- 1Agronomy Department, Plant Molecular and Cellular Biology Program, Genetics Institute, University of Florida, Institute of Food and Agricultural Sciences (IFAS), Gainesville, FL, United States
- 2DOE Center for Advanced Bioenergy and Bioproducts Innovation, Gainesville, FL, United States
Small heat shock protein (sHSP) promoters contain cis-regulatory elements that facilitate transcription in response to heat stress, making them valuable tools for functional studies through controlled gene expression and the precise regulation of gene-editing tools or morphogenic regulators. To evaluate their utility, GUS reporter gene expression driven by four plant-sourced HSP promoters (pGmHSP17.5, pHvHSP17, pZmHSP17.7, and pZmHSP26) was compared across various tissues of stably transformed sugarcane before and after heat treatment. At 22°C, all promoters showed minimal activity in leaves and roots, although pZmHSP17.7 and pHvHSP17 displayed moderate expression in stems. Following heat treatment, all promoters exhibited their highest activity in stems, followed by leaves and roots. In stem tissues, pGmHSP17.5 displayed heat-induced uidA expression comparable to the constitutive pZmUbi promoter. Notably, heat-induced reporter gene activity in stem middle sections of single-copy transgenic lines containing pZmHSP17.7, pHvHSP17, or pZmHSP26 exceeded pZmUbi-derived uidA activity by 9.7-fold, 3.8-fold, and 3.0-fold, respectively, with 346- to 3,672-fold induction compared to control conditions. Most promoters showed peak expression in the middle sections of the stem, while pHvHSP17 was the most active in the stem apices. Histochemical analysis revealed that pZmHSP17.7 and pHvHSP17 were active in both parenchyma cells and vascular bundles within sugarcane stems. Among leaf tissues, mature leaves exhibited greater expression than senescing or immature leaves, while root activity remained consistently minimal across all promoters. Temperature-course experiments identified distinct activation thresholds: 34°C–36°C for pZmHSP17.7, 36°C for pZmHSP26, 36°C–38°C for pHvHSP17, and 40°C–42°C for pGmHSP17.5. Drought stress also induced reporter gene transcription in stems under HSP promoters, although with lower fold induction than heat treatment. These findings provide valuable tools for gene function studies and biotechnology applications, including heat stress tolerance research, controlled transgene expression in metabolic engineering, precision gene editing, and developmental biology studies.
Introduction
Climate change, driven by increased human activity since the Industrial Revolution, poses an imminent and far-reaching threat. Consequently, heat stress has emerged as an increasingly frequent challenge in crop production (IPCC, 2022). High temperatures can significantly alter plant growth and development at morphological, physiological, and molecular levels (Fahad et al., 2017; Li et al., 2023). In response to heat stress, plants produce heat shock proteins (HSPs), which are essential for maintaining cellular homeostasis by preserving protein conformation and preventing non-functional protein aggregation (Vierling, 1991; Wang et al., 2004).
HSPs are well-conserved between species and can be identified by their characteristic heat shock domain (Helm et al., 1993). All plant HSPs can also be categorized into one of five families based on their molecular weight (HSP100, HSP90, HSP70, HSP60, and small HSPs). Among these, the small heat shock protein (sHSP) family acts as molecular chaperones to stabilize protein folding and degrade misfolding proteins (Haslbeck et al., 2004, 1999; Lee et al., 1997). sHSPs are regulated under a variety of abiotic stresses, including heat (Howarth, 1991), drought (Grigorova et al., 2011), heavy metals (Györgyey et al., 1991), and osmotic stress (Almoguera et al., 1993; Coca et al., 1996). The overexpression of sHSPs has been shown to improve abiotic stress tolerance in crops (Feng et al., 2019; Sato and Yokoya, 2008).
Over the last few decades, focus has also been placed on the promoters of HSP genes. HSP promoters harbor multiple heat shock factor (HSF) binding sites. These are short, highly conserved motifs (5′-nGAAn-3′), also known as heat shock elements (HSEs). The inducible nature of such promoters provides various applications for plant biotechnology, including promoter strength evaluation (Freeman et al., 2011; Lyznik et al., 1995; Rerksiri et al., 2013), gene functional characterization (Wu et al., 2009), the activation of expression for gene-editing components (Barone et al., 2020; Nandy et al., 2019), and the controlled excision of transgenes by activating site-specific recombination systems (Akbudak and Srivastava, 2011; Sheva et al., 2020; Zhao et al., 2019; Khattri et al., 2011).
A well-characterized plant HSP promoter is the Glycine max (L.) HSP17.5 promoter (pGmHSP17.5). pGmHSP17.5 was identified using insertion/deletion mutagenesis (Czarnecka et al., 1989) and has been shown to drive stronger transgene expression after heat induction than with the constitutive 35S promoter (Ainley and Key, 1990). pGmHSP17.5 has also been used to induce the CRISPR/Cas9 system for the generation of heritable mutations (Nandy et al., 2019) and to activate site-specific recombination in sugarcane (Zhao et al., 2019). Similar findings have also been demonstrated for the Hordeum vulgare (L.) HSP17 promoter (pHvHSP17). pHvHSP17 has two HSEs (Marmiroli et al., 1993; Raho et al., 1995) and was confirmed as heat-inducible in Nicotiana tabacum (Raho et al., 1996), Zea mays (Gullì et al., 2005), and Triticum aestivum L (Freeman et al., 2011). The ZmHSP26 protein has been identified (Nieto-Sotelo et al., 1990) and shown to be induced under heat stress in maize (Hu et al., 2015). The ZmHSP26 promoter (pZmHSP26) and ZmHSP17.7 promoter (pZmHSP17.7) have recently been documented to successfully activate a Cre-lox site-specific recombination system for the excision of selectable marker and morphogenic genes in Z. mays (Wang et al., 2020).
Sugarcane (Saccharum spp. hybrid) is the source of 40% of the global biofuel and 80% of the world’s table sugar production (Hoang et al., 2015). However, challenges associated with sugarcane’s polyploid genome make traditional breeding methods arduous, highlighting it as an ideal candidate for molecular improvement and research. While pGmHSP17.5 has previously been shown to induce FLPe/Frt for transgene excision in sugarcane (Zhao et al., 2019), the efficacy of pGmHSP17.5 has not been compared with that of other HSP promoters. The current study examined four different HSP promoters (pGmHSP17.5, pHvHSP17, pZmHSP17.7, and pZmHSP26) in stably transformed sugarcane using the uidA gene encoding β-glucuronidase (GUS) as a reporter gene. GUS is a commonly used reporter system in plant biotechnology studies. Its expression does not negatively impact plant growth and development and supports both histochemical and quantitative analysis (Vain, 2007). We evaluated the strengths of the four HSP promoters at the GUS activity level using histochemical GUS assays and quantitative MUG assays (Jefferson et al., 1987) in leaf, stem, and root tissues with and without heat induction, and we investigated their activating temperatures at the transcriptional level with qRT-PCR in sugarcane leaves. We also examined the efficacies of HSP promoters under drought stress in sugarcane stems. This study produced new quantitative knowledge on the temporal and spatial expression of HSP promoters in sugarcane under heat and drought, thus expanding the promoter toolbox for crop biotechnology.
Methods
Vector construction and gene transformation
Four vectors, each containing a uidA expression cassette and a reporter gene cassette, were constructed using the Golden Gate cloning method (Engler et al., 2014). The coding sequence of uidA was codon-optimized for sugarcane using custom gene synthesis (GenScript, NJ, USA). For the uidA expression cassettes, pGmHSP17.5, pHvHSP17, pZmHSP17.7, pZmHSP26, and pZmUbi (abbreviated as Ubi) were used to drive uidA in vectors QM134, QM127, QM125, QM126, and YR013, respectively (Figure 1A). In the reporter gene cassettes, neomycin phosphotransferase II (npt II) was used as a selectable marker driven by pZmUbi. Two nuclear matrix attachment regions (MARs) from N. tabacum were used as insulators to flank the linked expression cassettes (Allen et al., 1996; Xue et al., 2005). The vector backbone was removed via overnight restriction digestion using I-SceI (New England Biolabs, MA, USA). Transgene fragments were electrophoresed and purified using a GeneJET Gel Purification Kit (Thermo Fisher Scientific, MA, USA), coated onto gold microparticles, and delivered to callus cultures of sugarcane cultivar CP88–1762 using biolistics as described by Sandhu and Altpeter (2008).
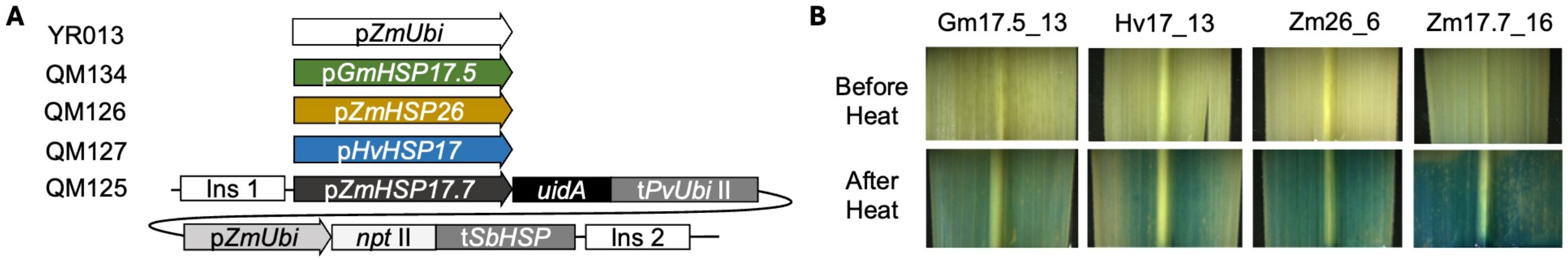
Figure 1. (A) Transgene cassettes used for transformation: pZmHSP17.7, promoter of Zea mays HEAT SHOCK PROTEIN 17.7 gene; pZmHSP26, promoter of Z. mays HEAT SHOCK PROTEIN 26 gene; pHvHSP17, promoter of Hordeum vulgare HEAT SHOCK PROTEIN 17 gene; pGmHSP17.5, promoter of Glycine max HEAT SHOCK PROTEIN 17.5 gene; pZmUbi, promoter of Z. mays UBIQUITIN gene; uidA, β-glucuronidase gene; tPvUbiII, Panicum virgatum ubiquitin terminator; pZmUbi, Z. mays ubiquitin promoter; npt II, neomycin phosphotransferase II; tSbHSP, Sorghum bicolor HEAT SHOCK PROTEIN terminator; Ins, insulator. (B) Representative leaf GUS staining results of different HSP lines before and after the heat treatment.
Plant material and conditions
The sugarcane tops of cultivar CP88–1762 were collected from field-grown sugarcane at the grand growth stage from the UF-IFAS Plant Research and Education Unit located near Citra, Florida. Callus induction was performed using immature leaf whorls for indirect embryogenesis as described by Kim et al. (2012). All culture media were prepared according to Kim et al. (2012). Once roots were established, regenerated plantlets (V0 generation; vegetative 0 generation) were transferred to soil and cultivated in a greenhouse setting. All plants under greenhouse conditions were grown in 22-cm-diameter pots, receiving 600 mL of irrigation per day via a drip irrigation system, with natural light and temperatures being controlled with air conditioning to 16°C to 20°C at night and 21°C to 25°C during the day. V0 plants (1m in height) were sampled for genomic DNA extraction.
Copy number assay
Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). Ten microliters of TaqMan® Gene Expression Master Mix, 1 μL of the customized TaqMan® probe (Applied Biosystems®, Thermo Fisher Scientific Inc., MA, USA), 7 μL of DNase/RNase-free water, and 20 ng of genomic DNA were used (20 μL total volume) to detect uidA copy number under the following conditions on a CFX connect system (Bio-Rad, CA, USA): denaturation for 10min at 95°C and 40 cycles of 15 s at 95°C and 60 s at 60°C. Sugarcane rust resistance gene (Bru1) was used for the normalization of the uidA gene copy number. Data were analyzed using the Applied Biosystems® CopyCaller® v2.1 software (Applied Biosystems, Thermo Fisher Scientific, MA, USA) following the manufacturer’s guidance. Primers used for copy number assay are listed in Supplementary Table S1.
Heat and drought stress treatments and sample collection
Heat treatments were conducted in a TPRB growth room (BioChambers Incorporated, MB, Canada) located at the UF/IFAS Growth Chamber Facility. Preliminary heat treatments consisted of heating 1-m-tall V0 plants for 2h (8:00 am to 10:00 am) at 40°C. Before and after heat treatment, two samples were collected from the middle of the first dewlap leaves of V0 plants. The mature node segments of V0 plants were used to generate the V1 generation under controlled greenhouse conditions as stated above.
Once exceeding 1.5m in height, V1 plants were sampled for fluorometric GUS assays (MUG assay) from the second dewlap leaves, stems (top of stem, middle of stem, and base of stem) of different tillers, and roots. A 4-day heat treatment was then completed, which contained four heat cycles, each 4 h long (from 8:00 am to 12:00 pm every day) at 40°C, 40% relative humidity, and 1,125 μmol m−2 s−1 Photosynthetic Photon Flux Density (PPFD) light intensity. During non-heat-treated hours, conditions were set to 22°C and 75% humidity. Day length was set for 15 h from 5:00 am to 8:00 pm with 1,125 μmol m−2 s−1 PPFD light intensity. After treatment, samples were collected from the middle sections of immature leaves, first dewlap leaves, third dewlap leaves, fifth dewlap leaves, stems (top of stem, middle of stem, and base of stem), and roots. Wild-type (WT) and HSP lines (V1 generation) were then vegetatively propagated from node cuttings to generate V2 progenies and biological replicates. V2 progenies were grown under controlled greenhouse conditions as stated above.
A subset of V2 plants was treated with drought stress by shutting off irrigation. Stem samples (top of stem, middle of stem, and base of stem) were collected when the relative water content (RWC %) in the potting soil reached 50% (mild drought) or 20% (severe drought). RWC % was measured using the FieldScout TDR 350 Soil Moisture Meter (Spectrum Technologies, Bridgend, UK).
A heat cycle (from 8:00 am to 10:00 am) was conducted to measure the minimal and optimal activation temperatures of HSP promoters. Another subset of V2 plants was split into six groups and received a 2-h heat treatment at 34°C, 36°C, 38°C, 40°C, 42°C, and 44°C, and the middle sections of the first dewlap leaves were sampled for RNA extraction.
Each treatment/genotype was sampled as three biological replicates, except for the V0 samples.
Histochemical GUS assay and counterstaining
A histochemical GUS assay was conducted to visualize GUS localization based on Jefferson et al. (1987). Tissues were immersed in GUS staining solution (Supplementary Table S2) and incubated for 48h at 37°C. The GUS-treated tissues were then incubated in 70% ethanol at room temperature to remove chlorophyll. Counterstaining was conducted following the procedures stated in Kim et al. (2002). Photos were captured using a ZEISS Axiocam 305 color microscope (ZEISS, Oberkochen, Germany).
Fluorometric GUS assay
Fluorometric GUS assays were conducted to quantify GUS activity based on Jefferson et al. (1987). A 4-methylumbelliferone (4-MU) standard curve was performed as follows: emissions of 1,000, 500, 150, 50, 20, 10, 5, and 1 nM 4-MU standards diluted in 0.2 M Na2CO3 were measured at 365 nm for excitation and 455 nm for emission using the BioTek Synergy H1 hybrid reader (Agilent Technologies, CA, USA). Protein quantification was conducted using Quick Start™ Bradford 1× Dye reagent (Bio-Rad, CA, USA) following the manufacturer’s instructions. A bovine serum albumin (BSA) standard curve was created with 2, 1.5, 1, 0.75, 0.5, 0.25, and 0.125 mg/mL BSA standards diluted in GUS extraction buffer (GEB) (Supplementary Table S2) and measured for absorbance at 595 nm. For MUG assays, mixtures of 20 μL of crude extract and 180 μL of AMB (Supplementary Table S2) were incubated at 37°C in the dark. After 30min of incubation, 100 μL of 0.2 M Na2CO3 was added to stop enzyme activity, and the mixtures were measured at 365 nm for excitation and 455 nm for emission. GUS activity was calculated in pmol 4-MU/(min·mg).
Quantitative RT-PCR analysis
RNA was extracted using TRIzol™ Reagent (Thermo Fisher Scientific, MA, USA), following the manufacturer’s protocol. One microgram of extracted RNA was used to obtain cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, MA, USA). Quantitative PCR (qPCR) was conducted with SsoAdvanced Universal SYBR green supermix (Bio-Rad, CA, USA) according to the manufacturer’s guidance using a CFX connect system (Bio-Rad, CA, USA). qPCR conditions were as follows: denaturation for 3min at 95°C, 40 cycles of 10 s at 95°C and 30 s at 60°C, and melting for 10 s at 95°C followed by 0.5°C increment temperature increase every 5 s from 65°C to 95°C. GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (GAPDH) was used to normalize uidA gene expression. uidA relative expression = 2{Ct (GAPDH)−Ct (transgene)}. Primers used for qPCR are listed in Supplementary Table S1.
In silico analysis of heat shock elements in the different HSP promoters
The sequences of the four HSP promoters were analyzed for HSE configurations using PlantPan3.0 (Chow et al., 2019), PlantCARE (Lescot et al., 2002), and PlantTFDB (PlantTFDB v5.0) (Tian et al., 2020). Putative HSEs were also called using motif pattern searches encompassing variants with two or three tandemly alternating repeats of nGAAn, allowing one to two nucleotide substitutions in the core GAA motif and/or one to two bp insertions between the repeats. Python codes used for HSE sequence calling were deposited at https://github.com/qiandemoni/HSE_sequence_finder. Motifs with two alternating repeats of nGAAn, no mismatches, and no insertions were considered minimal HSE. Detected imperfect HSEs were ranked according to the type and number of substitutions in the core motif, number of bp insertions between motifs, and distance from the transcription start site (TSS).
Statistical analyses
Statistical analysis was completed using ANOVA in GraphPad Prism (version 10.0.1). The least significant difference (LSD) method was used for the comparisons of means. Paired Student’s t-test was used to analyze the effect of mild/severe drought stresses within the same lines.
Results
Generation of low-copy-number GUS-expressing transgenic sugarcane lines
Linearized vectors containing HSP promoters driving uidA expression cassettes and npt II selectable marker cassettes flanked by insulators were delivered into sugarcane calli (Figure 1A). Thirty-one independent HSP promoter V0 transgenic lines were generated with seven to nine lines per construct (Supplementary Table S3). Twelve of the lines (two to six lines for each of the constructs) were identified to contain a single-copy uidA insertion, with the remaining 20 displaying between two and five copies (Supplementary Table S3). GUS assay was conducted in all lines before and after a preliminary 2-h heat treatment at 40°C (Supplementary Figure S1). For further analysis, five pGmHSP17.5 lines, five pHvHSP17 lines, five pZmHSP26 lines, and four pZmHSP17.7 lines were selected based on the before- and after-heat GUS staining. The selected HSP lines displayed growth and development similar to WT plants (Supplementary Figure S2). Histochemical GUS staining before and after heat treatment in the representative HSP lines also confirmed that all four promoters were sufficient to induce GUS expression in sugarcane leaves (Figure 1B). Two single-copy uidA lines driven by a ZmUbi promoter were used as constitutive controls.
Heat shock element analysis
Promoter analysis revealed substantial variation in HSE organization among the four HSP promoters. HSE motifs can be highly variable without compromising their function during the heat stress response. Neither PlantCARE nor PlantPAN3.0 provides a dedicated category for HSF-binding sites. PlantTFDB search included HSE motifs. However, it generated markedly fewer hits than those identified through a customized HSE motif search (Supplementary Tables S4, S5). Performing a customized HSE motif search revealed no canonical HSEs (nGAAnnTTCnnGAAn or nTTCnnGAAnnTTCn) for any of the four evaluated HSP promoters, but different groups of imperfect HSEs (Supplementary Figure S3). Allowing one to two nucleotide substitutions in the core GAA motif and/or one to two bp insertions between the three tandemly alternating repeats of the nGAAn motif will likely attenuate but not abolish the heat response. These criteria resulted in 23, 17, 10, and 10 imperfect HSEs for pGmHSP17.5, pZmHSP17.7, pHvHSP17.7, and pZmHSP26, respectively, in the customized motif search (Supplementary Table S5). The minimal partially functional HSE is represented by two alternating pentamers (nGAAnnTTCn or nTTCnnGAAn), for which two, one, two, and one perfect hits were identified in pGmHSP17.5, pZmHSP17.7, pHvHSP17.7, and pZmHSP26, respectively (Supplementary Table S5). Allowing one nucleotide substitution in the core GAA motif and/or one to two bp insertions between the two tandemly alternating repeats of the nGAAn motif resulted in 41, 69, 26, and 20 imperfect minimal HSEs in pGmHSP17.5, pZmHSP17.7, pHvHSP17.7, and pZmHSP26, respectively (Supplementary Table S5). Twelve, 13, two, and one of these minimal and imperfect HSE motifs were located within 100 bp upstream of the TSS of pGmHSP17.5, pZmHSP17.7, pHvHSP17.7, and pZmHSP26, respectively (Supplementary Table S5). Combining the different criteria, our custom motif search resulted in a total of 66, 87, 38, 31 non-canonical HSEs in pGmHSP17.5, pZmHSP17.7, pHvHSP17.7, and pZmHSP26, respectively, and were ranked according to their potential functionality in heat response (Supplementary Figure S3).
GUS activities in different leaf positions following heat induction
Trace amounts of GUS activity were detected in the second dewlap leaves of V1 WT plants, ranging from 0.01 to 0.03 and 0.00 to 0.02 pmol/(min·mg) before and after the 4-day heat treatment, respectively (Figure 2A, Supplementary Table S6). In the HSP lines, before-heat GUS activity was observed in the range of 0.00 to 0.10 pmol/(min·mg) (Supplementary Table S6), and this became elevated in all leaf tissues post-heat (Figure 2A). After heat treatment, the pGmHSP17.5, pZmHSP17.7, pZmHSP26, and pHvHSP17 lines exhibited 0.41, 3.87, 1.15, and 2.41 pmol/(min·mg) GUS activity levels per uidA copy, respectively, on average of all leaf positions (Figures 2A–D). The fold changes in GUS activity in leaves before and after heat treatment ranged from 4.2- to 4,665.9-fold for pGmHSP17.5, 3.0- to 172.8-fold for pZmHSP17.7, 1.6- to 1,137.2-fold for pZmHSP26, and 1.2- to 56.4-fold for pHvHSP17 lines (Figures 2A–D, Supplementary Table S6). In lines Gm17.5_13, Zm17.7_6, Zm26_6, and Hv17_13, the highest GUS activity was detected in the first dewlap leaf (Figures 2A–D). Conversely, in lines Zm17.7_16, Zm26_8, and Hv17_16, peak GUS activity was observed in the third dewlap leaves (Figures 2B–D). Lower GUS activity was observed in immature and fifth dewlap leaves for all the HSP lines (Figures 2A–D). In contrast, Ubi promoter lines showed consistent GUS activity levels across various leaf positions (Figure 2A). In lines Zm17.7_6, Zm26_6, and Hv17_13, GUS activity in the first dewlap leaves exceeded that of constitutive Ubi_15 and Ubi_16 controls (Figures 2B–D). However, for all leaf positions, pGmHSP17.5 lines showed lower GUS activity compared to Ubi promoter lines (Figure 2A).
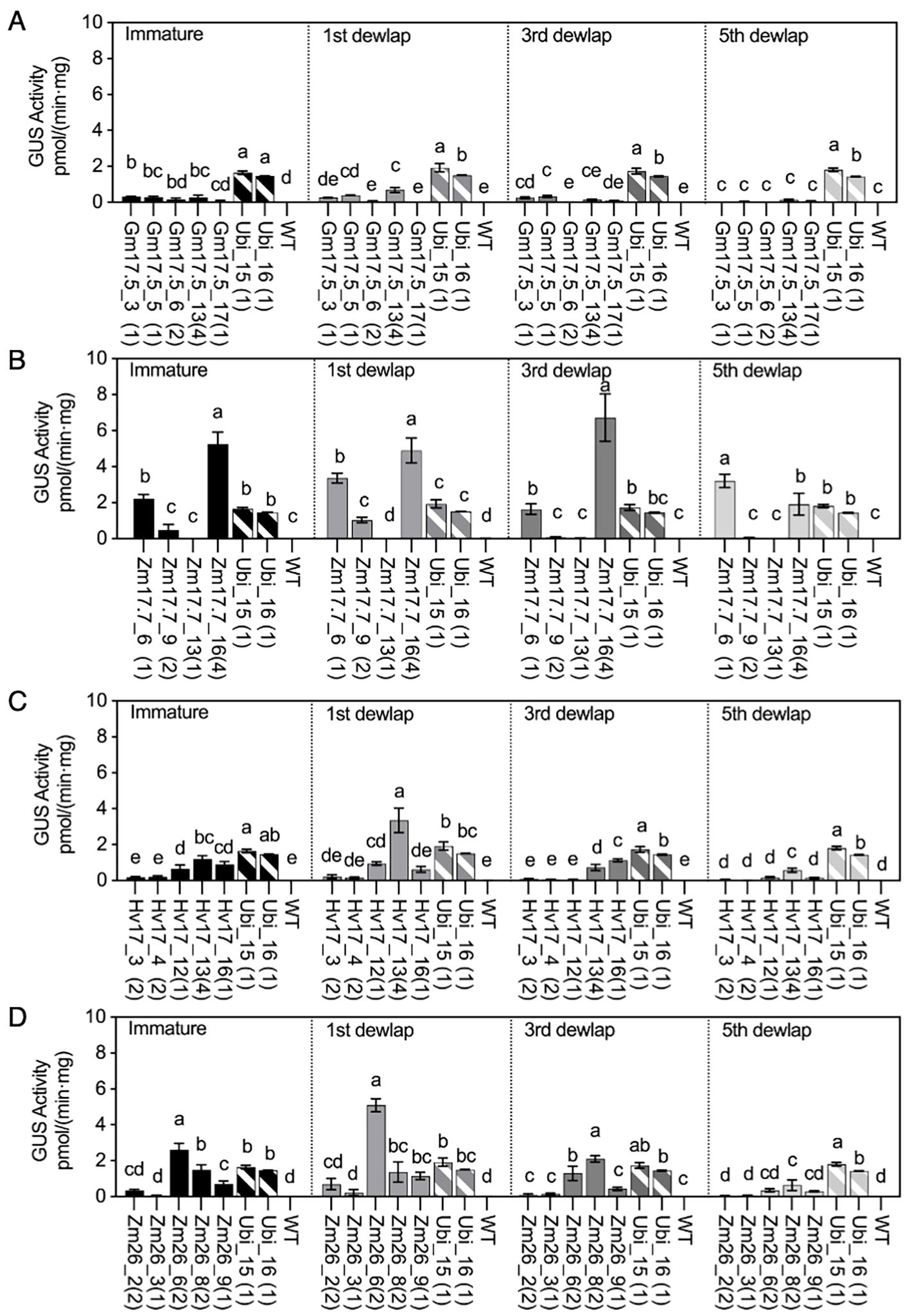
Figure 2. GUS activities in HSP lines and wild type (WT) after heat treatment compared with those in Ubi promoter lines under normal greenhouse conditions in different leaf positions (immature leaf, first dewlap leaf, third dewlap leaf, and fifth dewlap leaf). (A) pGmHSP17.5 lines, (B) pZmHSP17.7 lines, (C) pHvHSP17 lines, and (D) pZmHSP26 lines. uidA copy numbers are shown in parentheses following the line IDs. Solid bars indicate the HSP lines; shadowed bars indicate the Ubi promoter lines. Error bars indicate standard error. One-way ANOVA was conducted among different lines at same leaf positions, and different letters indicate significant difference at p < 0.05 according to Fisher’s least significant difference (LSD) comparison.
The gradient of GUS activity within the first dewlap leaves (tip, middle, and base) was also investigated (Figures 3A–D). Most lines, including all the pZmHSP17.7 lines, displayed a trend of leaf middle sections having the highest GUS activity (Figures 3A–D). However, for lines Gm17.5_3, Hv17_16, and Zm26_8, the highest GUS activity was observed at the tip of the leaf (Figures 3A, C, D).
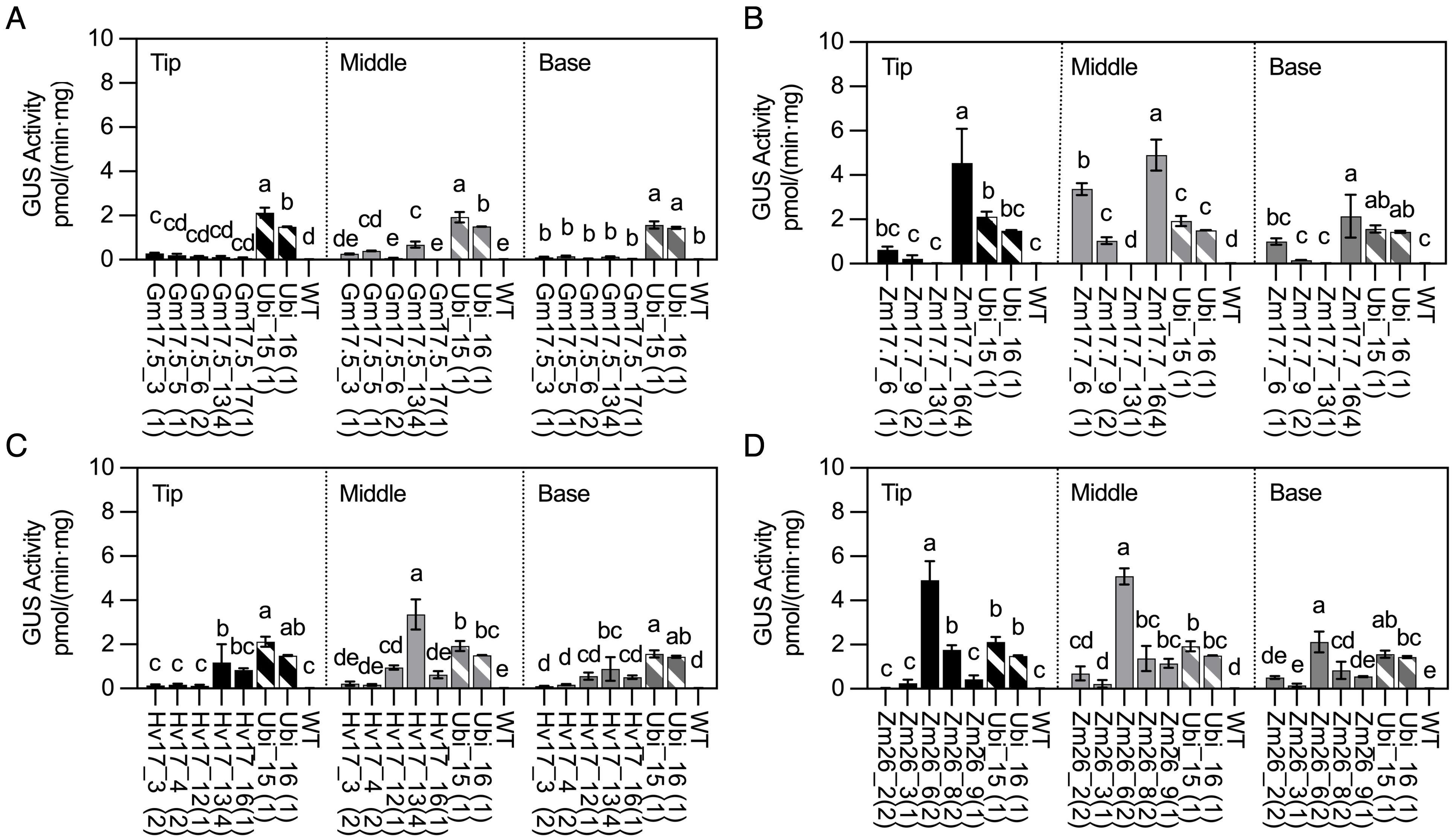
Figure 3. GUS activities in HSP lines and wild type (WT) after heat treatment compared with those in Ubi promoter lines under normal greenhouse conditions in first dewlap leaves (tip leaf section, middle leaf section, and base leaf section). (A) pGmHSP17.5 lines, (B) pZmHSP17.7 lines, (C) pHvHSP17 lines, and (D) pZmHSP26 lines. uidA copy numbers are shown in parentheses following the line IDs. Solid bars indicate the HSP lines; shadowed bars indicate the Ubi promoter lines. Error bars indicate standard error. One-way ANOVA was conducted among different lines at same leaf sections, and different letters indicate significant difference at p < 0.05 according to Fisher’s least significant difference (LSD) comparison.
GUS activity in different stem positions before and after heat induction
WT, HSP lines (V1 generation), and Ubi promoter lines were investigated for their GUS activity in the top, middle, and base positions of the stem. Negligible GUS activity was observed in any position of the stems in WT, pGmHSP17.5, and pZmHSP26 lines prior to heat treatment (Supplementary Figure S4). However, some of the pZmHSP17.7 and pHvHSP17 lines displayed GUS activity before heat treatment. In Hv17_13 and Zm17.7_6, the highest GUS activity levels before heat treatment were found in the middle and base of the stem, reaching 49.6% and 23.8% of that of the constitutive control line Ubi_16, respectively (Supplementary Figure S4). In WT, trace amounts of GUS activity, reaching up to 0.01 pmol/(min·mg), were detected in all stem positions after heat treatment (Figure 4). All HSP lines showed elevated GUS activity in the top, middle, and base of stem sections after heat (Figure 4). The before- and after-heat GUS activity fold changes were 32.3- to 3671.7-fold, 1.2- to 345.6-fold, 20.4- to 1567.6-fold, and 1.2- to 407.5-fold in all stem sections of all the pGmHSP17.5, pZmHSP17.7, pZmHSP26, and pHvHSP17 lines, respectively (Figure 4). All the pZmHSP17.7 and pZmHSP26 lines showed the highest GUS activity in the middle sections of the stems (Figures 4B, D), while pHvHSP17 lines displayed the highest activity at the top of the stems (Figure 4C). Gm17.5_6 showed peak GUS activity at the top of the stems, while GUS activity peaked in the middle of the stems for the rest of the pGmHSP17.5 lines (Figure 4A). Overall, the absolute GUS activity after heat treatment in the middle stem was up to 103.0-, 8.3-, 34.6-, and 63.2-fold greater than that in the first dewlap leaves in pGmHSP17.5, pZmHSP17.7, pZmHSP26, and pHvHSP17 lines, respectively (Figures 2, 4). Compared to the average of two Ubi promoter lines, after-heat GUS activity in the middle stem was up to 1.5-, 9.7-, 7.2-, and 4.6-fold greater in pGmHSP17.5, pZmHSP17.7, pZmHSP26, and pHvHSP17 lines, respectively (Figure 4).
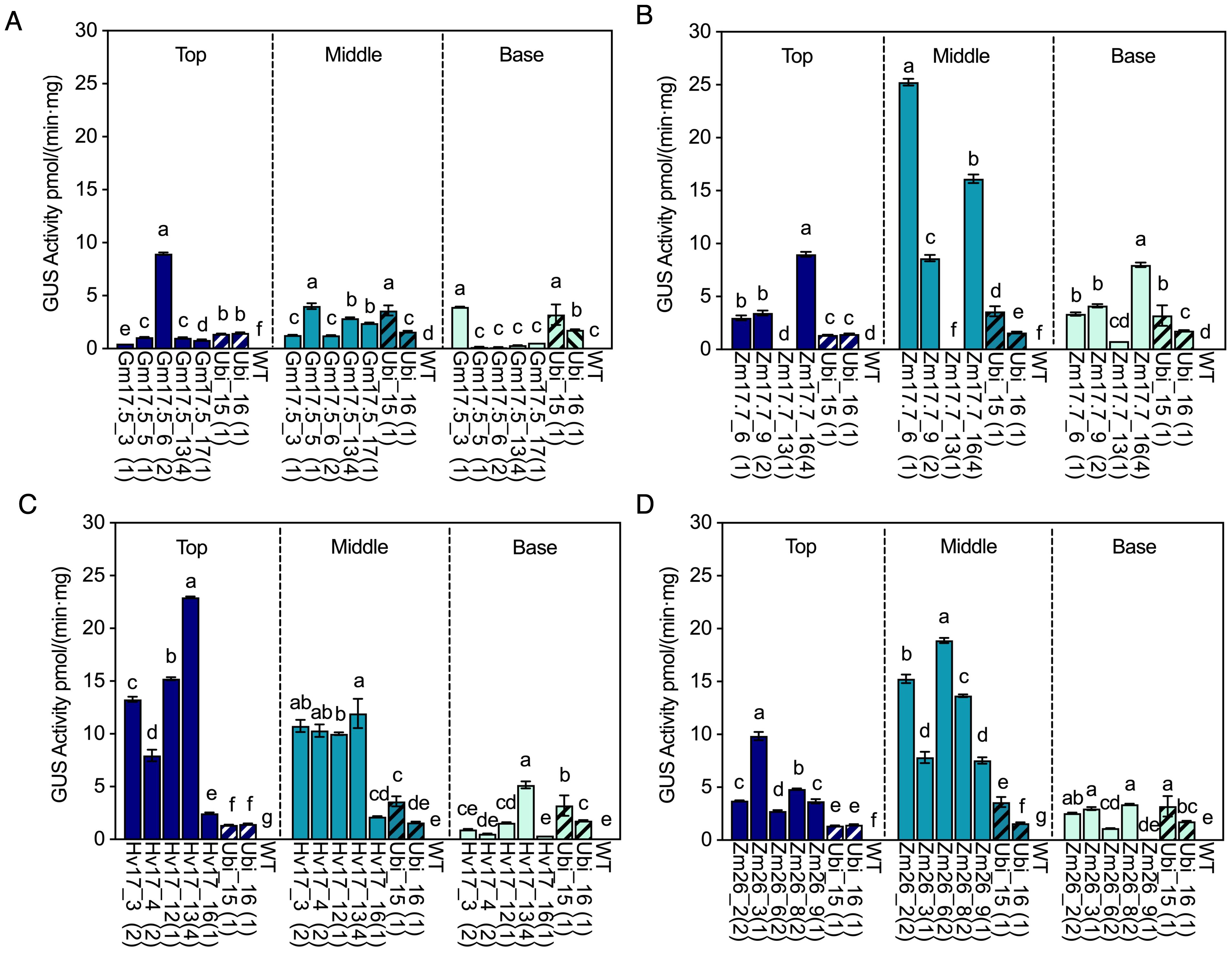
Figure 4. GUS activities in HSP lines and wild type (WT) after heat treatment compared with those in Ubi promoter lines under normal greenhouse conditions in different stem sections (top of stem, middle of stem, and base of stem). (A) pGmHSP17.5 lines, (B) pZmHSP17.7 lines, (C) pHvHSP17 lines, and (D) pZmHSP26 lines. UidA copy numbers are shown in parentheses following the line IDs. The solid bars indicate the HSP lines; the shadowed bars indicate the Ubi lines. Error bars indicate standard error. One-way ANOVA was conducted among different lines at same stem sections, and different letters indicate significant difference at p < 0.05 according to Fisher’s least significant difference (LSD) comparison.
Roots do not display elevated GUS expression following heat treatment
In WT roots, GUS activity was 0.07 and 0.06 pmol/(min·mg) before and after heat treatment, respectively (Supplementary Figure S5). GUS activity before heat treatment in the roots of transgenic HSP lines was slightly higher than that in the leaves and stems, ranging from 0.02 to 0.11 pmol/(min·mg) (Supplementary Figure S5, Supplementary Table S6). However, the after-heat root GUS activity ranged similarly to that before heat treatment between 0.05 and 0.12 pmol/(min·mg), which was significantly lower than that of the two Ubi lines, which ranged from 1.74 to 3.41 pmol/(min·mg) (Supplementary Figure S5). While an elevated trend was observed in root GUS activity for some lines following heat induction, the change was not significant, indicating a lack of significant reporter gene activation in roots with the applied heat treatment (Supplementary Figure S5).
Drought induces HSP promoters in sugarcane stem
After HSP lines from V2 generation reached a height of 1.5m, selected lines from pZmHSP17.7, pHvHSP17, and pZmHSP26 were subjected to drought, and samples were collected following mild and severe drought stress. Compared to before drought treatment, the increases in GUS activity after the severe drought stress were 2.3- to 54.7-fold, 0.2- to 27.4-fold, and 6.6- to 31.1-fold in pZmHSP17.7, pZmHSP26, and pHvHSP17 lines, respectively (Figure 5, Supplementary Figure S4). Notable increases of 3.5- and 7.5-fold were also observed between mild and severe drought stress in the top stem sections of Hv17_3 and the middle stem sections of Zm17.7_9 (Figure 5), yet for the rest of the lines, GUS activity measurements showed no significant differences between mild and severe drought stress (Figure 5, Supplementary Figure S6). The result of histochemical staining and counterstaining revealed that pZmHSP17.7 and pHvHSP17 were active in both vascular bundles and parenchyma cells in sugarcane stems, while pZmHSP26 was mostly active in vascular bundles (Supplementary Figure S7, Supplementary Figure S8).
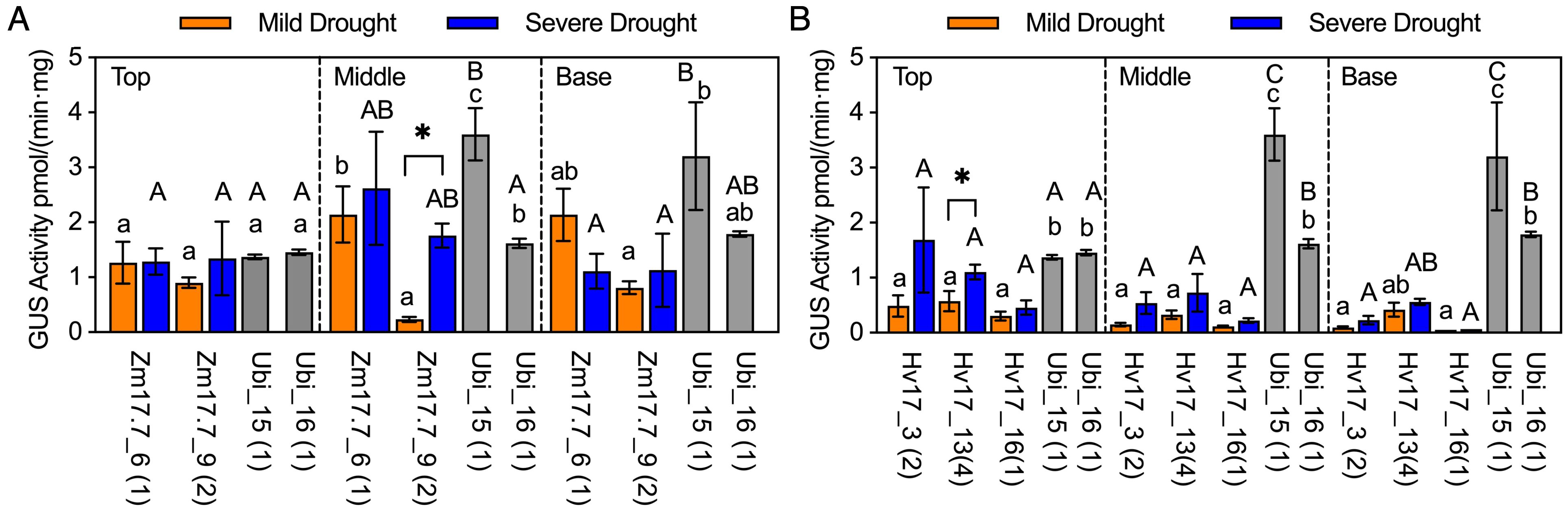
Figure 5. GUS activities in HSP lines after drought treatment compared with those in Ubi promoter lines under normal greenhouse conditions in different stem sections (top of stem, middle of stem, and base of stem). (A) pZmHSP17.7 lines and (B) pHvHSP17 lines. Gray bars indicate Ubi lines. uidA copy numbers are shown in parentheses following the line IDs. Error bars indicate standard error. One-way ANOVA was conducted among different lines (HSP lines and Ubi lines) at same stem sections, and different upper/lowercase letters indicate significant difference at p < 0.05 in mild/severe drought treatment according to Fisher’s least significant difference (LSD) comparison. Paired Student’s t-test was conducted to compare the values after mild or severe drought treatment within the same line, and the significance was indicated by * (p<0.05).
Activation temperatures for gene expression driven by different HSP promoters vary
The minimal and optimal induction temperatures for each HSP promoter were investigated using qRT-PCR at the transcriptional level in sugarcane leaf tissue. HSP lines from the V2 generation were heat-treated at temperatures ranging from 34°C to 44°C (2°C interval) for 2h in comparison to the 22°C control temperature. Significant transcription activation of uidA compared to the control temperature of 22°C was observed for the single-copy uidA lines of pZmHSP17.7 at 34°C to 36°C, pZmHSP26 at 36°C, for pHvHSP17 at 36°C to 38°C, and for pGmHSP17.5 at 42°C (Figures 6A–D). The highest uidA expression in these lines was detected between 40°C and 44°C (Figures 6A–D). The lines with four copies of pGmHSP17.5 or pZmHSP17.7 displayed significant uidA expression induction at lower temperatures and approximately two- to threefold higher maximum expression at 44°C than the corresponding single-copy lines (Figures 6A, B).
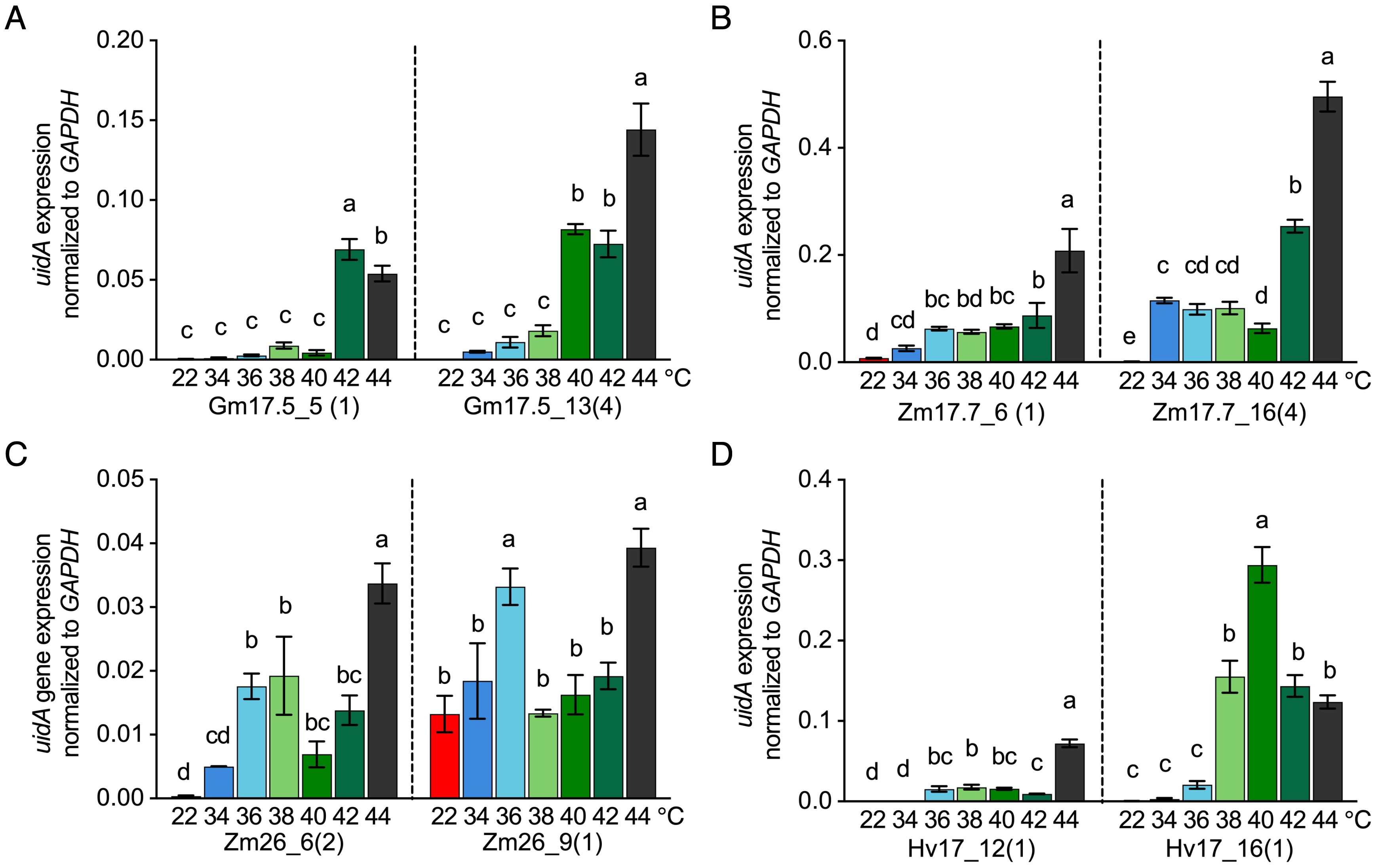
Figure 6. uidA expression normalized to housekeeping gene GAPDH after 2-h treatment at different temperatures (22°C, 34°C, 36°C, 38°C, 40°C, 42°C, and 44°C) in (A) pGmHSP17.5 lines, (B) pZmHSP17.7 lines, (C) pZmHSP26 lines, and (D) pHvHSP17 lines. uidA copy numbers are shown in parentheses following the line IDs. Values were derived from three biological replicates (n = 3). Error bars indicate standard error. One-way ANOVA was conducted, and different letters indicate significance at p<0.05 within the same line according to Fisher’s least significant difference (LSD) comparison.
Discussion
Inducible promoters provide remarkable utility when sustained transgene expression compromises plant development or agronomic performance. Well-characterized HSP promoters enable heat- and/or drought-inducible transgene expression for diverse applications, including metabolic engineering, site-specific recombination, gene editing with reduced off-target effects, and engineering enhanced heat and drought stress tolerance.
This study quantitatively and histochemically examined the spatial and temporal expression patterns of a GUS reporter gene under the transcriptional control of four HSP promoters in stably transformed sugarcane, comparing their performance to a constitutive maize ubiquitin promoter. The results demonstrate that HSP promoters induce stronger expression in mature leaf tissues compared to immature or senescing tissues.
Following heat treatment, leaves from multiple pZmHSP17.7, pZmHSP26, and pHvHSP17 transgenic lines displayed higher GUS activity than Ubi promoter lines, while all pGmHSP17.5 lines exhibited relatively weak GUS activity. These findings suggest that HSP promoters derived from monocotyledonous species (maize and barley) outperform those from dicotyledonous species (soybean) in sugarcane leaf tissues following heat induction.
Typically, leaves are expected to be more efficient at producing recombinant proteins than stems due to their distinct biochemical compositions. Sugarcane stems, for example, are rich in carbohydrates such as soluble sugars and lignocellulose and produce fewer proteins than sugarcane leaves (Palaniswamy et al., 2016). However, the results of this study indicate that differential regulation occurs in the leaves and stems of sugarcane following heat induction of HSP promoters.
Remarkably, sugarcane stems, which constitute approximately 70% of the plant’s biomass (Palaniswamy et al., 2016), displayed higher GUS activity than leaves after heat treatment. Similarly, a report in rice indicated that HSP-driven expression in the panicle was up to more than twofold higher than in the leaf (Rerksiri et al., 2013). Our findings have notable commercial value, as HSP promoters that enhance recombinant protein accumulation in sugarcane stems can be utilized for producing value-added proteins or expressing enzymes that catalyze hyper-accumulation of commercially important products, such as energy-dense lipids (Cao et al., 2023; Padilla et al., 2020; Parajuli et al., 2020). This approach also aligns with practical considerations, as existing sugar processing infrastructure for stem harvesting and processing can be readily adapted for these applications.
Prior to stress treatment, moderate levels of GUS activity were observed in the pZmHSP17.7 and pHvHSP17 lines in sugarcane stems, whereas GUS activity driven by pZmHSP26 and pGmHSP17.5 in stems was negligible. Most earlier reports have described that reporter gene activity or transcripts under the control of different HSP promoters are non-significant or undetectable prior to heat activation (Faralli et al., 2015; Freeman et al., 2011; Kuo et al., 2000; Pegoraro et al., 2011). However, several reports have described that some of the HSP promoters initiate transcripts or reporter gene activity under non-stress conditions, including during seed maturation (Prändl et al., 1995; Wehmeyer and Vierling, 2000) in the leaves of monocots (Harrington et al., 2020) and dicots (Bang et al., 2015; Khurana et al., 2013), the stigmas of monocots (Harrington et al., 2020), and the stems of dicots (Khurana et al., 2013).
Following drought treatment, the promoters pZmHSP17.7, pZmHSP26, and pHvHSP17 also activated GUS activity in sugarcane stems. GUS expression was localized in both the storage parenchyma cells and vascular bundles of pZmHSP17.7 and pHvHSP17 lines, and in the vascular bundles of pZmHSP26 lines. In comparison, in stable transgenic tobacco, GUS expression driven by pHvHSP17 was strictly restricted to the xylem tissues of stems and petioles after heat induction (Raho et al., 1996). Although GUS staining of some heat-shocked tissues, including leaves, glumes, and palea/lemma, in transgenic wheat lines was more intense in vascular bundles, GUS expression was not confined to vascular bundles in these tissues, with expression also observed in the internodes and nodes of stems. However, the relative expression difference between stem and leaf tissues was not quantified (Freeman et al., 2011; Raho et al., 1995). Similar to our study, Coca et al. reported higher reporter gene activity in stems than in leaves following heat activation when driven by the HaHSP17.7 promoter (Coca et al., 1996). However, in contrast to our observations, they found expression mainly in xylem and phloem rather than in parenchyma, and they did not observe upregulation by drought stress.
Fold inductions driven by HSP promoters following drought were substantially lower compared to those observed after heat treatment. This aligns with previous reports, which found that the expression of sHSPs, including HSP26, was more enhanced by heat than drought in maize seedlings (Hu et al., 2010). The HSP70 promoter from Oryza sativa also exhibited lower inducibility under drought compared to heat in rice (Rerksiri et al., 2013). In contrast, the GHSP26 gene was 100-fold more abundant in drought-stressed leaves, while only twofold more abundant in dehydrated stem and root compared to control tissues (Maqbool et al., 2007).
In a previous study, pHvHSP17 induced GUS activity in the roots of heat-treated wheat seedlings (Freeman et al., 2011). However, in mature sugarcane HSP lines, heat exposure did not significantly elevate GUS activity in roots. The lack of reporter gene activation in sugarcane roots may be due to decreased temperature exposure, as the thermal insulation of soil slows heat penetration (Katan, 1981). Freeman et al. (2011) used a temperature-controlled hydroponic system, which overcomes this limitation, but the hydroponic approach is less relevant for field performance than the approach we chose.
We also investigated the minimal and optimal activating temperatures of the four HSP promoters in sugarcane leaves. The results indicated that pZmHSP17.7 and pZmHSP26 were highly induced at approximately 44°C, requiring only a short heat pulse. This aligns with previous findings in maize (Wang et al., 2020), where site-specific transgene excision with Cre-lox under the transcriptional control of pZmHSP17.7 or pZmHSP26 was the most successful at 42–45°C. Similarly, pGmHSP17.5 was highly induced at 42–44°C. In contrast, pHvHSP17 was induced at a lower temperature (38°C–40°C), consistent with results found in wheat seedlings (Freeman et al., 2011). These findings could be invaluable for tailoring gene expression systems to specific thermal profiles, using different HSP promoters for genes that require activation under varied temperature conditions.
Comparing our in vivo results with in silico analyses of HSEs confirmed that the promoter performance of heat shock proteins cannot be reliably predicted from sequence-based HSE analysis alone. The abundance of canonical HSEs detected in silico within HSP promoters did not correspond to their heat-induced activation levels observed in vivo. For instance, pGmHSP17.5, which contained more high-confidence, non-canonical HSE motifs than the other promoters, did not exhibit the highest GUS induction following heat treatment. Despite the considerable degeneracy and variability among HSE motifs, many still support HSF binding with differing affinities, making it difficult for motif-based algorithms to distinguish functional from non-functional sites. Current in silico motif searches identify sequences in isolation, without considering chromatin context, cooperative HSF binding, epigenetic states, interactions with other regulatory elements, or nucleosome positioning—all of which strongly influence promoter activity (Huang et al., 2023; Abdulraheem et al., 2024; Fragkostefanakis et al., 2025) and may account for the comparatively high inducibility of pZmHSP17.7.
Heat-inducible promoters, such as HSP promoters, can open new avenues for breeding stress-resistant crop species. One notable example is that heat-inducible expression of miRNA398 enhanced the heat tolerance of Arabidopsis plants (Guan et al., 2013). HSP promoters are also particularly useful for complex metabolic engineering, where multiple transgenes need to be co-expressed to exert a synergistic impact on the desired phenotype. The inducible expression of specific transgenes allows the elucidation of their relative contribution to the phenotype in the context of constitutively co-expressed contributing factors by comparing phenotypes before and after induction. The availability of multiple well-characterized HSP promoters with similar induction profiles, such as in this study, also facilitates transgene stacking by decreasing risks associated with the repeated use of the same regulatory element, including unintended recombination and gene silencing (Matzke et al., 1994).
Conclusion
In this study, four different plant heat shock protein promoters were characterized in the vegetative tissues of stably transformed sugarcane to evaluate their efficacy and spatial expression profiles when directing the expression of a uidA reporter gene. Notably, pZmHSP17.7, pHvHSP17, and pZmHSP26 drove several-fold higher heat-induced reporter gene activity in stems compared to the constitutive pZmUbi promoter. The knowledge presented here will facilitate breeding for heat stress resilience and the development of traits requiring inducible transgene expression for complex metabolic engineering applications.
Data availability statement
The data presented in the study are deposited in the Zenodo repository under the following record locator: https://zenodo.org/records/17486962.
Author contributions
MQ: Investigation, Writing – original draft, Validation, Formal analysis, Visualization, Methodology. FA: Funding acquisition, Resources, Conceptualization, Writing – review & editing, Project administration, Supervision, Methodology.
Funding
The author(s) declared financial support was received for this work and/or its publication. This work was funded by the DOE Center for Advanced Bioenergy and Bioproducts Innovation (U.S. Department of Energy, Office of Science, Biological and Environmental Research Program under Award Number DE-SC0018420). Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Energy. This work was also supported by the USDA National Institute of Food and Agriculture, Hatch project 1020425.
Acknowledgments
The authors would like to thank Dr. Rajesh Yarra for providing transgenic sugarcane control lines harboring a single copy of vector YR 13, Dr. Eleanor Brant for her valuable contributions to the editing of this manuscript, and Franchesca Corral Dalugdug for her help with greenhouse work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1709171/full#supplementary-material
Supplementary Figure 1 | GUS staining of 1st dewlap leaves of all the transgenic HSP lines (V0) before and after 2h heat treatment at 40°C.
Supplementary Figure 2 | The selected HSP lines (V1) next to the wildtype sugarcane (WT) before any treatments.
Supplementary Figure 3 | In silico analysis of HSP promoter sequence revealing heat shock factor (HSF) binding sites containing two and three pentamers. Conservation levels of imperfect HSE motifs are “1”, “2A”, “2B”, “3”, “4A”, and “4B” as described below. 1: very likely to retain function (mildly reduced affinity/induction): no substitutions at conserved GAA or TTC positions of 2 pentamers (nGAAnnTTCn or nTTCnnGAAn) and may have a third pentamer with substitutions. No insertions between pentamers. 2A: likely to retain partial function (reduced affinity/induction): substitutions within a GAA where the substituted base is an A (i.e., one of the A’s but not the G) or single substitutions within a TTC where the substituted base is T (position 2) or C (position 3) with no insertions between pentamers. 2B: likely to retain partial function (reduced affinity/induction): no substitutions at conserved GAA or TTC positions of 2 pentamers (nGAAnnTTCn or nTTCnnGAAn) and may have a third pentamer with substitutions. 1–2 bp Insertions between pentamers. 3: less likely to retain partial function (more reduced affinity/induction): substitutions within a GAA where the substituted base is an A (i.e., one of the A’s but not the G) or single substitutions within a TTC where the substituted base is T (position 2) or C (position 3). 1–2 bp insertions between pentamers. 4A: least likely to retain partial function (most reduced affinity/induction): substitutions of the conserved G within a GAA (G→X at positions 2) or substitutions of the T in the TTC core (T→X at positions 2), without insertions between pentamers. 4B: least likely to retain partial function (most reduced affinity/induction): substitutions of the conserved G within a GAA (G→X at positions 2) or substitutions of the T in the TTC core (T→X at positions 2), with 1–2 bp insertions between pentamers. Data refers to Supplementary Table S5.
Supplementary Figure 4 | GUS activities in HSP lines and WT before heat treatment compared with that in Ubi promoter lines under normal greenhouse conditions in different stem sections (top of stem, middle of stem, and base of stem). uidA copy numbers were shown in parentheses following line IDs. The solid bars indicate the HSP lines; the shadowed bars indicate the Ubi lines. Error bars indicate standard error. One-way ANOVA was conducted among different lines at same stem sections, and different letters indicate significant difference at p < 0.05 according to Fisher’s least significant difference (LSD) comparison.
Supplementary Figure 5 | GUS activities in HSP lines and WT before and after heat treatment compared with that in Ubi promoter lines under normal greenhouse conditions in roots. uidA copy numbers were shown in parentheses following line IDs. The solid bars indicate the HSP lines; the shadowed bars indicate the Ubi lines. Error bars indicate standard error. One-way ANOVA was conducted among different lines at same stem sections, and different letters indicate significant difference at p < 0.05 according to Fisher’s least significant difference (LSD) comparison.
Supplementary Figure 6 | GUS activities in pZmHSP26 lines after drought treatment compared with that in Ubi lines under normal greenhouse conditions (gray bars) in different stem sections (top of stem, middle of stem, and base of stem). uidA copy numbers were shown in parentheses following line IDs. Error bars indicate standard error. One-way ANOVA was conducted among different lines (HSP lines and Ubi lines) at same stem sections, and different upper/lowercase letters indicate significant difference at p < 0.05 in mild/severe drought treatment according to Fisher’s least significant difference (LSD) comparison. Paired Student’s T test was conducted to compare the values after mild or severe drought treatment within the same line, and the significance was indicated by * (p < 0.05).
Supplementary Figure 7 | GUS staining in different stem sections of the selected HSP lines after mild and severe drought stress. uidA copy numbers are shown in parentheses following line IDs.
Supplementary Figure 8 | GUS counter-staining in different stem sections of the selected HSP lines after mild (50% RWC) and severe drought (20%RWC) stress. Pictures were taken at 5-10X magnitude and 40X magnitude. uidA copy numbers are shown in parentheses following line IDs.
References
Abdulraheem, M. I., Xiong, Y., Moshood, A. Y., Cadenas-Pliego, G., Zhang, H., and Hu, J. (2024). Mechanisms of plant epigenetic regulation in response to plant stress: Recent discoveries and implications. Plants 13, 163. doi: 10.3390/plants13020163
Ainley, W. M. and Key, J. L. (1990). Development of a heat shock inducible expression cassette for plants: Characterization of parameters for its use in transient expression assays. Plant Mol. Biol. 14, 949–967. doi: 10.1007/BF00019392
Akbudak, M. A. and Srivastava, V. (2011). Improved FLP recombinase, FLPe, efficiently removes marker gene from transgene locus developed by Cre-lox mediated site-specific gene integration in Rice. Mol. Biotechnol. 49, 82–89. doi: 10.1007/s12033-011-9381-y
Allen, G. C., Hall, G., Jr., Michalowski, S., Newman, W., Spiker, S., Weissinger, A. K., et al. (1996). High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell 8, 899–913. doi: 10.1105/tpc.8.5.899
Almoguera, C., Coca, M. A., and Jordano, J. (1993). Tissue-specific expression of sunflower heat shock proteins in response to water stress. Plant J. 4, 947–958. doi: 10.1046/j.1365-313X.1993.04060947.x
Bang, S. W., Park, S.-H., Kim, Y. S., Do Choi, Y., and Kim, J.-K. (2015). The activities of four constitutively expressed promoters in single-copy transgenic rice plants for two homozygous generations. Planta 241, 1529–1541. doi: 10.1007/s00425-015-2278-4
Barone, P., Wu, E., Lenderts, B., Anand, A., Gordon-Kamm, W., Svitashev, S., et al. (2020). Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol. Plant 13, 1219–1227. doi: 10.1016/j.molp.2020.06.008
Cao, V. D., Luo, G., Korynta, S., Liu, H., Liang, Y., Shanklin, J., et al. (2023). Intron-mediated enhancement of DIACYLGLYCEROL ACYLTRANSFERASE1 expression in energycane promotes a step change for lipid accumulation in vegetative tissues. Biotechnol. Biofuels Bioproducts 16, 1–15. doi: 10.1186/s13068-023-02393-1
Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G. Z., Tseng, K. C., Liu, Y. H., et al. (2019). Plantpan3.0: A new and updated resource for reconstructing transcriptional regulatory networks from chip-seq experiments in plants. Nucleic Acids Res. 47, D1155–D1163. doi: 10.1093/nar/gky1081
Coca, M. A., Almoguera, C., Thomas, T. L., and Jordano, J. (1996). Differential regulation of small heat-shock genes in plants: Analysis of a water-stress-inducible and developmentally activated sunflower promoter. Plant Mol. Biol. 31, 863–876. doi: 10.1007/BF00019473
Czarnecka, E., Key, J. L., and Gurley, W. B. (1989). Regulatory domains of the Gmhsp17.5-E heat shock promoter of soybean. Mol. Cell Biol. 9, 3457–3463. doi: 10.1128/mcb.9.8.3457-3463.1989
Engler, C., Youles, M., Gruetzner, R., Ehnert, T. M., Werner, S., Jones, J. D. G., et al. (2014). A Golden Gate modular cloning toolbox for plants. ACS Synth Biol. 3, 839–843. doi: 10.1021/sb4001504
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 8, 1147. doi: 10.3389/fpls.2017.01147
Faralli, M., Lektemur, C., Rosellini, D., and Gürel, F. (2015). Effects of heat shock and salinity on barley growth and stress-related gene transcription. Biol. Plant 59, 537–546. doi: 10.1007/s10535-015-0518-x
Feng, X. H., Zhang, H. X., Ali, M., Gai, W. X., Cheng, G. X., Yu, Q. H., et al. (2019). A small heat shock protein CaHsp25.9 positively regulates heat, salt, and drought stress tolerance in pepper (Capsicum annuum L.). Plant Physiol. Biochem. 142, 151–162. doi: 10.1016/j.plaphy.2019.07.001
Fragkostefanakis, S., Schleiff, E., and Scharf, K.-D. (2025). Back to the basics: the molecular blueprint of plant heat stress transcription factors. Biol. Chem. 406, 173–187. doi: 10.1515/hsz-2025-0115
Freeman, J., Sparks, C. A., West, J., Shewry, P. R., and Jones, H. D. (2011). Temporal and spatial control of transgene expression using a heat-inducible promoter in transgenic wheat. Plant Biotechnol. J. 9, 788–796. doi: 10.1111/j.1467-7652.2011.00588.x
Grigorova, B., Vaseva, I. I., Demirevska, K., and Feller, U. (2011). Expression of selected heat shock proteins after individually applied and combined drought and heat stress. Acta Physiol. Plant 33, 2041–2049. doi: 10.1007/s11738-011-0733-9
Guan, Q., Lu, X., Zeng, H., Zhang, Y., and Zhu, J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in A rabidopsis. Plant J. 74, 840–851. doi: 10.1111/tpj.12169
Gullì, M., Rampino, P., Lupotto, E., Marmiroli, N., and Perrotta, C. (2005). The effect of heat stress and cadmium ions on the expression of a small hsp gene in barley and maize. J. Cereal Sci. 42, 25–31. doi: 10.1016/j.jcs.2005.01.006
Györgyey, J., Gartner, A., Németh, K., Magyar, Z., Hirt, H., Heberle-Bors, E., et al. (1991). Alfalfa heat shock genes are differentially expressed during somatic embryogenesis. Plant Mol. Biol. 16, 999–1007. doi: 10.1007/BF00016072
Harrington, S. A., Backhaus, A. E., Fox, S., Rogers, C., Borrill, P., Uauy, C., et al. (2020). A heat-shock inducible system for flexible gene expression in cereals. Plant Methods 16, 1–16. doi: 10.1186/s13007-020-00677-3
Haslbeck, M., Braun, N., Stromer, T., Richter, B., Model, N., Weinkauf, S., et al. (2004). Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 23, 638–649. doi: 10.1038/sj.emboj.7600080
Haslbeck, M., Walke, S., Stromer, T., Ehrnsperger, M., White, H. E., Chen, S., et al. (1999). Hsp26: A temperature-regulated chaperone. EMBO J. 18, 6744–6751. doi: 10.1093/emboj/18.23.6744
Helm, K. W., LaFayette, P. R., Nagao, R. T., Key, J. L., and Vierling, E. (1993). Localization of small heat shock proteins to the higher plant endomembrane system. Mol. Cell Biol. 13, 238–247. doi: 10.1128/mcb.13.1.238-247.1993
Hoang, N. V., Furtado, A., Botha, F. C., Simmons, B. A., and Henry, R. J. (2015). Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front. Bioeng Biotechnol. 3, 182. doi: 10.3389/fbioe.2015.00182
Howarth, C. J. (1991). Molecular responses of plants to an increased incidence of heat shock. Plant Cell Environ. 14, 831–841. doi: 10.1111/j.1365-3040.1991.tb01446.x
Hu, X., Li, Y., Li, C., Yang, H., Wang, W., and Lu, M. (2010). Characterization of small heat shock proteins associated with maize tolerance to combined drought and heat stress. J. Plant Growth Regul. 29, 455–464. doi: 10.1007/s00344-010-9157-9
Hu, X., Yang, Y., Gong, F., Zhang, D., Zhang, L., Wu, L., et al. (2015). Protein sHSP26 improves chloroplast performance under heat stress by interacting with specific chloroplast proteins in maize (Zea mays). J. Proteomics 115, 81–92. doi: 10.1016/j.jprot.2014.12.009
Huang, Y., An, J., Sircar, S., Bergis, C., Lopes, C. D., He, X., et al. (2023). HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions. Nat. Commun. 14, 469. doi: 10.1038/s41467-023-36227-3
IPCC (2022). IPCC 2022: Climate Change 2022: Impacts, Adaptation and Vulnerability | Climate Change 2022: Impacts, Adaptation and Vulnerability (Cambridge, UK: Cambridge University Press).
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Katan, J. (1981). Solar heating (solarization) of soil for control of soilborne pests. Annu. Rev. Phytopathol. 19, 211–236. doi: 10.1146/annurev.py.19.090181.001235
Khattri, A., Nandy, S., and Srivastava, V. (2011). Heat-inducible Cre-lox system for marker excision in transgenic rice. J. Biosci. 36, 37–42. doi: 10.1007/s12038-011-9010-8
Khurana, N., Chauhan, H., and Khurana, P. (2013). Wheat chloroplast targeted sHSP26 promoter confers heat and abiotic stress inducible expression in transgenic Arabidopsis plants. PloS One 8, e54418–. doi: 10.1371/journal.pone.0054418
Kim, M. K., Choi, J. W., Jeon, J. H., Franceschi, V. R., Davin, L. B., and Lewis, N. G. (2002). Specimen block counter-staining for localization of GUS expression in transgenic arabidopsis and tobacco. Plant Cell Rep. 21, 35–39. doi: 10.1007/s00299-002-0475-7
Kim, J. Y., Gallo, M., and Altpeter, F. (2012). Analysis of transgene integration and expression following biolistic transfer of different quantities of minimal expression cassette into sugarcane (Saccharum spp. hybrids). Plant Cell Tiss. Organ Cult. 108, 297–302. doi: 10.1007/s11240-011-0043-3
Kuo, H., Tsai, Y., Young, L., and Lin, C. (2000). Ethanol treatment triggers a heat shock-like response but no thermotolerance in soybean (Glycine max cv. Kaohsiung No. 8) seedlings. Plant Cell Environ. 23, 1099–1108. doi: 10.1046/j.1365-3040.2000.00621.x
Lee, G. J., Roseman, A. M., Saibil, H. R., and Vierling, E. (1997). A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16, 659–671. doi: 10.1093/emboj/16.3.659
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Y., Ding, L., Zhou, M., Chen, Z., Ding, Y., and Zhu, C. (2023). Transcriptional regulatory network of plant cadmium stress response. Int. J. Mol. Sci. 24, 53–65. doi: 10.3390/ijms24054378
Lyznik, L. A., Hirayama, L., Rao, K. V., Abad, A., and Hodges, T. K. (1995). Heat-inducible expression of FLP gene in maize cells. Plant J. 8, 177–186. doi: 10.1046/j.1365-313X.1995.08020177.x
Maqbool, A., Zahur, M., Irfan, M., Qaiser, U., Rashid, B., Husnain, T., et al. (2007). Identification, characterization and expression of drought related alpha-crystalline heat shock protein gene (GHSP26) from Desi cotton. Crop Sci. 47, 2437–2444. doi: 10.2135/cropsci2007.03.0120
Marmiroli, N., Pavesi, A., Di Cola, G., Hartings, H., Raho, G., Conte, M. R., et al. (1993). Identification, characterization, and analysis of cDNA and genomic sequences encoding two different small heat shock proteins in Hordeum vulgare. Genome 36, 1111–1118. doi: 10.1139/g93-148
Matzke, A. J. M., Neuhuber, F., Park, Y. D., Ambros, P. F., and Matzke, M. A. (1994). Homology-dependent gene silencing in transgenic plants: epistatic silencing loci contain multiple copies of methylated transgenes. MGG Mol. Gen. Genet. 244, 219–229. doi: 10.1007/BF00285449
Murray, M. G. and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nandy, S., Pathak, B., Zhao, S., and Srivastava, V. (2019). Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 3(5), e00145. doi: 10.1002/pld3.145
Nieto-Sotelo, J., Vierling, E., and Ho, T. H. D. (1990). Cloning, sequence analysis, and expression of a cDNA encoding a plastid-localized heat shock protein in maize. Plant Physiol. 93, 1321–1328. doi: 10.1104/pp.93.4.1321
Padilla, C. S., Damaj, M. B., Yang, Z. N., Molina, J., Berquist, B. R., White, E. L., et al. (2020). High-level production of recombinant snowdrop lectin in sugarcane and energy cane. Front. Bioeng Biotechnol. 8. doi: 10.3389/fbioe.2020.00977
Palaniswamy, H., Syamaladevi, D. P., Mohan, C., Philip, A., Petchiyappan, A., and Narayanan, S. (2016). Vacuolar targeting of r-proteins in sugarcane leads to higher levels of purifiable commercially equivalent recombinant proteins in cane juice. Plant Biotechnol. J. 14, 791–807. doi: 10.1111/pbi.12430
Parajuli, S., Kannan, B., Karan, R., Sanahuja, G., Liu, H., Garcia-Ruiz, E., et al. (2020). Towards oilcane: Engineering hyperaccumulation of triacylglycerol into sugarcane stems. GCB Bioenergy 12, 476–490. doi: 10.1111/gcbb.12684
Pegoraro, C., Mertz, L. M., da Maia, L. C., Rombaldi, C. V., and de Oliveira, A. C. (2011). Importance of heat shock proteins in maize. J. Crop Sci. Biotechnol. 14, 85–95. doi: 10.1007/s12892-010-0119-3
Prändl, R., Kloske, E., and Schöffl, F. (1995). Developmental regulation and tissue-specific differences of heat shock gene expression in transgenic tobacco and Arabidopsis plants. Plant Mol. Biol. 28, 73–82. doi: 10.1007/BF00042039
Raho, G., Lupotto, E., Della Torre P, A., Hartings, H., Perrotta, C., and Marmiroli, N. (1995). Functional analysis of the temperature-dependent expression of the barley Hvhsp17 gene promoter in monocot and dicot cell systems. Plant Sci. 106, 63–69. doi: 10.1016/0168-9452(95)04064-2
Raho, G., Lupotto, E., Hartings, H., Della Torre P, A., Perrotta, C., and Marmiroli, N. (1996). Tissue-specific expression and environmental regulation of the barley Hvhsp17 gene promoter in transgenic tobacco plants. J. Exp. Bot. 47, 1587–1594. doi: 10.1093/jxb/47.10.1587
Rerksiri, W., Zhang, X., Xiong, H., and Chen, X. (2013). Expression and promoter analysis of six heat stress-inducible genes in rice. Sci. World J. 2013(1), 397401. doi: 10.1155/2013/397401
Sandhu, S. and Altpeter, F. (2008). Co-integration, co-expression and inheritance of unlinked minimal transgene expression cassettes in an apomictic turf and forage grass (Paspalum notatum Flugge). Plant Cell Rep. 27, 1755–1765. doi: 10.1007/s00299-008-0599-5
Sato, Y. and Yokoya, S. (2008). Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep. 27, 329–334. doi: 10.1007/s00299-007-0470-0
Sheva, M., Hanania, U., Ariel, T., Turbovski, A., Rathod, V. K. R., Oz, D., et al. (2020). Sequential genome editing and induced excision of the transgene in N. tabacum BY2 cells. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.607174
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., and Gao, G. (2020). PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Vain, P. (2007). Thirty years of plant transformation technology development: Review article. Plant Biotechnol. J. 5(2), 221–229. doi: 10.1111/j.1467-7652.2006.00225.x
Vierling, E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 579–620. doi: 10.1146/annurev.pp.42.060191.003051
Wang, N., Arling, M., Hoerster, G., Ryan, L., Wu, E., Lowe, K., et al. (2020). An efficient gene excision system in maize. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01298
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9(5), 244–252. doi: 10.1016/j.tplants.2004.03.006
Wehmeyer, N. and Vierling, E. (2000). The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance1. Plant Physiol. 122, 1099–1108. doi: 10.1104/pp.122.4.1099
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xue, H., Yang, Y.-T., Wu, C.-A., Yang, G.-D., Zhang, M.-M., and Zheng, C.-C. (2005). TM2, a novel strong matrix attachment region isolated from tobacco, increases transgene expression in transgenic rice calli and plants. Theor. Appl. Genet. 110, 620–627. doi: 10.1007/s00122-004-1880-9
Keywords: heat inducible promoter, sugarcane, vegetative tissue, uidA, stem, GUS activity, transgene expression, in silico analysis of heat shock elements
Citation: Qiande M and Altpeter F (2025) Comparing four heat-inducible promoters in stably transformed sugarcane regarding spatial and temporal control of transgene expression reveals candidates to drive stem-preferred transgene expression. Front. Plant Sci. 16:1709171. doi: 10.3389/fpls.2025.1709171
Received: 19 September 2025; Accepted: 28 October 2025;
Published: 03 December 2025.
Edited by:
Monalisa Sampaio Carneiro, Federal University of São Carlos, BrazilReviewed by:
Bin Xu, Nanjing Agricultural University, ChinaChakravarthi M, Sristi Biosciences Private Limited, India
Copyright © 2025 Qiande and Altpeter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fredy Altpeter, YWx0cGV0ZXJAdWZsLmVkdQ==
 Moni Qiande
Moni Qiande Fredy Altpeter
Fredy Altpeter