- College of Agriculture and Biology, Liaocheng University, Liaocheng, China
Agrobacterium tumefaciens-mediated genetic transformation has become an important method to study gene function and create new germplasm in plants. The classic genetic transformation process is time-consuming and laborious. In nature, some plants can propagate through detached leaves, which shows that the detached leaves can complete the dedifferentiation of leaf cell and bud differentiation through the intracellular hormone regulation of detached leaves without external hormones. Here, we report a simple leaf-cutting transformation (LCT) method without aseptic operation using Agrobacterium-mediated genetic transformation with detached leaves to complete the transgenic operation. Jonquil (Kalanchoe blossfeldiana), an ornamental plant with leaf propagation ability, was selected to transform via the LCT method using A. tumefaciens strain EHA105 and Agrobacterium rhizogenes K599. Without selection pressure, the visual reporter Ruby, a system for the biosynthesis of betalains, was used in the LCT method. New colorful germplasms of Jonquil accumulated with betalains were obtained. The transgenic Jonquils transformed by A. tumefaciens EHA105 grow normally, but these transgenic Jonquils transformed by A. rhizogenes K599 have abnormal growth, manifested as dwarf phenotype, and most Jonquils had protrusions like tentacles or polyps on their leaves. Stable transformation in Jonquil should be mediated by A. tumefaciens, not A. rhizogenes.
Introduction
Agrobacterium species are renowned for their role in creating genetically modified plants, thanks to their ability to integrate transfer DNA (T-DNA) into the nuclear chromosomes of plant hosts (Ziemienowicz, 2014). Agrobacterium tumefaciens and Agrobacterium rhizogenes are the most widely used Agrobacterium species in plant genetic engineering (Flores-Félix et al., 2020). The A. tumefaciens possesses a tumor-inducing (Ti) plasmid, which it employs to deliver its genetic material into the host’s chromosome. A. tumefaciens-mediated genetic transformation has become the predominant transformation approach in numerous plants. It is widely employed to introduce foreign genes into plants to study gene function or create new germplasm (Hiei et al., 2014; Yadava et al., 2017; Yu et al., 2024). The CRISPR/Cas9 editing technology has emerged as a powerful tool in gene function research and the development of novel plant germplasm (Gao, 2021). In the CRISPR/Cas9 gene editing process, the genetic transformation process is a critical step that involves effectively delivering the CRISPR/Cas9 system (including Cas9 protein and guide RNA) to target cells. A. rhizogenes contains a root-inducing (Ri) plasmid, and a cluster of rooting locus (rol) genes in the T-DNA of Ri can lead to produce hairy roots in recipient plants (Rugini et al., 1991; Bulgakov, 2008). Therefore, A. rhizogenes is often used to induce plants to produce hairy roots and form composite plants, becoming a powerful tool for studying rhizobium legume symbiosis, plant root systems, and other research.
In the process of plant genetic transformation, some plants need to induce callus first. For example, monocotyledonous plants such as rice, corn, and wheat induce callus through mature embryos or young embryos, and then infect the callus through A. tumefaciens-mediated transformation or particle bombardment. The infected callus is further induced to differentiate into buds, then the roots were produced at the base of the stem (Kausch et al., 2021; Wang et al., 2022). Many dicotyledonous plants, such as plants of legumes and Solanaceae, are directly infected with A. tumefaciens through leaves, cotyledons, or hypocotyls as explants. The infected explants are placed on the differentiation medium for callus induction and bud differentiation (Choudhury and Rajam, 2021). The whole transformation process using the A. tumefaciens-mediated genetic transformation method based on tissue culture is carried out in a sterile environment. For most crops, regeneration and genetic transformation are still arduous and highly dependent on species and genotype (Shin et al., 2020; Kausch et al., 2021; Maren et al., 2022; Wang et al., 2022). Some regulatory genes related to growth and development have been identified. Remarkably, these genes can notably enhance regeneration efficiency, curtail the transformation time, and even enable the transformation of recalcitrant genotypes (Maren et al., 2022; Wang et al., 2022; Yang et al., 2024). These studies still focused on how to improve the transformation efficiency and break through the restriction of genotype or species, but they do not reduce the process and steps of genetic transformation operation.
Traditional A. tumefaciens-mediated genetic transformation requires tissue culture in a sterile environment, which is cumbersome and time-consuming, and has high requirements in terms of professional skills and experience. Consequently, streamlining this approach has been regarded as a valuable objective. Moreover, achieving plant transformation without the need for tissue culture is extremely appealing. Floral dip is typically employed for genetic transformation in Arabidopsis thaliana, featuring a simple operation that does not require sterile procedures or tissue culture, aiding in its establishment as a model plant. Regeneration-dependent strategies for new plants can be carried out either within a living plant (in planta) or outside of a living plant (ex planta) (Bélanger et al., 2024), such as the Fast-TrACC (fast-treated agrobacterium co-culture) method that was developed in Nicotiana benthamiana and tomato (Maher et al., 2020). A simple and effective A. tumefaciens-mediated transformation method named regenerative activity-dependent in planta injection delivery (RAPID) was established in sweet potato. The RAPID method does not require tissue culture, overcoming the technical limitations of existing transformation strategies (Mei et al., 2024).
In nature, numerous plants possess the characteristic of root tillering, They are capable of spontaneously sprouting from their roots and subsequently developing stems and leaves. The cut–dip–budding (CDB) method was developed using A. rhizogenes-mediated genetic transformation in root-suckering plants (Cao et al., 2023). Similarly, the CDB delivery system has been constructed with A. rhizogenes-mediated genetic transformation and gene editing in succulent plants (Lu et al., 2024). The CDB method did not require an aseptic operation, but it was mediated by A. rhizogenes K599. However, when A. rhizogenes was used for stable genetic transformation, the resulting transgenic plants often exhibited defects. For instance, transgenic sweet potatoes transformed from A. rhizogenes had wrinkled leaves, and smaller storage roots with many fibrous roots (Otani et al., 1993). The expression of rolB2 gene derived from A. rhizogenes A4 led to a reduction in the growth of Jonquil (Kalanchoe blossfeldiana) plants and resulted in a more compact growth habit (Favero et al., 2021). Similarly, ornamental plants like Sinningia speciosa transformed with A. rhizogenes exhibited shortened peduncles, wrinkled leaves, and delayed flowering (Vereshchagina et al., 2023). The regenerated plants obtained with A. rhizogenes K599 showed wrinkled leaves and more branches when compared to the wild-type Chinese cabbage (Brassica rapa) (Wang et al., 2024). However, the CDB method did not specify whether the transgenic root-suckering plants derived from A. rhizogenes K599 grew normally.
To investigate whether genetic transformation using A. rhizogenes K599 in the ornamental plant Jonquil, which has a strong leaf propagation ability, would lead to abnormal growth, we employed a simple leaf-cutting transformation (LCT) method to transform it using A. rhizogenes K599 and A. tumefaciens EHA105. The LCT method does not require the aseptic operation to complete the transgenic operation. The visual Ruby reporter, which is composed of three key genes (CYP76AD1, DODA, and glucosyltransferase) involved in betalain biosynthesis, was used to monitor transgenic events in this study (He et al., 2020).
Results
Transgenic Jonquil produced with A. tumefaciens EHA105 grows normally
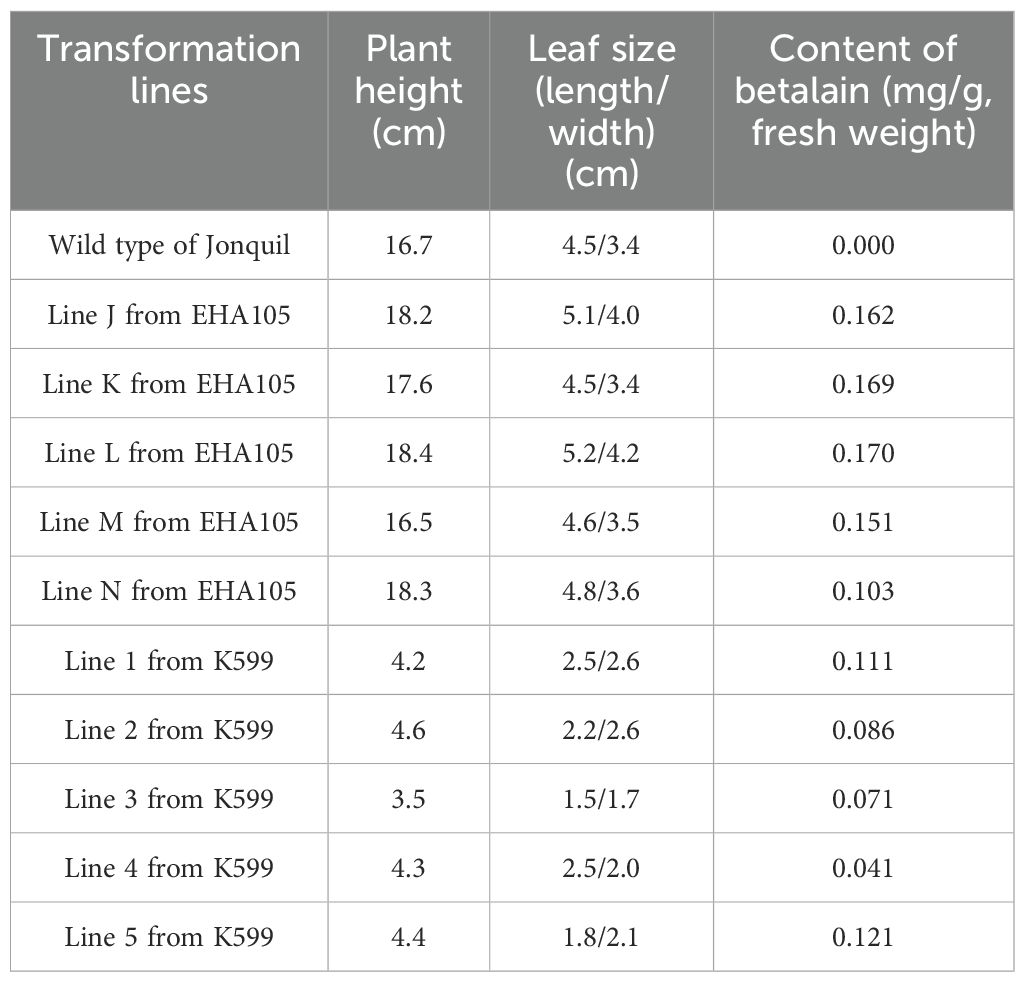
After infecting the propagated leaves with Agrobacterium for 12 weeks of culturing in vermiculite, the cluster of buds formed at the infected part of the petiole of Jonquil (Figure 1A). All explants produced buds, and the regeneration efficiency in Jonquil leaves was 100%, indicating that the Jonquil leaves have a strong reproductive ability. A total of 16 independent explants produced red buds with red roots (Figure 1A) and also green buds. After 15 weeks, these buds developed into Jonquil seedlings (Figure 1B) and then were transplanted into soil (Figures 1C–H). The leaves of red-colored Jonquil seedlings exhibited a variety of red color (Figures 1D–H).

Figure 1. The transgenic Jonquil produced via the LCT method using A tumefaciens strain EHA105. (A) Buds formed at the infected part of the petiole of Jonquil after 12 weeks of transformation. Black arrow: new red buds grow from the propagated leaves. White arrow: propagative leaf. (B) After approximately 15 weeks of transformation, the seedlings of Jonquil appeared with vividly red color. (C–H) One-week-old plants after transplant; (I–N) 8-week-old plants after transplant; (C, I) regenerated wild type. (D–H, J–N) Independent lines with red color. Bars = 1 cm.
After 8 weeks of transplantation, the red-colored Jonquil grew normally like the green plants (Figures 1I–N). The height of the red-colored Jonquil plant was approximately 18 cm (Table 1). Interestingly, there are both red- and green-colored leaves in one red-colored line, and even one leaf showed red and green colors (Figure 1L). The betalain measurement showed that the green part of the color mosaic leaf did not contain betalain, while the red part contained betalain in the color mosaic plant (line L in Figure 1) (Figure 2).
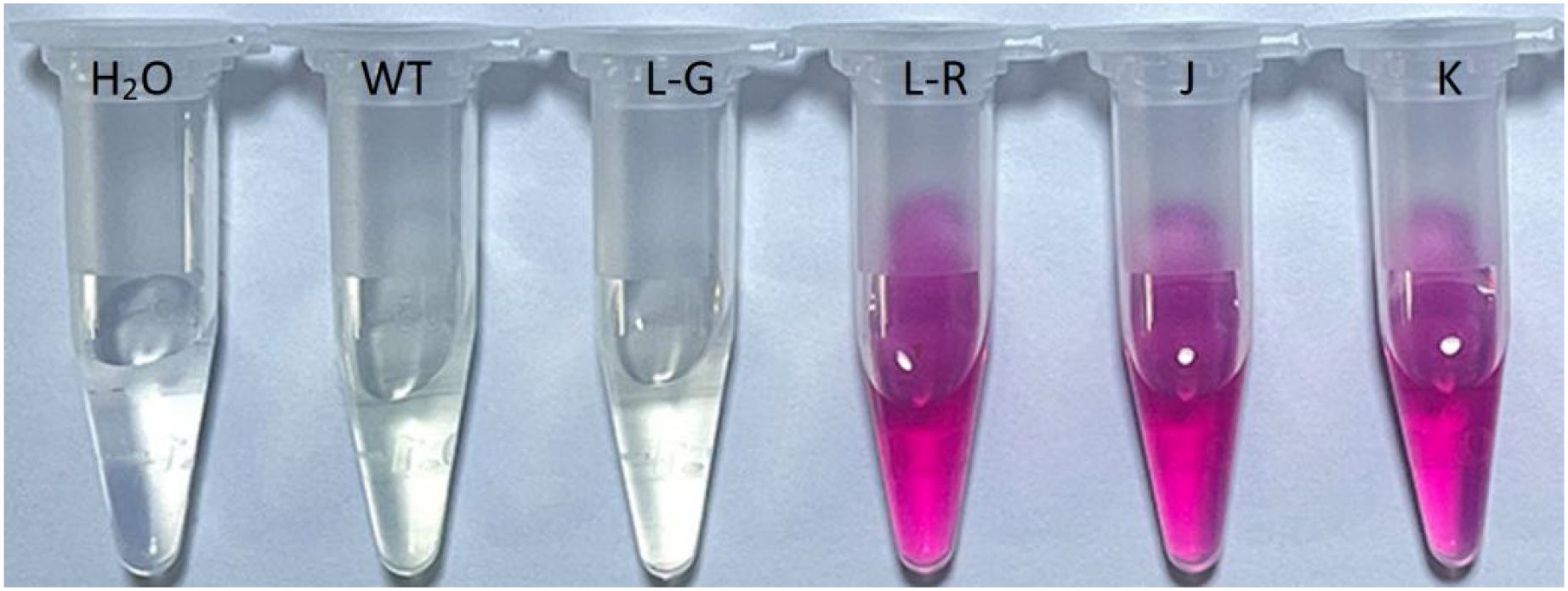
Figure 2. Betalain extract from leaves of red-colored Jonquil plants. H2O: water, WT: regenerated wild type, L-G: from the green part of one leaf showed red and green color in the independent line L in Figure 1, L-R: from the red part of one leaf showed red and green color in the independent line L in Figure 1, J: the independent line J in Figure 1, K: the independent line K in Figure 1.
Transgenic Jonquil produced with A. rhizogenes K599 grows slowly
New shoots were produced at the petiole incisions of all 60 explants of Jonquil after 12 weeks of culture (Figure 3A) and the regeneration efficiency of Jonquil leaves is 100%. Only six independent leaves have produced red color shoots with red roots from the petiole of Jonquil leaves (Figures 3A, B).

Figure 3. The transgenic Jonquil produced via the LCT method using A rhizogenes K599. (A) Buds and (B) seedlings formed at the infected part of the petiole of Jonquil after 12 and 15 weeks of transformation. Red arrows: new red buds grow from the propagated leaf. White arrows: propagative leaf. (C) Regenerated wild-type (WT) and transgenic (T) Jonquil. (C–G) Independent lines with red-colored Jonquils after 8 weeks of transplantation. White arrows: protrusions such as tentacles or polyps. (H) Leaves of regenerated Jonquil. WT: leaf of wild-type Jonquil; T: leaves of red-colored Jonquil. (I) Enlarged leaf part with protrusions such as tentacles or polyps. Bars = 1 cm.
After 8 weeks of transplantation, the height of the wild-type Jonquil was already 15 cm, while all these red-colored Jonquil plants grew slowly and the height of red-colored Jonquil was only 3–4 cm (Figures 3C–G). Among the six red-colored lines obtained with A. rhizogenes K599, five lines had protrusions like tentacles or polyps on their leaves. The number of protrusion structures such as tentacles or polyps differs in different leaves (Figures 3C–I). The length and width of the leaves obtained from K599 transformation are significantly reduced compared with wild-type Jonquil. Three mature leaves in the middle and lower parts of the plant were measured, and the average values were used to evaluate the size of the leaves (Table 1).
PCR identification of red-colored Jonquils
All of the red-colored Jonquils underwent amplification, resulting in the acquisition of an 818-bp fragment. In contrast, when using the propagative leaf as a negative control, no amplified fragment was detected (Figure 4A). The results showed that these red-colored Jonquils were transgenic plants. The genetic transformation efficiencies of A. tumefaciens EHA105 and A. rhizogenes K599 were 16% and 10%, respectively (Table 2).
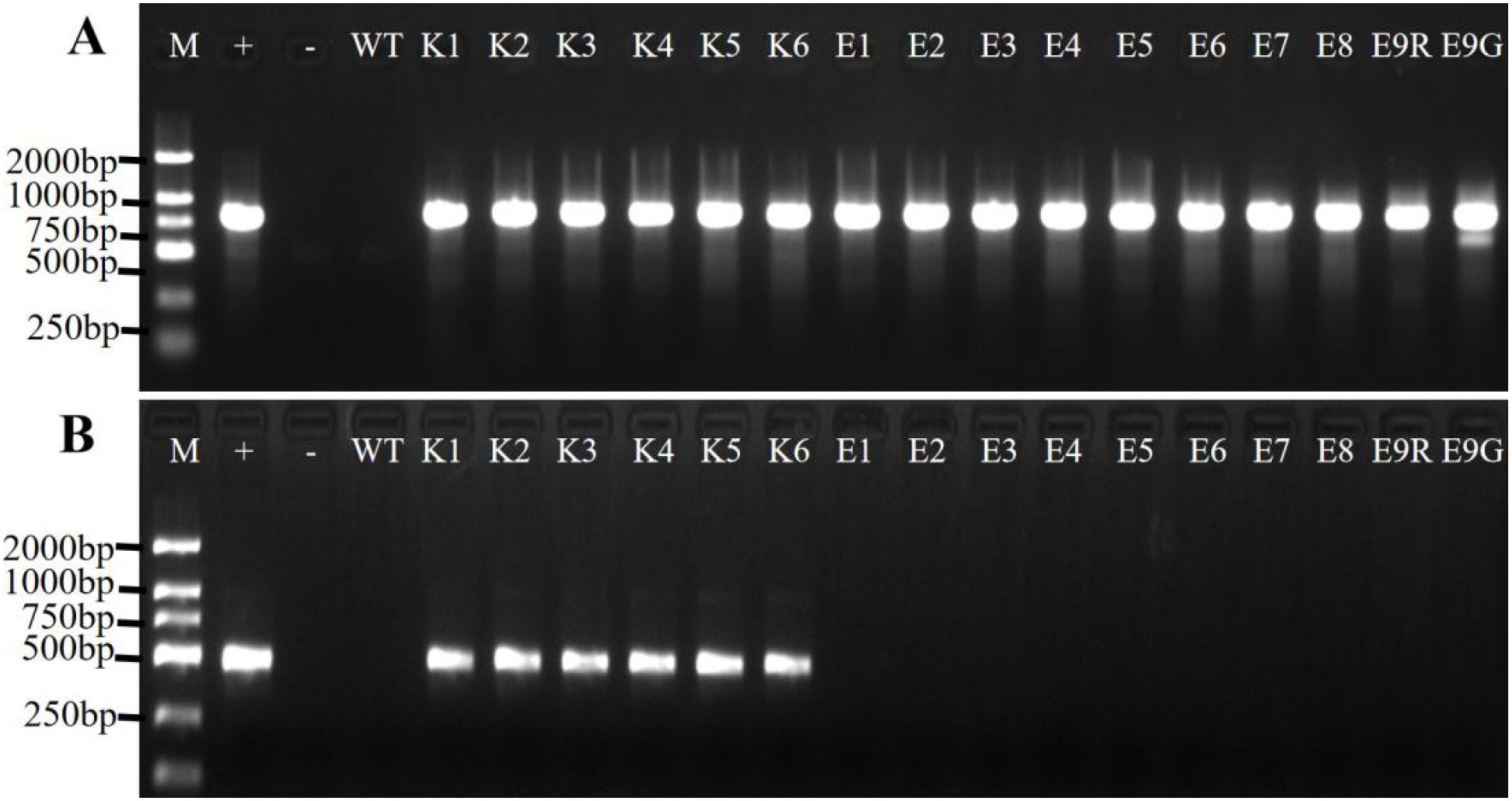
Figure 4. PCR validation with the transgenic Jonquil lines. (A) PCR validation of transformation lines with DoDF/R. (B) PCR validation of the RolB gene with the transgenic Jonquil lines. Lane M, DL2000 DNA marker; Lane +, p35RUBY plasmid; Lane -, negative control (water as template); WT, negative control (propagative leaf); K1–K6, independent transgenic Jonquil lines obtained from A rhizogenes K599; E1–E9, independent transgenic Jonquil lines obtained from A tumefaciens EHA105; E9R and E9G, red-colored and green-colored part form color chimera Jonquil, respectively.
One chimera Jonquil (line E9) with both red- and green-colored leaves was found (Figures 1F, L). Genomic DNAs were extracted from the red and green parts of one leaf, separately. DODA gene was amplified from both parts (Figure 4A), indicating that the color chimera Jonquil was not a transgenic chimera, which may be caused by the silencing of the Ruby gene.
PCR identification with RolB gene on the transgenic Jonquil plants produced with A. rhizogenes K599
The PCR amplification results showed that the 482-bp DNA fragment of RolB gene can be amplified from all the red-colored Jonquils obtained from A. rhizogenes K599, while there is no amplification product in the red-colored Jonquils obtained from A. tumefaciens EHA105 (Figure 4B). This indicated that these red-colored Jonquils obtained from A. rhizogenes K599 carried the RolB gene, which may have been introduced during the genetic transformation process mediated by A. rhizogenes K599.
Discussion
In nature, some plants can propagate by ex vivo leaves; that is to say, the ex vivo leaves of these plants can produce buds without exogenous hormones added. In this study, the regeneration efficiency of the ex vivo leaves of Jonquil is 100%; thus, Jonquil has a strong leaf propagation ability. The wild-type A. tumefaciens contains a Ti plasmid, and the T-DNA of Ti plasmid contains genes that can be integrated into the plant genome (Barton et al., 1983). These genes in T-DNA of the Ti plasmid can change the metabolic pathways of plant cells to produce phytohormones (such as auxin and cytokinin) and some amino acid derivatives, thereby promoting cell proliferation and forming crown galls (Baron and Zambryski, 1996). A. tumefaciens currently employed in plant genetic transformation has undergone artificial modification. Specifically, those genes within the T-DNA that are responsible for inducing crown galls have been deleted. By connecting the target gene into the plant binary vector and then transforming it into the modified A. tumefaciens, the target gene can be introduced into plant cells and stable inheritance can be achieved by infecting the explants with A. tumefaciens (Hoekema et al., 1983). Wild-type A. rhizogenes contains Ri plasmid and conveys its Ri plasmid to the host plant during infection and triggers the growth of a root mass characterized by excessive root proliferation at the infection site (Loyola-Vargas et al., 2024). The rol gene group on the T-DNA of Ri plasmid leads to the production of hairy roots by changing the hormone balance or the sensitivity of cells to auxin and cytokinin. These hairy roots have the characteristics of rapid growth (Rugini et al., 1991; Bulgakov, 2008). A. rhizogenes K599 contains pRi2659 and can induce hairy roots in transformation-recalcitrant plants, like soybean, and has higher transformation efficiency than other strains (Valdes Franco et al., 2016). Transgenic sweet potatoes were obtained with A. rhizogenes-mediated transformation, and the leaves developed from transgenic root tubers were wrinkled, the flower shape was changed, the apical dominance decreased, the internodes shortened, and the storage roots became smaller and grew a lot of fibrous roots; these plants had frequent branches and reduced geotropism (Otani et al., 1993). In this study, all these red-colored Jonquil plants grew slowly, and more than half of transgenic Jonquil plants had protrusions like tentacles or polyps on their leaves. The RolB gene was also amplified from all the red-colored Jonquils obtained from A. rhizogenes K599. Combined with the research results of A. rhizogenes-mediated genetic transformation in Jonquil, S. speciosa, and Chinese cabbage, the transgenic plants showed leaf curling and compact plant architecture (Favero et al., 2021; Vereshchagina et al., 2023; Wang et al., 2024). Therefore, we believe that the phenomena of dwarf symptoms and abnormal leaves are caused by the integration of the plasmid Ri of A. rhizogenes K599 into plants as well. Therefore, A. tumefaciens should be used in plant stable genetic transformation, while A. rhizogenes was usually used to produce composite plants.
There are numerous reports on the method of using hypocotyl injection to induce hairy roots (Kereszt et al., 2007; Meng et al., 2019). After the appearance of hairy roots at the injection site of the stem, the hairy roots are directly exposed to the air and do not come into contact with soil or other media. Therefore, when producing hairy roots by injecting hypocotyls, it is impossible to add antibiotics or herbicides to kill Agrobacterium or select transgenic positive roots. To prevent Agrobacterium from becoming an endophyte of the plant during Agrobacterium-mediated genetic transformation through propagative leaf, we recommend incorporating a cefotaxime cleaning phase. This step should be carried out 3 days after the plant has been infected with Agrobacterium. Nevertheless, we do not advocate the addition of selective antibiotics or herbicides. The underlying reason is that the process of shoot regeneration achieved through the propagation of leaves is highly dependent on the regulation of endogenous hormones within the propagated leaves. This endogenous hormone-mediated regulation plays a crucial role in triggering and facilitating the generation of regenerated shoots, and the introduction of selective antibiotics or herbicides may disrupt this delicate hormonal balance, potentially impeding the successful regeneration of shoots. There is no selection pressure using the LCT method, so the visual reporter gene is preferred. The visualized reporter gene can easily distinguish transgenic plants from non-transgenic plants. The genes that are frequently employed and can function as visualized reporter genes encompass β-glucuronidase (GUS), luciferin, fluorescent protein gene, anthocyanin synthesis regulatory factor (such as AtMYB75), and Ruby (He et al., 2020; Lyu et al., 2024). GUS staining assay is destructive to plant tissues and requires expensive chemical substrates (X-Gluc). Luciferin and fluorescent proteins stand out because of their ability to serve as a non-destructive, visual indicator, but they require optical equipment and/or substrates. Anthocyanin synthesis regulators are limited by the genotype of plant varieties. The reporter system Ruby overcomes the constraints of chemical substrates and optical instruments and can be used in all plant cells (He et al., 2020). Ruby is widely used as a reporter gene in many plants (Wang et al., 2023; Yu et al., 2023; Lyu et al., 2024). The reporter Ruby can also realize quantitative evaluation of gene expression level and promoter activity (Liu et al., 2024). Ruby was used to develop a splicing reporter, enabling the direct visual observation of pre-mRNA splicing (Prasad et al., 2024).
In this study, the genetic transformation efficiency is 16% and 10% with A. tumefaciens EHA105 and A. rhizogenes K599, respectively, with the LCT method using the Ruby reporter gene without selection pressure, while the transformation efficiency mediated by A. rhizogenes K599 was 74% with the CDB delivery system (Lu et al., 2024). The genotypes of the materials and infection solution used in these two methods are different, which may be the main reason for the difference in transformation efficiency. Using the CBD method, leaf segments used as explants were immersed in A. rhizogenes K599 infection solution [10 mM MgCl2, 10 mM 2-(N-morpholino) ethanesulfonic acid (MES), and 100 μM acetosyringone (AS), pH 6.0]. In this study, leaf segments were soaked with bacterial infection solution (OD600 = 0.6) (4.43 g/L M519 + 30 g/L sucrose + 2 mg/L 6-BA + 10 μmol/L AS; pH 5.8). There are differences in the infection solution used for these two transformation methods. MES and 100 μM AS were added in the CBD method, while only 10 μmol/L AS was used in the LCT method. Perhaps due to the absence of MES and the use of lower concentrations of AS, the transformation efficiency of the LCT method decreased. The genetic transformation efficiency of soybean is very low, and adding MES in the medium of genetic transformation in soybean can improve the transformation efficiency (Olhoft et al., 2003). In this study, an area of 32 cm × 45 cm in the culture room was occupied to obtain more than 10 independent transgenic lines; thus, the LCT method is suitable for high-throughput transgenic operations. The LCT method is very simple and does not require aseptic operation. There is no need to change the culture medium containing different hormones as required by the traditional transformation methods. The LCT method can be applied to the transgenic modification of plants that propagate asexually through leaves.
Conclusion
In summary, we have constructed a simple LCT method using Agrobacterium-mediated genetic transformation with detached leaves without aseptic operation to complete the transgenic operation. The LCT method is suitable for plants with leaves that have reproductive ability. The transgenic Jonquils transformed by A. tumefaciens EHA105 grew normally, but these transgenic Jonquils transformed by A. rhizogenes K599 had abnormal growth, manifested as a dwarf phenotype, and most Jonquils had protrusions like tentacles or polyps on their leaves. Consequently, stable transformation in Jonquil should be mediated by A. tumefaciens, not A. rhizogenes.
Methods
Plant materials and growth conditions
The Jonquil variety “Baishuijing” was purchased from the local flower market. These Jonquils were 2 years old and cultivated in a growth chamber (24–26°C, 16-h light/8-h dark cycle) with light intensity at 300 µmol·m−2·s−1. MS power medium (M519) was purchased from Phyto Technology laboratories (Shawnee Mission, Kansas).
Agrobacterium-mediated transformation
p35RUBY was constructed in our previous study (Lyu et al., 2024) and was transformed into A. tumefaciens strain EHA105 and A. rhizogenes K599 using electroporation. EHA105 and K599 harboring with p35RUBY were cultured in LB medium with 50 mg/L kanamycin and 50 mg/L rifampicin, and 50 mg/L kanamycin and 50 mg/L streptomycin, respectively, at 28°C. One clone was picked from Agrobacterium harboring with p35RUBY and cultured in liquid LB medium with the corresponding antibiotics. Bacterials were collected by centrifugation with 2,200g for 10 min at room temperature and resuspended with bacterial suspension solution (OD600 = 0.6) (4.43 g/L M519 + 30 g/L sucrose + 2 mg/L 6-BA + 10 μmol/L AS; pH 5.8).
The LCT method is the same as our previous one-step ARM hair root transformation (Fan et al., 2020). One- to two-month-old leaves as explants were cut from adult Jonquil plants. The incision of the petiole was soaked in the bacterial suspension solution of Agrobacterium harboring p35RUBY for 30 min (Supplementary Figure 1A) and then coated with the Agrobacterium bacterial harboring p35RUBY (Supplementary Figure 1B). The leaves were cultivated in sterile vermiculite wetted with sterile 1/10 MS solution (0.43 g/L M519) (Supplementary Figure 1C). All the explants were covered with transparent plastic bags to maintain humidity (Supplementary Figure 1D). Water was added every 10 days to maintain the humidity of vermiculite. There are 100 and 60 leaves used for the transformation of A. tumefaciens strain EHA105 and A. rhizogenes K599, respectively.
PCR identification with RUBY
To characterize the transgenic lines, the DODA gene, one gene of Ruby, was utilized for identifying the transgene events. The sequence of the DODA gene can be found in He et al. (2020). One leaf of independent line was subjected to DNA isolation. Genomic DNA was extracted according to a previously described method (Fan et al., 2020). The forward primer DoDF (5′-GATGAACGGCGAGGACG-3′) and the reverse primer DoDR (5′-CGGCGGAGGTGAACTTG-3′) were used for the PCR analysis. The PCR program was performed as follows: 94°C for 3 min, then 94°C for 30 s, 58°C for 30 s, and 72°C for 40 s, with 35 cycles, and finally 72°C for 5 min.
The fragment amplified with the primer pair DoDF/DoDR is expected to be 818 bp in size. All red-colored Jonquils were amplified with an expected 818-bp fragment, with the wild-type leaf of Jonquil as a negative control and p35RUBY plasmid as the positive control.
RolB gene identification with PCR amplification
A. rhizogenes K599 carries the pRi2659 plasmid. The sequencing of pRi2659 plasmid has been completed and the sequence was released in GenBank No. CP019703 (Mankin et al., 2007). The pRi2659 plasmid contains a cluster of rol genes in the T-DNA. RolA, RolB, RolC, and RolD in the cluster of rol genes are involved in auxin/cytokinin imbalance (Mankin et al., 2007; Kyndt et al., 2015). The primer set of the RolB gene, the forward primer rolB3 (5′-AGGGCGAGGTCGCTACGA-3′) and the reverse primer rolB4 (5′-GGCGGCGTACTTTAAATGGC-3′), was designed based on plasmid pRi2659, and the PCR fragment is expected to be 482 bp in size. PCR amplification was performed with the genomic DNA extracted from the red-colored Jonquil plants. The PCR program was performed as follows: 94°C for 3 min, then 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s, with 35 cycles, and finally 72°C for 5 min.
Betalain pigment measurement
Betalains were extracted based on the reported method with modification (Chen et al., 2023). A total of 0.5 g of fresh leaf was cut from Jonquil plants and then was immersed into 50 mL of 100% ethanol to remove chlorophyll pigments for 18 h at room temperature. The leaf was then transferred into 5 mL of water for 12 h at room temperature. The 5 mL extraction mixtures were used to measure. The light absorbance of the extraction mixtures was measured in a spectrophotometer at 538 nm wavelength. The calculation of betalain concentrations was detailed in the reported research (García-Cruz et al., 2013). Three biological replicates were performed for each independent line.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
WY: Methodology, Writing – original draft. HT: Writing – original draft, Methodology. YC: Methodology, Writing – original draft. SL: Writing – review & editing, Supervision. YF: Funding acquisition, Writing – original draft, Writing – review & editing, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Shandong province (No. ZR2023MC070) and the open project of Liaocheng University Landscape Architecture Discipline.
Acknowledgments
We thank Yubing He (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, China) for providing the pDR5:RUBY vector.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1594197/full#supplementary-material
References
Baron, C. and Zambryski, P. C. (1996). Plant transformation: A pilus in Agrobacterium T-DNA transfer. Curr. Biol. 6, 1567–1569. doi: 10.1016/S0960-9822(02)70773-2
Barton, K. A., Binns, A. N., Matzke, A. J. M., and Chilton, M.-D. (1983). Regeneration of intact tobacco plants containing full length copies of genetically engineered T-DNA, and transmission of T-DNA to R1 progeny. Cell 32, 1033–1043. doi: 10.1016/0092-8674(83)90288-X
Bélanger, J. G., Copley, T. R., Hoyos-Villegas, V., Charron, J.-B., and O’Donoughue, L. (2024). A comprehensive review of in planta stable transformation strategies. Plant Methods 20, 79. doi: 10.1186/s13007-024-01200-8
Bulgakov, V. P. (2008). Functions of rol genes in plant secondary metabolism. Biotechnol. Adv. 26, 318–324. doi: 10.1016/j.bioteChadv.2008.03.001
Cao, X., Xie, H., Song, M., Lu, J., Ma, P., Huang, B., et al. (2023). Cut–dip–budding delivery system enables genetic modifications in plants without tissue culture. Innovation 4, 100345. doi: 10.1016/j.xinn.2022.100345
Chen, J., Luo, M., Hands, P., Rolland, V., Zhang, J., Li, Z., et al. (2023). A split GAL4 RUBY assay for visual in planta detection of protein–protein interactions. Plant J. 114, 1209–1226. doi: 10.1111/tpj.16234
Choudhury, A. and Rajam, M. V. (2021). Genetic transformation of legumes: an update. Plant Cell Rep. 40, 1813–1830. doi: 10.1007/s00299-021-02749-7
Fan, Y., Zhang, X., Zhong, L., Wang, X., Jin, L., and Lyu, S. (2020). One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. BMC Plant Biol. 20, 208. doi: 10.1186/s12870-020-02421-4
Favero, B. T., Tan, Y., Lin, Y., Hansen, H. B., Shadmani, N., Xu, J., et al. (2021). Transgenic Kalanchoë blossfeldiana, Containing Individual rol Genes and Open Reading Frames Under 35S Promoter, Exhibit Compact Habit, Reduced Plant Growth, and Altered Ethylene Tolerance in Flowers. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.672023
Flores-Félix, J. D., Menéndez, E., Peix, A., García-Fraile, P., and Velázquez, E. (2020). History and current taxonomic status of genus Agrobacterium. Systematic Appl. Microbiol. 43, 126046. doi: 10.1016/j.syapm.2019.126046
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
García-Cruz, L., Valle-Guadarrama, S., Salinas-Moreno, Y., and Joaquín-Cruz, E. (2013). Physical, chemical, and antioxidant activity characterization of pitaya (Stenocereus pruinosus) fruits. Plant Foods Hum. Nutr. 68, 403–410. doi: 10.1007/s11130-013-0391-8
He, Y., Zhang, T., Sun, H., Zhan, H., and Zhao, Y. (2020). A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 7, 152. doi: 10.1038/s41438-020-00390-1
Hiei, Y., Ishida, Y., and Komari, T. (2014). Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00628
Hoekema, A., Hirsch, P. R., Hooykaas, P. J. J., and Schilperoort, R. A. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. doi: 10.1038/303179a0
Kausch, A. P., Wang, K., Kaeppler, H. F., and Gordon-Kamm, W. (2021). Maize transformation: history, progress, and perspectives. Mol. Breed. 41, 38. doi: 10.1007/s11032-021-01225-0
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kyndt, T., Quispe, D., Zhai, H., Jarret, R., Ghislain, M., Liu, Q., et al. (2015). The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: An example of a naturally transgenic food crop. Proc. Natl. Acad. Sci. U.S.A. 112, 5844–5849. doi: 10.1073/pnas.1419685112
Liu, J., Li, H., Hong, C., Lu, W., Zhang, W., and Gao, H. (2024). Quantitative RUBY reporter assay for gene regulation analysis. Plant Cell Environ. 47 (10), 3701–3711. doi: 10.1111/pce.14947
Loyola-Vargas, V. M., Méndez-Hernández, H. A., and Quintana-Escobar, A. O. (2024). “The history of agrobacterium rhizogenes: from pathogen to a multitasking platform for biotechnology,” in Plant cell culture protocols. Eds. Loyola-Vargas, V. and Ochoa-Alejo, N. (Springer US, New York, NY), 51–69. doi: 10.1007/978-1-0716-3954-2_4
Lu, J., Li, S., Deng, S., Wang, M., Wu, Y., Li, M., et al. (2024). A method of genetic transformation and gene editing of succulents without tissue culture. Plant Biotechnol. J. 22, 1981–1988. doi: 10.1111/pbi.14318
Lyu, K., Zhang, X., Yu, W., Lyu, S., and Fan, Y. (2024). Optimization of hairy root transformation and application of RUBY as a reporter in lotus corniculatus. Agronomy 14, 1335. doi: 10.3390/agronomy14061335
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., and Voytas, D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. doi: 10.1038/s41587-019-0337-2
Mankin, S. L., Hill, D. S., Olhoft, P. M., Toren, E., Wenck, A. R., Nea, L., et al. (2007). Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. In Vitro Cell. Dev. Biol.-Plant 43, 521–535. doi: 10.1007/s11627-007-9071-4
Maren, N. A., Duan, H., Da, K., Yencho, G. C., Ranney, T. G., and Liu, W. (2022). Genotype-independent plant transformation. Horticulture Res. 9, uhac047. doi: 10.1093/hr/uhac047
Mei, G., Chen, A., Wang, Y., Li, S., Wu, M., Hu, Y., et al. (2024). A simple and efficient in planta transformation method based on the active regeneration capacity of plants. Plant Commun. 5, 100822. doi: 10.1016/j.xplc.2024.100822
Meng, D., Yang, Q., Dong, B., Song, Z., Niu, L., Wang, L., et al. (2019). Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol. J. 17, 1804–1813. doi: 10.1111/pbi.13101
Olhoft, P. M., Flagel, L. E., Donovan, C. M., and Somers, D. A. (2003). Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method. Planta 216, 723–735. doi: 10.1007/s00425-002-0922-2
Otani, M., Mii, M., Handa, T., Kamada, H., and Shimada, T. (1993). Transformation of sweet potato (Ipomoea batatas (L.) Lam.) plants by Agrobacterium rhizogenes. Plant Sci. 94, 151–159. doi: 10.1016/0168-9452(93)90016-S
Prasad, K. V. S. K., Cheema, A., Scanlon, W., Matthews, A., Sharifova, S., Huq, E., et al. (2024). A simple method to visualize pre-mRNA splicing with the naked eye using a genetically encoded visual splicing reporter. Plant Physiol. 196 (2), 726–730. doi: 10.1093/plphys/kiae396
Rugini, E., Pellegrineschi, A., Mencuccini, M., and Mariotti, D. (1991). Increase of rooting ability in the woody species kiwi (Actinidia deliciosa A. Chev.) by transformation with Agrobacterium rhizogenes rol genes. Plant Cell Rep. 10 (6–7), 291–295. doi: 10.1007/BF00193144
Shin, J., Bae, S., and Seo, P. J. (2020). De novo shoot organogenesis during plant regeneration. J. Exp. Bot. 71, 63–72. doi: 10.1093/jxb/erz395
Valdes Franco, J. A., Collier, R., Wang, Y., Huo, N., Gu, Y., Thilmony, R., et al. (2016). Draft genome sequence of agrobacterium rhizogenes strain NCPPB2659. Genome Announc 4, e00746–e00716. doi: 10.1128/genomeA.00746-16
Vereshchagina, Y. V., Mironova, A. A., Bulgakov, D. V., and Bulgakov, V. P. (2023). Proteomic analysis of proteins related to defense responses in arabidopsis plants transformed with the rolB oncogene. IJMS 24, 1880. doi: 10.3390/ijms24031880
Wang, K., Shi, L., Liang, X., Zhao, P., Wang, W., Liu, J., et al. (2022). The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 8, 110–117. doi: 10.1038/s41477-021-01085-8
Wang, B., Wang, Y., Deng, Y., Yao, Q., and Xiong, A. (2023). Synthesis of betanin by expression of the core betalain biosynthetic pathway in carrot (Daucus carota L.). Hortic. Plant J. 10 (3), 732–742. doi: 10.1016/j.hpj.2023.05.012
Wang, Y., Yang, X., Wang, W., Wang, Y., Chen, X., Wu, H., et al. (2024). Efficient genetic transformation and gene editing of Chinese cabbage using Agrobacterium rhizogenes. Plant Physiol. 197 (2). doi: 10.1093/plphys/kiae543
Yadava, P., Abhishek, A., Singh, R., Singh, I., Kaul, T., Pattanayak, A., et al. (2017). Advances in maize transformation technologies and development of transgenic maize. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01949
Yang, W., Zhai, H., Wu, F., Deng, L., Chao, Y., Meng, X., et al. (2024). Peptide REF1 is a local wound signal promoting plant regeneration. Cell 187, 3024–3038.e14. doi: 10.1016/j.cell.2024.04.040
Yu, J., Deng, S., Huang, H., Mo, J., Xu, Z.-F., and Wang, Y. (2023). Exploring the potential applications of the noninvasive reporter gene RUBY in plant genetic transformation. Forests 14, 637. doi: 10.3390/f14030637
Yu, Y., Yu, H., Peng, J., Yao, W. J., Wang, Y. P., Zhang, F. L., et al. (2024). Enhancing wheat regeneration and genetic transformation through overexpression of TaLAX1. Plant Commun. 5, 100738. doi: 10.1016/j.xplc.2023.100738
Keywords: Jonquil (Kalanchoe blossfeldiana), leaf propagation, Agrobacterium tumefaciens, Agrobacterium rhizogenes, genetic transformation, leaf-cutting-transformation
Citation: Yu W, Tian H, Chen Y, Lyu S and Fan Y (2025) Stable transformation mediated by Agrobacterium tumefaciens in Jonquil is better than Agrobacterium rhizogenes. Front. Plant Sci. 16:1594197. doi: 10.3389/fpls.2025.1594197
Received: 15 March 2025; Accepted: 10 June 2025;
Published: 26 June 2025.
Edited by:
Phanikanth Jogam, Kakatiya Medical College, IndiaReviewed by:
Debee Prasad Sahoo, KIIT University, IndiaVijay Sheri, Texas Tech University, United States
Copyright © 2025 Yu, Tian, Chen, Lyu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinglun Fan, ZmFueWluZ2x1bkBsY3UuZWR1LmNu
 Wenjie Yu
Wenjie Yu Shanhua Lyu
Shanhua Lyu Yinglun Fan
Yinglun Fan