Arabidopsis cv Columbia (Col-0) plants were grown on half strength Murashige & Skoog (0.5 X MS) media agar plates or water culture pots containing 1.5% (w/v) sucrose under long day (16 h day cycle) growth conditions, with a light intensity of 100 µE m-2 s-1 at 22°C.
Biolistic transformation
GFP plasmids were biolistically transformed into 5-day old Arabidopsis PSBD wild-type cell culture (Menges and Murray, 2002) with either a mitochondrial RFP marker (AOX-RFP), or a plastid RFP marker (SSU-RFP) (Carrie et al., 2010) as described previously (Duncan et al., 2015). Biolistic transformation was carried out using the Bio-Rad Model PDS-1000/He Biolistic Particle Delivery System at a helium pressure of 1400 kPa, according to the manufacturer’s instructions. Transformed cells were placed in the dark at 26°C overnight prior to imaging. GFP and RFP fluorescence was visualized at 460-480 nm and 570-625 nm respectively using the BX61 Olympus confocal microscope. Images were captured using 100X magnification.
Generation of stable GFP lines
Stable transgenic C-terminal GFP fusions were created by Agrobacterium floral dip as described previously (Clough and Bent, 1998). For confirming plastid localisation, GFP fusion lines were crossed to a SSU-RFP stable transgenic line (Carrie et al., 2007). Root, guard, and epidermal cells were imaged as above.
Organelle isolation
Mitochondria were isolated from 14-day old Arabidopsis seedlings grown in 0.5 X MS liquid media as previously described (Duncan et al., 2015). Plastids were isolated from 14-day old seedlings grown on 0.5X MS plates as previously described (Aronsson and Jarvis, 2002)
Protein uptake assays
[35S]-methionine radiolabelled proteins were translated using the TNT® Coupled Reticulocyte system (Promega) according to manufacturer’s instructions. Protein uptake assays were carried out into either freshly isolated mitochondria or freshly isolated plastids as previously described (Duncan et al., 2015). To confirm the intra-mitochondrial localization of imported protein, the outer membrane was ruptured by osmotic shock following import and prior to the addition of Proteinase K (PK) as detailed previously (Duncan et al., 2015). Plastid protein uptake assays were carried out by incubating precursor protein with isolated plastids (30 ug) in 3 mM ATP, 10 mM Met, 10 mM Cys, 20 mM potassium gluconate, 10 mM NaHCO3, 3 mM MgSO4, 330 mM sorbitol, 50 mm HEPES-KOH, pH 8, 0.2% BSA, and 50 mM ascorbic acid) for 25°C for 5 min with gentle agitation. Plastids were and washed in cold 50 mM HEPES-KOH, pH 8, 0.3 m sorbitol, and 3 mm MgSO2 prior to treatment with thermolysin (10 μg) and CaCl2 (5 mm) for 15 min on ice. EDTA was added to stop the reaction and plastids pelleted. All import samples were resolved by SDS-PAGE (16% (w/v) Acrylamide SDS-PAGE gels or 16.5% (w/v) Acrylamide Tricine SDS-PAGE as described in (Schägger, 2006), dried, and exposed to a phosphor-imaging screen for 24 h.
Results
Phylogenetic analyses and protein characterization of the Arabidopsis PAM subunits
Pam16 and Pam18 of the PAM complex modulate mtHSP70 activity along with Tim15 and the nucleotide exchange factor, Mge1 required for the import of precursor proteins into the mitochondrial matrix to (Figure 1). The Arabidopsis genome encodes for two Pam16 (Pam16-1 and Pam16-2) and three Pam18 orthologues (Pam18-1, Pam18-2, and Pam18-3). Phylogenetic analysis indicates that Pam16 and Pam18 branch independently and are conserved across species (Supplementary Figure 1A). Arabidopsis encodes for three Tim15 orthologues (Tim15-1, Tim15-2 and Tim15-3), Tim15-2 (also known as ZR3) branches with human and yeast (Saccharomyces cerevisiae) Tim15 (also known as Hep1) whilst Tim15-1 and Tim15-3 branch distinctly to Tim15-2 and are conserved in the early land plant Selaginella moellendorffii, and monocots Zea mays and Oryza sativa (Supplementary Figure 1B) Pam16 and Pam18 orthologues contain the conserved intermembrane space (IMS) domain, J-domain, and a predicted N-terminal transmembrane region (Supplementary Figure 1C). Tim15 orthologues contain a zinc finger motif domain and a conserved C-C motif, present in human and yeast Tim15/HEP1 (Supplementary Figure 2). Structural predictions using Phyre2 (Kelley et al., 2015) show similarity for all Arabidopsis Pam16, Pam18, and Tim15 orthologues (green) when overlaid on their yeast counterpart (pink) (Supplementary Figure 1D).
Pam16 orthologues are targeted to the mitochondria whilst Pam18 orthologues are dual-targeted to mitochondria and plastid
The localization of C-terminal GFP-tagged Pam16 and Pam18 orthologues was examined using biolistic transformation of Arabidopsis cell suspensions. Full length GFP fusions were co-transformed with mitochondrial (AOX-RFP) or plastid (SSU-RFP) controls (Figure 2) (Carrie et al., 2010). The localization of Pam16-1 and Pam16-2 shows exclusive targeting to mitochondria as determined by co-localization with AOX-RFP (Figures 2A–D). Pam18-1-GFP fluorescence showed no overlay with AOX-RFP (Figure 2E) but did with SSU-RFP suggesting that Pam18-1 is targeted exclusively to the plastids (Figure 2F). Localization of Pam18-2 and Pam18-3 showed co-localization with both AOX-RFP and SSU-RFP suggesting dual-targeting to both mitochondria and plastid (Figures 2G–J).
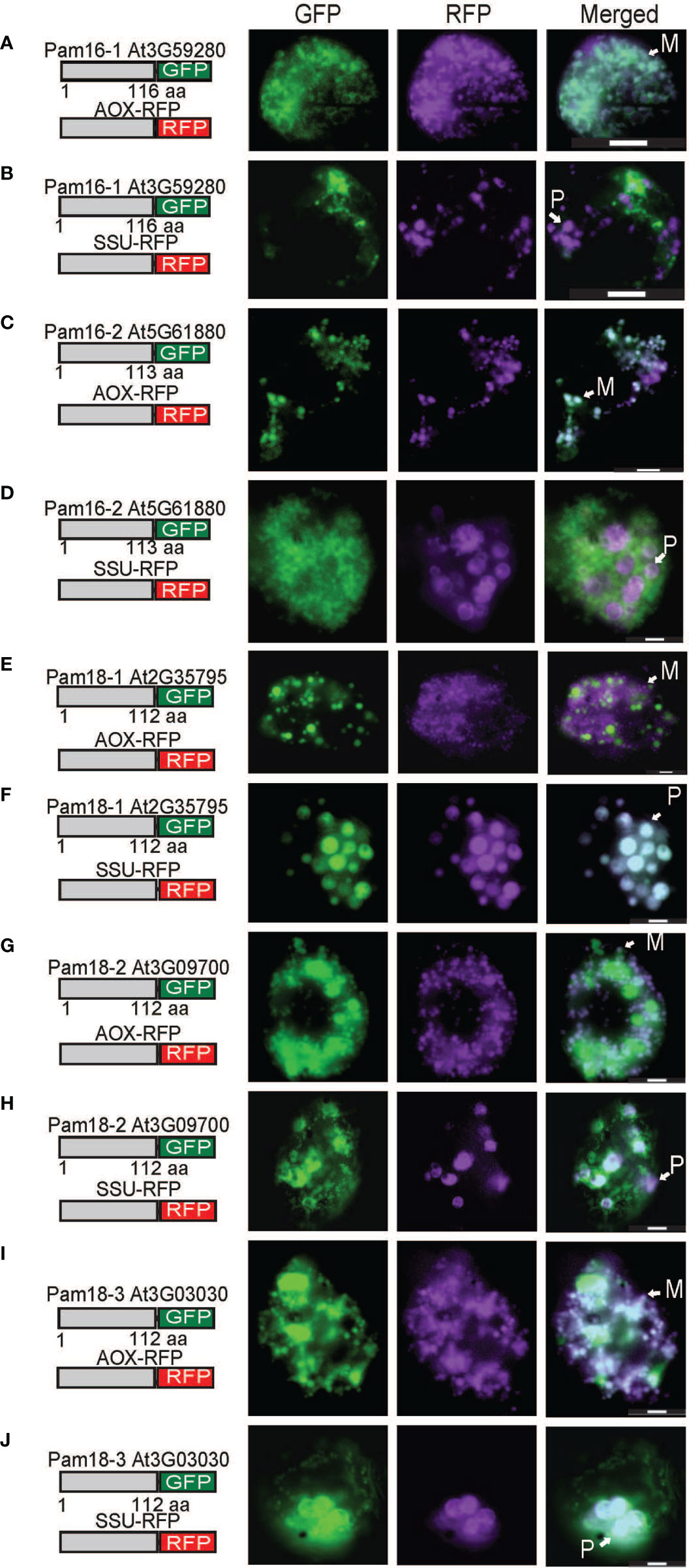
Figure 2 Biolistic transformation and localisation assays of Arabidopsis Pam16 and Pam18 GFP. C-terminal GFP fusions transformed into Arabidopsis cell suspensions alongside a mitochondrial targeted RFP (AOX-RFP) or plastid targeted RFP (SSU-RFP). Scale bar indicates 20 μm. M, mitochondria; P, plastid. Pam16-1 (A, B). Pam16-2 (C, D). Pam18-1 (E, F). Pam18-2 (G, H). Pam18-3 (I, J).
PAM interacting proteins Tim15 and Mge1 target to the mitochondria and plastids
As the plastid localization of Pam18 orthologues was unexpected, we tested the targeting of PAM Tim15, Mge2 and the plastid Mge1/2 counterparts, GrpE-like protein Cge1 and Cge2 (de Luna-Valdez et al., 2019). Tim15-1 exhibited exclusively plastid localization (Figures 3A,B), Tim15-2 co-localized to mitochondria (Figure 3C) and Tim15-3 co-localized to plastids and cytosol (Figure 3D). Multiple studies have comprehensively determined Mge1 to be dual-targeted to both mitochondria and plastids (Van Aken et al., 2009; Baudisch et al., 2014; Sharma et al., 2018) therefore we only tested Mge2 localization and determined it to target to the mitochondria (Figure 3E). Cge1 and Cge2 co-localized exclusively to the plastid (Figures 3F–I).
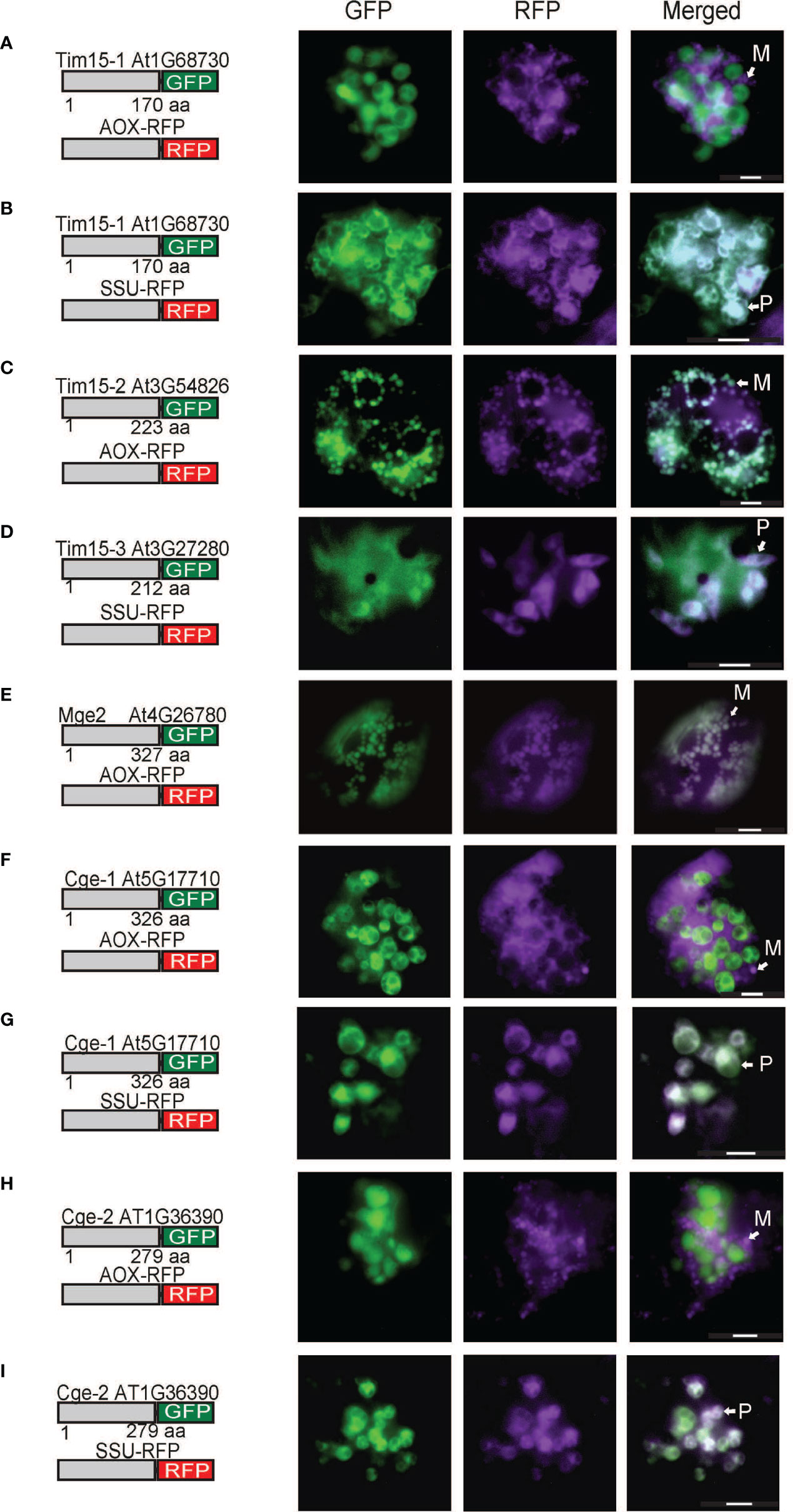
Figure 3 Biolistic transformation and localisation assays of Arabidopsis Tim15, Mge2 and Cge1/2 GFP. C-terminal GFP fusions transformed into Arabidopsis cell suspensions alongside a mitochondrial targeted RFP (AOX-RFP) or plastid targeted RFP (SSU-RFP). Tim15-1 (A, B). Tim15-2 (C). Tim15-3 (D). Mge2 (E). Cge1 (F, G). Cge2 (H, I). Scale bar indicates 20 μm. M, mitochondria; P, plastid.
Stable transgenic lines of Pam18 GFP show dual-targeting to the mitochondria and plastids
Due to the unexpected dual-targeting of Pam18 using transient GFP fusions, we generated stable transgenic Arabidopsis lines for all Pam16 and Pam18 orthologues fused to GFP at the C-terminus (Figure 4; Supplementary Figure 3). Analysis of Pam16-1GFP and Pam16-2GFP stable lines indicated a typical mitochondrial-like localization of protein in root cells, epidermis, and guard cells (Figures 4A, B). Pam18-1GFP was observed to be targeted to both the mitochondria and larger plastid-like organelles in root, epidermis, and guard cells (Figure 4C). Pam18-2GFP was observed to be targeted exclusively to mitochondria in root, epidermis, and guard cells (Figure 4D). Pam18-3GFP was observed to be targeted to both the mitochondria and larger plastid-like organelles in root, epidermis, and guard cells (Figure 4E).
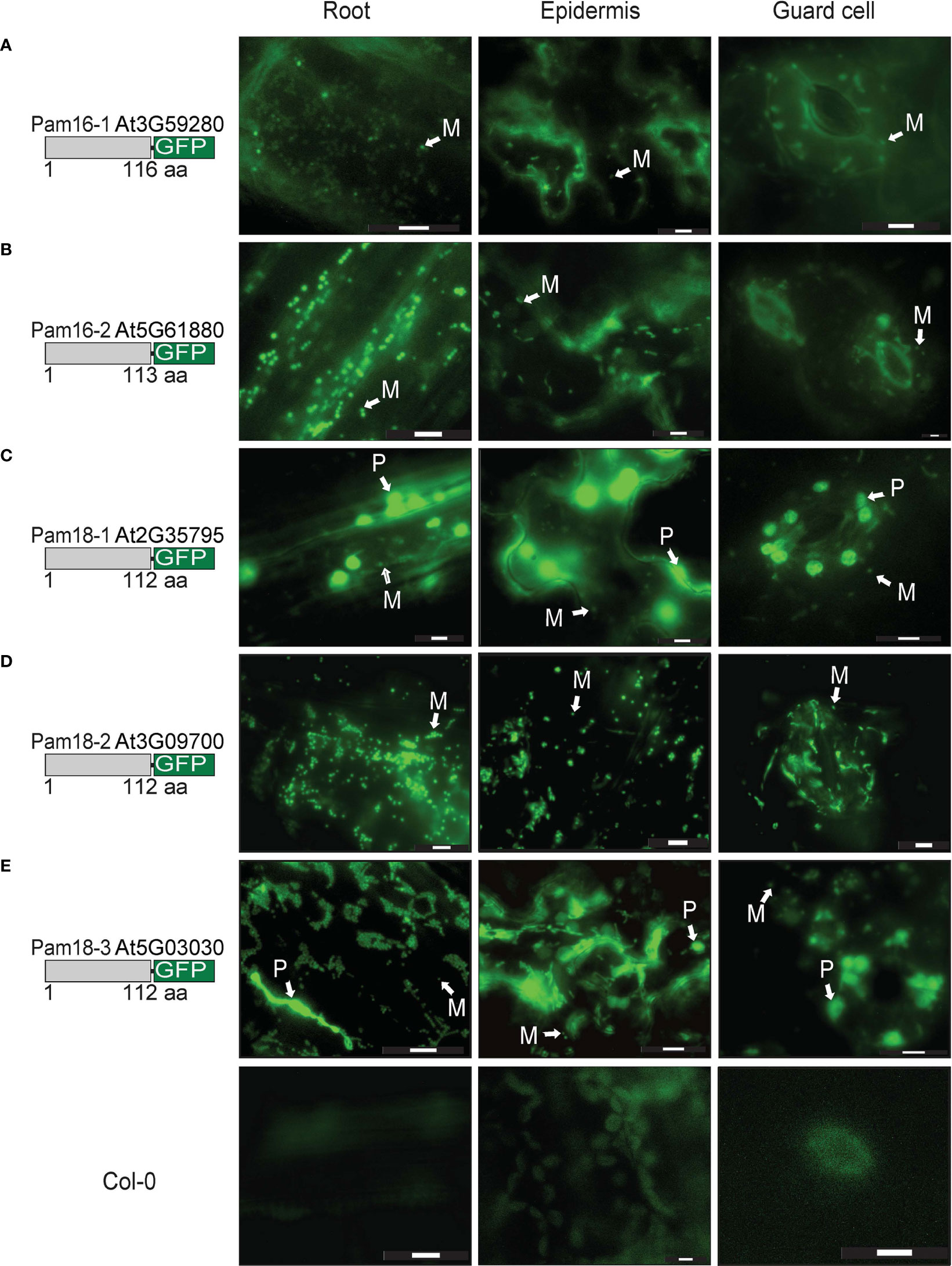
Figure 4 Pam16 and Pam18 GFP in stable transgenic Arabidopsis seedlings. Images taken from 3 weeks old Arabidopsis T2 transformants of root, epidermal, and guard cells Pam16-1 (A). Pam16-2 (B). Pam18-1 (C). Pam18-2 (D). Pam18-3 (E). Scale bar indicates 20 μM. M, mitochondria, P, plastid.
To confirm that the larger organellar structures that we observed targeting to were plastids, Pam18-1 and Pam18-3 GFP lines were crossed with a stable SSU-RFP line (Carrie et al., 2010) (Figure 5, Supplementary Figure 4). Imaging of these double GFP/SSU-RFP tagged lines showed mitochondrial-like GFP localization patterns and a clear overlay GFP/RFP overlay in root, epidermal and guard cells (Figures 5A–F), confirming that Pam18-1 and Pam18-3 are dual-targeted to mitochondria and plastids as observed with transient transformations Pam18-1 and Pam18-3FGFP fusions were also tested under their native promoters and crossed with the same SSU-RFP line. Under a native promoter, Pam18-1, was found to be exclusively mitochondrial-located in epithelial cells (Figure 6A, Supplementary Figure 4) and dual-targeted in guard cells and root cells (Figures 6B,C). Under a native promoter, Pam18-3 was observed to be targeted to mitochondria in all cell types investigated (Figures 6D–F). To confirm that the localization was to plastids, immunodetection was carried out against isolated plastids from 14-day old Pam18-1 GFP seedlings. Immunodetection with anti-GFP indicated a band ~38 kDa (Figure 7A) whilst immunodetection using RbCL, and Coomassie staining confirmed equally loading (Figures 7A, B).
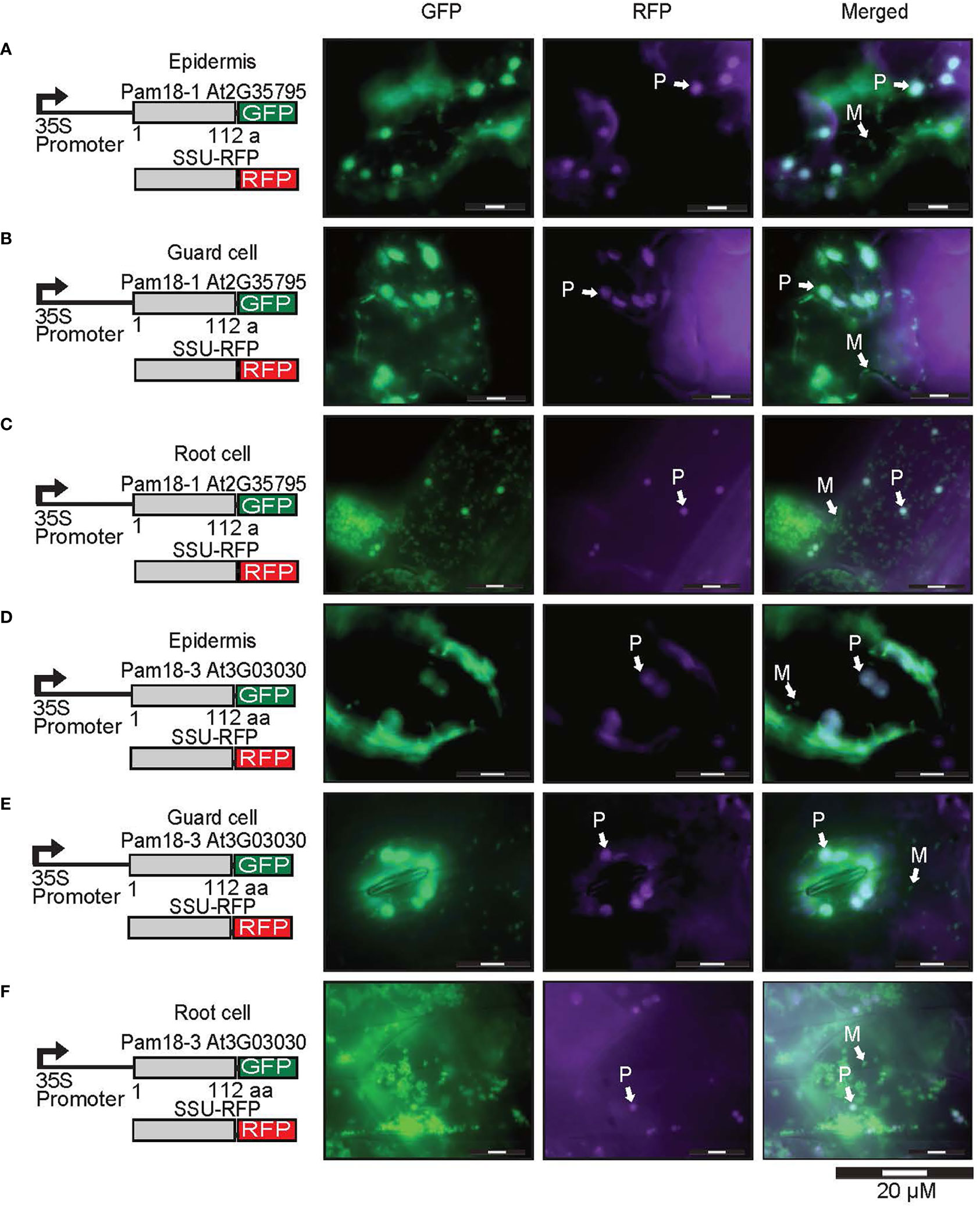
Figure 5 Pam18-1 and Pam18-3 GFP with SSU-RFP in stable transgenic Arabidopsis seedlings. Images taken from 3 weeks old Arabidopsis T2 transformants of root, epidermal, and guard cells Pam18-1 (A–C). Pam18-3 (D–F). Scale bar indicates 20 μM. M, mitochondria, P, plastid.
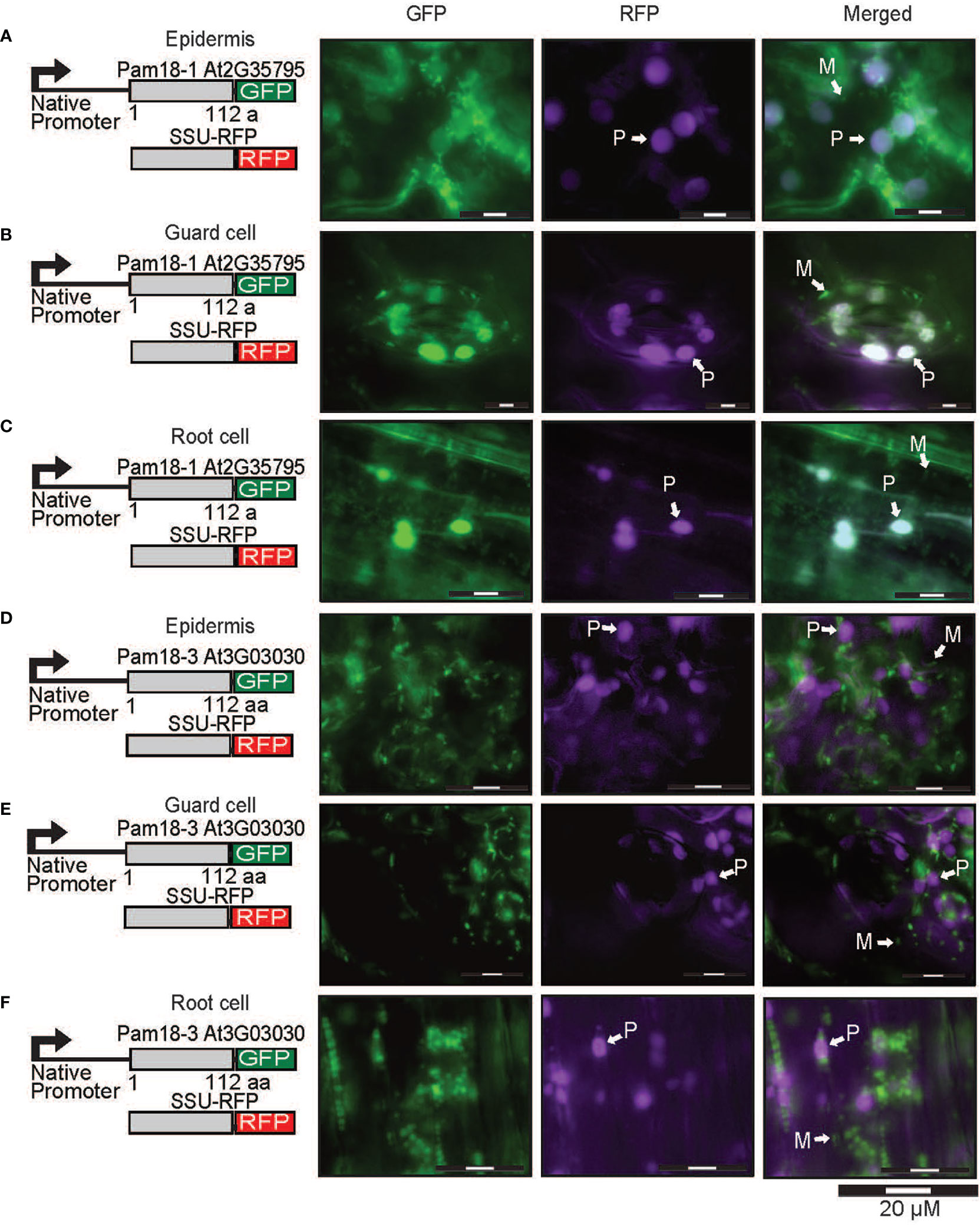
Figure 6 Pam18-1 and Pam18-3 GFP under endogenous promoters and plastid targeted-RFP (SSU-RFP) in stable transgenic Arabidopsis seedlings. Images taken from 3 weeks old Arabidopsis T2 transformants of root, epidermal, and guard cells Pam18-1 (A–C). Pam18-3 (D–F). Scale bar indicates 20 μM. M, mitochondria, P, plastid.
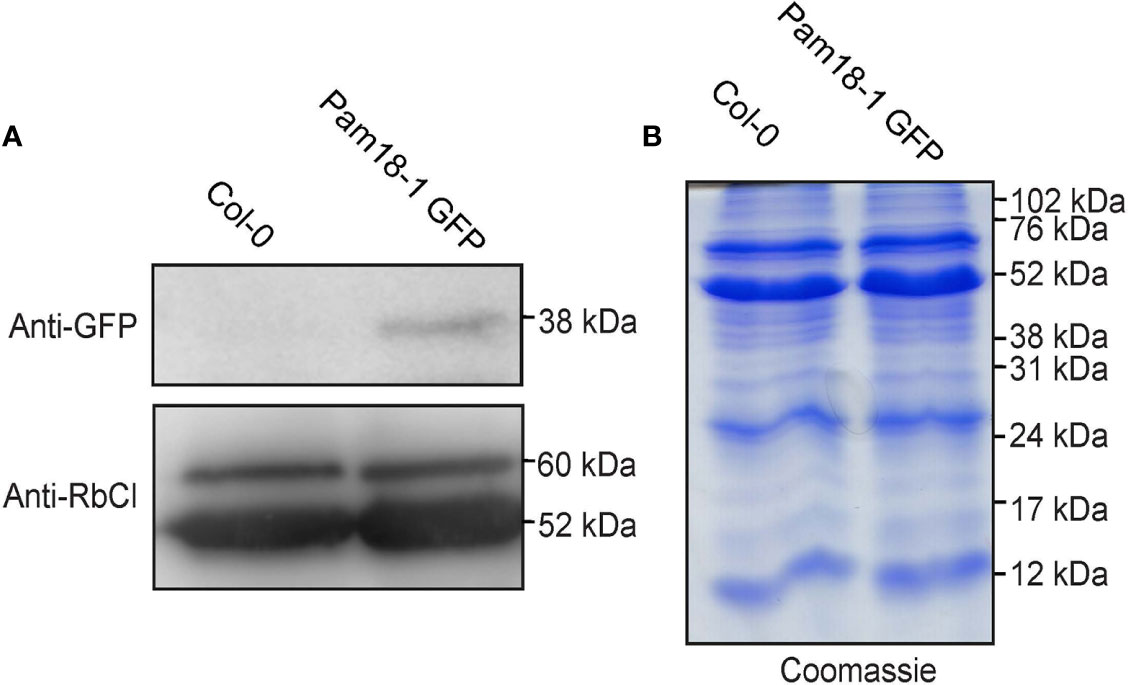
Figure 7 Immunodetection of Pam18-1 GFP under the expression of its endogenous promoter in isolated plastids. (A) Chloroplasts were isolated from 2 weeks old seedlings and immunodetected with anti-GFP and anti-RbCl. (B) Coomassie staining confirms equal protein loading and integrity.
Pam18 orthologues can be imported into isolated mitochondria and plastids
As the use of GFP fusion proteins can be error prone, (Duchêne et al., 2005; Sharma et al., 2018) we tested the targeting ability of PAM subunits without GFP tags. This was achieved by in vitro protein import assays using radiolabeled proteins imported into freshly isolated intact Arabidopsis mitochondria and plastids (Figures 8 and 9). For mitochondrial protein import assays, outer membranes were ruptured following protein uptake to confirm a proteins intra-mitochondrial localization, which is essential for proteins that do not contain a cleavable presequence (Murcha and Whelan, 2015). The Alternative Oxidase (AOX) and Tim23-2 proteins were used as controls to confirm protein uptake ability of the isolated mitochondria and to determine successful rupture of the outer membrane as previously described (Duncan et al., 2015). Import of the AOX precursor protein (AOX (p)) shows the generation of the mature protein (AOX (m)) upon successful import that is valinomycin sensitive (Figure 8A). Import of Tim23-2 reveals the generation of a ~14 kDa inner membrane PK protected fragment, confirming the successful import and rupture of the outer membrane (Figure 8B, lane 7). Import of Pam16 and Pam18 orthologues into isolated mitochondria showed the presence of a PK protected band in lane 3 and lane 7, confirming import through the inner membrane in a membrane potential dependent manner, with no processing observed (Figures 8C–G). Import of Tim15-1, and Tim15-3 did not suggest import into isolated mitochondria (Figures 8H, J), whilst Tim15-2 was imported and processed into a mature band of ~15 kDa (Figure 8I). Interestingly, a doublet band was visible suggesting an additional processing event may occur after presequence removal by MPP (Figure 8I). Import of Mge1 and Mge2 showed import and processing to mature proteins of ~25 and of 28 kDa respectively (Figures 8K, L). Both proteins show the presence of an intermediate processing product in lanes 2, 3 and 6, suggesting an additional processing event within the IMS (Figures 8K, L).
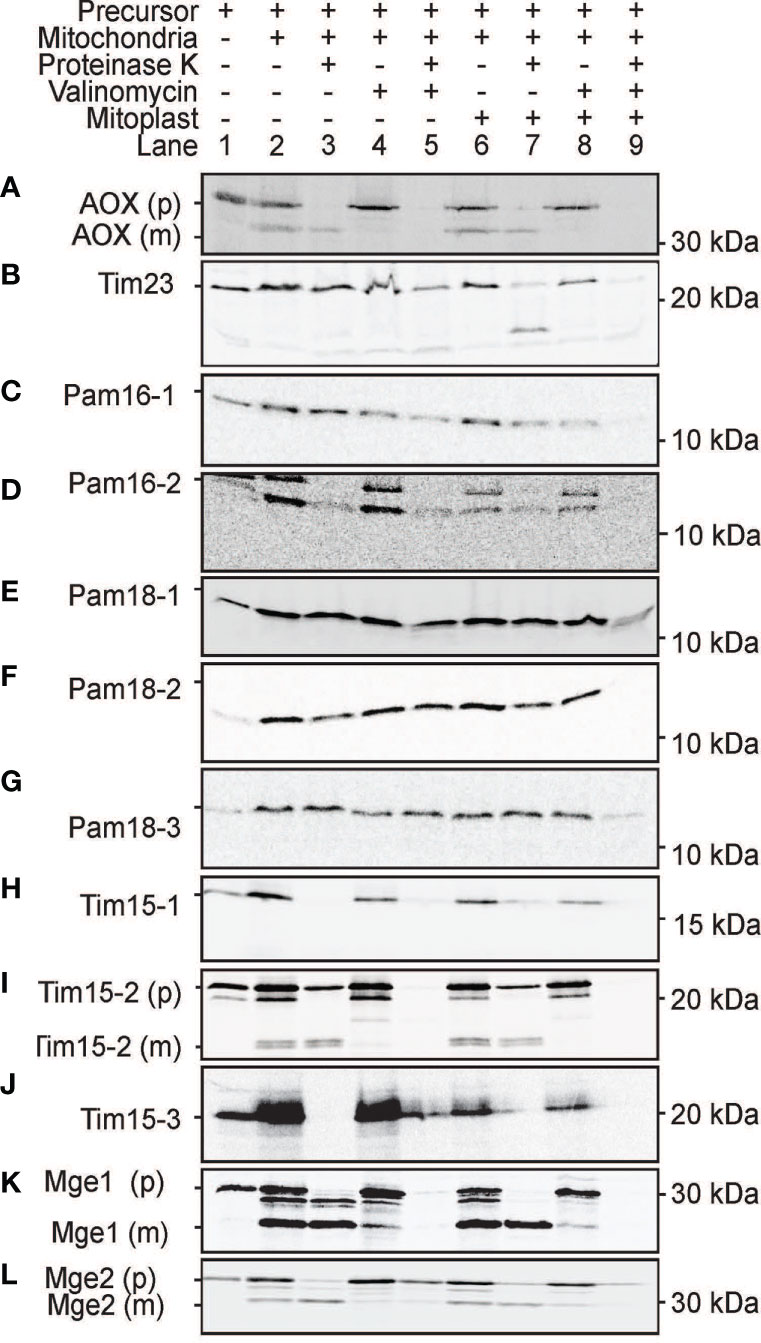
Figure 8 Protein import assays into isolated mitochondria. (A) Alternative Oxidase (AOX) was used as a control for successful import into the mitochondrial matrix while Tim23-2 (B) was used to test for the successful rupture of the outer membrane by osmotic shock following protein import. Pam16 (C, D). Pam18 (E–G). Tim15 (H–J). Mge (K, L). Lane 1 = precursor protein alone, lane 2 = precursor protein incubated with mitochondria under conditions that support import, lane 3 = as lane 2 with the addition of Proteinase K (PK) following import, lane 4 = same as lane 2 but with the addition of valinomycin prior to import, lane 5 = same as lane 4 but with the addition of PK following import, lane 6 = same as lane 2 but with outer membrane ruptured after import, lane 7 = same as lane 6 but with the addition of PK following import, lane 8 = same as lane 6 but with valinomycin prior to import, lane 9 = same as lane 8 but with PK treatment. (p) = precursor protein band. (i) = intermediate protein band. (m) = mature protein band. Position of protein size marker is shown on the right.
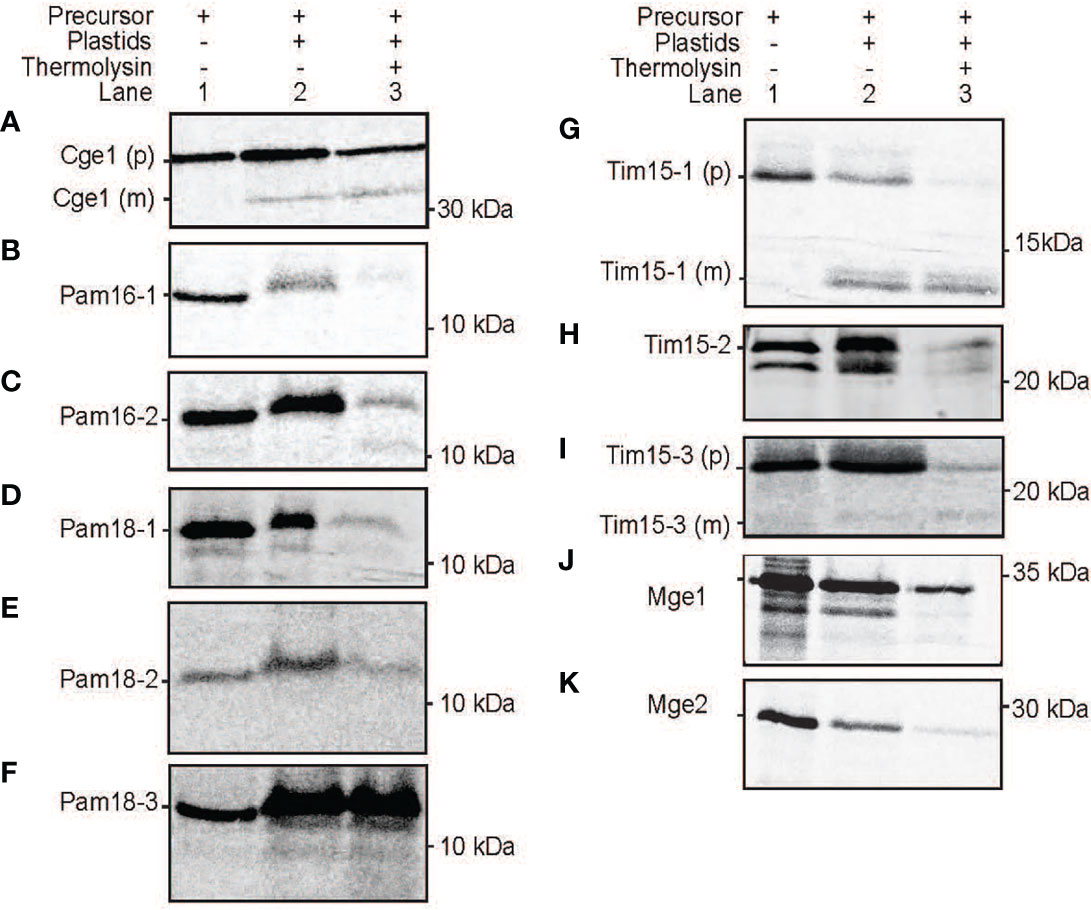
Figure 9 Protein import assays into isolated plastids. Import of radiolabelled proteins into isolated Arabidopsis plastids. Cge2 was used as a control for successful import (A). Pam16 (B, C), Pam 18 (D-F), Tim15 (G-I), Mge1 (J, K). Lane 1 = precursor protein alone, lane 2 = precursor protein incubated with chloroplasts under conditions that support import, lane 3 = as lane 2 with the addition of thermolysin following import. (p) = precursor protein band. (m) = mature protein band. Position of protein size marker is shown on the right.
All PAM orthologues were tested for import into isolated Arabidopsis plastids. The plastid Cge1 was used as a control, showing import and processing to the mature protein of ~ 30 kDa size (Figure 9A). The mitochondrial Pam16-1 did not show import into isolated chloroplasts with no band present in lane 3 following thermolysin treatment, though Pam16-2 exhibited some weak binding and import (Figures 9B,C). Import of Pam18-1 and Pam18-2 similarly showed weak import with radiolabeled bands present in lane 3 whilst Pam18-3 showed strong binding and import (Figures 9C–F). Tim15-1 and Tim15-3 showed import and to mature products of ~10 kDa (Figures 9G, I) whilst Tim15-2 did not show import into isolated chloroplasts (Figure 9H). The dual-targeted Mge1 exhibited import into isolated chloroplasts, though no processing to a mature protein was observed (Figure 9J). whilst the mitochondrial Mge2 did not exhibit import into isolated chloroplasts (Figure 9K). Therefore, in vitro protein import assays show that Pam16 and Pam18 orthologues have the ability to be imported into isolated mitochondria whilst Pam18 orthologues were targeted to isolated plastids.
Discussion
Pam16 and Pam18 are co-chaperones crucial for protein import into mitochondria. Plants contain multiple copies of Pam16 and Pam18 and whilst several studies have shown that Pam16 and Pam18 are targeted to mitochondria, none of these determined the localization of Pam18-1 (Tamadaddi et al., 2021). Furthermore, studies have shown that orthologues of PAM interacting partner proteins, Tim15 and Mge1, are localized to the mitochondria and plastids (Van Aken et al., 2009; Kluth et al., 2012; Langner et al., 2014). Therefore, we reexamined the targeting ability of these proteins utilizing multiple in vitro and in vivo approaches (Table 1). We found that Pam16-1 and Pam16-2 were targeted exclusively to the mitochondria using all methods tested such as transient biolistic transformation into cell suspensions, stable transgenic GFP-tagged lines, and in vitro protein import assays. We determined that Pam18 orthologues could be targeted to both mitochondria and plastids depending on the methodology used. For instance, Pam18-1 was exclusively targeted to the plastids in the biolistic transformation of cell suspensions, dual-targeted to plastids and mitochondria in the stable transgenic GFP lines using both the 35S promoter and native promoter and could be imported into isolated mitochondria and plastids. Pam18-2 was dual-targeted in transient biolistic transformations, exclusively targeted to mitochondria in stable transgenic GFP plants and could be imported into isolated mitochondria and plastids. Pam18-3 was found to be dual-targeted to mitochondria and chloroplasts, in both biolistic transformations and stable GFP lines but only localized to the mitochondria when using a native promoter (Table 1).
Whilst Pam18-2 and Pam18-3 localization has been investigated previously (Tamadaddi et al., 2021), localization was only tested in the root cells of stable GFP fusion lines suggesting that any potential plastid localization may not have been observed. Here, Pam18’s unexpected plastid localization was dependent on protein abundance and cell specific factors, highlighting the need for multiple and comprehensive experimental approaches. This has been previously shown with other dual-targeted proteins. The targeting of Mge1 was tested using various methods such as GFP-fusions delivered via biolistic transformation, protoplasts, agrobacterium transformation, and Mge1 showed both mitochondrial or dual-targeting ability (Sharma et al., 2018) as we confirmed here. Plastids are known to acquire specialized and distinct functions for photosynthesis, metabolism and storage and thus their proteomes differ in various cell types (Wise, 2007). Furthermore, recent studies have identified specialized stress sensing plastids known as sensory plastids (Dopp et al., 2021) and that the dual-targeted protein MutS Homolog1 (MSH1) involved in organelle genome functions (Xu et al., 2011) has been shown to reside exclusively in epidermal sensory plastids (Beltrán et al., 2018). This work also raises the possibility that Pam18 also plays a specialized role in plastids. Plastids, like mitochondria, evolved from an independent endosymbiotic event and similarly lost the majority of its genome, thus requiring import of the majority of its proteome to carry out its essential functions. Whilst each organelle possesses mostly unique and evolutionary distinct protein import machinery, the general mechanisms are strikingly similar (Flores-Pérez and Jarvis, 2013; Langner et al., 2014; Ghifari et al., 2018; Nakai, 2018). Both mitochondria and plastids utilize targeting peptides to deliver the cytosolic proteins to the organelle, which is cleaved upon import. Several of the proteases and aminopeptidases involved in the sequential degradation of the targeting signal have been shown to be dual-targeted to mitochondria and plastid. The Presequence Peptidase (PreP), Organellar oligopeptidase (OOP) and the M1 and M17 family of amino peptidase are all dual-targeted and play vital roles in plant homeostasis (Moberg et al., 2003; Kmiec et al., 2013; Kmiec et al., 2018; Ghifari et al., 2022). Whilst not a dual-targeted protein, the plant mitochondrial outer membrane receptor 64 (OM64) is paralogous to the plastid outer envelope receptor TOC64 (Lister et al., 2007). Both receptors function by binding to the cytosolic Hsp70 and Hsp90 via their TPR domains to initiate protein translocation across the outer membrane/envelope (Panigrahi et al., 2014; Nickel et al., 2019).
Both organelles utilize Hsp70 to translocate proteins in an ATP-dependent manner alongside co-chaperones and nucleotide exchange factors (Schilke et al., 2017; Clerico et al., 2019). The versatility of Hsp70 in such diverse mechanisms such as protein import, preventing protein aggregation, and folding of nascent polypeptide chains lies in its interaction with co-chaperones (Kampinga and Craig, 2010; Schilke et al., 2017). In plastids, cpHsp70s have been shown to interact with protein import components such as the inner envelope Tic40 and Tic110 (Nakai, 2018), both of which have shown to also associate with Cge1 (de Luna-Valdez et al., 2019). However, to date, no dedicated J-domain co-chaperones has been identified to interact with cpHsp70 or the plastid protein import systems (Flores-Pérez and Jarvis, 2013; Kikuchi et al., 2013). In Arabidopsis Mge1 is dual-targeted to mitochondria and plastids, and Mge2 is exclusively mitochondrial (Van Aken et al., 2009) (Hu et al., 2012). Whilst the specific role of Mge1 has yet to be determined the presence of two Mge homologs is conserved throughout plants suggesting diverged roles (Hu et al., 2012). Both Mge1 and 2 are highly stress responsive proteins and it has been suggested that Mge1 has a role in general protein import whilst the thermotolerant Mge2 plays a specific role when plants are exposed to high temperatures (Van Aken et al., 2009; Hu et al., 2012; Maziak et al., 2021). Though it remains to be seen what role Mge1 plays in the plastid.
Orthologues of Tim15 (HEP/ZR) another Hsp70 PAM complex co-chaperone has also been observed to be targeted to the mitochondria and plastid. Tim15-1 was shown to physically interact with mtHSP70 and was able to complement a yeast deletion strain (Kluth et al., 2012) with a likely role in preventing mtHSP70 aggregation (Nyakundi et al., 2016). Tim15-1 and Tim15-3 both have been confirmed as plastid proteins (Kluth et al., 2012) and similarly found to interact with cpHsp70 (Willmund et al., 2008). Plastid Tim15 was observed to be able to stimulate the ATPase activity of cpHSP70 required for protein import in moss (Liu et al., 2014) supporting this co-chaperones role in chloroplast protein import.
Both mtHsp70 and cpHsp70 show little divergence on a protein sequence level with their functions and specificity being regulated by their interactions with co-chaperones (Miernyk, 2001). The ability of Pam18 orthologues to be targeted to the plastid raises the possibility that the Pam18’s may also function as cpHSP70 co-chaperones alongside Mge and Tim15, to function in a developmental stress responsive and/or tissue specific manner. Pam18 transcript abundance profiles show that the dual-targeted Pam18-3 does not show high abundance in seed, a developmental stage where protein import components including Pam16’s are most abundant (Narsai et al., 2011; Chen et al., 2013). Furthermore, Pam18 homologues exhibit differential stress responses with regards to heat and drought (Chen et al., 2013) suggesting sub-functionalization. Recently, it has been shown that Pam18 has additional functions with regards to interacting with the respiratory chain in the mitochondrial inner membrane (Wiedemann et al., 2007; Priesnitz et al., 2022) promoting the assembly of Complex IV (Priesnitz et al., 2022). More interestingly, the function of mitochondrial Pam16 and Pam18 has been totally replaced by an unrelated protein Pam27 in trypanosomes (von Känel et al., 2020) which highlight the divergence and dynamic nature of protein import components and mechanisms across eukaryotes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MG-H and MWM; research design; MG-H, AW and MWM; conducting experiments and data analysis; MG-H, AW and MWM preparation of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
MWM is supported by an Australian Research Council Discovery Project (DP200101922). MG-H is supported by a University Postgraduate Award at the University of Western Australia. AW was supported by a Rowe Scientific Foundation Vacation Award.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.981552/full#supplementary-material
Supplementary Figure 1 | PAM complex subunits are conserved across species. (A, B) Phylogenetic analysis of Pam16, Pam18 and Tim15 orthologues from Saccharomyces cerevisiae (Sc), Arabidopsis thaliana (At), oryza sativa (Os), Caenorhabditis elegans (Ce), Zea mays (Zm), Homo sapiens (Hm), cyanobacteria (Cn), and Selaginella moellendorffii (Sm) PAM orthlogues with known localisation are highlighted in yellow (mitochondria) or green (chloroplast) respectively. (C) Protein features of PAM subunits; inner membrane space domain (IMS) domain (pink), transmembrane region (yellow), J-domain or J-like domain (purple, IPR001623) and a histidine-proline-aspartic acid (HPD) motif (red). The zinc ribbon superfamily domain present in Tim15 (green) and the zinc finger C-C motif (orange). (D) Predicted tertiary structures of the Arabidopsis Pam16, Pam18 and Tim15 orthologues overlaid over the experimentally determined structure of its putative yeast orthologue.
Supplementary Figure 2 | Conserved protein domains and motifs in Pam16, Pam18, Tim15, Mge1 and Cge orthologues from yeast and Arabidopsis. Domain mapping was done using InterPro (https://www.ebi.ac.uk/interpro/) and ScanProsite (https://prosite.expasy.org/scanprosite/). All Pam16s contain an intermembrane space domain (IMS domain, pink), and a transmembrane region (yellow), and a J-like domain (blue). The Pam18s additionally contain a J-domain (purple) instead of J-like domain distinguished by the presences of the HPD motif (red). The Tim15s all contain the zinc ribbon superfamily domain (green) with a C–C motif (orange). The Mge1 and Cge orthologues all contain a similar GrpE superfamily domain (yellow).
Supplementary Figure 3 | Bright field images corresponding to Figure 4. Scale bar indicates 20 mM.
Supplementary Figure 4 | Bright field images corresponding to Figures 5 and 6. Scale bar indicates 20 mM.
References
Aronsson, H., Jarvis, P. (2002). A simple method for isolating import-competent arabidopsis chloroplasts. FEBS Lett. 529, 215–220. doi: 10.1016/S0014-5793(02)03342-2
Baudisch, B., Langner, U., Garz, I., Klösgen, R. B. (2014). The exception proves the rule? dual targeting of nuclear-encoded proteins into endosymbiotic organelles. New Phytol. 201, 80–90. doi: 10.1111/nph.12482
Beltrán, J., Wamboldt, Y., Sanchez, R., LaBrant, E. W., Kundariya, H., Virdi, K. S., et al. (2018). Specialized plastids trigger tissue-specific signaling for systemic stress response in plants. Plant Physiol. 178, 672–683. doi: 10.1104/pp.18.00804
Carrie, C., Giraud, E., Duncan, O., Xu, L., Wang, Y., Huang, S., et al. (2010). Conserved and novel functions for arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285, 36138–36148. doi: 10.1074/jbc.M110.121202
Carrie, C., Murcha, M. W., Millar, A. H., Smith, S. M., Whelan, J. (2007). Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol. Biol. 63, 97–108. doi: 10.1007/s11103-006-9075-1
Chen, X., Ghazanfar, B., Khan, A. R., Hayat, S., Cheng, Z. (2013). Comparative analysis of putative orthologues of mitochondrial import motor subunit: Pam18 and Pam16 in plants. PloS One 8, e78400. doi: 10.1371/journal.pone.0078400
Clerico, E. M., Meng, W., Pozhidaeva, A., Bhasne, K., Petridis, C., Gierasch, L. M. (2019). Hsp70 molecular chaperones: multifunctional allosteric holding and unfolding machines. Biochem. J. 476, 1653–1677. doi: 10.1042/BCJ20170380
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
De Los Rios, P., Ben-Zvi, A., Slutsky, O., Azem, A., Goloubinoff, P. (2006). Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl. Acad. Sci. U. S. A. 103, 6166–6171. doi: 10.1073/pnas.0510496103
de Luna-Valdez, L. A., Villaseñor-Salmerón, C. I., Cordoba, E., Vera-Estrella, R., León-Mejía, P., Guevara-García, A. A. (2019). Functional analysis of the chloroplast GrpE (CGE) proteins from arabidopsis thaliana. Plant Physiol. Biochem. 139, 293–306. doi: 10.1016/j.plaphy.2019.03.027
Dopp, I. J., Yang, X., Mackenzie, S. A. (2021). A new take on organelle-mediated stress sensing in plants. New Phytol. 230, 2148–2153. doi: 10.1111/nph.17333
Duchêne, A. M., Giritch, A., Hoffmann, B., Cognat, V., Lancelin, D., Peeters, N. M., et al. (2005). Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 102, 16484–16489. doi: 10.1073/pnas.0504682102
Duncan, O., Carrie, C., Wang, Y., Murcha, M. W. (2015). In vitro and in vivo protein uptake studies in plant mitochondria. Methods Mol. Biol. 1305, 61–81. doi: 10.1007/978-1-4939-2639-8_5
Flores-Pérez, Ú., Jarvis, P. (2013). Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833, 332–340. doi: 10.1016/j.bbamcr.2012.03.019
Frazier, A. E., Chacinska, A., Truscott, K. N., Guiard, B., Pfanner, N., Rehling, P. (2003). Mitochondria use different mechanisms for transport of multispanning membrane proteins through the intermembrane space. Mol. Cell Biol. 23, 7818–7828. doi: 10.1128/mcb.23.21.7818-7828.2003
Ghifari, A. S., Gill-Hille, M., Murcha, M. W. (2018). Plant mitochondrial protein import: the ins and outs. Biochem. J. 475, 2191–2208. doi: 10.1042/BCJ20170521
Ghifari, A. S., Teixeira, P. F., Kmiec, B., Singh, N., Glaser, E., Murcha, M. W. (2022). The dual-targeted prolyl aminopeptidase PAP1 is involved in proline accumulation in response to stress and during pollen development. J. Exp. Bot. 73, 78–93. doi: 10.1093/jxb/erab397
Hennessy, F., Nicoll, W. S., Zimmermann, R., Cheetham, M. E., Blatch, G. L. (2005). Not all J domains are created equal: Implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci. 14, 1697–1709. doi: 10.1110/ps.051406805
Huang, Y., Chen, X., Liu, Y., Roth, C., Copeland, C., McFarlane, H. E., et al. (2013). Mitochondrial AtPAM16 is required for plant survival and the negative regulation of plant immunity. Nat. Commun. 4, 2558. doi: 10.1038/ncomms3558
Hu, C., Lin, S. Y., Chi, W. T., Charng, Y. Y. (2012). Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in arabidopsis. Plant Physiol. 158, 747–758. doi: 10.1104/pp.111.187674
Kampinga, H. H., Craig, E. A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. doi: 10.1038/nrm2941
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kikuchi, S., Bédard, J., Hirano, M., Hirabayashi, Y., Oishi, M., Imai, M., et al. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574. doi: 10.1126/science.1229262
Kluth, J., Schmidt, A., März, M., Krupinska, K., Lorbiecke, R. (2012). Arabidopsis zinc ribbon 3 is the ortholog of yeast mitochondrial HSP70 escort protein HEP1 and belongs to an ancient protein family in mitochondria and plastids. FEBS Lett. 586, 3071–3076. doi: 10.1016/j.febslet.2012.07.052
Kmiec, B., Branca, R. M. M., Murcha, M. W., Lehtiö, J., Glaser, E., Teixeira, P. F. (2018). A common peptidolytic mechanism for targeting peptide degradation in mitochondria and chloroplasts. Mol. Plant 11, 342–345. doi: 10.1016/j.molp.2017.11.008
Kmiec, B., Teixeira, P. F., Berntsson, R. P., Murcha, M. W., Branca, R. M., Radomiljac, J. D., et al. (2013). Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc. Natl. Acad. Sci. U. S. A. 110, E3761–E3769. doi: 10.1073/pnas.1307637110
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Langner, U., Baudisch, B., Klösgen, R. B. (2014). Organelle import of proteins with dual targeting properties into mitochondria and chloroplasts takes place by the general import pathways. Plant Signal Behav. 9, e29301. doi: 10.4161/psb.29301
Lister, R., Carrie, C., Duncan, O., Ho, L. H., Howell, K. A., Murcha, M. W., et al. (2007). Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19, 3739–3759. doi: 10.1105/tpc.107.050534
Liu, Y., Burch-Smith, T., Schiff, M., Feng, S., Dinesh-Kumar, S. P. (2004). Molecular chaperone Hsp90 associates with resistance protein n and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. doi: 10.1074/jbc.M310029200
Liu, Q., D'Silva, P., Walter, W., Marszalek, J., Craig, E. A. (2003). Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300, 139–141. doi: 10.1126/science.1083379
Liu, L., McNeilage, R. T., Shi, L. X., Theg, S. M. (2014). ATP requirement for chloroplast protein import is set by the km for ATP hydrolysis of stromal Hsp70 in physcomitrella patens. Plant Cell 26, 1246–1255. doi: 10.1105/tpc.113.121822
Maziak, A., Heidorn-Czarna, M., Weremczuk, A., Janska, H. (2021). FTSH4 and OMA1 mitochondrial proteases reduce moderate heat stress-induced protein aggregation. Plant Physiol. 187, 769–786. doi: 10.1093/plphys/kiab296
McFadden, G. I. (1999). Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 2, 513–519. doi: 10.1016/s1369-5266(99)00025-4
Menges, M., Murray, J. A. (2002). Synchronous arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. doi: 10.1046/j.1365-313x.2002.01274.x
Miernyk, J. A. (2001). The J-domain proteins of arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6, 209–218. doi: 10.1379/1466-1268(2001)0062.0.co;2
Moberg, P., Ståhl, A., Bhushan, S., Wright, S. J., Eriksson, A., Bruce, B. D., et al. (2003). Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J. 36, 616–628. doi: 10.1046/j.1365-313x.2003.01904.x
Mokranjac, D., Bourenkov, G., Hell, K., Neupert, W., Groll, M. (2006). Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 25, 4675–4685. doi: 10.1038/sj.emboj.7601334
Murcha, M. W., Narsai, R., Devenish, J., Kubiszewski-Jakubiak, S., Whelan, J. (2015). MPIC: a mitochondrial protein import components database for plant and non-plant species. Plant Cell Physiol. 56, e10. doi: 10.1093/pcp/pcu186
Murcha, M. W., Whelan, J. (2015). “Isolation of intact mitochondria from the model plant species arabidopsis thaliana and oryza sativa,” in Plant mitochondria. Eds. Murcha, M. W., Whelan, J. (New York, New York: Humana Press), 1–12. doi: 10.1007/978-1-4939-2639-8_1
Nakagawa, T., Ishiguro, S., Kimura, T. (2009). Gateway vectors for plant transformation. Plant Biotechnol. 26, 275–284. doi: 10.5511/plantbiotechnology.26.275
Nakai, M. (2018). New perspectives on chloroplast protein import. Plant Cell Physiol. 59, 1111–1119. doi: 10.1093/pcp/pcy083
Narsai, R., Law, S. R., Carrie, C., Xu, L., Whelan, J. (2011). In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in arabidopsis. Plant Physiol. 157, 1342–1362. doi: 10.1104/pp.111.183129
Neupert, W., Herrmann, J. M. (2007). Translocation of proteins into mitochondria. Annu. Rev. Biochem. 282, 18037-45. doi: 10.1146/annurev.biochem.76.052705.163409
Nickel, C., Horneff, R., Heermann, R., Neumann, B., Jung, K., Soll, J., et al. (2019). Phosphorylation of the outer membrane mitochondrial protein OM64 influences protein import into mitochondria. Mitochondrion 44, 93–102. doi: 10.1016/j.mito.2018.01.005
Nyakundi, D. O., Vuko, L. A., Bentley, S. J., Hoppe, H., Blatch, G. L., Boshoff, A. (2016). Plasmodium falciparum Hep1 is required to prevent the self aggregation of PfHsp70-3. PloS One 11, e0156446. doi: 10.1371/journal.pone.0156446
Pais, J. E., Schilke, B., Craig, E. A. (2011). Reevaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor. Mol. Biol. Cell 22, 4740–4749. doi: 10.1091/mbc.E11-08-0715
Panigrahi, R., Whelan, J., Vrielink, A. (2014). Exploring ligand recognition, selectivity and dynamics of TPR domains of chloroplast Toc64 and mitochondria Om64 from arabidopsis thaliana. J. Mol. Recognit 27, 402–414. doi: 10.1002/jmr.2360
Priesnitz, C., Böttinger, L., Zufall, N., Gebert, M., Guiard, B., van der Laan, M., et al. (2022). Coupling to Pam16 differentially controls the dual role of Pam18 in protein import and respiratory chain formation. Cell Rep. 39, 110619. doi: 10.1016/j.celrep.2022.110619
Sanjuán Szklarz, L. K., Guiard, B., Rissler, M., Wiedemann, N., Kozjak, V., van der Laan, M., et al. (2005). Inactivation of the mitochondrial heat shock protein zim17 leads to aggregation of matrix hsp70s followed by pleiotropic effects on morphology and protein biogenesis. J. Mol. Biol. 351, 206–218. doi: 10.1016/j.jmb.2005.05.068
Schilke, B. A., Ciesielski, S. J., Ziegelhoffer, T., Kamiya, E., Tonelli, M., Lee, W., et al. (2017). Broadening the functionality of a J-protein/Hsp70 molecular chaperone system. PloS Genet. 13, e1007084. doi: 10.1371/journal.pgen.1007084
Schiller, D., Cheng, Y. C., Liu, Q., Walter, W., Craig, E. A. (2008). Residues of Tim44 involved in both association with the translocon of the inner mitochondrial membrane and regulation of mitochondrial Hsp70 tethering. Mol. Cell Biol. 28, 4424–4433. doi: 10.1128/MCB.00007-08
Sharma, M., Bennewitz, B., Klösgen, R. B. (2018). Dual or not dual?–comparative analysis of fluorescence microscopy-based approaches to study organelle targeting specificity of nuclear-encoded plant proteins. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01350
Tamadaddi, C., Sagar, V., Verma, A. K., Afsal, F., Sahi, C. (2021). Expansion of the evolutionarily conserved network of J-domain proteins in the arabidopsis mitochondrial import complex. Plant Mol. Biol. 105, 385–403. doi: 10.1007/s11103-020-01095-8
Truscott, K. N., Voos, W., Frazier, A. E., Lind, M., Li, Y., Geissler, A., et al. (2003). A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163, 707–713. doi: 10.1083/jcb.200308004
Van Aken, O., Zhang, B., Carrie, C., Uggalla, V., Paynter, E., Giraud, E., et al. (2009). Defining the mitochondrial stress response in arabidopsis thaliana. Mol. Plant 2, 1310–1324. doi: 10.1093/mp/ssp053
van der Laan, M., Chacinska, A., Lind, M., Perschil, I., Sickmann, A., Meyer, H. E., et al. (2005). Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell Biol. 25, 7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005
von Känel, C., Muñoz-Gómez, S. A., Oeljeklaus, S., Wenger, C., Warscheid, B., Wideman, J. G., et al. (2020). Homologue replacement in the import motor of the mitochondrial inner membrane of trypanosomes. Elife 9, e52560. doi: 10.7554/eLife.52560
Wiedemann, N., van der Laan, M., Hutu, D. P., Rehling, P., Pfanner, N. (2007). Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179, 1115–1122. doi: 10.1083/jcb.200709087
Willmund, F., Hinnenberger, M., Nick, S., Schulz-Raffelt, M., Mühlhaus, T., Schroda, M. (2008). Assistance for a chaperone: Chlamydomonas HEP2 activates plastidic HSP70B for cochaperone binding. J. Biol. Chem. 283, 16363–16373. doi: 10.1074/jbc.M708431200
Wise, R. R. (2007). “The diversity of plastid form and function,” in The structure and function of plastids. Eds. Wise, R. R., Hoober, J.K. (Dordrecht: Netherlands: Springer), 3–26. doi: 10.1007/978-1-4020-4061-0_1
Keywords: mitochondria, plastids, dual-targeting, PAM, protein import apparatus
Citation: Gill-Hille M, Wang A and Murcha MW (2022) Presequence translocase-associated motor subunits of the mitochondrial protein import apparatus are dual-targeted to mitochondria and plastids. Front. Plant Sci. 13:981552. doi: 10.3389/fpls.2022.981552
Received: 29 June 2022; Accepted: 19 October 2022;
Published: 11 November 2022.
Edited by:
Yan Wang, La Trobe University, AustraliaReviewed by:
Oren Ostersetzer, Hebrew University of Jerusalem, IsraelMinu Chaudhuri, Meharry Medical College, United States
Copyright © 2022 Gill-Hille, Wang and Murcha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika W. Murcha, bW9uaWthLm11cmNoYUB1d2EuZWR1LmF1
 Mabel Gill-Hille
Mabel Gill-Hille Andre Wang
Andre Wang Monika W. Murcha
Monika W. Murcha