- Guangdong Provincial Key Laboratory of Plant Adaptation and Molecular Design, Guangzhou Key Laboratory of Crop Gene Editing, Innovative Center of Molecular Genetics and Evolution, School of Life Sciences, Guangzhou University, Guangzhou, China
FD gene family encodes transcription factors with a basic region/leucine zipper (bZIP) domain that play an essential role in floral transition regulation, which is vital for plants’ reproduction. Recent studies have uncovered additional functions for FD gene family in plant development, hormone signaling, and response to environmental cues. These pleiotropic roles make them promising targets for modern crops’ breeding. Here, we systematically review the diverse functions and regulation mechanisms of FD gene family in model plants and several crops, to provide important insights into their roles. By summarizing the current understanding of their molecular mechanisms, we aim to highlight their potential as key targets for improving crop yield, stress tolerance, and adaptation to changing climates. Furthermore, we propose future research directions, these efforts will pave the way for the effective utilization of them in modern crop breeding programs.
1 Introduction
Plant basic region/leucine zipper (bZIP) transcription factors function in many biological processes (Corrêa et al., 2008; Dröge-Laser et al., 2018; Yue et al., 2023). In the model species Arabidopsis thaliana, bZIP genes are classified into 13 groups (designated A-M), most of which display group-specific properties. FD and its homolog FD PARALOG (FDP) belong to group A and are involved in the control of floral transition, which is an important developmental process for angiosperms (Dröge-Laser et al., 2018; Romera-Branchat et al., 2020). Appropriate flowering time ensures reproduction success, seed set, and crop yield. Flowering time is regulated by signals from different pathways, such as age pathway, autonomous pathway, gibberellin pathway, photoperiod pathway, and vernalization pathway (Fornara et al., 2010; Wang et al., 2024). To govern flowering time by integrating signals from multiple pathways, FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1/AGL20), and LEAFY (LFY) are key floral integrators in promoting floral transition (Araki, 2001; Parcy, 2005; Lee and Lee, 2010; Hiraoka et al., 2013). FD, which is required for FT protein activity, also integrates flowering signals from different regulatory pathways (Abe et al., 2005; Wang et al., 2009; Benlloch et al., 2011; Seedat et al., 2013; Park et al., 2023). The FT protein is transported from the leaves to the SAM via the vascular tissue and interacts with FD to form the FT-FD complex, which in turn activates the expression of SOC1, APETALA1 (AP1), and LFY, thereby promoting plant flowering (Abe et al., 2005; Wigge et al., 2005; Park et al., 2023). Recent studies report that these floral integrators influence agronomic traits at the same time (Cai et al., 2020; Han et al., 2021; Kou et al., 2022). Therefore, understanding the mechanisms underlying flowering regulation by FD has significant implications for plant breeding and crop productivity.
The architecture of plants is tightly controlled by the identity and activity of meristems: during floral induction, the SAM transforms from a vegetative meristem to an inflorescence meristem (Zhu et al., 2021). The florigen FT promotes floral transition, whereas its homologous protein TERMINAL FLOWER 1 (TFL1) from the same family functions oppositely (Liu et al., 2021). FD interacts with either FT or TFL1, and as a weak activator, FD is converted into a strong activator by FT but into a repressor by TFL1 (Ahn et al., 2006). Recent studies have uncovered roles for FD gene family in inflorescence structure, stem growth, bud formation, and flower development (Tsuji et al., 2013; Sussmilch et al., 2015; Dutta et al., 2021). The morphogenetic effects induced by FD has a strong impact on plant architecture, thus FD homologs play crucial roles in biomass accumulation and plant production.
Crop yield is reduced when plants are exposed to extreme environmental conditions such as high salt, drought, cold, and heat. Plant bZIP transcription factors are considered as abiotic stress regulators, such as AtbZIP15 and AtbZIP35-AtbZIP38, which are involved in abscisic acid (ABA) and stress signaling (Choi et al., 2000; Uno et al., 2000). Similar functions have been reported in FD homologs, which provides a valuable basis for crop yield study in the future.
In this review, we analyze the phylogenetics and protein structures of members of the FD family, explore recent advances of the novel roles of FD in various species, comprehensively reveal the regulatory mechanisms of FD in floral transition, plant development, and environmental signal response. Ultimately, we provide perspectives for their further utilization in crop breeding.
2 Divergence of FD homologs
Full-length amino acid sequences of FD homologs were obtained from the Arabidopsis database TAIR, Phytozome database, and NCBI database. AtbZIP68 and AtbZIP16 (belong to group G) were used as outgroup proteins (Dröge-Laser et al., 2018) (Supplementary Table S1). We performed multiple sequence comparisons using MEGA7 software and conducted a phylogenetic analysis using the maximum likelihood method (Kumar et al., 2016). NCBI Batch CD-Search website (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) was used for structural domain prediction and MEME Motif Discovery (https://meme-suite.org/meme/tools/meme) was used to analyze the protein domains (Bailey et al., 2009). TBtools was used to visualize the results (Chen et al., 2020a).
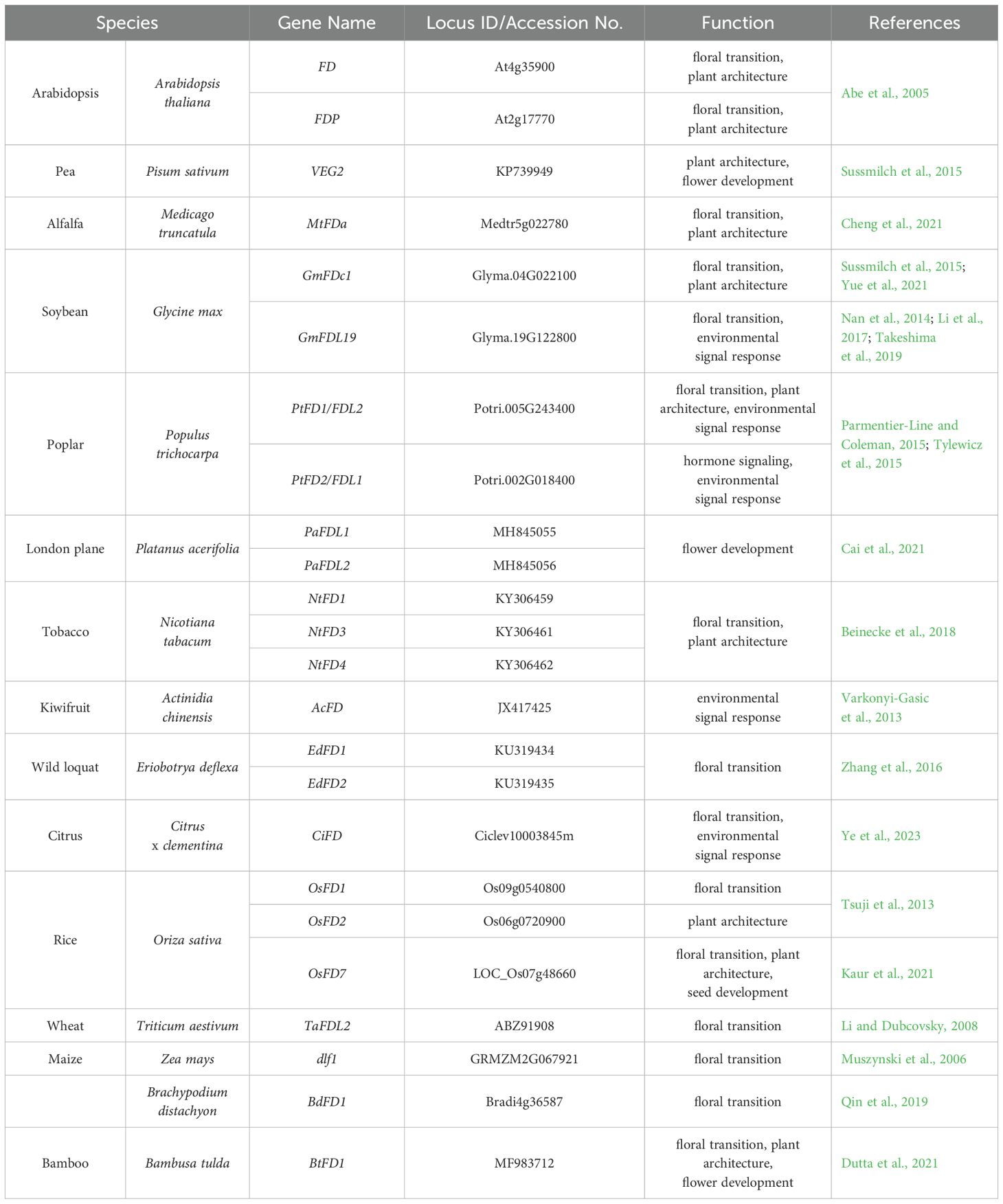
Based on the analysis, the FD homologs could be divided into three groups (Group I-III) (Figure 1). All FD homologs contain a bZIP domain; the basic region is responsible for binding to specific DNA sequences, and the leucine zipper motif is required for dimerization (Dröge-Laser et al., 2018). The SAP (C-terminal phosphorylation) motif, which is conserved in most FD-like proteins, is important for FD phosphorylation and dimeric 14-3–3 protein bridge binding (Tsuji et al., 2013). All FD proteins in Group I are derived from monocots and lack the A motif. It’s reported that OsFD2 regulates inflorescence architecture (Tsuji et al., 2013), but the exact functions of most members in Group I are unresolved. All FD proteins in Group III are clustered together with ABI5, AREBs, and ABFs, most of them lack the LSL motif. They might have similar functions with ABI5, AREBs, and ABFs. Although the SAP, A, and LSL motifs are conserved in most Group II FD homologs, the FD proteins from various eudicots species have divergent functions.
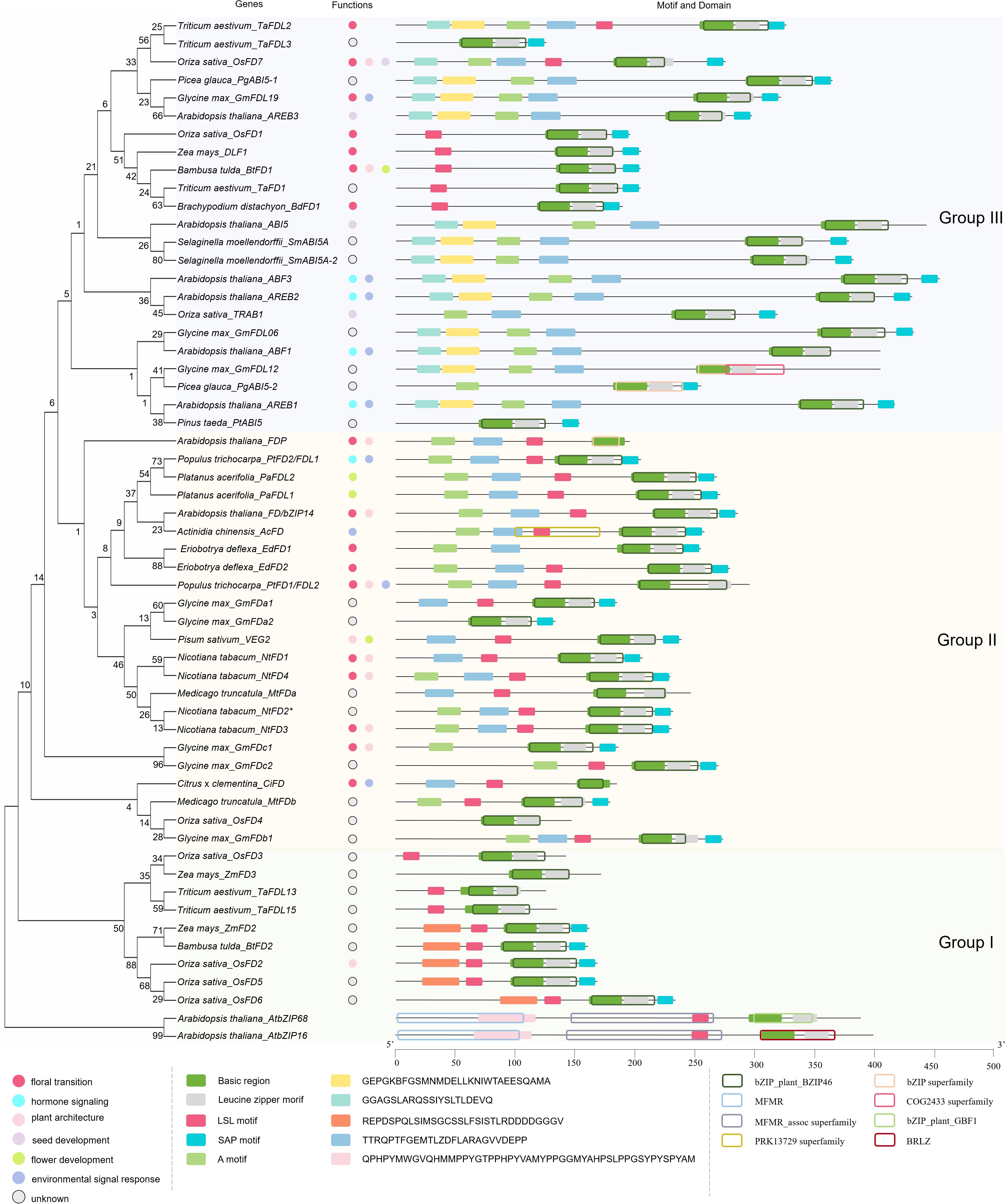
Figure 1. Phylogenetic analysis, protein structures, and functions of the FD proteins in plants. Basic region: DRRQKRMIKNRESAARSRARKQAYTNELE, leucine zipper motif: EVARLKEENARLKKQQEZLKE, SAP motif: LPKKKTLRRTSTAPF, A motif: TLPRTLSQKTVEEVWKDINLA, LSL motif: PPPATALSLNSGPGF.
3 FD gene family acts as a floral activator of the photoperiodic pathway
3.1 FD promotes flowering through florigen activation complex in Arabidopsis
The Arabidopsis FD gene encodes a bZIP protein of 285 amino acid residues, which is identified as AtbZIP14 (At4g35900). Arabidopsis FD mRNA is distributed in the shoot apex and leaves with consistent 24-h (hour) rhythms, and its expression is significantly upregulated after seedling emergence (Abe et al., 2005; Park et al., 2023). The results of functional studies on loss and gain of function mutants suggest that FD regulates floral transition (Abe et al., 2005). Arabidopsis fd-2 mutant has a late flowering phenotype (Wigge et al., 2005). Overexpression of FD results in early flowering, 35S:FD enhances 35S:FT phenotype; therefore, the amount of FD activity is one of the limiting factors for 35S:FT plants (Abe et al., 2005).
In Arabidopsis, 14-3–3 proteins interact with FD and FT to form the ‘florigen activation complex’ (FAC) complex (Ho and Weigel, 2014). 14-3–3 proteins act as adaptor proteins to recognize and interact with the phosphorylated FD. FAC formation is dependent on the phosphorylation at position 282 (T282) of FD. That is, only after FD has been phosphorylated can active FAC be formed to induce flowering (Collani et al., 2019) (Figure 2A). Kawamoto et al. (2015) found that the calcium-dependent protein kinases (CDPKs) CPK4, CPK6, and CPK33 are good candidates for FD kinases. FD binds DNA (a strong preference for binding to G-box motifs) but does not activate transcription. FT acts as a transcriptional coactivator, increasing the enrichment of FD on floral time and floral homeotic genes such as AP1, LFY, SOC1, FRUITFULL (FUL), SEPALLATA1 (SEP1), SEP2, and SEP3 (Wigge et al., 2005; Ahn et al., 2006; Collani et al., 2019). AP1 is a class A gene, SEP1, SEP2, and SEP3 are class E genes during flower development (Krizek and Fletcher, 2005). TWIN SISTER OF FT (TSF), a paralog of FT, promotes flowering by enhancing the binding of FD to DNA (Collani et al., 2019).
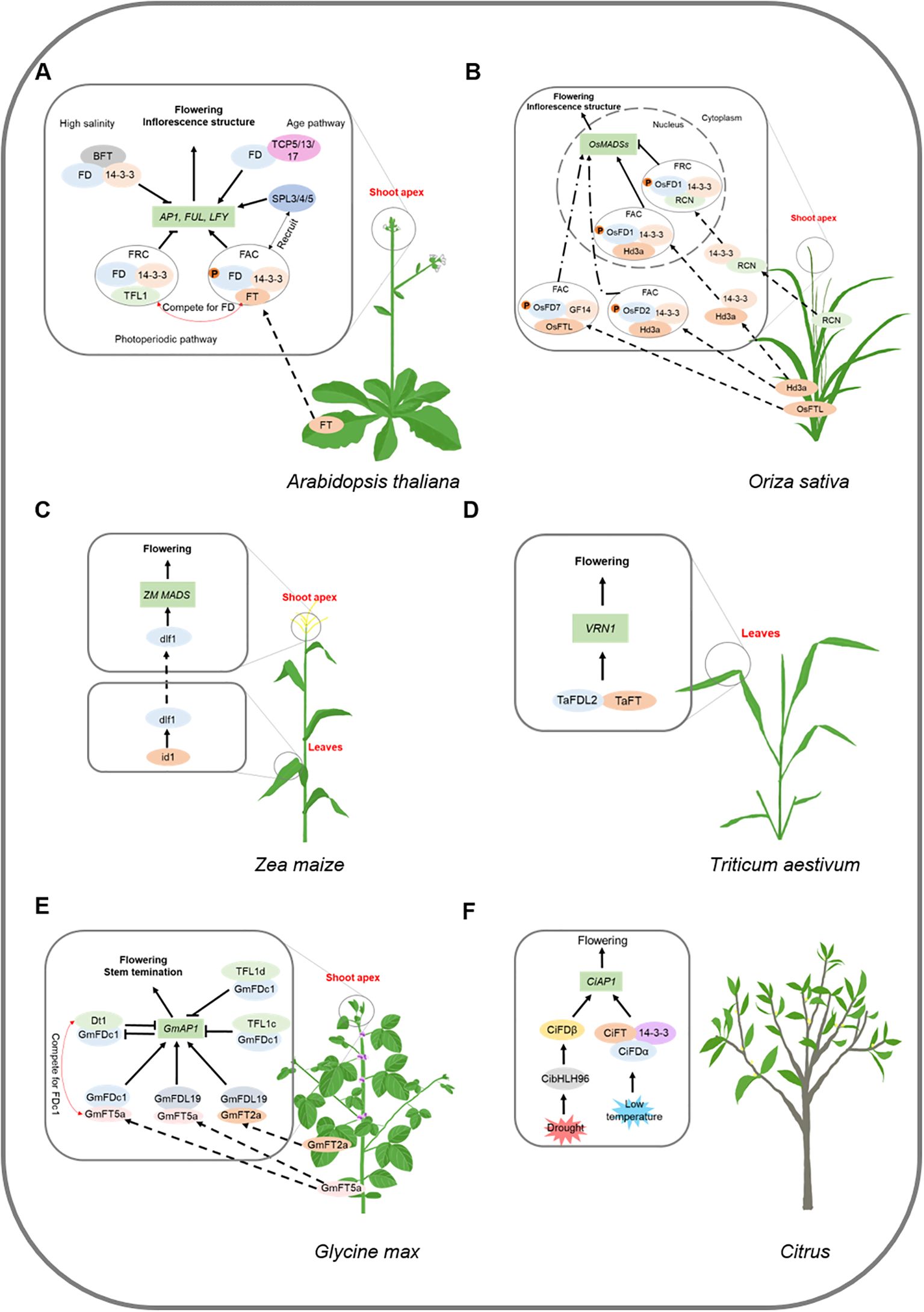
Figure 2. Regulation mechanisms of FD gene family in flowering pathways. Block letters represent proteins, italicized letters represent genes. Arrows represent facilitation and flathead arrows represent inhibition, dashed arrows represent translocation, dotted dashed arrows represent translocation and facilitation. P represents phosphorylation. IM represents inflorescence meristem, SAM represents shoot apical meristem, FM represents floral meristem, I1M represents primary inflorescence meristem, and I2M represents secondary inflorescence meristem. (A) Arabidopsis FT protein transports from leaves to shoot apex and work together with FD to promote floral transition, by activating the expression of AP1, FUL, LFY. FD is a floral integration factor that links photoperiod, age, and vernalization pathways. Under high salinity, BFT delays flowering, BFT protein competes with FT for FD binding. (B) At vegetative phase, FRC is formed. After Hd3a or OsFTL transports to the SAM, they compete with RCN for FAC formation. When the balance is shifted to the FAC, the reproductive program starts. (C) dlf1 mediates floral inductive signals transmitting from leaves to the shoot apex, activated by indeterminate1 (id1) in leaves. The targets of dlf1 may be maize MADS-box homologs (ZM MADS). (D) TaFDL2 interacts with TaFT and binds to the promoter of VRN1 to promote flowering. (E) Dt1 interacts with GmFDc1 and represses GmAP1a to repress flowering and stem termination. GmFT5a interferes with Dt1 for GmFDc1 binding and enhances the positive effect of GmFDc1 on GmAP1 expression. GmFDL19 interacting with GmFT2a and GmFT5a to promote flowering. TFL1c and TFL1d interact with GmFDc1 and binds to the promoter of GmAP1a to repress its activity. (F) CiFD forms two different proteins (CiFDα and CiFDβ) by low temperature and drought, respectively. Under low temperature, CiFDα can interact with CiFT and Ci14-3–3 to promote the expression of CiAP1. Under drought conditions, CibHLH96 activates the expression of CiFD and forms more CiFDβ. CiFDβ can bind directly to the CiAP1 promoter independently of CiFT and Ci14-3-3.
3.2 FD-like proteins in rice act differentially in FAC formation
In rice, FD homologs (OsFD1-OsFD7) share a conserved bZIP and SAP motif (Tsuji et al., 2013; Kaur et al., 2021). OsFD1-Hd3a-14-3–3 complex activates OsMADS15 (a homolog of AP1) and leads to early flowering (Tsuji et al., 2013). Silencing of OsFD7 correlates with late flowering and downregulation of MADS-box genes (e.g. OsMADS14, OsMADS15, and OsMADS18) involved in floral meristem development (Kaur et al., 2021) (Figure 2B). However, the regulation mechanism in rice is different from Arabidopsis. OsFD1 is located in the nucleus of shoot apex cells. The rice FT homolog, HEADING DATE 3a (Hd3a), translocates from the leaves to the shoot apex and binds 14-3–3 proteins in the cytoplasm. Then, the Hd3a-14-3–3 complex enters the nucleus and forms an FAC with OsFD1. The phosphorylated serine residue S192 in the OsFD1 SAP motif is recognized by 14-3–3 to facilitate the association between OsFD1 and Hd3a. FAC activates the transcription of OsMADS15, leading to floral induction (Taoka et al., 2011) (Figure 2B). Similar to OsFD1, OsFD7 is located in the nucleus of shoot apex cells. The rice FT homolog, OsFTL, translocates from the leaves to the shoot apex and binds OsGF14 proteins in the cytoplasm. Then, the OsFTL-OsGF14 complex enters the nucleus and forms an FAC with OsFD7. OsFD7 is phosphorylated by OsCDPK41 and OsCDPK49, and the interaction between OsGF14b and OsFD7 is dependent on this phosphorylation. FAC (OsFTL-OsGF14-OsFD7) activates the transcription of some floral meristem identity genes, leading to floral transition (Kaur et al., 2021) (Figure 2B). Unlike OsFD1 and OsFD7, OsFD2 shuttles between the cytoplasm and the nucleus. Normally, OsFD2 is restricted to the cytoplasm of shoot apex cells via its interaction with cytoplasmic 14-3–3 proteins. When Hd3a moves from the leaves to the shoot apex, the interaction between Hd3a and 14-3–3 initiates its nuclear translocation. The putative phosphorylation site, S164, within the SAP motif of OsFD2 is critical for the interaction between OsFD2 and 14-3-3 (Figure 2B). These results indicate that the FD function diverges among OsFD1, OsFD2, and OsFD7, but the formation of an FAC is essential for its function (Tsuji et al., 2013).
3.3 FD-like proteins have conserved functions in some important crops
Similar to Arabidopsis and rice, FD-like proteins in monocotyledonous plants such as maize, wheat, and bamboo also promote flowering (Table 1). Maize delayed flowering1 (dlf1) mediates floral inductive signals transmitted from the leaves to the shoot apex, activated by indeterminate1 (id1) in the leaves (Kozaki et al., 2004). The targets of dlf1 might be the maize MADS-box homologs (ZM MADS) (Muszynski et al., 2006) (Figure 2C). The id1 defined floral induction pathway may be unique to monocots, because no clear id1 orthologs exist in the Arabidopsis genome (Colasanti et al., 2006). In wheat, the regulatory mechanism of TaFDL2 is similar to that in Arabidopsis FD. TaFDL2 interacts with TaFT and binds to the ACGT elements in the promoter of VRN1 (homolog of Arabidopsis AP1) (Figure 2D). TaFT does not directly interact with the VRN1 promoter but interacts with TaFDL2 proteins (Li and Dubcovsky, 2008). Overexpressing bamboo BtFD1 in Arabidopsis leads to early flowering. BtFD1 binds to the ACGT motif of AtAP1’s promoter and upregulates the expression of AtAP1 (Dutta et al., 2021). In barley (Hordeum vulgare), HvFDL4 and HvFDL5 have been shown to physically interact with 14-3–3 proteins in a phosphorylation-dependent manner. Serine-to-alanine substitutions at critical residues (S333A in HvFDL4 or S216A in HvFDL5) abolish their binding to 14-3–3 proteins, suggesting that phosphorylation at these sites is essential for complex formation (Li et al., 2015). However, the biological functions of these interactions remain uncharacterized.
The FD homologs in dicotyledons also have consistent functions (Table 1). Overexpression of soybean GmFDc1 leads to early flowering, suggesting that GmFDc1 activates the floral transition (Yue et al., 2021). GmFT5a binds to GmFDc1 and enhances the positive effect of GmFDc1 on GmAP1 expression (Yue et al., 2021) (Figure 2E). Soybean GmFDL19-OE lines flower earlier than wild-type, which may be mediated by the direct up-regulation of GmAP1a. GmFDL19 also interacts with GmFT2a and GmFT5a to regulate flowering (Takeshima et al., 2019) (Figure 2E). GmFDL15 interacts with GmFT5b to promote flowering (Su et al., 2023). In poplar (Populus trichocarpa), the ectopic expression of PtFD1 (FDL2) results in early flowering (Tylewicz et al., 2015). Tobacco FD homologs participate in flowering regulation, NtFD1, NtFD3, and NtFD4 overexpression lines flower earlier than the wild-type (WT) (Beinecke et al., 2018). Overexpressing EdFD1 or EdFD2 in Arabidopsis results in early flowering. EdFT interacts with both EdFD1 and EdFD2 and regulates wild loquat flowering (Zhang et al., 2016).
4 FD-like proteins integrate endogenous and environmental stimuli
4.1 FD links the photoperiod, age, and vernalization pathways
SQUAMOSA PROMOTER BINDING LIKE (SPL) 3/4/5 are involved in the age pathway in Arabidopsis (Wang et al., 2009). SPL3/4/5 bind directly to the promoters of AP1, LFY, and FUL and recruit FD to these loci, mediating their activation by the FT-FD complex (Benlloch et al., 2011) (Figure 2A). SPL3/4/5 synergistically interact with the FT-FD module to induce flowering, linking the age and photoperiod pathways of flowering regulation (Jung et al., 2016). TCP5/13/17 (class II CIN TCP TFs) belonging to the Teosinte branched1/Cincinnata/proliferating cell factor (TCP) family act directly on the Telobox Motif cis-elements (GGACCA) of the AP1 promoter by interacting with FD (Figure 2A) (Li et al., 2019). They act synergistically and additively with the FT-FD module to positively regulate the initiation of flowering in Arabidopsis (Li et al., 2019). Chromatin remodeler HISTONE DEACETYLASE 19 (HDA19) preserves the identity of the inflorescence meristem (IM) in an age-dependent manner; in older hda19 inflorescence apices, floral organ identity genes are abnormally expressed, and the mutation of fd enhances the timing of these reproductive defects in hda19 (Gorham et al., 2018). FLOWERING H (FLH) is involved in the vernalization pathway of flowering. The early flowering Cape Verde Islands (CVI) allele of FLH requires the floral integrator FD to accelerate flowering (Seedat et al., 2013). These results confirm that FD is a central regulator of floral transition in the shoot meristem, which integrates signals from multiple pathways.
4.2 FD homologs integrate environment cues with flowering regulation
In citrus, CiFD forms two different proteins (CiFDα and CiFDβ) by alternatively splicing. Overexpressing CiFDα or CiFDβ in tobacco and trifoliate orange leads to early flowering (Ye et al., 2023). CiFDα and CiFDβ are induced by low temperatures and drought, respectively. Under low temperatures, CiFDα can interact with CiFT and Ci14-3–3 to form an FAC complex, which binds to the C-box element on the promoter of the floral meristem organization gene CiAP1, promoting its expression. In contrast, CiFDβ can bind directly to the CiAP1 promoter independently of CiFT and Ci14-3-3. The transcription factor CibHLH96 binds to the E-box of the CiFD promoter to promote CiFD expression. CibHLH96 is induced by drought but not at low temperatures. Under drought conditions, CibHLH96 activates the expression of CiFD and forms more CiFDβ (Figure 2F) (Ye et al., 2023). These results show that both CiFDα and CiFDβ are involved in the regulation of citrus flowering, but they have different mechanisms (Ye et al., 2023). In Kiwifruit, AcFD regulates cold signal response. It is downregulated in dormant buds in response to cold treatment (Varkonyi-Gasic et al., 2013). A member of soybean bZIP family group A, GmFDL19, is involved in abiotic stress tolerance and floral transition (Nan et al., 2014; Li et al., 2017; Yue et al., 2023). The tolerance to drought and salt stress is enhanced in GmFDL19-OE lines by upregulating ABA/stress-responsive genes and reducing the accumulation of Na+ ion content, and ectopic expression of GmFDL19 in soybean causes early flowering (Nan et al., 2014; Li et al., 2017).
5 FD gene family regulates plant morphogenesis
5.1 Inflorescence meristem identity and floral organ development
In pea, VEGETATIVE2 (VEG2), which is homologous to FD, plays a central role in regulating meristem identity throughout the development of the compound inflorescence. VEG2 interacts with FTb2 in the shoot apex to promote primary (I1) inflorescence meristem identity through DETERMINATE (DET), LATE FLOWERING (LF), FTa1, and FTc. VEG2 interacts with FTa1 at the shoot apex to promote secondary (I2) inflorescence meristem identity via VEG1 and FTc. veg2 mutant transforms I2 into I1 inflorescence meristems (Sussmilch et al., 2015). VEG2 is also involved in the regulation of floral architecture through the regulation of MADS-box genes such as PIM (AP1), AP3, and SEP1. veg2 mutant has defective sepals and petals, fused floral organs, reduced organ numbers, and malformed organs (Sussmilch et al., 2015) (Figure 3A).
Figure 3. Regulation mechanisms of FD gene family in environment signaling and plant development pathways. Block letters represent proteins, italicized letters represent genes. Arrows represent facilitation and flathead arrows represent inhibition, dashed arrows represent translocation. (A) VEG2 interacts with FTb2 in shoot apex to promote I1M identity through DET, LF, FTa1, and FTc. VEG2 interacts with FTa1 in shoot apex to promote I2M identity through VEG1 and FTc. VEG2 also involves in the regulation of floral architecture through regulating PIM. (B) PtFT- PtFD2 (FDL1) complex mediates photoperiodic growth by regulating LAP1. PtFD2 (FDL1) participates in the control of adaptive response and bud maturation pathways via interaction with ABI3. (C) EjFD interacts with EjTFL1s or EjFT1 to suppress the expression of EjAP1-1, which leads to the inhibition of loquat flower bud differentiation. EjFD-EjFT2 promotes floral bud formation by promoting the expression of EjAP1–1 and EjAP1-2, which is regulated by photoperiod and GA signals.
MtFDa, an ortholog of pea VEG2/PsFDa, plays a key role in inflorescence development in Medicago truncatula. mtfda has tertiary branches and bracts that transform into compound leaves, suggesting that MtFDa is required for I2 inflorescence meristem identity and development. mtfda and mtfta1 flower later than WT, the double mutant mtfda/mtfta1 never forms flowers, and no floral transition in mtfda/mtfta1 happens (Cheng et al., 2021). The phenotype of mtfda/mttfl1 double mutant phenotype is similar to that of mtfda. The I1 inflorescence shows indeterminate growth, indicating that MtFDa is epistatic to MtTFL1 for I1 indeterminacy. MtFDa and MtFULc co-determine I2 identity. The I2 inflorescence of the mtfda/mtfulc double mutant transforms into an I1-like vegetative structure, producing compound inflorescences, compound leaves, and indeterminate apices. The mtfda/mtfulc/mttfl1 triple mutant has a similar flowering time, inflorescence, and flower as mtfda, indicating that MtFDa has an epistatic effect on MtFULc (Cheng et al., 2018). Collectively, MtFDa plays a key role in inflorescence development, functions in coordination with MtFULc for I2 inflorescence meristem identification, and is epistatic to MtTFL1 for I1 indeterminacy (Cheng et al., 2018, 2021).
PaFDL1 and PaFDL2 are FD homologs of the London plane that participate in flower organ development. Overexpression of PaFDL1 and PaFDL2 in tobacco leads to extended stigmas, and curled petals at the tips (Cai et al., 2021).
5.2 Inflorescence structure and plant height
FD-like proteins participate in inflorescence development, stem growth, and seed development. The inflorescences of Nicotiana tabacum NtFD1, NtFD3, and NtFD4 overexpression lines are condensed and the pedicles, peduncles, and internodes are short, resulting in a bushy, bunch-like architecture. They flower earlier than the WT, fewer leaves are produced before flowering, and differentiation of axillary meristems is also premature compared with WT (Beinecke et al., 2018).
Rice OsFD7 RNAi lines have longer and denser panicles, more florets, elevated seed size and weight, and more seeds. The transcription levels of OsMADSs are down-regulated in OsFD7 RNAi lines (Kaur et al., 2021). OsFD2 inhibits the developmental shift from inflorescence branch meristem to floral (or spikelet) meristem in panicle branches, which leads to plentiful spikelets or secondary branches and a dense panicle phenotype with smaller leaves (Tsuji et al., 2013). The overexpression of bamboo BtFD1 in Arabidopsis also leads to dwarfism and an apparent reduction in the length of the flowering stalk and number of flowers per plant (Dutta et al., 2021). The overexpression of FD by the 35S promoter in Arabidopsis results in dwarf plants (Abe et al., 2005). Overexpression of FD and FDP (At2g17770/AtbZIP27) in rice causes a reduction in plant height and spikelet size with decreased expression of genes involved in cell elongation without significant flowering time alteration, which is linked to impaired gibberellin biosynthesis in plants (Jang et al., 2017). Romera-Branchat et al. (2020) reported that FD and FDP bind to genes involved in water deprivation and hormonal pathways, including gibberellic acid, ABA, and jasmonic acid. These results provide evidence of crosstalk between the regulation of plant morphogenesis and hormone signaling pathways.
In soybean, overexpression of GmFDc1 leads to fewer nodes (Yue et al., 2021). GmFT5a interferes with the binding of Dt1 to GmFDc1 and enhances the positive effect of GmFDc1 on GmAP1 expression (Yue et al., 2021) (Figure 2E). Dt1 controls stem growth habit and flowering time and strongly influences soybean grain yield (Liu et al., 2010; Yue et al., 2021). Mutations in the recessive alleles of gmft2a and gmft5a delay flowering and increase node number, branch number, and yield (Li et al., 2021). Thus, GmFDc1 appears to contribute significantly to soybean plant architecture and yield.
5.3 Photoperiod signal and plant growth
In poplar, overexpression of PtFD1 (FDL2) results in severe dwarfing under a LD photoperiod, however, SD-induced growth arrest and bud formation are lost in PtFD1 (FDL2)-overexpressing lines (Parmentier-Line and Coleman, 2015; Tylewicz et al., 2015). PtFD2 (FDL1) overexpression resulted in a delayed SD response compared to WT. FT- PtFD2 (FDL1) complex mediates photoperiodic growth by regulating Like AP1 (LAP1). PtFD2 (FDL1) also participates in controlling adaptive responses and bud maturation pathways by interacting with ABSCISIC ACID INSENSITIVE 3 (ABI3), a component of ABA signaling (Tylewicz et al., 2015) (Figure 3B). Loquat EjFD suppresses EjAP1–1 expression by interacting with EjTFL1s or EjFT1, inhibiting loquat flower bud differentiation. Conversely, EjFD-EjFT2 promotes floral bud formation under the regulation of photoperiod and GA signals (Jiang et al., 2020, 2024) (Figure 3C).
6 Competition of FT and TFL1 for FD binding
6.1 The balance between vegetative and reproductive stage
In contrast to FT and TSF, TERMINAL FLOWER 1 (TFL1) and BROTHER of FT and TFL1 (BFT) are floral repressors in the PEBP family. These PEBPs have conserved 14-3–3 binding motifs and interact with FD (Yoo et al., 2010; Hanano and Goto, 2011; Ryu et al., 2014). TFL1 competes with FT for FD binding and represses the transcription of floral meristem identity genes such as LFY and AP1 (Hanano and Goto, 2011) (Figure 2A). TFL1 interacts with unphosphorylated FD via 14-3–3 proteins, suggesting that the inactive FD/14-3-3/TFL1 ternary complex may be present in the basal state of the SAM. Only when FD is phosphorylated can FT form an active complex with the 14-3–3 proteins to induce flowering. The efficient phosphorylation of T282 in FD is calcium-dependent. This requirement may help prevent the premature induction of flowering (Kawamoto et al., 2015). Under high salinity conditions, BFT delays flowering. The relative transcript levels of BFT are higher than those of FT and the high-level BFT protein competes with FT for FD binding in the SAM (Ryu et al., 2014) (Figure 2A). Similar to FD, FDP is phosphorylated by CPK33, forming a complex with FT and TFL1 in a phosphorylation-dependent manner. The weak late-flowering phenotype of cpk33–1 may be due to the combined effect of the florigen and anti-florigen complex formation of FD and FDP (Kawamoto et al., 2015).
The rice TFL1-like protein RICE CENTRORADIALIS (RCN) competes with Hd3a for 14-3–3 binding. RCN protein transports from the vasculature to SAM to form the “florigen repression complex” (FRC) with 14-3–3 and OsFD1 and then represses florigenic activity. The balance between FRC and FAC depends on the ratio of Hd3a to RCN and regulates the development of SAM. In the vegetative phase, FRC are formed. Upon reaching the SAM, Hd3a competes with RCN for FAC formation. When the balance shifts to FAC, the reproductive program begins (Kaneko-Suzuki et al., 2018) (Figure 2B).In soybean, Dt1 and GmFT5a have opposite functions. Dt1 complementary lines produce more nodes and flower later than WT (Yue et al., 2021). gmft5a delays flowering and increases node number (Li et al., 2021). Dt1 interacts with GmFDc1 and binds to ACGT cis-elements in the promoter of GmAP1a to repress its activity. GmFT5a interferes with the binding of Dt1 to GmFDc1 and enhances the positive effect of GmFDc1 on GmAP1 expression (Chen et al., 2020b; Yue et al., 2021). TFL1c and TFL1d, homologs of Dt1, interact with GmFDc1 and binds to ACGT cis-elements in the promoter of GmAP1a to repress its activity (Wang et al., 2023) (Figure 2E). TFL1c and TFL1d might also compete with GmFT5a for GmFDc1 binding. EjTFL1s inhibit loquat flower bud differentiation through EjFD binding and suppression of EjAP1-1 (Jiang et al., 2020). In contrast, EjFT1 and EjFT2 interact with EjFD but have opposing effects: EjFT2-EjFD activates EjAP1–1 and EjAP1-2, while EjFT1-EjFD represses EjAP1-1. EjFT1 may resemble EjTFL1s in promoting vegetative growth. The competitive interaction between EjFT1 and EjFT2 with EjFD regulates floral bud differentiation, with EjFT2 promoting flowering and EjFT1 supporting vegetative growth (Jiang et al., 2024) (Figure 3C). Protein structural analysis of EjFT1 and EjFT2 suggests that differences in amino acid residues at Val123/Leu123, Ser157/Ala157, and Val158/Ala158 may be the reason for their functional differences (Jiang et al., 2024).
6.2 Environment cues and the antagonistic regulation
In Brachypodium distachyon, FTL9 and FD1 form an FAC that induces VRN1 and FUL2 expression, promoting flowering under SD conditions. Under LD conditions, however, FTL9 inhibits flowering. The FTL9-FD1 complex is less potent than the FT1-FD1 complex in inducing flowering. Overexpression of FTL9 disrupts FT1-FD1 complex formation by competing for FD1 binding, leading to reduced VRN1 expression and delayed flowering (Qin et al., 2019).
Kiwifruit CEN and FT interact with AcFD but exhibit distinct temporal expression patterns. FT is specifically induced in dormant buds during winter chilling, promoting dormancy release, whereas CEN transcripts accumulate in latent buds during summer but decrease in autumn prior to dormancy establishment. These contrasting expression patterns and functional roles suggest that CEN and FT may act as antagonistic regulators in kiwifruit development (Varkonyi-Gasic et al., 2013).
7 FD directly regulates genes related to flowering and endogenous signalling
The direct target genes of TFL1-FD and FT-FD complexes have been detected by ChIP-seq and RNA-seq experiments. The results reveal that the target genes have a prominent role in cell signaling, including flowering time genes (PRR7, CONSTANS (CO), and GIGANTEA (GI)) and floral identity genes (LFY, AP1, FUL, and LATE MERISTEM IDENTITY 2 (LMI2)). TFL1-FD represses while FT-FD activates the target genes (Collani et al., 2019; Goretti et al., 2020; Romera-Branchat et al., 2020; Zhu et al., 2021). CO and GI are regulators of flowering by activating FT in the photoperiodic pathway. CO and GI might act in feedback regulatory pathways with FT-FD complexes. The PRR7 gene is involved in the regulation of the plant biological clock and affects the circadian rhythmicity (Li et al., 2016). Current studies have not directly revealed the regulatory relationship between PRR7 and FT-FD. LFY, AP1, FUL, and LMI2 who are involving in the regulation of plant structure, floral meristem differentiation, and flowering time, have been proved to be the direct targets of TFL1-FD and FT-FD complexes in many plant species (Abe et al., 2005; Wigge et al., 2005; Ahn et al., 2006; Li and Dubcovsky, 2008; Taoka et al., 2011; Sussmilch et al., 2015; Collani et al., 2019; Takeshima et al., 2019; Yue et al., 2021; Dutta et al., 2021; Ye et al., 2023; Park et al., 2023).
TFL1-FD and FT-FD complexes also bind to genes linked to phytohormone biosynthesis, signaling, and response (auxin, ABA, brassinosteroid, cytokinin, jasmonic acid, and strigolactone) as well as genes linked to sugar signaling (Trehalose-6-phosphatases (TPP) genes) (Collani et al., 2019; Goretti et al., 2020; Zhu et al., 2021). The plant height, branch growth, bud growth, spikelet size, and tolerance to drought and salt stress influenced by FD gene family are reported to be related to these endogenous signals (Tylewicz et al., 2015; Jang et al., 2017; Li et al., 2017; Jiang et al., 2020, 2024).
8 Conclusions and perspectives
Flowering time of crops is essential for adaptation and yield. Early flowering enhances the efficiency of reproductive development, whereas delayed flowering facilitates the accumulation of materials through prolonged vegetative growth (Wang et al., 2024). Many genes regulating flowering time in crop species have been utilized in molecular breeding, such as Heading date 1 (Hd1) and Grain number, plant height, and heading date 7 (Ghd7) in rice (Yano et al., 2000; Xue et al., 2008), Vernalization 1 (VRN1) and Photoperiod-D1 (Ppd-D1) in wheat (Yan et al., 2003; Beales et al., 2007), J and SOC1 in soybean (Lu et al., 2017; Kou et al., 2022). However, FD genes act as a floral activator are seldom used. Perhaps FD’s function is less studied than FT. In the future, diverse FD alleles in crop germplasm resources should be utilized in modern breeding.
The bZIP transcription factor FD is a central regulator of yield traits, such as plant height, inflorescence structure, and seed development. In barley, bZIP transcription factors have been implicated in pre-anthesis tip degeneration (PTD), a programmed process critical for seed number and yield. However, the precise molecular functions and regulatory pathways remain unclear (Shanmugaraj et al., 2023). Interestingly, unlike in other species where TFL1 homologs control inflorescence determinacy, AP2L-5 (an AP2-family transcription factor) serves as the primary regulator of determinate/indeterminate inflorescence fate in barley (Zhong et al., 2021). This suggests that FD-like proteins may operate through distinct regulatory mechanisms in barley compared to other crops.
FD is essential for vegetative growth, overexpressing FD in Arabidopsis, rice, tobacco, bamboo, or poplar lead to dwarf phenotype. During the “Green revolution”, the utilization of “dwarfing genes” facilitated the breeding of novel cultivars in rice, wheat, and maize with enhanced resistance to lodging. The implementation of optimal plant densities contributed to a substantial augmentation in crop productivity (Hou et al., 2024). Therefore, FD might be a promising “dwarfing gene” in crop breeding and production increasing.
FD can respond to environmental factors such as photoperiod, temperature, phytohormones, and abiotic stresses (Zhu et al., 2021). This phenomenon suggests that FD may serve as a valuable genetic resource for enhancing plant adaptability to adverse environmental conditions. Whereas, FD’s potential has not been fully explored. It has been reported that other Group A members of bZIP transcription family, such as ABF1/AtbZIP35, ABF2/AREB1/AtbZIP36, ABF3/AtbZIP37, ABF4/AREB2/AtbZIP38, and ABI5/DPBF1/AtbZIP39 are associated with ABA and stress signaling or ABA-dependent seed maturation and germination (Choi et al., 2000; Uno et al., 2000; Skubacz et al., 2016). The functions and molecular mechanisms of FD homologs in relevant pathways might be a promising direction for future research.
Overall, a comprehensive overview of FD gene family will not only deepen our knowledge of the diverse roles executed by FD gene family in flowering, plant development, and environment signaling responses but will also facilitate the exploration of innovative strategies to improve crop productivity in challenging environments.
Author contributions
HY: Funding acquisition, Writing – original draft. MZ: Software, Writing – original draft. YL: Software, Writing – original draft. LC: Funding acquisition, Writing – review & editing. BL: Supervision, Writing – review & editing. FK: Supervision, Writing – review & editing. HL: Conceptualization, Writing – review & editing. LY: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32372158 to LY; 32372078 to HY; 32472164 to LC.); the Science and Technology Projects in Guangzhou, China (2023A04J1503 to LY); the Guangdong Basic and Applied Basic Research Foundation (2024A1515030288 to LC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1602756/full#supplementary-material
Supplementary Table 1 | List of genes in Figure 1.
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052. doi: 10.1126/science.1115983
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Lee, J. H., Yoo, S. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614. doi: 10.1038/sj.emboj.7600950
Araki, T. (2001). Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4, 63–68. doi: 10.1016/S1369-5266(00)00137-0
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Beales, J., Turner, A., Griffiths, S., Snape, J. W., and Laurie, D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115, 721–733. doi: 10.1007/s00122-007-0603-4
Beinecke, F. A., Grundmann, L., Wiedmann, D. R., Schmidt, F. J., Caesar, A. S., Zimmermann, M., et al. (2018). The FT/FD-dependent initiation of flowering under long-day conditions in the day-neutral species Nicotiana tabacum originates from the facultative short-day ancestor Nicotiana tomentosiformis. Plant J. 96, 329–342. doi: 10.1111/tpj.2018.96.issue-2
Benlloch, R., Kim, M. C., Sayou, C., Thévenon, E., Parcy, F., and Nilsson, O. (2011). Integrating long-day flowering signals: a LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J. 67, 1094–1102. doi: 10.1111/j.1365-313X.2011.04660.x
Cai, F., Shao, C., Zhang, Y., Shi, G., Bao, Z., Bao, M., et al. (2021). Two FD homologs from London plane (Platanus acerifolia) are associated with floral initiation and flower morphology. Plant Sci. 310, 110971. doi: 10.1016/j.plantsci.2021.110971
Cai, Y., Wang, L., Chen, L., Wu, T., Liu, L., Sun, S., et al. (2020). Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 18, 298–309. doi: 10.1111/pbi.13199
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., and He, Y. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Nan, H., Kong, L., Yue, L., Yang, H., and Zhao, Q. (2020b). Soybean AP1 homologs control flowering time and plant height. j. integr. Plant Biol. 62, 1–12. doi: 10.1111/jipb.v62.12
Cheng, X., Li, G., Krom, N., Tang, Y., and Wen, J. (2021). Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula. Plant Physiol. 185, 161–178. doi: 10.1093/plphys/kiaa005
Cheng, X., Li, G., Tang, Y., and Wen, J. (2018). Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 145, dev158766. doi: 10.1242/dev.158766
Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S. Y. (2000). ABFs, a family of ABA responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Colasanti, J., Tremblay, R., Wong, A. Y., Coneva, V., Kozaki, A., and Mable, B. K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7, 158. doi: 10.1186/1471-2164-7-158
Collani, S., Neumann, M., Yant, L., and Schmid, M. (2019). FT modulates genome-wide DNA-binding of the bZIP transcription factor FD. Plant Physiol. 180, 367–380. doi: 10.1104/pp.18.01505
Corrêa, L. G. G., Riaño-Pachón, D. M., Schrago, C. G., Vicentini dos Santos, R., Mueller-Roeber, B., and Vincentz, M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PloS One 3, e2944. doi: 10.1371/journal.pone.0002944
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP transcription factor family — an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Dutta, S., Deb, A., Biswas, P., Chakraborty, S., Guha, S., Mitra, D., et al. (2021). Identification and functional characterization of two bamboo FD gene homologs having contrasting effects on shoot growth and flowering. Sci. Rep. 11, 7849. doi: 10.1038/s41598-021-87491-6
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550.e552. doi: 10.1016/j.cell.2010.04.024
Goretti, D., Silvestre, M., Collani, S., Langenecker, T., Méndez, C., and Madueño, F. (2020). TERMINAL FLOWER 1 functions as a mobile transcriptional cofactor in the shoot apical meristem. Plant Physiol. 182, 2081–2095. doi: 10.1104/pp.19.00867
Gorham, S. R., Weiner, A. I., Yamadi, M., and Krogan, N. T. (2018). HISTONE DEACETYLASE 19 and the flowering time gene FD maintain reproductive meristem identity in an age-dependent manner. J. Exp. Bot. 69, 4757–4771. doi: 10.1093/jxb/ery239
Han, X., Wang, D., and Song, G. Q. (2021). Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering. Sci. Rep. 11, 12758. doi: 10.1038/s41598-021-92215-x
Hanano, S. and Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23, 3172–3184. doi: 10.1105/tpc.111.088641
Hiraoka, K., Yamaguchi, A., Abe, M., and Araki, T. (2013). The florigen genes FT and TSF modulate lateral shoot outgrowth in Arabidopsis thaliana. Plant Cell Physiol. 54, 352–368. doi: 10.1093/pcp/pcs168
Ho, W. and Weigel, D. (2014). Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26, 552–564. doi: 10.1105/tpc.113.115220
Hou, Z., Huang, H., Wang, Y., Chen, L., Yue, L., Liu, B., et al. (2024). Molecular regulation of shoot architecture in soybean. Plant Cell Environ., 1–14. doi: 10.1111/pce.15138
Jang, S., Li, H. Y., and Kuo, M. L. (2017). Ectopic expression of Arabidopsis FD and FD PARALOGUE in rice results in dwarfism with size reduction of spikelets. Sci. Rep. 7, 44477. doi: 10.1038/srep44477
Jiang, Y., Zhu, Y., Peng, Z., Su, W., Peng, J., Yuan, Y., et al. (2024). Two FT genes synergistically regulate the reproductive transition of loquat. Hortic. Plant J. 11, 548–563. doi: 10.1016/j.hpj.2023.08.003
Jiang, Y., Zhu, Y., Zhang, L., Su, W., Peng, J., Yang, X., et al. (2020). EjTFL1 genes promote growth but inhibit flower bud differentiation in loquat. Front. Plant Sci. 11, 576. doi: 10.3389/fpls.2020.00576
Jung, J.-H., Lee, H.-J., Ryu, J. Y., and Park, C.-M. (2016). SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 9, 1647–1659. doi: 10.1016/j.molp.2016.10.014
Kaneko-Suzuki, M., Kurihara-Ishikawa, R., Okushita-Terakawa, C., Kojima, C., Nagano-Fujiwara, M., Ohki, I., et al. (2018). TFL1-like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3–3 and FD. Plant Cell Physiol. 59, 458–468. doi: 10.1093/pcp/pcy021
Kaur, A., Nijhawan, A., Yadav, M., and Khurana, J. P. (2021). OsbZIP62/OsFD7, a functional ortholog of FLOWERING LOCUS D, regulates floral transition and panicle development in rice. J. @ Exp. Bot. 72, 7826–7845. doi: 10.1093/jxb/erab396
Kawamoto, N., Sasabe, M., Endo, M., Machida, Y., and Araki, T. (2015). Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci. Rep. 5, 8341. doi: 10.1038/srep08341
Kou, K., Yang, H., Li, H., Fang, C., Chen, L., Yue, L., et al. (2022). A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742. doi: 10.1016/j.cub.2022.02.046
Kozaki, A., Hake, S., and Colasanti, J. (2004). The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32, 1710–1720. doi: 10.1093/nar/gkh337
Krizek, B. A. and Fletcher, J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688–698. doi: 10.1038/nrg1675
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, J. and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Li, Y., Chen, Q., Nan, H., Li, X., Lu, S., Zhao, X., et al. (2017). Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PloS One 12, e0179554. doi: 10.1371/journal.pone.0179554
Li, C. X. and Dubcovsky, J. (2008). Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 55, 543–554. doi: 10.1111/j.1365-313X.2008.03526.x
Li, X., Fang, C., Yang, Y., Lv, T., Su, T., Chen, L., et al. (2021). Overcoming the genetic compensation response of soybean florigens to improve adaptation and yield at low latitudes. Curr. Biol. 31, 1–13. doi: 10.1016/j.cub.2021.06.037
Li, L., Li, X., Liu, Y., and Liu, H. (2016). Flowering responses to light and temperature. Sci. China-Life Sci. 59, 403–408. doi: 10.1007/s11427-015-4910-8
Li, C., Lin, H., and Dubcovsky, J. (2015). Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 84, 70–82. doi: 10.1111/tpj.2015.84.issue-1
Li, D., Zhang, H., Mou, M., Chen, Y., Xiang, S., Chen, L., et al. (2019). Arabidopsis class II TCP transcription factors integrate with the FT–FD module to control flowering. Plant Physiol. 181, 97–111. doi: 10.1104/pp.19.00252
Liu, B., Watanabe, S., Uchiyama, T., Kong, F., Kanazawa, A., Xia, Z., et al. (2010). The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 153, 198–210. doi: 10.1104/pp.109.150607
Liu, L., Xuan, L., Jiang, Y., and Yu, H. (2021). Regulation by FLOWERING LOCUS T and TERMINAL FLOWER 1 in flowering time and plant architecture. Small Struct. 2, 2000125. doi: 10.1002/sstr.202000125
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Muszynski, M. G., Dam, T., Li, B., Shirbroun, D. M., Hou, Z., Bruggemann, E., et al. (2006). Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142, 1523–1536. doi: 10.1104/pp.106.088815
Nan, H., Cao, D., Zhang, D., Li, Y., Lu, S., Tang, L., et al. (2014). GmFT2a and GmFT5a redundantly and differentially regulate flowering through interaction with and upregulation of the bZIP transcription factor GmFDL19 in soybean. PloS One 9, e97669. doi: 10.1371/journal.pone.0097669
Parcy, F. (2005). Flowering: a time for integration. Int. J. Dev. Biol. 49, 585–593. doi: 10.1387/ijdb.041930fp
Park, K. H., Kim, S. B., and Jung, J. H. (2023). Analysis of temperature effects on the protein accumulation of the FT–FD module using newly generated Arabidopsis transgenic plants. Plant Direct. 7, e552. doi: 10.1002/pld3.v7.12
Parmentier-Line, C. M. and Coleman, G. D. (2015). Constitutive expression of the Poplar FD-like basic leucine zipper transcription factor alters growth and bud development. Plant Biotechnol. J. 14, 260–270. doi: 10.1111/pbi.12380
Qin, Z., Bai, Y., Muhammad, S., Wu, X., Deng, P., Wu, J., et al. (2019). Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon. Nat. Commun. 10, 812. doi: 10.1038/s41467-019-08785-y
Romera-Branchat, M., Severing, E., Pocard, C., Ohr, H., Vincent, C., Nee, G., et al. (2020). Functional divergence of the Arabidopsis florigen-interacting bZIP transcription factors FD and FDP. Cell Rep. 31, 107717. doi: 10.1016/j.celrep.2020.107717
Ryu, J. Y., Lee, H. J., Seo, P. J., Jung, J. H., Ahn, J. H., and Park, C. M. (2014). The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol. Plant 7, 377–387. doi: 10.1093/mp/sst114
Seedat, N., Dinsdale, A., Ong, E. K., and Gendall, A. R. (2013). Acceleration of flowering in Arabidopsis thaliana by Cape Verde Islands alleles of FLOWERING H is dependent on the floral promoter FD. J. Exp. Bot. 64, 2767–2778. doi: 10.1093/jxb/ert120
Shanmugaraj, N., Rajaraman, J., Kale, S., Kamal, R., Huang, Y., Thirulogachandar, V., et al. (2023). Multilayered regulation of developmentally programmed pre-anthesis tip degeneration of the barley inflorescence. Plant Cell 35, 3973–4001. doi: 10.1093/plcell/koad164
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7, 1884. doi: 10.3389/fpls.2016.01884
Su, Q., Chen, L., Cai, Y., Wang, L., Chen, Y., Zhang, J., et al. (2023). The FLOWERING LOCUS T 5b positively regulates photoperiodic flowering and improves the geographical adaptation of soybean. Plant Cell Environ. 47, 246–258. doi: 10.1111/pce.14739
Sussmilch, F. C., Berbel, A., Hecht, V., Schoor, J. K. V., Ferrándiz, C., Madueño, F., et al. (2015). Pea VEGETATIVE2 is an FD homolog that is essential for flowering and compound inflorescence development. Plant Cell 27, 1046–1060. doi: 10.1105/tpc.115.136150
Takeshima, R., Nan, H., Harigai, K., Dong, L., Zhu, J., Lu, S., et al. (2019). Functional divergence between soybean FLOWERING LOCUS T orthologues FT2a and FT5a in post-flowering stem growth. J. Exp. Bot. 70, 3941–3953. doi: 10.1093/jxb/erz199
Taoka, K.-i., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3–3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Tsuji, H., Nakamura, H., Taoka, K., and Shimamoto, K. (2013). Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 54, 385–397. doi: 10.1093/pcp/pct005
Tylewicz, S., Tsuji, H., Miskolczi, P., Petterle, A., Azeez, A., Jonsson, K., et al. (2015). Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. P. Natl. Acad. Sci. U.S.A. 112, 3140–3145. doi: 10.1073/pnas.1423440112
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozake, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. P. Natl. Acad. Sci. U.S.A 97, 11632–11637. doi: 10.1073/pnas.190309197
Varkonyi-Gasic, E., Moss, S. M. A., Voogd, C., Wang, T., Putterill, J., and Hellens, R. P. (2013). Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytol. 198, 732–746. doi: 10.1111/nph.12162
Wang, J. W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, L., Lin, C., Li, B., Su, T., Li, S., Li, H., et al. (2023). Two soybean homologues of TERMINAL FLOWER 1 control flowering time under long day conditions. Crop J. 11, 704–712. doi: 10.1016/j.cj.2023.01.008
Wang, J., Xu, X., Wang, P., Zhang, L., Liu, L., Liu, L. P., et al. (2024). Floral-promoting GmFT homologs trigger photoperiodic after-effects: An important mechanism for early-maturing soybean varieties to regulate reproductive development and adapt to high latitudes. Plant Cell Environ. 47, 1656–1667. doi: 10.1111/pce.14833
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Xue, W., Xing, Y., Zhao, Y., Tang, W., Wang, L., Zhou, H., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., Dubcovsky, J., et al. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. 100, 6263–6268. doi: 10.1073/pnas.0937399100
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. doi: 10.1105/tpc.12.12.2473
Ye, L. X., Wu, Y. M., Zhang, J. X., Zhang, J. X., Zhou, H., Zeng, R. F., et al. (2023). A bZIP transcription factor (CiFD) regulates drought- and low-temperature-induced flowering by alternative splicing in citrus. J. Integr. Plant Biol. 65, 674–691. doi: 10.1111/jipb.13390
Yoo, S. J., Chung, K. S., Jung, S. H., Yoo, S. Y., Lee, J. S., and Ahn, J. H. (2010). BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 63, 241–253. doi: 10.1111/j.1365-313X.2010.04234.x
Yue, L., Li, X., Fang, C., Chen, L., Yang, H., Yang, J., et al. (2021). FT5a interferes with the Dt1-AP1 feed-back loop to control flowering time and shoot determinacy in soybean. J. Integr. Plant Biol. 63, 1004–1020. doi: 10.1111/jipb.13070
Yue, L., Pei, X., Kong, F., Zhao, L., and Lin, X. (2023). Divergence of functions and expression patterns of soybean bZIP transcription factors. Front. Plant Sci. 14, 1150363. doi: 10.3389/fpls.2023.1150363
Zhang, L., Yu, H., Lin, S., and Gao, Y. (2016). Molecular characterization of FT and FD homologs from Eriobotrya deflexa Nakai forma koshunensis. Front. Plant Sci. 7, 8. doi: 10.3389/fpls.2016.00008
Zhong, J., wan Esse, G. W., and Bi, X. (2021). INTERMEDIUM-M encodes an HvAP2L-H5 ortholog and is required for inflorescence indeterminacy and spikelet determinacy in barley. Proc. Natl. Acad. Sci. 118, e2011779118. doi: 10.1073/pnas.2011779118
Keywords: FD, floral transition, plant development, adaptation, crop breeding
Citation: Yang H, Zhong M, Liu Y, Chen L, Liu B, Kong F, Li H and Yue L (2025) Multifaceted roles of FD gene family in flowering, plant architecture, and adaptation. Front. Plant Sci. 16:1602756. doi: 10.3389/fpls.2025.1602756
Received: 30 March 2025; Accepted: 05 June 2025;
Published: 20 June 2025.
Edited by:
Dilip R. Panthee, North Carolina State University, United StatesReviewed by:
Dionysia A. Fasoula, Agricultural Research Institute, CyprusMichela Osnato, University of Urbino Carlo Bo, Italy
Copyright © 2025 Yang, Zhong, Liu, Chen, Liu, Kong, Li and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Li, bGlob25nMTAwNUBnemh1LmVkdS5jbg==; Lin Yue, eXVlbGluQGd6aHUuZWR1LmNu
†These authors have contributed equally to this work
 Hui Yang
Hui Yang Meiling Zhong†
Meiling Zhong† Fanjiang Kong
Fanjiang Kong Hong Li
Hong Li Lin Yue
Lin Yue