- College of Life Sciences, Zhejiang Normal University, Jinhua, China
Bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae (Xoo), is a widespread and destructive disease in rice production. Previously, we cloned an executor R gene, Xa7, which confers durable and broad-spectrum resistance to BB. Here, we further confirmed that the transcription activator-like effector (TALE) AvrXa7 in Xoo strains could directly bind to the effector-binding element (EBE) in the promoter of the Xa7 gene. Other executor R genes (Xa7, Xa10, Xa23, and Xa27) driven by the promoter of the Xa7 gene could be activated by AvrXa7 and trigger the hypersensitive response (HR) in tobacco leaves. When the expression of the Xa23 gene was driven by the Xa7 promoter, the transgenic rice plants displayed a similar resistance spectrum as the Xa7 gene, demonstrating that the disease resistance characteristics of executor R genes are mainly determined by their induction patterns. Xa7 gene is induced locally by Xoo in the infected leaves, and its induction not only inhibited the growth of incompatible strains but also enhanced the resistance of rice plants to compatible strains, which overcame the shortcomings of its race-specific resistance. Transcriptome analysis of the Xa7 gene constitutive expression in rice plants displayed that Xa7-mediated disease resistance was related to the biosynthesis of lignin and thus enhanced resistance to Xoo. Overall, our results provided novel insights and important resources for further clarifying the molecular mechanisms of the executor R genes.
Introduction
Xanthomonas oryzae pv. oryzae (Xoo), the causal agent of bacterial blight (BB) disease in rice (Oryza sativa L.), is a notorious plant pathogenic bacteria that greatly affects global food security (Jiang et al., 2020). The interaction between Xoo strains and rice plants has been regarded as a classic model for studying bacteria–plant interaction (NIÑO-LIU et al., 2006). Xoo strains inject transcription activator-like effectors (TALEs) into rice plant cells through their type III secretion system (T3SS), which is capable of activating the transcription of specific genes. Among them are the susceptibility genes, such as the SWEETs (sugars will eventually be exported transporter) family members, benefiting their own reproduction and invasion, a phenomenon referred to as effector-triggered susceptibility (ETS) (Boch et al., 2014). To combat the attack of Xoo, plants evolved several types of resistance (R) genes that can trap Xoo TALEs and trigger immunity in host cells, which is known as effector-triggered immunity (ETI) (Gu et al., 2005). A typical TALE usually has a secretion signal motif in its N-terminal, a binding domain for target recognition, a transcription factor binding (TFB) domain, highly conserved nuclear localization signals (NLSs), and acidic transcription activator-like domains (AADs) (Szurek et al., 2001). Distinctively, the central repeat domain of TALEs contains a variable number of tandemly arranged 33 to 34 amino acid repeats, and the 12th and 13th, two residues in each repeat called repeat variable di-residues (RVDs), can recognize a specific nucleotide (Boch et al., 2009). Therefore, TALEs can specifically bind to the promoter of R or S genes by recognizing the effector-binding elements (EBEs) and then activating their expression (Schornack et al., 2006)
Executor R genes are regarded as a unique type of disease resistance genes in plants, which can specifically trap TALEs of plant pathogens and trigger strong hypersensitive response (HR) and directly programmed cell death (PCD) in the host. Up to now, six genes belonging to this type have been cloned; among them, Bs3 (Römer et al., 2007) and Bs4C (Strauss et al., 2012) were isolated from pepper (Capsicum frutescens), while Xa27 (Gu et al., 2005), Xa10 (Tian et al., 2014), Xa23 (Wang et al., 2015), and Xa7 (Chen et al., 2021; Luo et al., 2021; Wang et al., 2021) were cloned from rice. Interestingly, each identified executor R gene (except Xa10) has an identical coding sequence (CDS) with its susceptible allele but only differs in the promoter, which has an EBE sequence specifically recognized by a cognate TALE of pathogens, such as AvrBs3, AvrBs4, AvrXa27, AvrXa10, AvrXa23, or AvrXa7 (Ballvora et al., 2001; Szurek et al., 2002; Ji et al., 2022b). Therefore, the executor R genes are usually “silent” and only activated to trigger rapid host-cell death when the invading pathogens are present, effectively preventing further pathogen attacks (Nowack et al., 2022). Evidence has shown that the expression of the executor R genes could trigger cell death in both plant and animal cells (Strauss et al., 2012; Tian et al., 2014; Chen et al., 2021), suggesting that the defense pathway of these executor R genes may be conserved. However, only the executor R gene Bs3 has been known to encode a putative flavin-containing monooxygenase (FMO), which may be involved in the metabolism of reactive oxygen species (Krönauer et al., 2019). Unlike the Bs3 gene, the other five executor R genes encode small unknown proteins, which have 113 to 164 amino acids with two to four transmembrane domains (Ji et al., 2022b; Nowack et al., 2022). In addition, those executor R genes have low similarities in nucleotide sequence and no homology with other R proteins that have been identified. To date, little is known about the molecular mechanisms of the executor R genes.
Recently, we cloned the rice executor R gene Xa7 (Chen et al., 2021; Luo et al., 2021; Wang et al., 2021), which has been confirmed to be a broad-spectrum and durable BB resistance gene (Vera Cruz et al., 2000; White and Yang, 2009). We found that the expression of Xa7 was strongly induced by the incompatible Xoo strains, and a putative EBE sequence (named EBEAvrXa7) in the promoter of Xa7 could be perfectly matched by the RVDs of AvrXa7 (Chen et al., 2021). AvrXa7 has been reported to play dual roles as both virulence factor and avirulence effector in Xoo, which makes the Xa7 gene a broad-spectrum resistance against BB disease (Zhang et al., 2015). To study the disease resistance mechanism of Xa7 gene, here, we confirmed the interaction between AvrXa7 and the EBEAvrXa7, constructed the transgenic plants with different executor R genes driven by the Xa7 promoter to test BB resistance, analyzed the relationship between the expression pattern of Xa7 gene and the infection of Xoo, and explored the defense response pathway of Xa7 gene by RNA-seq. Our results will be helpful and provide novel insights for future research on the molecular mechanisms of the executor R genes.
Materials and methods
Plant materials and growth conditions
IR24 is a susceptible indica rice variety without the Xa7 gene, while IRBB7 is a near-isogenic line of IR24 containing the Xa7 gene. A japonica rice variety Zhonghua11 (ZH11) does not contain the Xa7 gene locus was selected as the wild-type control and transgenic recipient. Transgenic lines constitutively expressing the Xa7 gene (Xa7-OE-1 and Xa7-OE-2) were obtained in our previous research (Chen et al., 2021). The detailed information on rice varieties used in this study is listed in Supplementary Table S3. All rice plants were grown in natural paddy fields during summer in southern China. Tobacco (Nicotiana benthamiana) plants were cultured in a growth chamber at 25°C (16-hour light/8-hour dark).
Yeast one-hybrid assay
A synthetic DNA fragment containing three tandem repeats of the predicted EBEAvrXa7 sequence was inserted into the pLacZ vector (digested with NcoI) via homologous recombination. Subsequently, the engineered pLacZ construct was linearized with NcoI and used for the transformation of the yeast strain YM4271. Positive colonies were able to grow on a nutrient-deficient medium (SD/-Ura) (Breton et al., 2016). A vector expressing the fusion protein of the GAL4 activation domain (AD) and the RVDs of AvrXa7 was constructed using the pGADT7 vector and then transformed into YM4271 harboring the corresponding pLacZ vector. Positive colonies were screened on a nutrient-deficient plate (SD/-Ura/-Leu). β-Galactosidase activity was assessed using a commercially available luminescent β-galactosidase substrate Beta-Glo, which was cleaved and then released d-luciferin for detection using firefly luciferase.
HR assay in tobacco leaves
Constructs of 35S::avrXa7, Xa7pro::Xa7, Xa7pro::Xa10, Xa7pro::Xa23, and Xa7pro::Xa27 were transformed into Agrobacterium tumefaciens strain EHA105 using the heat-shock method. The bacteria were cultured in 20 mL of Lysogeny Broth (LB) liquid medium with 50 mg/L kanamycin and 25 mg/L rifampin until the density reached OD600 = 0.6. The qualified bacterial cells were then collected by centrifugation at 4°C for 10 min at 3,000 rpm and re-suspended in an inoculation medium containing 10 mM MgCl2, 10 mM MES, and 200 μM AS. Leaves of 4-week-old tobacco were inoculated using needleless syringes following the method described by Kay et al. (2007). The inoculated leaves were harvested after 2 days of inoculation and stained using 3,3′-diaminobenzidine (DAB), as described by Daudi and O'Brien (2012).
Vector construction and genetic transformation
The PrimeSTAR HS DNA Polymerase Kit (TaKaRa, Mountain View, CA, USA) was used to amplify the fragments of interest with specific primer pairs listed in Supplementary Table S4. The PCR products were then inserted into the frame vector pCAMBIA1300 using an In-Fusion HD Cloning Kit (TaKaRa). The verified vectors were transformed into the rice japonica variety Zhonghua11 using the Agrobacterium-mediated method, as described previously (Nishimura et al., 2006). DNA sequences of used constructs are shown in Supplementary 5.
Inoculation of Xoo strains
The Xoo strains PXO86 and PXO99 were obtained from the International Rice Research Institute (IRRI) in the Philippines. The engineered strain PXO99avrXa7 was created by our group in the previous research, which contained an avrXa7 overexpression construct in the PXO99 strain (Chen et al., 2021). Among them, PXO86 and PXO99avrXa7 are incompatible strains of the Xa7 gene in the rice variety IRBB7, while PXO99 is a compatible strain of the Xa7 gene in IRBB7. The strains were initially cultured on agar medium containing 20 g/L sucrose, 5 g/L peptone, 0.5 g/L Ca(NO3)2, 0.43 g/L Na2HPO4, and 0.05 g/L FeSO4 at 28°C for 2 days. The bacterial colonies were then eluted with sterile water and diluted to a concentration of OD600 = 1.0 for subsequent inoculation. The leaf-tip clipping method (Kauffman et al., 1973) was used for inoculation of the Xoo strains, and lesions on the inoculated leaves were measured 2 weeks after inoculation to evaluate BB resistance (Yin et al., 2000).
GUS-staining assay
During the amplification of the Xa7 promoter, we obtained a mutated Xa7 promoter, which has two cytosine bases deleted; as a result, the Xa7 gene driven by the mutated Xa7 promoter could not induced by AvrXa7. Thus, we chose this promoter as a native control. We inoculated the leaves of Xa7pro::Gus and Xa7mpro::Gus transgenic plants with PXO86 at the early stage of tillering, and we detected Gus activity by submerging the infected leaves in GUS-staining solution (Zhang et al., 2018).
RT-qPCR assay
Total RNA was extracted from rice leaves using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and was purified with the RNase-Free DNase Set (Qiagen) in accordance with the manufacturer’s instructions. Synthesis of first-strand cDNAs from the extracted RNA was performed using the M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Real-time PCR (qRT-PCR) was carried out using the Fast SYBR Green Master Mix Reagent (Applied Biosystems, Foster City, CA, USA) on an ABI7500 Real-time PCR System. For qRT-PCR in rice, the housekeeping gene Actin was used as the internal control, while in N. benthamiana, the expression of the NbGAPDH was used as the internal control. The expression of target genes was quantified using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Standard errors were calculated based on a minimum of three biological replicates. The primers used are listed in Supplementary Table S4.
Compatible and incompatible strain treatment assay
The leaves of IRBB7 were inoculated with the positive control strain PXO86 (incompatible), the negative control strain PXO99 (compatible), and a mixed control strain (PXO86+PXO99) at the same time, while the remaining treatments were first inoculated with PXO86 strain simultaneously to induce the expression of the Xa7 gene and then inoculated with PXO99 strain at 1 day, 2 days, and 3 days after the initial inoculation, separately. One-month-old rice plants after transplantation were inoculated with Xoo strains at approximately 1 × 109 cfu/mL using the leaf-clipping method described above.
RNA-seq analysis
Leaves of the transgenic plants of Xa7-OE-1 and Xa7-OE-2, as well as the wild-type control ZH11, were harvested with three biological replicates at the tillering stage (2 months old) and immediately frozen in liquid nitrogen. All samples were sent to the Beijing Genomics Institute (BGI) for RNA-seq using a HiSeq 2500 sequencing system (Illumina, San Diego, CA, USA). The raw data were processed to remove the adaptor sequences and low-quality reads by SOAPnuke (Kawahara et al., 2013). Then, the cleaning reads were mapped onto the reference genome sequence of Nipponbare (http://rice.plantbiology.msu.edu/) by Bowtie2 (Cock et al., 2010; Langmead and Salzberg, 2012). The transcriptional level of expression genes was calculated using RSEM (Li and Dewey, 2011). Differentially expressed genes (DEGs) were identified using DEGseq (Wang et al., 2010) with the Log2FC ≥1 or ≤−1 and significance adjusted at p ≤ 0.05. Enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the DEGs was performed on the BGI website (https://report.bgi.com). The raw sequencing data were deposited in the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) with the BioProject accession number PRJNA954312.
Determination of lignin content
The leaves of ZH11, Xa7-OE-1, and Xa7-OE-2 at the early stage of tillering were sampled and then ground into powder after fully drying for 96 hours. The powder was filtered using a 40 mesh sieve, and then 5 mg was weighed for the determination of lignin content. The measurement method was carried out according to the handbook of the Lignin Content Assay Kit (JC2203-S, China). The samples were mixed with acetic acid, and absorbance was measured at 280 nm. The content of lignin (%) was calculated using a computed formula with an extinction coefficient of 17.75 mL·mg−1·cm−1 for rice.
Statistical methods and significance analysis
Data plotting and statistical analysis were performed using GraphPad Prism 8.0 software (https://www.graphpad.com/). Data are shown as the means ± SD, and significance analysis was conducted using a two-tailed Student’s t-test or a one-way analysis of variance (ANOVA) with least significant difference (LSD) multiple comparisons test. Asterisks represent statistical significance as *p < 0.05 and **p < 0.01.
Results
AvrXa7 directly binds to the EBEAvrXa7 in the promoter of the Xa7 gene
In a previous study, a 26-bp putative EBEAvrXa7 sequence (ATAACCCCCCCCCCCCCAGATAACCA) was predicted in the promoter of the Xa7 gene, which matched perfectly with the 25.5 RVDs of AvrXa7 based on the recognition principle between TALEs and EBEs (Chen et al., 2021). To confirm that AvrXa7 can indeed interact with EBEAvrXa7, a yeast one-hybrid assay was carried out. In the reporter vector, three tandem repeats of both Xa7-EBEAvrXa7 and SWEET14-EBEAvrXa7 were inserted upstream of the LacZ gene, while the effector vector (AD-RVD) expressed the RVDs of AvrXa7 fused with the GAL4 activation domain (Figure 1A). The yeast strains co-transformed with Xa7-EBEAvrXa7 and AD-RVD grew on a nutrient-deficient medium and showed significant β-gal activity, as detected by blue staining (Figure 1B). These results indicate that AvrXa7 can directly bind to the EBEAvrXa7 sequence in the promoter of the Xa7 gene.
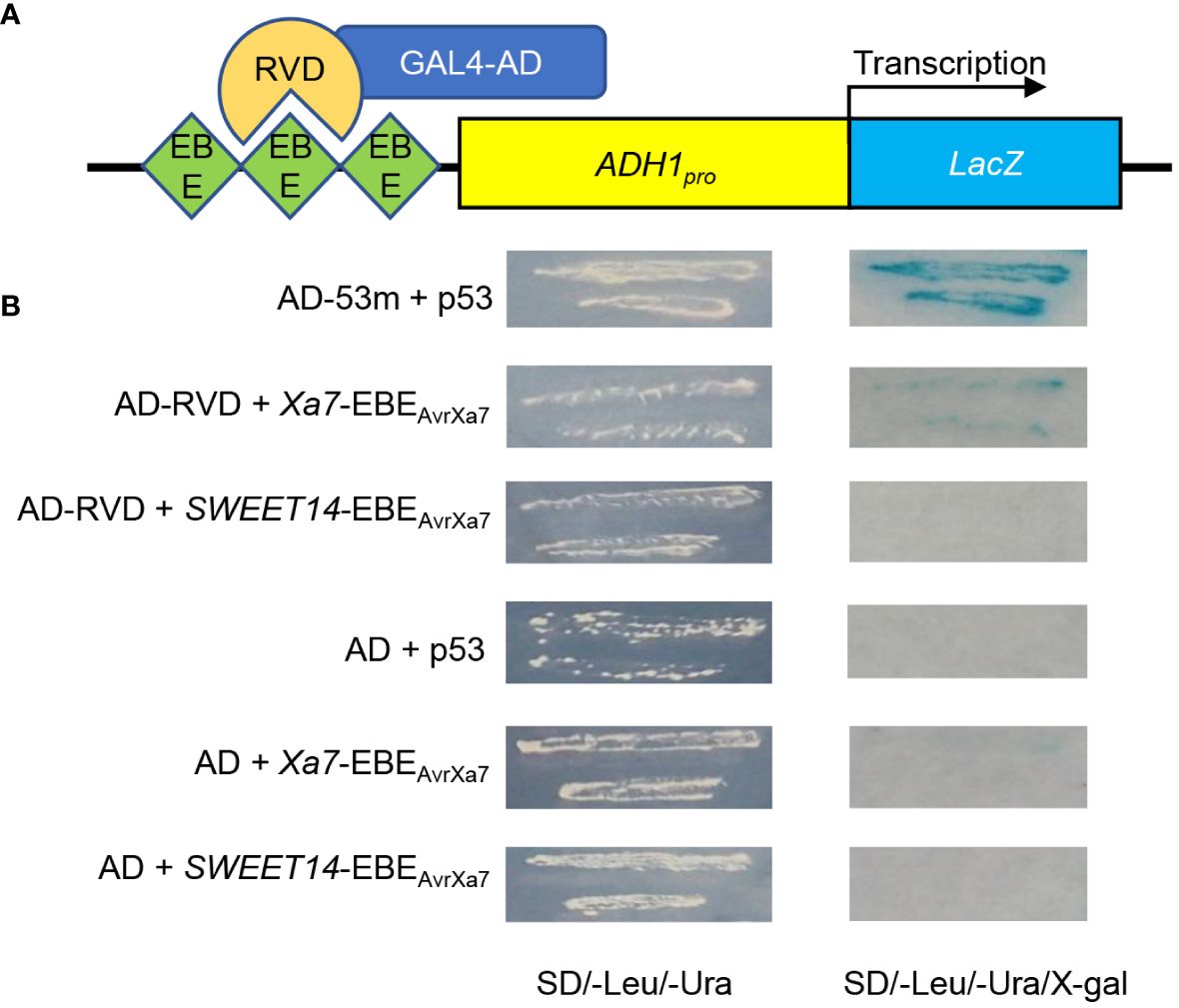
Figure 1 Identification of the interaction between AvrXa7 and Xa7-EBEAvrXa7. (A) Schematic diagram of the yeast one-hybrid system used in this study. Three tandem repeats of Xa7-EBEAvrXa7 and SWEET14-EBEAvrXa7 were inserted upstream of the LacZ gene. The repeat variable di-residues (RVDs) of AvrXA7 (AD-RVD) were fused with the activation domain (AD) of GAL4. ADH1pro refers to the constitutive ADH1 promoter, which drives the expression of the LacZ gene. (B) The β-gal activity test results of different transformants. The interaction between p53Blue and AD-53m was used as the positive control. pLacZ-EBEAvrXa7 represents the reporter vector, while AD-RVD represents the effector vector.
AvrXa7 activates the executor R genes driven by the Xa7 promoter in tobacco
To investigate whether the promoter of the Xa7 gene can activate other executor R genes by binding with AvrXa7, six constructs were created (Figure 2A) and transiently transferred into tobacco for HR analysis—a plant-specific basic immunity that causes infected cell death via the bursts of reactive oxygen species (ROS; Lorrain et al., 2003). Among them, four constructs containing the executor R gene Xa7, Xa10, Xa23, and Xa27 were driven by the native promoter of Xa7 (Xa7pro). The remaining two constructs were driven by the CaMV35S promoter that constitutively expressed Xa7 or GFP (Figure 2A). The empty vector (EV) was used as a negative control. The results showed that significant cell death and excessive ROS accumulation occurred only when transforming the 35S::Xa7 or co-transforming the Xa7pro::executor (Xa7, Xa10, Xa23, or Xa27) with 35S::avrXa7 in the tobacco leaves. No or only slight (Xa7pro::Xa10) HR phenotype was observed in other situations (Figures 2B–E; Supplementary Figure S1). The above results suggested that the promoter of the Xa7 gene can activate other executor R genes to trigger HR in plants by inoculation with AvrXa7.
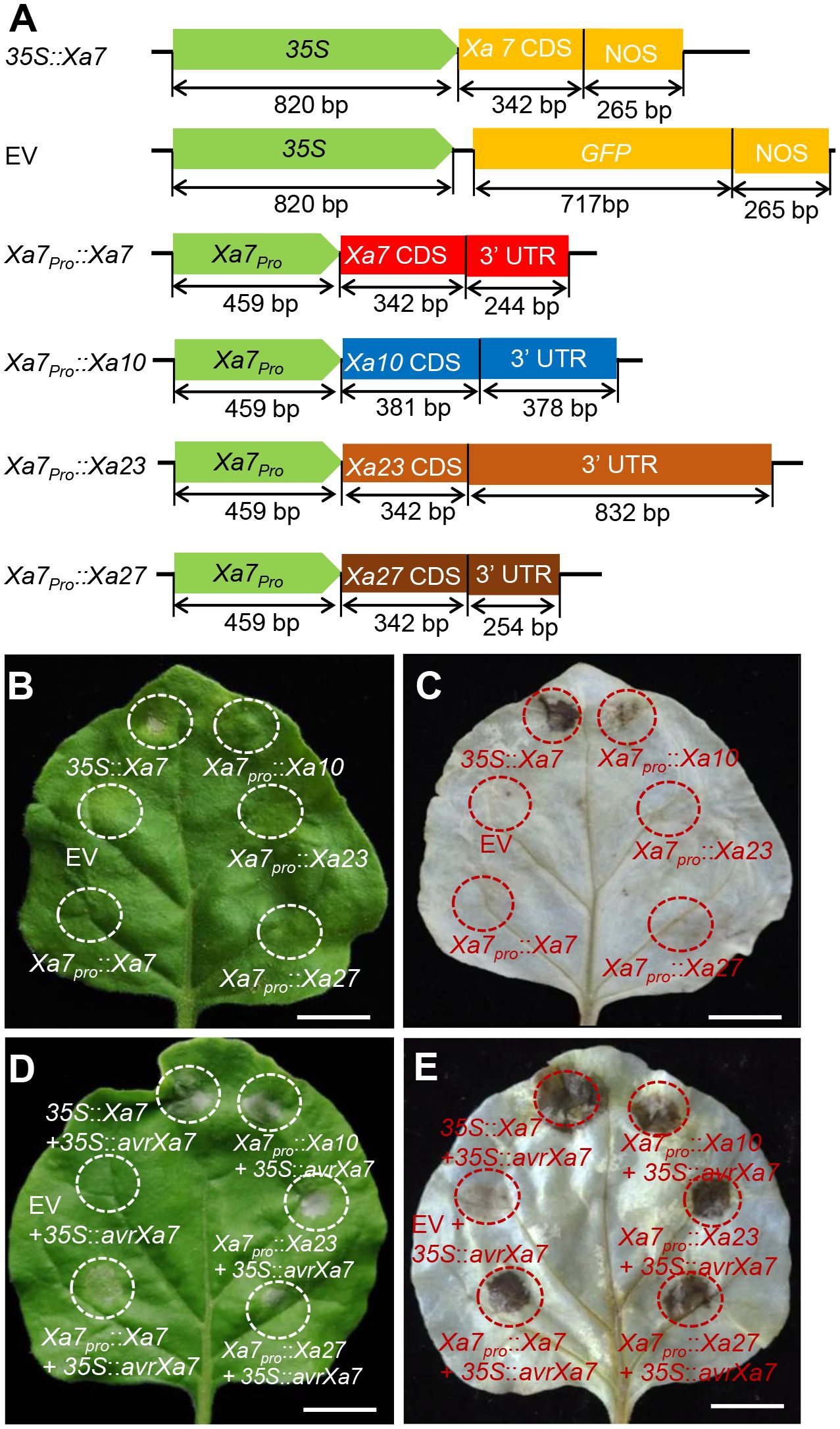
Figure 2 Executor R genes activated by the Xa7 promoter triggered hypersensitive response (HR) in tobacco leaves. (A) Vector diagrams of positive control 35S::Xa7, negative control empty vector (EV), and four executor R genes driven by the Xa7 promoter, named Xa7pro::Xa7, Xa7pro::Xa10, Xa7pro::Xa23, and Xa7pro::Xa27. (B) HR assays in tobacco leaves. Agrobacterium tumefaciens containing different vectors were injected into tobacco leaves using needleless syringes. 35S::Xa7 was selected as the positive control, while EV as the negative control. The infiltrated areas are outlined with dashed circles. (C) The results of 3,3′-diaminobenzidine (DAB) staining assay. The corresponding leaf from (B) was stained with DAB. (D) Phenotype of vectors co-injected with the 35S::AvrXa7 in tobacco leaves. (E) The DAB staining result of corresponding leaves in (D).
The Xa7 gene promoter can endow Xa23 with similar disease resistance characteristics
To further verify whether the executor R gene driven by the Xa7 promoter can enhance the disease resistance of rice plants, the mentioned construct Xa7pro::Xa23 was transformed into a BB-susceptible rice cultivar Zhonghua11 (ZH11). Twenty-five stable transgenic T2 lines were obtained and used for the analysis of the BB resistance (Figure 3A). The transgenic recipient ZH11 was highly susceptible to BB, while the transgenic plants with Xa7pro::Xa23 exhibited a similar disease resistance spectrum of Xa7, instead of Xa23. The Xa7pro::Xa23 transgenic plants were resistant to the Xa7-incompatible strain PXO86 and susceptible to the Xa7-compatible strain PXO99, while rice cultivar CBB23 containing the native Xa23 gene was resistant to both stains (Figures 3B, C). Additionally, the expression of Xa23 gene in those transgenic lines was significantly induced at different time points after PXO86 inoculation (Figure 3D). The above results showed that the executor R gene driven by the promoter of Xa7 gene enhanced host resistance to BB and indicated that the disease resistance characteristics of the executor R genes mainly depend on their promoters.
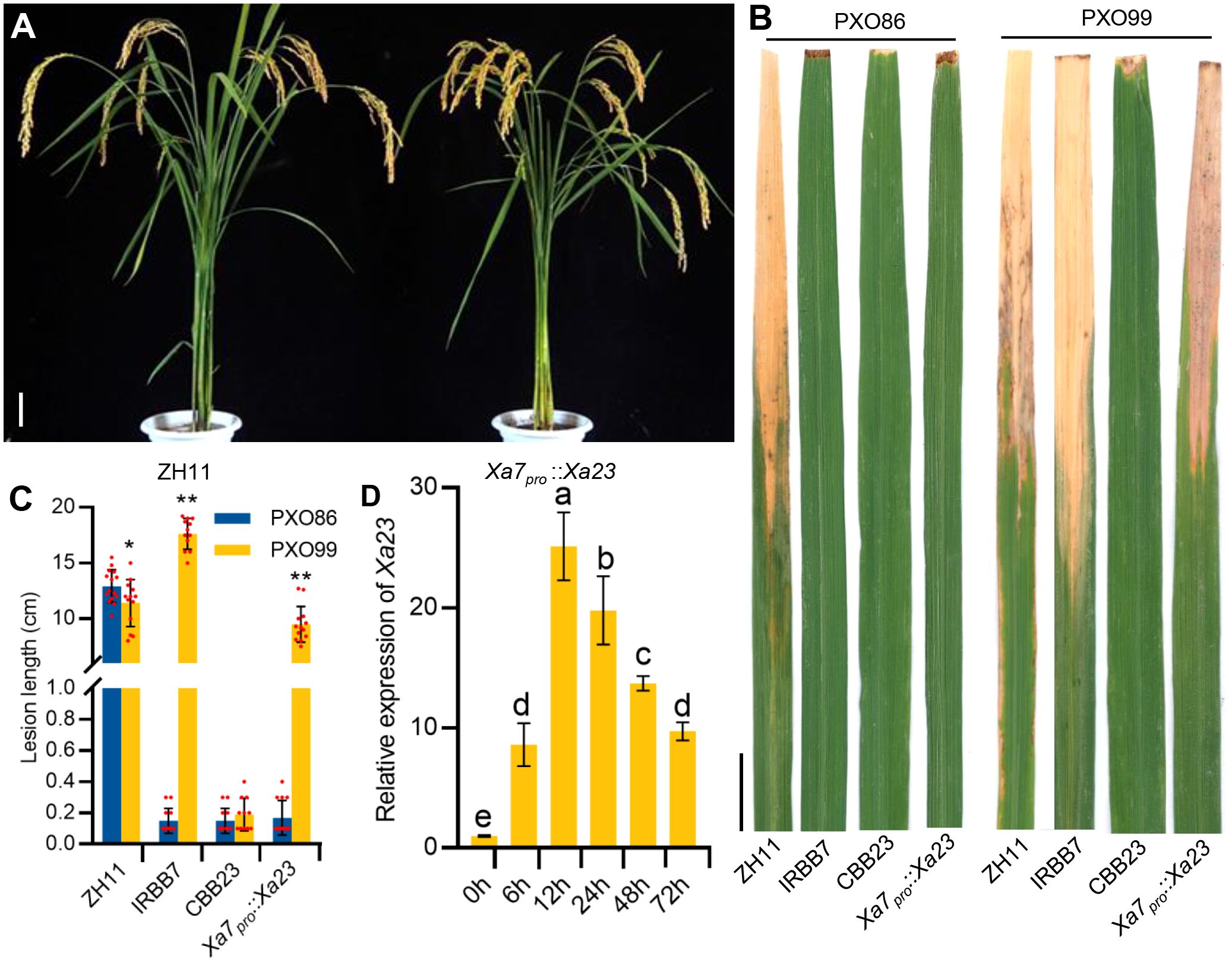
Figure 3 The resistance and expression characteristics of Xa7pro::Xa23 transgenic plants. (A) Phenotypes of ZH11 and Xa7pro::Xa23 transgenic plants at mature stage. Bar = 10 cm. (B) Lesions of ZH11, IRBB7, CBB23, and the Xa7pro::Xa23 transgenic plants inoculated with Xoo strains PXO86 and PXO99. Photographs were taken 2 weeks after inoculation with Xoo strains. Bar = 2 cm. (C) Lesion lengths (cm) of ZH11, IRBB7, CBB23, and the Xa7pro::Xa23 transgenic plants. Data are shown as means ± SD; significance analysis was conducted using a two-tailed Student’s t-test. (D) The induced expression characteristics of Xa23 gene in the Xa7pro::Xa23 transgenic plants. Leaves were sampled at 0 hours, 6 hours, 12 hours, 24 hours, 48 hours, and 72 hours after inoculated with PXO86. Data are shown as means ± SD; significance analysis was conducted using ordinary one-way ANOVA with least significant difference (LSD) multiple comparisons test. * represents significance differences exist at p < 0.05; ** represents significance differences exist at p < 0.01.
Expression of the Xa7 gene is induced locally by its incompatible Xoo strain
To explore the mechanism of Xa7 gene-mediated disease resistance, the expression pattern of the Xa7 gene after inoculation with Xoo strains was analyzed. A native promoter of the Xa7 gene (abbreviated as Xa7pro) and a mutated promoter of the Xa7 gene (abbreviated as Xa7mpro) with two base deletions in the EBEAvrXa7 sequence were constructed upstream of the Gus gene and obtained corresponding transgenic rice plants designated as Xa7pro::Gus and Xa7mpro::Gus. After inoculation with the Xa7 incompatible Xoo strain PXO86, which contains an AvrXa7, GUS activity was significantly detected in leaves of the Xa7pro::Gus transgenic plants, and it was localized around the infection site, while no induced GUS activity was detected in the leaves of the Xa7mpro::Gus transgenic plants (Figure 4A).
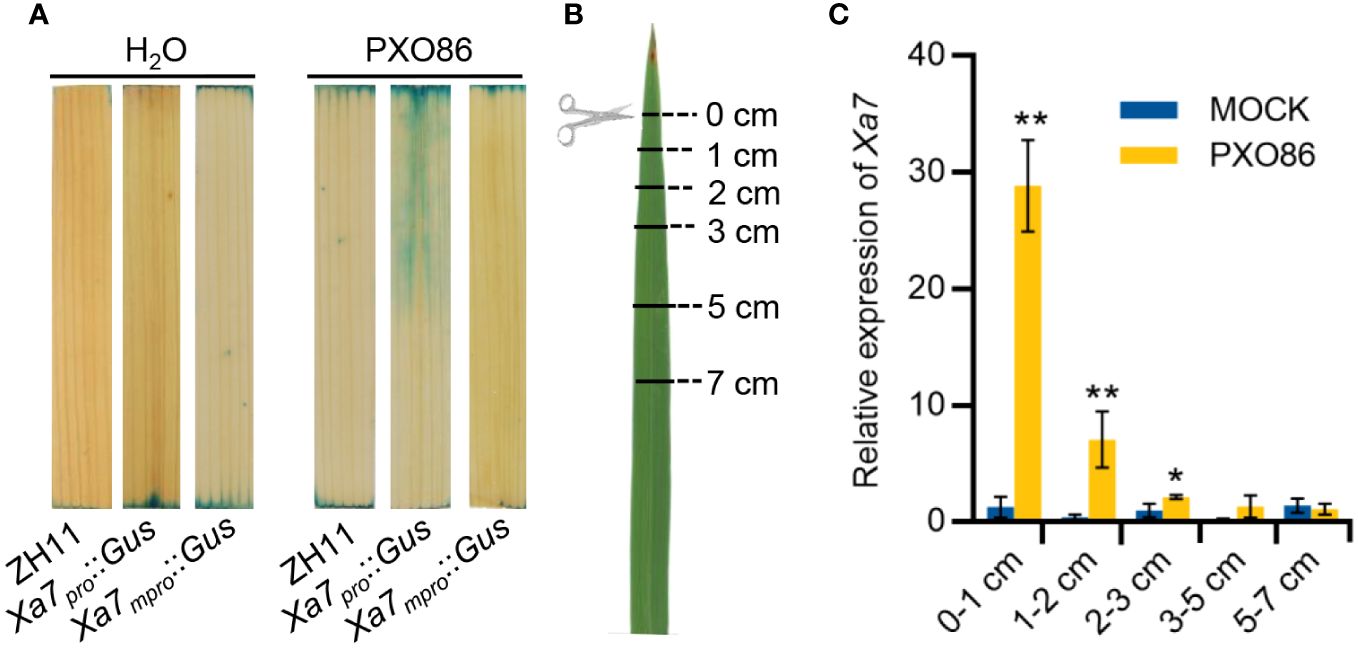
Figure 4 Induction expression characteristics of the Xa7 gene. (A) The GUS staining results of leaves from transgenic plants Xa7pro::Gus and Xa7mpro::Gus. Leaves were stained after inoculated with H2O and Xoo strain PXO86 for 24 hours. (B) Leaves of IRBB7 were inoculated with PXO86 by clipping the leaf tips, and the infected leaves were divided into five different regions away from the clipping site (0–1 cm, 1–2 cm, 2–3 cm, 3–5 cm, and 5–7 cm). (C) Expression of the Xa7 gene in different leaf regions after inoculation with PXO86 and H2O for 24 hours. Gene expression was measured by qRT-PCR, and data are presented as the means of three replicates ± SD; significance analysis was conducted using a two-tailed Student’s t-test; * represents significance differences exist at p < 0.05; ** represents significance differences exist at p < 0.01.
Additionally, leaves from rice cultivar IRBB7 that contain the Xa7 gene were inoculated with PXO86 by clipping the leaf tips. After 24 hours, the infected leaves were collected and divided into five different regions based on distance from the clipping site (0–1 cm, 1–2 cm, 2–3 cm, 3–5 cm, and 5–7 cm) (Figure 4B). The expression level of the Xa7 gene was then analyzed via RT-qPCR. The results showed that expression of the Xa7 gene was strongly induced near the infection site of the leaves (<1 cm) and gradually decreased as the distance increased (Figure 4C). These results suggested that the Xa7 gene was locally induced in the host cells by Xoo infection.
Activation of the Xa7 gene inhibits the growth of incompatible Xoo strain
As the induced expression of the Xa7 gene is localized, it is likely that the pathogen would be controlled locally in the host. To investigate this, the leaves of IR24 and its near-isogenic line IRBB7 were inoculated with PXO86 via leaf-tip clipping, and the growth of Xoo strains was analyzed in the five different regions of the inoculated leaves (Figure 4B) at 1 day, 3 days, 5 days, 9 days, and 14 days after inoculation (DAI) (Figures 5A, B). At 1 DAI, the growth of Xoo was only detected in the 0–1-cm region of the IRBB7 leaves, whereas it was detected in the 0–3-cm region of the IR24 leaves. Subsequently, the growth of Xoo spread on the leaves from the tips of both IRBB7 and IR24. However, the pathogen was limited to the 0–5-cm region of IRBB7 from 5 DAI, while it had spread to areas beyond 7 cm in IR24. Moreover, the amount of Xoo strains in IRBB7 was at least 10-fold lower than that in IR24 over 0–1-cm regions. These results indicated that the induced expression of the Xa7 gene could retard and restrict the growth of Xoo strains.
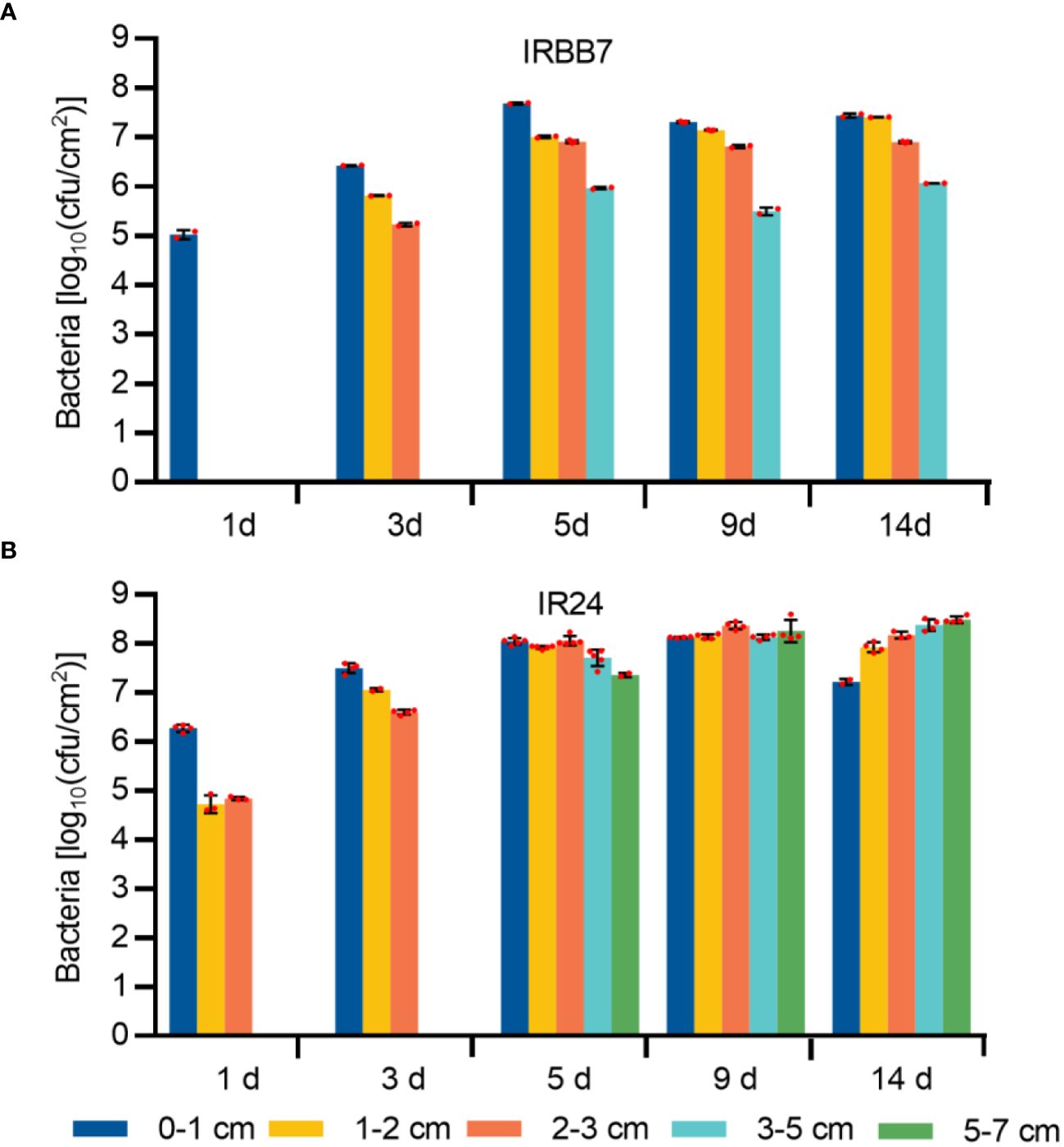
Figure 5 Growth quantity of Xoo strains in the leaves of IRBB7 and IR24 after inoculation with PXO86. (A, B) Population number of Xoo strains after inoculation with PXO86 for 1 day, 3 days, 5 days, 9 days, and 14 days in IRBB7 and IR24, respectively. Leaves were sampled from the regions 0–1 cm, 1–2 cm, 2–3 cm, 3–5 cm, and 5–7 cm after inoculated with PXO86.
The induced expression of the Xa7 gene enhances resistance of rice plants to compatible strains
As the induced expression of Xa7 gene inhibited the growth of incompatible strain PXO86, we hypothesized that the activated expression of the Xa7 gene may also enhance the host’s defense against compatible Xoo strains. To test this hypothesis, a frequently used Xoo strain PXO99, which does not contain an AvrXa7 and cannot activate the expression of the Xa7 gene, was selected as a compatible strain of the Xa7 gene (Chen et al., 2021). First, the leaves of IRBB7 were infected with the incompatible strain PXO86 to activate the expression of the Xa7 gene and then further inoculated with the compatible strain PXO99 by clipping closely to the PXO86-infected site at 1 day, 2 days, and 3 days after PXO86 inoculation (<0.5 cm). The lesion lengths on leaves were measured 2 weeks later after Xoo inoculation. The results showed that the lesions of leaves inoculated with combined PXO86 and PXO99 strains were significantly shorter than those only inoculated with PXO99 (Figures 6A, B). To further verify that the observed effect was solely due to AvrXa7-mediated induction of Xa7, the leaves of IRBB7 were infected with PXO99avrXa7, PXO99, and combined strains (PXO99avrXa7 and PXO99 mixed). The lesion lengths were measured 2 weeks later after Xoo inoculation (Supplementary Figure S2), and the results were consistent with those shown in Figure 6. Thus, the induced expression of the Xa7 gene could significantly increase the resistance of host plants to the compatible strains.
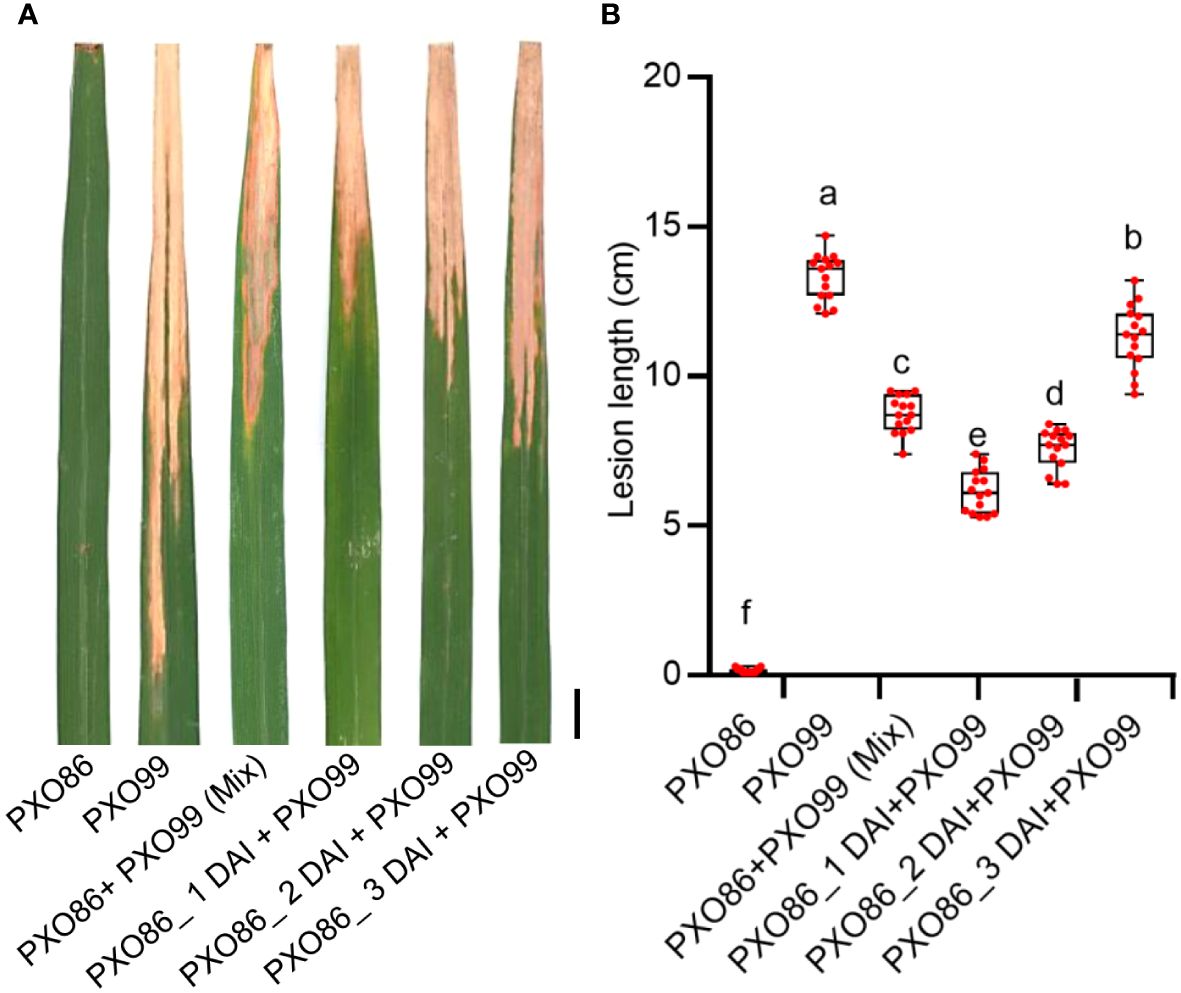
Figure 6 Evaluation of disease resistance to compatible strain PXO99 after induced expression of the Xa7 gene. (A) The lesion phenotype of IRBB7 after inoculation with different races of Xoo strains for 2 weeks. Bar = 1 cm. (B) Statistics of lesion length in panel (A) From left to right, they represent leaves of IRBB7 infected with PXO86, PXO99, PXO86+PXO99 (Mix), and combined inoculation of PXO99 after inoculated with PXO86 for 1 day, 2 days, and 3 days. Data are shown as means ± SD; significance analysis was conducted using ordinary one-way ANOVA with least significant difference (LSD) multiple comparisons test.
Xa7 gene mediates transcriptional reprogramming during disease resistance
To further explore the molecular basis of cell death and defense response mediated by the Xa7 gene, transgenic rice plants constitutively expressing the Xa7 gene that was constructed in our previous study were applied (Chen et al., 2021). Two different lines, named Xa7-OE-1 and Xa7-OE-2, were selected. Compared to the wild-type control ZH11, Xa7-OE-1 plants display normal phenotypes and have low expression levels of the Xa7 gene, while the growth and development of the Xa7-OE-2 plants are obviously inhibited and have significantly higher expression of the Xa7 gene (Figures 7A, B). RNA-seq analysis was performed on both transgenic lines and the wild-type ZH11. Compared to ZH11, 897 and 2,561 DEGs were detected in Xa7-OE-1 and Xa7-OE-2, respectively, and among them, 617 DEGs were shared by the two transgenic lines (Figure 7C), in which 491 genes were collectively upregulated and 87 genes were downregulated (Supplementary Table S1, S2).
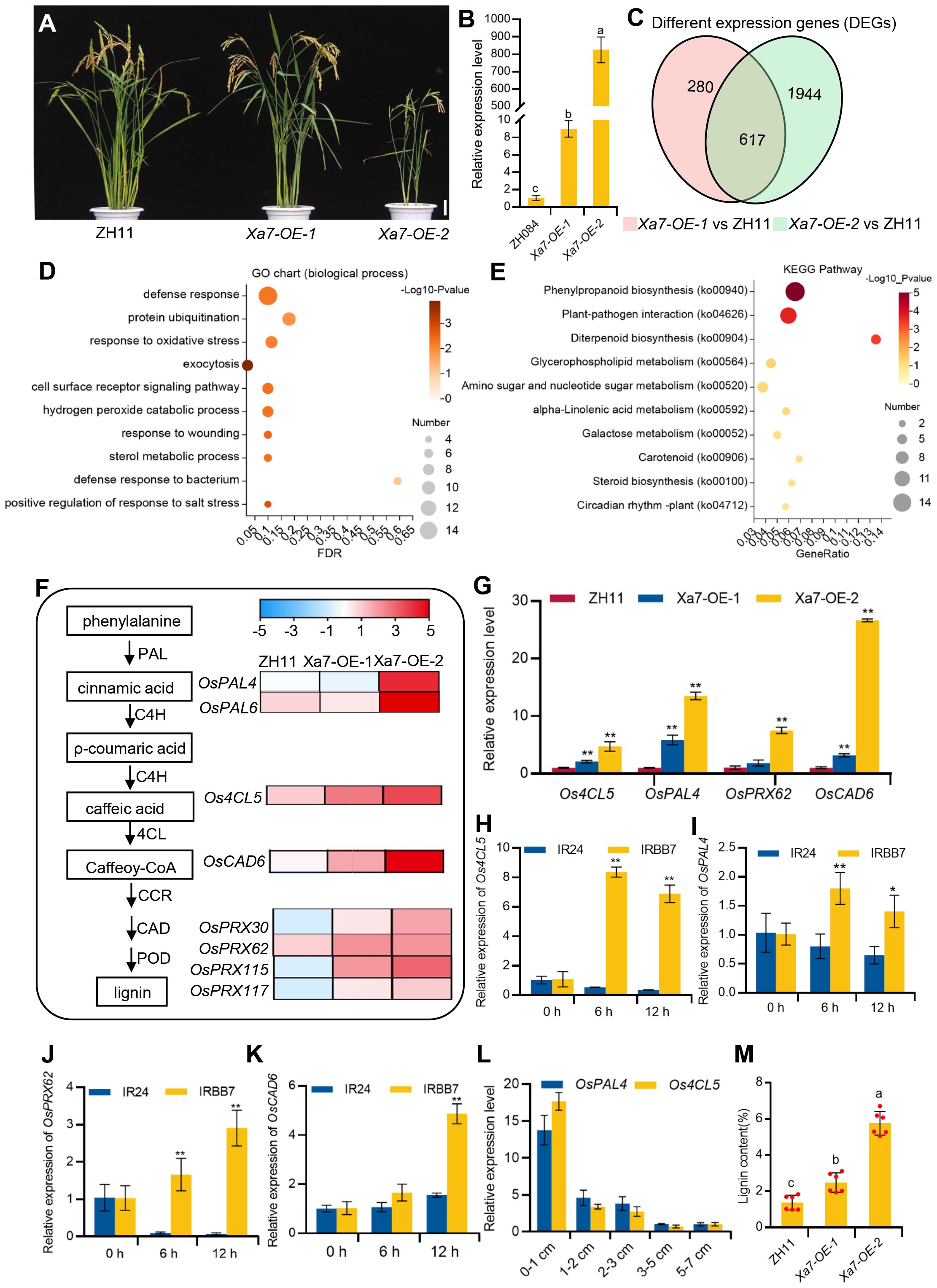
Figure 7 Transcriptome analysis and verification of the Xa7-constitutive expression lines. (A) Phenotypic characterization of ZH11, Xa7-OE-1, and Xa7-OE-2. Bar = 10 cm. (B) qRT-PCR analysis of the relative transcript levels of the Xa7 gene in ZH084, Xa7-OE-1, and Xa7-OE-2 with the absence of Xoo. (C) Venn diagram illustrating the number of differentially expressed genes (DEGs) identified in Xa7-OE-1 and Xa7-OE-2 compared to ZH11. (D) Gene Ontology (GO) enrichment analysis of the DEGs in Xa7-OE-1 and Xa7-OE-2 when compared to ZH11. (E) The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment of the upregulated DEGs between Xa7-OE-1 and Xa7-OE-2 when compared to ZH11. The value on the right side of the column indicates the reliability of the enrichment degree of the pathways (p-value). (F) The heat map of DEGs associated with the phenylpropanoid pathway. Log values of the FPKM of DEGs were used for the display of the heat map. (G) Verification of the relative expression levels of DEGs in rice plants ZH11, Xa7-OE-1, and Xa7-OE-2. Data are shown as means (three independent replicates) ± SD. (H–K) The expression levels of Os4CL5, OsPAL4, OsPRX62, and OsCAD6 in IRBB7 and IR24 at different time points after inoculation with PXO86, respectively. Data are shown with means ± SD. (L) The expression of OsPAL4 and Os4CL5 in different leaf regions 24 hours after IRBB7 inoculated with PXO86. (M) The quantitative determination of lignin content in rice plants ZH11, Xa7-OE-1, and Xa7-OE-2. In panels (B, G–K, M), statistical significance was determined by ordinary one-way ANOVA with least significant difference (LSD) multiple comparisons test. * represents significance differences exist at p < 0.05; ** represents significance differences exist at p < 0.01.
The Gene Ontology (GO) enrichment of the upregulated DEGs showed that genes involved in biological processes and defense response were prevalent (Figure 7D), such as the transcription factor OsWRKY19, which can activate the transcription of PR gene and play an important role in the defense response against pathogens (Du et al., 2021). To further explore Xa7-mediated pathways, KEGG enrichment analysis of the DEGs was performed, and the most enriched pathway was phenylpropanoid biosynthesis (ko00940) (Figure 7E). It has been reported that this pathway is involved in lignin biosynthesis and plays a key role in plant protection under biotic stress (Hamberger et al., 2007; Vogt, 2010). Enzymes such as phenylalanine ammonia-lyase (PAL), 4-coumarate–CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD) are involved in the biosynthesis of monolignol from phenylalanine (Vanholme et al., 2010; Tianpei et al., 2015), and then laccases (Lac) or peroxidases (POD) oxidize monolignols to monolignol radicals in the cell wall, which subsequently combine in a combinatorial fashion to form lignin (Miedes et al., 2014). In the Xa7-OE lines, genes encoding the enzyme involved in lignin biosynthesis, such as PAL (OsPAL4 and OsPAL6), 4CL (Os4CL5), CAD (OsCAD6), and POD (OsPRX30, OsPRX62, OsPRX115, and OsPRX117), were found to be significantly upregulated (Figure 7F).
The transcription levels of selected genes, including OsPAL4, Os4CL5, OsPRX62, and OsCAD6, were verified via qRT-PCR, and the results were consistent with the RNA-seq data (Figure 7G). Furthermore, the expression of these genes was significantly induced in the leaves of IRBB7 but not in IR24 after inoculation with the PXO86 strain (Figures 7H–K). Moreover, qRT-PCR was selected to detect the expression level of two lignin-related genes in the different regions of IRBB7 after inoculation with PXO86 24 hours, and the results showed that the expression pattern was similar to that of the Xa7 gene. Subsequently, quantitative determination of lignin content in the leaves from ZH11, Xa7-OE-1, and Xa7-OE-2 proved that with the increased expression of Xa7, the lignin content was significantly enriched (Figure 7M). The above results suggested that the executor R gene Xa7 may enhance the resistance of rice plants to Xoo pathogens by increasing lignin synthesis.
Discussion
The identification and cloning of executor R genes from rice and pepper that can specifically trap TALEs of pathogens and trigger defense reactions in host plants have revolutionized our understanding of plant–pathogen interactions (Ji et al., 2022b; Nowack et al., 2022). Among the six executor R genes cloned to date, Xa7 is a newly identified executor R gene of rice that contains a putative AvrXa7-target sequence EBEAvrXa7 in its promoter (Chen et al., 2021; Luo et al., 2021; Wang et al., 2021). Here, we experimentally confirmed that AvrXa7 can directly bind to the EBEAvrXa7, suggesting that Xa7 can be activated by the binding of AvrXa7 to its promoter via EBEAvrXa7 (Figure 1).
In fact, the specific recognition between the EBEs of target genes in the host plant and the RVDs of TALEs plays a critical role in determining disease susceptibility or resistance in plants. Variations in the EBE sequences of S genes in plants decreased their susceptibility to pathogen attack (Antony et al., 2010). Conversely, the TALE-dependent R genes of plants have evolved to specifically interact with TALEs to activate immunity. To date, four executor genes, Xa7, Xa10, Xa23, and Xa27, have been cloned from rice, which confer race-specific disease resistance against Xoo strains carrying TALEs AvrXa7, AvrXa10, AvrXa23, and AvrXa27, respectively (Ji et al., 2022b). In this study, we proved that the disease resistance spectrum of Xa7 is largely determined by its promoter, as the executor R gene Xa23 driven by the Xa7 promoter was found to confer similar disease resistance characteristics to Xa7 rather than Xa23 (Figure 3). Promoter modification of R genes has been proposed as an effective strategy for engineering novel recognition specificity between TALEs and EBEs to gain broad-spectrum resistance to pathogens (Li et al., 2016). For instance, in the transgenic rice plants, six EBE sequences specifically recognized by cognate TALEs of PthXo1, PthXo6, Tal9a, Tal4c, Tal2g, and Tal4a were introduced into the promoter of Xa27 gene to achieve broad-spectrum resistance to both Xoo and Xoc (X. oryzae pv. oryzicola) strains (Hummel et al., 2012). Similarly, an additional 17-bp EBE inserted into the promoter of Xa23 in rice plants also specifically trapped conserved TALEs from multiple Xoc strains, providing an alternative strategy to effectively utilize the executor R genes in disease resistance (Ji et al., 2022a). In the future, as more R genes and TALEs are identified, pyramiding the EBEs of major TALEs from pathogens, such as AvrXa7, AvrXa23, PthXo3, and Tal2g, will be a promising strategy for breeding crops with broad-spectrum disease resistance.
In this research, we were surprised to find that Xa7 was locally activated in infected leaves by the incompatible strain PXO86 (Figure 4), and compared to the rice variety IR24, the bacteria grew to similar levels in the infiltration area and significantly deceased outside of the infection site, finally limited inside of the 5-cm region in IRBB7 (Figure 5). This response was similar to the localized acquired resistance (LAR), which did not suppress pathogen growth at the local site but rather halted pathogen propagation into adjacent regions (Jacob et al., 2023). The mechanism of LAR is distinct from that of systemic acquired resistance (SAR), which primes defense in distal tissues to prevent secondary infections (Ross, 1961). Contrary to SAR, LAR is acute and short-lasting, and the HR triggered by an elicitor was found to activate defense in the surrounding cells without direct contact with the elicitor (Dorey et al., 1997). Moreover, Xa7 expression appeared to occur earlier than the onset of cell death (Figure 4). This raises a possibility that complete cell death may not be necessary to generate cell non-autonomous danger signals that activate LAR. Additionally, both the induced and constitutive expression of Xa7 gene enhanced the resistance of rice plants to Xa7-compatible strain PXO99 (Figure 6; Chen et al., 2021), indicating that the LAR activated by the executor genes has no race specificity. An open question is to identify the immunogenic molecules triggering LAR. Candidates like ROS or calcium signaling released from infection cells are worth investigating.
Groundbreaking research on the Xa10 provided exciting insights into how Xa7 initiated immune signaling. Similar to XA7, XA10 was a compact protein featuring four transmembrane α-helices and localized to the endoplasmic reticulum (ER) membrane of plants, which was coincidental with the ER Ca2+ depletion and the onset of XA10-induced cell death (Tian et al., 2014). The ER is one of the important intracellular Ca2+ stores in eukaryotic cells, and the reduction of ER Ca2+ levels can induce the unfolded protein response, an evolutionarily conserved stress response that can trigger apoptosis (Shore et al., 2011). Additionally, the formation of hexamers of XA10 on the ER membrane was reinforced by the WTS protein, which formed pentameric architecture with a central pore and functioned as a new type of Ca2+ permeable cation-selective channel on the ER membrane (Wang et al., 2023). Based on the results obtained in the study of Xa10 and WTS, we propose that Xa7 may form a calcium channel on the ER membrane and transport Ca2+ from the ER to the cytosolic when the pathogens are present; however, this hypothesis remains to be validated further.
In order to investigate the disease resistance mechanism conferred by the Xa7 gene, RNA-seq was performed. In the present study, several genes encoding key enzymes of lignin synthesis were induced in both the Xa7-overexpression lines and the Xa7-containing rice variety IRBB7 after Xoo inoculation, resulting in the accumulation of lignin (Figure 7). Lignin is the second most abundant component in the cell walls of vascular tissues (Neutelings, 2011) and is often rapidly deposited through the induction of lignin biosynthesis genes in responses to pathogen infection in plants (Malinovsky et al., 2014; Yoon et al., 2015). The lignin-mediated vascular defense was also used by other race-specific R genes against vascular pathogens in the rice–Xoo pathosystem. For example, the Xa21-mediated Xoo resistance was related to lignin accumulation (Shamsunnaher et al., 2020). Similarly, Xa10 stimulated the accumulation of lignin-like phenolic compounds (Reimers and Leach, 1991). Based on these findings, it is possible to speculate that Xa7 may enhance the resistance to Xoo by increasing the biosynthesis of lignin, which could change the mechanical strength of the cell wall and enhance resistance against Xoo. This raises the following question: how does Xa7 gene activate lignin-based vascular-specific immune responses upon perceiving Xoo in rice? This will be worthy of further investigation. All in all, the discovery of the disease resistance mechanism of Xa7 opens the door to investigating how the other executor genes confer resistance to Xoo.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA954312.
Author contributions
LH: Writing – original draft, Writing – review & editing. PL: Funding acquisition, Writing – original draft, Writing – review & editing. LM: Writing – review & editing. HL: Writing – review & editing. TB: Writing – review & editing. XC: Writing – review & editing, Funding acquisition. BM: Writing – review & editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (32071987, 32101697, and 32272096) and the Natural Science Foundation of Zhejiang Province (LZ23C130004, LQ22C130005, and LTGN24C13002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1365989/full#supplementary-material
References
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 22, 3864–3876. doi: 10.1105/tpc.110.078964
Ballvora, A., Pierre, M., van den Ackerveken, G., Schornack, S., Rossier, O., Ganal, M., et al. (2001). Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol. Plant Microbe Interact. 14, 629–638. doi: 10.1094/MPMI.2001.14.5.629
Boch, J., Bonas, U., Lahaye, T. (2014). TAL effectors–pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. doi: 10.1111/nph.13015
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-Type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Breton, G., Kay, S. A., Pruneda-Paz, J. L. (2016). Identification of Arabidopsis transcriptional regulators by Yeast One-Hybrid screens using a transcription factor ORFeome. Methods Mol. Biol. 1398, 107–118. doi: 10.1007/978-1-4939-3356-3_10
Chen, X., Liu, P., Mei, L., He, X., Chen, L., Liu, H., et al. (2021). Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacteria-blight disease in rice. Plant Commun. 2, 100143. doi: 10.1016/j.xplc.2021.100143
Cock, P. J., Fields, C. J., Goto, N., Heuer, M. L., Rice, P. M. (2010). The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38, 1767–1771. doi: 10.1093/nar/gkp1137
Daudi, A., O'Brien, J. A. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc. 2, e263. doi: 10.21769/BioProtoc.263
Dorey, S., Baillieul, F., Pierrel, M. A., Saindrenan, P., Fritig, B., Kauffmann, S. (1997). Spatial and temporal induction of cell death, defense genes, and accumulation of salicylic acid in tobacco leaves reacting hypersensitively to a fungal glycoprotein elicitor. Mol. Plant Microbe Interact. 10, 646–655. doi: 10.1094/MPMI.1997.10.5.646
Du, D., Zhang, C., Xing, Y., Lu, X., Cai, L., Yun, H., et al. (2021). The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19 (5), 1052–1064. doi: 10.1111/pbi.13530
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
Hamberger, B., Ellis, M., Friedmann, M., Clarice, D. A. S., Barbazuk, B., Douglas, C. J. (2007). Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The populus lignin toolbox and conservation and diversification of angiosperm gene families. Botany 85, 1182–1201. doi: 10.1139/B07-098
Hummel, A. W., Doyle, E. L., Bogdanove, A. J. (2012). Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195, 883–893. doi: 10.1111/j.1469-8137.2012.04216.x
Jacob, P., Hige, J., Dangl, J. L. (2023). Is localized acquired resistance the mechanism for effector-triggered disease resistance in plants? Nat. Plants. 9, 1184–1190. doi: 10.1038/s41477-023-01466-1
Ji, Z., Guo, W., Chen, X., Wang, C., Zhao, K. (2022b). Plant executor genes. Int. J. Mol. Sci. 23, 1524. doi: 10.3390/ijms23031524
Ji, Z., Sun, H., Wei, Y., Li, M., Wang, H., Xu, J., et al. (2022a). Ectopic Expression of executor gene Xa23 enhances resistance to both bacterial and fungal diseases in rice. Int. J. Mol. Sci. 23, 6545. doi: 10.3390/ijms23126545
Jiang, N., Yan, J., Liang, Y., Shi, Y., He, Z., Wu, Y., et al. (2020). Resistance genes and their interactions with Bacterial Blight/Leaf Streak pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.). Rice 13, 3. doi: 10.1186/s12284-019-0358-y
Kauffman, H. E., Reddy, A., Hsieh, S. P. Y., Merca, S. D. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Kay, S., Hahn, S., Marois, E., Hause, G., Bonas, U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651. doi: 10.1126/science.1144956
Kawahara, Y., de la Bastide, M., Hamilton, JP., Kanamori, H., McCombie, WR., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6 (1), 4. doi: 10.1186/1939-8433-6-4
Krönauer, C., Kilian, J., Strauß, T., Stahl, M., Lahaye, T. (2019). Cell death triggered by the YUCCA-like Bs3 protein coincides with accumulation of Salicylic Acid and Pipecolic Acid but not of Indole-3-Acetic Acid. Plant Physiol. 180, 1647–1659. doi: 10.1104/pp.18.01576
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, J., Meng, X., Zong, Y., Chen, K., Zhang, H., Liu, J., et al. (2016). Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants. 2, 16139. doi: 10.1038/nplants.2016.139
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lorrain, S., Vailleau, F., Balagué, C., Roby, D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. doi: 10.1016/S1360-1385(03)00108-0
Luo, D., Huguet-Tapia, J. C., Raborn, R. T., White, F. F., Brendel, V. P., Yang, B. (2021). The Xa7 resistance gene guards the rice susceptibility gene SWEET14 against exploitation by the bacterial blight pathogen. Plant Commun. 2, 100164. doi: 10.1016/j.xplc.2021.100164
Malinovsky, F. G., Fangel, J. U., Willats, W. G. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5, 178. doi: 10.3389/fpls.2014.00178
Miedes, E., Vanholme, R., Boerjan, W., Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5, 358. doi: 10.3389/fpls.2014.00358
Neutelings, G. (2011). Lignin variability in plant cell walls: contribution of new models. Plant Sci. 181, 379–386. doi: 10.1016/j.plantsci.2011.06.012
NIÑO-LIU, D. O., Ronald, P. C., Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Nishimura, A., Aichi, I., Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Nowack, M. K., Holmes, D. R., Lahaye, T. (2022). TALE-induced cell death executors: an origin outside immunity? Trends Plant Sci. 27, 536–548. doi: 10.1016/j.tplants.2021.11.003
Reimers, P. J., Leach, J. E. (1991). Race-specific resistance to Xanthomonas oryzae pv. oryzae conferred by bacterial blight resistance gene Xa10 in rice (Oryza Sativa) involves accumulation of a lignin-like substance in host tissues. Physiol. Mol. Plant Pathol. 38, 39–55. doi: 10.1016/S0885-5765(05)80141-9
Römer, P., Hahn, S., Jordan, T., Strauss, T., Bonas, U., Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645–648. doi: 10.1126/science.1144958
Ross, A. F. (1961). Systemic acquired resistance induced by localized virus infections in plants. Virology 14, 340–358. doi: 10.1016/0042-6822(61)90319-1
Schornack, S., Meyer, A., Romer, P., Jordan, T., Lahaye, T. (2006). Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 163, 256–272. doi: 10.1016/j.jplph.2005.12.001
Shamsunnaher, Chen, X., Zhang, X., Wu, X., Huang, X., Song, W. (2020). Rice immune sensor XA21 differentially enhances plant growth and survival under distinct levels of drought. Sci. Rep. 10, 16938. doi: 10.1038/s41598-020-73128-7
Shore, G. C., Papa, F. R., Oakes, S. A. (2011). Signaling cell death from the endoplasmic reticulum stress response. Curr. Opin. Cell Biol. 23, 143–149. doi: 10.1016/j.ceb.2010.11.003
Strauss, T., Van Poecke, R. M., Strauss, A., Römer, P., Minsavage, G. V., Singh, S., et al. (2012). RNA-seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc. Natl. Acad. Sci. U S A. 109, 19480–19485. doi: 10.1073/pnas.1212415109
Szurek, B., Marois, E., Bonas, U., Van den Ackerveken, G. (2001). Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26, 523–534. doi: 10.1046/j.0960-7412.2001.01046.x
Szurek, B., Rossier, O., Hause, G., Bonas, U. (2002). Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46, 13–23. doi: 10.1046/j.1365-2958.2002.03139.x
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The rice TAL effector–dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 26, 497–515. doi: 10.1105/tpc.113.119255
Tianpei, X., Li, D., Qiu, P., Luo, J., Zhu, Y., Li, S. (2015). Scorpion peptide LqhIT2 activates phenylpropanoid pathways via jasmonate to increase rice resistance to rice leafrollers. Plant Sci. 230, 1–11. doi: 10.1016/j.plantsci.2014.10.005
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Vera Cruz, C. M., Bai, J., Ona., I., Leung, H., Nelson, R. J., Mew, T. W., et al. (2000). Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. U S A. 97, 13500–13505. doi: 10.1073/pnas.250271997
Wang, C., Chen, S., Feng, A., Su, J., Wang, W., Feng, J., et al. (2021). Xa7, a small orphan gene harboring promoter trap for AvrXa7, leads to the durable resistance to Xanthomonas oryzae pv. oryzae. Rice 14, 1–16. doi: 10.1186/s12284-021-00490-z
Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, W., Qin, L., Zhang, W., Tang, L., Zhang, C., Dong, X., et al. (2023). WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance. Cell 186, 2656–2671.e18. doi: 10.1016/j.cell.2023.05.023
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
White, F. F., Yang, B. (2009). Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. doi: 10.1104/pp.109.139360
Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G. S., et al. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 13, 869–876. doi: 10.1094/MPMI.2000.13.8.869
Yoon, J., Choi, H., An, G. (2015). Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 57, 902–912. doi: 10.1111/jipb.12422
Zhang, Z., Li, J., Tang, Z., Sun, X., Zhang, H., Yu, J., et al. (2018). Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3-OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J. Exp. Bot. 69, 4723–4737. doi: 10.1093/jxb/ery256
Keywords: rice, bacterial blight, executor R genes, Xa7, rice-Xoo interaction
Citation: He L, Liu P, Mei L, Luo H, Ban T, Chen X and Ma B (2024) Disease resistance features of the executor R gene Xa7 reveal novel insights into the interaction between rice and Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 15:1365989. doi: 10.3389/fpls.2024.1365989
Received: 05 January 2024; Accepted: 14 March 2024;
Published: 03 April 2024.
Edited by:
Edmund Kozieł, Warsaw University of Life Sciences, PolandReviewed by:
Benjamin J. Spears, Butler University, United StatesTom Schreiber, Leibniz Institute of Plant Biochemistry, Germany
Copyright © 2024 He, Liu, Mei, Luo, Ban, Chen and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xifeng Chen, eGZjaGVuQHpqbnUuY24=; Bojun Ma, bWJqQHpqbnUuY24=
 Lumei He
Lumei He Pengcheng Liu
Pengcheng Liu