- State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China
Dendrobium nobile is a prized orchid species with both medicinal and ornamental values, known for accumulating flavonoids that contribute to its bioactivity and flower pigmentation. Chalcone isomerase (CHI), a key enzyme in the flavonoid biosynthetic pathway, catalyzes the conversion of chalcone to naringenin, thereby promoting the production of anthocyanins and other flavonoids. In this study, we present the first systematic analysis of the CHI gene family in D. nobile, classifying them into two subfamilies. Expression profiling revealed that DnCHI1, a type IV CHI, is highly expressed in petals, suggesting a potential role in anthocyanin metabolism. Heterologous transient overexpression assays showed that DnMYB90 significantly downregulated endogenous PeCHI expression and reduced anthocyanin accumulation, whereas DnCHI1 overexpression resulted in a 15-fold increase in anthocyanin content. Dual-luciferase reporter and yeast one-hybrid assays further confirmed that DnMYB90 acts as a transcriptional repressor of DnCHI1. These results provide new insights into the regulatory module in anthocyanin biosynthesis in D. nobile, highlighting the functional divergence of CHI genes and their interaction with MYB transcription factors.
1 Introduction
Dendrobium nobile Lindl. (Orchidaceae) is a perennial herbaceous species in the genus Dendrobium. The Chinese Pharmacopoeia records its rich content of bioactive compounds, including polysaccharides, alkaloids, and flavonoids, underpinning its pharmacological effects. Its vibrant floral coloration and distinctive morphology also make it an attractive ornamental species (Xu et al., 2022a) and an important parental line in Dendrobium hybrida breeding programs (Meng et al., 2025).
Flavonoids, a major class of plant secondary metabolites, are widely distributed across plant organs. In flowering plants, floral color, a key ornamental trait, is determined primarily by pigments such as carotenoids, flavonoids, and alkaloids (Qiu et al., 2023). Among these, anthocyanins, a subclass of flavonoids, are the predominant determinants of floral color diversity (Sun et al., 2015). The flavonoid biosynthetic pathway originates from the phenylpropanoid pathway, in which phenylalanine is sequentially converted to p-coumaroyl-CoA, a central intermediate that can enter either the flavonoid biosynthetic pathway or the monolignol biosynthetic pathway. Within the flavonoid pathway (Ge et al., 2023), chalcone synthase (CHS) and chalcone isomerase (CHI) catalyze the first two committed steps, generating essential precursors for downstream anthocyanin biosynthesis (Chao et al., 2021). In Hevea brasiliensis, for example, the overexpression of HbCHI2 significantly upregulates downstream genes such as anthocyanin synthase (ANS), flavonoid 3′-hydroxylase (F3′H), and leucoanthocyanidin reductase (LAR), thereby enhancing flavonoid accumulation (Wu et al., 2025a).
CHI is the second enzyme in the flavonoid biosynthetic pathway (Yin et al., 2019; Nan et al., 2022). It catalyzes the intramolecular cyclization of 2′,4,4′,6′-tetrahydroxychalcone (produced by CHS) into (2S)-flavanone (Kang et al., 2014). While this reaction can occur spontaneously under physiological conditions, CHI accelerates it by approximately 107-fold, greatly boosting flavonoid output in plant cells. Phylogenetic and functional analyses classify plant CHIs into four types (I–IV) (Luo et al., 2025). Types I and II exhibit true CHI activity, catalyzing chalcone to naringenin, which is essential for flavonoid biosynthesis and anthocyanin biosynthesis (Ni et al., 2020). Type I CHIs are widely distributed across vascular plants, including Aquilaria sinensis (Ding et al., 2021), bryophytes, ferns, and most angiosperms (Ni et al., 2020). Type II CHIs are largely confined to legumes, although some occur in basal land plants (Cheng et al., 2018). Type III and IV CHIs lack catalytic residues and therefore demonstrate no chalcone-cyclizing activity (Dastmalchi and Dhaubhadel, 2015). Type IV CHIs, also known as chalcone isomerase-like proteins (CHILs), instead regulate flavonoid biosynthesis (Zhao et al., 2020; Wolf-Saxon et al., 2023). CHILs are unique to land plants (Jiang et al., 2015) and remain among the least characterized components of the Arabidopsis flavonoid pathway. For instance, eggplant SmCHI2 is preferentially expressed in pigmented tissues and promotes anthocyanin accumulation (Zhou et al., 2023), while DcCHI4 from Dracaena cambodiana enhances flavonoid production when overexpressed in Nicotiana benthamiana (Zhu et al., 2021). Such findings highlight the regulatory importance of CHIL proteins.
In orchids, flavonoid biosynthesis is tightly controlled by transcription factors, particularly the MYB family, which modulates structural gene expression and thereby floral pigmentation. MYB proteins are classified into four groups based on the number and arrangement of MYB repeats (R) in their conserved domains: 1R-MYB, R2R3-MYB, R1R2R3-MYB, and 4R-MYB. Of these, R2R3-MYB proteins are predominant regulators of anthocyanin biosynthesis (Yan et al., 2021; Yang et al., 2022; Li et al., 2015). In Phalaenopsis-type Dendrobium, DhMYB2 and DhbHLH1 enhance petal anthocyanin accumulation by activating DhDFR and DhANS (Li et al., 2017). Similarly, the transient overexpression of R2R3-MYB genes RcPAP1 and RcPAP2 from Cattleya in Phalaenopsis induced purplish-red pigmentation in previously white tepals (Li et al., 2020). In Dendrobium officinale, DoMYB5 and DobHLH24 directly bind to the promoters of DoCHS and DoDFR, positively regulating anthocyanin biosynthesis (Yang et al., 2023). A recent genomic analysis of D. nobile identified 125 MYB genes, classified into 26 subgroups, with DnMYB90 assigned to subgroup S4, suggesting a potential role in flavonoid regulation (Wu et al., 2024). In addition, the systematic characterization of the DnCHI gene family revealed two type IV CHIs (DnCHI1 and DnCHI3). Subsequent qRT-PCR analysis revealed that DnCHI1 expression was the highest in petals. Under dual-luciferase reporter and yeast one-hybrid assays, we confirmed that DnMYB90 binds to the DnCHI1 promoter, implicating it in the transcriptional regulation of anthocyanin metabolism. These findings advance our understanding of the regulatory network governing floral pigmentation in D. nobile and provide a framework for further research into flavonoid biosynthesis in orchids.
2 Materials and methods
2.1 Identification of CHI gene family members from D. nobile
The genome assembly, protein sequences, and annotation files of D. nobile were obtained from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/, accessed on April 7, 2025) under accession number GCA_022539455.1. CHI genes in D. nobile were identified by performing a BlastP search against the D. nobile protein database using characterized Arabidopsis thaliana CHI amino acid sequences from the TAIR database (https://www.arabidopsis.org/) as queries (Zhang et al., 2017). Candidate CHI genes were selected with an e-value cutoff of 1e−5. The CHI family-specific Pfam accession (PF02431) was retrieved from the Pfam database (http://pfam.xfam.org/), and its HMM profile was downloaded (Potter et al., 2018). Using the Simple HMM Search tool in TBtools, the D. nobile genome was systematically screened for potential CHI family members. Candidate sequences were validated via protein domain analysis using NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART (http://smart.embl-heidelberg.de), and those lacking the conserved CHI domain were excluded. Six CHI genes were ultimately identified and named DnCHI1–DnCHI6 according to their chromosomal locations. The physicochemical properties of each protein were estimated using ProtParam (http://web.expasy.org/protparam/), and their subcellular localization was predicted using Plant-mPloc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (Chou and Shen, 2010; Artimo et al., 2012).
2.2 Phylogenetic analysis of the DnCHI genes
A phylogenetic tree was constructed using MEGA 11.0 with the neighbor-joining (NJ) algorithm, analyzing CHI protein sequences from A. thaliana, Oryza sativa, Glycine max, and D. nobile to investigate the evolutionary relationships among CHI genes. Bootstrap analysis was conducted using 1,000 replicates under default parameters. The resulting tree was exported in Newick format and visualized using the iTOL online tool (https://itol.embl.de/).
2.3 Conserved motif and gene structure analysis
The conserved motifs of the DnCHI gene family were analyzed using the MEME suite (http://meme-suite.org/) with the following parameters: Maximum motifs = 10 and optimum motif width = 6–50 amino acids (Bailey et al., 2009). Gene structures (exon–intron organization) were determined by aligning coding sequences (CDS) with their corresponding genomic DNA sequences in TBtools. Motif distribution and gene structure were visualized using TBtools visualization modules to enable comparative analysis among DnCHI members.
2.4 Analysis of cis-active elements of the DnCHI genes
The promoter regions (2,000 bp upstream of the ATG start codon) of all DnCHI gene family members were extracted using TBtools and analyzed for cis-acting elements using the PlantCARE online platform (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The results were visualized using TBtools.
2.5 Plant materials and sample preparation
Mature D. nobile plants were collected from Chishui City, Guizhou, China, and cultivated under natural light conditions in the Chinese Academy of Forestry greenhouse (Beijing, China). Root, stem, leaf, and flower tissues were harvested at full bloom; immediately frozen in liquid nitrogen for 1 hour; and stored at −80°C. Three biological replicates were prepared for each tissue type.
2.6 Tissue-specific expression analysis
The frozen tissues of D. nobile were ground in liquid nitrogen, and total RNA was extracted using the DP441 RNA Extraction Kit (Tiangen, Beijing, China). RNA concentration and purity were measured using a NanoDrop™ One/OneC spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and integrity was assessed using agarose gel electrophoresis. First-strand cDNA was synthesized using the One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China). Gene-specific primers (Supplementary Table 1) were designed using NCBI Primer-BLAST and validated using the TBtools Primer Check tool, with all primers synthesized by Sangon Biotech (Shanghai, China). The DnUBQ1 gene was used as an internal reference based on a prior evaluation of housekeeping genes in D. nobile (Guo et al., 2012). qRT-PCR reactions were prepared according to the manufacturer’s protocol of TB Green® Premix Ex Taq™ (Takara, Tokyo, Japan) and run on a LightCycler 480 System (Roche, Basel, Switzerland). Relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Each sample analysis included three biological replicates and four technical replicates.
2.7 Cloning of DnCHI1 and DnMYB90
The reference sequences of DnCHI1 (LOC110099164) and DnMYB90 (KAI0514214.1) were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov). Gene-specific primers were designed using SnapGene 8.0 (Supplementary Table 1). Target fragments were amplified from petal cDNA templates using 2× TransStart® FastPfu Fly PCR SuperMix (-dye) (TransGen Biotech, Beijing, China), and amplification products were verified using agarose gel electrophoresis. Positive bands were excised and purified using a gel extraction kit (GenBetter, Beijing, USA) and subsequently ligated into the pMD19-T cloning vector (Takara, Japan). The recombinant plasmids were transformed into Escherichia coli DH5α competent cells (Huayueyang, Beijing, China) using the freeze–thaw method. Positive clones were screened using colony PCR and confirmed by sequencing (Sangon Biotech, Shanghai, China).
2.8 Transient heterologous expression of DnCHI1 and DnMYB90 in Phalaenopsis
Given the lack of a reliable transient overexpression system for D. nobile, Phalaenopsis sogo Yukidian ‘V3’, which has a well-established transformation system, was used for heterologous transient expression (Li et al., 2020). The binary vector pCAMBIA1302 was linearized with NcoI and purified. Using SnapGene 8.0, primer pairs for vector construction (Supplementary Table 1) were used to construct recombinant plasmids p1302-DnCHI1 and p1302-DnMYB90, which were transformed into E. coli DH5α, and single clones were selected for sequencing at Sangon Biotech (Shanghai, China). The sequencing-verified plasmids were introduced into Agrobacterium tumefaciens GV3101 (Huayueyang, Beijing, China). Positive colonies were selected using PCR and cultured in Luria-Bertani (LB) medium containing 20 mg/L rifampicin and 50 mg/L kanamycin. Bacterial cells were collected by centrifugation at optimal density and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 µM acetosyringone). The suspensions were syringe-infiltrated into P. sogo Yukidian ‘V3’ petals. Following visible phenotype development, the relative expression levels of ABP genes were determined via qRT-PCR using published primers (Supplementary Table 1), with PeActin as the reference gene. All measurements included six biological replicates, each with four technical replicates. Anthocyanins were extracted from 0.5 g of petal tissue using 10 mL of acidic methanol containing 1% (v/v) HCl and incubated overnight at 4°C. After centrifugation, the supernatant was collected, and absorbance was measured at 530 and 657 nm using a spectrophotometer. Anthocyanin content (AT) was calculated using the formula AT (U/g) = A530 − 0.25 × A657 (Zhou et al., 2024). All assays were performed in triplicate.
2.9 Subcellular localization of DnMYB90
The coding sequence of DnMYB90 (without a stop codon) was inserted into the GFP-tagged pCAMBIA1302 vector using seamless cloning (primers listed in Supplementary Table 1) to generate the 35S::DnMYB90-GFP construct (Jiang et al., 2023). Both the empty vector and 35S::DnMYB90-GFP were transformed into the A. tumefaciens strain GV3101. Positive transformants were cultured, resuspended, and infiltrated into the abaxial side of N. benthamiana leaves. After 48 hours of incubation, fluorescence signals were observed using a confocal laser scanning microscope (Zeiss LSM 880 Meta, Jena, Germany). Three independent biological replicates were performed for each construct.
2.10 Dual-luciferase assay
Genomic DNA was extracted using a rapid DNA extraction kit (Tiangen, Beijing, China). The DnCHI1 promoter was amplified by nested PCR using genomic DNA and inserted into the pGreenII 0800-LUC reporter vector via homologous recombination. The DnMYB90 coding sequence was cloned into the pGreenII 62-SK effector vector (primer sequences in Supplementary Table 1). Recombinant vectors were transformed into A. tumefaciens GV3101 (pSoup) and verified by bacterial PCR. The bacterial suspensions were co-infiltrated into different sites of the same N. benthamiana leaf to ensure internal comparability. After 48 hours under low-light conditions, LUC/REN ratios were quantified using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) on a multimode plate reader, and fluorescence activity was visualized using the NightSHADE LB 985 imaging system (Berthold, Bad Wildbad, Germany). All assays included three biological replicates, each with three technical replicates.
2.11 Yeast one-hybrid assay
The DnCHI1 promoter sequence was truncated into 150–350-bp fragments containing specific binding sites to avoid potential auto-activation. The truncated promoter fragments and the DnMYB90 coding sequence were inserted into the pLacZi-2μ bait vector and pB42AD prey vector, respectively, via seamless cloning (primers listed in Supplementary Table 1). To preclude autoactivation and ensure experimental rigor, two control groups were established: Control 1 (DnCHI1pro::LacZi + Empty pB42AD) and Control 2 (empty pLacZi + AD-DnMYB90). Two independent constructs were generated for each combination and co-transformed into Saccharomyces cerevisiae strain EGY48 and plated on SD dropout medium (SD/-Trp/-Ura). After 2–3 days of inverted incubation at 28°C, single colonies were diluted, serially diluted, and spotted onto SD/-Trp/-Ura/Gal/Raf/X-Gal indicator plates. Both colony growth and blue coloration were subsequently recorded.
3 Results
3.1 Identification and physicochemical property analysis of the DnCHI genes
Six CHI genes were systematically identified from the D. nobile genome and designated as DnCHI1 to DnCHI6 according to their chromosomal locations (Figure 1A). The CHI gene family members displayed considerable variation in physicochemical properties, with protein lengths ranging from 209 to 425 amino acids (DnCHI5 being the longest and DnCHI3 the shortest), molecular weights varying between 23,682.12 and 47,187.5 Da, and theoretical isoelectric points (pI) from 4.99 to 9.28. Instability indices varied from 35.1 to 45.23, aliphatic indices from 92.8 to 98.2, and the grand average of hydropathicity values from −0.161 to 0.047, with four members predicted as hydrophilic proteins. Subcellular localization predictions indicated predominant chloroplast localization for most DnCHI proteins, additional nuclear localization for DnCHI2 and DnCHI5, and unique membrane localization for DnCHI6 (Supplementary Table 2).
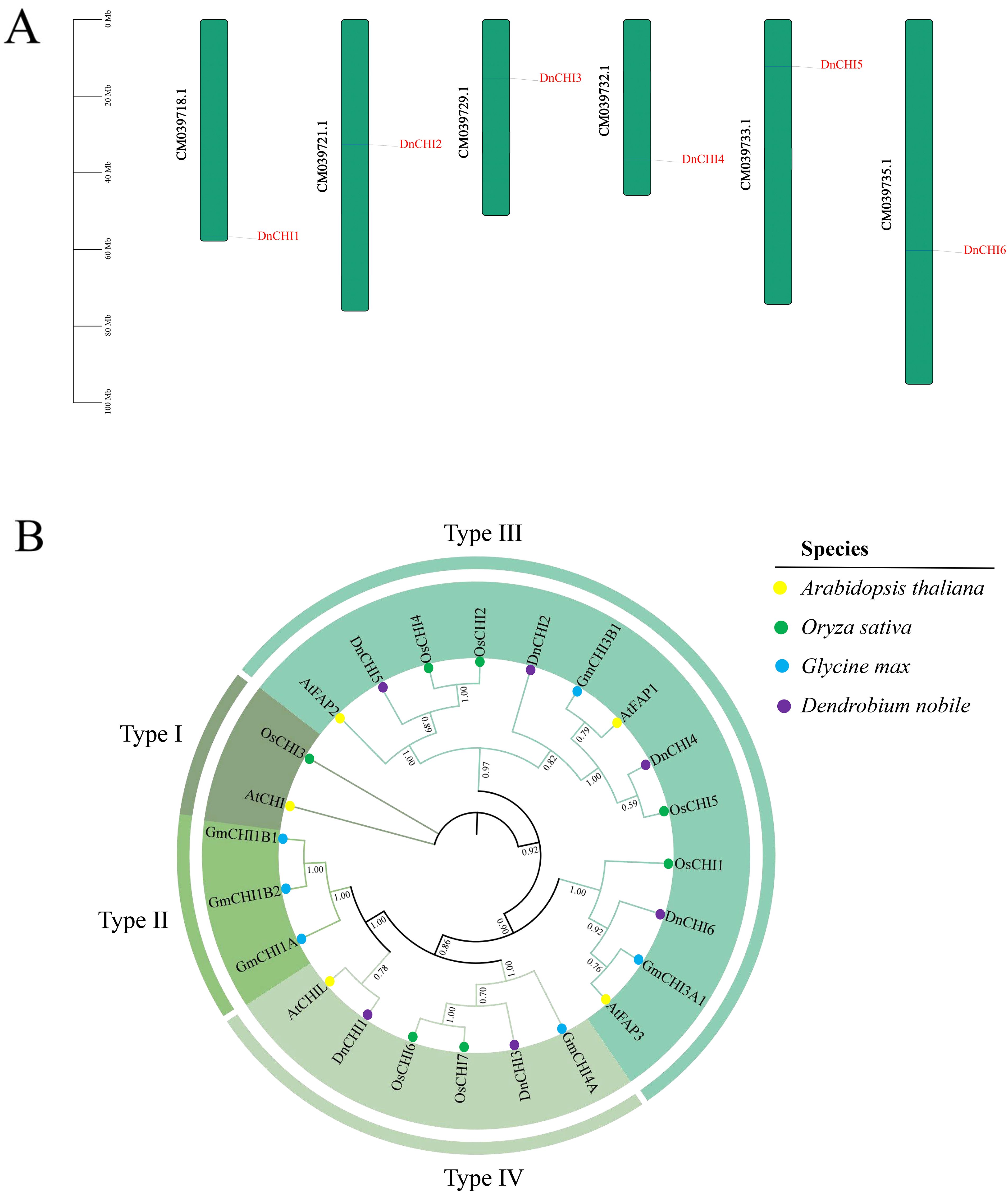
Figure 1. Chromosomal localization and phylogenetic analysis of DnCHI genes. (A) Distribution of DnCHI genes from Dendrobium nobile across six chromosomes. The scale on the left indicates chromosome length (in megabases, Mb). (B) Phylogenetic analysis of CHI genes among Arabidopsis thaliana (yellow circle), Oryza sativa (green circle), Glycine max (blue circle), and D. nobile (purple circle). The unrooted phylogenetic tree was constructed using the neighbor-joining (NJ) method in the MEGA X software with 1,000 bootstrap replicates.
3.2 Phylogenetic analysis of the DnCHI genes
A phylogenetic tree was constructed using five A. thaliana, seven O. sativa, six G. max, and six DnCHI genes, revealing four distinct subfamilies (Types I–IV), with Type III containing the highest number of members (Figure 1B). Four DnCHI genes (DnCHI2, DnCHI4, DnCHI5, and DnCHI6) clustered within Type III, which, according to previous studies, can be further subdivided into three subgroups (FPA1, FPA2, and FPA3) involved in fatty acid metabolism. The Type IV subfamily included two DnCHI genes (DnCHI1 and DnCHI3) that may encode key enzymes for flavonoid biosynthesis (Wang et al., 2022). No D. nobile CHI genes were classified within Type I or Type II subfamilies.
3.3 Conserved motif and gene structure analysis
MEME-based analysis identified 10 conserved motifs (motifs 1–10) with copy numbers ranging from 2 to 9 per protein (Figure 2A). Motif 3 was universally present across all DnCHI, while motif 4 was exclusively found in Type III subfamily proteins (Figure 2C). The Type III subfamily exhibited significantly higher motif abundance compared with the two Type IV members, accompanied by distinct motif compositions and exon positional arrangements between subfamilies, suggesting higher motif abundance compared with Type IV members, accompanied by distinct motif compositions and exon positional arrangements between subfamilies, suggesting greater structural conservation within Type III and potential functional divergence during evolution. Within the Type III, despite quantitative differences in motif numbers, similar compositional patterns and spatial distributions were observed, indicating conserved functional roles among these evolutionarily related proteins.
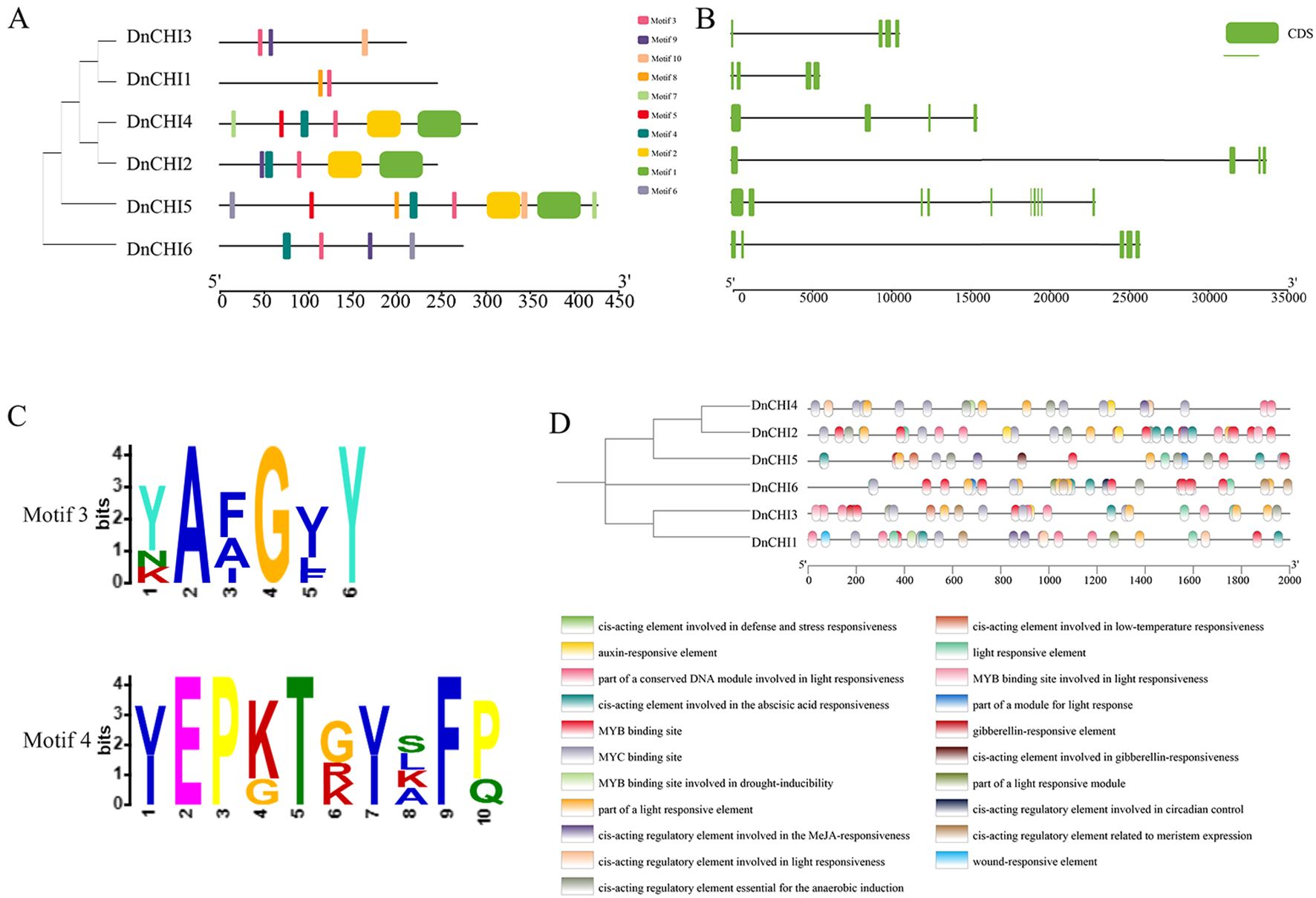
Figure 2. Conserved motif, gene structure, conserved domains, and cis-acting regulatory elements of the DnCHI genes. (A) Ten conserved motifs in the DnCHI genes are represented by different colored squares. (B) Exon and intron structures of DnCHI genes in Dendrobium nobile. Exons are shown as green squares and introns as gray lines. (C) Conserved motifs motif3 and motif4. (D) Promoter element sites, with colored rectangles representing distinct cis-acting regulatory elements.
During gene family evolution, introns and exons play distinct roles in gene expression and regulation. The six DnCHI family members in D. nobile contained 4–10 exons, with DnCHI5 possessing the highest number (10 exons). Four genes (DnCHI3, DnCHI1, DnCHI4, and DnCHI2) each contain four exons, while DnCHI6 has five exons (Figure 2B). Genes within the same subgroup generally displayed similar exon numbers, with intron positions remaining relatively conserved throughout evolution.
3.4 cis-regulatory element analysis of DnCHI promoters
cis-Acting elements within gene promoter regions are key regulators of transcriptional activity and participate extensively in diverse biological processes (Wittkopp and Kalay, 2012). An analysis of the promoter regions of the DnCHI genes revealed cis-acting elements grouped into four major functional categories: regulation of plant growth and development, hormone responsiveness, stress responses, and transcription factor binding (Figure 2D). Most DnCHI promoters contained elements responsive to abscisic acid, auxin, methyl jasmonate, gibberellin, and light signaling, as well as binding sites for MYB and MYC transcription factors. A notable exception was DnCHI4, whose promoter lacked abscisic acid-responsive elements and MYB binding sites, which were conserved in all other members.
3.5 Tissue-specific expression analysis of DnCHI1 and DnCHI3 genes
qRT-PCR analysis revealed distinct expression profiles of DnCHI1 and DnCHI3 across roots, stems, leaves, and flowers of D. nobile (Figure 3). DnCHI1 showed the highest transcript accumulation in petals, followed by roots, with minimal expression in stems and leaves. In contrast, DnCHI3 exhibited predominant expression in leaves. The distinct tissue-specific expression patterns, together with a positive correlation between transcript abundance and secondary metabolite accumulation, suggest the potential involvement of type IV DnCHI1 in floral pigment biosynthesis via secondary metabolic pathways.
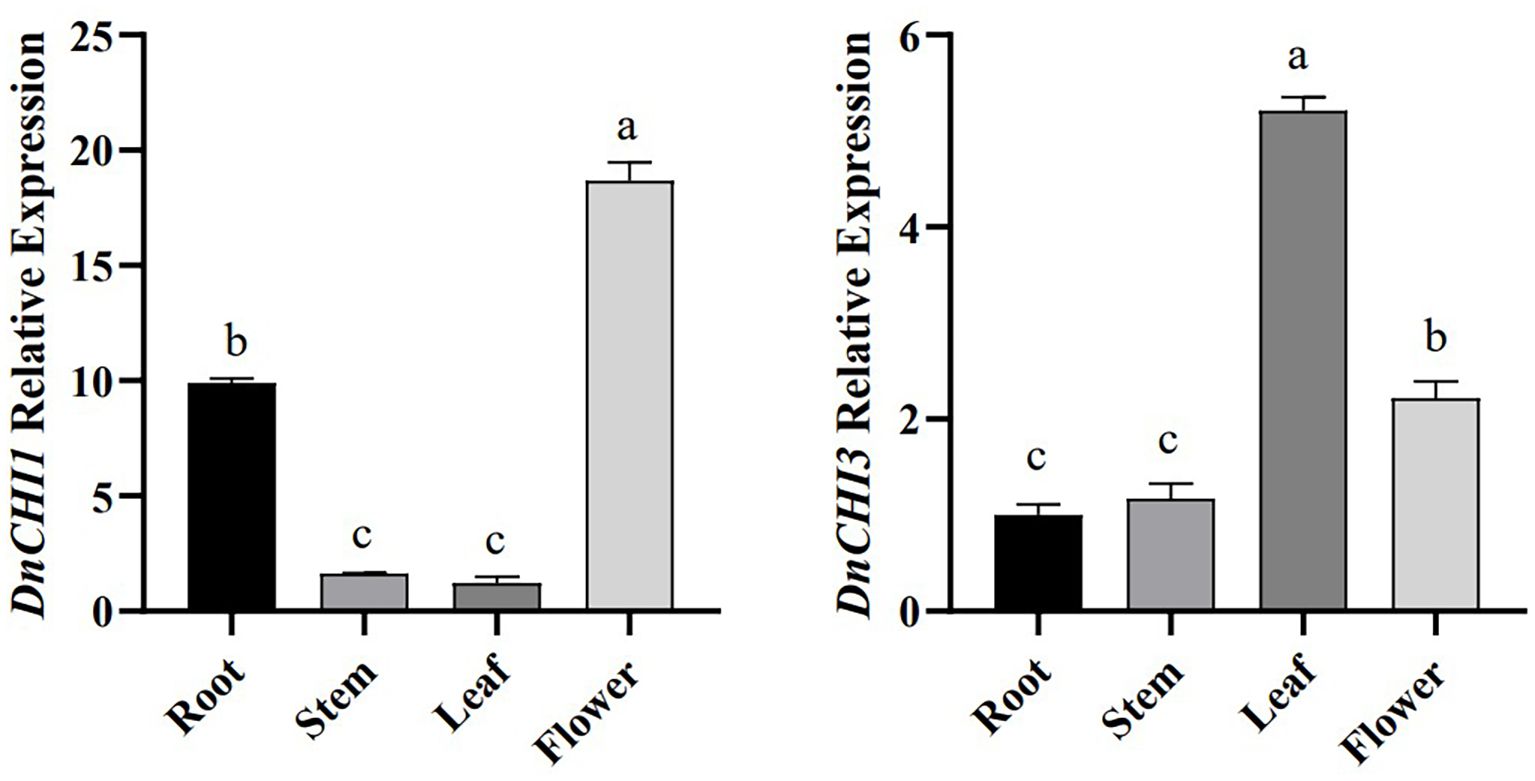
Figure 3. Relative expression levels of DnCHI1 and DnCHI3 in different tissues of Dendrobium nobile. Different lowercase letters indicate significant differences (p < 0.05).
3.6 Functional analysis of DnCHI1 and DnMYB90 through heterologous overexpression
The heterologous overexpression of DnCHI1 and DnMYB90 in Phalaenopsis petals induced distinct phenotypic changes. DnCHI1-overexpressing lines exhibited significant yellow pigmentation relative to controls, whereas DnMYB90-overexpressing plants displayed coloration comparable to empty-vector (pCAMBIA1302) controls (Figure 4A). Anthocyanin quantification revealed substantially higher pigment accumulation in DnCHI1-transformed petals compared with wild type, whereas DnMYB90 overexpression resulted in moderate but significantly lower anthocyanin increases (Figure 4B).
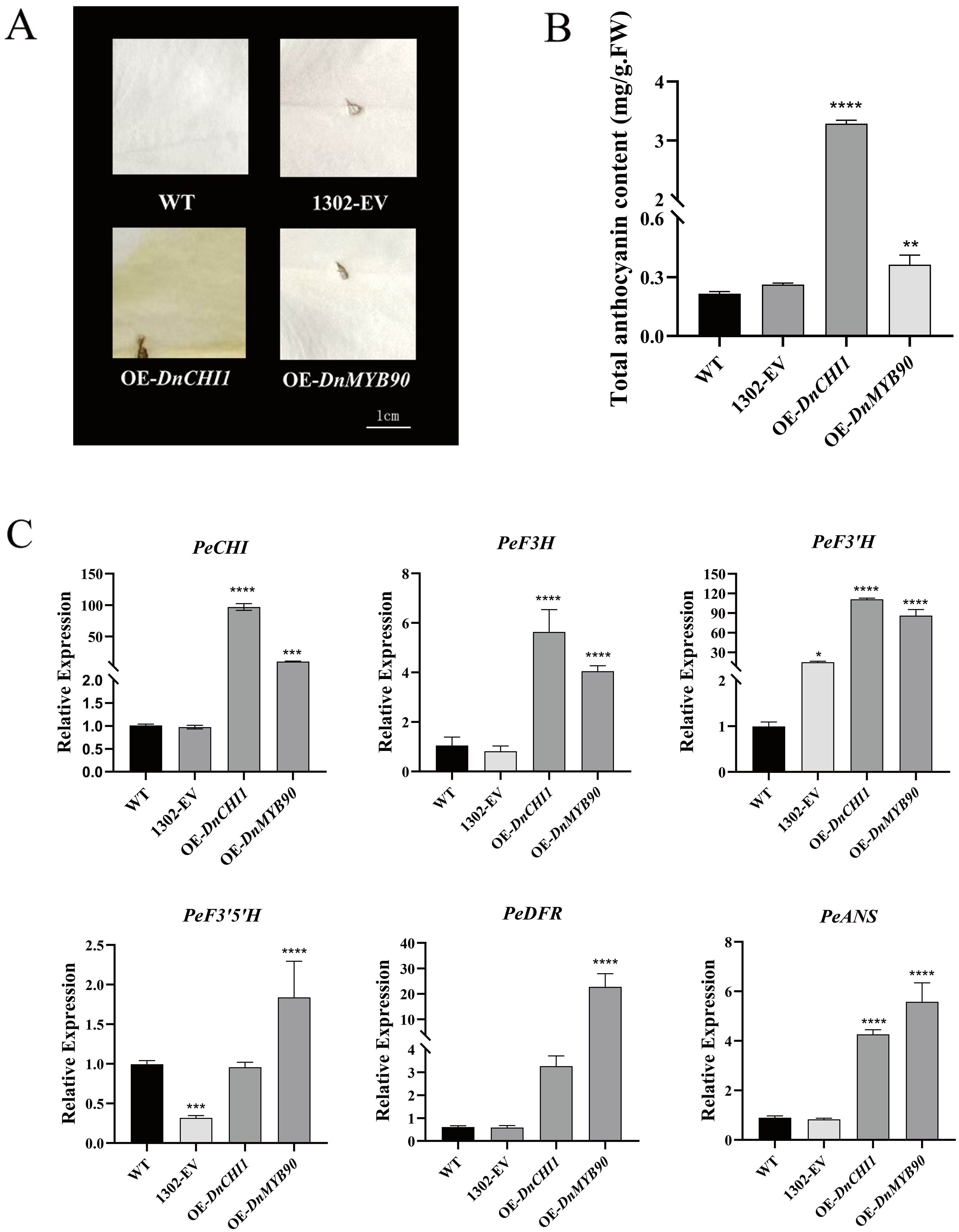
Figure 4. Transient overexpression of DnCHI1 and DnMYB90 in Phalaenopsis petals. (A) Flower phenotypes. WT, wild-type; 1302-EV, empty-vector control; OE-DnCHI1 and OE-DnMYB90, overexpression lines. (B) Total anthocyanin content in transgenic lines. (C) Expression levels of anthocyanin biosynthesis-related genes. Error bars represent SEM of six biological replicates. Asterisks indicate statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Scale bar = 1.0 cm.
qRT-PCR analysis confirmed effective transgene expression, with wild-type and empty-vector controls showing comparable expression levels. DnCHI1 overexpression markedly upregulated flavonoid biosynthesis genes (PeCHI, PeF3H, PeF3′H, and PeANS), whereas DnMYB90 enhanced PeF3′H, PeF3′5′H, and PeANS expression (111-fold and 85-fold increases in PeF3′H for DnCHI1 and DnMYB90 overexpressors, respectively), despite lower PeCHI expression in DnMYB90 transformations (Figure 4C). Both transgenic systems consistently upregulated DnANS expression.
3.7 Nuclear localization of DnMYB90
Subcellular localization analysis provided insights into the functional properties of DnMYB90. In N. benthamiana leaf cells, GFP-tagged empty vectors exhibited fluorescence in both the nucleus and cytoplasm, whereas the 35S::GFP-DnMYB90 fusion protein localized exclusively to the nucleus. These findings indicate that DnMYB90 functions primarily within the nuclear compartment (Figure 5).
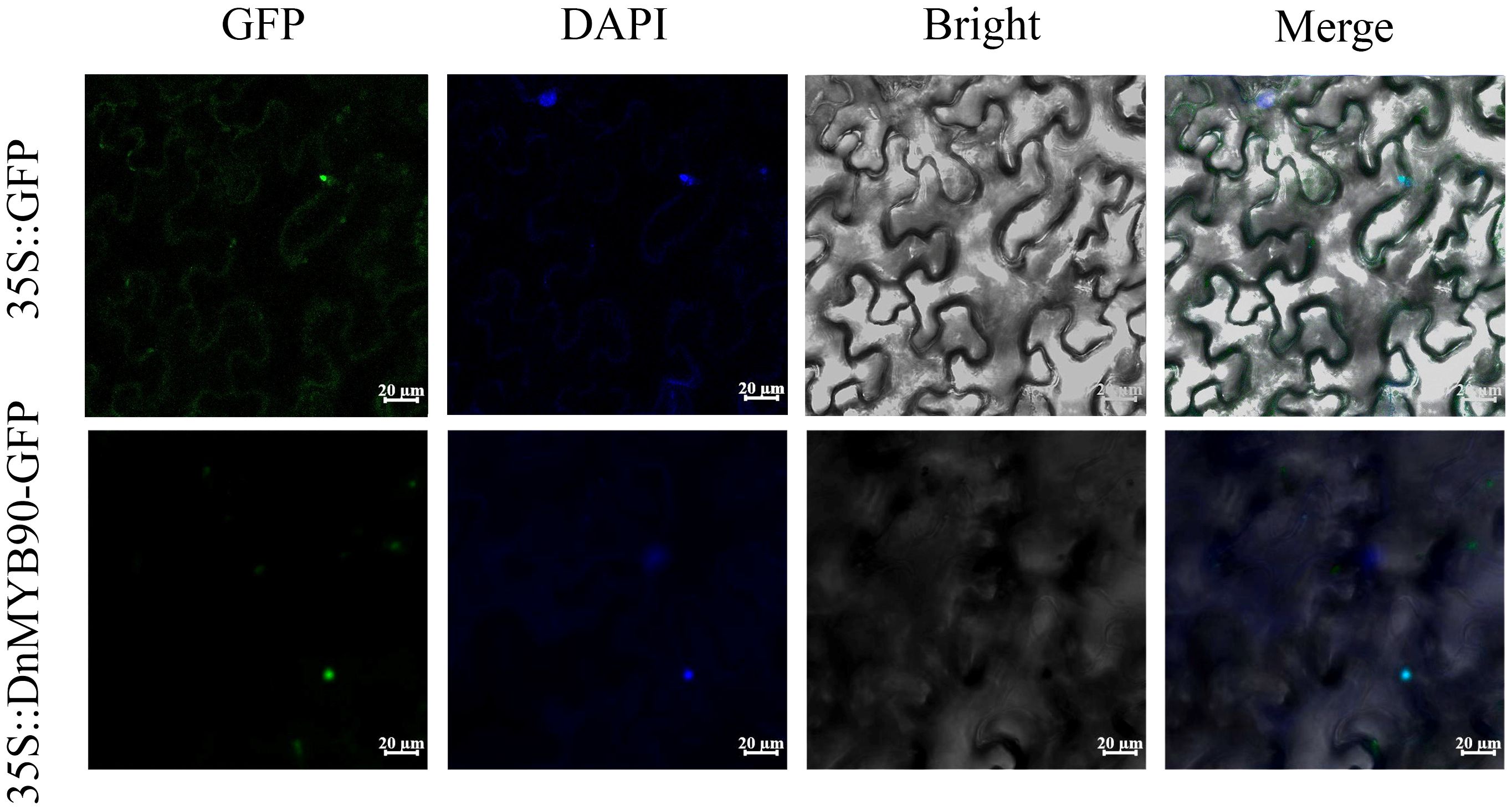
Figure 5. Subcellular localization of DnMYB90. GFP, green fluorescent protein fluorescence; DAPI, blue nucleus fluorescence; Merge: the superposition of green and blue fields. Bar = 20 μm.
3.8 DnMYB90 bound and inhibited the DnCHI1 promoter
The bioinformatics analysis of the DnCHI1 promoter sequence (Figure 2D) identified conserved MYB transcription factor binding sites. Yeast one-hybrid assays confirmed a direct interaction between DnMYB90 and the DnCHI1 promoter region (Figure 6A). Dual-luciferase reporter assays (Figures 6B–D) demonstrated that DnMYB90 significantly repressed DnCHI1 promoter activity, thereby establishing a regulatory mechanism in which DnMYB90 binds to and negatively regulates DnCHI1.
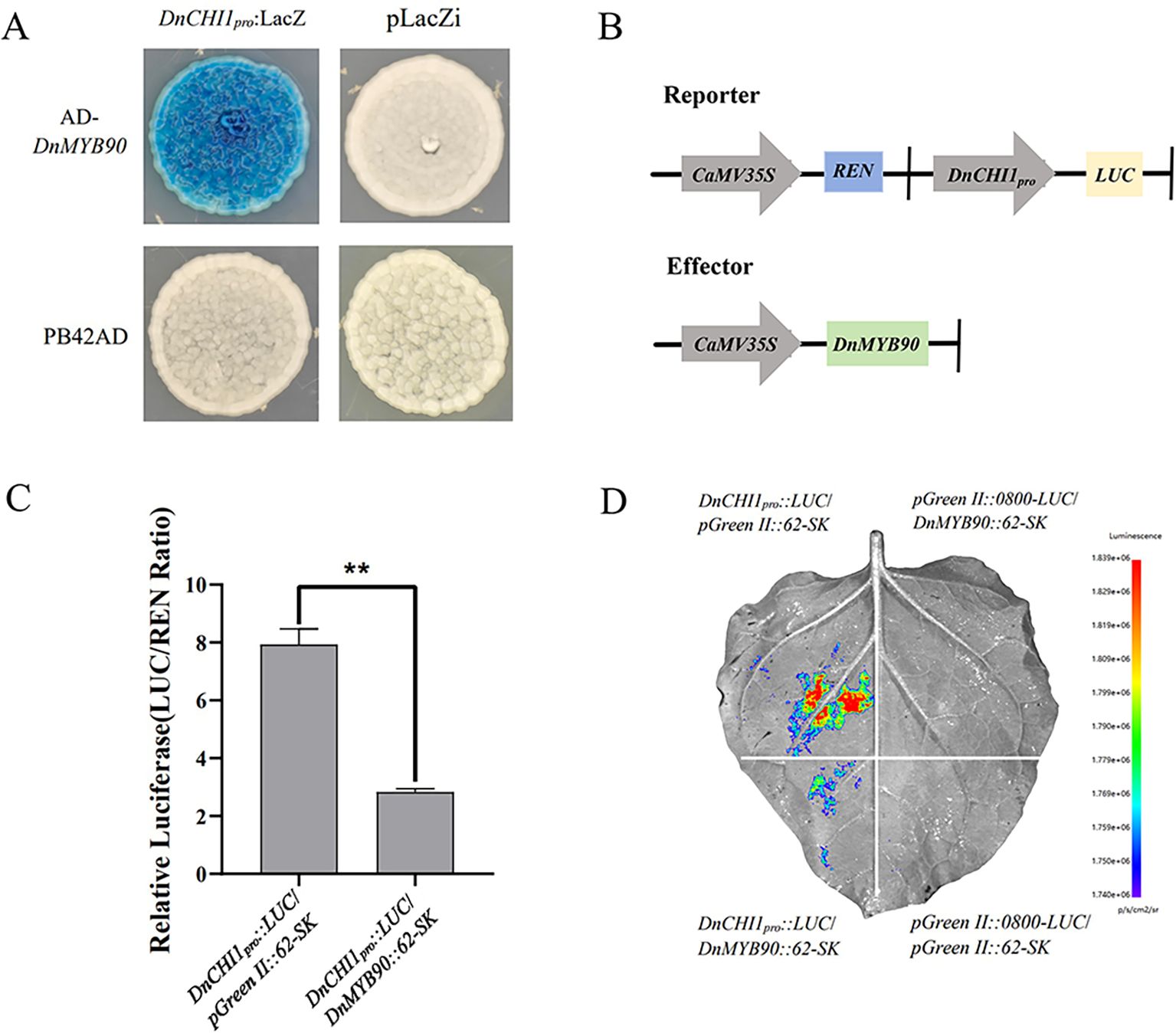
Figure 6. Regulatory effect of DnMYB90 on DnCHI1. (A) Yeast one-hybrid analyses of the interaction between DnMYB90 and the promoter of DnCHI1. (B) Schematic of reporter and effector constructs. (C) Dual-luciferase reporter assay showing repression of DnCHI1 promoter activity by DnMYB90. Data represent mean ± SE (n = 3). Statistical significance was determined by Student’s t-test (**p < 0.01). (D) Representative image of a Nicotiana benthamiana leaf 3 days after co-infiltration.
4 Discussion
CHI is a key enzymatic regulator in the flavonoid biosynthetic pathway, influencing both the rate and yield of flavonoid production in plants. In this study, six DnCHI genes with putative roles in flavonoid biosynthesis were identified in D. nobile. A classification system for G. max CHI genes (Type I–IV), first established by Dastmalchi and Dhaubhadel (2015), has since been applied to model plants such as A. thaliana, in which AtCHI represents Type I, AtFAP1/2/3 belong to Type III, and AtCHIL is a Type IV gene (Ngaki et al., 2012). Subsequent studies have expanded CHI gene characterization across diverse species. For example, Wu et al. identified five IbCHI genes in Ipomoea batatas (Wu et al., 2025b) and reported 33 CHI family members across four Gossypium species, with expression patterns responsive to Fusarium wilt race 7 infection (Zu et al., 2019). In Allium fistulosum, Xu et al. (2022) demonstrated that Type IV AfCHIL enhances naringenin synthesis efficiency via functional interaction with AfCHS (Xu et al., 2022b). Phylogenetic analysis in this present study classified the six identified DnCHI genes in D. nobile into two distinct subfamilies: Type III (DnCHI4/5/6) and Type IV (DnCHI1/3), with no Type I or II members detected. Functional divergence was evident among subfamilies, with Type II members occurring exclusively in leguminous plants and Type III members typically associated with fatty acid biosynthesis. Although Type I CHIs functionally overlap with Type IV CHIs, they generally exhibit lower catalytic efficiency. In contrast, Type IV CHIs often act as enhancer-flavonoid producers (EFPs), promoting flavonoid biosynthesis (Luo et al., 2025; Ngaki et al., 2012). The absence of Type I and Type II CHI genes in D. nobile may reflect limitations in the current genome assembly or annotation; however, it more plausibly represents a distinct evolutionary trajectory unique to this species. The conserved structural features of these DnCHI genes suggest functional conservation with established orthologs, although experimental validation is required to confirm these predicted roles.
Distinct expression profiles were observed for DnCHI1 and DnCHI3: DnCHI1 was predominantly expressed in petals, while DnCHI3 showed higher transcript abundance in leaves. Similar spatial expression trends have been documented in other species, such as root-preferential CHI expression in Scutellaria baicalensis (Lam et al., 2021) and the leaf-dominant expression of Camellia sinensis CsF3′H (Wang et al., 2017). Given the established correlation between flavonoid content and biosynthetic gene activity, the findings suggest that DnCHI1 is a strong candidate gene involved in petal-specific metabolic processes.
Flavonoids represent one of the most important classes of plant secondary metabolites, and CHI expression has been shown to enhance flavonoid content in multiple systems (Yin et al., 2019). The heterologous expression of onion CHI in Del/Ros1-overexpressing tomatoes substantially increased anthocyanin content in both peel and pulp (Lim and Li, 2017), and the overexpression of CnCHI4 from camellia markedly promoted flavonoid biosynthesis in transgenic tobacco and camellia plants (Yu et al., 2022). In this study, a qRT-PCR analysis of DnCHI1-overexpressing and DnMYB90-overexpressing lines revealed markedly lower PeCHI expression in OE-DnMYB90 compared with OE-DnCHI1, whereas downstream ABP genes (PeF3′5′H, PeDFR, and PeANS) exhibited higher expression in OE-DnMYB90 plants. Results showed that OE-DnMYB90 plants accumulated less anthocyanin than OE-DnCHI1 plants. This apparent paradox suggests that the negative effect of the DnMYB90-mediated transcriptional repression of the upstream key gene DnCHI1 may outweigh the positive effect of its partial upregulation of downstream genes. Supporting this view, Kriangphan et al. (2015) proposed that the low color intensity observed in the pink hybrid orchid D. ‘Sirinclassic’ can be attributed to the reduced expression of upstream ABP genes CHI1 and CHI2, resulting in limited naringenin production and consequently lower pigment accumulation and color intensity. Furthermore, the yellow pigmentation observed in OE-DnCHI1 plants suggests that the overexpression of DnCHI1 may promote flavanol accumulation, potentially enhancing the species’ medicinal value. Future studies could use Liquid Chromatography-Mass Spectrometry (LC–MS) to quantify metabolite levels in both OE-DnMYB90 and OE-DnCHI1 plants to validate these hypotheses. The preliminary evidence from yeast one-hybrid and dual-luciferase assays supports the hypothesis that DnMYB90 directly represses DnCHI1. However, we cannot exclude the possibility that DnMYB90 also directly or indirectly regulates other genes in the anthocyanin biosynthetic pathway (Dubos et al., 2010), with these multifaceted regulations collectively shaping the observed phenotype.
The R2R3-MYB subfamily, closely associated with anthocyanin regulation, has been well characterized in orchids, including Cymbidium goeringii (104 members), Cymbidium faberi (102 members), D. officinale (101 members), and Phalaenopsis equestris (99 members) (Wang et al., 2018; Fan et al., 2020; Ke et al., 2021; Chen et al., 2022). SG4-clade MYB transcription factors in particular often act as repressors of the ABP via the downregulation of key biosynthetic genes (Chen et al., 2022). Based on a genome-wide phylogenetic and conserved motif analysis of the R2R3-MYB family in D. nobile, Wu et al. (2024) classified DnMYB90 within this subgroup, suggesting that it may function analogously as a repressor. In A. thaliana, R2R3-MYBs such as MYB11 and MYB12 have been shown to activate CHI gene expression (Pandey et al., 2015; Liu et al., 2015), and the overexpression of EsAN2 in tobacco upregulated the expression of key flavonoid biosynthetic genes, including CHS, CHI, and ANS (Huang et al., 2016). However, in D. nobile, the regulatory mechanisms of SG4-clade R2R3-MYBs on CHI expression remain poorly understood, and no prior study has provided direct evidence of MYB–type IV CHI promoter interaction. This study provides preliminary evidence through dual-luciferase and yeast one-hybrid assays, indicating that DnMYB90 directly binds the DnCHI1 promoter and represses its transcription in heterologous systems. These findings offer new insight into the regulation of anthocyanin biosynthesis in D. nobile. Nevertheless, given the complexity of the ABP regulatory network, DnMYB90-mediated repression of DnCHI1 alone is unlikely to represent the sole control point. Further studies are needed to evaluate its broader effects on ABP genes, as well as to functionally characterize additional SG4-clade DnMYBs to determine whether such repressive regulation is a conserved feature in this orchid species.
5 Conclusions
Six CHI gene family members were identified in the D. nobile genome, which are clustered into two distinct phylogenetic subfamilies. Expression profiling revealed the predominant accumulation of type IV DnCHI1 transcripts in petals, indicating a potential key role in anthocyanin metabolism. Heterologous transient overexpression in Phalaenopsis demonstrated that DnMYB90 overexpression led to significantly lower PeCHI expression and reduced anthocyanin accumulation compared with DnCHI1 transformants. Dual-luciferase reporter and yeast one-hybrid experiments confirmed that DnMYB90 acts as a transcriptional repressor of DnCHI1, providing new mechanistic insights into the regulation of anthocyanin biosynthesis in orchids. These findings establish a framework for the further elucidation of the complex transcriptional networks governing floral pigmentation. To further elucidate the mechanistic role of DnMYB90, subsequent studies could employ electrophoretic mobility shift assays (EMSAs) to quantify its binding affinity to the DnCHI1 promoter, together with chromatin immunoprecipitation (ChIP) assays to comprehensively identify target genes involved in anthocyanin biosynthesis. In addition, yeast two-hybrid assays may be applied to screen for transcription factors that interact with DnMYB90 and potentially co-regulate anthocyanin biosynthesis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
L-LC: Software, Writing – original draft, Visualization, Methodology, Investigation, Validation, Data curation. Y-XZ: Validation, Investigation, Writing – original draft. L-RW: Validation, Writing – original draft. LL: Software, Writing – review & editing. Z-JL: Resources, Writing – review & editing, Supervision, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by Germplasm Innovation of Excellent Forage and Breeding of New Varieties (2024RCYJ04004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1687738/full#supplementary-material
References
Artimo, P., Jonnalagedda, M., Arnold, M., Baratin, D., Csardi, C., Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, 597–603. doi: 10.1093/nar/gks400
Bailey, T. L., Mikael, B., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Chao, N., Wang, R., Hou, C., Yu, T., Miao, K., Cao, F. Y., et al. (2021). Functional characterization of two chalcone isomerase (CHI) revealing their responsibility in anthocyanins accumulation in mulberry. Plant Physiol. Biochem. 161, 65–73. doi: 10.1016/j.plaphy.2021.01.044
Chen, J., Bi, Y. Y., Wang, Q. Q., Liu, D. K., Zhang, D. Y., Ding, X. Q., et al. (2022). Genome-wide identification and analysis of anthocyanin synthesis-related R2R3-MYB genes in Cymbidium goeringii. Front. Plant Science. 13. doi: 10.3389/fpls.2022.1002043
Cheng, A. X., Zhang, X. B., Han, X. J., Zhang, Y. Y., Gao, S. A., Liu, C. J., et al. (2018). Identification of chalcone isomerase in the basal land plants reveals an ancient evolution of enzymatic cyclization activity for the synthesis of flavonoids. New Phytologist. 217, 909–924. doi: 10.1111/nph14852
Chou, K. C. and Shen, H. B. (2010). A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: Euk-mPLoc 2.0. PLoS One 5, e9931. doi: 10.1371/journal.pone.0009931
Dastmalchi, M. and Dhaubhadel, S. (2015). Soybean chalcone isomerase: evolution of the fold, and the differential expression and localization of the gene family. Planta. 241, 507–523. doi: 10.1007/s00425-014-2200-5
Ding, N., Hai, Y., Wang, X. H., Tu, P. F., Gao, B. W., and Shi, S. P. (2021). Cloning and expression analysis of chalcone isomerase from Aquilaria sinensis. Acta Pharm. Sin. 56, 630–638. doi: 10.16438/j.0513-4870.2020-1588
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Science. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fan, H. H., Cui, M. L., Li, N. H., Li, X. J., Liang, Y. X., Liu, L., et al. (2020). Genome-wide identification and expression analyses of R2R3-MYB transcription factor genes from two Orchid species. PeerJ. 8, e9781. doi: 10.7717/peerj.9781
Ge, S. B., Zhang, X. N., Han, W. Y., Li, Q. Y., and Li, X. (2023). Research progress on plant flavonoid biosynthesis and its anti-stress mechanism. Acta Hortic. Sinica. 50, 16. doi: 10.16420/j.issn.0513-353x.2021-1186
Guo, W. X., Li, H. Q., and Liang, S. (2012). Sequence and expression analysis of five housekeeping genes in Dendrobium nobile: selection of reference genes for gene expression quantification. J. Mianyang Normal University. 31, 59–66. doi: 10.16276/j.cnki.cn51-1670/g
Huang, W., Khaldun, A. B. M., Lv, H. Y., Du, L. W., Zhang, C. J., and Wang, Y. (2016). Isolation and functional characterization of an R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum. Plant Cell Rep. 35, 883–894. doi: 10.1007/s00299-015-1929-z
Jiang, B. X., Wu, Z. H., Yang, G. X., Lv, S. J., Jia, Y. H., Wu, Y. Y., et al. (2023). Cloning and functional analysis of flavanone 3-hydroxylase gene in Rhododendron hybridum Hort. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 39, 653–669. doi: 10.13345/j.cjb.220581
Jiang, W., Yin, Q., Wu, R., Zheng, G. S., Liu, J. Y., Dixon, R. A., et al. (2015). Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 66, 7165–7179. doi: 10.1093/jxb/erv413
Kang, Y. L., Pei, J., Cai, W. L., Liu, W., Luo, J., and Wu, Q. H. (2014). Research progress on flavonoid metabolic synthesis pathway and related function genes in medicinal plants. Chin. Traditional Herbal Drugs 45, 6. doi: 10.7501/j.issn.0253-2670.2014.09.026
Ke, Y. J., Zheng, Q. D., Yao, Y. H., Ou, Y., Chen, J. Y., Wang, M. J., et al. (2021). Genome-wide identification of the MYB gene family in Cymbidium ensifolium and its expression analysis in different flower colors. Int. J. Mol. Sci. 22, 13245. doi: 10.3390/ijms222413245
Kriangphan, N., Vuttipongchaikij, S., Kittiwongwattana, C., Suttangkakul, A., Pinmanee, P., Sakulsathaporn, A., et al. (2015). Effects of sequence and expression of eight anthocyanin biosynthesis genes on gloral coloration in four Dendrobium Hybrids. Horticulture J. 84, 83–92. doi: 10.2503/hortj.MI-020
Lam, P. Y., Wang, L., Lui, A. C. W., Liu, H. J., TakeaKimura, Y., Chen, M. X., et al. (2021). Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiol. 188, 1993–2011. doi: 10.1093/plphys/kiab606
Li, C., Ng, K. Y., and Fan, L. M. (2015). MYB transcription factors are active players in abiotic stress signaling. Environ. Exp. Botany. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014
Li, C. H., Qiu, J., Ding, L., Huang, M. Z., Huang, S. R., Yang, G. S., et al. (2017). Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 112, 335–345. doi: 10.1016/j.plaphy.2017.01.019
Li, B. J., Zheng, B. Q., Wang, J. Y., Tsai, W. C., Lu, H. S., Zou, L. H., et al. (2020). New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun. Biol. 3, 89–102. doi: 10.1038/s42003-020-0821-8
Lim, W. and Li, J. (2017). Co-expression of onion chalcone isomerase in Del/Ros1-expressing tomato enhances anthocyanin and flavonol production. Plant Cell Tissue Organ Culture (PCTOC) 128, 113–124. doi: 10.1007/s11240-016-1090-6
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Livak, K. J. and Schmittgen, T. D. L. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, K. Y., Wang, S. P., Yang, L., Luo, S. L., Cheng, J., Dong, Y., et al. (2025). Evolutionary landscape of plant chalcone isomerase-fold gene families. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1559547
Meng, H. X., Li, J. W., Gong, J. Y., and Wang, H. X. (2025). Physiological and transcriptome response analysis of Dendrobium nobile flowers under high temperature stress. Chin. J. Trop. Crops. 46, 266–277. doi: 10.3969/j.issn.1000-2561.2025.02.003
Nan, G., Wu, H., Wu, Q., Liu, L. S., Liao, Q. C., Li, C. L., et al. (2022). Cloning, identification, and functional analysis of the chalcone isomerase gene and its promoter from Tartary buckwheat. Acta Physiologiae Plantarum 14, 1400. doi: 10.1007/s11738-022-03410-w
Ngaki, M. N., Louie, G. V., Philippe, R. N., Manning, G., Pojer, F., Bowman, M. E., et al. (2012). Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature. 485, 530–533. doi: 10.1038/nature11009
Ni, R., Zhu, T. T., Zhang, X. S., Wang, P. Y., Sun, C. J., Qiao, Y. N., et al. (2020). Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. J. Exp. Botany. 71, 290–304. doi: 10.1093/jxb/erz425
Pandey, A., Misra, P., and Trivedi, P. K. (2015). Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 34, 1515–1528. doi: 10.1007/s00299-015-1803-z
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Qiu, Y. J., Cai, C. C., Mo, X., Zhao, X. Y., Wu, L. J., Liu, F., et al. (2023). Transcriptome and metabolome analysis reveal the effect of flavonoids on flower color variation in Dendrobium nobile Lindl. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1220507
Sun, Y., Bao, J. Z., Liu, C. G., Li, F. T., and Chen, X. L. (2015). Progresses on genetic engineering in Orchid floral color. J. Nucl. Agric. Sci. 29, 1701–1710. doi: 10.11869/j.issn.100-8551.2015.09
Wang, J. Y., Jiang, Y. F., Sun, T., Zhang, C. H., Liu, X. H., and Li, Y. S. (2022). Genome-wide classification and evolutionary analysis reveal diverged patterns of chalcone isomerase in plants. Biomolecules. 12, 961. doi: 10.3390/biom12070961
Wang, X. J., Liang, L. X., Li, L. B., and Wang, T. (2018). Genome-wide analysis of R2R3-MYB transcription factors in Phalaenopsis equestris. For. Res. 31, 104–113. doi: 10.13275/j.cnki.lykxyj.2018.03.014
Wang, W. L., Wu, Z. J., Liu, Z. W., Wang, Y. X., Li, H., Cui, X., et al. (2017). Cloning and expression analysis of the gene encoding flavonoid 3′-hydroxylase in tea plant (Camellia sinensis). J. Tea Science. 37, 108–118. doi: 10.13305/j.cnki.jts.2017.01.012
Wittkopp, P. J. and Kalay, G. (2012). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wolf-Saxon, E. R., Moorman, C. C., Castro, A., Ruiz-Rivera, A., Mallari, J. P., and Burke, J. R. (2023). Regulatory ligand binding in plant chalcone isomerase–like (CHIL) proteins. J. Biol. Chem. 299, 104804. doi: 10.1016/j.jbc.2023.104804
Wu, L. P., Fan, J. Z., Su, X. L., Peng, D. Y., and Xing, S. H. (2024). Genome-wide identification of R2R3-MYB family genes and their response to stress in Dendrobium nobile. Front. Bioscience-Landmark. 29, 1. doi: 10.21203/rs3.rs-2749425/v1
Wu, Y. Q., Jin, X. J., Wang, L. J., Wang, C., Lei, J., Chai, S. S., et al. (2025a). Bioinformatics and expression analysis of the CHI Gene Family in sweet potato. Plants 14, 752. doi: 10.3390/plants14050752
Wu, Q. Q., Xia, L., Yu, F., Liang, X. Y., Wang, M., and Zhang, Y. (2025b). Identification and preliminary verification of gene function of chalcone isomerase HbCHI gene family in Hevea brasiliensis. Chin. J. Trop. Crops. 46, 247–255. doi: 10.3969/j.issn.1000-2561.2025.02.001
Xu, Q., Niu, S., Li, K., Zheng, X., Jia, Y., Liu, Y., et al. (2022a). Chromosome-scale assembly of the Dendrobium nobile genome provides insights into the molecular mechanism of the biosynthesis of the medicinal active ingredient of Dendrobium. Front. Genet. 13. doi: 10.3389/fgene.2022.844622
Xu, H. H., Lan, Y. P., Xing, J. Y., Li, Y., Liu, L. C., and Wang, Y. Q. (2022b). AfCHIL, a type IV chalcone isomerase, enhances the biosynthesis of naringenin in metabolic engineering. Front. Plant Science. 13. doi: 10.3389/fpls.2022.891066
Yan, H., Pei, X., Zhang, H., Li, X., Zhang, X. X., Zhao, M. G., et al. (2021). MYB-Mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 22, 3103. doi: 10.3390/ijms22063103
Yang, J. F., Chen, Y. Z., Xiao, Z. H., Shen, H. L., Li, Y. H., and Wang, Y. (2022). Multilevel regulation of anthocyanin-promoting R2R3-MYB transcription factors in plants. Front. Plant Science. 13. doi: 10.3389/fpls.2022.1008829
Yang, K., Hou., Y. B., Wu, M., Pan, Q. Y., Xie, Y. L., Zhang, Y. S., et al. (2023). DoMYB5 and DobHLH24, transcription factors involved in regulating anthocyanin accumulation in Dendrobium officinale. Int. J. Mol. Sci. 24, 7552. doi: 10.3390/ijms24087552
Yin, Y. C., Zhang, X. D., Gao, Z. Q., Hu, T., and Liu, Y. (2019). The research progress of chalcone isomerase (CHI) in plants. Mol. Biotechnol. 61, 32–52. doi: 10.1007/s12033-018-0130-3
Yu, S. H., Li, J. Y., Peng, T., Ni, S., Feng, Y., Wang, Q. S., et al. (2022). Identification of chalcone isomerase family genes and roles of CnCHI4 in flavonoid metabolism in Camellia nitidissima. Biomolecules. 13, 41. doi: 10.3390/biom13010041
Zhang, C., Wang, D., Yang, C., Kong, N., Shi, Z., Zhao, P., et al. (2017). Genome-wide identification of the potato WRKY transcription factor family. PLoS One 12, e0181573. doi: 10.1371/journal.pone.0181573
Zhao, C., Liu, X. J., Gong, Q., Cao, J. P., Shen, W. X., Yin, X. R., et al. (2020). Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 19, 671–688. doi: 10.1111/pbi.13494
Zhou, L., Cui, Q. X., Liu, T. J., Ban, Q. Y., Yuan, Y. H., Ning, K., et al. (2023). Cloning and analysis of the type IV chalcone isomerase gene SmCHI2 in eggplant. Southwest China J. Agric. Sci. 36, 1475–1481. doi: 10.16213/j.cnki.scjas.2023.7.015
Zhou, Y. W., Xu, Y. C., Tan, J. J., Huang, L. S., Zhu, G. F., and Ye, Y. J. (2024). Role of anthocyanin metabolic diversity in bract coloration of Curcuma alismatifolia varieties. Plant Physiol. Biochem. 216, 109156. doi: 10.1016/j.plaphy.2024.109156
Zhu, J. H., Zhao, W., Li, R. S., Guo, D., Li, H. L., Wang, Y., et al. (2021). Identification and characterization of chalcone isomerase genes involved in flavonoid production in dracaena Cambodiana. Front. Plant Science. 12, 616396. doi: 10.3389/fpls.2021.616396
Keywords: Dendrobium nobile, chalcone isomerase, flavonoid biosynthesis, DnCHI1, MYB
Citation: Chang L-l, Zhou Y-x, Wang L-r, Liu L and Li Z-j (2025) Identification of chalcone isomerase family genes and roles of DnCHI1 in flavonoid metabolism in Dendrobium nobile. Front. Plant Sci. 16:1687738. doi: 10.3389/fpls.2025.1687738
Received: 18 August 2025; Accepted: 26 October 2025;
Published: 14 November 2025.
Edited by:
Surendra Pratap Singh, Chhatrapati Shahu Ji Maharaj University, IndiaReviewed by:
Shihai Xing, Anhui University of Chinese Medicine, ChinaSrivani Mallela, Tata Memorial Hospital, India
Copyright © 2025 Chang, Zhou, Wang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-jian Li, emhlbmppYW5saUAxNjMuY29t
 Ling-ling Chang
Ling-ling Chang Yu-xin Zhou
Yu-xin Zhou Ling-rong Wang
Ling-rong Wang Lei Liu
Lei Liu Zhen-jian Li
Zhen-jian Li