- 1College of Horticulture and Landscape Architecture, Northeast Agricultural University, Harbin, China
- 2Horticultural Research Institute, Hangzhou Academy of Agricultural Sciences, Hangzhou, China
A sudden cooling in the early spring or late autumn negatively impacts the plant growth and development. Although a number of studies have characterized the role of the transcription factors (TFs) of plant R2R3-myeloblastosis (R2R3-MYB) in response to biotic and abiotic stress, plant growth, and primary and specific metabolisms, much less is known about their role in Rosa multiflora under chilling stress. In the present study, RmMYB108, which encodes a nuclear-localized R2R3-MYB TF with a self-activation activity, was identified based on the earlier published RNA-seq data of R. multiflora plants exposed to short-term low-temperature stress and also on the results of prediction of the gene function referring Arabidopsis. The RmMYB108 gene was induced by stress due to chilling, salt, and drought and was expressed in higher levels in the roots than in the leaves. The heterologous expression of RmMYB108 in Arabidopsis thaliana significantly enhanced the tolerance of transgenic plants to freezing, water deficit, and high salinity, enabling higher survival and growth rates, earlier flowering and silique formation, and better seed quantity and quality compared with the wild-type (WT) plants. When exposed to a continuous low-temperature stress at 4°C, transgenic Arabidopsis lines–overexpressing RmMYB108 showed higher activities of superoxide dismutase and peroxidase, lower relative conductivity, and lower malondialdehyde content than the WT. Moreover, the initial fluorescence (Fo) and maximum photosynthetic efficiency of photosystem II (Fv/Fm) changed more dramatically in the WT than in transgenic plants. Furthermore, the expression levels of cold-related genes involved in the ICE1 (Inducer of CBF expression 1)-CBFs (C-repeat binding factors)-CORs (Cold regulated genes) cascade were higher in the overexpression lines than in the WT. These results suggest that RmMYB108 was positively involved in the tolerance responses when R. multiflora was exposed to challenges against cold, freeze, salt, or drought and improved the cold tolerance of transgenic Arabidopsis by reducing plant damage and promoting plant growth.
Introduction
Freezing temperatures induce cellular dehydration and limit the plant growth by inhibiting water uptake. Furthermore, chilling stress is fatal to plants, as cold thermodynamically lowers the membrane fluidity and directly inhibits several vital metabolic reactions (Mehrotra et al., 2020). In winter-hardy plants, the genes comprising a cryogenic response network mediate the physiological and biochemical changes and transcriptional modifications to maintain the cell integrity and plant survival at low temperatures (Zhao et al., 2015). We previously described that Rosa multiflora, a creeping thorny plant with medicinal and ornamental values, has a strong ability to withstand subzero temperatures (up to –40°C) in winter. The transcriptomic analysis of R. multiflora leaves exposed to different temperatures (25, 4, and –20°C) revealed that genes encoding APETALA2/ethylene-responsive factor (AP2/ERF), myeloblastosis (MYB), basic helix-loop-helix (bHLH), zinc finger protein (ZFP), NAC [NAM (NO APICAL MERISTEM), ATAF (Arabidopsis ACTIVATION FACTOR) and CUC (CUP-SHAPED COTYLEDON)] and WRKY transcription factors (TFs) actively participate in the response to cold treatment (Zhang et al., 2016). In plants, MYB TFs form one of the largest TF families, which are characterized by 1–4 incomplete conserved repeat (R)-containing DNA-binding domain located near the N-terminus (Klempnauer et al., 1982). Each R is composed of approximately 51–53 conserved amino acid residues with three α-helices (Dubos et al., 2010). The MYB family contains four types of TFs (i.e., 1R, R2R3, 3R, and 4R), depending on the number of MYB domains (Rogers and Campbell, 2004). The R2R3-MYB TFs form the largest clade and participate in the plant growth, especially in cell differentiation, development of floral organ, specific metabolisms, and response to environmental stress (Albert et al., 2014; Li et al., 2019; Liu et al., 2021).
In Arabidopsis thaliana, the R2R3-MYB TFs, such as AtMYB15, AtMYB30, AtMYB44, AtMYB96, and AtMYB108, are associated with stress responses (Dubos et al., 2010). The AtMYB15 protein interacts with the Inducer of C-repeat binding factor (CBF) Expression 1 (ICE1) and binds to Myb recognition sequences in the promoters of CBF genes to repress cold tolerance (Agarwal et al., 2006). In Rosa chinensis, anthocyanidin synthase, flavonol synthase, orcinol O-methyltransferase 1 (RcOOMT1), and RcOOMT2 genes are highly expressed in pink petals of flower buds and open flowers (Han et al., 2019). The silencing of RcMYB84/RcMYB123 increases the susceptibility of R. chinensis to Botrytis cinerea and reduces the protective effects of treatment with jasmonic acid (JA; Ren et al., 2020). Moreover, the structural genes of proanthocyanidins and flavonoid have a high expression level in RrMYB5- and RrMYB10-overexpressed Rosa rugosa and tobacco (Shen et al., 2019). However, a complete understanding of how R2R3-MYB TFs respond to low-temperature stress in Rosa species is lacking.
Here, we identified the R2R3-MYB genes of R. multiflora that respond to low-temperature stress from its transcriptome data (Zhang et al., 2016). The functions of these genes were predicted based on the phylogenetic analysis of the homologs of R. multiflora and Arabidopsis R2R3-MYB. We further investigated the RmMYB108 gene after combination of the RNA-seq data and prediction of the gene function. The overexpression (OE) of RmMYB108 in Arabidopsis confirmed the role of RmMYB108 in improving the tolerance against plant stress. This study provides a theoretical basis for cold-tolerance breeding in Rosa species.
Materials and Methods
Plant Materials and Growth Conditions
The cuttings of R. multiflora were obtained from the forest botanical garden of Heilongjiang in China (45.0°N, 128.4°E) and were grown under a 16-h light/8-h dark cycle at 25°C [control (CK)]. The 1-year-old cutting seedlings of R. multiflora were exposed to 4°C (CT1) in an artificial climate room or to −20°C (CT2) in a freezer to induce cold stress. Additionally, plants were either not watered or watered with 150 mM NaCl to imitate drought and salt stress, respectively. The leaves and roots of R. multiflora were collected and immediately frozen in liquid nitrogen.
Arabidopsis thaliana ecotype Columbia-0 was used for stable genetic transformation, and tobacco (Nicotiana benthamiana) was used for transient transformation. Arabidopsis plants were grown at 23°C under 12-h light/12-h dark cycle during the vegetative period and under 14-h light/10-h dark cycle during the reproductive phase.
Bioinformatics Analysis of R2R3-MYB Proteins in R. multiflora
Putative R2R3-RmMYB proteins, which were queried with the MYB DNA-binding protein PF00249 in the Linux system HMMER 3.3.1 (<1E−10), were identified from a published RNA-seq data (SRA accession no.: PRJNA698412) and the whole genome sequence of R. multiflora1 (Jung et al., 2019). The coding sequences (CDSs) of R2R3-MYB genes in A. thaliana and amino acid sequences of the encoded proteins were downloaded from The Arabidopsis Information Resource (TAIR2). The sequence of AtICE1 promoter was downloaded from the National Center for Biotechnology Information3, and the cis-acting elements of AtICE1 promoter were predicted using PlantCARE4. The conserved MYB domains were defined by using SMART5 (Letunic and Bork, 2018). The sequence alignments were performed using DNAMAN. A phylogenetic analysis was carried out using the maximum likelihood method with 1,000 bootstrap replicates in MEGA X. The heatmap was generated by using the Toolbox for Biologists (TBtools) software; the value in the row scale was normalized by the equation: , X is fragments per kilobase per million (FPKM), μ is mean, and σ is standard deviation of all FPKM in the row (Chen C. et al., 2020).
Quantitative Real-Time Polymerase Chain Reaction
The levels of gene expression were determined by using the quantitative real-time polymerase chain reaction (qRT-PCR), as described earlier (Zhang et al., 2016). The comparative Ct method was used to analyze the level of gene expression (Schmittgen and Livak, 2008). Primers used for qRT-PCR are listed in Supplementary Table 1. RmUBC (Klie and Debener, 2011) and AtActin2 (Zhou et al., 2018) were selected as reference genes in R. multiflora and Arabidopsis, respectively. All experiments were performed in three biological replicates, each containing three technical repeats.
Subcellular Localization of RmMYB108
The RmMYB108 CDS without the stop codon was amplified by PCR and cloned into the pBI121-green fluorescent protein (GFP) vector using ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) to generate 35S::RmMYB108-GFP construct. The primers used for plasmid construction are listed in Supplementary Table 1. The 35S::RmMYB108-GFP plasmid (Addgene: 173181) and 35S::GFP [positive control (PC)] plasmid were introduced into Agrobacterium tumefaciens strain GV3101, which was then injected into the leaves of 1-month-old N. benthamiana plants. The subcellular localization of 35S::RmMYB108-GFP and 35S::GFP was visualized by confocal laser scanning microscopy (TCS SP8, Wetzlar, Germany) 3 days post infiltration.
Transcriptional Activation of RmMYB108
The full-length CDS of RmMYB108 was introduced into the pGBKT7 vector using NdeI and BamHI restriction endonucleases and T4 ligase (Thermo Fisher Scientific, Waltham, MA, United States). The primers that were used for plasmid construction are listed in Supplementary Table 1. pGBT9 was used as a PC that could activate HIS3, ADE2, and MEL1 reporter genes and X-α-Gal activity in the transformed yeast cells on the medium lacking tryptophan (Trp) and histidine (His). pGBKT7 was used as a negative control (NC). The NC, PC, and pGBKT7-RmMYB108 plasmids (Addgene: 173183) were transformed into the yeast strain AH109 and cultivated on synthetic-defined (SD)/-Trp or SD/-Trp/-His medium with or without X-α-Gal.
Vector Construction and Plant Transformation
To construct 35S::RmMYB108, the RmMYB108 CDS was amplified using the primers listed in Supplementary Table 1, and then the PCR product was ligated into the KpnI and BamHI sites of the pCAMBIA1301 vector downstream to the 35S promoter of the cauliflower mosaic virus (Addgene: 173180). The resulting vector was introduced into A. tumefaciens strain GV3101.
To generate RmMYB108-overexpressing lines, Arabidopsis plants were transformed with the 35S::RmMYB108 construct using the floral dipping method (Clough and Bent, 1998). A semiquantitative reverse-transcription PCR (RT-PCR) assay was performed to select T2 generation, which was obtained from healthy T1 plants grown on the half-strength Murashige and Skoog (1/2 MS)–agar medium containing 30 mg/L Hygromycin. Homozygous RmMYB108 OE lines (#7, #9, and #11) in the T3 generation were further identified by screening of Hygromycin resistance and PCR.
Stress Treatments of Arabidopsis
For induce stress as oxidation, dehydration, and salt, sterilized seeds or 1-week-old seedlings of Arabidopsis were grown in 1/2 MS medium containing 3% sugar and 0.75% agar, which was further supplemented with or without 1.2 mM hydrogen peroxide (H2O2), 150 mM mannitol, or 150 mM NaCl. The plates were incubated under 12-h light/12-h dark cycle at 23°C vertically. To induce cold stress, the sterilized seeds and 1-week-old seedlings in plates without any supplement were incubated in a growth chamber that was maintained at 15 or 4°C vertically. The seed germination and seedling growth were observed and measured after cultivating for 1 and 2 weeks, respectively. To test the freezing tolerance of OE lines, the 20-day-old Arabidopsis plants were cold-acclimated at 4°C for 12 h and then subjected to −10°C for 2.5 or 4 h. Subsequently, the plants were subjected again to 4°C for 12 h and then grown at 23°C.
Physiological Measurements of Plants
The leaves of 20-day old wild-type (WT) plants and RmMYB108 OE lines were collected after exposure to 4°C for 0, 1, 3, 6 12, and 24 h. The relative conductivity (RC), superoxide dismutase (SOD) activity, peroxidase (POD) activity, and malondialdehyde (MDA) content of plant leaves were measured as described earlier (Miao et al., 2010).
Measurement of Chlorophyll Fluorescence
The 20-days-old WT and transgenic Arabidopsis plants were exposed to 4°C for 0, 1, 3, 6, 12, and 24 h; the rosette leaves of Arabidopsis were used for measuring chlorophyll fluorescence. The initial fluorescence (Fo) and the highest electronic efficiency (Fv/Fm) of photosystem II (PS II) were measured using IMAGING-PAM (Walz, Germany), according to the instructions of the manufacturer. The same leaf position was used for measurements in both plant groups.
Statistics and Analysis
All data were subjected to ANOVA, followed by the least significant difference test. All statistical analyses were performed using IBM SPSS (New York, NY, United States) software. The results were displayed by graphs and charts using GraphPad Prism 8.0.
Results and Analysis
R2R3-RmMYBs Potentially Regulate Plant Growth, Development, and Stress Tolerance in R. multiflora
The analysis of R. multiflora RNA-seq data (SRA accession no.: PRJNA698412) revealed many differentially expressed genes, which encoded TFs involved in metabolism, transcription, transport, and signal transduction in response to cold stress. For example, some genes belonging to the AP2/ERF, MYB, and NAC families were expressed in higher levels at both 4°C (CT1) and −20°C (CT2) than at 25°C (CK); however, the specific functions of most of these genes are unknown. In this study, we focused on how the R2R3-RmMYBs respond to low-temperature stress in R. multiflora. Based on the FPKM data, 37 R2R3-RmMYB genes (Supplementary Table 2 and Supplementary Figure 1) exhibited three expression trends in response to cold treatment. Genes including RmMYB018 and RmMYB44b were upregulated in the CT1 and CT2 treatments, while genes such as RmMYB308c and RmMYB24a were downregulated in both treatments. Several genes including RmMYB35 and RMMYB106 were induced only at CT1.
To understand the possible function as well as identifying potential chilling stress-responsive genes in this family, 119 R2R3-MYBs with complete sequences, which were extracted from 2R subgroup (Supplementary Table 3), were identified and named by referring to the protein BLAST results and the naming principle of R2R3-MYB in R. chinensis (Han et al., 2019; Supplementary Figure 2). Notably, the TFs with identical names were differentiated by lowercase letters; these MYB TFs showed extremely similar sequences and blastp results (Supplementary Table 4). According to the function of 134 Arabidopsis R2R3-MYBs (Supplementary Table 5), 114 R2R3-MYBs in R. multiflora were divided and clustered to 30 function-annotated and six function-unknown subgroups, with the exception of 5 R2R3-MYBs, which shared low sequence similarity with AtMYBs in the ML phylogenetic tree (Figure 1). Based on their functional annotation, RmMYBs were divided into three classes. RmMYBs in class I were further divided into eight subgroups (i.e., S9, S10, S16, S24, S25, S26, S29, and S33) and were involved in specific metabolisms, such as the regulation of lignin, anthocyanin, and flavonol biosynthesis. For example, RmMYB8e clustered closely with AtMYB11 and AtMYB12, which modulate flavonoid biosynthesis in favor of flavonol accumulation (Pandey et al., 2015; Wang et al., 2016). Class II contained 14 subgroups (i.e., S2, S3, S4, S11, S13, S14, S15, S19, S20, S21, S23, S30, S31, and S35), and RmMYBs in these subgroups played important roles in the fate of the plant cell, especially in the regulation of axillary meristem and stamen development. Class III RmMYBs, which were divided into eight clades (i.e., S1, S6, S8, S18, S22, S32, S34, and S36), were involved in the response to biotic and abiotic stress. In class III, some RmMYBs showed a close relationship with Arabidopsis R2R3-MYBs involved in stress tolerance, such as AtMYB21 (Zhang et al., 2021), AtMYB44 (Jung et al., 2007), or AtMYB108 (Cui et al., 2013). Furthermore, some R2R3-MYB genes involved in the response to abiotic and biotic stress were also involved in the regulation of stamen development or specific metabolisms. These results suggest that R. multiflora R2R3-MYBs perform a wide range of functions including the regulation of growth and development of rose plants, as well as its tolerance to harsh environments.
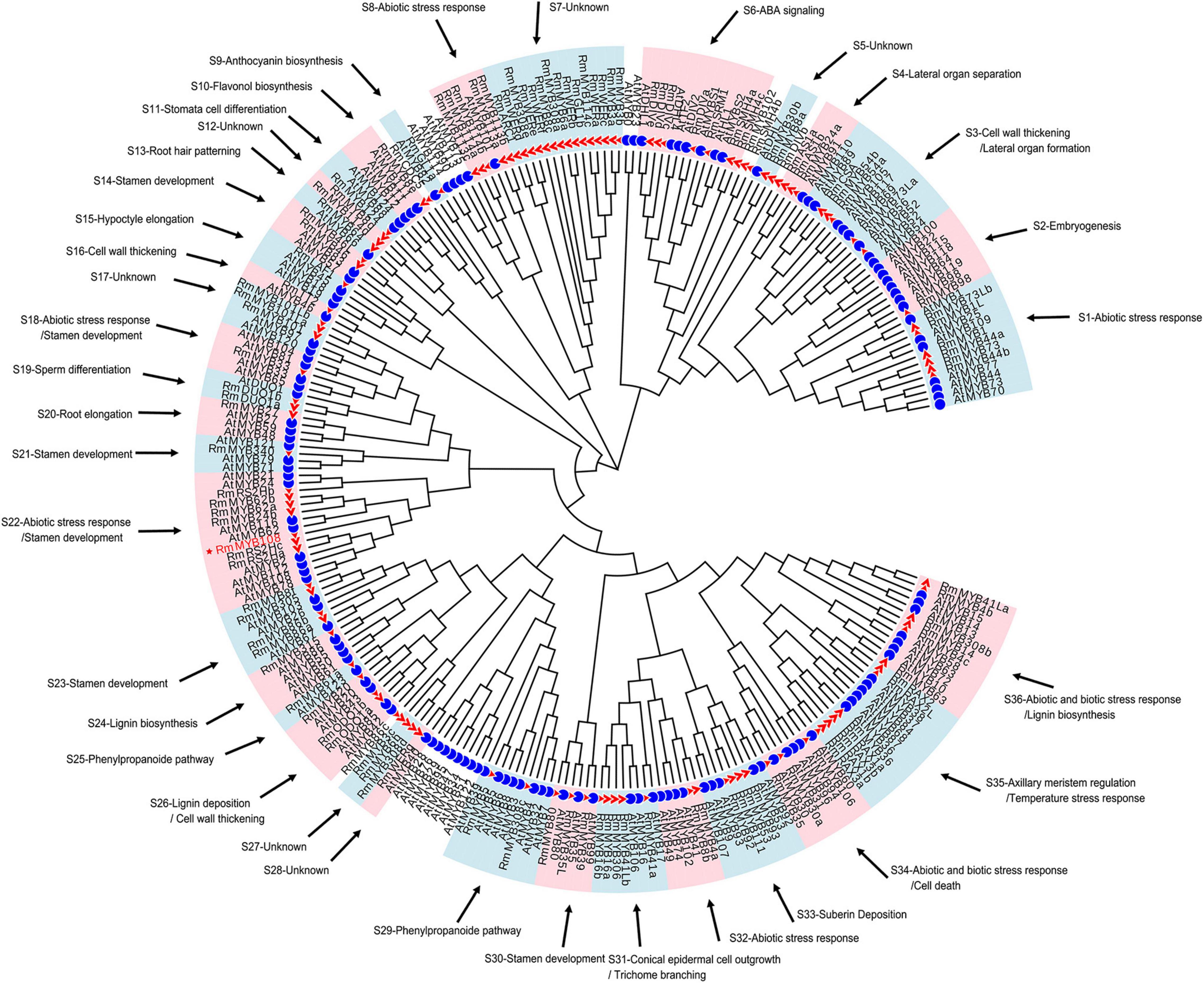
Figure 1. Putative functions of R2R3-MYB genes in Rosa multiflora based on the cluster results between R. multiflora and Arabidopsis thaliana in the phylogenetic tree. Red triangle marked R2R3-MYBs in R. multiflora and blue circle represented R2R3-MYBs in A. thaliana. The circular unrooted tree was generated by maximum likelihood method. Different subgroups were marked with pink and light blue background, and the function of each subgroup referred the genes with confirmed functions in A. thaliana.
Rosa multiflora R2R3-MYB Genes Response to Chilling Stress
The R. multiflora RNA-seq data analysis and prediction of gene function revealed a total of 15 stress-responsive R2R3-MYB genes (Figure 2). The gene expression analysis by using the qRT-PCR confirmed that RmMYB108, RmMYB44b, and RmMYB44a were upregulated in response to chilling stress, while RmMYB106, RmMYB35, RmMYB114b, and RmMYB308c were upregulated at 4°C and downregulated at –20°C. Moreover, the expression trends of the abovementioned genes showed a strong correlation with the RNA-seq results. However, while some other R. multiflora genes such as DIVARICATA (RmDIVa), RmMYB308d, and Salt-Related MYB1 were differentially expressed under low-temperature treatment, their correlation between qRT-PCR and RNA-seq data was not significant (Figures 2A–O). The alignments of the amino acid sequence of the R domains of these 15 R2R3-MYBs showed that the Myb domains of R2 and R3 were evolutionarily highly conserved, with spacer sequences [-W-(X19)-W-(X19)-W-…-F/I-(X18)-W-(X18)-W-] (Figures 2P,Q).
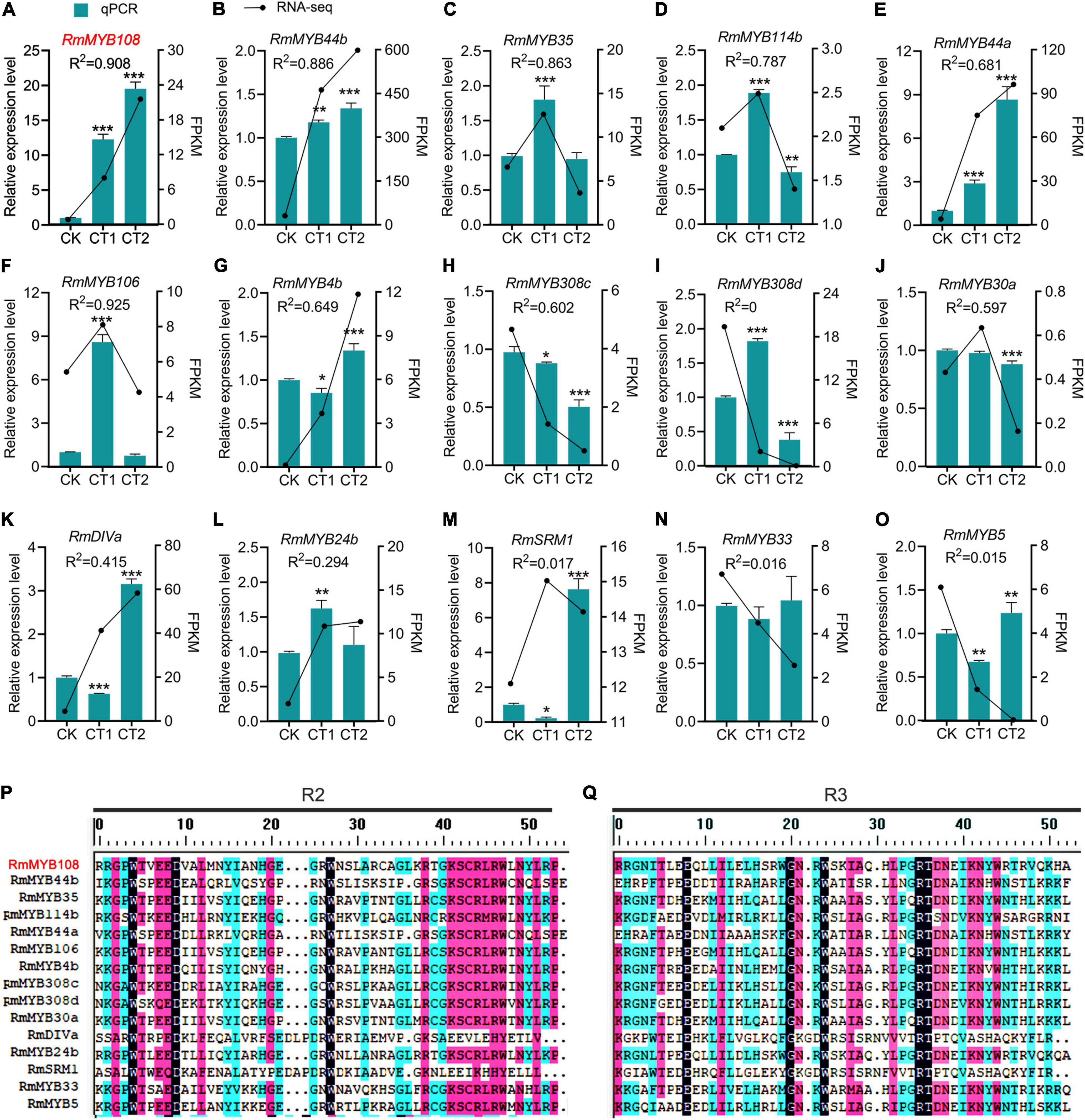
Figure 2. R2R3-MYBs in R. multiflora respond to chilling stress. Relative expression level and fragments per kilobase per million (FPKM) of (A) RmMYB108, (B) RmMYB44b, (C) RmMYB35, (D) RmMYB114b, (E) RmMYB44a, (F) RmMYB106, (G) RmMYB4b, (H) RmMYB308c, (I) RmMYB308d, (J) RmMYB30a, (K) RmDIVa, (L) RmMYB24b, (M) RmSRM1, (N) RmMYB33, (O) RmMYB5 under 25°C (CK), 4°C (CT1) and −20°C (CT2). The correlation coefficient of each gene between FPKM from RNA-seq and relative expression level from quantitative real-time polymerase chain reaction (qRT-PCR) was represented by R2 (coefficient of determination). *p < 0.05; **p < 0.01; and ***p < 0.001 in statistics. The aligned sequences of both R2 (P) and R3 (Q) domains of 15 R2R3-MYB from R. multiflora were evolutionarily highly conserved.
Thus, the results of the prediction of gene function together with RNA-seq data revealed that the 15 R. multiflora R2R3-MYB genes are responsive to the cold treatment. Among those genes, RmMYB108 showed the highest relative expression level, and its expression trend was fitted with the transcriptome result under low-temperature treatment. In addition, the effect of RmMYB108 on plant cold tolerance has not been investigated to date although the phylogenetic tree showed that RmMYB108 has a close relationship with R2R3-MYBs of Arabidopsis which are involved in stress resistance. Therefore, we further explored the potential role of RmMYB108 in cold tolerance in Arabidopsis.
Molecular Characterization of RmMYB108
The expression of RmMYB108 was first investigated under stress due to cold, salt, and drought using the qRT-PCR. At 4°C, RmMYB108 expression in rose leaves was induced within 0.5–2 h of the treatment, peaked at 1 h, and then decreased to its original level at 2 h. Furthermore, under cold stress, RmMYB108 showed a greater increase in the expression in roots over time, with the highest value at 8 h (Figure 3A). Notably, the expression pattern of RmMYB108 under salt stress (150 mM NaCl) was similar to that under cold stress (Figure 3B). In the drought treatment, the RmMYB108 expression peaked at 6 h in roots and at 12 h in leaves (Figure 3C). Thus, the expression of RmMYB108 in rose leaves and roots was affected by stress due to cold, salt, and drought at different time points. Interestingly, these results suggest that rose leaves sense environmental signals earlier than roots, although roots potentially exhibited a greater tolerance to stress than leaves.
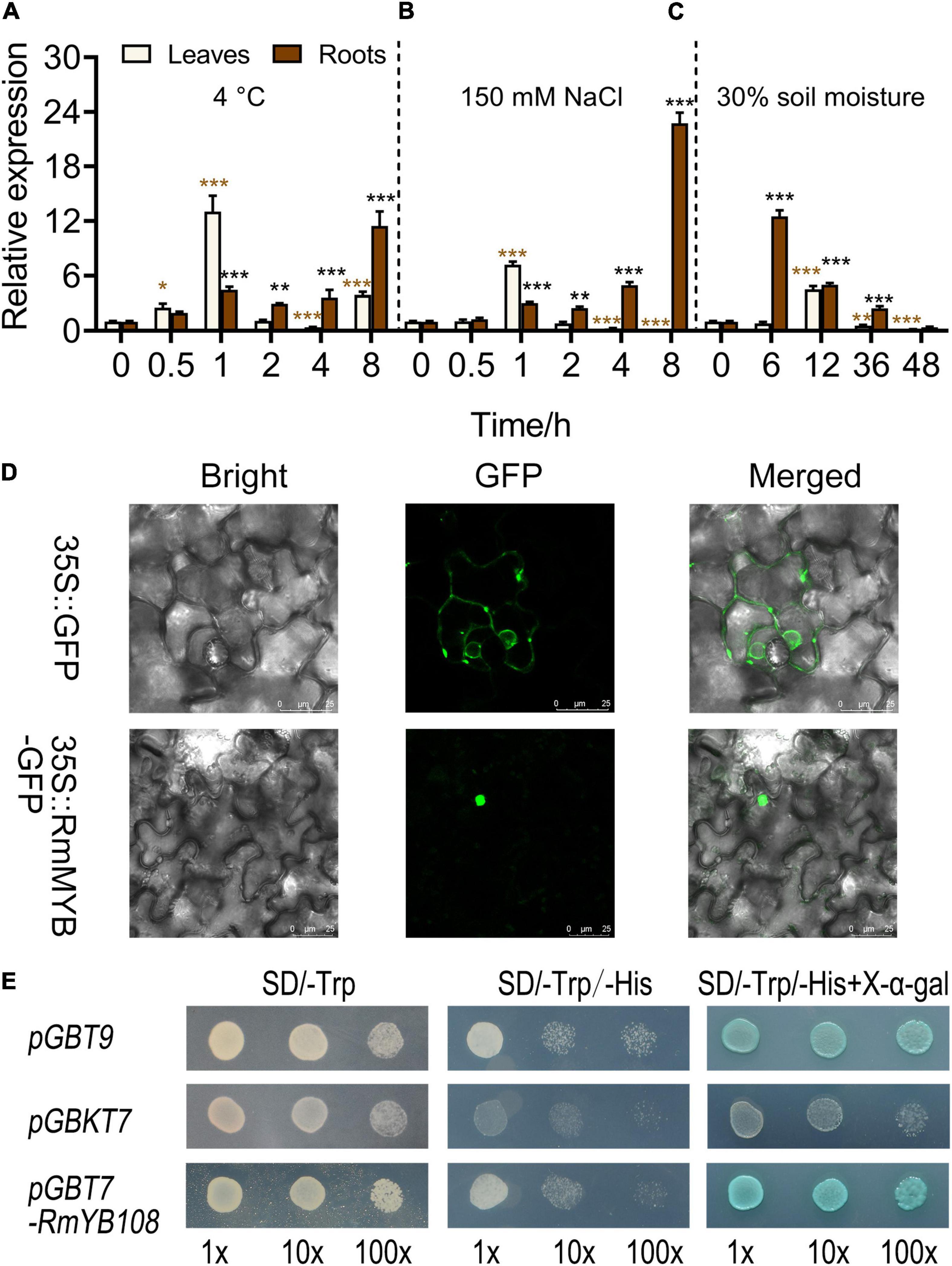
Figure 3. Analysis of gene expression, localization, and transactivation assay of the RmMYB108 transcription factor. RmMYB108 expression patterns in R. multiflora leaves and roots responding abiotic stress under continuous 4°C (A), 150 mM NaCl (B), and drought (C) treatment. (D) The positive control 35S::GFP and 35S::RmMYB108 constructs were expressed in tobacco epidermal cells, showing the nuclear localization of RmMYB108. (E) Transcriptional activation assay of RmMYB108. Growth of yeast cells (strain AH109) transformed with pGBT9 vector (a positive control), pGBKT7 vector (a negative control), or pGBKT7-RmMYB108 vector on SD/-Trp or SD/-Trp/-His with or without X-α-gal. 10×, 10×, and 100× represent yeasts were diluted with original concentration (OD600 = 0.4), 10–, 100-folds, respectively. *p < 0.05; **p < 0.01; ***p < 0.001 in statistics.
To determine the subcellular localization of the RmMYB108 protein, the RmMYB108 CDS was fused with the N-terminus of the GFP gene. As expected, RmMYB108 was localized to the nucleus in tobacco epidermal cells (Figure 3D). In addition, the experiments conducted in yeast confirmed that the full-length RmMYB108 exhibited self-activation, as the yeast AH109 cells transformed with the pGBKT7-RmMYB108 and pGBT9 (PC) grew well on the SD/-Trp/-His medium and turned blue on the selective medium containing X-α-Gal (Figure 3E).
Overexpression of RmMYB108 Enhanced the Cold Tolerance of Arabidopsis at the Early Growth Stage
To characterize the function of RmMYB108, pCAMBIA1301-RmMYB108 was transformed into Arabidopsis via the Agrobacterium-mediated transformation. Three RmMYB108 OE lines (#7, #9, and #11) were identified by using the semiquantitative RT-PCR (Figure 4A). We then compared the growth and cold tolerance of WT and RmMYB108 OE lines at the stages of germination and seedling. The rate of seed germination of the WT was similar to that of the OE lines under normal (no stress) conditions but reduced significantly at 15 and 4°C. While 98% of the transgenic seeds stayed alive, more than 40% WT seeds showed no germination at 4°C (Figures 4B,C). Interestingly, the growth vigor of seedlings (determined by their fresh weight and primary root length) was weaker in the WT than in OE lines even at 23°C, and the growth potential of WT seedlings was affected more dramatically than that of OE lines with the decrease in temperature (Figures 4D–F). At 14 days after treatment, the weight of WT seedlings decreased by 31.78% at 15°C and by 90.27% at 4°C, whereas that of OE lines decreased by 13.6 and 82.78%, respectively. These results suggest that the OE of RmMYB108 in Arabidopsis accelerated the plant growth as well as enhanced tolerance to chilling temperatures by alleviating stress-induced inhibition of seed germination and plant growth.
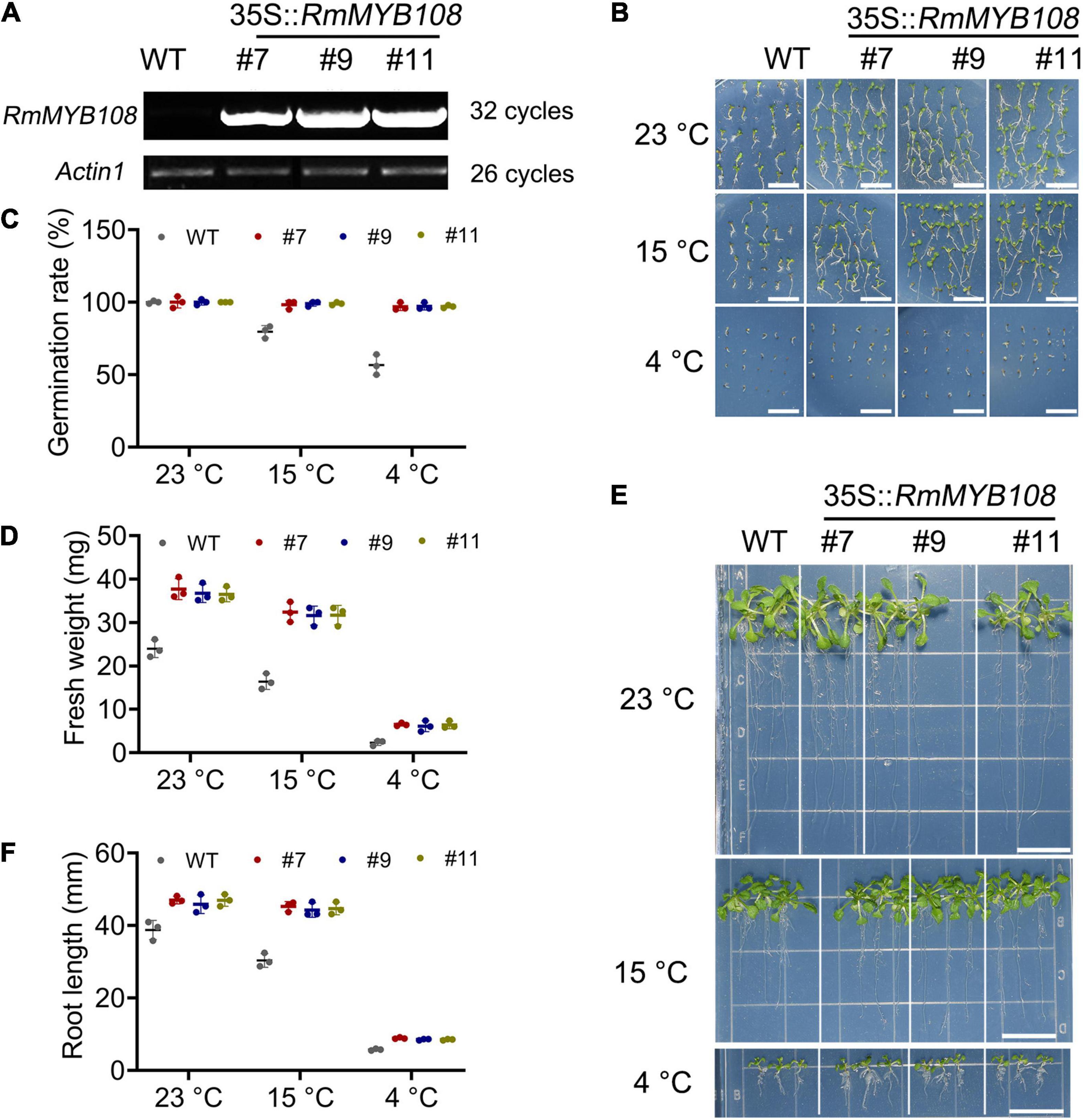
Figure 4. Identification of RmMYB108-overexpressing Arabidopsis and cold tolerance contrast between wild type (WT) and #7, #9, and #11 OE lines at early growth stage of plants. (A) semiquantitative RT-PCR analysis of RmMYB108 and Actin1 in WT and overexpression (OE) lines showed the overexpression of RmMYB108 in transgenic plants. The 7-days seed germination phenotype (B), seed germination rate (C), 21-day seedlings fresh weight (D), growth (E), and root length (F) of WT and OE lines under 23, 15, and 4°C, respectively, indicated OE lines had higher cold tolerance than WT. Data represent mean value + SD (n = 30). *p < 0.05; **p < 0.01; ***p < 0.001 in statistics.
Overexpression of RmMYB108 Enhanced Cold Tolerance at Maturity
The cold tolerance assays were further carried out in mature plants of the WT and OE lines. When exposed to –10°C for 4 h, transgenic plants showed a higher survival rate and greater tolerance against freezing stress than WT plants, with several yellow leaves (Figure 5A), whereas WT plants showed the most withered leaves and less than 15% survival (Figure 5B). When the duration of freezing stress was shortened to 2.5 h, the survival rate of WT plants increased to 20%, while that of OE lines was maintained at approximately 80% (Figures 5B,C). Forty-day-old transgenic plants were more robust than WT plants with or without cold treatment. A short exposure to freezing temperatures greatly harmed WT plants, as evident from the sharp reduction in the plant height from 19 to 8 cm, whereas OE lines suffered less damage, as the plant height decreased only from 31.22 to 21.27 cm (Figure 5D). In addition, flowering and silique formation occurred earlier in OE lines than in the WT under CK and cold conditions. The first silique appeared at 29 days in the WT and 22.5 days in OE lines under normal conditions but at 41.33 and 28.43 days, respectively, under cold conditions (Figure 5E). Furthermore, after freezing, silique length of the WT was only 5.95 cm, which is much shorter than that of OE lines with 13.00 cm at average (Figures 5F,G). These results suggest that the OE of RmMYB108 in Arabidopsis improved cold tolerance of the plant and shortened the plant growth cycle even after freezing.
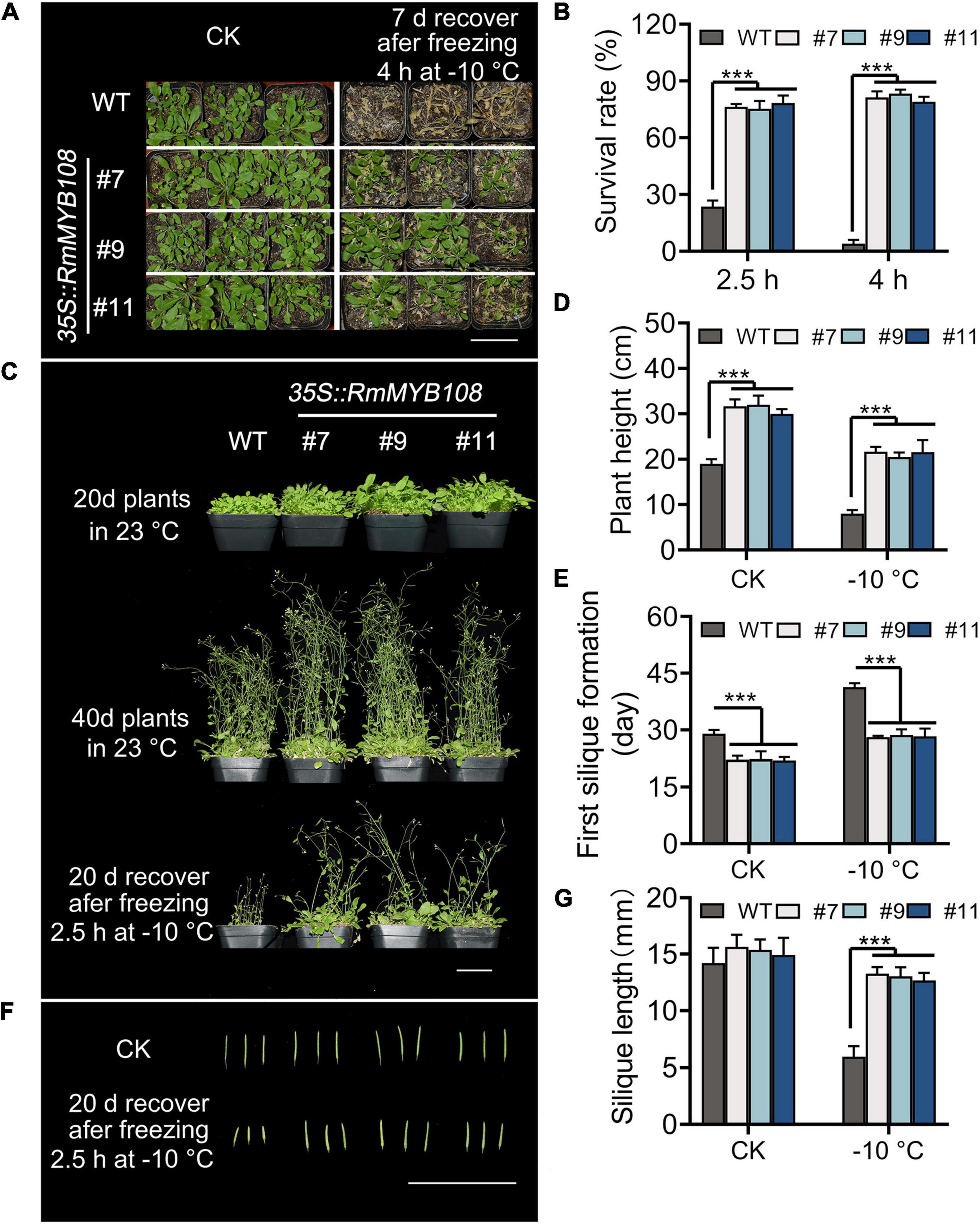
Figure 5. Phenotype of transgenic RmMYB108 plants after shortly freezing at –10°C at vegetative and reproductive growth stages of plants. (A) Phenotype of 20-days-old WT and OE lines that were treated at –10°C for 4 h and then recovered for 7 days. (B) Survival rate of recovered plants after being frozen at –10°C. The 20-day WT and OE lines were pictured after freezing 2.5 h at –10°C and recovering after 20 days (C), the plant height (D), first silique formation time (E), and silique length (F,G) were measured to show that the overexpression of RmMYB108 enhanced cold tolerance at maturity. Data represent mean value + SD (n = 20). *p < 0.05; **p < 0.01; ***p < 0.001 in statistics.
Overexpression of RmMYB108 Induced Physiological Changes in Arabidopsis
The healthy WT and OE lines were grown under the treatment at 4°C for 1 week, while the leaves of these plants were green, with no indication of wilting or dehydration. The physiological changes were observed in plants within 24 h of treatment at 4°C and indicated that all the physiological indexes of WT plants and OE lines showed the same variational trend under cold treatment in the vegetative phase. The SOD and POD activities of Arabidopsis peaked at 6 h after chilling stress, and their values were higher in OE lines than in WT plants at each time point, except at 0 h. For example, compared with the control at 6 h, SOD activity increased by 128.08% in the WT and by 284.20, 291.96, and 281.87% in OE lines #7, #9, and #11, respectively (Figures 6A,B). In addition, MDA content and RC, which exhibit a negative correlation with cold tolerance, showed a greater increase in WT plants than in OE lines. At 24 h, the RC of WT lines increased by 20%, whereas that of OE lines increased by an average of 15.54% (Figures 6C,D). Furthermore, the value of Fo, which shows a linear relationship with the content of photosynthetic pigment, increased more in WT plants than in OE lines under chilling stress (Figure 6E). The value of Fv/Fm decreased over time from 0.795 to 0.490 in the WT, 0.806 to 0.570 in line #7, 0.809 to 0.579 in line #9, and 0.805 to 0.577 in line #11 (Figure 6F).
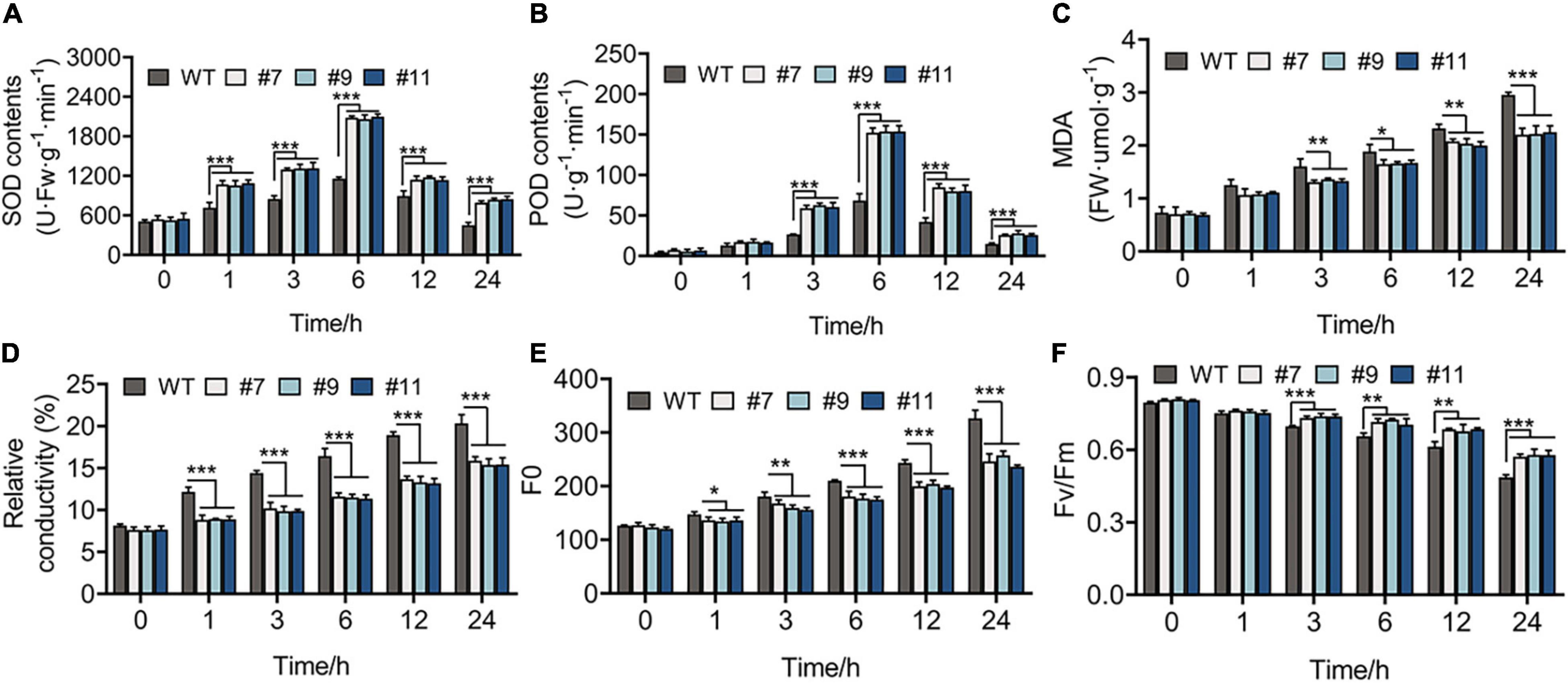
Figure 6. Physiological indicators of 20-days-WT and OE lines that were treated under 4°C for 0, 1, 3, 6, 12, and 24 h, respectively. (A) Superoxide dismutase (SOD) activity, (B) peroxidase (POD) activity, (C) malondialdehyde (MDA) content, (D) relative conductivity, (E) initial fluorescence (Fo), (F) the biggest electronic efficiency (Fv/Fm) changes between WT and OE lines provided an evidence for the growth of OE lines was less restricted than WT under chilling stress. *p < 0.05; **p < 0.01; ***p < 0.001 in statistics.
RmMYB108 Improves Plant Cold Tolerance by Upregulating the CBF Cascade
The expression of some marker genes involved in cold tolerance was tested in the WT and OE lines under chilling stress. Genes, such as AtICE1, AtCOR47A, AtCBF3, AtCOR15B, and AtRD29A (Zhou et al., 2011; Thalhammer and Hincha, 2014), in the ICE1-CBF-COR cascade showed a similar expression in the WT and OE lines under normal conditions. Under chilling stress, all genes were upregulated, reaching peak values at 3 or 6 h (Figures 7A–E), with more drastic changes in OE lines than in the WT. No phenotypic difference was apparent between WT plants and OE lines at continuous 4°C; however, the accumulation of oxides and loss of permeability of the cell membrane were observed in plants of all genotypes. Notably, compared with WT plants, all OE lines showed a minimal injury with a similar trend in physiological indexes and expression of genes related to cold resistance.
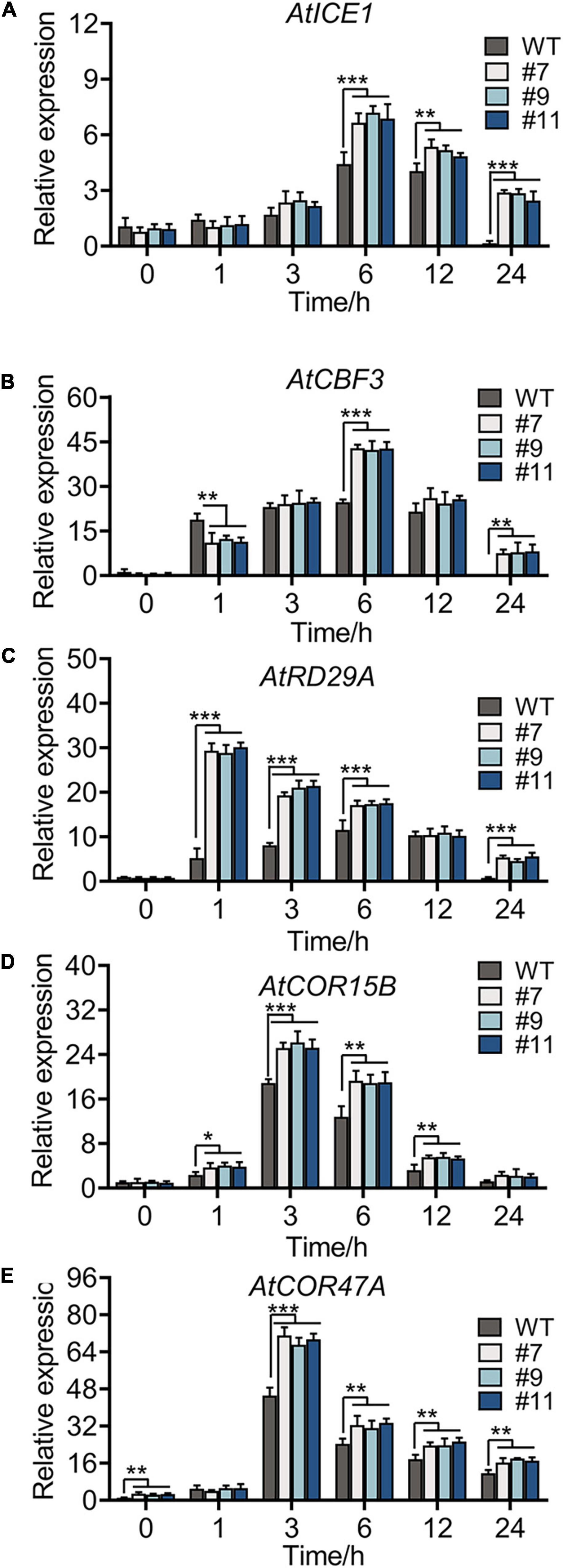
Figure 7. Cold-responsive marker genes expression level of WT and OE lines under 4°C for 0, 1, 3, 6, 12, and 24 h, respectively. Relative expression level of AtICE1 (A), AtCBF3 (B), AtRD29A (C), AtCOR15B (D), and AtCOR47A (E) were measured to reveal that RmMYB108 probably improved plant tolerance by CBF-dependent under cold treatment. *p < 0.05; **p < 0.01; ***p < 0.001 in statistics.
Overexpression of RmMYB108 Decreases Sensitivity to Other Stress
The seed germination and seedling growth of Arabidopsis were investigated under other stress to exemplify the essential role of RmMYB108 in abiotic stress tolerance. In deionized water, WT and transgenic seeds showed the same germination rate (Figure 8A). However, in the presence of 1.2 mM H2O2, 150 mM mannitol, or 150 mM NaCl, the germination rate of WT seeds decreased to 50–70%, while that of OE lines #7, #9, and #11 remained over 95% (Figure 8B). Furthermore, the vigor of WT seedlings was lower than that of OE lines in all treatments. The fresh weight and primary root length of WT seedlings were significantly lower than those of OE lines in the treatment of stress due to oxidation, drought, and salt (Figure 8C). Under normal conditions, the average primary root length of WT seedlings was 38.79 mm, while that of OE lines was approximately 46.63 mm; however, when treated with 1.2 mM H2O2, 150 mM mannitol, and 150 mM NaCl, the average primary root length decreased to 11, 7.47, and 13 mm in the WT and averaged to 14.59, 13.27, and 18.82 mm in the OE lines, respectively (Figures 8D,E). Additionally, water deficiency or 200 mM NaCl treatments for 10 days significantly damaged the growth and development of Arabidopsis plants (Figure 8F). The survival rate of WT was less than 20% in both salt and drought treatments after returning to normal growth conditions, while that of OE lines was maintained at 60–80% (Figures 8G,H). These phenomena revealed that RmMYB108 enhanced plant tolerance to stress due to dehydration and oxidation.
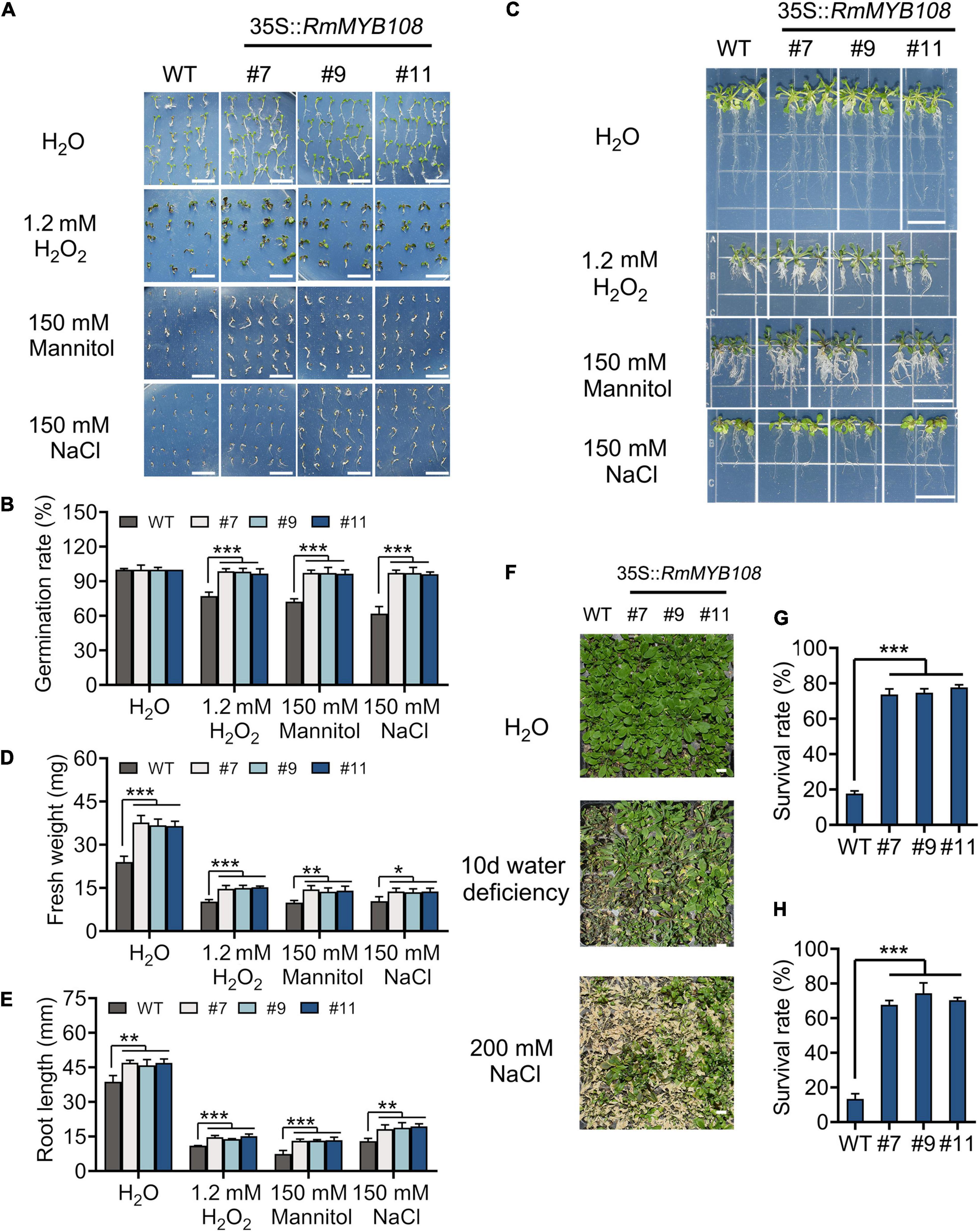
Figure 8. Effects of stress due to simulated oxidation, drought, and salt on plant growth. Seed germination phenotype (A) and germination rate (B), seedlings growth (C), fresh weight (D), and root length (E) of WT and OE lines were observed and measured under deionized water (H2O), 1.2 mM H2O2, 150 mM mannitol, and 150 mM NaCl, respectively. The 20-days WT and #7, #9, and #11 were not watered or watered with 200 mM NaCl for 10 days (F); the survival rate (G,H) were calculated and revealed that RmMYB108 enhanced plant tolerance against stress due to dehydration and oxidation. Data represent mean value + SD (n = 30). *p < 0.05; **p < 0.01; ***p < 0.001 in statistics.
Discussion
The R2R3-MYBs play essential roles in responses of the plant to abiotic stress. MYB genes in wheat [Triticum aestivum; TaMYB31 and TaMpc1-D4 (Myb protein colorless 1 located on chromosome D)] and cotton (Gossypium hirsutum; GaMYB85) respond to drought stress (Zhao et al., 2018; Li et al., 2020). Similarly, the Salt Stress Regulator 1 (PtrSSR1) gene in poplar (Populus trichocarpa), AcoMYB4 in pineapple (Ananas comosus), and SiMYB305 in sesame (Sesamum indicum) mediate the tolerance against stress due to salt or drought through abscisic acid (ABA) signaling (Fang et al., 2017; Chen H. et al., 2020; Dossa et al., 2020). In apple (Malus domestica), MdMYB308L, MdMYB23, MdMYB88, and MdMYB124 regulated cold hardiness via CBF-dependent and CBF-independent pathways (An et al., 2018; Xie et al., 2018; An et al., 2020). In this study, we found that RmMYB108, RmMYB44a, and RmMYB44b participated in the cold tolerance response of R. multiflora (Figure 1). The ICE1-CBF-COR gene cascade has been shown to contribute to cold acclimation by protecting plants from freezing damage in Arabidopsis (Zhao et al., 2015). AtICE1, AtCBF3, AtRD29A, AtCOR15B, and AtCOR47A were upregulated in Arabidopsis within 24 h under chilling stress, and RmMYB108 OE lines showed the higher expression levels of these genes than those in WT plants (Figure 7). Furthermore, the promoter of ICE1 harbors MYB-binding sequences (Supplementary Table 6). Therefore, we predicted that RmMYB108 enhances cold tolerance probably via the CBF-dependent pathway in Arabidopsis (Supplementary Figure 3).
RmMYB108 was upregulated in the transcriptome of R. multiflora in response to low-temperature stress, improved the ability to resist abiotic stress, and accelerated the growth of Arabidopsis plants by regulating changes in proteins in the cytoplasm or by indirectly maintaining the integrity of the cell membrane under stress. The stress due to low temperature, drought, and salt limits the availability of water to plant cells (Khan et al., 2018). This stress can cause membrane rupture and the outward flow of ions due to difference in osmotic pressure between the intra- and extracellular spaces, leading to plasmolysis and even cell death (Djemal and Khoudi, 2016; Ritonga and Chen, 2020). RmMYB108, which acts as a TF in the nucleus, regulates the expression of downstream genes probably by binding to their promoter regions on receiving the signals due to abiotic stress transmitted by the receptors. These genes generally participate in the minimization of oxidative damage, ion and water balance, osmotic stress response, and specific metabolisms to protect plant cells from water-related stress (Figure 6; Khan et al., 2018). Once the culture conditions were appropriate, the OE lines recovered and blossomed faster than WT obviously.
The recent studies showed that MYB108 orthologs are associated with plant development, specific metabolisms, and stress response. MYB108 is believed to be a JA-responsive TF involved in the development of stamen and pollen and defense signaling in plants (Mandaokar and Browse, 2009; Cheng et al., 2016; Xu et al., 2019). RmMYB108 has a close relationship with AtMYB112, AtMYB78, and AtMYB108 in Arabidopsis (Figure 1 and Supplementary Figure 2); AtMYB112 responds to salinity and high-light stress (Lotkowska et al., 2015), AtMYB78 belongs to ABA-related genes (Sun et al., 2020), and AtMYB108 acts as a negative regulator of ABA-induced cell death, (Cui et al., 2013). In Prunus mume, PmMYB108 (90% sequence similarity with RmMYB108) is positively associated with organ color (Zhang et al., 2018). In R. chinensis, RcMYB108, which is homologous to RmMYB108 (94.17% sequence similarity), was upregulated during petal senescence and shedding, and silencing of RcMYB108 altered the expression of senescence-associated genes and blocked ethylene- and JA-induced petal senescence (Zhang et al., 2019). Interestingly, when Arabidopsis seeds were cultured at 4°C for 6 months, we found that the speed of seed germination and plant growth was greatly reduced (data not shown). The germination rate of WT seeds was very low, whereas the seeds of most of the OE lines germinated well, and the resulting seedlings subsequently bloomed under harsh environments. After transferring to normal conditions, the transgenic plants grew well and quickly formed siliques. By contrast, WT seedlings showed a very low survival rate. Based on the evaluation of stress tolerance of OE lines and WT plants, we speculated that the RmMYB108 gene promotes cell division and differentiation to facilitate plant senescence, shortens the plant growth cycle, and enhances tolerance against abiotic stress.
In addition to an increasing plant yield, plant breeders focus on how to shorten the breeding period and increase the environmental stress resistance of plants (Bhatta et al., 2021). These breeding goals can generally be achieved through the genetic modification of plants by targeting genes encoding TFs (Hoang et al., 2017; Khan et al., 2018; Nowicka et al., 2018; Baillo et al., 2019). In this study, OE of RmMYB108 in Arabidopsis promoted the tolerance against stress due to drought, salt, and freezing of transgenic plants, manipulated their growth cycle, and increased their biomass and seed yield under cold stress. This approach can be used to accelerate wood growth, reduce the length of the plant breeding cycle, enhance stress tolerance, and improve land utilization by developing more productive and resilient crops that can feed the global population, which is predicted to reach 10 billion by 2050 (Bhatta et al., 2021).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JD designed and performed the experiments, interpreted the data, and wrote the article. DC, JZ, and LC designed the experiments and edited the article. XZ performed the experiments. WZ and TY analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31971700) and Joint Guiding Project of Natural Science Foundation of Heilongjiang Province (No. LH2020C014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Deguo Han in Northeast Agricultural University for providing the plasmids of pCAMBIA1301 and pBI121.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.696919/full#supplementary-material
Supplementary Figure 1 | The heatmap indicated row-normalized fragments per kilobase per million (FPKM) of 37 R2R3-MYBs from the RNA-seq database under 25°C (CK), 4°C (CT1), –20°C (CT2) in Rosa multiflora.
Supplementary Figure 2 | The phylogenetic tree of R2R3-MYB genes among Rosa multiflora, Rosa chinensis, and Arabidopsis thaliana. Red triangle marked R2R3-MYBs in R. multiflora, green star marked R2R3-MYBs in R. chinensis, and blue circle represented R2R3-MYBs in A. thaliana.
Supplementary Figure 3 | The putative regulation network of RmMYB108 after encountering chilling in overexpressed RmMYB108 Arabidopsis.
Supplementary Table 1 | The primers names and sequences (5′–3′) for quantitative real-time polymerase chain reaction (qRT-PCR), reverse-transcriptase polymerase chain reaction (RT-PCR), cloning, overexpression, subcellular localization, and self-activation.
Supplementary Table 2 | The name, raw fragments and fragments per kilobase per million (FPKM) of R2R3-MYBs that respond to low temperature from transcriptome sequencing of Rosa multiflora.
Supplementary Table 3 | The subgroups and sequences of 185 RmMYBs from the whole genome of Rosa multiflora.
Supplementary Table 4 | The names, nucleotide sequences, and encoded amino acid sequences of 119 R2R3-MYBs in Rosa multiflora.
Supplementary Table 5 | The nucleotide and encoded amino acid sequences of R2R3-MYBs in Arabidopsis thaliana from The Arabidopsis Information Resource (TAIR).
Supplementary Table 6 | The prediction of sequence and cis-acting elements from PlantCARE of AtICE1 promoter.
Footnotes
- ^ https://www.rosaceae.org/
- ^ http://www.arabidopsis.org/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://smart.embl-heidelberg.de/
References
Agarwal, M., Hao, Y. J., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X. W., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.m605895200
Albert, N., Griffiths, A., Cousins, G., Verry, I., and Williams, W. (2014). Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 205, 882–893. doi: 10.1111/nph.13100
An, J.-P., Li, R., Qu, F.-J., You, C.-X., Wang, X.-F., and Hao, Y.-J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
An, J. P., Wang, X. F., Zhang, X. W., Xu, H. F., Bi, S. Q., You, C. X., et al. (2020). An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 18, 337–353. doi: 10.1111/pbi.13201
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10:771. doi: 10.3390/genes10100771
Bhatta, M., Sandro, P., Smith, M. R., Delaney, O., Voss-Fels, K. P., Gutierrez, L., et al. (2021). Need for speed: manipulating plant growth to accelerate breeding cycles. Curr. Opin. Plant Biol. 60:101986. doi: 10.1016/j.pbi.2020.101986
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Lai, L., Li, L., Liu, L., Jakada, B. H., Huang, Y., et al. (2020). AcoMYB4, an ananas comosus L. MYB transcription factor, functions in osmotic stress through negative regulation of ABA signaling. Int. J. Mol. Sci. 21:5727. doi: 10.3390/ijms21165727
Cheng, H.-Q., Han, L.-B., Yang, C.-L., Wu, X.-M., Zhong, N.-Q., Wu, J.-H., et al. (2016). The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Botany 67, 1935–1950. doi: 10.1093/jxb/erw016
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cui, F., Brosché, M., Sipari, N., Tang, S., and Overmyer, K. (2013). Regulation of ABA dependent wound induced spreading cell death by MYB108. New Phytol. 200, 634–640. doi: 10.1111/nph.12456
Djemal, R., and Khoudi, H. (2016). TdSHN1, a WIN1/SHN1-type transcription factor, imparts multiple abiotic stress tolerance in transgenic tobacco. Environ. Exp. Botany 131, 89–100. doi: 10.1016/j.envexpbot.2016.07.005
Dossa, K., Mmadi, M. A., Zhou, R., Liu, A., Yang, Y., Diouf, D., et al. (2020). Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB Plants 12:lz081.
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581.
Fang, Q., Jiang, T., Xu, L., Liu, H., Mao, H., Wang, X., et al. (2017). A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiol. Biochem. 114, 100–110. doi: 10.1016/j.plaphy.2017.02.018
Han, Y., Yu, J. Y., Zhao, T., Cheng, T. R., Wang, J., Yang, W. R., et al. (2019). Dissecting the genome-wide evolution and function of R2R3-MYB transcription factor family in rosa chinensis. Genes 10:16.
Hoang, X. L. T., Nhi, D. N. H., Thu, N. B. A., Thao, N. P., and Tran, L. P. (2017). Transcription factors and their roles in signal transduction in plants under abiotic stresses. Curr. Genomics 18, 483–497.
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2007). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic arabidopsis. Plant Physiol. 146, 323–324. doi: 10.1104/pp.107.110981
Jung, S., Lee, T., Cheng, C. H., Buble, K., Zheng, P., Yu, J., et al. (2019). 15 years of GDR: new data and functionality in the genome database for rosaceae. Nucleic Acids Res. 47, D1137–D1145.
Khan, S. A., Li, M. Z., Wang, S. M., and Yin, H. J. (2018). Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 19:29.
Klempnauer, K.-H., Gonda, T. J., and Michael Bishop, J. (1982). Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31, 453–463. doi: 10.1016/0092-8674(82)90138-6
Klie, M., and Debener, T. (2011). Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res. Notes 4:518. doi: 10.1186/1756-0500-4-518
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496.
Li, J., Han, G., Sun, C., and Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 14:1613131. doi: 10.1080/15592324.2019.1613131
Li, X., Tang, Y., Li, H., Luo, W., Zhou, C., Zhang, L., et al. (2020). A wheat R2R3 MYB gene TaMpc1-D4 negatively regulates drought tolerance in transgenic Arabidopsis and wheat. Plant Sci. 299:110613. doi: 10.1016/j.plantsci.2020.110613
Liu, Z.-Y., Li, X.-P., Zhang, T.-Q., Wang, Y.-Y., Wang, C., and Gao, C.-Q. (2021). Overexpression of ThMYB8 mediates salt stress tolerance by directly activating stress-responsive gene expression. Plant Sci. 302:110668. doi: 10.1016/j.plantsci.2020.110668
Lotkowska, M. E., Tohge, T., Fernie, A. R., Xue, G.-P., Balazadeh, S., and Mueller-Roeber, B. (2015). The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169, 1862–1880.
Mandaokar, A., and Browse, J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in arabidopsis. Plant Physiol. 149, 851–862. doi: 10.1104/pp.108.132597
Mehrotra, S., Verma, S., Kumar, S., Kumari, S., and Mishra, B. N. (2020). Transcriptional regulation and signalling of cold stress response in plants: an overview of current understanding. Environ. Exp. Bot. 180:104243. doi: 10.1016/j.envexpbot.2020.104243
Miao, B. H., Han, X. G., and Zhang, W. H. (2010). The ameliorative effect of silicon on soybean seedlings grown in potassium-deficient medium. Ann. Bot. 105, 967–973. doi: 10.1093/aob/mcq063
Nowicka, B., Ciura, J., Szymańzska, R., and Kruk, J. (2018). Improving photosynthesis, plant productivity and abiotic stress tolerance – current trends and future perspectives. J. Plant Physiol. 231, 415–433. doi: 10.1016/j.jplph.2018.10.022
Pandey, A., Misra, P., and Trivedi, P. K. (2015). Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 34, 1515–1528. doi: 10.1007/s00299-015-1803-z
Ren, H., Bai, M., Sun, J., Liu, J., Ren, M., Dong, Y., et al. (2020). RcMYB84andRcMYB123mediate jasmonate-induced defense responses againstBotrytis cinereain rose (Rosa chinensis). Plant J. 103, 1839–1849. doi: 10.1111/tpj.14871
Ritonga, F. N., and Chen, S. (2020). Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 9:560. doi: 10.3390/plants9050560
Rogers, L. A., and Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protocols 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shen, Y. X., Sun, T. T., Pan, Q., Anupol, N., Chen, H., Shi, J. W., et al. (2019). RrMYB5-and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 17, 2078–2095. doi: 10.1111/pbi.13123
Sun, L., Liu, L. P., Wang, Y. Z., Yang, L., Wang, M. J., and Liu, J. X. (2020). NAC103, a NAC family transcription factor, regulates ABA response during seed germination and seedling growth in Arabidopsis. Planta 252:95.
Thalhammer, A., and Hincha, D. K. (2014). A mechanistic model of COR15 protein function in plant freezing tolerance: integration of structural and functional characteristics. Plant Signal. & Behav. 9:e977722. doi: 10.4161/15592324.2014.977722
Wang, F., Kong, W., Wong, G., Fu, L., Peng, R., Li, Z., et al. (2016). AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics 291, 1545–1559. doi: 10.1007/s00438-016-1203-2
Xie, Y. P., Chen, P. X., Yan, Y., Bao, C. N., Li, X. W., Wang, L. P., et al. (2018). An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 218, 201–218.
Xu, X. F., Wang, B., Feng, Y. F., Xue, J. S., Qian, X. X., Liu, S. Q., et al. (2019). AUXIN RESPONSE FACTOR17 directly regulates MYB108 for anther dehiscence. Plant Physiol. 181, 645–655. doi: 10.1104/pp.19.00576
Zhang, Q., Zhang, H., Sun, L., Fan, G., Ye, M., Jiang, L., et al. (2018). The genetic architecture of floral traits in the woody plant Prunus mume. Nat. Commun. 9:1702.
Zhang, S., Zhao, Q., Zeng, D., Xu, J., Zhou, H., Wang, F., et al. (2019). RhMYB108, an R2R3-MYB transcription factor, is involved in ethylene- and JA-induced petal senescence in rose plants. Hortic Res. 6:131.
Zhang, X., Wu, S., Liu, S., and Takano, T. (2021). The Arabidopsis sucrose non-fermenting-1-related protein kinase AtSnRK2.4 interacts with a transcription factor, AtMYB21, that is involved in salt tolerance. Plant Sci. 303:110685. doi: 10.1016/j.plantsci.2020.110685
Zhang, X. Y., Zhang, J. Z., Zhang, W. W., Yang, T., Xiong, Y., and Che, D. D. (2016). Transcriptome sequencing and de novo analysis of Rosa multiflora under cold stress. Acta Physiol. Plantarum 38:13.
Zhao, C., Lang, Z., and Zhu, J.-K. (2015). Cold responsive gene transcription becomes more complex. Trends Plant Sci. 20, 466–468. doi: 10.1016/j.tplants.2015.06.001
Zhao, Y., Cheng, X. Y., Liu, X. D., Wu, H. F., Bi, H. H., and Xu, H. X. (2018). The wheat MYB transcription factor TaMYB31 is involved in drought stress responses in arabidopsis. Front. Plant Sci. 9:1426. doi: 10.3389/fpls.2018.01426
Zhou, A., Sun, H., Feng, S., Zhou, M., Gong, S., Wang, J., et al. (2018). A novel cold-regulated gene from Phlox subulata, PsCor413im1, enhances low temperature tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 495, 1688–1694. doi: 10.1016/j.bbrc.2017.12.042
Keywords: Rosa multiflora, R2R3 MYB transcription factors, MYB108, cold tolerance, transgenic, Arabidopsis
Citation: Dong J, Cao L, Zhang X, Zhang W, Yang T, Zhang J and Che D (2021) An R2R3-MYB Transcription Factor RmMYB108 Responds to Chilling Stress of Rosa multiflora and Conferred Cold Tolerance of Arabidopsis. Front. Plant Sci. 12:696919. doi: 10.3389/fpls.2021.696919
Received: 18 April 2021; Accepted: 30 June 2021;
Published: 27 July 2021.
Edited by:
Rosalyn B. Angeles-Shim, Texas Tech University, United StatesReviewed by:
M. A. Qibin, South China Agricultural University, ChinaBoas Pucker, University of Cambridge, United Kingdom
Mei Zhang, South China Botanical Garden, Chinese Academy of Sciences, China
Aiping Song, Nanjing Agricultural University, China
Copyright © 2021 Dong, Cao, Zhang, Zhang, Yang, Zhang and Che. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinzhu Zhang, amluemh1emhhbmdAbmVhdS5lZHUuY24=; Daidi Che, ZGFpZGljaGVAbmVhdS5lZHUuY24=
 Jie Dong
Jie Dong Lei Cao
Lei Cao Xiaoying Zhang2
Xiaoying Zhang2 Jinzhu Zhang
Jinzhu Zhang Daidi Che
Daidi Che