- 1College of Environmental Science and Engineering, Yangzhou University, Yangzhou, China
- 2Key Laboratory of Arable Land Quality Monitoring and Evaluation, Ministry of Agriculture and Rural Affairs, Yangzhou University, Yangzhou, China
- 3Key Laboratory of Saline-Alkali Soil Reclamation and Utilization in Coastal Areas, The Ministry of Agriculture and Rural Affairs of China, Yangzhou University, Yangzhou, China
- 4Jiangsu Key Laboratory of Crop Genetic and Physiology, Yangzhou University, Yangzhou, China
- 5Jiangsu Key Laboratory of Crop Cultivation and Physiology, Yangzhou University, Yangzhou, China
- 6Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops, Yangzhou University, Yangzhou, China
- 7Research Institute of Rice Industrial Engineering Technology, Yangzhou University, Yangzhou, China
Introduction: Salt stress represents a critical abiotic constraint that severely impedes plant growth and agricultural productivity. While plant growth-promoting rhizobacteria (PGPR) demonstrate potential in enhancing plant salt tolerance, their precise mechanisms remain incompletely elucidated. This study systematically investigates the mechanistic basis by which PGPR inoculation ameliorates salt stress in rice seedlings through modulation of rhizosphere microbiota and root exudate profiles.
Methods: We inoculated rice seedlings with five monocultures (Bacillus sp., Providencia sp., etc.) and a synthetic consortium (T6) under salt stress conditions. Growth parameters, rhizobacterial communities (via 16S rRNA sequencing), and root exudates (untargeted metabolomics) were comparatively analyzed against uninoculated controls (CK).
Results: PGPR inoculation significantly promoted rice seedling growth under salt stress. Treatments T2-T6 exhibited substantial increases in key biomass parameters—including root length, plant height, and dry weight—relative to the CK control. Concurrently, elevated chlorophyll content and enhanced photosynthetic efficiency were observed. Inoculated plants also displayed significantly higher activities of antioxidant enzymes (Superoxide dismutase, Peroxidase, Catalase activity) (SOD, POD, CAT) and proline (Pro) accumulation in both leaves and roots, coupled with a marked reduction in Malondialdehyde, indicating effective mitigation of oxidative damage. PGPR inoculation altered rhizosphere bacterial community composition, reducing overall alpha-diversity. Notably, the relative abundance of dominant bacterial phyla (e.g., Proteobacteria, Acidobacteriota) and beneficial genera (e.g., Subgroup_7, Lysobacter) increased significantly. These microbial shifts showed positive correlations with improved plant physiological status, suggesting a synergistic role in promoting seedling growth under salt stress. Root exudate metabolomics revealed a substantial number of differentially abundant metabolites in inoculated plants compared to CK, encompassing lipids, hormones, and signaling molecules. Crucially, the production of these specific exudates correlated with the enrichment of dominant bacterial taxa in the rice rhizosphere. Metabolic pathway analysis indicated significant enrichment primarily within Nucleotide metabolism and Purine metabolism pathways (belonging to the Metabolism superclass) and ABC transporter pathways (within Environmental Information Processing). The T6 consortium treatment induced enrichment across a significantly greater number of key metabolic pathways compared to single-strain inoculations.
Discussion: PGPR inoculation enhances rice seedling growth and salt tolerance by: (1) optimizing rhizosphere microbiota (enriching dominant phyla and beneficial genera); (2) recruiting stress-mitigating microbial consortia; and (3) stimulating root exudates enriched in nucleotide/purine metabolism and ABC transporters. The superior efficacy of the T6 consortium underscores the advantage of synergistic microbial interactions. Collectively, these findings reveal plant-microbe-metabolite mechanisms underlying PGPR-mediated salt tolerance, providing a foundation for developing salinized soil remediation strategies.
1 Introduction
Statistics from the Food and Agriculture Organization (FAO) of the United Nations show that the global area of salinized land has reached 1.381 billion hectares (representing approximately 10.7% of the global land area) (Yu, 2025). As an important existing and potential arable resource, saline-alkali soil has significant potential for development and utilization. Traditional saline-alkali land management mainly relies on physical methods (such as irrigation for salt leaching) and chemical improvement, yet these are constrained by methodological limitations, high costs, and high recurrence risk. Contemporary research has shifted toward bio-agronomic synergistic strategies, establishing sustainable ecological reclamation systems through revegetation (e.g., halophyte cultivation) integrated with microbial remediation (e.g., PGPR inoculation) (Yang et al., 2016; Luo et al., 2021; Wang et al., 2024; Wang, 2024).
PGPR are widely distributed and diverse, encompassing groups such as bacteria, fungi, and actinomycetes, with Pseudomonas, Bacillus, and Rhizobium as dominant groups (Lugtenberg and Kamilova, 2009; Luo et al., 2021). Due to their dual functions of promoting plant growth and controlling pests/diseases, as well as advantages such as ecological environmental protection and strong environmental compatibility, PGPR have been widely applied and attracted significant attention in soil remediation (Arora and Kumar, 2019; Luo et al., 2021). As core components of plant-microbe interaction systems, PGPR participate in stress resistance regulation of host plants through multiple mechanisms such as metabolic interaction and signal transduction. For example, nutrient deficiency in saline-affected soils constitutes a major constraint limiting plant growth and yield enhancement. Rhizosphere microorganisms play a pivotal role in enhancing plant nutrient acquisition through multifaceted mechanisms, Bacillus aquimaris can increase nitrogen content in wheat leaves under salt stress (Upadhyay and Singh, 2015); Azospirillum brasilense and Pseudomonas fluorescens synergistically promote soil nitrogen mineralization and significantly enhance rice biomass. Inoculation with salt-tolerant PGPR elevates phytohormone levels in plant roots, consequently mitigating stress-imposed impairments under saline conditions. B. cereus regulates the plant cytokinin (CTK) signal transduction system under drought stress, coordinating CTK transport in root-shoot communication (Liu et al., 2015); Bacillus licheniformis increases the relative biomass of chrysanthemums by 35-42% and reduces root Na+ accumulation by 28-33% under 200 mM NaCl stress by synthesizing ABA (Cheng et al., 2017). Additionally, PGPR can enhance plant salt-alkali resistance by synthesizing osmolytes. Studies have shown that Bacillus subtilis improved proline synthesis capacity through overexpression of proB and proA genes, simultaneously enhancing its own salt tolerance and plant osmotic tolerance (Wu et al., 2018). These studies have revealed the multiple action mechanisms of PGPR in plant stress resistance regulation and their important application value in agricultural ecology.
Recent breakthroughs in functional research on Plant Growth-Promoting Rhizobacteria (PGPR) have revealed their significant biocontrol potential in crop cultivation and soil remediation. PGPR orchestrate host plant responses to abiotic stresses through multifaceted metabolic pathways, demonstrating versatile roles in plant-microbe interactions. Substantial evidence confirms that diverse PGPR taxa—including Bacillus, Chryseobacterium, Pseudomonas, Azospirillum, Achromobacter, Aeromonas, and Acetobacter (Etesami and Maheshwari, 2018)—enhance plant salinity tolerance via synergistic mechanisms. Under saline soil conditions, PGPR inoculation improves stress resilience metrics, promotes biomass accumulation, optimizes nutrient cycling efficiency, and stabilizes agricultural ecosystems. Metabolomic profiling by Ali et al (Ali et al., 2022). demonstrated that Enterobacter cloacae PM23 upregulates the proline synthesis gene ProDH, triggering osmolyte accumulation (e.g., proline, glycine betaine) in maize seedlings. This process maintains cellular turgor under moderate salt stress (100 mM NaCl). Complementary transcriptomic analysis (Girma et al., 2022) revealed that Klebsiella sp. KBG6.2 mitigates sodium chloride-induced oxidative damage (150 mM) in rice by activating indole-3-acetic acid (IAA) signaling while suppressing reactive oxygen species (ROS) burst in root apices.
This study integrates metagenomic sequencing and plant untargeted metabolomics technologies to systematically clarify the mechanism by which salt-tolerant PGPR inoculation promotes rice growth in salinized soils by regulating rhizospheric microbial community structure and root exudates. On the one hand, it enhances our theoretical understanding of PGPR enhancing crop salt tolerance; on the other hand, it provides experimental evidence for developing salinized soil improvement technologies based on salt-tolerant rice-microbe synergistic systems.
2 Materials and methods
2.1 Pot experiments and preparation of inoculants
The pot experiments with rice were conducted in a rooftop experimental facility at Yangzhou University (Yangzhou, China). Air-dried and sieved soil was filled into plastic pots (15 cm height × 15 cm diameter) at 2 kg per pot. Seven treatments were set up: CK (non-inoculated control), T1 (Bacillus sp.), T2 (Providencia sp.), T3 (Planococcus sp.), T4 (Pseudoclavibacter sp.), T5 (Dietzia sp.), and T6 (a mixture of the above five strains), with three replicates per treatment (21 pots in total). Rice seedlings with uniform growth were selected and transplanted into the pots (two hills per pot, four plants per hill). Seven days later, bacterial suspensions were evenly irrigated around the rice roots at an inoculation concentration of 108 CFU/g soil, while CK was watered with an equal volume of sterile water. A water layer depth of 2–3 cm was maintained by daily observation and replenishment with deionized water (Li et al., 2025). Physiological and biochemical indices of rice were measured after four weeks.
Typical single colonies were selected and inoculated into 5 mL of LB liquid medium, then cultured in a 28°C shaking incubator at 180 rpm for 12 h to prepare primary seed solutions. Subsequently, the primary culture was transferred to 100 mL of LB medium at a volume ratio of 1:100 and continuously cultured at 30°C with the same rotation speed for 12 h. After cultivation, bacterial pellets were collected by centrifugation (4°C, 8000×g, 10 min), and the supernatant was discarded. The pellets were resuspended in sterile water to an appropriate concentration.
2.2 Determination of physiological and biochemical indices
After four weeks of growth, intact rice plants were harvested, and root soils were rinsed with tap water. Residual water was blotted dry with filter paper, and the plant height and main root length were measured and recorded. Roots and shoots were separated, blanched in a constant-temperature drying oven at 115°C for 30 min, and then dried at 80°C to constant weight. Dry weights of roots and shoots were weighed using an electronic balance.
Chlorophyll content in rice leaves was determined by ethanol extraction colorimetry (Li, 2000). For antioxidant enzyme activity assays (Wang and Huang, 2015), SOD activity was measured by the nitroblue tetrazolium method, CAT activity by the guaiacol method, and POD activity by the UV absorption method in rice tissues (roots and leaves). MDA content was determined by the thiobarbituric acid colorimetry, and free proline content was measured by sulfosalicylic acid extraction–acid ninhydrin colorimetry (Wang and Huang, 2015) in roots and leaves.
2.3 Collection and analysis of root exudates
Root exudates of each treatment were collected in the fourth week of the experiment. Roots were first rinsed with sterile water, transferred to a 1 L beaker wrapped with tin foil, and immersed in sterile water. After continuous culture for 24 h, the solution was filtered through a sterile 0.45 μm microporous membrane. The collected solution was lyophilized by a freeze dryer to obtain dry powder of root exudates, which was stored in a sterile airtight container at -20°C. Subsequent LC-MS untargeted metabolomics analysis was commissioned to Majorbio Bio-Pharm (Shanghai, China), and the obtained data were further processed.
Metabolites that passed quality control were functionally annotated using the KEGG database, categorizing them based on their involvement in biological pathways or molecular functions. Orthogonal partial least squares-discriminant analysis (OPLS-DA) was conducted to statistically compare metabolic profiles among six inoculation treatments (T1–T6) and the control group (CK), systematically elucidating the regulatory effects of plant growth-promoting rhizobacteria (PGPR) on the rice root exudome. Significantly differential metabolites (top 30 ranked by variable importance in projection (VIP) scores with VIP ≥ 1) were selected for hierarchical clustering analysis using Euclidean distance and complete linkage algorithms. A composite visualization integrating a heatmap with VIP bar plots was generated to display metabolite distribution patterns, expression profiles, and statistical parameters (VIP values and P-values) across experimental groups.
Comparative metabolomic profiling between inoculated and non-inoculated groups identified differentially abundant metabolites exhibiting significant up- and down-regulation. Core metabolites were structurally classified using the Human Metabolome Database (HMDB), and KEGG pathway enrichment analysis was performed to identify significant pathways (false discovery rate [FDR] < 0.05) containing ≥2 mapped metabolites. Finally, Spearman’s rank correlation analysis was employed to elucidate association networks between dominant microbial taxa and root-secreted metabolites.
2.4 Processing and sequencing analysis of rhizospheric soil samples
During plant harvest, rhizospheric soil samples were collected using standardized methods. After intact root systems were removed, large soil clumps were shaken off, and soils tightly attached to root surfaces were collected and immediately stored in a -80°C freezer.
After DNA extraction, PCR amplification was performed for the V1-V9 variable regions of the bacterial 16S rRNA gene using the forward primer 27F (5’-[Barcode]AGAGTTTGATCMTGGCTCAG-3’) and reverse primer 1492R (5’-ACCTTGTTACGACTT-3’). The PCR reaction system contained 15 μL of 2× Phanta Max Master Mix (Vazyme), 0.5 μM of each primer, 10–30 ng of template DNA, and ddH2O up to 30 μL. The thermal cycling parameters were: initial denaturation at 95°C for 5 min; 30 cycles of 94°C for 30s (denaturation), 56°C for 30 s (annealing), and 72°C for 45s (extension); final extension at 72°C for 10 min; and storage at 4°C. The amplified products were verified by 2% agarose gel electrophoresis (120V, 40 min), and specific bands (450–550 bp) were excised, purified using the AxyPrep DNA Gel Extraction Kit (Axygen), and eluted with 30 μL of sterile EB buffer (10 mM Tris-HCl, pH 8.5). Microbiome data analysis was performed based on the QIIME2 platform (version 2022.11). In the sequence processing stage, demultiplexing assigned paired-end reads to sample-specific barcodes, and primer sequences were excised using Cutadapt (v3.7). The DADA2 pipeline performed quality filtering (Q-score ≥ 20), denoising with removal of non-overlapping sequences, paired-end merging requiring a minimum 12-bp overlap, and chimera detection against the SILVA 138 reference database. High-quality sequences were clustered at 100% similarity to generate amplicon sequence variants (ASVs), and a feature table (ASV-sample abundance matrix) was constructed. For taxonomic annotation and filtering, ASVs were classified via BLASTn alignment (E-value ≤ 1e−5) against the Greengenes 13_8 database (99% identity threshold). Low-abundance ASVs (relative abundance < 0.001%, equivalent to < 10 reads per million) were removed to generate a filtered feature table. In community profiling, taxonomic composition was visualized using stacked bar plots (phylum/class levels) generated with ggplot2 (v3.4.2) in R. Rarefaction curves confirmed adequate sequencing depth (Good’s coverage > 99%). Features were subsampled to 8,500 sequences per sample (5th percentile of minimum sequencing depth) to standardize library sizes.
For diversity analysis: Alpha diversity indices (Chao1 for richness; Shannon for evenness) were computed in QIIME2; intergroup differences were visualized via boxplots;Beta diversity analysis used weighted UniFrac distances for principal coordinate analysis (PCoA). Intergroup dissimilarities were assessed by Bray-Curtis-based PERMANOVA (999 permutations; P < 0.05). For differential analysis and integration: LEfSe identified differentially abundant taxa across groups. Spearman’s rank correlation revealed associations between dominant microbiota and rice physiological indices. Metabolic pathways with significant alterations (adj. P < 0.05, |log2FC| > 1) were characterized with key contributing taxa.
3 Results
3.1 Effects of PGPR inoculation on rice growth under salt stress
As shown in Figures 1A, B, compared with CK, all inoculated treatments promoted rice growth. Root length significantly increased (p<0.05) in all T1-T6 treatments compared to CK, with the largest increase (51.77%) observed in T6 (Figures 1B, C). Plant height significantly increased (p<0.05) in T1-T4 and T6 treatments, but not in T5 (10.76% increase, not significant). The T4 treatment showed the greatest increase in plant height (23.43%) (Figure 1B). In the T1 treatment, there were no significant changes in the dry weight of roots and shoots. However, the dry weight of shoots significantly increased in T2-T6 treatments, with the T3 treatment showing the greatest increase (133.33%) (Figure 1D). The dry weight of roots in the T4 treatment was 1.9 times that of CK (Figure 1D).
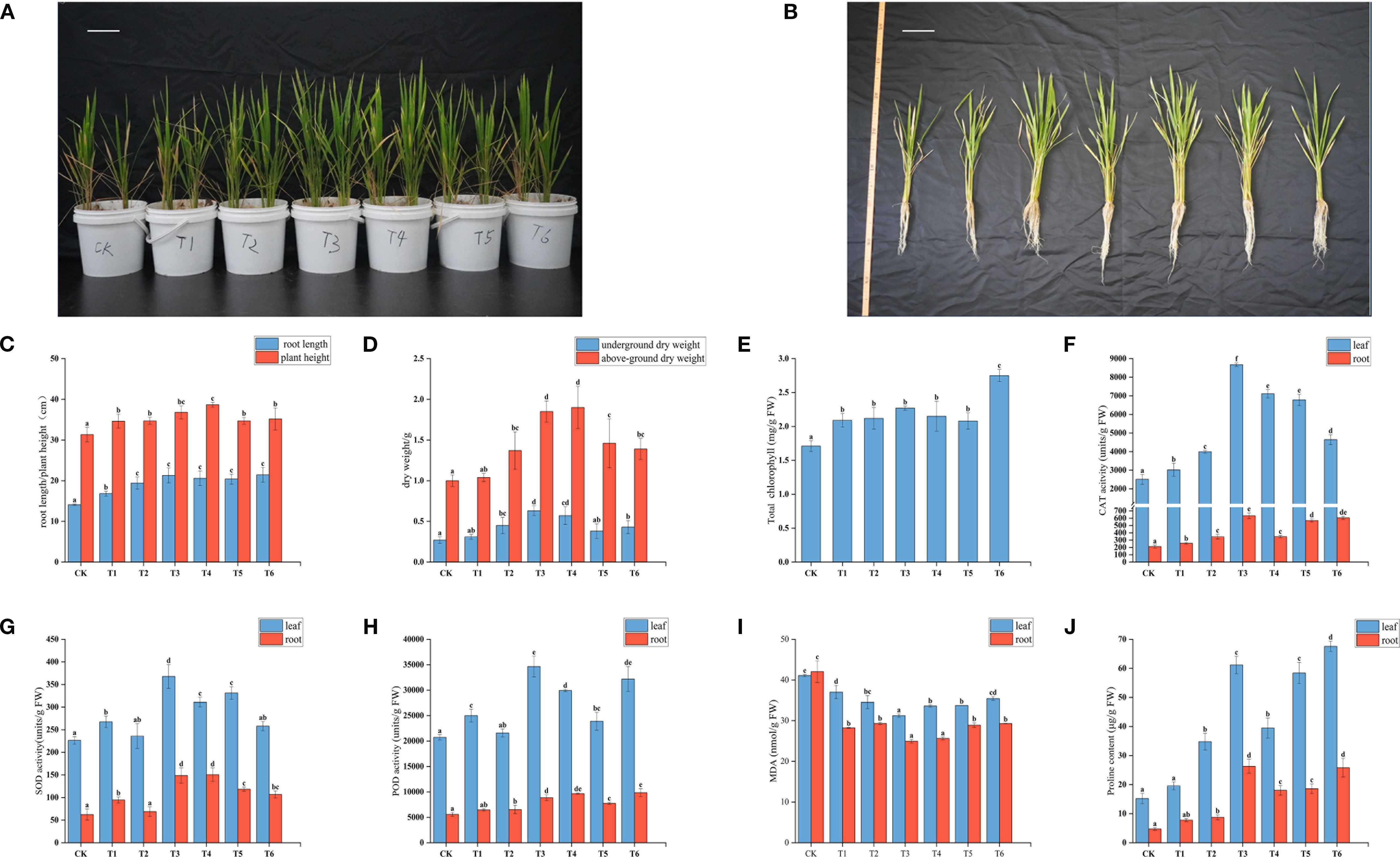
Figure 1. Effects of different treatment groups on (A, B) rice seedling growth, (C) root length and plant height,(D) above-ground and below-ground dry weight, (E) total chlorophyll content, (F-H) SOD, CAT, and POD content in rice leaves and roots, (I, J) MDA and proline content in rice leaves and roots.
Chlorophyll, a key catalyst for plant photosynthesis, is an important indicator of photosynthetic efficiency and growth status. The chlorophyll content in all T1-T6 treatments was significantly higher than in CK (p < 0.05). The T6 treatment showed the most significant effect, with an increase of 60.82% compared to CK (Figure 1E), indicating that mixed inoculation with PGPR has a clear advantage in improving total chlorophyll content in rice leaves under salt stress.
To assess the effects of different treatments on rice defense capacity, the activities of antioxidant enzymes (POD, SOD, CAT), MDA content, and Pro content were measured in rice leaves and roots. The antioxidant enzyme activities in both leaves and roots of all treatment groups were higher than in CK (Figures 1F–J), indicating that PGPR inoculation enhances the antioxidant capacity of rice seedlings under salt stress. Overall, most treatments showed significant increases compared to CK. The largest increase in SOD activity in leaves was observed in T3 (62.40%), while in roots, T4 showed the greatest increase (141.10%). For CAT activity, T3 had the greatest increase in both leaves (245.66%) and roots (199.15%). For POD activity, T3 showed the greatest increase in leaves (67.16%), and T4 in roots (73.26%).
MDA content, a key indicator of oxidative stress, increases when plants are subjected to stress due to intensified lipid peroxidation. The MDA content in both leaves and roots of T1-T6 treatments was significantly reduced (p<0.05) compared to CK (Figure 1I), indicating that PGPR inoculation effectively alleviates salt stress in rice. The inhibitory effects on MDA content varied among treatments, with T3 showing the most prominent effect: MDA content in leaves and roots decreased by 24.04% and 40.65%, respectively. The reduction in MDA content in roots was significantly greater than in leaves for all T1-T6 treatments (p<0.05), indicating that PGPR inoculation provides more significant protective effects on roots.
Free Pro is an important osmolyte in plant responses to abiotic stress, and its accumulation is closely associated with stress tolerance. The Pro content in both leaves and roots of T1-T6 treatments increased compared to CK, with significant increases in T2-T6 (p<0.05) (Figure 1J), indicating that PGPR inoculation enhances rice salt tolerance by promoting Pro accumulation. The T3 and T6 treatments showed the most significant increases: leaf Pro content increased by 302.10% and 344.31%, respectively, and root Pro content by 456.03% and 445.24%, respectively. The T2, T4, and T5 treatments also exhibited significant increases in Pro content in both leaves and roots (p<0.05).
3.2 Effects of PGPR inoculation on the rhizosphere bacterial community of rice under salt stress
Multi-omics analysis of the rhizosphere soil microbial community of rice using 16S rRNA amplicon sequencing showed that after rigorous quality control procedures, the total number of raw sequencing data reached 522,660, with the number of raw sequencing data per sample ranging from 73,120 to 75,989. After chimera filtering and sequence optimization, the total number of high-quality sequences was 492,998, with individual sample valid sequence counts ranging from 68,807 to 71,759. Sequencing read lengths, after quality assessment, ranged from 681 to 1,471 base pairs, meeting the requirements for subsequent analysis. At the same time, we uploaded the sequencing sequences to the NCBI database with the accession number PRJNA1304747.
Principal Coordinates Analysis (PCoA) based on Bray-Curtis dissimilarity (Figure 2A) revealed clear separation between inoculated (T1-T6) and CK groups. PC1 (21.38%) and PC2 (12.86%) collectively explained 34.24% of community variance, confirming significant PGPR-induced restructuring of the rhizosphere microbiome under salt stress.
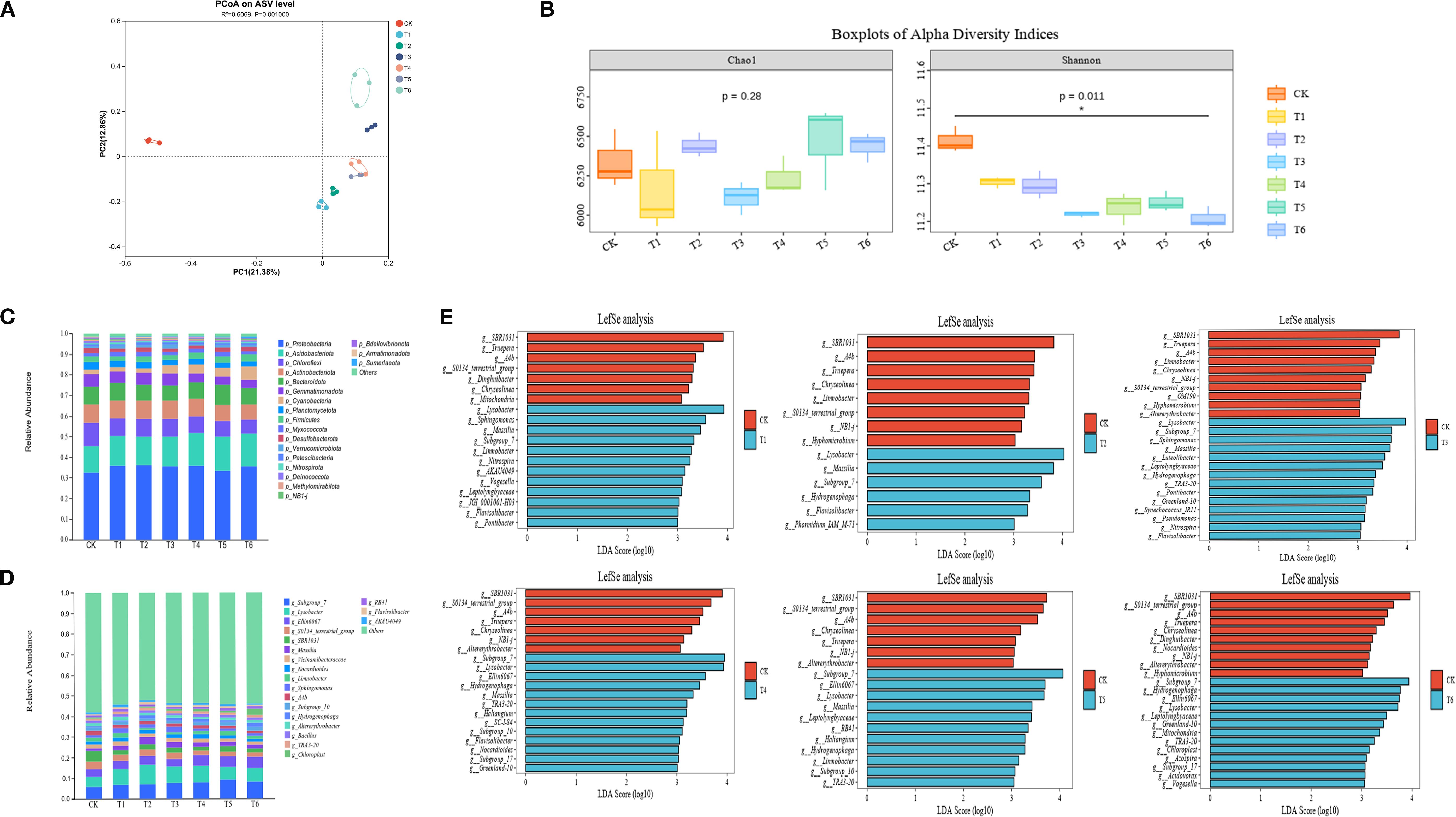
Figure 2. Rice rhizosphere bacterial community (A) PcoA plot and (B) Alpha diversity box plot, [(C) phylum level; (D) genus level] rice rhizosphere bacterial community composition and (E) genus level LefSe analysis.
Alpha diversity analysis (Figure 2B) showed significantly reduced Shannon indices in all inoculated treatments (T1-T6) versus CK (p < 0.05), indicating decreased species diversity and evenness. The Chao1 index showed no significant differences among treatments (p > 0.05), demonstrating that PGPR inoculation primarily affects community evenness rather than species richness.
Community composition analysis identified dominant bacterial phyla across all treatments (Figure 2C): Proteobacteria, Acidobacteriota, Chloroflexi, Actinobacteriota, Bacteroidota, and Gemmatimonadota. At genus level (Figure 2D), core taxa included: Subgroup_7 (6.07–9.23%), Lysobacter (4.63–9.52%), Ellin6067 (3.49–5.70%), S0134_terrestrial_group (2.06–3.65%), and SBR1031 (1.79–4.98%). PGPR inoculation consistently enriched beneficial genera (Subgroup_7, Lysobacter, Ellin6067, Massilia, Sphingomonas) while suppressing S0134_terrestrial_group, SBR1031, Limnobacter, and A4b relative to CK.
Linear discriminant analysis ffect Size (LEfSe) (LDA > 3; Figure 2E) identified treatment-specific biomarkers, with shared enrichment of beneficial taxa (Subgroup_7, Lysobacter, Massilia, Hydrogenophaga, Ellin6067, Sphingomonas) across multiple inoculated treatments.
Finally, OTU abundance profiles derived from high-throughput sequencing were mapped to the KEGG database to characterize functional attributes of rhizobacterial communities across treatment groups. At KEGG Pathway Level 3, PGPR inoculation elevated the abundances of key bacterial metabolic pathways in saline-stressed rhizosphere soils, including:Ansamitocin biosynthesis (ko01051); Vancomycin antibiotic biosynthesis (ko01055); C5-Branched dibasic acid metabolism (ko00660); Valine, leucine and isoleucine biosynthesis (ko00290); Fatty acid biosynthesis (ko00061); Pantothenate and CoA biosynthesis (ko00770); Synthesis and degradation of ketone bodies (ko00072); Lipoic acid metabolism (ko00785).
(Table 1).
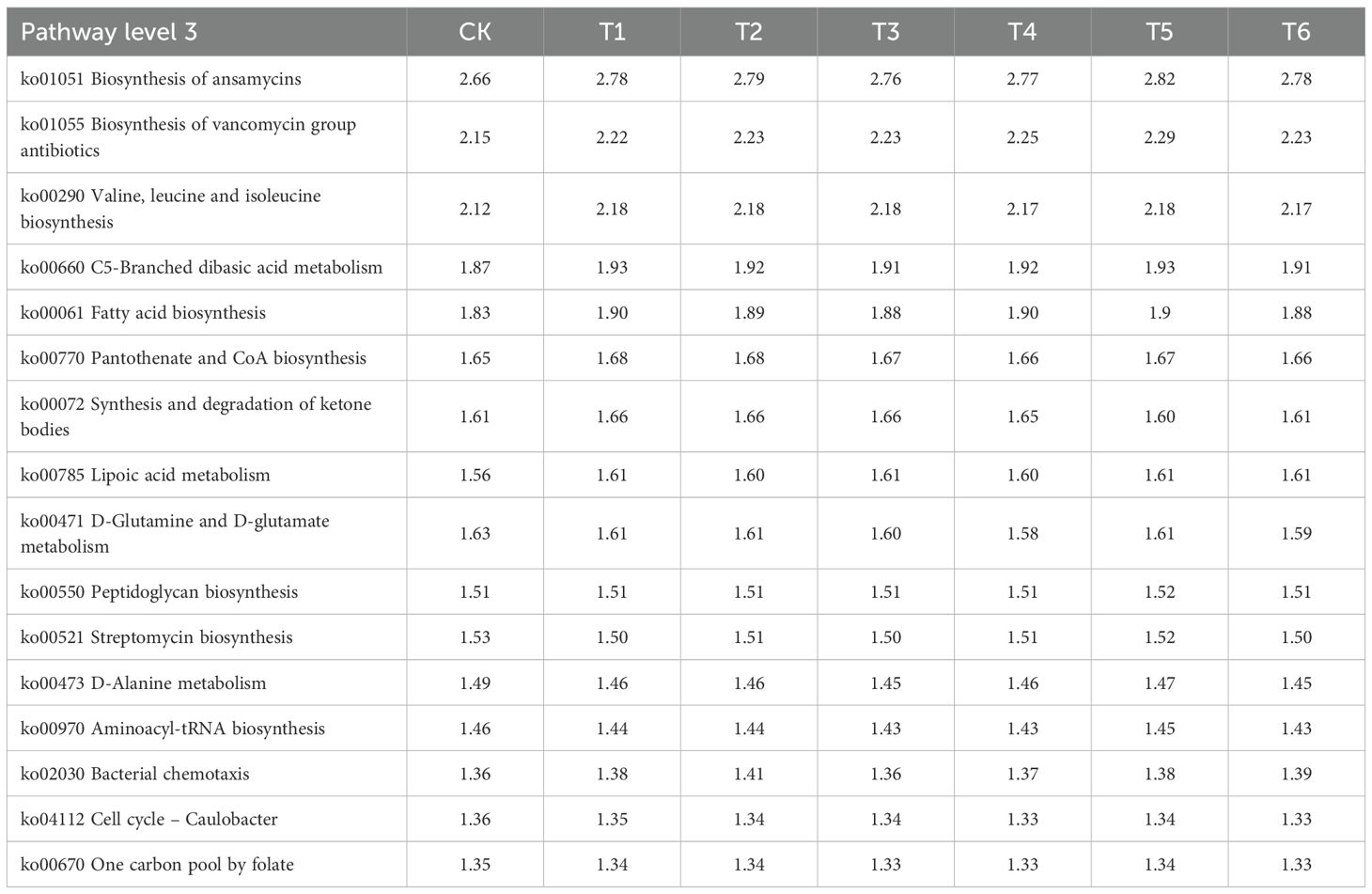
Table 1. Functional annotation of KEGG pathway 3 for rhizosphere soil bacterial communities of rice in different treatment groups.
3.3 Differences in the composition of rice root exudates
To investigate the effects of PGPR inoculation on rice root exudates under salt stress and their association with rhizosphere microbiome functional regulation, we analyzed exudates from six treatments. We identified 2,029 metabolites, comprising 1,379 cationic and 650 anionic metabolites. Partial Least Squares Discriminant Analysis (PLS-DA) revealed significant intergroup differences in exudate composition (Figure 3A).
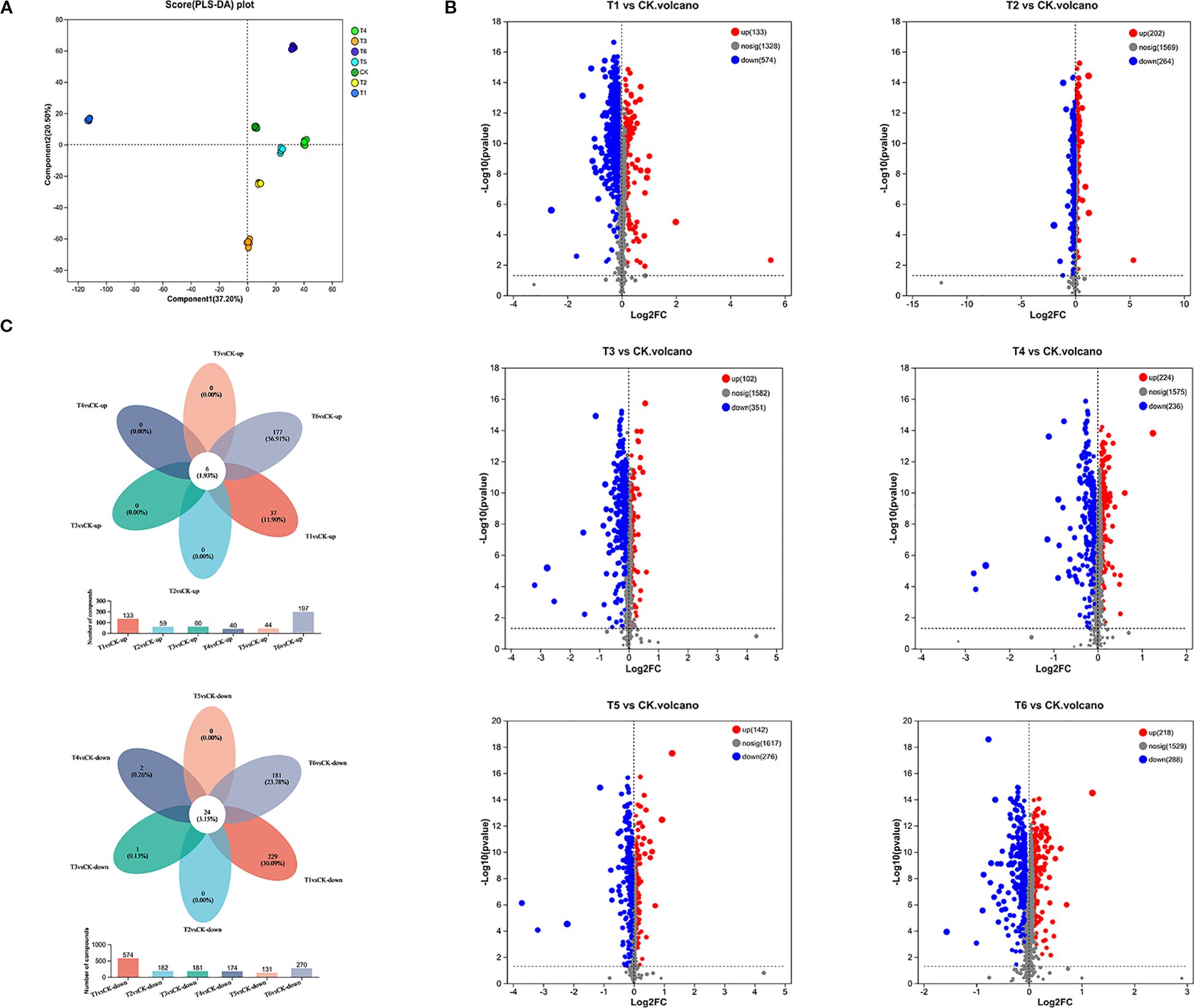
Figure 3. (A) PLS-DA of root exudates, (B) Total ion model differential metabolite volcano plot for each differential grouping, (C) Venn diagram of differentially expressed exudates in rice under different treatment groups.
Volcano plots of differentially expressed metabolites (DEMs) indicated a predominant downregulation trend in all treatments relative to CK (Figure 3B). T1 exhibited the highest number of downregulated metabolites (n=574). These results confirm that PGPR inoculation significantly alters root exudate composition, with downregulation being the dominant response.
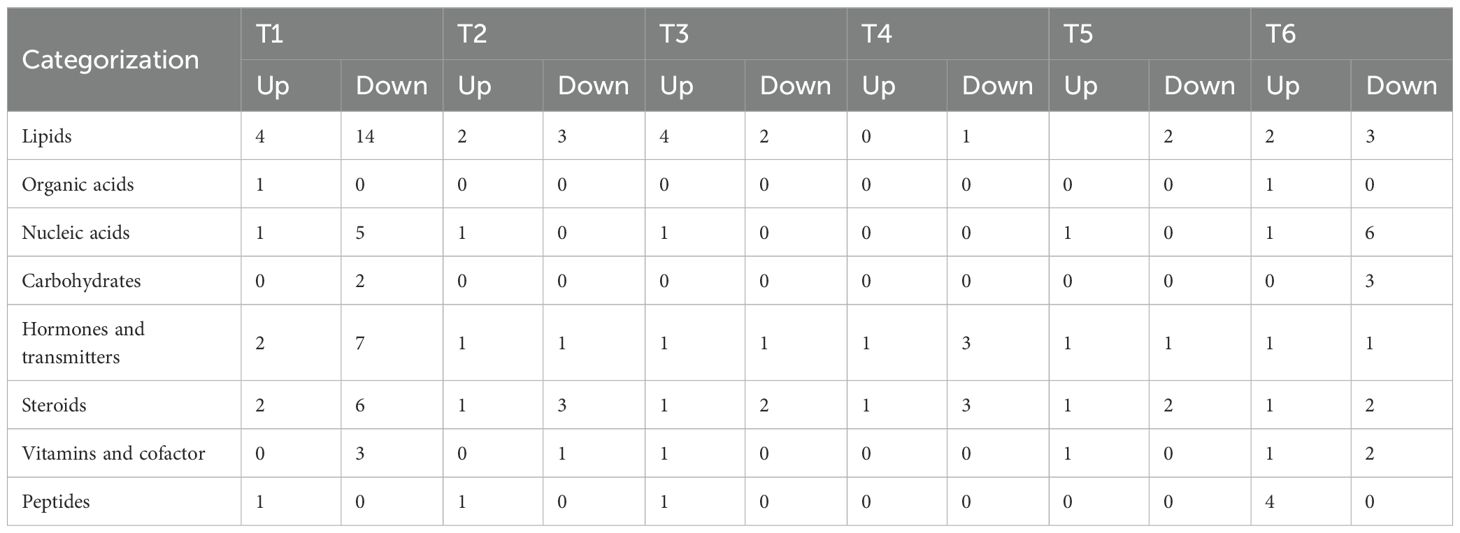
Through Venn diagram analysis of common and unique differential exudates among different comparison treatment groups, the results were shown as follows: Six common upregulated DEMs, with 37 and 177 unique to T1 and T6 (vs. CK), respectively;Twenty-four common downregulated DEMs, with 229 and 181 unique to T1 and T6 (vs. CK), respectively. (Figure 3C). Functional annotation classified these DEMs into lipids, hormones and transmitters, steroids, vitamins and cofactors, and nucleic acids (Table 2). Downregulated DEMs in T1-T6 (vs. CK) were predominantly lipids, hormones and transmitters, and steroids, while upregulated DEMs were primarily nucleic acids and peptides.
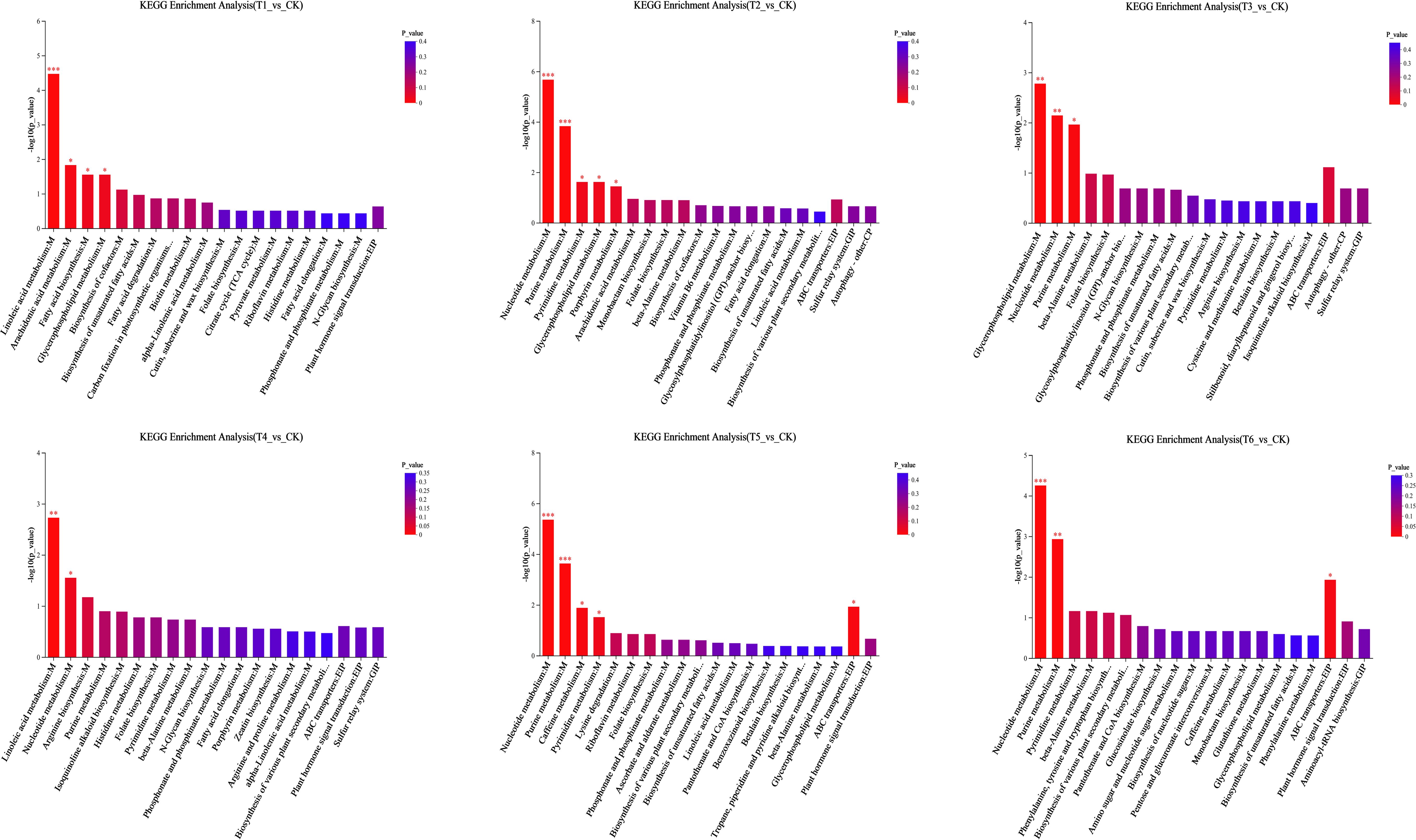
Pathway enrichment analysis indicated that key DEMs in T1-T6 were primarily enriched in nucleotide metabolism, purine metabolism (Metabolism), and ABC transporters (Environmental Information Processing) (Figure 4). Specifically:T1 exhibited significant enrichment (p < 0.001) in linoleic acid metabolism (Metabolism).T2 and T3 were enriched in glycerophospholipid metabolism (Metabolism).The mixed inoculation (T6) showed significantly more enriched pathways than single strains, including tryptophan metabolism, phenylalanine/tyrosine/tryptophan biosynthesis (Metabolism), and plant hormone signal transduction (Environmental Information Processing).
3.4 Correlation analysis between rice rhizosphere soil bacterial communities and physiological indicators, root exudates
Correlation analysis revealed significant associations between key soil bacterial groups and rice physiological indicators and root exudates. We concluded that:
Beneficial taxa (Subgroup_7, Subgroup_17, Hydrogenophaga, Haliangium, Leptolyngbyaceae) showed significant positive correlations (p < 0.05) with growth parameters (root length, plant height, shoot/root biomass, chlorophyll) and stress markers (SOD, CAT, POD, Proline), while exhibiting negative correlations with MDA (Figure 3A).Conversely, SBR1031, A4b, Altererythrobacter, Arcicella, and Rhodobacter displayed inverse correlation patterns (Figure 5A).
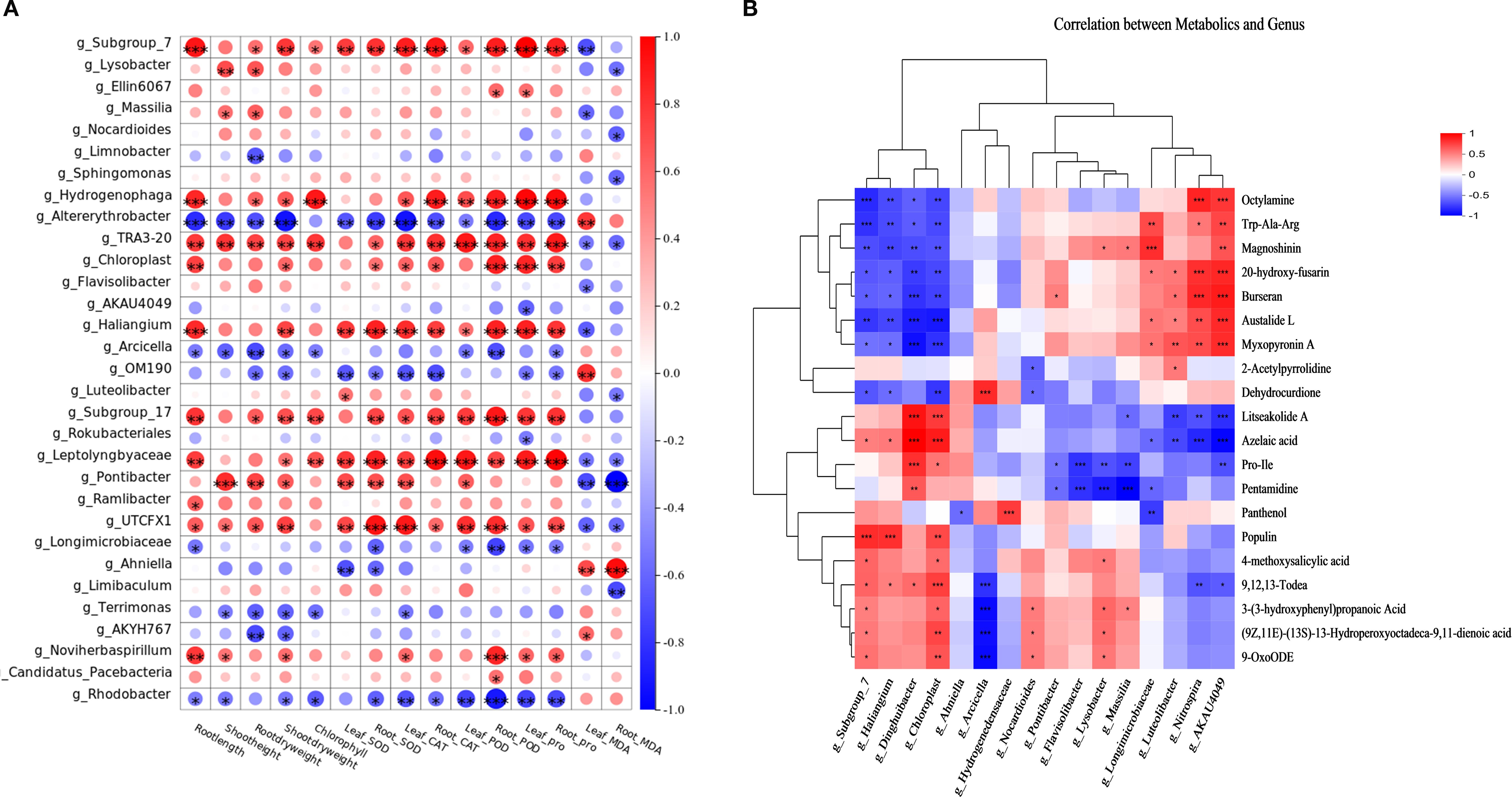
Figure 5. (A) Heat map of correlation between genus-level enriched taxa of rice rhizosphere bacteria and rice physiological indicators;(B) Heatmap of correlation analysis between rice root secretion and rhizosphere bacterial genus level enrichment taxa.
Subgroup_7, Subgroup_17, Hydrogenophaga, TRA3-20, Haliangium, Leptolyngbyaceae, etc., showed significant positive correlations with growth parameters (root length, plant height, above- and below-ground dry weight, chlorophyll content) and stress markers (SOD, CAT, POD, Pro), and significant. negative correlations with MDA. In contrast, SBR1031, A4b, Altererythrobacter, Arcicella, Rhodobacter, exhibited inverse correlations (Figure 5A).
Metabolite-specific relationships revealed three distinct microbial functional groups:
Protective Taxa: Subgroup_7 and Haliangium exhibited significant negative correlations with stress-associated metabolites (octylamine, tryptophyl-alanyl-arginine, magnoshinin, 20-hydroxyfusarin, burseran, austalide L, myxopyronin A, dehydrocurdione) while showing positive correlations with beneficial compounds (litseakolide A, azelaic acid, populin, 9,12,13-trihydroxy-10E-octadecenoic acid). Stress-Associated Taxa: Longimicrobiaceae, AKAU4049, Luteolibacter, and Nitrospira demonstrated inverse patterns - positively correlating with the aforementioned stress metabolites but negatively correlating with litseakolide A, azelaic acid, prolyl-isoleucine, and pentamidine. Dual-Role Taxa: Lysobacter, Massilia, and Flavisolibacter displayed selective associations: positive correlations with magnoshinin and 3-(3-hydroxyphenyl)propanoic acid, yet negative correlations with prolyl-isoleucine and pentamidine (Figure 5B).
4 Discussion
Salt stress is an important environmental factor that severely limits global crop productivity. The use of appropriate beneficial PGPR represents a greener, cleaner approach to improving crop production and environmental resilience under salt stress. This study systematically investigated the effects of PGPR inoculation on rice seedling growth, rhizosphere microbial communities, and root exudate metabolism under salt stress. The results align with existing research and provide multidimensional evidence for elucidating the salt tolerance regulatory mechanisms of PGPR.
4.1 Effects of different treatments on rice growth
We found that PGPR alleviated salt stress damage by enhancing the antioxidant system and osmotic regulation function, consistent with the theory proposed by Fiodor et al (Fiodor et al., 2021). that “PGPR inoculation may increase nutrient absorption and accumulation by promoting root development.” In the experiment, different PGPR inoculation treatments significantly increased SOD, CAT, and POD activities in rice leaves and roots, consistent with the results reported by Jha et al (Jha and Subramanian, 2013) and Chen et al (Chen et al., 2024). Meanwhile, MDA content was significantly reduced, alleviating oxidative stress caused by salt stress, in line with the findings of Han et al (Han et al., 2014). and Chauhan et al (Chauhan et al., 2019), indicating that PGPR activated the enzymatic defense network to remove excess ROS. Notably, under mild salt stress (e.g., soil salinity of 0.2–2.2‰), PGPR significantly enhanced chlorophyll content more effectively than under severe stress, possibly because the low-salt environment did not exceed the metabolic regulatory threshold of PGPR. Overall, PGPR inoculation improved chlorophyll content in rice leaves under salt stress (Xin et al., 2011; Gao, 2014). In this experiment, proline content in rice leaves and roots increased significantly across all treatment groups, consistent with the findings of Jha et al (Jha and Subramanian, 2013). As a core component of osmotic regulation, proline accumulation was enhanced by PGPR through the activation of the OsP5CS1/2 gene, aligning with the molecular mechanisms observed in transgenic rice studies (Wang et al., 2021).
4.2 Differences in rhizosphere bacteria under salt stress
Our study also observed that PGPR inoculation led to a decrease in rhizosphere bacterial alpha diversity (Shannon index), consistent with the community clustering results from PCoA. Specifically, under salt stress, PGPR preferentially promoted the colonization of beneficial bacteria, leading to enhanced community structure selectivity and a reduction in overall α-diversity (Tahir et al., 2015; Jing et al., 2018). The survival of exogenous bacteria depends on environmental compatibility; when soil conditions such as pH and temperature are suboptimal, even inoculants with high viable cell counts may exhibit poor survival rates and consequently fail to outcompete indigenous microbial populations. In most cases, inoculation enhances plant nutrient acquisition and promotes growth—both directly and indirectly—by altering rhizosphere soil pH and modulating microbiome composition and interactions (da Cunha et al., 2024).
At the phylum level, Proteobacteria, Acidobacteriota, Chloroflexi, and Actinobacteriota were the four dominant phyla across all treatments, similar to the findings of Sarathambal et al (Sarathambal et al., 2022), confirming that PGPR inoculation does not recruit dominant bacterial groups in a host plant-specific manner. The enrichment of these dominant bacterial groups carries clear functional implications. α- and γ-Proteobacteria—exemplified by nitrogen-fixing genera such as Rhizobium (α-class) and Pseudomonas (γ-class)—convert atmospheric nitrogen into plant-available forms, significantly enhancing plant growth through biological nitrogen fixation. In saline-alkaline soils, these Proteobacteria facilitate rapid rhizospheric organic matter degradation, exhibiting competitive dominance in carbon-rich environments. Their abundance correlates positively with soil organic matter content, and their metabolic plasticity contributes to plant stress tolerance under adverse conditions (Rilling et al., 2018; Das et al., 2020; Anzalone et al., 2022). Acidobacteriota abundance frequently exhibits negative correlations with soil nitrogen/phosphorus availability. This phylum likely employs oligotrophic strategies to outcompete copiotrophs under nutrient-depleted conditions, contributing to carbon cycling through decomposition of recalcitrant organic compounds (e.g., cellulose) and thereby enhancing soil fertility sustainability (Zhang et al., 2023). Chloroflexi contributes to phytoremediation of contaminated soils under heavy metal-polluted or hypersaline conditions through sulfur cycling and anaerobic metabolic pathways. In contrast, Actinobacteriota plays a role in enhancing soil nutrient content, inhibiting the growth of pathogenic microorganisms, and promoting plant growth (Bhattacharyya et al., 2018; Guarino et al., 2020). Proteobacteria and Actinobacteriota dominate nitrogen and phosphorus transformation, while Chloroflexi and Acidobacteriota participate in carbon cycling, collectively maintaining root zone nutrient balance and resisting environmental fluctuations. This functional division confirms that PGPR exhibits cross-host conservation in recruiting dominant microbial communities.
At the genus level, genera such as Subgroup_7, Lysobacter, Massilia, and Hydrogenophaga were significantly enriched in the rice rhizosphere. This directed regulation formed a specific microbial community architecture that was more conducive to the adaptive growth of rice seedlings under salt stress. Among these, Subgroup_7, as the core group of Acidobacteriota, prefers acidic environments, participates in the decomposition of recalcitrant organic matter (such as cellulose), regulates soil carbon cycling, and stabilizes soil aggregates by secreting extracellular polysaccharides, thereby improving the rhizosphere microenvironment (Yang et al., 2021). Lysobacter, Massilia, and Hydrogenophaga (all belonging to Proteobacteria) construct the defense and nutritional network of the rhizosphere microbiome through functions such as decomposing recalcitrant carbon sources, inhibiting pathogens, promoting nutrient cycling, and enhancing plant resistance (Ruth et al., 2015). Additionally, the relative abundance of these groups showed a significant positive correlation with rice physiological and biochemical indicators, promoting seedling growth under salt stress.
Functional gene analysis further revealed enrichment of the pantothenate and CoA biosynthesis pathway (ko00770) at KEGG Level 3, indicating PGPR may enhance microbial oxidative stress adaptation by modulating redox cofactor metabolism (Ilangumaran and Smith, 2017). The upregulation of ansamitocin (ko01051) and vancomycin-type antibiotic biosynthesis (ko01055) genes suggests PGPR inoculation induces antagonistic interactions. These antibiotics not only suppress phytopathogens but also reinforce the competitive dominance of beneficial taxa (e.g., Bacillus, Pseudomonas) through rhizosphere microbiome restructuring (Khanghahi et al., 2025). Notably, although salinity stress typically reduces Actinobacteriota abundance, PGPR-secreted antibiotics may preserve functional guild equilibrium—a mechanism corroborated in rapeseed rhizospheres by Świątczak et al (Swiatczak et al., 2023).
4.3 Differences in the composition of rice root exudates under salt stress
Analysis using the OPLS-DA model indicated that PGPR treatment significantly altered the composition of root exudates, with the number of DEMs ranging from 418 to 707 across treatment groups—primarily downregulated, suggesting that PGPR reshapes the rhizosphere microenvironment by inhibiting specific metabolic pathways. The chemical classification of differentially secreted compounds showed that lipids and steroids were predominantly downregulated, while peptides, vitamins, and cofactors were predominantly upregulated. Nucleic acids were predominantly downregulated in T1 and T6 but upregulated in T2–T5. Peptides (Trp-Ala-Arg) play critical roles in plant root secretions as signal regulators, growth promoters, stress resistance enhancers, and influencers of rhizosphere microorganisms. For example, brassinosteroids collaborate with other plant hormones (such as auxins) to regulate growth and development, while steroid components in root exudates act as signaling molecules, influencing rhizosphere microbial composition and regulating plant–microbe interactions.
Furthermore, heat map analysis showed that the secretion of lipids (e.g., Austalide L, Myxopyronin A) reduced the enrichment of bacterial colonies such as Subgroup_7 and Haliangium, while Litseakolide A and 9,12,13-Todea enhanced their enrichment. Organic acids (e.g., Azelaic acid, 3-(3-hydroxyphenyl)propanoic acid) exhibited the opposite effect. KEGG enrichment analysis revealed significant enrichment of nucleotide metabolism, purine metabolism, and the ABC transporter pathway (within Environmental Information Processing) across treatment groups. Rhizosphere microorganisms support proliferation through nucleotide synthesis (e.g., adenosine and guanosine nucleotides)—for instance, D-ribose transport, mediated by the RbsA enzyme, represents a critical step in microbial reproduction (Abulfaraj et al., 2024), while secreted ribose functions as a plant symbiosis signal (Keren et al., 2024). Purine metabolism generates cAMP via the cascade AMP → ADP → ATP, directly influencing microbial energy metabolism efficiency (Kai Wang et al., 2016). ABC transporters participate in nutrient transmembrane transport (e.g., amino acids) and microbial toxin efflux—e.g., Aeromonas hydrophila alters outer membrane phospholipid transport through MlaF gene mutations to enhance antibiotic resistance (Powers and Trent, 2018). Furthermore, ABC transporters are implicated in plant hormone signaling (e.g., IAA). This association potentially underpins a mutualistic relationship characterized by bidirectional resource exchange: microbes secrete auxins, while plants provide carbon sources. This establishes a cyclical “microbe-secreted auxin–plant-provided carbon source” relationship (Wang, 2017).
The T6 treatment (mixed strains) enriched more pathways—including nucleotide metabolism and glycerophospholipid metabolism—than single strains. Nucleotide synthesis supports rapid microbial proliferation, while adjustments in membrane lipid composition (e.g., upregulated phosphatidylethanolamine) enhance environmental adaptability (Zhao et al., 2021; Abulfaraj et al., 2024), providing metabolic evidence for why mixed bacterial agents outperform single strains.
5 Conclusions
This study demonstrates that PGPR inoculation significantly enhances root exudate production in rice, and these exudates subsequently recruit beneficial microbiota (Subgroup_7, Lysobacter, Massilia) that promote plant growth and development (Figure 6).
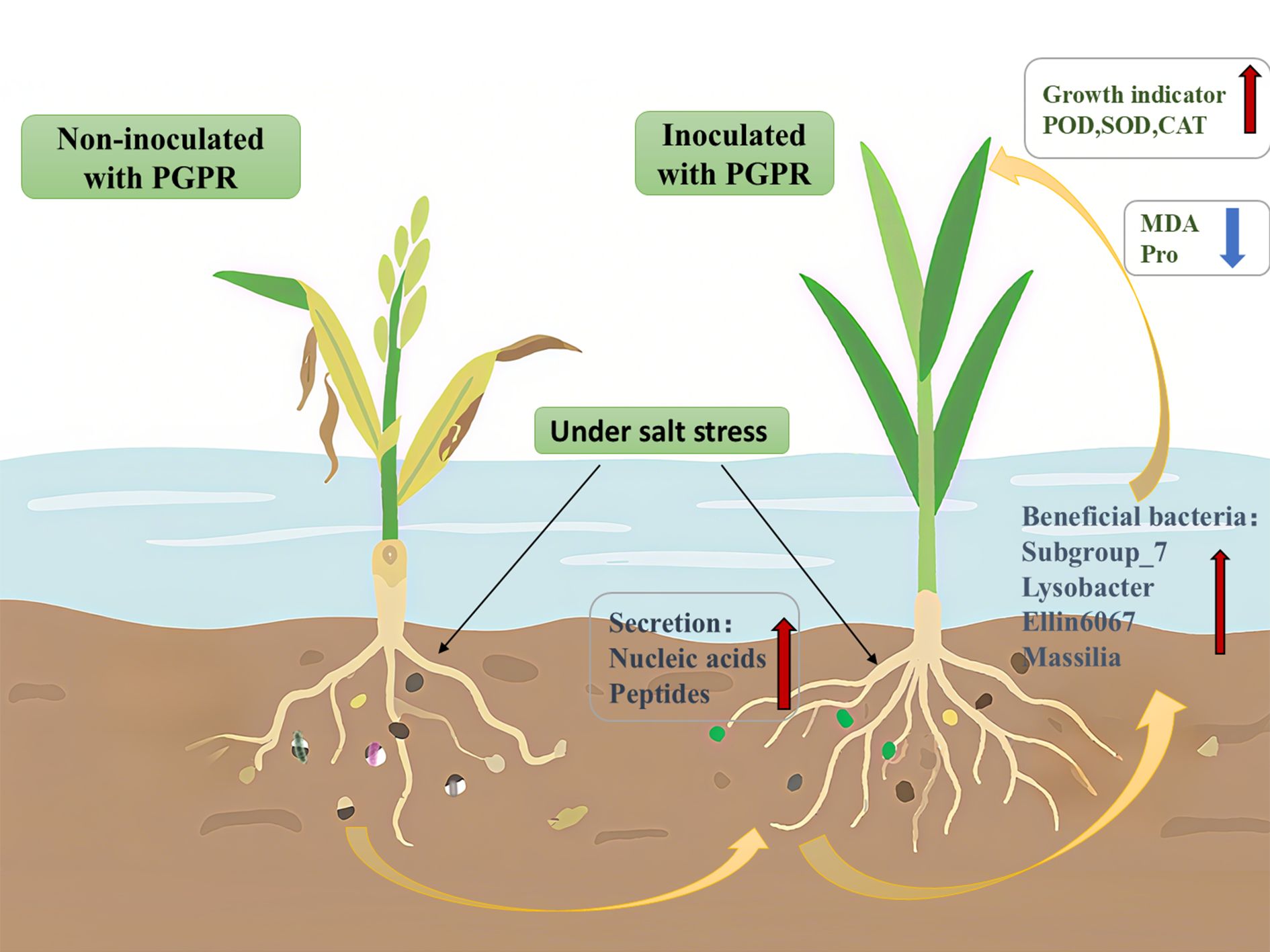
Figure 6. Effect of Plant Growth-promoting Rhizobacteria on the Growth of Rice Seedlings under Salt Stress and Its Microbiological Mechanism. This illustration demonstrates the effects of PGPR inoculation on rice cultivated in saline-stressed soil. The left panel depicts untreated rice plants and their root systems, where the native microbial community remains unaffected by exogenous microorganisms under chronic salt stress, resulting in significant growth inhibition. In contrast, the right panel shows PGPR-inoculated rice and associated roots. Inoculation stimulated the secretion of specific rhizosphere metabolites, which effectively recruited beneficial microbiota and restructured the microbial community composition. Consequently, soil microbial diversity was enhanced, concomitant with marked improvements in rice growth indices. These synergistic effects collectively promoted plant development and stress adaptation.
These findings establish a PGPR-rice metabolic mutualism model wherein PGPR recruit stress-protective microbiomes while synchronously tailoring root exudates to optimize nutrient acquisition and ROS scavenging. The enhanced performance of the T6 consortium highlights the potential of synthetic microbial communities for reclaiming saline soils. Future research should validate these mechanisms in field trials and integrate transcriptomics to elucidate underlying gene regulatory networks.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JW: Conceptualization, Writing – review & editing, Data curation, Investigation. XG: Data curation, Writing – original draft. MY: Methodology, Formal Analysis, Writing – review & editing. YZ: Writing – review & editing, Supervision, Investigation. YC: Project administration, Supervision, Writing – review & editing, Investigation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research of the thesis was supported by the National Key Research and Development Program (2022YFD1900704-05), the National Natural Science Foundation of China (81873877, 42307443) and Jiangsu Agriculture Science and Technology Innovation Fund (CX (23) 1020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1661074/full#supplementary-material
References
Abulfaraj, A. A., Shami, A. Y., Alotaibi, N. M., Alomran, M. M., Aloufi, A. S., Al-Andal, A., et al. (2024). Exploration of genes encoding KEGG pathway enzymes in rhizospheric microbiome of the wild plant Abutilon fruticosum. Amb Express 14, 2–21. doi: 10.1186/s13568-024-01678-4
Ali, B., Wang, X., Saleem, M. H., Sumaira, A., Hafeez, A., Afridi, M. S., et al. (2022). PGPR-Mediated Salt Tolerance in maize by modulating plant physiology, antioxidant defense, compatible solutes accumulation and bio-surfactant producing genes. Plants-Basel 11, 2–26. doi: 10.3390/plants11030345
Anzalone, A., Mosca, A., Dimaria, G., Nicotra, D., Tessitori, M., Privitera, G. F., et al. (2022). Soil and soilless tomato cultivation promote different microbial communities that provide new models for future crop interventions. Int. J. Mol. Sci. 23, 2–19. doi: 10.3390/ijms23158820
Arora, N. K. and Kumar, N. (2019). “Plant growth-promoting rhizospheric microbes for remediation of saline soils,” in Phyto and Rhizo Remediation. Eds. Arora, N. K. and Kumar, N. Springer-Verlag, Singapore, 121–146. doi: 10.1007/978-981-32-9664-0_5
Bhattacharyya, D., Duta, S., Yu, S.-M., Jeong, S. C., and Lee, Y. H. (2018). Taxonomic and functional changes of bacterial communities in the rhizosphere of kimchi cabbage after seed bacterization with Proteus vulgaris JBLS202. Plant Pathol. J. 34, 286–296. doi: 10.5423/ppj.Oa.03.2018.0047
Chauhan, P. S., Lata, C., Tiwari, S., Chauhan, A. S., Mishra, S. K., Agrawal, L., et al. (2019). Transcriptional alterations reveal Bacillus amyloliquefaciens-rice cooperation under salt stress. Sci. Rep. 9, 11912. doi: 10.1038/s41598-019-48309-8
Chen, J., Yang, C., Qi, W., Lin, X., Li, S., and Liu, S. (2024). Effects of PGPR biofertilizer on the physiological characteristics of ryegrass plants under salt stress. Beifang Yuanyi., 55–60. doi: 10.11937/bfyy.20233451
Cheng, Z., Lin, Z., Yue, X., Feiyue, L., Xin, X., Zhongyou, M., et al. (2017). Bacillus licheniformis SA03 confers increased saline–alkaline tolerance in chrysanthemum plants by induction of abscisic acid accumulation. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01143
da Cunha, I. D. A. M., da Silva, A. V. O. R., Boleta, E. H. M., Pellegrinetti, T. A., Zagatto, L. F. G., Zagatto, S. D. S., et al. (2024). The interplay between the inoculation of plant growth-promoting rhizobacteria and the rhizosphere microbiome and their impact on plant phenotype. Microbiol. Res 283, 127706. doi: 10.1016/j.micres.2024.127706
Das, A., Chaudhari, D., Rangappa, K., Layek, J., and Rahi, P. (2020). Pea (Pisum sativum l.) plant shapes its rhizosphere microbiome for mutrient uptake and stress amelioration in acidic aoils of the North-East region of India. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00968
Etesami, H. and Maheshwari, D. K. (2018). Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 156, 225–246. doi: 10.1016/j.ecoenv.2018.03.013
Fiodor, A., Singh, S., and Pranaw, K. (2021). The contrivance of plant growth promoting microbes to mitigate climate change impact in agriculture. Microorganisms 9, 2–36. doi: 10.3390/microorganisms9091841
Gao, X. (2014). Effects of different concentrations of saline and alkaline stress on the growth and physiological and ecological characteristics of rice. Jilin Agric. University.
Girma, B., Panda, A. N., Roy, P. C., Ray, L., Mohanty, S., and Chowdhary, G. (2022). Molecular, biochemical, and comparative genome analysis of a rhizobacterial strain Klebsiella Sp. KBG6.2 imparting salt stress tolerance to Oryza sativa L. Environ. Exp. Bot. 203, 105066. doi: 10.1016/j.envexpbot.2022.105066
Guarino, F., Improta, G., Triassi, M., Cicatelli, A., and Castiglione, S. (2020). Effects of zinc pollution and compost amendment on the root microbiome of a metal tolerant poplar clone. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01677
Han, Y., Liu, R., and Li, M. (2014). Effect of combination of AM fungi and PGPR mycorrhizal agents on growth and defense enzyme activities in cucumber under low temperature stress. Chin. vegetables 07, 35–39.
Ilangumaran, G. and Smith, D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: A Systems Biology Perspective. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01768
Jha, Y. and Subramanian, R. B. (2013). Paddy plants inoculated with PGPR show better growth physiology and nutrient content under saline conditions. Chil. J. Ag. Res 73, 213–219. doi: 10.4067/S0718-58392013000300002
Jing, D., Ma, H., Liu, F., Du, Z., Jia, H., Ma, B., et al. (2018). Effect of inoculation with inter-root facultative bacteria on microenvironmental characteristics of inter-root soil of alkali pontica under salt stress environment. Soil Fer. Sci. China, 34–39. doi: 10.11838/sfsc.20180406
Kai Wang, X. J., Xu, H., Qiu, S., and Zou, S. (2016). Integration of overexpression of key enzyme genes of purine metabolism pathway to increase adenosine cyclic phosphate production in Saccharomyces cerevisiae strains. Food Ferment. Ind. 42, 6. doi: 10.13995/j.cnki.11−1802/ts.201608005
Keren, G., Yehezkel, G., Satish, L., Adamov, Z., Barak, Z., Ben-Shabat, S., et al. (2024). Root-secreted nucleosides: signaling chemoattractants of rhizosphere bacteria. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1388384
Khanghahi, M. Y., Spagnuolo, M., Filannino, P., Minervini, F., and Crecchio, C. (2025). Genetic and ecological inheritance of plant growth-promoting rhizobacteria. Plant Soil. 509, 15–33. doi: 10.1007/s11104-024-06852-y
Li, H. (2000). Principles and techniques of plant physiology and biochemistry experiments (Beijing: Higher Education Press).
Li, Q., Zhang, B., Zhang, Y., Hu, J., and Dai, Q. (2025). Bacterial communities and tetracycline resistance genes in the soil-rice continuum upon tetracycline exposure. App. Soil Ecol. 205, 105762. doi: 10.1016/j.apsoil.2024.105762
Liu, F., Ma, H., Du, Z., and Ma, B. (2015). Physiological response of honeysuckle containerized seedlings to drought stress inoculated with inter-root growth-promoting bacteria. Ecol. Lett. 35, 7003–7010. doi: 10.5846/stxb201402180282
Lugtenberg, B. and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Ann. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Luo, J., Xun, D., Song, Q., Wang, J., Wang, Z., and Song, R. (2021). Effects of inter-root growth promoting bacteria on the growth and soil physicochemical indexes of 1-year-old camphor pine. J. Northeast For. Univ. 49, 110–116. doi: 10.13759/j.cnki.dlxb.2021.10.019
Powers, M. J. and Trent, M. S. (2018). PNAS Plus: Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. P. Nat. Acad. Sci. U.S.A. 115, E8518–E8527. doi: 10.1073/pnas.1806714115
Rilling, J. I., Acuna, J. J., Sadowsky, M. J., and Jorquera, M. A. (2018). Putative nitrogen-fixing bacteria associated with the rhizosphere and root endosphere of wheat plants grown in an andisol from southern Chile. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02710
Ruth, G. E., Joeke, P., Raaijmakers, J. M., and Irene, D. B. (2015). Diversity and activity of lysobacter species from disease suppressive soils. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01243
Sarathambal, C., Dinesh, R., Srinivasan, V., Sheeja, T. E., Jeeva, V., and Manzoor, M. (2022). Changes in bacterial diversity and composition in response to Co-inoculation of Arbuscular Mycorrhizae and Zinc-Solubilizing Bacteria in turmeric rhizosphere. Curr. Microbiol. 79, 3–9. doi: 10.1007/s00284-021-02682-8
Swiatczak, J., Kalwasinska, A., Szabó, A., and Brzezinska, M. (2023). 2RO45 inoculation alters the taxonomic structure and functioning of the canola rhizosphere microbial community. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1168907
Tahir, M., Mirza, M. S., Hameed, S., Dimitrov, M. R., and Smidt, H. (2015). Cultivation-Based and molecular assessment of bacterial diversity in the rhizosheath of wheat under different crop rotations. PloS One 10, e0130030. doi: 10.1371/journal.pone.0130030
Upadhyay, S. K. and Singh, D. P. (2015). Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 17, 288–293. doi: 10.1111/plb.12173
Wang, Y. (2017). Study on the uric acid-lowering effect of chicory extract modulating intestinal transporter protein ABCG2. Beijing Univ. Chin. Med.
Wang, X. (2024). Research on plant adaptation and planting technology based on saline and alkaline land management. Cott. Sci. 46, 155–157.
Wang, X. and Huang, J. (2015). Principles and techniques of plant physiological biochemical experiment.
Wang, N., Huang, D., Zhang, Y., Xu, W., and Feng, X. (2024). Patent status of key technologies for comprehensive management and utilization of saline and alkaline land. China Sci. Technol. Inf. 46, 31–34.
Wang, F., Niu, H., Xin, D., Long, Y., Wang, G., Liu, Z., et al. (2021). OsIAA18, an Aux/IAA transcription factor gene, is involved in salt and drought tolerance in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.738660
Wu, Z., Li, Y., and Gu, Z. (2018). Exploration of the functions of glnA, proB, and proA genes in the L-proline synthesis pathway of Bacillus subtilis. J. Microbiol. 58, 39–50. doi: 10.13343/j.cnki.wsxb.20170011
Xin, S., Wang, G., Gao, Y., and Zhao, J. (2011). Effectiveness of plant-promoting bacteria CS2, CC9, and charcoal soil additions on rice-planting in saline soils. Guangdong Agric. Sci. 38, 82–85. doi: 10.16768/j.issn.1004-874x.2011.07.041
Yang, Y., Chen, J., Chen, X., Jiang, Q., and Xie, S. (2021). Cyanobacterial bloom induces structural and functional succession of microbial communities in eutrophic lake sediments. Environ. pollut., 117157. doi: 10.1016/j.envpol.2021.117157
Yang, J., Yao, R., Wang, X., and Xie, W. (2016). Ecological management and ecological industry development model of saline-alkaline land in the river-loop plains. Ecol. Lett. 36, 7059–7063. doi: 10.5846/stxb201610182130
Yu, Z. (2025). Efficient management of saline soils through land reclamation and water conservation. Qual. Agric. Prod., 39–41.
Zhang, Z., Sun, J., Li, T., Shao, P., Ma, J., and Dong, K. (2023). Plants changed the response of bacterial community to the nitrogen and phosphorus addition ratio. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1168111
Keywords: PGPR, salt stress, rice seedlings, rhizobacterial community, root exudates
Citation: Wang J, Gao X, Yang M, Zhang Y and Chen Y (2025) Alleviative effects of plant growth-promoting rhizobacteria on salt-stressed rice seedlings: mechanisms mediated by rhizosphere microbiota and root exudates. Front. Plant Sci. 16:1661074. doi: 10.3389/fpls.2025.1661074
Received: 07 July 2025; Accepted: 22 September 2025;
Published: 08 October 2025.
Edited by:
Shixiao Yu, Sun Yat-Sen University, ChinaReviewed by:
Anukool Vaishnav, GLA University, IndiaLei Ren, Guangdong Ocean University, China
Ziming Ma, Max Planck Institute of Molecular Plant Physiology, Germany
Copyright © 2025 Wang, Gao, Yang, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinjun Wang, amluanVud2FuZ0B5enUuZWR1LmNu; Yinglong Chen, Y2hlbnlpbmdsb25nQHl6dS5lZHUuY24=
 Jinjun Wang
Jinjun Wang Xinru Gao
Xinru Gao Yang Zhang
Yang Zhang Yinglong Chen
Yinglong Chen